ABSTRACT
The tripartite motif (TRIM) proteins constitute a family of ubiquitin E3 ligases involved in a multitude of cellular processes, including protein homeostasis and autophagy. TRIM32 is characterized by six protein–protein interaction domains termed NHL, various point mutations in which are associated with limb-girdle-muscular dystrophy 2H (LGMD2H). Here, we show that TRIM32 is an autophagy substrate. Lysosomal degradation of TRIM32 was dependent on ATG7 and blocked by knockout of the five autophagy receptors p62 (also known as SQSTM1), NBR1, NDP52 (also known as CALCOCO2), TAX1BP1 and OPTN, pointing towards degradation by selective autophagy. p62 directed TRIM32 to lysosomal degradation, while TRIM32 mono-ubiquitylated p62 on lysine residues involved in regulation of p62 activity. Loss of TRIM32 impaired p62 sequestration, while reintroduction of TRIM32 facilitated p62 dot formation and its autophagic degradation. A TRIM32LGMD2H disease mutant was unable to undergo autophagic degradation and to mono-ubiquitylate p62, and its reintroduction into the TRIM32-knockout cells did not affect p62 dot formation. In light of the important roles of autophagy and p62 in muscle cell proteostasis, our results point towards impaired TRIM32-mediated regulation of p62 activity as a pathological mechanisms in LGMD2H.
KEY WORDS: TRIM32, p62/SQSTM1, LGMD2H, Autophagy, BBS11, Ubiquitylation
Highlighted Article: Mutations in the E3 ligase TRIM32 associated with the muscular dystrophy disease LGMD2H, impair its autophagic degradation and its ability to activate the autophagy receptor p62/SQSTM1.
INTRODUCTION
Protein quality control is crucial for cellular health, especially in tissues containing long-lived cells like the muscles and nervous system. Dysfunctional quality control leads to protein aggregation and development of diseases with increasing prevalence related to age (Dikic and Elazar, 2018). The two most important biological machineries controlling protein degradation are autophagy and the ubiquitin-proteasome system (UPS), where protein ubiquitylation mediated by E3 ubiquitin ligases provide substrate specificity. Based on their structural properties, E3 ubiquitin ligases are classified as RING, HECT or RBR ligases (Morreale and Walden, 2016). TRIM family proteins belong to the RING E3s, which constitute the most-abundant family of ubiquitin ligases (Hatakeyama, 2017). TRIMs are characterized by the highly conserved RING finger–B-box–coiled-coil domains at their N-termini, generally constituting their E3 ligase and oligomerization activity. Human TRIM family proteins comprise 82 members that are classified into 12 different subfamilies according to the domain structure of their C-terminal region (Fig. 1A). They have important functions in development, differentiation, immune responses and carcinogenesis (reviewed in Hatakeyama, 2017). Furthermore, certain TRIM proteins act as regulators of autophagy activity (Pizon et al., 2013; Tomar et al., 2012) and as autophagy receptors (Chauhan et al., 2016; Kimura et al., 2015, 2016; Mandell et al., 2016, 2014). TRIM32 is characterized by six repeats of NHL (NCL-1, HT2A and LIN-41) motifs in its C-terminus, which are thought to mediate protein–protein interactions (Slack and Ruvkun, 1998). TRIM32 is expressed throughout the body, but with high expression levels in the brain and heart (Frosk et al., 2002). The biological roles of TRIM32 are multi-faceted, impacting muscle physiology, cancer and immunity. This is reflected in the many substrates reported to be targeted by its E3 ligase activity, such as the muscular relevant proteins actin, α-actinin, desmin, tropomysin and dysbindin, the cell cycle regulators c-Myc, MYCN and p53, and the innate immunity adaptor STING (also known as TMEM173) (reviewed in Lazzari and Meroni, 2016). The finding that mutations in the NHL domains of TRIM32 can lead to the hereditary muscular disease limb-girdle muscular dystrophy 2H (LGMD2H), identified TRIM32 as an important player in muscular physiology (Frosk et al., 2002). Whether the pathogenic effect of LGMD2H is due to a role for TRIM32 in muscle atrophy (Cohen et al., 2012) or more in muscle homeostasis and regrowth after atrophy (Kudryashova et al., 2012, 2005; Nicklas et al., 2012; Servián-Morilla et al., 2019) is not well understood. However, it has been shown that the LGMD2H mutations lead to impaired self-oligomerization and auto-ubiquitylation, and reduced TRIM32 expression level (Albor et al., 2006; Kudryashova et al., 2011; Locke et al., 2009; Servián-Morilla et al., 2019). Moreover, recent work analyzing muscle biopsies and primary myoblasts from LGMD2H patients, described reduced proliferation and differentiation capacity, diminished satellite cell pool, and enhanced senescence and autophagy activity (Servián-Morilla et al., 2019). In addition, another very recent publication reported that TRIM32 is important for induction of autophagy in atrophic muscle cells, while the LGMD2H mutant was unable to activate autophagy in these cells (Di Rienzo et al., 2019). All mutations associated with LGMD2H are either deletions or point mutations located in the NHL domains, while a point mutation in the B-box of TRIM32 is associated with a completely unrelated disease, multisystemic disorder Bardet–Biedl syndrome 11 (BBS11) (Chiang et al., 2006). The pathogenic mechanism for the disease is not understood.
Fig. 1.
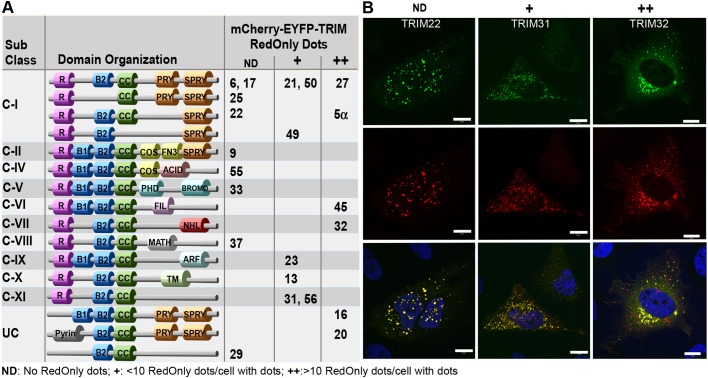
TRIM proteins from various subclasses are degraded in the lysosome. (A) Subclasses and domain organization of the 22 TRIM proteins analyzed in the double-tag screen. The ability of each mCherry–EYFP–TRIM protein to form RedOnly structures is indicated by ND (no RedOnly structures detected),+(<10 RedOnly structures in cells with fluorescent puncta), ++ (many RedOnly structures in cells with fluorescent puncta). R, Ring finger B1/B2: Bbox1/Bbox2; CC, Coiled coil. (B) Confocal images of mCherry–EYFP-tagged TRIM proteins representing each of the categories ND, + and ++. HeLa cells were transfected with mCherry–EYFP-tagged TRIM22, TRIM31 or TRIM32 1 day before fixation and imaged using a Zeiss780 confocal microscope and the Zen software. Blue, DAPI staining. Scale bars: 10 µm.
Autophagy is a cellular renovation system that uses lysosome-mediated pathways to degrade and recycle almost any type of cell content (Mizushima and Komatsu, 2011). Autophagy is described to act both as a non-selective bulk degradation pathway, and as a selective elimination of components such as aberrant protein aggregates, RNA bodies, lipid droplets, unwanted organelles and invading pathogens in a process termed selective autophagy (reviewed in Rogov et al., 2014). Selective autophagy provides quality control of cellular components, degrading damaged and harmful material during normal cellular conditions. When cells are stressed by hypoxia, ischemia, starvation or infection, selective autophagy is stimulated and specifically targets excess or toxic structures (Johansen and Lamark, 2011; Rogov et al., 2014). This allows the cell to adapt to the unfavorable conditions. Autophagy receptors are instrumental in the process. They bind specifically to the cargo and to ATG8 family proteins (such as LC3B, also known as MAP1LC3B) at the forming autophagosome, directing the cargo for degradation in the lysosome. The first selective autophagy receptor to be identified was p62, also known as sequestosome-1 (SQSTM1) (Bjørkøy et al., 2005). Ubiquitin labeling is one of the signals for cargo degradation via selective types of autophagy. Ubiquitylated protein aggregates containing autophagy receptors are evident in a range of human diseases. p62 contains a C-terminal ubiquitin-binding domain (UBA), a LC3-interacting region (LIR) and an N-terminal Phox and Bem1 (PB1) domain with an oligomerization property (Pankiv et al., 2007). The oligomerization activity of p62 is important for sequestering its cargo and for scaffolding phagophore membrane development (Bjørkøy et al., 2005; Itakura and Mizushima, 2011), described as a p62-mediated phase separation process dependent on ubiquitin (Sun et al., 2018; Zaffagnini et al., 2018). Recent studies have revealed that the autophagy activity of p62 itself is regulated by ubiquitylation (Heath et al., 2016; Jongsma et al., 2016; Lee et al., 2017; Pan et al., 2016; Peng et al., 2017). The UBA domain of unmodified p62 has a strong tendency to form an intermolecular dimer that spatially occludes ubiquitin binding (Isogai et al., 2011; Long et al., 2010). Ubiquitylation of several residues both inside and outside the UBA domain is reported to enhance the ubiquitin-binding activity and autophagy activity of p62 (Lee et al., 2017; Peng et al., 2017). Interestingly, several studies implicate p62 as an important player of protein homeostasis in muscle cells (Arndt et al., 2010; Lee et al., 2018; Rodriguez-Muela et al., 2018). Furthermore, proper regulation of autophagy is fundamental for muscle homeostasis, and dysregulation of autophagy has a pathogenic role in several forms of muscle diseases (Jiao and Demontis, 2017).
By employing a fluorescence-based double-tag screen of 22 TRIM proteins representing each of the TRIM subfamilies, we identified TRIM32 as a potential autophagy cargo protein. Interaction analysis in vitro and in cells revealed direct interaction and colocalization of TRIM32 and p62, while autophagy assays showed that p62 was able to mediate autophagic degradation of TRIM32. Conversely, ubiquitylation assays and proteomic analysis identified p62 as a TRIM32 substrate. TRIM32 mediated mono-ubiquitylation of p62 at residues previously shown to be important for the ubiquitin-binding activity of p62. By establishment of TRIM32-knockout (KO) and reconstituted cells, we show that TRIM32 facilitates p62 sequestration and autophagic degradation. Introduction of the LGMD2H disease mutation in TRIM32 inhibited its autophagic degradation, and also its ability to regulate p62 activity. In contrast, introduction of the BBS11 mutation in TRIM32 strongly facilitated p62 sequestration and degradation. Our results demonstrate a dual role for TRIM32 in autophagy, acting both as a substrate and as a positive regulator of p62. Importantly, the inactivity of the TRIM32 LGMD2H mutant points toward dysfunctional TRIM32 mediated regulation of p62 as a pathological mechanism in LGMD2H.
RESULTS
TRIM proteins from various subclasses are degraded in the lysosome
Recent studies have shown that certain TRIM proteins are implicated in the autophagy process, as regulators and as receptors in selective autophagy (reviewed in Di Rienzo et al., 2019; Hatakeyama, 2017; Kimura et al., 2017, 2016; van Gent et al., 2018). Furthermore, a few TRIM proteins seemingly are degraded by autophagy themselves, including TRIM50 (Fusco et al., 2012), TRIM30 (Choi et al., 2015) and TRIM5α (Mandell et al., 2016). Here, we employed the double-fluorescence-tag strategy (Pankiv et al., 2007) to identify TRIM proteins that could be degraded by autophagy, and hence that are potential as autophagy regulators and receptors. A total of 22 different TRIM proteins, representing 11 subclasses of the TRIM family, were fused to the double fluorescence tag mCherry–EYFP and expressed in HeLa cells. Since EYFP is unstable in acidic milieus with a pH below 6, while mCherry is stable, double-tagged proteins will only have red florescence when they are sequestered in the lysosome (denoted ‘RedOnly’ structures), which has a pH of ∼4.7. At 24 h after transfection, the cells were exposed to normal medium or were starved for 2 h in Hanks’ balanced salt solution (HBSS), before fixation and confocal microscopy imaging. To verify that the RedOnly structures represented lysosomal compartments, we analyzed, in parallel, cells treated with the lysosomal inhibitor Bafilomycin A1 (BafA1) for 4 h before fixation. BafA1 impairs the acidification of the lysosomes, and hence the quenching of EYFP localized in the lysosome. As presented in Fig. 1, 13 of the 22 TRIM proteins tested formed some RedOnly structures. Nine of these have previously been linked to autophagy, namely, TRIM20 and TRIM21 (Kimura et al., 2015), TRIM50 (Fusco et al., 2012), TRIM23 (Sparrer et al., 2017), TRIM13 (Tomar et al., 2012), TRIM31 (Ra et al., 2016), TRIM5α (Mandell et al., 2014), TRIM32 (Di Rienzo et al., 2019; Yang et al., 2017) and TRIM16 (Chauhan et al., 2016; Kimura et al., 2017). The observation that not all TRIM proteins form RedOnly structures may indicate that this is not a general trait of the conserved N-terminal RING finger–B-box–coiled-coil domains, or that degradation of certain TRIMs by autophagy is dependent on factors not present in HeLa cells. Furthermore, RedOnly structures were detected among TRIMs from many different subclasses (Fig. 1A), suggesting that it is not dependent on any specific domains in the C-terminal. However, four of the six TRIM proteins that gave a substantial amount of RedOnly structures both in normal medium and upon starvation conditions contain a SPRY domain in their very C-terminal end. Three of these, TRIM5α, TRIM16 and TRIM20, have been previously identified as autophagy receptors (Chauhan et al., 2016; Kimura et al., 2017; Mandell et al., 2014) and hence confirm our screening strategy. mCherry–EYFP–TRIM32 displayed a strong and reproducible formation of RedOnly dots in both normal and starved conditions (Fig. 1B). Interestingly, TRIM32 is reported to interact with the autophagy receptor TAX1BP1, and thereby mediate autophagic degradation of the TLR3/4 adaptor protein TRIF (Yang et al., 2017) and a recent report suggests autophagic degradation of the TRIM32 LGMD2H disease mutants (Servián-Morilla et al., 2019). However, whether TRIM32 is a substrate for autophagic degradation is unresolved.
