ABSTRACT
Human cell reprogramming remains extremely inefficient and the underlying mechanisms by different reprogramming factors are elusive. We found that NANOG and LIN28 (NL) synergize to improve OCT4, SOX2, KLF4 and MYC (OSKM)-mediated reprogramming by ∼76-fold and shorten reprogramming latency by at least 1 week. This synergy is inhibited by GLIS1 but reinforced by an inhibitor of the histone methyltransferase DOT1L (iDOT1L) to a ∼127-fold increase in TRA-1-60-positive (+) iPSC colonies. Mechanistically, NL serve as the main drivers of reprogramming in cell epithelialization, the expression of Let-7 miRNA target LIN41, and the activation of canonical WNT/β-CATENIN signaling, which can be further enhanced by iDOT1L treatment. LIN41 overexpression in addition to OSKM similarly promoted cell epithelialization and WNT activation in reprogramming, and a dominant-negative LIN41 mutation significantly blocked NL- and iDOT1L-enhanced reprogramming. We also found that NL- and iDOT1L-induced canonical WNT activation facilitates the initial development kinetics of iPSCs. However, a substantial increase in more mature, homogeneous TRA-1-60+ colony formation was achieved by inhibiting WNT activity at the middle-to-late-reprogramming stage. We further found that LIN41 can replace LIN28 to synergize with NANOG, and that the coexpression of LIN41 with NL further enhanced the formation of mature iPSCs under WNT inhibition. Our study established LIN41 and canonical WNT signaling as the key downstream effectors of NL for the dramatic improvement in reprogramming efficiency and kinetics, and optimized a condition for the robust formation of mature human iPSC colonies from primary cells.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Induced pluripotent stem cell (iPSC), LIN28, LIN41, WNT, Epithelialization, Reprogramming efficiency
Summary: Robust human iPSC reprogramming through the synergy of NANOG, LIN28, and inhibition of DOT1L in modulation of LIN41 expression and the canonical WNT pathway.
INTRODUCTION
Two gene cocktails, OCT4, SOX2, KLF4, and MYC (OSKM) (Takahashi et al., 2007; Takahashi and Yamanaka, 2006) and OCT4, SOX2, NANOG and LIN28A (OSNL) (Yu et al., 2007b), can reprogram somatic cells to embryonic stem cell (ESC)-like induced pluripotent stem cells (iPSCs). The reprogramming of mouse somatic cells involves two major waves of transcriptional changes (Hussein et al., 2014). The first transcriptional change occurs at the early reprogramming stage, with cells undergoing mesenchymal-to-epithelial transition (MET) for iPSC colony formation (Hussein et al., 2014; Li et al., 2010; Samavarchi-Tehrani et al., 2010). This stage is followed by the second wave that occurs during maturation and stabilization, when the pluripotency regulatory network is activated and stabilized in reprogrammed cells (Buganim et al., 2012; Golipour et al., 2012; Hussein et al., 2014; Polo et al., 2012; Samavarchi-Tehrani et al., 2010). In human cells, the early-to-middle reprogramming stages are characterized by multiple waves of lineage-related gene activation in the order of developmental reversal, with MET occurring at the middle-to-late-reprogramming stage along with pluripotent network activation (Cacchiarelli et al., 2015). This transcriptional alteration in reprogramming is accompanied by epigenomic modifications that suppress somatic gene expression/reactivation and maintain the active pluripotency regulatory network (Cacchiarelli et al., 2015; Hussein et al., 2014; Xu et al., 2016). However, the exact molecular mechanism that ensures successful human cell reprogramming is still poorly defined.
Thus far, induced pluripotency in humans remains a very inefficient and lengthy process. The reprogramming efficiency for human iPSC generation is generally at the low end of the reported range (0.00002–∼1%) in different laboratories, and it usually takes between 3 and 5 weeks for the induced iPSC colonies to appear (Malik and Rao, 2013; Rao and Malik, 2012). Additional reprogramming factors have been reported to enhance the reprogramming efficiency induced by OSKM (Hanna et al., 2009; Maekawa et al., 2011; Silva et al., 2009; Tanabe et al., 2013; Worringer et al., 2014; Yu et al., 2007b; Zhang et al., 2016). NANOG is a key gene required for pluripotency maintenance (Pan and Thomson, 2007) and is thought to stabilize reprogramming at the late iPSC induction stage (Hanna et al., 2009; Silva et al., 2009; Yu et al., 2007b). GLIS1 promotes human iPSC generation and activates Foxa2 in mouse cell reprogramming to promote MET and to reinforce the activity of the core pluripotent gene network (Maekawa et al., 2011). LIN28 is exclusively expressed in completely but not partially reprogrammed human iPSCs (Zhang et al., 2016) and promotes the maturation of reprogrammed cells, a major roadblock for successful human iPSC generation (Tanabe et al., 2013). The best known function of LIN28 is to inhibit Let-7 miRNA maturation to promote the expression of HMGA2, KRAS, MYC (Viswanathan et al., 2009) and HRAS in cancer cells (Cai et al., 2013; Yu et al., 2007a). However, unlike the ectopic expression of MYC (Takahashi et al., 2007; Takahashi and Yamanaka, 2006), ectopically expressed HMGA2, KRAS or HRAS failed to improve human iPSC generation (Worringer et al., 2014). Thus, the exact mechanisms by which these reprogramming factors regulate human cell reprogramming remain elusive.
The canonical WNT/β-CATENIN pathway signals through the T cell factor (TCF)/lymphoid enhancer factor and exerts pleiotropic effects on pluripotency establishment and maintenance. WNT maintains naïve-pluripotent mouse ESCs by suppressing the negative effector TCF7L1 (formally known as TCF3), and stimulating WNT/β-CATENIN activity facilitates mouse iPSC induction (Lluis et al., 2011; Zhang et al., 2014). However, WNT also inhibits mouse ESC proliferation via the effectors TCF7 and TCF7L2 (formally known as TCF1 and TCF4, respectively) (Cole et al., 2008; De Jaime-Soguero et al., 2017; Martello et al., 2012). In humans, WNT/β-CATENIN activity is needed for the self-renewal of primed-state human ESCs (Fernandez et al., 2014) or the generation of human iPSCs (Cevallos et al., 2018; Ross et al., 2014). However, enhancing WNT/β-CATENIN and TCF7 signaling promotes differentiation of ESCs or the reprogrammed cells (Cevallos et al., 2018; Davidson et al., 2012; Dravid et al., 2005; Jiang et al., 2013). In addition, the WNT negative regulator TCF7L1 is needed for the generation of human ESC-like, primed-state pluripotent mouse cells (Hoffman et al., 2013) and maintains human ESC pluripotency by inhibiting primitive streak commitment (Sierra et al., 2018). Thus, WNT activity needs to be carefully controlled in reprogramming. However, how different reprogramming factors regulate canonical WNT signaling for successful reprogramming remains unclear.
In the current study, we used primary human mesenchymal stem cells (MSCs) with very low efficiency in OSKM-mediated reprogramming to study the iPSC induction mediated by OSKM and the reprogramming factors GLIS1, NANOG and LIN28 (GNL). We used TRA-1-60, one of the best markers for primed-state pluripotency (Andrews et al., 1984; Chan et al., 2009) and successful iPSC generation (Onder et al., 2012; Tanabe et al., 2013), to monitor the reprogramming process. We found that NANOG and LIN28 (NL), but not GLIS1, synergize to stimulate the expression of the Let-7 target LIN41 and to enhance canonical WNT activity for human iPSC generation. The synergistic effects can be re-enforced by the inhibition of the histone 3 lysine 79 (H3K79) methyltransferase DOT1L, resulting in a more than ∼127-fold increase in TRA-1-60 positive (+) iPSC colonies. Furthermore, we discovered that although the elevated canonical WNT activity facilitates initial reprogramming kinetics, the inhibition of WNT signaling at the middle-to-late-reprogramming stage dramatically enhances the maturation of reprogrammed cells.
RESULTS
NL is more efficient than GNL in reprogramming
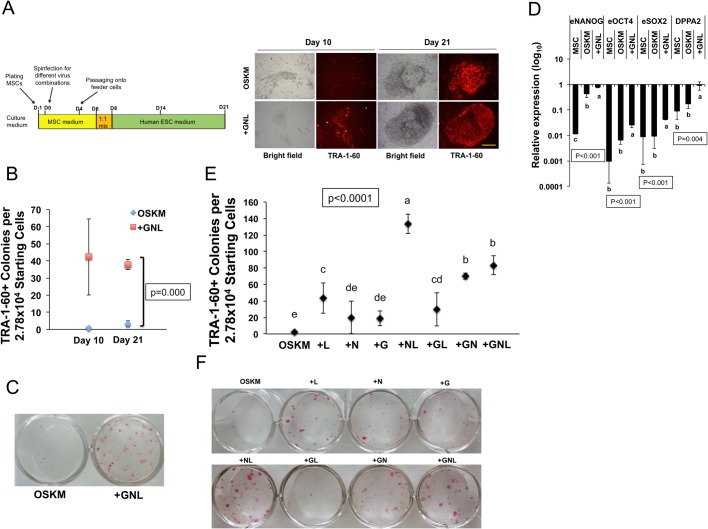
We first hypothesized that coexpressing GLIS1, NANOG and LIN28 would greatly enhance OSKM-mediated reprogramming based on their reported individual effects (Hanna et al., 2009; Lee et al., 2017; Maekawa et al., 2011; Yu et al., 2007b; Zhang et al., 2016). Primary MSCs were transduced with OSKM or OSKM+GNL expressed in a retroviral pMXs-vector (Fig. 1A). In the OSKM reprogramming condition, few TRA-1-60+ cell aggregates were observed on day 10 of viral infection and the development of TRA-1-60+ colonies appeared 1–2 weeks later (Fig. 1A,B). In contrast, many ESC-like TRA-1-60+ colonies readily appeared in the GNL condition on day 10 (Fig. 1A,B). The difference in the number of TRA-1-60+ colonies was also correlated with the alkaline phosphatase (AP)-staining of reprogrammed cells (Fig. 1C). Quantitative-reverse transcription PCR (qRT-PCR) analysis on reprogrammed cells at day 14 showed that the GNL combination significantly stimulated the expression of the endogenous (e) pluripotent genes OCT4, SOX2, NANOG and DPPA2 (Fig. 1D).
