Abstract
Human cortical organoids (hCOs), derived from human embryonic stem cells (hESCs), provide an excellent platform to study human brain development and diseases in complex 3D tissue. However, current hCOs lack microvasculature, resulting in limited oxygen and nutrient delivery to the inner-most parts of hCOs. Here, we engineered hESCs to ectopically express human ETS variant 2 (hETV2) to create in vitro vasculature in hCOs, namely vhCOs (vascularized hCOs). hETV2-expressing cells in hCOs contributed to forming a complex vascular-like network in hCOs. Importantly, the presence of vascularization resulted in enhanced functional maturation of organoids. We found that vhCOs acquired several blood-brain barrier (BBB) characteristics, including an increase in the expression of tight junctions, nutrient transporters, and trans-endothelial electrical resistance. Finally, hETV2-induced endothelium supported the formation of perfused blood vessels in vivo. These vhCOs form vasculature that resemble the early prenatal brain, and present a robust model to study brain disease in vitro.
The human brain is composed of multiple cell types, including neurons, astrocytes, oligodendrocytes, and microglia. Understanding human brain development and neurological disease mechanisms has been challenging due to the paucity of available tissues. Pluripotent stem cell-derived three-dimensional (3D) cortical organoids (hCOs) recapitulate the developing human cortex in both architecture and function. The hCOs have provided an exclusive opportunity to study the initial brain development and neurologic disorders 1-5. Although the advent of 3D brain organoids has offered innovative avenues, there are several unresolved limitations: the lack of functional blood vessels, the limited formation of microglia and the six distinct cortical layers 6. Notably, the absence of vascular system brain is deemed detrimental to organoids, and millimeter-scale organoids under long-term culture consistently exhibit apoptotic cell death at the inner-most regions 7,8. It is well-known that neuron progenitor differentiation is impaired without functional vasculature 9.
Recently, efforts have been made to implement vascular structure in human brain organoids. Mansour et al.10 showed that human brain organoids transplanted onto the cortex of the mouse brain induced the outgrowth of murine vessels into the human tissue, which increased cell survival and maturation. Beyond this, there is no systematic approach to produce robust vascularization in brain organoids, which limits their application to the study in normal or pathogenic development in vitro. Here, we present a unique method to generate in vitro functional vasculature-like network in hCOs from human embryonic stem cells (hESCs).
Our recent study found that the expression of the transcription factor human ETS variant 2 (hETV2) reprograms human dermal fibroblasts into endothelial cells (EC) 11. Here, we demonstrated that the expression of hETV2 induces EC differentiation in hESC under three conditions (Supplementary Note 1). These data suggest that overexpression of hETV2 induces the EC formation regardless of differentiation conditions and in the absence of growth factors (e.g., VEGF) essential for differentiating and maintaining mature endothelial cells in culture 12,13.
We hypothesized that the hETV2 expression induces the formation of ECs and vascular-like structures in hCOs. We tested the hETV2 dosage and induction time to determine the optimal stage of hCO development for the formation of vascular-like structure in hCOs. We identified hCOs with 20% hETV2 induced at day 18 start to form vascular-like structures, characterized by EC markers (Supplementary Note 2), which were used in subsequent analysis. We named the hCOs with the vascular-like structure, as vascularized hCOs (vhCOs).
To examine whether the hETV2-expressing organoids have functional vasculature-like structures, we evaluated the endothelial characteristics on day 30, 70 and 120. The formation of ventricular (VZ)-like zones, sub-ventricular (SVZ)-like zones, and cortical layers is a distinct feature of cortical organoids 1,2. Both control hCOs and vhCOs exhibited well-organized SOX2 positive VZ and TBR1 positive SVZ around a lumen (Supplementary Fig. 1a, left panel). Moreover, the separation of early-born CTIP2+ and late-born SATB2+ neurons became distinct and formed deep and upper cortical layers in a similar manner in control and vhCOs at day 120 (Supplementary Fig. 1a, right panel). Following neuroectoderm commitment, both control hCOs and vhCOs demonstrated a down-regulation of pluripotency genes and a concomitant up-regulation of neural genes (Supplementary Fig. 1b). Overall, these results show that both control hCOs and vhCOs differentiated at a similar extent and acquired the regional and morphological features of organized cortical structures.
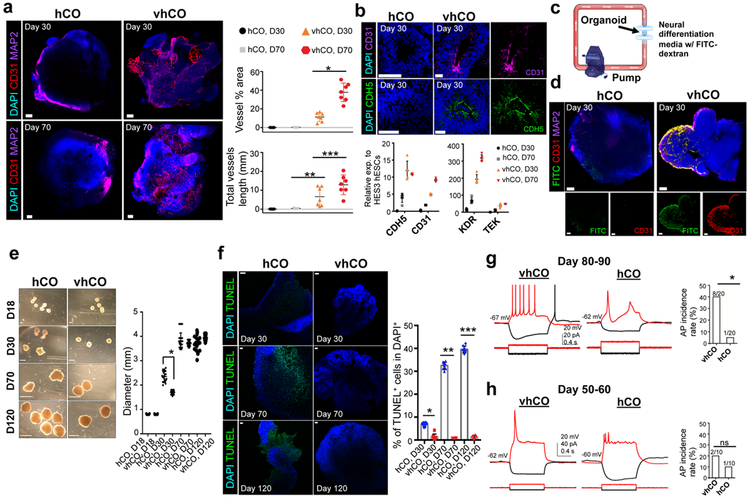
We next investigated the organization of the endothelium and vascular networks. Whole-mount immunostaining demonstrated that enhanced CD31+ endothelial tubes were formed in vhCOs at day 30 while control hCOs lacked the formation of these tubes (Fig. 1a and Supplementary video 1 and 2). Importantly, we observed that the vhCOs formed more complex network of hETV2-derived CD31+ vascular-like structure at 70 day old (Fig. 1a). The quantification of vascularization via ImageJ (AngioTool) demonstrated that vhCOs had a significantly more vessel area and vessel length compared with control hCOs (Fig. 1a and Supplementary Fig. 1c). When we sectioned organoids, the lumens of VZ in hETV2-expressing vhCOs were stained strongly for EC markers CD31 and CDH5, whereas lumens of VZ in control hCOs lacked these EC markers (Fig. 1b). qPCR results supported our immunostaining analysis, revealing an elevated expression of CD31 and CDH5 in vhCOs compared to control hCOs at day 30 (Fig. 1b). When control hCOs were cultured up to 70 days, expression of TEK and KDR genes increased compared to day 30 control hCOs. However, vhCOs demonstrated earlier and much higher induction of EC genes compared to control hCOs (Fig. 1b). Co-immunostaining indicated the presence of endothelial cells, i.e., CD31+ or CDH5+, derived from mCherry+ cells, in vhCOs at day 70, whereas control hCOs failed to generate these endothelial cells (Supplementary Fig. 1dand e). Additionally, electron microscopy confirmed the presence of endothelial cells in vhCOs (Supplementary Fig. 1f). When repeated with different hESC line (H1), a similar EC staining pattern was observed in H1-derived vhCOs (Supplementary Fig. 2a). Overall, hETV2 induction leads to consistent generation of organoids with vascular-like architectures.
Figure 1. Characterization of vasculature in vhCOs.