TRIM32 interacts and colocalizes with the autophagy marker proteins LC3B and p62
The double tag assay clearly indicated lysosomal degradation of exogenous TRIM32. In order to determine whether endogenous TRIM32 is degraded via lysosomal pathways or via the proteasome, the lysosomal inhibitor BafA1 and the proteasomal inhibitor MG132 were applied on HEK293 cells grown in full medium or starved in HBSS for 4 h. The protein levels of endogenous TRIM32 during the various conditions was analyzed by western blotting and the band intensities quantitated and correlated to the level of PCNA (Fig. 2A). Treatment with the lysosomal inhibitor and the proteasomal inhibitor increased the amount of TRIM32 to a similar degree under starvation conditions, while the proteasomal pathway seems to be most important under normal cellular conditions. The increase of the LC3B II band (representing lipidated LC3B) upon BafA1 treatment, and the phosphorylation status of the mTOR-regulated translation initiation factor 4E (p4E-BP1) verified that the cells responded as expected to the lysosomal inhibitor and to the starvation medium. These data suggest that TRIM32 is degraded via the proteasomal and lysosomal pathways to a similar extent during starvation conditions, while the proteasomal pathway is dominant under normal conditions.
Fig. 2.
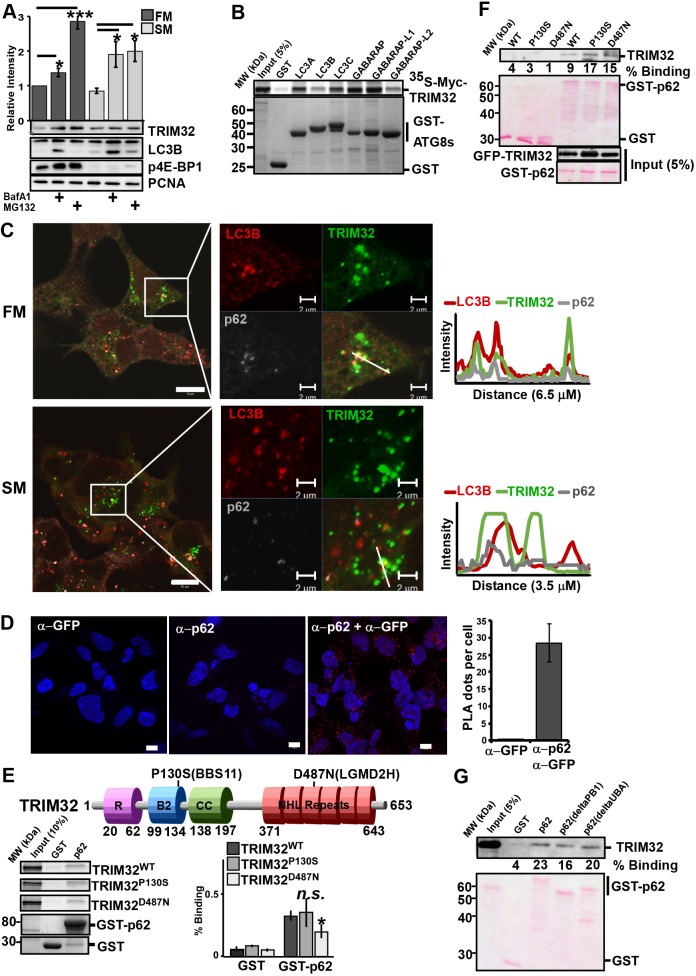
TRIM32 interacts and colocalizes with the autophagy marker proteins LC3B and p62. (A) HEK293 FlpIn cells were treated with BafA1 (200 nM) or MG132 (10 µM) for 4 h in HBSS (starvation medium, SM) or full medium (FM), before cell extracts were harvested in 1× SDS. 10 µg of the various cell extracts were separated on SDS-PAGE gels and blotted against the indicated antibodies. The bar graphs on top panel represent the mean± s.e.m. relative band intensities estimated by ImageJ from three independent experiments (n=3). *P<0.05, ***P<0.0005 (Student's t-test). (B) GST-pulldown assay using 35S-labeled Myc–TRIM32 and recombinant GST and GST–ATG8 proteins. The amount of Myc–TRIM32 bound to the various ATG8 proteins was detected by autoradiography. The assay was repeated with similar results. (C) HEK293 FlpIn EGFP-TRIM32 cells in full medium or starved in HBSS (2 h) were fixed and stained with antibodies for LC3B and p62. Images were obtained using a ZEISS780 confocal laser scanning microscope, and the colocalization monitored using the ZEN software. Scale bars: 10 µm (main images), 2 µm (magnifications). Intensity profiles along the indicated line are shown to the right. (D) Representative images of the Flp-In EGFP-TRIM32 cells analyzed by the proximation ligation assay DuoLink using antibodies against GFP and p62. The graph represents the mean±s.d. of colocalization dots using the Volocity software, from three independent experiments. Each experiment includes z-stack images of n=100 cells per condition. Scale bars: 10 µm. (E) The top panel shows a schematic of the TRIM32 domain organization with the localization of the disease point mutations P130S and D487N indicated. R, Ring finger; B2, Bbox2; CC, Coiled coil. The lower panels show GST-pulldown assays using 35S-labeled Myc–TRIM32WT, Myc–TRIM32P130S or Myc–TRIM32D487N and recombinant GST and GST–p62 immobilized on glutathione–Sepharose beads. Quantifications of the binding of wild-type and mutant constructs to GST–p62 are presented as percentage binding relative to input. The bars represent the mean±s.d. band intensities compared to input as quantified using ImageJ of three independent experiments. *P=0.025; n.s., not significant (Student's t-test). (F) GST-pulldown assay using cell extracts from FlpIn EGFP-TRIM32WT, EGFP-TRIM32P130S or EGFP-TRIM32D487N cells and GST–p62 immobilized on glutathione–Sepharose beads. EGFP–TRIM32 was detected using an anti-GFP antibody. GST proteins were visualized by Ponceau staining. Quantifications of the band intensities of the bound proteins compared to 5% input are indicated below the blot. (G) GST-pulldown assays using cell extracts from the FlpIn EGFP–TRIM32 cells and recombinant GST, GST-p62, or various GST-p62 deletion constructs immobilized on glutathione–Sepharose beads. Bound EGFP–TRIM32 was detected using anti-GFP antibody, while the GST proteins were visualized by Ponceau staining. Quantifications of the band intensities of the bound proteins compared to 5% input using ImageJ are indicated below the blot.
Several TRIM proteins are reported to interact with core components of the autophagy machinery, and to function as autophagy receptors and regulators of autophagosome formation (Kimura et al., 2015, 2016; Mandell et al., 2016, 2014). We applied in vitro GST-pulldown assays to determine whether TRIM32 has the ability to bind directly to the ATG8 family proteins. Fig. 2B shows that TRIM32 binds to LC3A (MAP1LC3A), LC3C (MAP1LC3C), GABARAP and GABARAP-L1, and weakly to LC3B and GABARAP-L2. However, we were not able to find any candidate LC3-interacting regions (LIRs) in TRIM32 with bioinformatic tools, and GST-pulldown assays with the N-terminal part of TRIM32 (the RING finger–B-box–coiled-coil domains) and the C-terminal part (NHL-domains) both displayed binding to GST–LC3B and GST–GABARAP (Fig. S1A). Further attempts to identify LIRs in TRIM32 did not give any consistent results (data not shown). We next tried to identify whether the interaction between TRIM32 and the ATG8 protein family was mediated by the newly characterized ubiquitin-interacting motif (UIM) docking site (Marshall et al., 2019). However, we were not able to identify a UIM motif in TRIM32. This suggests that TRIM32 can interact directly with ATG8 proteins in a non-LIR- and non-UIM-dependent way, suggesting that other or more complex interfaces are involved.
To further investigate the lysosomal degradation of the TRIM32 mutants, Flp-In T-Rex 293 cell lines with tetracycline-inducible expression of EGFP–TRIM32P130S and EGFP–TRIM32D487N were established (Fig. S1B). In accordance with previous reports, TRIM32 is normally a cytoplasmic protein forming distinct perinuclear bodies or aggregates (Fig. 2C; Fig. S1B) (Locke et al., 2009). Furthermore, western blots of the expressed EGFP fusion proteins revealed a slower-migrating form of EGFP–TRIM32 and the BBS11 mutant EGFP–TRIM32P130S, but not the LMGD2H mutant EGFP–TRIM32D487N (Fig. S1B). Co-staining the FlpIn EGFP–TRIM32 cells with antibodies against the autophagy marker protein LC3B and the autophagy receptor protein p62 showed that certain TRIM32 bodies colocalized with LC3B-positive p62 bodies (Fig. 2C). This indicates that a subpopulation of the TRIM32 bodies represent autophagosomes. Such bodies were observed both in full medium and in starvation conditions. To further evaluate whether TRIM32 interacts with p62 in cells, the DuoLink proximity ligation assay was employed to detect close colocalization of EGFP–TRIM32 and endogenous p62 in the Flp-In EGFP–TRIM32 cell line. The DuoLink assay revealed close to 30 distinct spots per cell (Fig. 2D). Together, these data show that TRIM32 is recruited to LC3B-positive p62 bodies, and is stabilized by treating cells with the lysosomal inhibitor BafA1. This strongly indicates that a subpopulation of TRIM32 proteins is degraded by autophagy.
Seven various mutations in the NHL repeat region of TRIM32 are associated with the recessive muscular dystrophy disease LGMD2H, with the TRIM32D487N mutation being the most common (Fig. 2E) (Frosk et al., 2002), while a point mutation in the B-Box2, TRIM32P130S, leads to the pleiotropic disorder BBS11 (Fig. 2E) (Chiang et al., 2006). We applied the in vitro GST-pulldown assay to verify the direct interaction between p62 and wild-type TRIM32 and its two disease mutants. p62 expressed in E.coli and immobilized on glutathione beads was added to in-vitro-translated Myc–TRIM32WT, Myc–TRIM32P130S or Myc–TRIM32D487N. The amount of bound proteins was analyzed by SDS-PAGE (Fig. 2E). Wild-type TRIM32 and both its disease mutants bound weakly to p62. The interaction between TRIM32 and p62 in cells was further confirmed by GST-pulldown assays using cell extracts from HEK293 cell lines expressing EGFP-tagged wild-type TRIM32 and its disease mutants (Fig. 2F), and by co-immunoprecipitation of endogenous p62 (Fig. S1D).
The PB1 domain and the UBA domain of p62 is important for p62 oligomerization and for binding to ubiquitylated proteins (Bjørkøy et al., 2005; Lee et al., 2017). In order to identify whether the PB1 domain or the UBA domain of p62 is implicated in the interaction with TRIM32, we performed GST-pulldown assays using p62 PB1 and UBA deletion constructs fused to GST and cell extracts from cells expressing exogenous EGFP–TRIM32. We found that deletion of the UBA domain or the PB1 domain did not affect the interaction with TRIM32, indicating that the interaction is not mediated by ubiquitin (Fig. 2G). These results show that TRIM32 binds directly to the autophagy receptor p62, and this interaction is not impaired by the LGMD2H and BBS11 disease mutations.