Fig. 1.
Effects of LIN28, NANOG and GLIS1 on promoting the reprogramming of human MSCs. (A, left) Schematic diagram of the timeline of human MSC reprogramming. (Right) Representative images of TRA-1-60 immunofluorescence in OSKM- and +GNL-induced colonies of human MSCs on days 10 and 21. Scale bar: 250 μm. (B) Number of TRA-1-60+ colonies in OSKM- and +GNL-mediated reprogramming conditions on days 10 and 21. Scatter plots represent the mean±s.d., n=3. (C) Representative images of putative iPSC colonies in OSKM- and +GNL-mediated reprogramming stained with AP on day 21. (D) qRT-PCR results of the pluripotent gene expression in parental MSCs and cells transduced with OSKM or +GNL on reprogramming day 14 relative to the pluripotent gene expression in H9-ESCs. e, endogenous genes. Bars represent the mean±s.d., n=3. (E) Number of TRA-1-60+ colonies in different reprogramming conditions on days 12 and 18. +G, +N and +L represent the addition of GLIS1, NANOG and LIN28, respectively, to OSKM for reprogramming; +NL, +GL, +GN and +GNL represent the respective combinations added to OSKM for reprogramming. Scatter plots represent the mean±s.d., n=3. (F) Representative images of putative iPSC colonies under different reprogramming conditions stained with AP on day 21. In all graphs, conditions with different letters are significantly different.
We then asked which factor(s) in GLIS1, NANOG and LIN28 most effectively promoted reprogramming. We applied the factors individually or in two-factor combinations to the OSKM condition. On day 12, TRA-1-60+ colonies were evident in all other conditions except for the OSKM alone (Fig. S1). The applications of GLIS1, NANOG or LIN28 each improved the reprogramming efficiency of human MSCs compared with OSKM, albeit with less efficiency than the GNL combined (Fig. 1E,F). Furthermore, while the GLIS1 and NANOG (GN) combination produced similar reprogramming efficiency to GNL, NL together increased TRA-1-60+ colonies by ∼1.6-fold over that of GNL (Fig. 1E). This result was also correlated with an increase in AP-stained colonies (Fig. 1F). No synergistic effect was observed for the GLIS1 and LIN28 (GL) combination (Fig. 1E,F). Thus, among the three additional reprogramming factors, the NL combination most dramatically enhanced OSKM-mediated reprogramming and shortened reprogramming latency by more than 1 week compared with the OSKM condition.
NL co-stimulate LIN41 to promote cell epithelialization in reprogramming
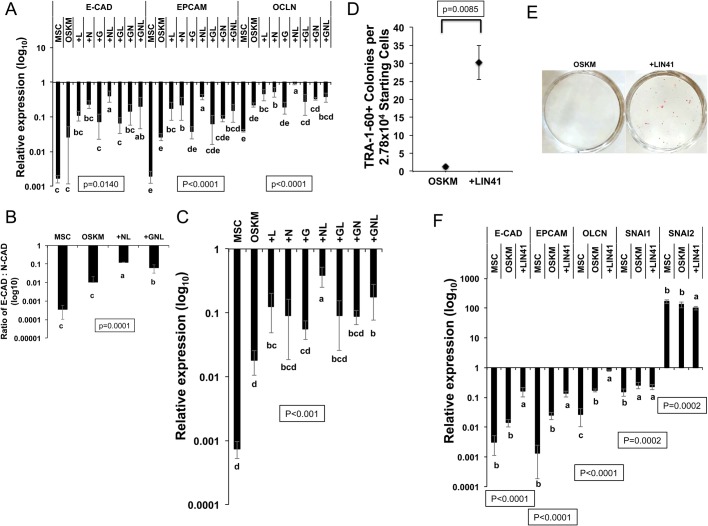
To identify a possible mechanism for the NL-enhanced reprogramming, we evaluated the gene expression in reprogrammed cells on day 14. Compared with the OSKM condition, both the addition of NL and GNL significantly improved the expression of core pluripotent genes, including endogenous NANOG, OCT4 and SOX2, with no significant difference between the two conditions (Fig. S2). We then asked if NL and GNL differentially regulate the MET process in reprogramming. Compared with OSKM alone or with GLIS1, NANOG or LIN28, NL but not GNL significantly increased the expression of the epithelial markers E-CADHERIN (E-CAD), EPCAM and OCLN (Fig. 2A). Moreover, GNL resulted in more decreased EPCAM and OCLN expression than NL (Fig. 2A). In addition, the ratio of the epithelial marker E-CAD versus the mesenchymal marker N-CAD (Nakajima et al., 2004; Wang et al., 2016) was increased more significantly in NL than in GNL compared with the OSKM-alone condition (Fig. 2B). No obvious difference was observed in the expression of mesenchymal markers among different reprogramming conditions (Fig. S3). These data indicate that NL synergize to promote cellular epithelialization in reprogramming, while the addition of GLIS1 reduces this synergy.
Fig. 2.
NL synergize to activate LIN41 and promote the reprogramming of human MSCs. (A) qRT-PCR results of epithelial gene expression in MSCs and reprogrammed cells on day 14 under different conditions relative to the epithelial gene expression in H9-ESCs. Bars represent the mean±s.d., n=3. (B) Ratio of E-CAD:N-CAD mRNA expression in MSCs and reprogrammed cells on day 14 under different conditions relative to the expression in H9-ESCs. Bars represent the mean±s.d., n=3. (C) qRT-PCR results of LIN41 expression in MSCs and reprogrammed cells on day 14 under different conditions relative to H9-ESC expression. Bars represent the mean±s.d., n=3. (D) Number of TRA-1-60+ colonies under the OSKM- and OSKM+LIN41 (+LIN41)-mediated reprogramming conditions on day 12. Scatter plots represent the mean±s.d., n=3. (E) Representative images of putative iPSC colonies in OSKM- and +LIN41-mediated reprogramming stained with AP on day 18. (F) qRT-PCR results of mesenchymal and epithelial marker expression in parental MSCs and human MSCs reprogrammed with OSKM or +LIN41 on day 14 relative to H9-ESC expression. Bars represent the mean±s.d., n=3. In all graphs, conditions with different letters are significantly different.
The mRNA of the ubiquitin ligase LIN41 is targeted by Let-7 miRNAs in Caenorhabditis elegans, mice and humans (Ecsedi et al., 2015; Nguyen et al., 2017; Slack et al., 2000; Worringer et al., 2014), and LIN41 plays an important role in overcoming the Let-7 barrier for OSKM-mediated reprogramming from fibroblasts (Worringer et al., 2014). However, although the RNA-binding protein LIN28 directly inhibits the maturation of Let-7 miRNAs (Viswanathan et al., 2008), whether it regulates LIN41 expression to promote successful reprogramming is not known. We asked if LIN41 is a downstream target of LIN28 in reprogramming. Compared with the OSKM condition, the addition of LIN28 significantly stimulated LIN41 expression, and this stimulatory effect was synergistically enhanced by NL but not by GL or GN (Fig. 2C). The addition of GNL also exhibited less LIN41 stimulation than NL (Fig. 2C). Thus, NL co-stimulate the expression of LIN41 in reprogramming whereas GLIS1 reduces this effect. We also questioned whether LIN41 overexpression could improve the OSKM-mediated reprogramming from human MSCs as previously reported from fibroblasts (Worringer et al., 2014). Similar to LIN28 overexpression (Fig. 1E), ectopic LIN41 significantly improved OSKM-mediated reprogramming efficiency (Fig. 2D,E; Fig. S4). We further questioned whether LIN41 regulates MET in reprogramming. Indeed, the overexpression of LIN41 significantly stimulated the expression of the epithelial markers E-CAD, EPCAM and OCLN (Fig. 2F). Additionally, LIN41 did not affect the expression of the mesenchymal marker SNAI1 and only slightly reduced (<25%) the expression of SNAI2 (Fig. 2F). Thus, our data indicate that LIN41 functions as a downstream target and effector of LIN28 and is co-stimulated by NL to promote reprogramming, at least partially by enhancing cellular epithelialization.