(a) Left, immunostaining of whole mount vhCOs and control hCOs at the different time point (30-day and 70-day) for CD31 and MAP2. Right, AngioTool analysis indicating the abundance and type of vasculature in hCOs. Data represent the mean ± SEM (n=7, from three independent batches). (*p=0.00003699, and **p=0.00064, ***p=0.0403) (b) Top, immunostaining for CD31 and CDH5 reveals the production of endothelial cells in sectioned-vhCOs at day 30. CD31 and CDH5 were present at lumens of ventricular zone in sectioned vhCOs while they were not found in control hCOs. Bottom, expression of endothelial genes from organoids at day 30 and day 70 was measured relative to HES3 hESCs. Data represent the mean ± SEM (n=5, from three independent batches). (c) Illustration of FITC-dextran perfusion into organoids via bioreactor with the flow rate of 0.88 ml/min. (d) Immunostaining of whole mount FITC-dextran perfused vhCOs and control hCOs for CD31. Representative images were shown (N=5, from three independent batches). (e) Left, morphology and size of the control hCOs and vhCOs after 120-day culture. Right, quantification of diameter (mm) from organoids at different stages (n=20, *p=0.000045, from four independent batches) (Mean values of hCO at day 18, 30, 70, 120 are 0.803, 2.354, 3.802 and 3.731 mm, respectively, and mean values of vhCO at day 18, 30, 70, 120 are 0.831, 1.695, 3.697 and 3.938 mm, respectively). Error bar represents the ± SEM. (f) Left, TUNEL staining of organoids after 30-, 70- and 120-day culture. Right, quantification of TUNEL+/DAPI+ cells indicated that the increase in cell death at the center of control hCOs at day 70 and 120 was dramatically reduced in vhCOs. Data represent the mean ± SEM (n=8, from three independent batches). (D30: T=9.97 DF=4 and *p=0.000096, D70: T=26.02 DF=4 and **p=0.000012, D120: T=34.78 DF=4 and ***p=00000408). (g-h) Left, voltage traces of current-clamp recordings of a cell in control hCOs and in vhCOs at day 80-90 (g) and day 50-60 (h) in response to hyperpolarizing (−10 pA) and depolarizing (+5 pA or +10 pA) current steps. Right, bar graph shows the difference in AP incidence rate between control hCOs and vhCOs. *p<0.05 in g and p<0.5 in h. The scale bar represents 100 μm in a, d, and 50 μm in f, b, and 1, 2 and 4 mm at day 18, 30, and 70 and 120, respectively in e. The unpaired two-tail t-test was used for all comparisons.
We further examined the function of vascular structures in vhCOs by assessing in vitro capillary networks using a bioreactor (Fig. 1c and Supplementary Fig. 2b). Perfusion of fluorescein isothiocyanate (FITC)-dextran (100 nm) was performed by placing the organoids between two syringe filters under constant flow rate (Fig. 1c). To determine the extent of perfusable vasculature in vhCOs at day 30, we performed whole-mount immunostaining for both CD31 and MAP2. We observed the presence of FITC-dextran in hETV2 organoid CD31+ vasculature, indicating the existence of perfusable vascular-like network in vhCOs (Fig. 1d and Supplementary video 3). Similarly, FITC-positive lumens were observed only in vhCOs while control hCOs failed to show FITC-dextran filling in the lumens of VZ (Supplementary Fig. 2c). FITC-dextran filled regions were co-stained with endothelial marker CDH5, and around 8% of lumens in vhCOs were found FITC-dextran filled (n=5, p<0.0001, Supplementary Fig. 2c). Overall, the incorporation of FITC-dextran in CD31+ vascular networks supports the presence of functional vascular-like openings inside vhCOs.
Next, we investigated the function of the vascular-like system in supporting the growth and viability of organoids. Both control hCOs and vhCOs were similar in size at day 18 before hETV2 induction. At day 30, when hETV2 was induced for 12 days, vhCOs were significantly smaller than control organoids (Fig. 1e, right, n=20, p<0.0001). By day 70 and 120, the growth rate of vhCOs exceeded that of control hCOs, although the size of both organoid types similarly reached 3.5–4mm in diameter (Fig. 1e). This early lag in growth followed by a late increase appears to be temporally correlated with organoid vascularization; this phenomenon was also noted by others while evaluating hCOs transplanted into mouse brains 10. As the organoid size increased, control hCOs exhibited significantly higher apoptotic signals (TUNEL+) at day 70 (around 35%) compared to day 30 (n=5, p<0.00001, Fig. 1f). On the other hand, no significant cell death was found in vhCOs, even at day 120 (Fig. 1f). Similarly, the analysis of hypoxia-inducible factor 1-α (HIF-1α) staining demonstrated the significantly elevated (~42%) regions of hypoxic cells in control hCOs compared to vhCOs at day 120 (~2.5%) (Supplementary Fig. 2d). Collectively, these results suggest that hETV2-induced vascular-like architectures support the gradual increase in the size of hCOs by supporting the diffusion of oxygen, thus preventing cell death inside of vhCOs.
The murine central nervous system (CNS) starts to form vascularization at embryonic day 7.5–9.5 14,15, whereas human brain vascular components are identified in 6- to 7-week old human embryos 16. The formation of ECs was shown to be critical for the maturation of cortical neurons 9,17,18. Thus, we performed whole-cell patch-clamp recordings to characterize neuronal activities in control hCOs and vhCOs. 8 of 20 cells from vhCOs (day 80–90) produced multiple action potentials (APs) with spike frequency adaptation in response to 1 s-long depolarizing current pulses (from +5 to +20 pA with 5 pA increments; cells were held at approximately −60 mV) (Fig. 1g, Left) whereas the remaining 12 cells did not produce APs. In sharp contrast, only 1 of 20 cells from control hCOs (day 80–90) produced single low-amplitude APs (Fig. 1g, Left) while the remaining 19 cells did not produce any APs. The incidence rate of obtaining neurons that were able to produce APs was significantly higher in vhCOs compared to that in control hCOs (Fig. 1g, Right, p<0.0197, Fisher’s exact test). In 50–60 day organoids, there was no difference in the incidence rate of obtaining neurons producing APs. Specifically, 2 out of 10 cells and 1 out of 10 cells produce APs in vhCOs and control hCOs, respectively (Fig. 1h, p<0.5, Fisher’s exact test). An earlier study identified the spontaneous firing of neurons in 8-month old organoids while 4-month old organoids failed to contain these neuronal activities 19. Strikingly, we observed neurons with generating APs from 3-month old vhCOs. Moreover, vhCOs expressed dramatically higher levels of pre-synaptic marker synapsin 1 (SYN1), compared to control hCOs at day 70 (Supplementary Fig. 2e). When organoids were cultured longer, SYN1 expression in control hCOs became increased at 120-day, although that in vhCOs was higher (Supplementary Fig. 2e). Thus, these results suggest that functional vessels-like structures in hCOs play a critical role in neuronal maturation observed in vhCOs.