TRIM32 is targeted to autophagic degradation by p62
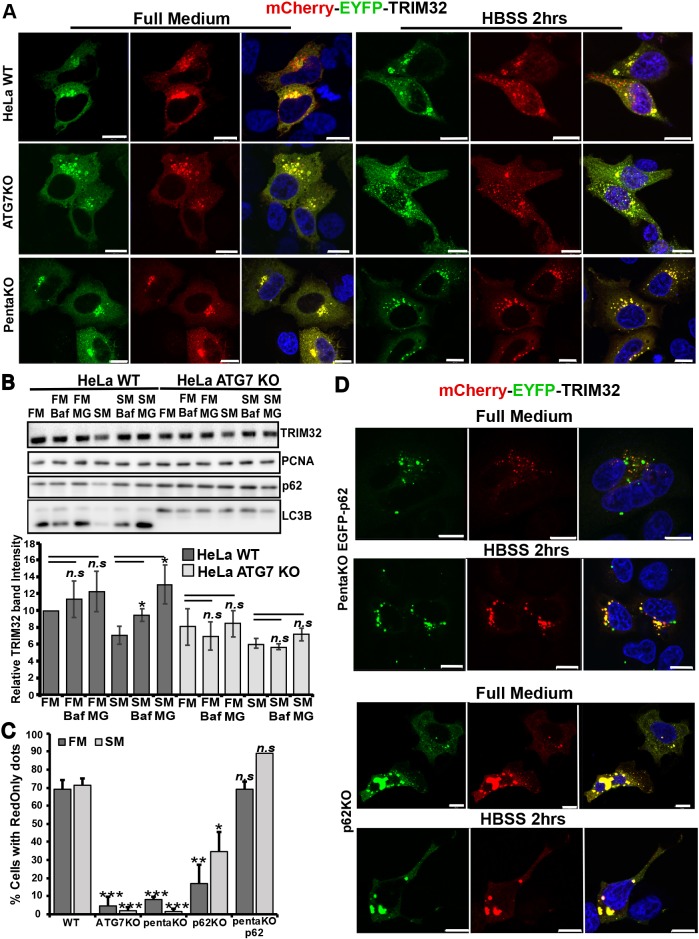
Next, we evaluated whether the lysosomal degradation of TRIM32 is mediated by macroautophagy. For this purpose, we applied the mCherry–EYFP–TRIM32 double-tag assay in the autophagy-deficient ATG7-KO cells (Mejlvang et al., 2018). ATG7 is an ubiquitin E1-like activating enzyme that is essential for assembly and function of the ubiquitin-like conjugation systems (Nakatogawa et al., 2009). mCherry–EYFP–TRIM32 did not form any RedOnly dots in the ATG7-KO cells (Fig. 3A,C). Furthermore, degradation of endogenous TRIM32 was not stabilized by BafA1 treatment in ATG7-KO cells (Fig. 3B). This clearly indicates that lysosomal degradation of TRIM32 is mediated by the macroautophagy pathway. Next question was whether TRIM32 is degraded by selective autophagy. To this end, the degradation of TRIM32 was assayed in a cell line lacking the five autophagy receptors p62, NBR1, NDP52, TAX1BP1 and OPTN (denoted the ‘pentaKO’) (Lazarou et al., 2015). Only a few cells with mCherry–EYFP–TRIM32 RedOnly dots were observed in pentaKO cells grown under normal and starvation conditions (Fig. 3A,C). This suggests that TRIM32 is dependent on the autophagy receptors for efficient degradation by autophagy, pointing towards degradation by selective autophagy.
Fig. 3.
TRIM32 is targeted to autophagic degradation by p62. (A) Normal HeLa cells and cells that were genetically knocked out for ATG7 or the five autophagy receptors p62, NBR1, NDP52, OPTN or TAX1BP1 (pentaKO) as indicated, were transfected with mCherry–EYFP–TRIM32. At 1 day post transfection, half of the cells were treated with HBSS for 2 h (starvation medium, SM) and half were maintained in full medium (FM), before all cells were fixed, stained with DAPI and imaged using a Zeiss780 confocal microscope. Scale bars: 10 µm. (B) Western blot analysis of endogenous TRIM32 in wild-type HeLa cells and HeLa ATG7-KO cells in normal medium (FM) in HBSS (SM), and treated or not with BafA1 (Baf) and MG132 (MG) for 4 h. The graph represents the mean±s.d. TRIM32 band intensities from three independent experiments quantitated using ImageJ. P-values are obtained using Student's t-test. n.s.=not significant (P-value>0.05). PCNA represents the loading control, while p62 and LC3B are controls of autophagy flux. (C) The graphs represent the amount of mCherry-EYFP-TRIM32 transfected cells that contain RedOnly dots compared to the amount of cells that display mCherry-EYFP-TRIM32 dots (representative images shown in A and D) under normal and starvation (HBSS 2 h) conditions. Each graph represents the mean±s.e.m. of at least three independent experiments (n=25–50 cells). *P<0.05, **P<0.005, ***P<0.0005, n.s., not significant (Student's t-test). (D) PentaKO cells reconstituted with GFP–p62 (green dots) and HEK293 FlpIn cells genetically knocked out for p62 as indicated to the right, were transfected and treated as described in A. Scale bars: 10 µm.
The interaction and colocalization of TRIM32 with p62 prompted us to analyze the degradation of TRIM32 in pentaKO cells reconstituted with EGFP–p62. mCherry–EYFP–TRIM32 formed several RedOnly dots both under normal and starvation conditions in this cell line, indicating that p62 is able to mediate autophagic degradation of TRIM32 (Fig. 3C,D). To evaluate if p62 is essential for autophagic degradation of TRIM32, we employed the double tag assay in a HEK293 FlpIn cell line knocked out for p62. The T-REx HEK293 p62−/− cell line was established by CRISPR/Cas9 as described in the Materials and Methods section (Fig. S1C). Ablation of p62 strongly reduced the number of RedOnly dots in the mCherry–EYFP–TRIM32-transfected cells compared to wild-type cells (Fig. 3C,D), but did not completely abolish lysosomal degradation of mCherry–EYFP–TRIM32. This indicates that p62 can mediate autophagic degradation of TRIM32. However, other autophagy receptors beside p62 may also be involved.
The LGMD2H disease mutant of TRIM32 is not degraded by autophagy
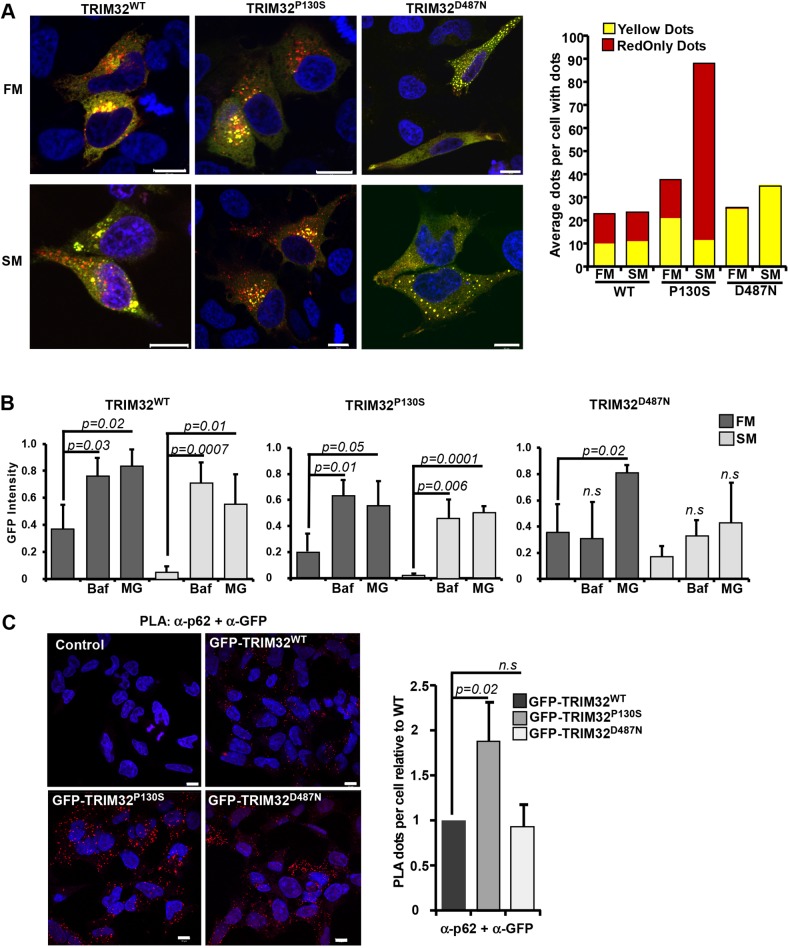
To investigate whether the TRIM32P130S and TRIM32D487N disease mutants were degraded by autophagy, they were fused to the mCherry–EYFP double tag and transfected into HeLa cells. Interestingly, we found that mCherry–EYFP–TRIM32P130S displayed a strong ability to form RedOnly dots, especially under starvation conditions (Fig. 4A). TRIM32D487N on the other hand, was mainly diffuse except from in some cells where it formed several round dots distributed throughout the cell. However, none of these dots were found to be RedOnly (Fig. 4A). Similar results were obtained when we transfected the double-tag constructs into the myoblast cell line C2C12 (Fig. S2A). Around 75% of the C2C12 cells expressing mCherry–EYFP–TRIM32WT and mCherry–EYFP–TRIM32P130S contained RedOnly dots, compared to only 12% of the C2C12 cells expressing mCherry–EYFP–TRIM32D487N. Moreover, double-tag analyses in the C2C12 cells expressing the mutants TRIM32R394H and TRIM32V591M, both of which are within the NHL repeats region and are associated with LGMD2H, revealed that they did not localize in acidic compartments (Fig. S2A). These results clearly indicate that TRIM32 undergoes autophagic degradation in muscle cells, and that this degradation is strongly inhibited by mutations associated with LGMD2H.
Fig. 4.
The LGMD2H disease mutant of TRIM32 is not degraded by autophagy. (A) HeLa cells were transfected with mCherry–EYFP–TRIM32WT, mCherry–EYFP–TRIM32P130S or mCherry–EYFP–TRIM32D487N as indicated, kept in normal medium (full medium, FM) or starved with HBSS for 2 h (starvation medium, SM) at 1 day post transfection and imaged by a Zeiss780 confocal microscope. The graphs to the right display manual quantification of RedOnly dots and yellow dots in n=30 cells, and are representative of three independent experiments. Scale bars: 10 µm. (B) GFP fluorescence intensities (arbitrary units) of EGFP–TRIM32WT, EGFP–TRIM32P130S and EGFP–TRIM32D487N as measured by flow cytometry 24 h after promoter shut off in full medium (FM) or HBSS for 6 h (SM), supplemented with BafA1 2 µm (Baf) or MG132 20 µM (MG) as indicated. Bars show average mean±s.d. GFP intensity from three independent experiments (n=10,000 cells). P-values are as indicated; n.s., not significant (Student's t-test). (C) Representative images of the Flp-In EGFP-TRIM32 cells analyzed by the proximation ligation assay DuoLink using antibodies against GFP and p62. The graph represents the mean±s.d. relative number of colocalization dots with from three independent experiments. Each experiment includes z-stack images of n≈100 cells per condition, and was quantified using the Volocity software. P-values are as indicated; n.s., not significant (Student's t-test). Scale bars: 10 µm.
Next, a flow-based reporter cell system (Larsen et al., 2010) was applied on the HEK293 FlpIn EGFP–TRIM32 cell lines, to monitor degradation of EGFP–TRIM32 and the two disease mutants under normal conditions and starvation. Expression of EGFP–TRIM32 or the disease mutants was induced by treatment with tetracycline for 24 h. Thereafter, the expression was shut off by tetracycline wash-out. The cells were starved in HBSS for 6, 12 and 24 h, and the degradation measured by flow cytometry using the mean GFP intensity in the cell population as a read out. BafA1 was applied to determine degradation by lysosomal pathways, while MG132 was included to measure proteasomal degradation. In full medium, EGFP–TRIM32 is stabilized to a similar extent by BafA1 and MG132, implicating both autophagy and the proteasome system in TRIM32 degradation under normal conditions (Fig. 4B). This is in line with the data presented in Fig. 2A, showing that endogenous TRIM32 is degraded both by autophagy and by the proteasome. This shows that EGFP–TRIM32 inducibly expressed in FlpIn cell lines is degraded similarly to the endogenous protein. Upon 6 h starvation, EGFP–TRIM32 expression is reduced by 85% compared to normal conditions. The degradation is inhibited both by BafA1 and MG132, pointing towards involvement of both autophagy and the proteasomal system also under starved conditions. Similar results were obtained when the starvation period was extended to 12 and 24 h (data not shown). However, the expression level of EGFP–TRIM32 did not decline further, indicating that the expression of TRIM32 is stabilized after 6 h starvation. Applying the flow reporter assay on the BBS11 disease mutant EGFP–TRIM32P130S indicated a higher rate of degradation compared to the wild-type protein, with a 90% reduction in the expression after 6 h of starvation (Fig. 4B). The LGMD2H disease mutant EGFP–TRIM32D487N, however, was not stabilized by BafA1 treatment under either normal or starvation conditions (Fig. 4B). The expression level of EGFP–TRIM32D487N upon starvation is reduced by 50%, which indicates a dramatically lower turnover than the wild-type protein and the BBS11 mutant. Hence, as indicated by the double-tag assay in Fig. 4A, EGFP–TRIM32D487N is not an autophagic substrate, suggesting that it shows dependence on its oligomerization and/or auto-mono-ubiquitylation abilities to be recognized as an autophagic substrate. This is in contrast to a recent report showing BafA1-mediated stabilization of a TRIM32 LGMD2H mutant in primary myoblasts isolated from a patient (Servián-Morilla et al., 2019). This discrepancy can be explained by a TRIM32V591M mutation in this patient, while we have analyzed the TRIM32D487N mutation.