Canonical WNT signaling is synergistically stimulated by NL in reprogramming
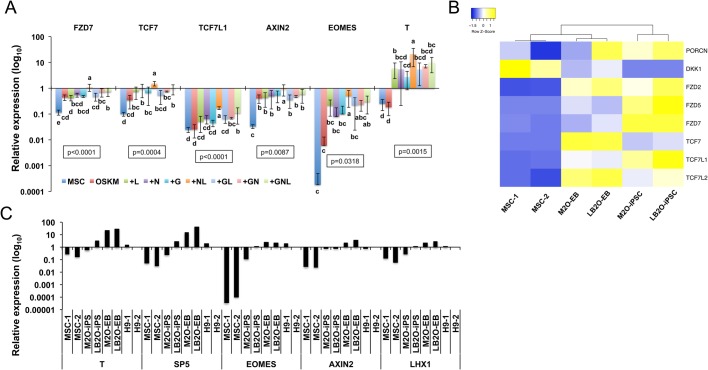
The overexpression of the canonical WNT/β-CATENIN signaling effector TCF7 initially promoted reprogramming but induced differentiation at late-reprogramming stage (Cevallos et al., 2018). We asked whether WNT activity is modulated by GLIS1, NANOG or LIN28 in reprogramming. Among all conditions, NL substantially stimulated the expression of FZD7, the most abundant WNT receptor specific to human ESCs and necessary for pluripotency maintenance (Fernandez et al., 2014). In addition, TCF7 and the canonical WNT signaling targets AXIN2, EOMES and T (Huggins et al., 2017; Yan et al., 2001) were also greatly activated by NL (Fig. 3A). GNL exerted a smaller stimulatory effect on WNT activity than NL (Fig. 3A). These findings indicate that NL synergistically stimulates canonical WNT activity in reprogramming, while GLIS1 mitigates this stimulatory effect. Meanwhile, NL also moderately but significantly promoted the expression of TCF7L1 (Fig. 3A), the WNT antagonist and pluripotent marker necessary to prevent hyperactive WNT signaling-induced primitive streak differentiation in human ESCs/iPSCs (Cevallos et al., 2018; Sierra et al., 2018).
Fig. 3.
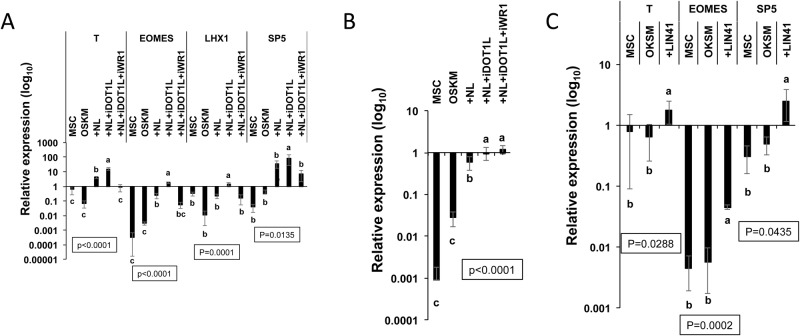
Activation of canonical WNT signaling in reprogrammed cells and pluripotent stem cells. (A) qRT-PCR results of the expression of WNT/β-CATENIN pathway components and target genes in MSCs and reprogrammed cells on day 14 under different conditions relative to H9-ESC expression. Bars represent the mean±s.d., n=3. Conditions with different letters are significantly different. (B) Heatmap showing the expression of canonical WNT/β-CATENIN pathway regulatory genes in MSCs, two iPSC lines (M2O and LB2O) and the day-5 EBs differentiated from these iPSCs. (C) qRT-PCR results of the expression levels of WNT/β-CATENIN target genes in MSCs, two iPSC lines (M2O and LB2O), the day-5 EBs differentiated from these iPSCs and H9-ESCs.
We also evaluated whether WNT signaling is elevated in human iPSCs by comparing two previously reported human iPSC lines (Wang et al., 2017) with their parental MSCs and the day 5 embryoid bodies (EBs) differentiated from these iPSCs. Although no obvious difference in the expression of eight canonical WNT ligands (Staal et al., 2008) was found between iPSCs and MSCs (Fig. S5), increased expression of PORCN, a membrane bound O-acetyltransferase necessary for WNT ligand secretion (Barrott et al., 2011; Biechele et al., 2011; Proffitt and Virshup, 2012), and decreased expression of DKK1, an inhibitor of canonical WNT signaling (Cruciat and Niehrs, 2013) were evident in human iPSCs compared with human MSCs (Fig. 3B). Furthermore, the three WNT receptors reported to enrich in human ESCs – FZD2/5/7 (Fernandez et al., 2014), and the WNT effectors TCF7 and TCF7L2 – were all increased in iPSCs compared with MSCs (Fig. 3B). We further found that the primitive streak/mesoendoderm markers targeted by canonical WNT signaling, including T, SP5, EOMES, AXIN2 and LHX1 (Huggins et al., 2017; Yan et al., 2001), were all highly or moderately upregulated in human iPSCs and ESCs compared with MSCs (Fig. 3C). Taken together, these results indicate that canonical WNT signaling is more active in human pluripotent stem cells than in MSCs and is synergistically stimulated by NL in reprogramming. Additionally, consistent with the known differentiation-stimulating function of fully activated WNT signaling (Sierra et al., 2018), we noticed that compared with iPSCs and MSCs, EBs exhibited markedly elevated WNT ligands (Fig. S5) and WNT effectors TCF7/TCF7L2 only moderately increased WNT antagonist TCF7L1 (Fig. 3B).
Inhibiting H3K79 methyltransferase enhances NL-stimulated reprogramming, while blocking WNT signaling promotes iPSC maturation
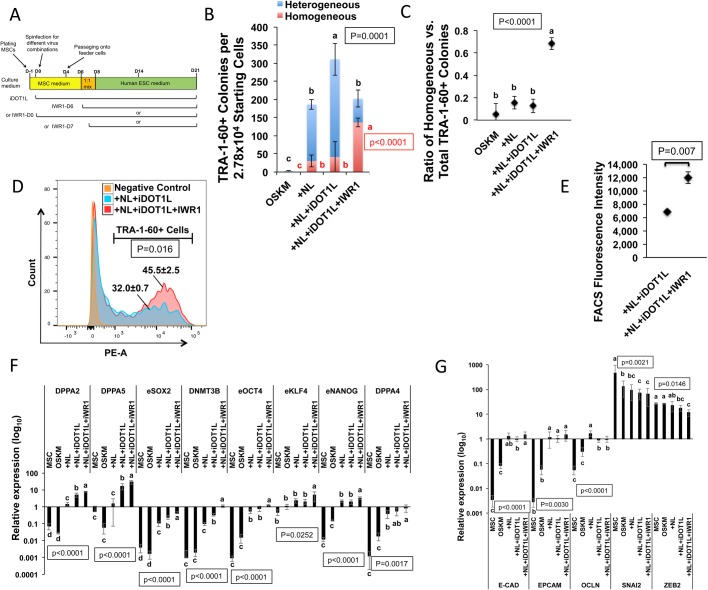
H3K79 dimethylation (H3K79me2) is a barrier of reprogramming from human fibroblasts (Onder et al., 2012). We asked if inhibiting H3K79me2 would further enhance the NL-mediated improvement in reprogramming efficiency. An inhibitor of the H3K79 methyltransferase DOT1L (iDOT1L) (Onder et al., 2012) was added at day 0 of reprogramming (Fig. 4A). The addition of iDOT1L enhanced OSKM-mediated reprogramming (Figs S6 and S7). Similarly, iDOT1L also enhanced the reprogramming mediated by OSKM plus a polycistronic NL expression (used hereafter in all +NL conditions), resulting in an ∼127-fold increase in total TRA-1-60+ colonies compared with the OSKM condition, in contrast to the ∼76-fold increase in the NL condition with no iDOT1L (Fig. 4B).
Fig. 4.