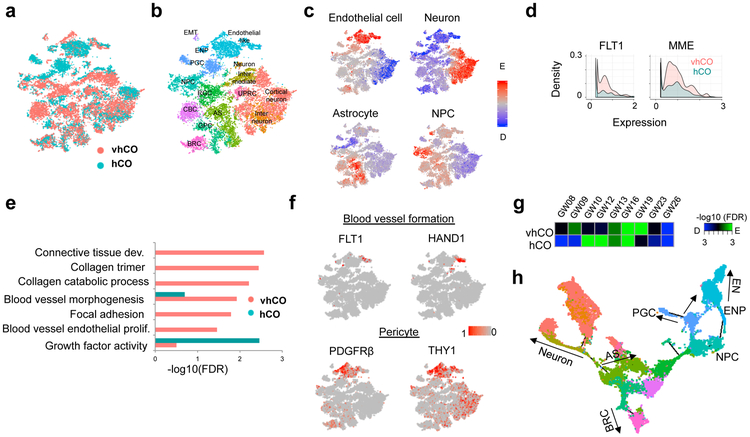
To examine the global differences between control and vascularized cortical organoids, we performed the single-cell transcriptome analysis of a total of 9,748 and 10,278 cells derived from vhCOs and control hCOs, respectively, at day 70 (Fig. 2a). A total of 26 clusters were identified by unbiased K-mean clustering from a combination of all cells from both organoids, which were systematically assigned to 15 distinct cell types, including cortical neuron (CN), interneuron (IN), astrocyte (AS), radial glia cell (RGC), glial progenitor cell (GPC) and neuronal progenitor cell (NPC) (Fig. 2b and Supplementary Fig. 3a-c) 20,21. Our scRNA-seq indicated a minimal doublet rate and a limited bias of unique molecular index (UMI) among clusters (Supplementary Fig. 3d and e). We detected an endothelial-like cell cluster (EN), which is mainly composed of both vhCO and control hCO-derived cells. These clusters display significant enrichment for endothelial cell-specific gene signatures (Fig. 2c) 20,22, and several neuroepithelial markers (VIM and HES1) 23 which also act as vascular remodelers (Supplementary Fig. 3c). Analysis of cells within EN cluster revealed that vhCOs showed dramatic expression of several vasculogenesis factors (FLT1, HAND1, MME, and VTN), pericyte markers (PDGRFβ and THY1), collagens and cell adhesion-related genes; control hCOs lacked the expression of these markers (Fig. 2d-f and Supplementary Fig. 3f) 24. Although some control hCO-derived cells are close to the endothelium, these results suggest that ETV2 is critical for vascular endothelial cell maturation and vascular morphogenesis in vhCOs.
Figure 2. Single cell analysis of vhCOs.
(a-b) t-distributed Stochastic Neighbor Embedding (tSNE) plot of single cells distinguished by a. organoid type and b. cell annotation. RGC: radial glia cell, GPC: glia progenitor cell, NPC: neuronal progenitor cell, CBC: Cilium-bearing cell, BRC: BMP signal-related cell, EN: endothelial-like cell, ENP: endothelial-like progenitor, PGC: proteoglycan-expressing cell, EMT: epithelial-mesenchymal transition-related cell, UPRC: unfolded protein response-related cell. Data are representative of 20026 cells. (c) Enrichment of gene signatures for endothelial cells, neuron, NPC, astrocyte and oligodendrocyte. Data are representative of 20026 cells. (d) Histogram of gene expression related to vasculogenesis. Two-sided T test p-value=2.96e-6 (FLT1) and 4.68e-14 (MME). Statistical significance was calculated by hypergeometric test and adjusted by Benjamini-Hochberg procedure. Data are representative of 2257 cells. (e) GO enrichment for differentially-expressed genes in endothelial-like clusters between vhCO and control hCO. Data are representative of 2257 cells. (f) tSNE plot showing gene expression related to blood vessel formation and pericyte. Data are representative of 20026 cells. (g) Estimation of neurodevelopmental stage by enrichment of vhCO- and control hCO-specific genes. (h) Monocle-based trajectory analysis of vascularized organoid, vhCO.
To further address whether organoid vascularization promotes neuronal maturation, we compared the single-cell transcriptome profiles of the neuronal clusters from vascularized organoids with those from developing human brains (GW08–23) 25. Gene Set Enrichment Analysis revealed that vhCOs-derived neurons resemble in vivo neurons corresponding to gestational week 16–19 (Fig. 2g). Critically, control hCOs-derived neurons from the same timepoint resemble an earlier stage (GW10–12) than vhCOs-derived neurons (Fig. 2g). In combination with the above immunostaining and electrophysiological analysis (Fig. 1g and Supplementary Fig. 2e), the scRNA-seq results further support our findings that vascularization accelerates the functional maturation of neurons. In addition, we constructed a differentiation trajectory from the single-cell transcriptome profile, and proteoglycan-expressing cells (PGC) were grouped into one branch with endothelial-like cells suggesting their potential to differentiate toward the endothelial lineage (Fig. 2h and Supplementary Note 3).
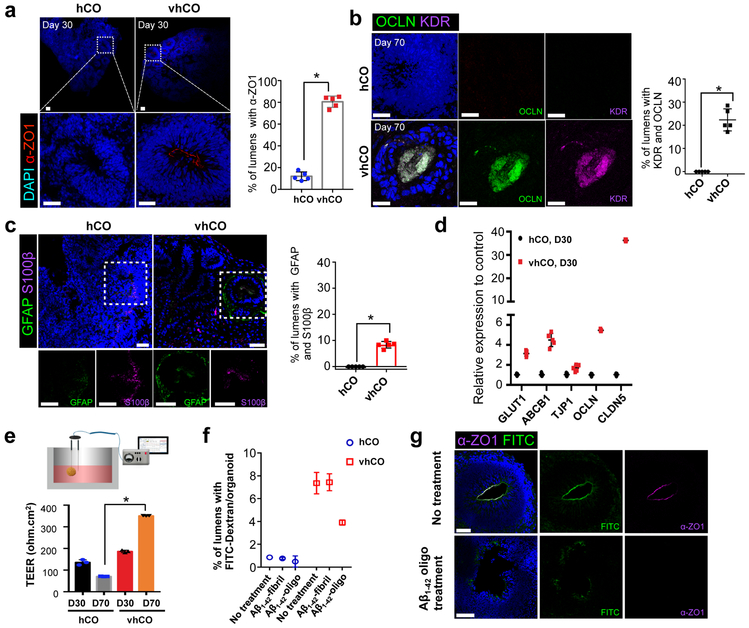
The human embryonic brain starts to develop blood-brain barrier (BBB) via specialized endothelial cells that generate the perineural vascular plexus and connect cerebral capillaries by tight junctions, the major characteristics of BBB 26,27. In addition to the expression of EC markers (Fig. 1b), we noted that most vhCOs are positive for the tight junction marker α-ZO1 in the lumens (Fig. 3a). Around 90% of lumens of vhCOs were stained for α-ZO1 compared to 18% in control hCOs (n=4, p<0.001, Fig. 3a). At day 70, lumens of VZ in vhCOs were stained for α-ZO1 while control hCOs lacked this unique tight junction staining at the lumens (Supplementary Fig. 4a). Similar α-ZO1 staining at the lumens was observed in hETV2-expressing H1-derived hCOs (Supplementary Fig. 2a). Both control hCOs and vhCOs showed a similar expression pattern of β-catenin at the lumens of VZ (Supplementary Fig. 4b). In addition to α-ZO1, occludin (OCLN) and KDR were co-stained in vhCOs at day 70 (Fig. 3b). The vhCOs showed clear but sparse OCLN and KDR co-staining at day 30 (Supplementary Fig. 4c). Control hCOs failed to express OCLN and KDR (Fig. 3b). Furthermore, glial cells marked by glial fibrillary acidic protein (GFAP) and glial-specific calcium binding protein B (S100β) were present at the lumen of VZ in vhCOs at day 70 (Fig. 3c and Supplementary Fig. 4d). Pericytes are an essential part of a neurovascular unit and contribute to vessel stability in BBB 28. The interesting BBB immunostaining results prompted us to determine if pericytes were produced in these vhCOs. Co-staining of pericyte marker, PDGFRβ, and astrocytic marker, GFAP, identified pericytes, which surrounded astrocytes but did not colocalize with them. Control hCOs did not contain any pericyte staining (Supplementary Fig. 4e). Lastly, vhCOs demonstrated increased expression of the other tight junction-related genes, including claudin-5 (CLDN5), TJP1 and efflux transporter (ABCB1) and BBB glucose transporter (GLUT1) (Fig. 3d). Collectively, hETV2 induction gave rise to the distinct expression of tight junction markers, astrocytic and pericytic proteins, and transporters, mimicking a BBB-like phenotype, albeit a structural difference from naturally formed BBB.