Since we found that TRIM32 colocalizes with p62 in punctate structures, and that reintroduction of p62 into the pentaKO cells was sufficient to direct TRIM32 to autophagic degradation, the next question was whether EGFP–TRIM32D487N had lost its ability to interact with endogenous p62. To this end, the Duolink proximation ligation assay was performed on the FlpIn EGFP–TRIM32 wild-type and mutant cell lines (Fig. 4C). Importantly, EGFP–TRIM32D487N colocalized with p62 to a similar extent as EGFP–TRIM32WT. Hence, interaction with p62 is not sufficient to target TRIM32 to autophagic degradation. Interestingly, the BBS11 disease mutant interacted strongly with p62, forming around twice as many dots in the Duolink assay as wild-type TRIM32 (Fig. 4C). This suggests that the P130S mutation in TRIM32 may modulate the colocalization of TRIM32 and p62 in cells.
Wild-type TRIM32, but not the LGMD2H disease mutant, mono-ubiquitylates p62
In order to investigate the role of TRIM32 in selective autophagy, we established HEK293 FlpIn TRIM32-KO cell lines by CRISPR/Cas9. Knockout of TRIM32 expression in the cell lines was verified by western blotting (Fig. S2B) and genomic sequencing (Fig. S2C). Measuring the proliferation curve of the TRIM32-KO cell lines revealed that they had impaired proliferation compared to the mother HEK293 FlpIn cell line (Fig. 2D). This is in line with the reported roles of TRIM32 in tumorigenesis (Liu et al., 2014; Wang et al., 2018; Yin et al., 2018; Zhao et al., 2018), and thus validates the TRIM32-KO cell lines further.
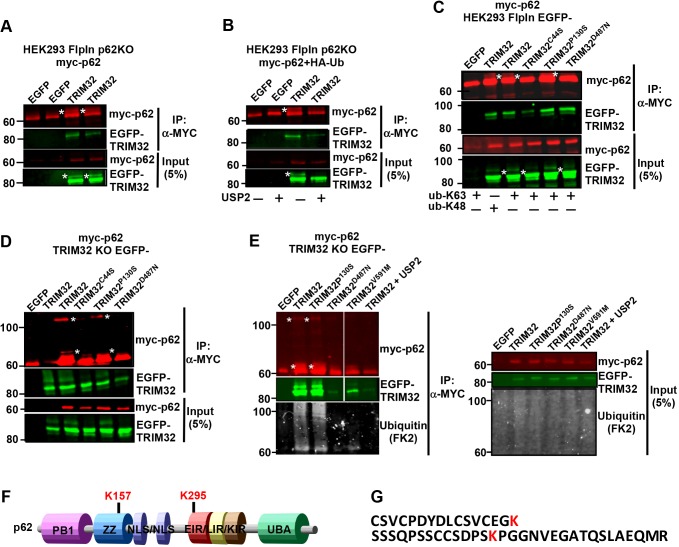
Recent research connects the upregulation of TRIM proteins in tumor cells to enhanced protein quality control and antioxidant defense (Chen et al., 2017). p62 plays important roles both in protein quality control and in antioxidant defense via NRF2 (also known as NFE2L2) (Jain et al., 2010; Pankiv et al., 2007), and ubiquitylation is an important signaling event in these processes. Moreover, reports describing ubiquitin modifications of p62 as a modulator of its activity are emerging (Heath et al., 2016; Jongsma et al., 2016; Lee et al., 2017; Lin et al., 2017; Pan et al., 2016). This prompted us to investigate whether TRIM32 has the ability to ubiquitylate p62. Myc–p62 was immunoprecipitated from a HEK293 p62-KO cell line co-expressing Myc–p62 and EGFP–TRIM32. Western blotting revealed a slow-migrating band both for Myc–p62 and EGFP–TRIM32 (Fig. 5A), indicative of mono-ubiquitylation of both proteins. Whether these slower migrating bands indeed represented ubiquitin modification was verified by coexpressing the de-ubiquitylase USP2 (Fig. 5B). Expressing USP2 removed the slow-migrating band of both p62 and TRIM32. Furthermore, such slower-migrating bands were not detected when Myc–p62 was precipitated from HEK293 cells expressing EGFP–TRIM32C44S (catalytic dead) or EGFP–TRIM32D487N (an LGMD2H disease mutation). This indicates that TRIM32 mediates mono-ubiquitylation of p62, and that this is impaired by mutations in the NHL domains involved in the LGMD2H disease. Mutation in the BBox2 (P130S, a BBS11 disease mutation), however, did not affect mono-ubiquitylation of p62. Both K63- and K48-linked ubiquitylation were responsible for the p62 modification to a similar extent (Fig. 5C). Both wild-type and mutated forms of EGFP–TRIM32 co-precipitated with Myc–p62, which is in line with the GST-pulldown (Fig. 2) and Duolink assay (Fig. 4) results. Hence, the disease mutations do not impair the p62–TRIM32 interaction in cells. Similar ubiquitylation of p62 was obtained when co-transfecting Myc–p62 and wild-type or P130S EGFP–TRIM32 constructs in the HEK293 FlpIn TRIM32-KO cells (Fig. 5C). Myc–p62 also formed a high-molecular-mass band in the SDS gel when co-transfected with the active forms of TRIM32 (Fig. 5D), suggesting that TRIM32-mediated ubiquitylation of p62 leads to the formation of stable p62 oligomers. To identify whether impaired ubiquitylation of p62 is a general trait of LGMD2H disease mutations, the recently described V591M mutant of TRIM32 (Di Rienzo et al., 2019) was co-transfected with Myc–p62 in the HEK293 FlpIn TRIM32-KO cells (Fig. 5E). Introduction of the V591M mutation strongly impaired the p62 ubiquitylation, clearly pointing towards aberrant ubiquitylation of p62 as a general phenotype of LGMD2H. Western blotting of the Myc–p62 precipitates using an antibody against mono-and poly-ubiquitin (FK2) verified that p62 ubiquitylation was strongly reduced upon introduction of the LGMD2H disease mutations D487N and V591M (Fig. 5E).
Fig. 5.
TRIM32 wild-type, but not the LGMD2H disease mutant, mono-ubiquitylates p62. (A) Western blot of two independent experiments from lysates of HEK293 p62-KO cells co-expressing Myc–p62 with EGFP–TRIM32. Myc–p62 was immunoprecipitated (IP) using a Myc-trap and precipitated p62 detected using an anti-Myc antibody. The EGFP–TRIM proteins were detected using an anti-GFP antibody. * indicates mono-ubiquitylated Myc–p62 and EGFP–TRIM32. (B) Myc–p62 was co-expressed with EGFP–TRIM32 and HA–ubiquitin, and with mCherry–USP2, where indicated, in the HEK293 p62-KO cell line. Myc–p62 was immunoprecipitated using a Myc-trap and precipitated p62 detected using an anti-Myc antibody. The EGFP–TRIM proteins were detected using an anti-GFP antibody. * indicates mono-ubiquitylated Myc–p62 and EGFP–TRIM32. (C) Myc–p62 was co-transfected with HA–Ub-K48 or HA–Ub-K63 in the HEK293 EGFP–TRIM32WT, EGFP–TRIM32P130S and EGFP–TRIM32D487N cell lines and expression induced by tetracyclin (1 µM) for 24 h. Myc–p62 was immunoprecipitated using a Myc-trap and precipitated p62 detected using an anti-Myc antibody. The EGFP–TRIM proteins were detected using an anti-GFP antibody. * indicates mono-ubiquitylated p62 and mono-ubiquitylated EGFP–TRIM32. (D) Myc–p62 was co-transfected with EGFP–TRIM32WT, EGFP–TRIM32P130S or EGFP–TRIM32D487N in the HEK293 TRIM32-KO (#1) cell line. Myc-p62 was immunoprecipitated using a Myc-trap and precipitated p62 detected using an anti-Myc antibody. The EGFP–TRIM proteins were detected using an anti-GFP antibody. * indicates mono-ubiquitylated p62 and a high-molecular-mass form of p62. (E) Myc–p62 was co-transfected with EGFP–TRIM32WT, EGFP–TRIM32P130S, EGFP–TRIM32D487N, EGFP–TRIM32V591M, or EGFP–TRIM32 and mCherry–USP2 in the HEK293 TRIM32-KO (#1) cell line. Myc–p62 was immunoprecipitated using a Myc-trap and precipitated p62 detected using an anti-Myc antibody. Ubiquitin modification of p62 was detected using an antibody recognizing mono- and poly-ubiquitin (FK2) (lower panels). The EGFP–TRIM proteins were detected using an anti-GFP antibody. * indicates mono-ubiquitylated p62 and a high-molecular-mass p62. In all blots, EGFP indicates a control plasmid expressing only EGFP. (F) Schematic of p62 domain organization with location of the two lysine residues mapped to be ubiquitylated by TRIM32 indicated above. ZZ, Zinc finger; NLS, Nuclear localization signal; EIR, E2-interacting region; LIR, LC3-interacting region; KIR, Keap1-interacting region; UBA, Ubiquitin-binding domain. (G) Sequence of the p62 peptides found to be ubiquitylated by TRIM32 by MS analyses.
Next, we undertook mass spectrometry (MS) analysis to identify which lysine residues in p62 were modified by TRIM32. MS analysis of Myc–p62 immunoprecipitated from HEK293 TRIM32-KO cells co-transfected with Myc–p62 and EGFP–TRIM32, or the TRIM32 disease mutants EGFP–TRIM32P130S and EGFP–TRIM32D487N, revealed that co-expression of wild-type TRIM32 and the BBS11 disease mutant lead to ubiquitylation of p62 at two lysine residues, K157 and K295 (Fig. 5F,G). No lysine residue was found to be ubiquitylated when the LGMD2H disease mutant was co-expressed with p62. To verify ubiquitylation on these two residues, we introduced K157R or K295R single and double mutations into p62. However, these mutations did not abolish TRIM32 mediated mono-ubiquitylation (Fig. S3A,B). Mono-ubiquitylation of endogenous p62 under ubiquitin stress is reported to occur on p62K157 and p62K295, together with p62K313 and p62K420 (Peng et al., 2017); in that study, mono-ubiquitylation of p62 at those residues was proposed to inhibit formation of inactive p62 dimers with ‘closed’ UBA domains, and thereby promote p62–polyubiquitin interactions and autophagy activity. Hence, next we established a p62K157R/K295R/K420R triple mutant and subjected it to TRIM32, but still TRIM32 was able to mediate ubiquitylation of p62 (Fig. S3B). Neither did introduction of the single K165R or K313R mutations, or the four mutations p62K157R/K295R/K313R/K420R impair ubiquitylation (Fig. S3B). These results indicate that TRIM32-mediated ubiquitylation of p62 may happen at several lysine residues. Interestingly, introducing the D69A mutation in p62, which impairs its ability to undergo oligomerization (Lamark et al., 2003), abolished the TRIM32-mediated mono-ubiquitylation (Fig. S3A,B). This indicates that TRIM32 recognize and ubiquitylate oligomerized p62, but not monomeric p62 molecules.