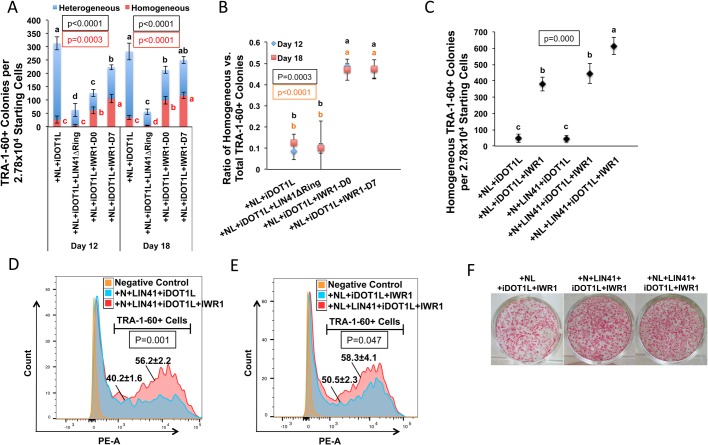
Effects of inhibiting H3K79 methylation and the WNT signaling pathway on reprogramming. (A) Schematic diagram showing the timeline of iDOT1L and IWR1 administration in reprogramming. (B) Numbers of homogeneous (red), heterogeneous (blue) and total (red plus blue) TRA-1-60+ colonies in the OSKM condition and the other reprogramming conditions on day 12. +NL, +NL+iDOT1L and +NL+iDOT1L+IWR1 represent the addition of NL alone or NL plus inhibitor(s) to OSKM for reprogramming. Bars represent the mean±s.d., n=3. Letters and P-values shown in red and black represent the statistics for the numbers of homogeneous and total TRA-1-60+ colonies, respectively. (C) Ratio of homogeneous versus total TRA-1-60+ colonies in the OSKM condition and the other reprogramming conditions as indicated in B on day 12. Scatter plots represent the mean±s.d., n=3. (D) FACS analysis of cellular TRA-1-60 immunofluorescence on reprogramming day 14 with or without WNT inhibition. The percentage of TRA-1-60+ cells out of the total reprogrammed cells is shown as the mean±s.d., n=3. (E) Median fluorescence intensity of TRA-1-60+ cells as determined by FACS analysis on reprogramming day 14 with or without WNT inhibition. Scatter plots represent the mean±s.d., n=3. (F) qRT-PCR results of pluripotent marker gene expression in MSCs and reprogrammed cells on day 14 under the conditions described in B relative to H9-ESC expression. Bars represent the mean±s.d., n=3. (G) qRT-PCR results of epithelial and mesenchymal gene expression in MSCs and reprogrammed cells on day 14 under the conditions described in B relative to H9-ESC expression. Bars represent the mean±s.d., n=3. In all graphs, conditions with different letters are significantly different.
As we found that NL stimulate canonical WNT signaling in reprogramming, and hyperactive WNT causes human iPSC/ESC differentiation (Cevallos et al., 2018; Sierra et al., 2018), we wondered if inhibiting WNT would improve the NL-enhanced reprogramming to a greater extent. A canonical WNT inhibitor IWR1 (Chen et al., 2009) has been shown to improve the maintenance of human ESC self-renewal (Kim et al., 2013). We added IWR-1 at day 6 of reprogramming, when iPSC colony formation was evident (Fig. 4A). Interestingly, the addition of IWR1 produced flat-shaped iPSC colonies that more morphologically resemble human ESCs than the other conditions on day 12 (Fig. S8). Furthermore, the colonies observed with the addition of IWR1 exhibited brighter and more homogeneous TRA-1-60 fluorescence (Fig. S9). We therefore counted both the homogeneous and heterogeneous TRA-1-60+ colonies in reprogramming. Although the number of total (homogenous and heterogeneous) TRA-1-60+ colonies was greatest in the NL+iDOT1L condition (Fig. 4B), the ratio of homogeneous versus total TRA-1-60+ colonies remained low (<20%) (Fig. 4C). However, compared with the NL+iDOT1L condition, the NL+iDOT1L+IWR1 reprogramming condition exhibited striking increase in the number and ratio of homogeneous TRA-1-60+ colonies (Fig. 4B,C). The positive effects of iDOT1L and IWR1 on NL-enhanced TRA-1-60+ colony formation were also correlated with the AP-staining of induced colonies at reprogramming day 18 (Fig. S10). The fluorescence-activated cell sorting (FACS) further confirmed that compared with the NL+iDOT1L condition, the NL+iDOT1L+IWR1 condition significantly increased the percentage of TRA-1-60+ cells on day 14 (Fig. 4D); the median fluorescence intensity of TRA-1-60+ cells was also increased ∼1.8-fold in the NL+iDOT1L+IWR1 condition (Fig. 4E).
Consistent with the increase in total TRA-1-60+ colonies, compared with the NL condition, NL+iDOT1L increased the expression of the pluripotency markers DPPA2/5 (Qian et al., 2016; Tung et al., 2013) and some late-reprogramming stage markers, including endogenous SOX2 and DNMT3B (Buganim et al., 2012; Cacchiarelli et al., 2015; Takahashi et al., 2014) (Fig. 4F). However, compared with the NL and NL+iDOT1L conditions, the addition of IWR1 not only further enhanced the expression of these genes mentioned above, but also increased the expression of more core pluripotency markers, including endogenous OCT4, NANOG, KLF4 and DPPA4 (Cacchiarelli et al., 2015) (Fig. 4F). This was correlated with the increased TRA-1-60+ cell population as well as the enhanced TRA-1-60 fluorescence intensity in the NL+iDOT1L+IWR1 condition, and indicates a reinforcement of pluripotency network activity for the NL+iDOT1L enhanced reprogramming by inhibiting WNT.
We asked whether the addition of iDOT1L or IWR1 would impact MET in reprogramming. While NL stimulated dramatic epithelial marker expression compared with OSKM, it exhibited no obvious effect on mesenchymal markers similarly as we had observed (Fig. 4G; Fig. S3). However, compared with the OSKM condition, the NL+iDOT1L condition significantly decreased the expression of the mesenchymal markers SNAI2 (∼46%) and ZEB2 (∼33%), and the addition of IWR1 further reduced ZEB2 expression (∼57%) (Fig. 4G). These data indicate that NL are the main driving forces underlying cell epithelialization in reprogramming. Additionally, iDOT1L could enhance reprogramming by suppressing the expression of mesenchymal markers, which can be further enhanced by the addition of IWR1. All these underpin the activation of pluripotency network and promote the maturation of reprogrammed cells.
To verify the pluripotency of putative iPSCs, we picked the homogeneous TRA-1-60+ colonies on reprogramming days 18–21 from different conditions (NL, NL+iDOT1L, and NL+iDOT1L+IWR1). These cells readily expanded in a mTeSR1 feeder-free condition (Ludwig et al., 2006a,b). iDOT1L and IWR1 were removed during the expansion. These iPSC lines exhibited silencing of all transgenes at passage 11 (Fig. S11) and expressed pluripotent genes/proteins at similar levels as human ESCs (Figs S11 and S12). To confirm their differentiation capacity, iPSCs established from different conditions were subjected to EB differentiation (Fig. S13). qRT-PCR and immunostaining analyses of EBs at day 5 demonstrated significant activation of lineage markers for three germ layers (Figs S14 and S15).
iDOT1L treatment enhances NL-stimulated WNT and LIN41 activities, and LIN41 expression contributes to WNT activation in reprogramming
We asked if inhibiting H3K79me2 by iDOT1L would affect the NL-stimulated WNT activity. Interestingly, we found that iDOT1L treatment further enhanced the expression of WNT target genes induced by NL in reprogramming (Fig. 5A). As expected, IWR1 inhibited the WNT activity co-stimulated by NL and iDOT1L (Fig. 5A).
Fig. 5.
iDOT1L treatment enhances WNT and LIN41 activities in reprogramming. (A) qRT-PCR results showing the expression of WNT/β-CATENIN target genes in MSCs and reprogrammed cells by OSKM, +NL, +NL+iDOT1L and +NL+iDOT1L+IWR1 on day 14 under the conditions described in Fig. 4B relative to H9-ESC expression. Bars represent the mean±s.d., n=3. (B) qRT-PCR results of NL- and iDOT1L-induced LIN41 expression on reprogramming day 14 under the conditions described in A relative to H9-ESC expression. Bars represent the mean±s.d., n=3. (C) qRT-PCR results of the OSKM- and OSKM+LIN41 (+LIN41)-mediated reprogramming conditions WNT target genes on reprogramming day 14 relative to H9-ESC expression. Bars represent the mean±s.d., n=3. In all graphs, conditions with different letters are significantly different.
We have shown that NL synergistically stimulate LIN41 expression and that LIN41 enhances the OSKM-mediated reprogramming of MSCs (Fig. 2C–E). We further questioned if LIN41 expression is regulated by iDOT1L treatment and WNT inhibition. Compared with the NL condition, the addition of iDOT1L further enhanced LIN41 expression in reprogramming (Fig. 5B). IWR1, however, did not significantly alter LIN41 expression level (Fig. 5B). We also asked whether LIN41 could regulate WNT activity by analyzing the reprogrammed cells in OSKM and OSKM+LIN41 conditions (Fig. 2D,E). Compared with the OSKM condition, the OSKM+LIN41 condition exhibited significantly enhanced expression of canonical WNT targets, including T, EOMES and SP5 (Fig. 5C). These results indicate that NL and iDOT1L co-stimulate LIN41 expression, which is independent of WNT signaling, and LIN41 participates in the activation of canonical WNT signaling in reprogramming, which is consistent with what we had observed for NL (Fig. 3A).