Figure 3. The vhCOs demonstrate BBB characteristics.
(a) Left, immunostaining showed that α-ZO1 was present in most lumens of vhCOs. Right, quantification of lumens with α-ZO1. Data represent the mean ± SEM (n=5, from three independent batches). Unpaired two-tail t-test was used for comparison (T=10 DF=2 and *p<0.0001). (b) Top, co-immunostaining for OCLN and KDR indicates that they are co-localized at the lumen of vhCOs day 70. Bottom, quantification of lumens stained with both OCLN and KDR. Data represent the mean ± SEM (n=5, from three independent batches). Unpaired two-tail t-test was used for comparison (T=9.77 DF=4 and *p=0.0006). (c) Left, co-immunostaining for GFAP and S100β indicates that they are co-localized at the lumen of vhCOs at day 70. Right, quantification of lumens stained with both GFAP and S100β. Data represent the mean ± SEM (n=5, from three independent batches). Unpaired two-tail t-test was used for comparison (T=16 DF=4 and *p=0.00009). (d) Expression of markers for tight junction and transporters from control hCOs and vhCOs at day 30. Data represent the mean ± SEM (n=5, from three independent batches). (e) Left, depiction of TEER analysis from hCOs. Right, TEER was measured in hCOs at day 30 and 70. TEER was sharply increased from vhCOs at day 70. Data represent the mean ± SEM (n=3, from three independent batches). (f) Aβ1-42 treatment of hCOs induces a decrease in FITC-dextran filled lumens. Organoids were incubated with 10 μM Aβ1-42 (Aβ1-42-fibril and Aβ1-42-oligo) for 48h, and then FITC-dextran perfusion was performed. FITC-dextran filled lumens were significantly decreased in vhCOs treated with Aβ1–42-oligo. Data represent the mean ± SEM (n=6, from three independent batches, *p<0.000001). (g) Immunostaining for tight junction marker α-ZO1 in the FITC-dextran perfused vhCOs with or without Aβ1-42-oligo treatment. Data are representative of three independent experiments. The scale bar represents 50 μm in a, b, c and g.
As noted, one critical characteristic of the BBB is a tight junction barrier formed by the connected endothelial cells with tight junction proteins. To measure tight junction integrity, we used trans-endothelial electrical resistance (TEER) analysis. We measured TEER in hCOs by locating micro-electrode probes in three different regions of spheres (Fig. 3e, left panel). At 30 days, TEER in vhCOs (186 ±4 Ω cm2) was significantly increased compared to control hCOs (135 ±10 Ω cm2) (Fig. 3e, right panel). The vhCOs grown for 70 days in culture exhibited much higher TEER (351 ±10 Ω cm2) than controls (71 ±1 Ω cm2) (Fig. 3e, right panel). The elevated expression of tight junction markers and the continuous cell contact via α-ZO1 stain observed in vhCOs likely (Fig. 3a and Supplementary Fig. 4a) resulted in higher TEER. The TEER measurement we observed is lower than monolayer-derived BBB models reported before (~1500 Ω cm2)29. However, 3D constructed BBB models30 showed similar TEER values (~50 Ω cm2) to those measured in our system. Collectively, vhCOs not only exhibited vascular-like phenotypes and the expression of endothelial and BBB markers, but they also contain tight junctions comparable to previously described 3D BBB models.
In addition to the physical features, we further evaluated the biological function of vascular-like structures in vhCOs. We developed a neurotoxicity model in cortical organoids by assessing the deposition of amyloid-β (Aβ) peptide species. The accumulation of Aβ1–42 leads to malformation in tight junctions and the subsequent disruption of BBB through matrix metalloproteinases (MPPs) 31-33. We examined the effects of Aβ1–42 species, including Aβ1–42-oligo and Aβ1–42-fibril, on vascular structure in hCOs using the FITC-dextran perfusion assay. The Aβ1–42-oligo treatment led to decreased FITC-dextran filling and depletion of α-ZO1 signal in lumens of vhCOs, whereas Aβ1–42-fibril treatment did not affect FITC-dextran perfusion (Fig. 3f and g). Similar to an earlier study 31, Aβ1–42-fibril did not damage tight junctions as effectively as Aβ1–42-oligo, and this is supported by FITC-dextran leakage in vhCOs (Fig. 3f and Supplementary Fig. 4f). Collectively, these results suggest that hETV2 induction facilitates the formation of functional endothelial tight junctions and BBB-like characteristics.