TRIM32 enhances the formation and turnover of p62 dots
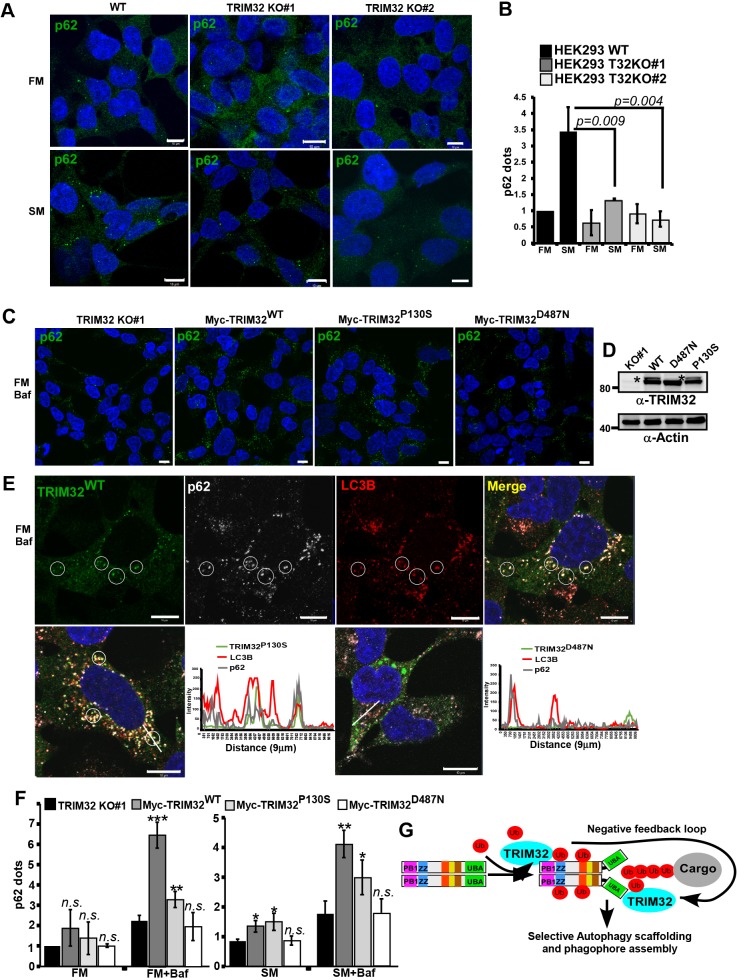
To assess the impact of TRIM32 on the early autophagy events, we assayed WIPI-dot formation in the TRIM32-KO cell lines compared to wild-type cells in normal and starvation conditions. The TRIM32-KO cells did not display any strong effect on the formation of WIPI dots upon starvation (Fig. S3C). However, confocal analysis of p62 in the TRIM32-KO cells showed that the ability of p62 to form dots was significantly reduced (Fig. 6A,B). This suggests that TRIM32-mediated mono-ubiquitylation of p62 stimulates the formation of p62 dots and its autophagic activity, in line with previous studies of the roles of p62 ubiquitylation (Conway and Kirkin, 2017; Lee et al., 2017; Peng et al., 2017). In order to verify this further, we reconstituted the TRIM32-KO cell lines with stably expressing Myc–TRIM32WT, Myc–TRIM32P130S and Myc–TRIM32D487N, and monitored the formation of p62 dots under normal conditions and upon inhibition of autophagy activity by BafA1 treatment (Fig. 6C–F). Conclusively, we found that reintroduction of TRIM32 in the TRIM32-KO cell line strongly facilitated the formation of p62 dots. The number of p62 dots was enhanced upon BafA1 treatment, indicating that TRIM32 augments the autophagic degradation of p62. Importantly, reconstitution with the TRIM32P130S mutant facilitated p62 dot formation nearly as efficiently as TRIM32WT, while reconstitution with the TRIM32D487N mutant had no effect on p62 dot formation (Fig. 6F). This clearly indicates that the E3 ligase activity of TRIM32 is important for its regulatory role on p62. Interestingly, in the TRIM32-KO cells reconstituted with Myc–TRIM32, most cells displayed a diffuse cytoplasmic localization of TRIM32 – with only a few cells containing TRIM32 dots. This was in contrast to the HEK293 FlpIn cells stably overexpressing EGFP–TRIM32. Here, wild-type TRIM32 and the BBS11 disease mutant were found to form cytoplasmic dots in most cells. The more diffuse localization pattern in the reconstituted TRIM32-KO cells suggests that high expression levels of TRIM32 leads to the formation of cytoplasmic dots. However, the dot formation seems to be dependent on TRIM32 ubiquitylation, since the catalytic dead TRIM32 and the LGMD2H disease mutation were mainly diffuse. Importantly, BafA1 treatment of the reconstituted TRIM32-KO cells resulted in formation of several cytoplasmic dots in the cells expressing wild-type Myc–TRIM32 and the BBS11 disease mutant. Myc–TRIM32WT and Myc–TRIM32P130S colocalized with p62 and LC3B in these dots (Fig. 6E). In addition, Myc–TRIM32D497N formed large cytoplasmic dots in some cells. However, these did not colocalize with p62 and LC3B (Fig. 6E). To conclude, our data show that TRIM32 can act as a positive regulator of p62 sequestration and degradation. Concomitantly, TRIM32 itself is a p62 cargo, and thus will be directed to degradation by an active p62. This creates a negative-feedback loop in the TRIM32–p62 interaction pathway (Fig. 6G). Hence, TRIM32-mediated ubiquitylation of p62 seems to regulate its autophagic activity, and thereby cellular proteostasis. One mechanism for the LGMD2H disease can thus be explained by diminished p62 mono-ubiquitylation and lower p62-mediated autophagy activity, resulting in impaired proteostasis.
Fig. 6.
TRIM32 enhances the formation and turnover of p62 dots. (A) Immunostaining of endogenous p62 in HEK293 FlpIn cells and the two TRIM32 KO clones under normal (FM) and starvation (SM) conditions (HBSS 2 h). Scale bars: 10 µm. (B) Quantification of p62 dots in the two TRIM32 KO clones and the HEK293 FlpIn wild type cell line under normal (FM) and starvation (SM) (HBSS 2 h) conditions using the Volocity software (PerkinElmer). Each graph represents the mean±s.d. of the relative number of p62 dots per cell (normalized to the value for FM in wild-type cells). The results are from three independent experiments each including ∼100 cells per condition. P-values were obtained using Student's t-test. (C) Immunostaining of endogenous p62 in the TRIM32 KO (#1) cell line and TRIM32 KO (#1) cells reconstituted with stable expression of Myc–TRIM32WT, Myc–TRIM32P130S or Myc–TRIM32D487N as indicated. The cells are grown in FM with BafA1 (2 µM, 7 h). Scale bars: 10 µm. (D) Western blot showing the expression of Myc–TRIM32, Myc–TRIM32P130S and Myc–TRIM32D487N in the reconstituted HEK293 TRIM32-KO cell lines, grown in FM. * indicates mono-ubiquitylated TRIM32. (E) Upper panels show immunostaining of Myc–TRIM32WT, p62 and LC3B in the reconstituted HEK293 TRIM32-KO cells grown in normal medium supplemented with BafA1 (2 µM) for 7 h. The circles exemplify colocalization of all three proteins in BafA1 dots. The lower panels show merged images of similar immunostainings in the TRIM32-KO cell line reconstituted with Myc–TRIM32P130S or Myc–TRIM32D487N. Images were obtained using a ZEISS780 confocal laser scanning microscope, and the colocalization line plots to the right obtained using the ZEN software. Scale bars: 10 µm. (F) Quantification of p62 dots in the cell lines represented in C, under normal (FM) and normal plus BafA1 (7 h) conditions, and starvation (SM) (HBSS 4 h) and starvation plus BafA1 conditions. The graphs represent the mean±s.d. of the relative number of p62 dots per cell (normalized to the value for FM in TRIM32-KO cells) quantified using Volocity software (PerkinElmer). The results are from three independent experiments each including n>100 cells per condition. *P<0.05, **P<0.005, ***P<0.0005; n.s., not significant (Student's t-test). (G) A model depicting the roles of TRIM32 both as a p62 activator and a p62 substrate in selective autophagy. ZZ, zinc finger; UBA, ubiquitin-binding domain; Ub, Ubiquitin.
DISCUSSION
Owing to constant mechanical stress, striated muscle proteins are particularly prone to wear and tear, and require several protein quality-control mechanisms to coordinate protein turnover and removal of damaged proteins. Among these, ubiquitin signaling and the autophagy-lysosomal pathways are important players (reviewed in Dong and Cui, 2018). Here, we identified the E3 ubiquitin ligase TRIM32 as a substrate for the autophagy receptor p62, which targets TRIM32 for degradation by selective autophagy. Interestingly, TRIM32 was identified as one of the proteins rapidly degraded upon starvation together with the autophagy receptor proteins and certain other proteins (Mejlvang et al., 2018). However, the disease mutant of TRIM32 associated with LGMD2H was not degraded by autophagy. Furthermore, we found TRIM32 to bind directly to p62 and mediate mono-ubiquitylation on several lysine residues, among them p62K157 and p62K295. In addition, the TRIM32 LGMD2H disease mutant D487N bound directly to p62. However, TRIM32D487N did not mediate ubiquitylation of p62. Mono-ubiquitylation of p62 is proposed to enhance its autophagy receptor potential, which is critical for maintaining proteostasis during cellular stress (Peng et al., 2017). We found the formation of p62 bodies to be reduced in TRIM32-KO cells, while it was strongly facilitated when TRIM32WT or TRIM32P130S was reintroduced into the knockout cells. Importantly, the LGMD2H disease mutation was not able to rescue p62 dot formation in the TRIM32-KO cells. Together, this implicates TRIM32 as a positive regulator of p62-mediated selective autophagy, and suggests that one mechanism for development of LGMD2H in TRIM32D487N muscle cells is due to reduced p62 autophagy activity leading to dysregulated cellular proteostasis.
Several TRIM proteins have been shown to modulate the selective autophagy process (Chauhan et al., 2016; Fusco et al., 2018; Jena et al., 2018; Kimura et al., 2015, 2016; Mandell et al., 2014), and during the completion of this paper Di Rienzo and co-workers (2019) identified TRIM32 as a regulator of ULK1 activity in atrophic muscle cells. Similarly to ULK1, p62 activity during selective autophagy is regulated by ubiquitylation. TRIM21 and NEDD4 are reported to mediate ubiquitylation of p62K7 leading to suppressed protein sequestration and induced inclusion body autophagy (Lin et al., 2017; Pan et al., 2016). The E3 ligase RNF166 ubiquitylates p62 at K91 and K189, facilitating the role of p62 in xenophagy (Heath et al., 2016), while RNF26 ubiquitylates p62 within the UBA domain facilitating TOLLIP interaction and vesicular cargo sorting (Jongsma et al., 2016). Keap1–Cullin3 poly-ubiquitylates p62K420 leading to diminished p62 sequestration and degradation activity (Lee et al., 2017). The E2 conjugating enzymes UBE2D2 and UBE2D3 are found to bind directly to p62 via the E2-interacting region (EIR) (Fig. 5E), and mediate mono-ubiquitylation of p62 at several lysine residues, among them K157, K295 and K420, upon ubiquitin stress signaling (Peng et al., 2017). This mono-ubiquitylation is proposed to relieve the inhibition of the UBA domain in the p62 UBA dimer, leading to enhanced binding and tethering of ubiquitylated cargoes to ATG8 proteins conjugated to the phagophore (Conway and Kirkin, 2017). Here, we mapped a direct interaction between TRIM32 and p62 that was independent of the PB1 domain and the UBA domain, similar to the p62 interaction with Keap1–Cullin3. However, Keap1–Cullin3 poly-ubiquitylated p62 at K420 in the UBA domain, while we found TRIM32 to mediate mono-ubiquitylation of p62 at K157 and K295. The Keap1–Cullin3-mediated ubiquitylation of p62 increases p62 inclusion body formation and subsequent degradation, which is proposed to be a mechanism for Keap1 to release the antagonistic role of p62 and thereby dampen the NRF2-mediated response to oxidative stress (Lee et al., 2017). On the other hand, mono-ubiquitylation of p62 at several residues, including K157, K295 and K420, enhances p62 body formation and p62-mediated autophagy activity. This is proposed to be critical for homeostasis during various cellular stresses.
We show that introduction of the LGMD2H disease mutation in TRIM32 makes it unable to mono-ubiquitylate p62. The D487N mutation located in the evolutionarily conserved NHL region of TRIM32 does not inhibit binding to p62. Previously, the LGMD2H mutant is reported to have impaired oligomerization and E3 ligase activity and to display a more diffuse cytoplasmic localization than TRIM32 WT and TRIM32P130S that are enriched in puncta/aggregates in most cells (Locke et al., 2009). Also, the LGMD2H disease mutants seem to be less expressed in muscle tissues from affected patients (Servián-Morilla et al., 2019). Apart from that, the molecular mechanisms of TRIM32 mutants causing the LGMD2H and BBS11 diseases are far from being understood. It is not known if the self-mono-ubiquitylation is important for activation of TRIM32. Our results suggest that this may be the case, and also that this mono-ubiquitylation is important for degradation of TRIM32 via selective autophagy. Several reports connect TRIM32 ubiquitylation activity to substrates whose deregulation might influence the onset and progression of muscular dystrophy, including actin, tropomyosin, troponins, α-actinin, dysbindin and c-Myc (reviewed in Tocchini and Ciosk, 2015). Interestingly, p62 is strongly implicated in chaperone-assisted selective autophagy (CASA), which is essential for muscle maintenance (Arndt et al., 2010). Dysregulated CASA resulted in Z-disk disintegration and progressive muscle weakness. Here, we found that the TRIM32 mutation implicated in muscle dystrophy is unable to ubiquitylate p62 at lysine residues that are linked to p62-mediated substrate sequestration and autophagy activity. Furthermore, TRIM32-KO cells displayed a reduced capacity for p62 sequestration upon cellular stress induced by starvation. Hence, LGMD2H mutations in TRIM32 lead to impaired substrate ubiquitylation and impaired degradation via p62-mediated selective autophagy, both important players in cellular proteostasis. These findings contribute to the understanding of the pathological mechanisms of LMGD2H.