The activities of WNT and LIN41 are critical for NL- and iDOT1L-mediated reprogramming
We wondered how the LIN41 and WNT activities contribute to the enhanced reprogramming by the NL and iDOT1L addition. A dominant-negative LIN41 mutant with an N-term RING domain deletion (pMXs-LIN41ΔRing) (Worringer et al., 2014) was added to the NL+iDOT1L condition. Additionally, IWR1 was added to the NL+iDOT1L condition from initial (day 0) or middle-to-late-reprogramming (day 7) to evaluate the effect of WNT signaling on reprogramming (Fig. 4A). The numbers of homogenous/heterogeneous TRA-1-60+ colonies were counted on reprogramming days 12 and 18 (Fig. 6A). Compared with the NL+iDOT1L condition, LIN41ΔRing reduced the total TRA-1-60+ colonies to only ∼20% of the NL+iDOT1L condition on both days 12 and 18 (Fig. 6A). This result also correlated with the reduced number of AP-stained colonies in the LIN41ΔRing condition on day 18 (Fig. S16). These data demonstrate that LIN41 plays a critical role in NL-induced iPSC colony formation. For WNT inhibition during reprogramming, we found that on day 12, the addition of IWR1 from day 0 reduced the number of total TRA-1-60+ colonies to ∼41% of those in the NL+iDOT1L condition, in contrast to the reduction to ∼71% when IWR1 was added from day 7 (Fig. 6A). However, on day 18, the total number of TRA-1-60+ colonies increased to ∼75% and ∼89% of the NL+iDOT1L condition for IWR1 treatments from day 0 and 7, respectively (Fig. 6A). Additionally, the ratio of homogeneous versus total TRA-1-60+ colonies was similar regardless whether IWR1 was applied from day 0 or 7, and was significantly greater than the NL+iDOT1L condition on days 12 and 18 (Fig. 6B). These data indicate that the activated WNT signaling by NL and iDOT1L plays a significant role in facilitating the kinetics of initial iPSC colony development. However, the subsequent maturation of reprogrammed cells in these colonies requires the inhibition of WNT activity.
Fig. 6.
LIN41 and WNT play critical roles in the enhancement of human cell reprogramming induced by NL and iDOT1L. (A) Effects of dominant-negative LIN41 overexpression and the WNT inhibition applied from reprogramming day 0 or day 7 on the numbers of homogeneous (red), heterogeneous (blue) and total (red plus blue) TRA-1-60+ colonies. Bars represent the mean±s.d., n=3. Letters and P-values shown in red and black represent the statistics for the number of homogeneous and total TRA-1-60+ colonies, respectively. (B) Ratio of homogeneous versus total TRA-1-60+ colonies under the different reprogramming conditions indicated in C. Scatter plots represent mean±s.d., n=3. Letters and P-values shown in black and red represent the day 12 and day 18 statistics, respectively. (C) Effects of LIN41 overexpression and IWR1 addition beginning on reprogramming day 6 on the formation of homogeneous TRA-1-60+ colonies on reprogramming day 12. Scatter plots represent the mean±s.d., n=3. (D) FACS analysis of cellular TRA-1-60 immunofluorescence on reprogramming day 14 in the +N+LIN41+iDOT1L condition with or without WNT inhibition. The percentage of TRA-1-60+ cells out of total reprogrammed cells is shown as the mean±s.d., n=3. (E) FACS analysis of cellular TRA-1-60 immunofluorescence on reprogramming day 14 in the +NL+iDOT1L+IWR1 and +NL+LIN41+iDOT1L+IWR1 conditions. The percentage of TRA-1-60+ cells out of total reprogrammed cells is shown as the mean±s.d., n=3. (F) Representative pictures of putative iPSC colonies stained with AP on reprogramming day 21. In all graphs, conditions with different letters are significantly different.
As we found that LIN41 is a critical downstream effector of LIN28 in reprogramming, we asked whether LIN41 could replace LIN28 in synergizing with NANOG (N+LIN41) for reprogramming. In striking similarity to the NL+iDOT1L condition, when IWR1 was added (from day 6), the replacement of LIN28 with LIN41 (N+LIN41+iDOT1L) induced a ∼10-fold increase in the homogeneous TRA-1-60+ colonies compared with the condition without IWR1 on day 12 (Fig. 6C). FACS analysis further revealed a significant increase in the TRA-1-60+ cell population and fluorescence intensity when IWR1 was added to the N+LIN41+iDOT1L condition (Fig. 6D; Fig. S17). These results indicate that LIN41 can replace LIN28 to synergize with NANOG in reprogramming. Furthermore, when LIN41 was coexpressed with NL, the homogeneous TRA-1-60+ colonies further increased by >1.6-fold over the NL+iDOT1L+IWR1 condition (Fig. 6C). The increase in the TRA-1-60+ cell population by NL+LIN41+iDOT1L+IWR1 condition was also confirmed by FACS analysis (Fig. 6E). These results correlated with the number of AP-stained colonies at 3 weeks of reprogramming, showing a dramatic generation of AP+ colonies (Fig. 6F). Thus, the efficiency of establishing homogeneous TRA-1-60+ colonies from the initial MSCs by combined NL and LIN41 overexpression was ∼2% (Fig. 6C), in contrast to the ∼0.0004% efficiency under the OSKM condition (Fig. 4B), representing a 1000-fold increase in reprogramming efficiency. Taken together, the results in our study demonstrated that NL and iDOT1L promote reprogramming efficiency and kinetics via mechanisms that include LIN41 stimulation, MET and canonical WNT activation, and that the inhibition of WNT at the middle-to-late-reprogramming stage dramatically facilitates the maturation of reprogrammed cells (Fig. 7).
Fig. 7.
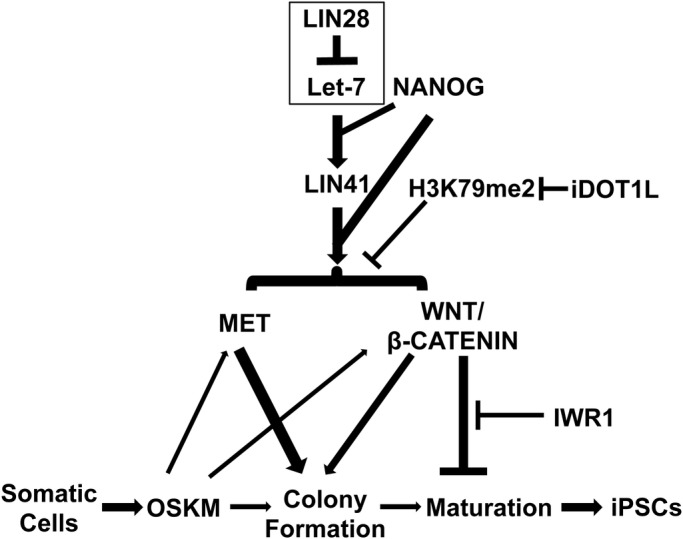
Schematic model for the enhanced reprogramming of human somatic cells by LIN28, NANOG and iDOT1L. Proposed model: the enhanced human somatic cell reprogramming by NANOG and LIN28 involves their synergy in activation of LIN41, which is a target of LIN28/Let-7 pathway. LIN41 can replace LIN28 to synergize with NANOG, achieving the same amplification of reprogramming efficiency as the NANOG and LIN28 combination. This synergy can be further enhanced with the inhibition of H3K79 methyltransferase DOT1L. The strong stimulation of MET and activation of the canonical WNT signaling pathway contribute to the massive colony formation in the optimized reprogramming system. For the WNT pathway, despite its positive role in promoting colony formation, hyperactivation of WNT triggers the differentiation of the emerging presumptive colonies. Hence, inhibition of the WNT pathway by IWR1 at late stage of reprogramming can promote the maturation of the emerging colony, without compromising the induced iPSC colony numbers. A thicker line within the graph indicates a stronger stimulation or inhibition compared with a thinner line toward the same biological effect.
DISCUSSION
Human somatic cell reprogramming by OSKM or OSNL remains highly inefficient. This inefficiency is likely due to the required coordination of many cellular events to overcome the reprogramming roadblocks, including the activation of the cell cycle and MET, the silence of lineage gene expression, metabolic resetting, and the complete activation of the pluripotent regulatory network (Brouwer et al., 2016; Xu et al., 2016). We found that among the combinations of GLIS1, NANOG and LIN28, OSKM-mediated reprogramming is synergistically stimulated by the NL combination, while GLIS1 mitigates this synergy. ‘The pioneering model’ of OSKM-mediated reprogramming showed that OSK factors bind to the shared genomic targets at the initial reprogramming stage to remodel chromatin with the assistance of MYC. This gradually enhances the binding of reprogramming factors to the genomic loci and the activation of the endogenous pluripotent network (Soufi et al., 2012). However, how NL factors induce iPSCs or improve reprogramming efficiency is not completely understood (González and Huangfu, 2016). Let-7 miRNAs promote differentiation by inhibiting the genes targeted by the core reprogramming factors OCT4, SOX2 and NANOG, and the inhibition of Let-7 increased reprogramming efficiency in mice (Melton et al., 2010) and humans (Worringer et al., 2014). However, whether Lin28 can regulate LIN41 in reprogramming has not been demonstrated. We demonstrated that in reprogramming, LIN28 significantly stimulates the expression of LIN41, the direct target of Let-7, consistent with the demonstrated inhibitory function of LIN28 protein for Let-7 miRNA maturation (Heo et al., 2008; Newman et al., 2008; Rybak et al., 2008; Viswanathan et al., 2009). We also found that NL synergize in LIN41 activation and that this effect can be further augmented by inhibiting the H3K79 methyltransferase DOT1L. Similar to NL, LIN41 overexpression significantly promoted epithelial gene expression in reprogramming. We further showed that a dominant-negative mutation of LIN41 greatly suppressed the enhanced reprogramming by NL+iDOT1L. Finally, we showed that the combination of NANOG and LIN41 resulted in similar reprogramming efficiency to NL. Thus, for the first time, our findings established LIN41 as a key downstream effector of the LIN28- and NANOG-mediated enhancement in human iPSC generation and indicate that this mechanism occurs, at least partially, by promoting cellular epithelialization.