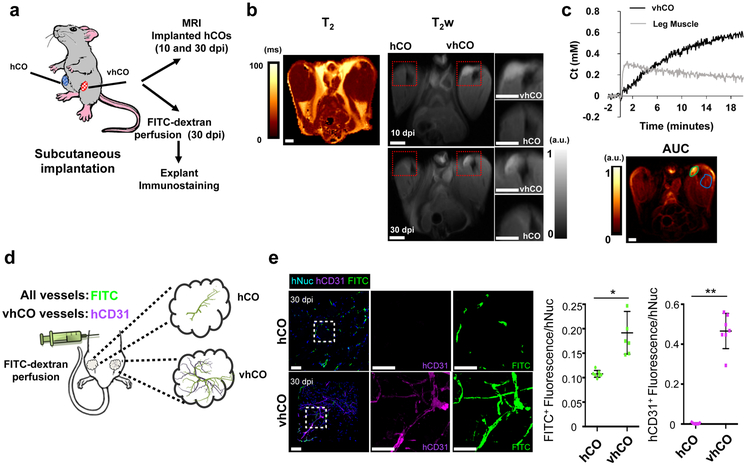
Lastly, we examined whether control hCOs and vhCOs form functional vasculature in vivo. We implanted hCOs subcutaneously into the hind limbs of immune-deficient mice, and subsequently performed magnetic resonance imaging (MRI) on the xenograft (Fig. 4a). The transplanted vhCOs relative to the surrounding muscle and skin tissues showed a high contrast, weighted by the transverse relaxation time (T2), while the control hCO was not clearly defined both at 10 and 30 dpi (Fig. 4b). Since vascularization is critical for the survival of implanted organoids, control organoids, lacking in vitro generated vasculature, exhibited faint contrast relative to the surrounding muscle and skin tissues. Additionally, using dynamic contrast enhancement with contrast weighted by the longitudinal relaxation time (T1) shortening induced by gadolinium to assess permeability through the vasculature. The vhCOs exhibited slow but irreversible uptake, indicating a highly vascularized, but less permeable tissue compared to the surrounding hind limb muscle (rapid uptake with backflow; Fig. 4c). At 30 day after organoid transplantation, mice were perfused with FITC-dextran to validate whether hCOs formed functional blood vessels in vivo (Fig. 4d). Immunostaining for human-specific CD31 and FITC revealed that the functional vasculature in vhCOs were formed and connected to the mouse vasculature; this was absent in control hCOs (Fig. 4e and Supplementary video 4). In addition, vhCOs not only contained significantly elevated hCD31+ vascular networks, but also a higher percentage of FITC-dextran filled vessels in total (Fig. 4e and Supplementary videos 5-7). Control organoids exhibited limited FITC-dextran filled host vessels, which corroborated the MRI data and indicated that control hCOs lacked a functional vascular connection to the host mouse (Fig. 4b and e). Overall, in vitro-generated vasculature-like network in hETV2 organoids was essential for blood flow and connected with host blood network.
Figure 4. The vhCOs possess functional vascular system.
(a) Depiction of subcutaneous implantation of control hCOs and vhCOs in the right and left leg of immune-deficient mice. (b) Left, in vivo T2 map of the implanted control hCOs and vhCOs. Right, anatomical image of hCOs after 10- and 30-day post-implantation (dpi). Both images detected the vhCO region, but area of implanted control hCO was not apparent. Data are representative of three independent experiments. (c) Top, tissue concentration of gadolinium contrast agent as a function of time in the left leg muscle (gray trace) and implanted vhCOs (black trace). Muscle tissue indicates a rapid uptake and backflow into the vasculature, but vhCOs showed a slower and irreversible uptake. Bottom, map of the area under the curve (AUC) of the concentration curve, with ROIs outlined for the vhCOs (green) and muscle (blue). Data are representative of three independent experiments. (d) Schematic of the method for FITC-dextran perfusion. Host blood vessels are filled with FITC-dextran, shown green, and endogenous vessels in vhCOs were shown as magenta. (e) Explanted organoids from FITC-perfused mice were stained for human-specific CD31 and hNuclei at day 30 dpi. The scale bar represents 50 μm, n=3 animals and 3 organoids from three independent batches for MRI and n=7 animals and 7 organoids from four independent batches for FITC-perfusion (*p=0.00412, **p=0.000183, the unpaired two-tail t-test was used for all comparisons). Mean ± SEM are shown.
In conclusion, hETV2 induced reprogramming of EC cells in organoids is a robust method to generate vhCOs with functional vasculature-like network, thereby offering a valuable platform to investigate brain development and disease mechanisms. These vhCOs are a new model system that may facilitate a more accurate physiological representation of the brain by exhibiting the interplay of neural and endothelial cells and by reducing the apoptotic and hypoxic condition of the interior tissues that damages typical, avascular organoids.
METHODS
Animals
The Rag2−/− GammaC−/− mice were purchased from Jackson Laboratories. All animal experiments described in this study were approved by the Institutional Animal Care & Use Committee (IACUC) of Yale University.
hESCs culture
HES-3 NKX2–1GFP/w and BC4 human ESCs were cultured on Matrigel (BD Biosciences) coated dishes with mTeSR1 media (Stem Cell Technologies) and passaged every week by treatment with 0.83 U/ml Dispase (Stem Cell Technologies). All experiments including hESCs were approved by Yale Embryonic Stem Cell Research Oversight (ESCRO).
Generation and differentiation of BC4 hESCs
As described previously 34, doxycycline-inducible (BC4) dCAS9-mCherry and rTTA were introduced into the AAVS1 locus of HES-3 NKX2–1GFP/w. Briefly, 2 million HES-3 NKX2–1GFP/w cells were electroporated with 8 μg donor plasmid, 1 μg AAVS1 TALEN-L and 1 μg AAVS1 TALEN-R by using the Amaxa Nucleofector device (AAB-1001, Lonza) and seeded in mTeSR1 plus Y-27632 (10 μM). After 3 days, G-418 (Thermo Fisher Scientific) was applied for 7 days (400 μg/ml for the first 3 days and 300 μg/ml for the next 4 days) to obtain stable colonies. A single isogenic colony was picked and expanded. BC4 was differentiated with the induction of hETV2 in differentiation conditions: 1) EB differentiation media (IMDM containing 15%FBS, 200 μg/ml transferrin, 0.05 ng/ml ascorbic acid, 1 mM sodium pyruvate, 100 U/ml penicillin/streptomycin, 2 mM glutamine and 450 μM monothioglycerol), 2) Neuronal differentiation media (DMEM-F12, 15% (v/v) KSR, 5% (v/v) heat-inactivated FBS (Life Technologies),1% (v/v) Glutamax, 1% (v/v) MEM-NEAA, 100 μM β-Mercaptoethanol, 10 μM SB-431542, 100 nM LDN-193189, and 2 μM XAV-939), and 3) EC differentiation protocol (first cells differentiated to mesoderm in N2B27 supplemented with 8 μM CHIR99021 and 25 ng/ml BMP4, and then cells were converted into EC cells in StemPro-34, 200 ng/ml VEGF-A and 2 μM forskolin) as described previously 35.
Electric cell-substrate impedance sensing (ECIS) analysis
ECIS assay was utilized to evaluate 2D-endothelial differentiated cells barrier function, as described previously 36. The wells of ECIS chamber slides were functionalized with 1 mM cysteine and coated with Matrigel at 37 °C for 1.5 hours, respectively. Chamber slides were inserted into the ECIS device (8W10E+, Applied Biophysics, Troy, NY) that was situated within a cell culture incubator. BC4 hESCs were inoculated into the chambers, and impedance was recorded in real-time during the 3 different differentiation protocols described above.
Generation of human cortical organoids (hCOs) with hETV2 induction
For the generation of endothelial cell containing hCOs, BC4 hESCs were infected with the lentivirus containing the FUW-tetO-hETV2 as described previously 11. The lentivirus containing inducible hETV2 was transduced for 7 days in the absence of dox. As we described earlier 20, hCOs were generated by mixing hETV2-infected BC4 and non-infected parental HES3 hESCs. Briefly, hESCs were dissociated into single cells via Accutase and 9000 cells, containing 5%, 10% or 20% hETV2 infected BC4 hESCs, were plated into a well of U-bottom ultra-low-attachment 96-well plate in neural induction media (DMEM-F12, 15% (v/v) KSR, 5% (v/v) heat-inactivated FBS (Life Technologies),1% (v/v) Glutamax, 1% (v/v) MEM-NEAA, 100 μM β-Mercaptoethanol, 10 μM SB-431542, 100 nM LDN-193189, 2 μM XAV-939 and 50 μM Y27632). Basal activation of hETV2 was started on day 2 by adding 0.5 μM dox, and FBS and Y27632 were removed from day 2 and 4, respectively. At day 10, organoids were transferred to ultra-low-attachment 6-well plate in hCO media with minus vitamin A (1:1 mixture of DMEM-F12 and Neurobasal media, 0.5% (v/v) N2 supplement, 1% (v/v) B27 supplement without vitamin A, 0.5% (v/v) MEM-NEAA, 1% (v/v) Glutamax, 50 μM β-Mercaptoethanol, 1% (v/v) Penicillin/Streptomycin and 0.025% Insulin) for spinning culture and the media was changed every other day. After day 18, the media was changed to hCO media with vitamin A (the same composition as described above except B27 with vitamin A was used and 20 ng/ml BDNF and 200 μM ascorbic acid were supplemented). The media was changed every 4 days after day 18. Activation of hETV2 was performed beginning on either day 10 or day 18 by adding 2 μM dox continuously in the media.