The TRIM32-mediated ubiquitylation site p62K157 is located in the zinc-finger (ZZ) domain (Fig. 5E), and this lysine residue is evolutionarily conserved from zebrafish to human. Recently the small non-coding RNA molecule Vault RNA1-1 was found to bind to the ZZ domain of p62 and thereby modulate its dimerization and autophagy activity (Horos et al., 2019). Hence, post-translational modifications in the ZZ domain could impact such interactions and thereby p62-mediated autophagy activity. The p62K295 site lies at the border of the EIR and of a previously predicted PEST sequence. Thus, it can be speculated that this modification can regulate interactions with ubiquitin E2 enzymes, but also have an impact on p62 stability. The latter can thereby explain the reduced expression level of p62 detected in muscle tissue isolated from LGMD2H patients (Servián-Morilla et al., 2019).
In addition to being a positive regulator of p62 activity, this study revealed that TRIM32 is a p62 substrate in selective autophagy (Fig. 6G). TRIM32 first facilitates p62-mediated autophagy activity. This, after a certain period, leads to a reduced TRIM32 expression caused by p62-mediated degradation. Diminished TRIM32 expression will in turn lead to declined p62 activity, creating a negative-feedback loop. This negative-feedback mechanism may be a potential target for therapeutic intervention.
MATERIALS AND METHODS
Antibodies and reagents
The following primary antibodies were used: rabbit polyclonal antibody for TRIM32 (Proteintech, 10326-1-AP; 1:2000); rabbit polyclonal anti-GFP (Abcam, ab290; 1:5000); mouse monoclonal Myc-Tag (9B11) [Cell Signaling Technologies, #2276; 1:2000 for western blotting (WB), 1:200 for immunofluorescence (IF)]; rabbit polyclonal anti-LC3B (Sigma, L7543; 1:1000 for WB, 1:500 for IF); rabbit polyclonal antibody for phospho-4E-BP1 (Cell Signaling Technologies, #9451; 1:1000); rabbit polyclonal anti-actin (Sigma, A2066; 1:1000); mouse monoclonal anti-p62 lck ligand (BD Biosciences, 610833; 1:2000); guinea pig polyclonal anti-p62 (Progen, GP62-C; 1:2000); mouse monoclonal anti-ubiquitin (FK2) (BIOMOL, PW8810, 1:1000) and mouse monoclonal anti-PCNA (DAKO, M0879; 1:1000). The following secondary antibodies were used: horseradish-peroxidase (HRP)-conjugated goat anti-rabbit-IgG (BD Biosciences, 554021; 1:2000); HRP-conjugated goat anti-mouse-Ig (BD Biosciences, 554002; 1:2000); and HRP-conjugated anti-biotin antibody (Cell Signaling Technologies, #7075; 1:2000). The following fluorescent secondary antibodies were used: Alexa Fluor® 488-conjugated goat anti-mouse-IgG (Life Technologies, A-11029; 1:1000); Alexa Fluor® 488-conjugated goat anti-rabbit-IgG (Life Technologies, A-11008; 1:1000); Alexa Fluor® 555-conjugated goat anti-rabbit-IgG (Life Technologies, A-11008; 1:5000); Alexa Fluor® 555-conjugated goat anti-guinea-pig-IgG (Life Technologies, A-21435; 1:1000); Alexa Fluor® 555-conjugated goat anti-mouse-IgG (Life Technologies, A-21424; 1:1000); Alexa Fluor® 647-conjugated goat anti-guinea pig IgG (Life Technologies, A-21450; 1:1000). The reagents used were: BafA1 (Sigma, B1793); MG132 (Sigma, C2759); tetracycline (Sigma, #87128); doxycycline (Sigma, D9891); and Hanks balanced salt solution (Sigma, H8264).
Cell culture and transfections
HeLa (ATCC, CCL2), HEK293 (ATCC, CRL-1573) and Hek293 T-Rex (Thermo Fisher Scientific, R714-07) cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Sigma, D6046) with 10% fetal bovine serum and 1% streptomycin-penicillin (Sigma, P4333). Hek FlpIn T-Rex cells with integrated TRIM32, TRIM32 C44S, TRIM32 P130S or TRIM32 D487N were grown in the same medium with additional selection marker antibiotics, 200 µg/ml hygromycin B (Invitrogen, #10687010) and 7.5 µg/ml blasticidin (Gibco, A1113903). Sub-confluent cells were transfected using TransIT-LT1 (Mirus, MIR2300) or Metafectene Pro (Biontex, T040) following the manufacturer's instructions. All cell lines were routinely tested for mycoplasma contamination.
Construction of plasmids
All plasmids used in this study are listed in Table S1. Plasmids were made by conventional restriction enzyme-based cloning or by use of the Gateway recombination system (Thermo Fisher Scientific). Gateway LR reactions were performed as described in the instruction manual. Point mutation was carried out using the site-directed-mutagenesis kit from Stratagene. PCR and sequencing oligonucleotides (Table S2) were ordered from Thermo Fisher Scientific. All plasmids were verified by restriction enzyme digestion and DNA sequencing (BigDye, Applied Biosystems, 4337455). All TRIM proteins were cloned into the pDest mCherry-EYFP (Bhujabal et al., 2017) double tag vector.
Recombinant protein production and GST-pulldown analysis
GST or GST-tagged proteins were expressed in Escherichia coli strain SoluBL21 (Genlantis, #C700200). Protein expression was induced by treating overnight bacterial culture with 50 µg/ml isopropyl β-D-1-thiogalactopyranoside (IPTG). GST or GST fusion proteins were purified and immobilized on glutathione–Sepharose 4 Fast Flow beads (GE Healtcare, 17-5132-01). Myc-tagged proteins were in vitro translated using the TNT T7 reticulocyte Lysate system (Promega, #14610) in the presence of [35S]methionine. In vitro translated protein or total cell lysate was pre-incubated with 10 µl glutathione–Sepharose beads and 100 µl of NETN buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM EDTA and 0.5% Nonidet P-40) with cOmplete Mini EDTA-free protease inhibitor mixture tablets (Roche Applied Science, 11836170001) for 1 h at 4°C to reduce unspecific binding. Pre-incubated lysate was then incubated with the immobilized GST fusion protein for 2 h at 4°C. Beads were washed five times with NETN buffer, boiled with 2× SDS gel loading buffer (125 mM Tris-HCl pH 7.5, 4% SDS, 0.04% Bromophenol Blue, 8% sucrose and 100 mM dithiolthreitol) and subjected to SDS-PAGE. Gels were stained with Coomassie Brilliant Blue R-250 Dye (Thermo Fisher Scientific, #20278) to visualize GST fusion proteins and then vacuum dried. Signals from 35S-labeled proteins were detected with a Fujifilm bioimaging analyzer BAS-5000 (Fujifilm).
Immunoblotting and immunoprecipitation
Cells were seeded in 6-cm plates and treated as indicated. Cells were lysed in 1× SDS buffer (50 mM Tris-HCl pH 7.4, 2% SDS and 10% glycerol) supplemented with 200 mM dithiothreitol (DTT, Sigma, #D0632) and heated at 100°C for 10 min. Protein concentration was measured using the Pierce BCA protein assay kit (Thermo Fisher Scientific, #23227). Equal amounts of protein were resolved by SDS-PAGE and transferred to nitrocellulose membrane (Sigma, GE10600003). The membrane was stained with Ponceau S (Sigma, P3504), blocked with 5% non-fat dry milk in 1% TBS-T [0.2 M Tris-HCl pH 8, 1.5 M NaCl and 0.05% Tween 20 (Sigma, P9416)] and then incubated with indicated primary antibodies for 24 h. The membrane was washed three times for 10 min each with TBS-T followed by incubation with secondary antibody for 1 h. The membrane was washed three times for 10 min and analyzed by enhanced chemiluminescence using the ImageQuant LAS 4000 (GE Lifescience). For immunoprecipitation, cells were lysed in modified radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris pH 7.5; 150 mM NaCl; 1 mM EDTA; 1% NP40; 0.25% Triton-X-100) supplemented with cOmplete Mini EDTA-free protease inhibitor cocktail tablets (Roche, #11836170001) and phosphatase inhibitor cocktail (Merck Millipore, #524625) by shaking at 4°C for 30 min. The cell lysate was centrifuged at 10,000 g for 10 min. The resulting supernatant was incubated with antibody to endogenous protein for endogenous immunoprecipitation or with Myc-TRAP (Chromotek, yta-20) for cells stably expressing, or transiently transfected, with Myc-tagged proteins. They were washed five times in RIPA buffer before boiling in 2× SDS gel loading buffer. This was followed by protein identification by immunoblotting as previously described but on Immobilon-FL PVDF membrane (Millipore, IPFL00010), blocked with Odyssey® blocking buffer (PBS) (LI-COR Biosciences, #927-40000) and scanned on Odyssey CLx Imager (LI-COR).
Mass spectrometry
Cells were directly lysed in lysis buffer (Myc-TRAP®_A; Chromotek) supplemented with cOmplete Mini EDTA-free protease inhibitor (Roche), 5 mM NAM (N-Arachidonylmaleimide; Sigma) and 10 mM NEM (N-ethylmaleimide; Sigma). Immunoprecipitation of Myc-tagged proteins was performed according to the supplier's protocol (Chromotek). The samples were fractionated by SDS-PAGE followed by SimplyBlue™ SafeStain (Thermo Fisher Scientific) staining. The lanes representing proteins of molecular masses within 55–80 kDa were cut from the gel. In-gel trypsin digestion was performed before analysis by high-performance liquid chromatography–tandem mass spectrometry (HPLC-MS/MS). Gel pieces were subjected to in-gel reduction, alkylation and tryptic digestion using 6 ng μl−1 trypsin (V511A; Promega). OMIX C18 tips (Varian) were used for sample cleanup and concentration. Peptide mixtures containing 0.1% formic acid were loaded onto a Thermo Fisher Scientific EASY-nLC1200 system. Samples were injected to a trap column (Acclaim PepMap 75 μm×2 cm, C18, 3 μm, 100 Å; Thermo Fisher Scientific) for desalting before elution to the separation column (EASY-Spray column, C18, 2 μm, 100 Å, 50 μm, 50 cm; Thermo Fisher Scientific). Peptides were fractionated using a 4–40% gradient of increasing amounts of 80% acetonitrile in water over 60 min at a flow rate of 300 nl/min. The mobile phases contained 0.1% formic acid. Separated peptides were analyzed using an Orbitrap Fusion Lumos mass spectrometer. The mass spectrometer was operated in a data-dependent mode with the precursor scan in the orbitrap over the range m/z 350–1500. The most-intense ions were selected for electron-transfer dissociation (ETD) or collision-induced dissociation (CID) fragmentation using 3 s between each master scan. Dynamic exclusion was set to 30 s. The Orbitrap AGC target was set to 4E5, and the MS2 scans in the Ion Trap were set to 1E4 with maximum injection times 50 and 100 ms, respectively. Precursor ions with charge 3+ in the m/z range 350–650 and 4+ or 5+ ions in the m/z range 350–900 were fragmented with ETD. All ions with 6+ or higher were also fragmented using ETD. The rest of the precursor ions were fragmented using CID. Protein identification and post-translational modification mapping was performed using the Proteome Discoverer 2.2 software (Thermo Fisher Scientific).
Immunostaining and fluorescence confocal microscopy
Subconfluent cells grown in 24-well plates on coverslips (VWR, #631-0150) coated with fibronectin (Sigma, F1141) and treated as indicated. They were fixed in 4% paraformaldehyde for 20 min to allow visualization of acidic structures after the staining procedure. The cells were then permeabilized with methanol at room temperature for 5 min, blocked in 5% goat serum in PBS or 5% BSA in PBS and incubated at room temperature with a specific primary antibody followed by Alexa Fluor 488-, 555- or 647-conjugated secondary antibody and DAPI. Confocal images were obtained using a 63× NA 1.4 oil immersion objective on an LSM780/LSM800 system. Quantification of p62 dots were performed using the Volocity software (Perkin Elmer) on 10 images per condition, in three independent experiments. Each image contained 10–20 cells, and was generated using the z-stack function in the ZEN software (Zeiss). The line-profile tool in the ZEN software was exploited to plot colocalization of EGFP–TRIM32 and immunostained p62 and LC3B. Quantification of cells containing red only dots in the double-tagged screen was done manually by two independent researchers in three independent experiments.