The effect of the canonical WNT/β-CATENIN pathway on reprogramming remains contradictory. A recent report showed that in OSKM-mediated human cell reprogramming, the hyperactivation of WNT at early stages promoted iPSC colony formation, while it stimulated differentiation at late stages (Cevallos et al., 2018). However, in mouse cell reprogramming, WNT inhibited early-stage reprogramming but promoted late-stage reprogramming (Ho et al., 2013). Both GLIS1 and NANOG have been indicated to activate certain components of the WNT pathway in reprogramming (Maekawa et al., 2011; Marucci et al., 2014), and WNT and LIN28 co-amplify the expression of their target genes in cancer cells (Tu et al., 2015). However, how canonical WNT activity is regulated by the reprogramming factors for iPSC generation is unclear. We found that NL factors exert synergistic effects in the stimulation of WNT/β-CATENIN activity and that this stimulation can be further enhanced by inhibiting DOT1L. Additionally, we found that NL- and iDOT1L-activated canonical WNT signaling contributes to the kinetics of initial iPSC colony development. We also found that similar to LIN28, LIN41 plays a positive role in stimulating WNT activity in reprogramming. Furthermore, we found that the inhibition of WNT activity from the middle-to-late-reprogramming stage dramatically improved the homogeneity of TRA-1-60+ colonies and the population/intensity of TRA-1-60 expression in reprogrammed cells. This finding correlates with the enhanced expression of late-reprogramming stage markers in reprogrammed cells (Fig. 4G). Our study thus unveiled a mechanism of the synergistic stimulation of LIN41 and canonical WNT activities by NL and the inhibition of H3K79me2 to ensure highly efficient reprogramming from human primary somatic cells; moreover, the suppression of WNT signaling further improved the maturation of reprogrammed cells (Fig. 7). Exactly how LIN41 works with NANOG to activate MET and WNT activities in reprogramming warrants further investigation. The robust reprogramming system we described here would be of great value to study reprogramming mechanisms using primary cell culture and to rapidly establish the appropriate quality and quantity of mature human iPSCs for differentiation studies as well as for further translational research and applications.
MATERIALS AND METHODS
Chemicals and DNA constructs
The DOT1L inhibitor EPZ004777 (iDOT1L) was purchased from AOBIOUS Inc. (Gloucester, MA, USA). WNT inhibitor IWR1 was purchased from Selleckchem (Houston, TX, USA). The constructs pMXs-OCT4, NANOG, LIN28A and GLIS1 were purchased from Addgene (Cambridge, MA, USA). Construction of the polycistronic vector pMXs-KLF4, MYC and SOX2 (KMS) was described in our previous study (Wang et al., 2017). To clone the pMXs-GNL or NL polycistronic vector, the coding sequences for human NANOG, LIN28A and GLIS1 were PCR-amplified from the above-mentioned Addgene constructs. The amplified DNA sequences for each gene were then inserted into linearized pMXs vectors (Cell Biolabs, San Diego, CA, USA) using an In-Fusion kit (Clontech Inc., Mountain View, CA, USA). 2A sequences (Carey et al., 2009; Ryan and Drew, 1994; Ryan et al., 1991) were inserted between each gene.
Retrovirus packaging with 293T cells
293T cells were plated onto six-well plates at 2.5×106 cells/plate. The next day, pMXs constructs, PUMVC and pCMV-VSVG (Addgene) plasmids were co-transfected into 293T cell using Fugene 6 reagent (Promega, Madison, WI, USA). Cell culture media containing retroviruses were harvested at 48 and 72 h post-transfection and filtered through a 0.8 μm filter. The viruses were stored in −70°C before use.
Human somatic cell reprogramming
Primary human umbilical cord-derived MSCs from ATCC (Manassas, VA, USA) were used to carry out the reprogramming experiments. MSCs were maintained with low serum mesenchymal stem cell growth kit (ATCC). For reprogramming, on day −1, MSCs at passages 5–6 were plated onto six-well tissue-culture plates at a density of 5×105 cells/plate. On day 0, retroviruses carrying OSKM and other reprogramming factors were added to the cell culture with 10 μg/ml polybrene and spinfected at 650 g for 45 min. The infected cells on day 4 were passaged onto mitomycin C-treated mouse embryonic fibroblast (MEF) feeders in the presence of 10 μM Y-27632 (Selleckchem) ROCK inhibitor. On day 4, the medium was changed to a 1:1 mix of UC-MSCs medium and human ESC medium. Starting from day 6, the cells were maintained in complete human ESC medium, which contains 20% knockout serum replacement (KSR) in DMEM/F12, supplemented with 1× NEAA, 1× Glutamax, 0.5× penicillin and streptomycin, 4 ng/ml human FGF2 (all from Thermo Fisher Scientific, Waltham, MA, USA) and 1× β-mercaptoethanol (Merck Millipore, Billierica, MA, USA). iDOT1L (3.3 μM) and IWR1 (2.5 μM) were added in reprogramming as specified in the main text and maintained thereafter. For iPSC line characterization, TRA-1-60+ colonies were picked on days 18–21 of reprogramming and grown in human ESC medium on MEF feeders. The colonies were dispatched by 1 mg/ml dispase (Thermo Fisher Scientific) at passage 2, transferred to a Matrigel (Corning Inc., NY, USA) feeder-free system and then cultured in mTeSR1 medium (STEMCELL Technologies, Inc., Vancouver, Canada) for expansion.
EB formation
EB formation experiments were carried out with human iPSC lines at passage 11. When growing to 70–80% confluency with mainly middle-size colonies, the cells were treated with freshly prepared 1 mg/ml dispase for 30 min and removed from the plate by pipetting. After three washes with DMEM/F12, the cells were then plated onto low-adhesive petri dishes in EB formation medium, which is human ESC medium without FGF2. EBs at day 5 were harvested for RNA isolation and gene expression analysis. For immunofluorescence analysis, EBs were treated by TrypLE (Thermo Fisher Scientific) on day 4 and plated onto gelatin-coated plates. The cells were subjected to immunofluorescence staining on day 14.
Immunofluorescence and TRA-1-60 live staining
Putative iPSC lines at passage 11 were subjected immunofluorescence-staining for pluripotent marker expression. The cells from EB differentiation were studied for lineage differentiation markers. For immunofluorescence, the cells were first fixed in 4% PFA for 15 min at room temperature. Following fixation, the cells were treated with 0.5% Triton X-100 in PBS for 15 min at room temperature for cell membrane permeabilization. After blocking, the cells were incubated in primary antibodies for 2 h at 37°C, followed by secondary antibodies at room temperature for 1 h. Cells were counter-stained with DAPI and imaged under a Nikon fluorescence microscope. Primary antibodies including rabbit anti-OCT4 (Merck Millipore), rabbit anti-SOX2 (Abcam, San Francisco, CA, USA), rabbit anti-NANOG (Merck Millipore), NL-557 conjugated OTX2, NL-493 conjugated GATA4 (R&D Systems, Minneapolis, MN, USA) and mouse anti-SMA (Sigma-Aldrich, St. Louis, MO, USA) were used at 1:100 dilution. Alexa Fluro 488 conjugated goat anti-rabbit or goat anti-mouse secondary antibody (Cell Signaling Technology, Danvers, MA, USA) was used in 1:500 dilution.
For TRA-1-60 live staining, the cells in different reprogramming conditions were stained with GloLIVE TRA-1-60 live-stain antibodies (R&D Systems) according to the manufacturer's protocol. Briefly, the cells were incubated in reprogramming media containing TRA-1-60 antibodies at 1:100 dilution for 30 min. The cells were then washed with DPBS and continued to be cultured in reprogramming media. For colony counting, the stained colonies were visualized under a Nikon fluorescence microscope, with homogenous and heterogeneous TRA-1-60+ colony numbers counted. For FACS analysis, cells were treated with TrypLE and resuspended in reprogramming media. Stained cells were then analyzed with a BD LSRFortessa flow cytometer with fluorescence excitation at 557 nm (BD Biosciences, San Jose, CA, USA). FlowJo software was used for data analysis.
qRT-PCR analysis
Total RNAs were isolated from parental MSCs, reprogrammed MSCs, or putative iPSCs, or human H9 ESCs with RNeasy mini kits (Qiagen, Hilden, Germany). Genomic DNAs were removed by DNase I (Qiagen) incubation. 0.5 μg total RNAs were then reverse transcribed into cDNA using iScript reverse transcription supermix (Bio-Rad Laboratories, Hercules, CA, USA). qPCR reactions were performed with SYBR Green supermix (Bimake, Houston, TX, USA) using the ABI 7500 Fast platform (Thermo Fisher Scientific). GAPDH was used as the housekeeping gene for gene expression normalization. Data were processed with the software associated with ABI 7500. Heatmap based on the qRT-PCR data were generated using Heatmapper (Babicki et al., 2016) (www.heatmapper.ca).