Live Imaging of hCOs
Live images were captured in hCOs at day 30 and day 70 to visualize the mCherry expressing cells and structures. The Leica TCS SP5 confocal microscope, equipped with a controlled cell chamber possessing 37 °C temperature and 5% CO2, was used to generate z stack images. 3D reconstruction of images was attained by using Leica Las-X software.
FITC-Dextran Perfusion
In vitro capillary network is developed by using 2 Cole Palmer silicone tubing (size L/S160 connected to Master-flex peristaltic pump. Organoids were located in between two EZFlow Syringe filters (MedLab) that were attached to the silicone tube. Perfusion of fluorescein isothiocyanate (FITC)-dextran (Sigma, average molecular weight) was performed for 1hour under constant flow rate (0.88 ml/min). All experiments were performed in the dark and organoids were fixed as described below for characterizing the FITC localization.
Electrophysiological recordings
The control hCOs and vhCOs (D80-D90) were cut into two halves in chilled oxygenated slicing medium containing (in mM) 85 NaCl, 75 sucrose, 2.5 KCl, 25 glucose, 1.25 NaH2PO4, 4 MgCl2, 0.5 CaCl2, and 24 NaHCO3. They were kept in the same solution at room temperature until recording. We performed whole-cell patch-clamp recordings using artificial cerebrospinal fluid containing (in mM) 126 NaCl, 2.5 KCl, 26 NaHCO3, 2 CaCl2, 2 MgCl2, 1.25 NaH2PO4, and 10 glucose) gasses with 95% O2/5% CO2. Cortical cells in hCOs were visualized using an upright microscope (Eclipse FN1; Nikon Instruments Inc.: Melville, NY, U.S.A.) with infrared differential interference contrast optics and water immersion 60X objective. Borosilicate glass pipettes (5–7 MΩ) were filled with intracellular solution containing (in mM) 126 K-gluconate, 4 KCl, 10 HEPES, 4 ATP-Mg, 0.3 GTP-Na, and 10 phosphocreatine (pH 7.2 and osmolality of 290 mOsm). They were used to record activities from cortical neurons. MultiClamp700B amplifier and Digidata 1440A digitizer (Molecular Devices: San Jose, CA, U.S.A.) were used for recordings. Voltage signals were filtered at 3 kHz using a Bessel filter and digitized at 10 kHz. We used the Clampfit 10 software (Molecular Devices: San Jose, CA, U.S.A.) to analyze the data.
Electron Microscopy
The cultured organoids were fixed in 2% glutaraldehyde and 0.1 M Na-cacodylate (pH 7.4), at different time points, for 1 hour at RT. The organoids were post-fixed via incubation in 1% osmium tetroxide for 1 h, staining en bloc in 2% aqueous uranyl acetate, dehydration in ethanol and propylene oxide and infiltration with Embed 812. After the hardening of the blocks, 60 nm sections were generated, post-stained with uranyl acetate and lead citrate, and analyzed by EM.
Transendothelial electrical resistance (TEER) analysis
The TEER 3-D organoids were measured by locating micro-electrode probes in three different regions of the spheres. TEER measurements were performed by using an electrochemical impedance analyzer (Z100, eDAQ). For each control hCO and vhCO (day 30 and day 70, n=3), three different measurements were performed and Z navigator was used to produce EIS data from the organoids. ZMAN software, both metal and battery supercapacitor circuits, was used to analyze the data. Finally, resistivity is calculated as follow:
TEER value (Ω cm2) = TEERaverage (Ω, ZMAN software) ⅹ hCO surface area (cm2, 4πr2organoid)
Library Preparation of ScRNA-seq
hCOs at 70-days old were randomly obtained from 3 different culture dishes (10 hCOs pooled together). As described previously 20, organoids were first dissociated via the papain dissociation system according to the manufacturer’s instruction. After washing with HBSS, hCOs were dissected into small pieces in papain solution and oxygenated with 95% O2 :5% CO2 for 5 min. Then, the tissue was oxygenated for 5 min and incubated at 37 °C for 1 hour. After generating single-cell suspension via trituration, single cells were suspended in 1% BSA/PBS supplemented with 10 μM Y27632 and stained for propidium iodide (PI). FACS sorted PI cells were re-suspended in 0.04% BSA/PBS (128 cell/μl) and used to generate cDNA libraries by using the Single Cell 3’ Reagent Kits. In short, cells were partitioned into nanoliter-scale Gel Bead-In-Emulsions (GEMs), and microfluidic cells were streamed at limiting dilution into a stream of Single Cell 3’ Gel Beads and then a stream of oil. After cell lysis, primers, an Illumina P7 and R2 sequence, a 14 bp 10xBarcode, a 10 bp randomer, and a poly-dT primer were released and mixed with the cell lysate and a bead-derived Master mix. In each individual bead, full-length cDNA from poly-adenylated mRNA was generated. After individual droplets were broken down, homogenized and cleaned from the remaining non-cDNA, the libraries were size-selected followed addition of R2, P5, P7 sequences to each selected cDNA. Finally, each library was sequenced using the Illumina HiSeq4000 2×150 bp in Rapid Run Mode.
Data processing of ScRNA-seq
Alignment to human (hg19) genome, initial quality control and UMI counting per Ensembl genes were processed by CellRanger with the count function by default parameters (v2.1.0). Then, all libraries were normalized to the same sequencing depth and analyzed for dimensional reduction and K-means clustering with “aggr” function of CellRanger by default parameters. Batch effect was then minimized by Seurat (v2.2.1)37. Each organoid type’s raw UMI count was normalized to total UMI with log-normalization and scaled by a factor of 10,000. Highly variable genes were identified with more than one dispersion and an average expression of 0.1–8. Canonical correlation analysis (CCA) was then performed with common variable genes from both types of the organoid. Single-cell transcriptome profiles from vascularized and non-vascularized organoids were merged into CCA subspace using the 1st to 20th CCs. For dimensional reduction, tSNE was implemented with the 1st to 20th CCs. Finally, cell clusters were identified by a shared nearest neighbor method by FindClusters function with “k.param=30, resolution=1.5, reduction.type=”cca”” options. Differentially-expressed genes in each cluster were identified by 1.5- fold change and p<0.05 with two-sided T-test. Gene Ontology analysis was implemented to the differentially-expressed genes by GOstats (v2.36.0) in Bioconductor package. Multiple statistical tests were adjusted by the Benjamini-Hochberg method using the p.adjust function in R. A false discovery rate (FDR) < 0.05 was used as statistical significance. Cell type labels were assigned to each cluster by cell-type specific gene signatures, significant GO terms, genes involved in specific cellular events, neurotransmitter transporter and synthetic enzymes as described previously (Supplementary Fig. 3b) 20,21. Further scRNA-seq analyses were described detail in Supplementary Note 4.