Generation of TRIM32- and p62-KO HEK293 FlpIn cell lines
To generate a knockout cell line for TRIM32, the CRISPR/Cas9 system was exploited. The knockout was generated as described by Ran et al. (2013). Guide RNAs (gRNA) as described in Table S2 were annealed and ligated into the vector pSpCas9(BB)-2A-GFP (PX458) (Addgene, #48138) using BbsI restriction sites. Subconfluent Hek293 FlpIn T-Rex cells were transfected with the targeting plasmids using Metafectene Pro (Biontex, T020). EGFP-positive cells were sorted by FACS and plated into 96-well plates at 3 days post transfection. Single colonies were expanded up to 12-well plates and knockout validated by immunoblotting. Confirmed knockout clones were further screened by genomic sequencing. The targeted genomic regions were amplified by PCR using the primers TRIM32 PCRfw and TRIM32 PCRrv, or p62 PCRfw and p62PCRrv (Table S2) and the resulting PCR products ligated into the pGEM-T-EASy vector (Promega, A3600). Sequencing were conducted for at least three clones for each PCR product.
Reconstitution of TRIM32-KO cell lines
The TRIM32-KO cell lines were transduced using the lentiviral-based GATEWAY destination vector pCDH-EF1a-Myc-IRES conferring puromycine resistance. This vector enables constitutive expression of Myc–TRIM32WT, Myc–TRIM320P130S or Myc–TRIM32D487N under control of an EF1α promoter.
Generation of stable cell lines
Stable cell lines were generated using the FlpIn T-Rex system (Thermo Fisher Scientific, R71407). cDNAs were PCR amplified and ligated into the inducible FlpIn expression vector, pDest-EGFP-Flp-In (Alemu et al., 2012). FlpIn T-Rex cells were then co-transfected with the cDNA-containing FlpIn expression vector and the FlpIn recombinase vector pOG44 in the ratio of 1:3. Cells were selected by treatment with 200 µg/ml hygromycin B (Invitrogen, #10687010) and 7.5 µg ml−1 blasticidin (Gibco, A1113903), and protein expression verified by induction with tetracycline (Sigma, #87128) or doxycycline (Sigma, D9891).
Flow cytometry
Hek293 FlpIn TRIM32 WT, P130S and D487N cells in 12-well plates were trypsinized (Sigma, T4049) and passed through cell strainer caps (BD Biosciences, 352235) to obtain single-cell suspensions. Cells were analyzed on a FACSAria cell sorter running FACSDiva software version 5.0 (BD Biosciences) using the blue laser for excitation of GFP. GFP fluorescence was collected through a 530/30 nm bandpass filter in the E detector. Data was collected from a minimum of 10,000 singlet events per tube, and the median GFP-A value was used for quantification. All bar graphs show the mean±s.d. of the GFP-A median values from three independent experiments.
Proximity ligation assay
Subconfluent HEK293 FlpIn EGFP-TRIM32 cells were fixed with 4% paraformaldehyde. Colocalization of EGFP–TRIM32 and endogenous p62 was detected by performing a Duolink® PLA (Sigma; DUO92101) according to the manufacturer's protocol. The primary antibodies rabbit polyclonal anti-GFP (1:2000, Abcam, ab290) and mouse monoclonal anti-p62 lck ligand (1:200, BD Biosciences, 610833) were incubated at 4°C overnight. The signals were detected using a 63× NA 1.4 oil immersion objective on an LSM780 system. Quantification of dots was performed using the Volocity software (Perkin Elmer) on 10 images per condition, in three independent experiments. Each image contained 10–20 cells, and was generated using the z-stack function in the ZEN software (Zeiss).
Cell proliferation assay
Hek FlpIn TRIM32 KO Clone #1, KO Clone #2 and FlpIn T-Rex control cells were seeded with four different concentrations (2000, 4000, 6000 and 8000 cells) in 100 μl DMEM (Sigma, D6046) on E-Plate L16 PET readers (ACEA Biosciences Inc, #2801185). xCELLigence® Real-Time Cell Analysis (RTCA) (ACEA Biosciences) were used to measure the cell proliferation over a time of 96 h with recording at 1 h intervals.
Statistics
All experiments were repeated at least three times, unless otherwise specified. Error bars represent the s.d. or s.e.m. as indicated in figure legends, and two-sided unpaired, homoscedastic Student's t-tests were performed to assess significant differences between populations. Replicates were not pooled for statistical analyses. *P<0.05, **P<0.005 and ***P<0.0005; n.s. denotes not signififcant (P>0.05). Sample sizes are denoted in the figure legends.
Supplementary Material
Acknowledgements
We thank Richard Youle, National Institutes of Health, Bethesda, MD 20892, USA, for the generous gift of the pentaKO cell line, and Yakubu P. Abudu, UiT, The Arctic University of Norway, for the gift of the EGFP–p62, reconstituted pentaKO cell line. Thanks to Jack-Ansgar Bruun at the Tromsø University Proteomics Platform for help with mass spectrometry analysis, and the Advanced Microscopy Core Facility at UiT, The Arctic University of Norway, for the use of instrumentation.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: T.J., T.L., E.S.; Methodology: K.S.O., J.G.G., Z.B., E.S.; Validation: J.G.G., E.S.; Formal analysis: E.S.; Investigation: K.S.O., J.G.G., Z.B., A.J., A.Ø., K.B.L., E.S.; Writing - original draft: K.S.O., E.S.; Writing - review & editing: V.D., T.J., T.L., E.S.; Visualization: K.S.O., E.S.; Supervision: T.J., T.L., E.S.; Project administration: E.S.; Funding acquisition: V.D., T.J., E.S.
Funding
This work was supported by PhD grants to K.S.O. and J.G.G. from UiT – The Arctic University of Norway, by the Research Council of Norway (Norges Forskningsråd) (TOPPFORSK program grant #249884 to T.J.), and by the National Institutes of Health (R37AI042999, R01AI042999, and NIH center grant P20GM121176 to V.D.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.236596.supplemental
References
- Albor A., El-Hizawi S., Horn E. J., Laederich M., Frosk P., Wrogemann K. and Kulesz-Martin M. (2006). The interaction of Piasy with Trim32, an E3-ubiquitin ligase mutated in limb-girdle muscular dystrophy type 2H, promotes Piasy degradation and regulates UVB-induced keratinocyte apoptosis through NFkappaB. J. Biol. Chem. 281, 25850-25866. 10.1074/jbc.M601655200 [DOI] [PubMed] [Google Scholar]
- Alemu E. A., Lamark T., Torgersen K. M., Birgisdottir A. B., Larsen K. B., Jain A., Olsvik H., Øvervatn A., Kirkin V. and Johansen T. (2012). ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region (LIR) motifs. J. Biol. Chem. 287, 39275-39290. 10.1074/jbc.M112.378109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt V., Dick N., Tawo R., Dreiseidler M., Wenzel D., Hesse M., Fürst D. O., Saftig P., Saint R., Fleischmann B. K. et al. (2010). Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr. Biol. 20, 143-148. 10.1016/j.cub.2009.11.022 [DOI] [PubMed] [Google Scholar]
- Bhujabal Z., Birgisdottir A. B., Sjøttem E., Brenne H. B., Øvervatn A., Habisov S., Kirkin V., Lamark T. and Johansen T. (2017). FKBP8 recruits LC3A to mediate Parkin-independent mitophagy. EMBO Rep. 18, 947-961. 10.15252/embr.201643147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Øvervatn A., Stenmark H. and Johansen T. (2005). p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171, 603-614. 10.1083/jcb.200507002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S., Kumar S., Jain A., Ponpuak M., Mudd M. H., Kimura T., Choi S. W., Peters R., Mandell M., Bruun J.-A. et al. (2016). TRIMs and galectins globally cooperate and TRIM16 and galectin-3 co-direct autophagy in endomembrane damage homeostasis. Dev. Cell 39, 13-27. 10.1016/j.devcel.2016.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Brewer M. D., Guo L., Wang R., Jiang P. and Yang X. (2017). Enhanced degradation of misfolded proteins promotes tumorigenesis. Cell Rep. 18, 3143-3154. 10.1016/j.celrep.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang A. P., Beck J. S., Yen H.-J., Tayeh M. K., Scheetz T. E., Swiderski R. E., Nishimura D. Y., Braun T. A., Kim K.-Y. A., Huang J. et al. (2006). Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet-Biedl syndrome gene (BBS11). Proc. Natl. Acad. Sci. USA 103, 6287-6292. 10.1073/pnas.0600158103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi U. Y., Choi W. Y., Hur J. Y. and Kim Y.-J. (2015). Polyubiquitin chain-dependent protein degradation in TRIM30 cytoplasmic bodies. Exp. Mol. Med. 47, e159 10.1038/emm.2015.12 [DOI] [PubMed] [Google Scholar]
- Cohen S., Zhai B., Gygi S. P. and Goldberg A. L. (2012). Ubiquitylation by Trim32 causes coupled loss of desmin, Z-bands, and thin filaments in muscle atrophy. J. Cell Biol. 198, 575-589. 10.1083/jcb.201110067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway O. and Kirkin V. (2017). Love laughs at Locksmiths: ubiquitylation of p62 unlocks its autophagy receptor potential. Cell Res. 27, 595-597. 10.1038/cr.2017.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rienzo M., Antonioli M., Fusco C., Liu Y., Mari M., Orhon I., Refolo G., Germani F., Corazzari M., Romagnoli A. et al. (2019). Autophagy induction in atrophic muscle cells requires ULK1 activation by TRIM32 through unanchored K63-linked polyubiquitin chains. Sci. Adv. 5, eaau8857 10.1126/sciadv.aau8857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I. and Elazar Z. (2018). Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19, 349-364. 10.1038/s41580-018-0003-4 [DOI] [PubMed] [Google Scholar]
- Dong Z. and Cui H. (2018). The autophagy-lysosomal pathways and their emerging roles in modulating proteostasis in tumors. Cells 8, 4 10.3390/cells8010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosk P., Weiler T., Nylen E., Sudha T., Greenberg C. R., Morgan K., Fujiwara T. M. and Wrogemann K. (2002). Limb-girdle muscular dystrophy type 2H associated with mutation in TRIM32, a putative E3-ubiquitin-ligase gene. Am. J. Hum. Genet. 70, 663-672. 10.1086/339083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco C., Micale L., Egorov M., Monti M., D'Addetta E. V., Augello B., Cozzolino F., Calcagni A., Fontana A., Polishchuk R. S. et al. (2012). The E3-ubiquitin ligase TRIM50 interacts with HDAC6 and p62, and promotes the sequestration and clearance of ubiquitinated proteins into the aggresome. PLoS ONE 7, e40440 10.1371/journal.pone.0040440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco C., Mandriani B., Di Rienzo M., Micale L., Malerba N., Cocciadiferro D., Sjøttem E., Augello B., Squeo G. M., Pellico M. T. et al. (2018). TRIM50 regulates Beclin 1 proautophagic activity. Biochim. Biophys. Acta Mol. Cell Res. 1865, 908-919. 10.1016/j.bbamcr.2018.03.011 [DOI] [PubMed] [Google Scholar]
- Hatakeyama S. (2017). TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem. Sci. 42, 297-311. 10.1016/j.tibs.2017.01.002 [DOI] [PubMed] [Google Scholar]
- Heath R. J., Goel G., Baxt L. A., Rush J. S., Mohanan V., Paulus G. L. C., Jani V., Lassen K. G. and Xavier R. J. (2016). RNF166 determines recruitment of adaptor proteins during antibacterial autophagy. Cell Rep. 17, 2183-2194. 10.1016/j.celrep.2016.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horos R., Büscher M., Kleinendorst R., Alleaume A.-M., Tarafder A. K., Schwarzl T., Dziuba D., Tischer C., Zielonka E. M., Adak A. et al. (2019). The small non-coding vault RNA1-1 acts as a riboregulator of autophagy. Cell 176, 1054-1067.e1012. 10.1016/j.cell.2019.01.030 [DOI] [PubMed] [Google Scholar]
- Isogai S., Morimoto D., Arita K., Unzai S., Tenno T., Hasegawa J., Sou Y.-S., Komatsu M., Tanaka K., Shirakawa M. et al. (2011). Crystal structure of the ubiquitin-associated (UBA) domain of p62 and its interaction with ubiquitin. J. Biol. Chem. 286, 31864-31874. 10.1074/jbc.M111.259630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E. and Mizushima N. (2011). p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J. Cell Biol. 192, 17-27. 10.1083/jcb.201009067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A., Lamark T., Sjøttem E., Larsen K. B., Awuh J. A., Øvervatn A., McMahon M., Hayes J. D. and Johansen T. (2010). p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 285, 22576-22591. 10.1074/jbc.M110.118976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jena K. K., Kolapalli S. P., Mehto S., Chauhan S. and Chauhan S. (2018). TRIM16 controls turnover of protein aggregates by modulating NRF2, ubiquitin system, and autophagy: implication for tumorigenesis. Mol. Cell. Oncol. 5, e1532251 10.1080/23723556.2018.1532251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J. and Demontis F. (2017). Skeletal muscle autophagy and its role in sarcopenia and organismal aging. Curr. Opin. Pharmacol. 34, 1-6. 10.1016/j.coph.2017.03.009 [DOI] [PubMed] [Google Scholar]