Statistical analysis
Unless specifically indicated, all experiments were performed at least three times and data were shown as mean±standard deviations (s.d.) of the mean. Statistical analysis was carried out using either two-sample t-test with Minitab 18, or ANOVA with Randomized Complete Block design (RCB) and LSD post hoc test with SAS 9.4. P<0.05 was considered to be significant.
Supplementary Material
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: L.W., Y.T.; Validation: Y.S., C.H., Y.Y., A.C., A.K.; Formal analysis: L.W., Y.T.; Investigation: L.W., Y.S., C.H., Y.Y., A.C., A.K.; Writing - original draft: L.W., Y.T.; Writing - review & editing: L.W., Y.T.; Supervision: Y.T.; Project administration: Y.T.; Funding acquisition: Y.T.
Funding
This work was supported by the Agriculture and Food Research Initiative Competitive Grant no. 2016-67016-24894 and 2019-67015-29413 to Y.T. from the United States Department of Agriculture (USDA)/National Institute of Food and Agriculture (NIFA), and by a USDA/NIFA W3171/W4171 regional project to Y.T.
Supplementary information
Supplementary information available online at http://bio.biologists.org/lookup/doi/10.1242/bio.047225.supplemental
References
- Andrews P. W., Banting G., Damjanov I., Arnaud D. and Avner P. (1984). Three monoclonal antibodies defining distinct differentiation antigens associated with different high molecular weight polypeptides on the surface of human embryonal carcinoma cells. Hybridoma 3, 347-361. 10.1089/hyb.1984.3.347 [DOI] [PubMed] [Google Scholar]
- Babicki S., Arndt D., Marcu A., Liang Y., Grant J. R., Maciejewski A. and Wishart D. S. (2016). Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 44, W147-W153. 10.1093/nar/gkw419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrott J. J., Cash G. M., Smith A. P., Barrow J. R. and Murtaugh L. C. (2011). Deletion of mouse Porcn blocks Wnt ligand secretion and reveals an ectodermal etiology of human focal dermal hypoplasia/Goltz syndrome. Proc. Natl. Acad. Sci. USA 108, 12752-12757. 10.1073/pnas.1006437108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biechele S., Cox B. J. and Rossant J. (2011). Porcupine homolog is required for canonical Wnt signaling and gastrulation in mouse embryos. Dev. Biol. 355, 275-285. 10.1016/j.ydbio.2011.04.029 [DOI] [PubMed] [Google Scholar]
- Brouwer M., Zhou H. and Nadif Kasri N. (2016). Choices for induction of pluripotency: recent developments in human induced pluripotent stem cell reprogramming strategies. Stem Cell Rev. 12, 54-72. 10.1007/s12015-015-9622-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y., Faddah D. A., Cheng A. W., Itskovich E., Markoulaki S., Ganz K., Klemm S. L., van Oudenaarden A. and Jaenisch R. (2012). Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell 150, 1209-1222. 10.1016/j.cell.2012.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacchiarelli D., Trapnell C., Ziller M. J., Soumillon M., Cesana M., Karnik R., Donaghey J., Smith Z. D., Ratanasirintrawoot S., Zhang X. et al. (2015). Integrative analyses of human reprogramming reveal dynamic nature of induced pluripotency. Cell 162, 412-424. 10.1016/j.cell.2015.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W.-Y., Wei T.-Z., Luo Q.-C., Wu Q.-W., Liu Q.-F., Yang M., Ye G.-D., Wu J.-F., Chen Y.-Y., Sun G.-B. et al. (2013). The Wnt-beta-catenin pathway represses let-7 microRNA expression through transactivation of Lin28 to augment breast cancer stem cell expansion. J. Cell Sci. 126, 2877-2889. 10.1242/jcs.123810 [DOI] [PubMed] [Google Scholar]
- Carey B. W., Markoulaki S., Hanna J., Saha K., Gao Q., Mitalipova M. and Jaenisch R. (2009). Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc. Natl. Acad. Sci. USA 106, 157-162. 10.1073/pnas.0811426106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevallos R. R., Rodríguez-Martínez G. and Gazarian K. (2018). Wnt/beta-catenin/TCF pathway is a phase-dependent promoter of colony formation and mesendodermal differentiation during human somatic cell reprogramming. Stem Cells 36, 683-695. 10.1002/stem.2788 [DOI] [PubMed] [Google Scholar]
- Chan E. M., Ratanasirintrawoot S., Park I.-H., Manos P. D., Loh Y.-H., Huo H., Miller J. D., Hartung O., Rho J., Ince T. A. et al. (2009). Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat. Biotechnol. 27, 1033-1037. 10.1038/nbt.1580 [DOI] [PubMed] [Google Scholar]
- Chen B., Dodge M. E., Tang W., Lu J., Ma Z., Fan C.-W., Wei S., Hao W., Kilgore J., Williams N. S. et al. (2009). Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat. Chem. Biol. 5, 100-107. 10.1038/nchembio.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole M. F., Johnstone S. E., Newman J. J., Kagey M. H. and Young R. A. (2008). Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 22, 746-755. 10.1101/gad.1642408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciat C.-M. and Niehrs C. (2013). Secreted and transmembrane wnt inhibitors and activators. Cold Spring Harb. Perspect. Biol. 5, a015081 10.1101/cshperspect.a015081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson K. C., Adams A. M., Goodson J. M., McDonald C. E., Potter J. C., Berndt J. D., Biechele T. L., Taylor R. J. and Moon R. T. (2012). Wnt/beta-catenin signaling promotes differentiation, not self-renewal, of human embryonic stem cells and is repressed by Oct4. Proc. Natl. Acad. Sci. USA 109, 4485-4490. 10.1073/pnas.1118777109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jaime-Soguero A., Aulicino F., Ertaylan G., Griego A., Cerrato A., Tallam A., Del Sol A., Cosma M. P. and Lluis F. (2017). Wnt/Tcf1 pathway restricts embryonic stem cell cycle through activation of the Ink4/Arf locus. PLoS Genet. 13, e1006682 10.1371/journal.pgen.1006682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dravid G., Ye Z., Hammond H., Chen G., Pyle A., Donovan P., Yu X. and Cheng L. (2005). Defining the role of Wnt/beta-catenin signaling in the survival, proliferation, and self-renewal of human embryonic stem cells. Stem Cells 23, 1489-1501. 10.1634/stemcells.2005-0034 [DOI] [PubMed] [Google Scholar]
- Ecsedi M., Rausch M. and Großhans H. (2015). The let-7 microRNA directs vulval development through a single target. Dev. Cell 32, 335-344. 10.1016/j.devcel.2014.12.018 [DOI] [PubMed] [Google Scholar]
- Fernandez A., Huggins I. J., Perna L., Brafman D., Lu D., Yao S., Gaasterland T., Carson D. A. and Willert K. (2014). The WNT receptor FZD7 is required for maintenance of the pluripotent state in human embryonic stem cells. Proc. Natl. Acad. Sci. USA 111, 1409-1414. 10.1073/pnas.1323697111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golipour A., David L., Liu Y., Jayakumaran G., Hirsch C. L., Trcka D. and Wrana J. L. (2012). A late transition in somatic cell reprogramming requires regulators distinct from the pluripotency network. Cell Stem Cell 11, 769-782. 10.1016/j.stem.2012.11.008 [DOI] [PubMed] [Google Scholar]
- González F. and Huangfu D. (2016). Mechanisms underlying the formation of induced pluripotent stem cells. Wiley Interdiscip. Rev. Dev. Biol. 5, 39-65. 10.1002/wdev.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J., Saha K., Pando B., van Zon J., Lengner C. J., Creyghton M. P., van Oudenaarden A. and Jaenisch R. (2009). Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 462, 595-601. 10.1038/nature08592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I., Joo C., Cho J., Ha M., Han J. and Kim V. N. (2008). Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell 32, 276-284. 10.1016/j.molcel.2008.09.014 [DOI] [PubMed] [Google Scholar]
- Ho R., Papp B., Hoffman J. A., Merrill B. J. and Plath K. (2013). Stage-specific regulation of reprogramming to induced pluripotent stem cells by Wnt signaling and T cell factor proteins. Cell Rep. 3, 2113-2126. 10.1016/j.celrep.2013.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman J. A., Wu C.-I. and Merrill B. J. (2013). Tcf7l1 prepares epiblast cells in the gastrulating mouse embryo for lineage specification. Development 140, 1665-1675. 10.1242/dev.087387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins I. J., Bos T., Gaylord O., Jessen C., Lonquich B., Puranen A., Richter J., Rossdam C., Brafman D., Gaasterland T. et al. (2017). The WNT target SP5 negatively regulates WNT transcriptional programs in human pluripotent stem cells. Nat. Commun. 8, 1034 10.1038/s41467-017-01203-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein S. M. I., Puri M. C., Tonge P. D., Benevento M., Corso A. J., Clancy J. L., Mosbergen R., Li M., Lee D.-S., Cloonan N. et al. (2014). Genome-wide characterization of the routes to pluripotency. Nature 516, 198-206. 10.1038/nature14046 [DOI] [PubMed] [Google Scholar]