Aβ1-42 peptide generation and treatment
Amyloid-β protein fragment 1–42 (Aβ1–42, Sigma) was used to prepare fibrils (Aβ1–42-fibril) and oligomers (Aβ1–42-oligo) as described previously 31. Briefly, 2 mM stock was prepared by dissolving Aβ1–42 in dimethylsulfoxide (DMSO, Sigma). For generating fibrils, 2 mM Aβ1–42 was diluted to 100 μM by adding 10 mM HCl and vortexed for 30 seconds followed by 24h incubation at 37 °C with continuous shaking. For oligomer conditions, 2mM Aβ1–42 was diluted in neural differentiation media to 100 μM vortexed for 30 seconds followed by 24h incubation at 4 °C. After generating fibrils and oligomers, 10 μM Aβ1–42 peptide variants were incubated with control hCOs and vhCOs for 2 days. Then, the effects of Aβ1-42 on FITC-Dextran perfusion and tight junction marker expression were analyzed.
Subcutaneous implantation of hCOs
Control and hETV2-infected hCOs grown for 40–50 days were embedded with Matrigel and used for implantation. Mice were placed into a chamber by providing 2% isoflurane for anesthetization. Then, mice were fixed in a laminar hood to cut a small incision at each back leg. Matrigel-embedded control hCOs and vhCOs were subcutaneously placed into the incision of the right and left back legs, respectively. Once organoids were inserted, the wound was closed with sutures and buprenorphine (0.1 mg/kg) was administered for pain relief. Subsequently, implanted organoids were analyzed via MRI analysis and immunofluorescence staining.
FITC-dextran perfusion into mice containing subcutaneous implanted hCOs
After 30-days of organoid implantation, mice were injected with 15 mg/ml FITC-dextran via perfusion device. Briefly, the perfusion device was inserted in the left ventricle, and then we punctured the right ventricle. FITC-dextran was immediately injected into mice with a 5ml/min flow rate via a peristaltic pump. After monitoring the color change in the blood vessels of mice (around 3–5 min), the mice brain, vhCO and control hCOs were explanted and imaged for FITC via confocal microscopy. Then, explanted tissues were fixed and sliced for further immunofluorescence staining.
Immunofluorescence staining
For explanted organoids, residual Matrigel was removed by washing with PBS. As described earlier 20, all organoids were fixed in 4% paraformaldehyde (PFA) at 4°C overnight following 3 washes with PBS at RT. Then, organoids were transferred to a 30% sucrose solution for 2 days at 4°C. For FITC-Dextran perfused hCOs, organoids were fixed in 4% PFA at RT for 15 minutes, and incubated in 30% sucrose at 4°C for 1 day in dark. Organoids were equilibrated with O.C.T compound at RT for 15 min, transferred to base molds, and embedded in O.C.T compound on dry ice. 40-μm cryosections were generated and organoid blocks were stored at −80°C. Slides were dried for 2 hours at RT and incubated with 0.1% Triton-100 for 15 min at RT in a humidified chamber. 3% bovine serum albumin (BSA) was used for blocking at RT for 2 hr, and then organoids were incubated with a primary antibody diluted in 3% BSA overnight at 4°C. After 2 washing steps, organoids were incubated with Alexa Fluor Dyes (1:1000) for 1 hr following nuclei staining with DAPI (1:1000). Finally, slides were mounted with ProLong Gold Antifade Reagent and images were taken with Leica TCS SP5 confocal microscope. The tunnel assay (C100247, Invitrogen) was performed to detect apoptotic cells following the manufacturer’s protocol. A list of antibodies is presented in Supplementary Table 1.
Whole-mount Immunostaining of organoids
We performed whole-mount immunostaining followed by confocal microscopy to examine the localization and organization of endothelial cell networks within the cortical organoids. Organoids were washed with PBS and fixed overnight in 4% paraformaldehyde (PFA) at 4°C. After washing the organoids with PBS for 5 hours, the organoids were blocked overnight at RT in 0.5% BSA and 0.125% Triton-100 in PBS. Organoids were incubated in primary antibodies (anti-Cd31 1:100, anti-MAP2 1:200) and diluted in 0.5% BSA and 0.125% Triton-100 in PBS for 2-days at 4°C. Unbound antibodies were removed via washing with PBS for one day at RT. Then, organoids were incubated with Alexa Fluor Dyes (1:500) for 4 h following nuclei staining with DAPI (1:1000) for 2 hours. After washing with PBS for several hours, clearing of the organoids was performed as described previously 38. The organoids were applied to a sequential dehydration series of 30%, 50%, 70% and 99% 1-Propanol (diluted in PBS pH adjusted to 9.5 via triethylamine) for 4 hours at 4°C. After dehydration, the organoids were incubated with ethyl cinnamate for 1h at RT in light protected and air-sealed tubes.
Flow Cytometry
For flow cytometry analysis, EC cells generated via induction of hETV2 were dissociated with Accutase. Single cells were incubated with CD31 (Abcam, Ab119339, 0.5 μq/5*105 cells) for 30 min on ice. The cells were washed and subsequently incubated with secondary FITC-labelled antibody for 30 min on ice. Finally, the cells were washed, filtered and analyzed on the BD LSRII instrument.
MRI data acquisition and analysis
Images were acquired using an 11.74T horizontal-bore spectrometer with Bruker console, using a dual-coil setup (volume and surface coils in transmit-receive configuration; RAPID Biomedical GmbH). The animal was anesthetized using 1.5% isoflurane in oxygen, and body temperature was maintained at 35–37°C throughout the procedure using a circulating warm water pad. First, anatomical images were acquired using a T2-weighted turboRARE sequence with 4 averages and a RARE factor of 4 (effective echo time (TE) = 15 ms). Then, an intrinsic T1 map was fitted from a multi-recycle time (TR) acquisition (300, 700, 1000, 2000, 4000 & 8000 ms). Finally, a spoiled-gradient sequence was used for the dynamic contrast enhancement (flip angle = 15°, TR = 41.6ms, dual TE = 2.5/5ms), acquired every 4 seconds for 22 minutes, with a bolus injection of Gadobutrol in the tail vein (0.1 mmol/Kg) after 2 minutes. The dynamic signal change was converted to contrast agent concentration calibrated from voxel-level intrinsic T1, and an additional apparent T2 (T2*) correction from the second echo due to magnetic susceptibility of Gadobutrol, where the relaxivity of Gadobutrol (r1) was previously measured in vitro. The DCMRSoft is available on demand.