- Johansen T. and Lamark T. (2011). Selective autophagy mediated by autophagic adapter proteins. Autophagy 7, 279-296. 10.4161/auto.7.3.14487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongsma M. L. M., Berlin I., Wijdeven R. H. M., Janssen L., Janssen G. M. C., Garstka M. A., Janssen H., Mensink M., van Veelen P. A., Spaapen R. M. et al. (2016). An ER-associated pathway defines endosomal architecture for controlled cargo transport. Cell 166, 152-166. 10.1016/j.cell.2016.05.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T., Jain A., Choi S. W., Mandell M. A., Schroder K., Johansen T. and Deretic V. (2015). TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J. Cell Biol. 210, 973-989. 10.1083/jcb.201503023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T., Mandell M. and Deretic V. (2016). Precision autophagy directed by receptor regulators - emerging examples within the TRIM family. J. Cell Sci. 129, 881-891. 10.1242/jcs.163758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T., Jia J., Kumar S., Choi S. W., Gu Y., Mudd M., Dupont N., Jiang S., Peters R., Farzam F. et al. (2017). Dedicated SNAREs and specialized TRIM cargo receptors mediate secretory autophagy. EMBO J. 36, 42-60. 10.15252/embj.201695081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudryashova E., Kudryashov D., Kramerova I. and Spencer M. J. (2005). Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J. Mol. Biol. 354, 413-424. 10.1016/j.jmb.2005.09.068 [DOI] [PubMed] [Google Scholar]
- Kudryashova E., Struyk A., Mokhonova E., Cannon S. C. and Spencer M. J. (2011). The common missense mutation D489N in TRIM32 causing limb girdle muscular dystrophy 2H leads to loss of the mutated protein in knock-in mice resulting in a Trim32-null phenotype. Hum. Mol. Genet. 20, 3925-3932. 10.1093/hmg/ddr311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudryashova E., Kramerova I. and Spencer M. J. (2012). Satellite cell senescence underlies myopathy in a mouse model of limb-girdle muscular dystrophy 2H. J. Clin. Invest. 122, 1764-1776. 10.1172/JCI59581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamark T., Perander M., Outzen H., Kristiansen K., Øvervatn A., Michaelsen E., Bjørkøy G. and Johansen T. (2003). Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J. Biol. Chem. 278, 34568-34581. 10.1074/jbc.M303221200 [DOI] [PubMed] [Google Scholar]
- Larsen K. B., Lamark T., Øvervatn A., Harneshaug I., Johansen T. and Bjørkøy G. (2010). A reporter cell system to monitor autophagy based on p62/SQSTM1. Autophagy 6, 784-793. 10.4161/auto.6.6.12510 [DOI] [PubMed] [Google Scholar]
- Lazarou M., Sliter D. A., Kane L. A., Sarraf S. A., Wang C., Burman J. L., Sideris D. P., Fogel A. I. and Youle R. J. (2015). The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524, 309-314. 10.1038/nature14893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzari E. and Meroni G. (2016). TRIM32 ubiquitin E3 ligase, one enzyme for several pathologies: From muscular dystrophy to tumours. Int. J. Biochem. Cell Biol. 79, 469-477. 10.1016/j.biocel.2016.07.023 [DOI] [PubMed] [Google Scholar]
- Lee Y. J., Chou T.-F., Pittman S. K., Keith A. L., Razani B. and Weihl C. C. (2017). Keap1/Cullin3 modulates p62/SQSTM1 activity via UBA domain ubiquitination. Cell Rep. 20, 1994 10.1016/j.celrep.2017.08.019 [DOI] [PubMed] [Google Scholar]
- Lee Y. J., Jonson P. H., Sarparanta J., Palmio J., Sarkar M., Vihola A., Evilä A., Suominen T., Penttilä S., Savarese M. et al. (2018). TIA1 variant drives myodegeneration in multisystem proteinopathy with SQSTM1 mutations. J. Clin. Invest. 128, 1164-1177. 10.1172/JCI97103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Dai Q., Meng H., Sun A., Wei J., Peng K., Childress C., Chen M., Shao G. and Yang W. (2017). The HECT E3 ubiquitin ligase NEDD4 interacts with and ubiquitylates SQSTM1 for inclusion body autophagy. J. Cell Sci. 130, 3839-3850. 10.1242/jcs.207068 [DOI] [PubMed] [Google Scholar]
- Liu J., Zhang C., Wang X. L., Ly P., Belyi V., Xu-Monette Z. Y., Young K. H., Hu W. and Feng Z. (2014). E3 ubiquitin ligase TRIM32 negatively regulates tumor suppressor p53 to promote tumorigenesis. Cell Death Differ. 21, 1792-1804. 10.1038/cdd.2014.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke M., Tinsley C. L., Benson M. A. and Blake D. J. (2009). TRIM32 is an E3 ubiquitin ligase for dysbindin. Hum. Mol. Genet. 18, 2344-2358. 10.1093/hmg/ddp167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J., Garner T. P., Pandya M. J., Craven C. J., Chen P., Shaw B., Williamson M. P., Layfield R. and Searle M. S. (2010). Dimerisation of the UBA domain of p62 inhibits ubiquitin binding and regulates NF-kappaB signalling. J. Mol. Biol. 396, 178-194. 10.1016/j.jmb.2009.11.032 [DOI] [PubMed] [Google Scholar]
- Mandell M. A., Kimura T., Jain A., Johansen T. and Deretic V. (2014). TRIM proteins regulate autophagy: TRIM5 is a selective autophagy receptor mediating HIV-1 restriction. Autophagy 10, 2387-2388. 10.4161/15548627.2014.984278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell M. A., Jain A., Kumar S., Castleman M. J., Anwar T., Eskelinen E.-L., Johansen T., Prekeris R. and Deretic V. (2016). TRIM17 contributes to autophagy of midbodies while actively sparing other targets from degradation. J. Cell Sci. 129, 3562-3573. 10.1242/jcs.190017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall R. S., Hua Z., Mali S., McLoughlin F. and Vierstra R. D. (2019). ATG8-binding UIM proteins define a new class of autophagy adaptors and receptors. Cell 177, 766-781.e724. 10.1016/j.cell.2019.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejlvang J., Olsvik H., Svenning S., Bruun J.-A., Abudu Y. P., Larsen K. B., Brech A., Hansen T. E., Brenne H., Hansen T. et al. (2018). Starvation induces rapid degradation of selective autophagy receptors by endosomal microautophagy. J. Cell Biol. 217, 3640-3655. 10.1083/jcb.201711002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. and Komatsu M. (2011). Autophagy: renovation of cells and tissues. Cell 147, 728-741. 10.1016/j.cell.2011.10.026 [DOI] [PubMed] [Google Scholar]
- Morreale F. E. and Walden H. (2016). Types of ubiquitin ligases. Cell 165, 248-248.e241. 10.1016/j.cell.2016.03.003 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H., Suzuki K., Kamada Y. and Ohsumi Y. (2009). Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458-467. 10.1038/nrm2708 [DOI] [PubMed] [Google Scholar]
- Nicklas S., Otto A., Wu X., Miller P., Stelzer S., Wen Y., Kuang S., Wrogemann K., Patel K., Ding H. et al. (2012). TRIM32 regulates skeletal muscle stem cell differentiation and is necessary for normal adult muscle regeneration. PLoS ONE 7, e30445 10.1371/journal.pone.0030445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J.-A., Sun Y., Jiang Y.-P., Bott A. J., Jaber N., Dou Z., Yang B., Chen J.-S., Catanzaro J. M., Du C. et al. (2016). TRIM21 ubiquitylates SQSTM1/p62 and suppresses protein sequestration to regulate redox homeostasis. Mol. Cell 62, 149-151. 10.1016/j.molcel.2016.03.015 [DOI] [PubMed] [Google Scholar]
- Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J.-A., Outzen H., Øvervatn A., Bjørkøy G. and Johansen T. (2007). p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131-24145. 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Peng H., Yang J., Li G., You Q., Han W., Li T., Gao D., Xie X., Lee B.-H., Du J. et al. (2017). Ubiquitylation of p62/sequestosome1 activates its autophagy receptor function and controls selective autophagy upon ubiquitin stress. Cell Res. 27, 657-674. 10.1038/cr.2017.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizon V., Rybina S., Gerbal F., Delort F., Vicart P., Baldacci G. and Karsenti E. (2013). MURF2B, a novel LC3-binding protein, participates with MURF2A in the switch between autophagy and ubiquitin proteasome system during differentiation of C2C12 muscle cells. PLoS ONE 8, e76140 10.1371/journal.pone.0076140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ra E. A., Lee T. A., Won Kim S., Park A., Choi H. J., Jang I., Kang S., Hee Cheon J., Cho J. W., Eun Lee J. et al. (2016). TRIM31 promotes Atg5/Atg7-independent autophagy in intestinal cells. Nat. Commun. 7, 11726 10.1038/ncomms11726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A. and Zhang F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281-2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Muela N., Parkhitko A., Grass T., Gibbs R. M., Norabuena E. M., Perrimon N., Singh R. and Rubin L. L. (2018). Blocking p62-dependent SMN degradation ameliorates spinal muscular atrophy disease phenotypes. J. Clin. Invest. 128, 3008-3023. 10.1172/JCI95231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov V., Dötsch V., Johansen T. and Kirkin V. (2014). Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell 53, 167-178. 10.1016/j.molcel.2013.12.014 [DOI] [PubMed] [Google Scholar]
- Servián-Morilla E., Cabrera-Serrano M., Rivas-Infante E., Carvajal A., Lamont P. J., Pelayo-Negro A. L., Ravenscroft G., Junckerstorff R., Dyke J. M., Fletcher S. et al. (2019). Altered myogenesis and premature senescence underlie human TRIM32-related myopathy. Acta Neuropathol. Commun. 7, 30 10.1186/s40478-019-0683-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack F. J. and Ruvkun G. (1998). A novel repeat domain that is often associated with RING finger and B-box motifs. Trends Biochem. Sci. 23, 474-475. 10.1016/S0968-0004(98)01299-7 [DOI] [PubMed] [Google Scholar]
- Sparrer K. M. J., Gableske S., Zurenski M. A., Parker Z. M., Full F., Baumgart G. J., Kato J., Pacheco-Rodriguez G., Liang C., Pornillos O. et al. (2017). TRIM23 mediates virus-induced autophagy via activation of TBK1. Nat. Microbiol. 2, 1543-1557. 10.1038/s41564-017-0017-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Wu R., Zheng J., Li P. and Yu L. (2018). Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 28, 405-415. 10.1038/s41422-018-0017-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocchini C. and Ciosk R. (2015). TRIM-NHL proteins in development and disease. Semin. Cell Dev. Biol. 47-48, 52-59. 10.1016/j.semcdb.2015.10.017 [DOI] [PubMed] [Google Scholar]
- Tomar D., Singh R., Singh A. K., Pandya C. D. and Singh R. (2012). TRIM13 regulates ER stress induced autophagy and clonogenic ability of the cells. Biochim. Biophys. Acta 1823, 316-326. 10.1016/j.bbamcr.2011.11.015 [DOI] [PubMed] [Google Scholar]
- van Gent M., Sparrer K. M. J. and Gack M. U. (2018). TRIM proteins and their roles in antiviral host defenses. Annu. Rev. Virol. 5, 385-405. 10.1146/annurev-virology-092917-043323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Xu J., Fu H., Zhang Y., Zhang X., Yang D., Zhu Z., Wei Z., Hu Z., Yan R. et al. (2018). TRIM32 promotes cell proliferation and invasion by activating beta-catenin signalling in gastric cancer. J. Cell. Mol. Med. 22, 5020-5028. 10.1111/jcmm.13784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Liu T.-T., Lin H., Zhang M., Wei J., Luo W.-W., Hu Y.-H., Zhong B., Hu M.-M. and Shu H.-B. (2017). TRIM32-TAX1BP1-dependent selective autophagic degradation of TRIF negatively regulates TLR3/4-mediated innate immune responses. PLoS Pathog. 13, e1006600 10.1371/journal.ppat.1006600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Li Z., Chen J. and Hu X. (2018). Expression and the potential functions of TRIM32 in lung cancer tumorigenesis. J. Cell. Biochem. 120, 5232-5243. 10.1002/jcb.27798 [DOI] [PubMed] [Google Scholar]
- Zaffagnini G., Savova A., Danieli A., Romanov J., Tremel S., Ebner M., Peterbauer T., Sztacho M., Trapannone R., Tarafder A. K. et al. (2018). Phasing out the bad-How SQSTM1/p62 sequesters ubiquitinated proteins for degradation by autophagy. Autophagy 14, 1280-1282. 10.1080/15548627.2018.1462079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T.-T., Jin F., Li J.-G., Xu Y.-Y., Dong H.-T., Liu Q., Xing P., Zhu G.-L., Xu H., Yin S.-C. et al. (2018). TRIM32 promotes proliferation and confers chemoresistance to breast cancer cells through activation of the NF-kappaB pathway. J. Cancer 9, 1349-1356. 10.7150/jca.22390 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.