Real-time quantitative PCR (qPCR)
The whole organoids were used for total RNA isolation via RNeasy Mini Kit (Qiagen). 1 μg RNA was converted to cDNA using iScript Select cDNA Synthesis Kit. For the quantification of gene expression, qPCR was carried out on the CFX96 Real-Time PCR system (Biorad) using the SsoFast EvaGreen Supermix (Biorad). The PCR conditions were: 95 °C for 15 min, followed by 40 two-step cycles at 94 °C for 10 s and 60 °C for 45 s. A list of primers used in this study is presented in Supplementary Table 2.
Statistics
Data are presented as mean ± SEM. The paired or unpaired two-tail t-test (GraphPad Prism software version 7.0), hypergeometric test adjusted by Benjamini-Hochberg procedure, and two-sided t-test (R version 3.5.0 software) were used to determine the statistical significance. Statistical tests and biological replicates for each experiment are presented in the figure legends.
Data availability
Single-cell transcriptome data are available in the Gene Expression Omnibus under accession code: GSE134049. The data that support the findings of this study are available on request from corresponding author.
Reporting Summary
More information in detail on research design, antibodies, and cell lines is available in Life Sciences Reporting Summary linked to this article.
Supplementary Material
Acknowledgements
I.-H. P. was partly supported by NIH (GM111667–01, R01AA025080–01, R01CA203011–2), CSCRF (14-SCC-YALE-01, 16-RMB-YALE-04), Kavli Foundation, Simons Foundation, KRIBB/KRCF research initiative program (NAP-09–3). This work was supported by the College of Medicine, University of Arkansas for Medical Sciences to S.-H. L., Core Facilities of the Center for Translational Neuroscience, Award P30 GM110702 from the IDeA program at NIGMS, and by NIH (R01HL127759, DP3DK108245) and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), Republic of Korea (HI15C2782, HI16C2211) . Computation time was provided by Yale University Biomedical High Performance Computing Center.
Footnotes
Competing interests I.-H.P. serves on the scientific advisory boards of ELGEN Therapeutics and INSTEMCARE with financial interest. However, the work presented in this manuscript is not related to ELGEN Therapeutics or INSTEMCARE.
REFERENCES
- 1.Lancaster MA et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paşca AM et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nature methods 12, 671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qian X et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165, 1238–1254 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pașca SP The rise of three-dimensional human brain cultures. Nature 553, 437 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Quadrato G, Brown J & Arlotta P The promises and challenges of human brain organoids as models of neuropsychiatric disease. Nature medicine 22, 1220 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Heide M, Huttner WB & Mora-Bermudez F Brain organoids as models to study human neocortex development and evolution. Curr Opin Cell Biol 55, 8–16 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Lancaster MA & Knoblich JA Generation of cerebral organoids from human pluripotent stem cells. Nature protocols 9, 2329 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin X et al. Engineering stem cell organoids. Cell stem cell 18, 25–38 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen Q et al. Endothelial cells stimulate self-renewal and expand neurogenesis of neural stem cells. Science 304, 1338–1340 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Mansour AA et al. An in vivo model of functional and vascularized human brain organoids. Nature biotechnology 36, 432 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S et al. Direct reprogramming of human dermal fibroblasts into endothelial cells using ER71/ETV2 Circulation research, CIRCRESAHA; 116.309833 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee S, Kim JE, Johnson BA, Andukuri A & Yoon Y-S (Future Medicine, 2017). [DOI] [PMC free article] [PubMed]
- 13.Morita R et al. ETS transcription factor ETV2 directly converts human fibroblasts into functional endothelial cells. Proceedings of the National Academy of Sciences 112, 160–165 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelhardt B & Liebner S Novel insights into the development and maintenance of the blood–brain barrier. Cell and tissue research 355, 687–699 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hogan KA, Ambler CA, Chapman DL & Bautch VL The neural tube patterns vessels developmentally using the VEGF signaling pathway. Development 131, 1503–1513 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Norman MG & O’kusky JR The growth and development of microvasculature in human cerebral cortex. Journal of Neuropathology & Experimental Neurology 45, 222–232 (1986). [PubMed] [Google Scholar]
- 17.Paredes I, Himmels P & de Almodóvar CR Neurovascular Communication during CNS Development. Developmental cell 45, 10–32 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Jin K et al. Vascular endothelial growth factor (VEGF) stimulates neurogenesis in vitro and in vivo. Proceedings of the National Academy of Sciences 99, 11946–11950 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quadrato G et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature 545, 48 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiang Y et al. Fusion of Regionally Specified hPSC-Derived Organoids Models Human Brain Development and Interneuron Migration. Cell Stem Cell 21, 383–398 e387, doi: 10.1016/j.stem.2017.07.007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiang Y et al. hESC-Derived Thalamic Organoids Form Reciprocal Projections When Fused with Cortical Organoids. Cell Stem Cell 24, 487–497 e487, doi: 10.1016/j.stem.2018.12.015 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darmanis S et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A 112, 7285–7290, doi: 10.1073/pnas.1507125112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu B et al. The endothelial cell-specific antibody PAL-E identifies a secreted form of vimentin in the blood vasculature. Mol Cell Biol 24, 9198–9206, doi: 10.1128/MCB.24.20.9198-9206.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morikawa Y & Cserjesi P Extra-embryonic vasculature development is regulated by the transcription factor HAND1. Development 131, 2195–2204, doi: 10.1242/dev.01091 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Zhong S et al. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555, 524–528, doi: 10.1038/nature25980 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Obermeier B, Daneman R & Ransohoff RM Development, maintenance and disruption of the blood-brain barrier. Nature medicine 19, 1584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daneman R, Zhou L, Kebede AA & Barres BA Pericytes are required for blood–brain barrier integrity during embryogenesis. Nature 468, 562 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winkler EA, Bell RD & Zlokovic BV Central nervous system pericytes in health and disease. Nature neuroscience 14, 1398 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lippmann ES et al. Derivation of blood-brain barrier endothelial cells from human pluripotent stem cells. Nature biotechnology 30, 783 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho C-F et al. Blood-brain-barrier spheroids as an in vitro screening platform for brain-penetrating agents. Nature communications 8, 15623 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wan W et al. Aβ1–42 oligomer‐induced leakage in an in vitro blood–brain barrier model is associated with up‐regulation of RAGE and metalloproteinases, and down‐regulation of tight junction scaffold proteins. Journal of neurochemistry 134, 382–393 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Almutairi MM, Gong C, Xu YG, Chang Y & Shi H Factors controlling permeability of the blood–brain barrier. Cellular and molecular life sciences 73, 57–77 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deane R et al. RAGE mediates amyloid-β peptide transport across the blood-brain barrier and accumulation in brain. Nature medicine 9, 907 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Mandegar MA et al. CRISPR interference efficiently induces specific and reversible gene silencing in human iPSCs. Cell stem cell 18, 541–553 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patsch C et al. Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nature cell biology 17, 994 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan Y, Altalhi WA, Ng JJ & Courtman DW Derivation of human peripheral blood derived endothelial progenitor cells and the role of osteopontin surface modification and eNOS transfection. Biomaterials 34, 7292–7301 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Macosko EZ et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214, doi: 10.1016/j.cell.2015.05.002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masselink W et al. Broad applicability of a streamlined ethyl cinnamate-based clearing procedure. Development 146, doi: 10.1242/dev.166884 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell transcriptome data are available in the Gene Expression Omnibus under accession code: GSE134049. The data that support the findings of this study are available on request from corresponding author.