- Jiang W., Zhang D., Bursac N. and Zhang Y. (2013). WNT3 is a biomarker capable of predicting the definitive endoderm differentiation potential of hESCs. Stem Cell Rep. 1, 46-52. 10.1016/j.stemcr.2013.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Wu J., Ye S., Tai C.-I., Zhou X., Yan H., Li P., Pera M. and Ying Q.-L. (2013). Modulation of beta-catenin function maintains mouse epiblast stem cell and human embryonic stem cell self-renewal. Nat. Commun. 4, 2403 10.1038/ncomms3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.-Y., Noh H. B., Kim H.-T., Lee K.-I. and Hwang D.-Y. (2017). Glis family proteins are differentially implicated in the cellular reprogramming of human somatic cells. Oncotarget 8, 77041-77049. 10.18632/oncotarget.20334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q. et al. (2010). A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7, 51-63. 10.1016/j.stem.2010.04.014 [DOI] [PubMed] [Google Scholar]
- Lluis F., Ombrato L., Pedone E., Pepe S., Merrill B. J. and Cosma M. P. (2011). T-cell factor 3 (Tcf3) deletion increases somatic cell reprogramming by inducing epigenome modifications. Proc. Natl. Acad. Sci. USA 108, 11912-11917. 10.1073/pnas.1017402108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig T. E., Bergendahl V., Levenstein M. E., Yu J., Probasco M. D. and Thomson J. A. (2006a). Feeder-independent culture of human embryonic stem cells. Nat. Methods 3, 637-646. 10.1038/nmeth902 [DOI] [PubMed] [Google Scholar]
- Ludwig T. E., Levenstein M. E., Jones J. M., Berggren W. T., Mitchen E. R., Frane J. L., Crandall L. J., Daigh C. A., Conard K. R., Piekarczyk M. S. et al. (2006b). Derivation of human embryonic stem cells in defined conditions. Nat. Biotechnol. 24, 185-187. 10.1038/nbt1177 [DOI] [PubMed] [Google Scholar]
- Maekawa M., Yamaguchi K., Nakamura T., Shibukawa R., Kodanaka I., Ichisaka T., Kawamura Y., Mochizuki H., Goshima N. and Yamanaka S. (2011). Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature 474, 225-229. 10.1038/nature10106 [DOI] [PubMed] [Google Scholar]
- Malik N. and Rao M. S. (2013). A review of the methods for human iPSC derivation. Methods Mol. Biol. 997, 23-33. 10.1007/978-1-62703-348-0_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G., Sugimoto T., Diamanti E., Joshi A., Hannah R., Ohtsuka S., Göttgens B., Niwa H. and Smith A. (2012). Esrrb is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell 11, 491-504. 10.1016/j.stem.2012.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marucci L., Pedone E., Di Vicino U., Sanuy-Escribano B., Isalan M. and Cosma M. P. (2014). beta-catenin fluctuates in mouse ESCs and is essential for Nanog-mediated reprogramming of somatic cells to pluripotency. Cell Rep. 8, 1686-1696. 10.1016/j.celrep.2014.08.011 [DOI] [PubMed] [Google Scholar]
- Melton C., Judson R. L. and Blelloch R. (2010). Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 463, 621-626. 10.1038/nature08725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima S., Doi R., Toyoda E., Tsuji S., Wada M., Koizumi M., Tulachan S. S., Ito D., Kami K., Mori T. et al. (2004). N-cadherin expression and epithelial-mesenchymal transition in pancreatic carcinoma. Clin. Cancer Res. 10, 4125-4133. 10.1158/1078-0432.CCR-0578-03 [DOI] [PubMed] [Google Scholar]
- Newman M. A., Thomson J. M. and Hammond S. M. (2008). Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14, 1539-1549. 10.1261/rna.1155108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D. T. T., Richter D., Michel G., Mitschka S., Kolanus W., Cuevas E. and Wulczyn F. G. (2017). The ubiquitin ligase LIN41/TRIM71 targets p53 to antagonize cell death and differentiation pathways during stem cell differentiation. Cell Death Differ. 24, 1063-1078. 10.1038/cdd.2017.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder T. T., Kara N., Cherry A., Sinha A. U., Zhu N., Bernt K. M., Cahan P., Mancarci B. O., Unternaehrer J., Gupta P. B. et al. (2012). Chromatin-modifying enzymes as modulators of reprogramming. Nature 483, 598-602. 10.1038/nature10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G. and Thomson J. A. (2007). Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 17, 42-49. 10.1038/sj.cr.7310125 [DOI] [PubMed] [Google Scholar]
- Polo J. M., Anderssen E., Walsh R. M., Schwarz B. A., Nefzger C. M., Lim S. M., Borkent M., Apostolou E., Alaei S., Cloutier J. et al. (2012). A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 151, 1617-1632. 10.1016/j.cell.2012.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proffitt K. D. and Virshup D. M. (2012). Precise regulation of porcupine activity is required for physiological Wnt signaling. J. Biol. Chem. 287, 34167-34178. 10.1074/jbc.M112.381970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Kim J. K., Tong W., Villa-Diaz L. G. and Krebsbach P. H. (2016). DPPA5 supports pluripotency and reprogramming by regulating NANOG turnover. Stem Cells 34, 588-600. 10.1002/stem.2252 [DOI] [PubMed] [Google Scholar]
- Rao M. S. and Malik N. (2012). Assessing iPSC reprogramming methods for their suitability in translational medicine. J. Cell. Biochem. 113, 3061-3068. 10.1002/jcb.24183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J., Busch J., Mintz E., Ng D., Stanley A., Brafman D., Sutton V. R., Van den Veyver I. and Willert K. (2014). A rare human syndrome provides genetic evidence that WNT signaling is required for reprogramming of fibroblasts to induced pluripotent stem cells. Cell Rep. 9, 1770-1780. 10.1016/j.celrep.2014.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M. D. and Drew J. (1994). Foot-and-mouth disease virus 2A oligopeptide mediated cleavage of an artificial polyprotein. EMBO J. 13, 928-933. 10.1002/j.1460-2075.1994.tb06337.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M. D., King A. M. Q. and Thomas G. P. (1991). Cleavage of foot-and-mouth disease virus polyprotein is mediated by residues located within a 19 amino acid sequence. J. Gen. Virol. 72, 2727-2732. 10.1099/0022-1317-72-11-2727 [DOI] [PubMed] [Google Scholar]
- Rybak A., Fuchs H., Smirnova L., Brandt C., Pohl E. E., Nitsch R. and Wulczyn F. G. (2008). A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 10, 987-993. 10.1038/ncb1759 [DOI] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P., Golipour A., David L., Sung H.-K., Beyer T. A., Datti A., Woltjen K., Nagy A. and Wrana J. L. (2010). Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7, 64-77. 10.1016/j.stem.2010.04.015 [DOI] [PubMed] [Google Scholar]
- Sierra R. A., Hoverter N. P., Ramirez R. N., Vuong L. M., Mortazavi A., Merrill B. J., Waterman M. L. and Donovan P. J. (2018). TCF7L1 suppresses primitive streak gene expression to support human embryonic stem cell pluripotency. Development 145, dev161075 10.1242/dev.161075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Nichols J., Theunissen T. W., Guo G., van Oosten A. L., Barrandon O., Wray J., Yamanaka S., Chambers I. and Smith A. (2009). Nanog is the gateway to the pluripotent ground state. Cell 138, 722-737. 10.1016/j.cell.2009.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack F. J., Basson M., Liu Z., Ambros V., Horvitz H. R. and Ruvkun G. (2000). The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5, 659-669. 10.1016/S1097-2765(00)80245-2 [DOI] [PubMed] [Google Scholar]
- Soufi A., Donahue G. and Zaret K. S. (2012). Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151, 994-1004. 10.1016/j.cell.2012.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staal F. J. T., Luis T. C. and Tiemessen M. M. (2008). WNT signalling in the immune system: WNT is spreading its wings. Nat. Rev. Immunol. 8, 581-593. 10.1038/nri2360 [DOI] [PubMed] [Google Scholar]
- Takahashi K. and Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663-676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K. and Yamanaka S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861-872. 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Sasaki A., Yamamoto M., Nakamura M., Sutou K., Osafune K. and Yamanaka S. (2014). Induction of pluripotency in human somatic cells via a transient state resembling primitive streak-like mesendoderm. Nat. Commun. 5, 3678 10.1038/ncomms4678 [DOI] [PubMed] [Google Scholar]
- Tanabe K., Nakamura M., Narita M., Takahashi K. and Yamanaka S. (2013). Maturation, not initiation, is the major roadblock during reprogramming toward pluripotency from human fibroblasts. Proc. Natl. Acad. Sci. USA 110, 12172-12179. 10.1073/pnas.1310291110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu H.-C., Schwitalla S., Qian Z., LaPier G. S., Yermalovich A., Ku Y.-C., Chen S.-C., Viswanathan S. R., Zhu H., Nishihara R. et al. (2015). LIN28 cooperates with WNT signaling to drive invasive intestinal and colorectal adenocarcinoma in mice and humans. Genes Dev. 29, 1074-1086. 10.1101/gad.256693.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung P.-Y., Varlakhanova N. V. and Knoepfler P. S. (2013). Identification of DPPA4 and DPPA2 as a novel family of pluripotency-related oncogenes. Stem Cells 31, 2330-2342. 10.1002/stem.1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan S. R., Daley G. Q. and Gregory R. I. (2008). Selective blockade of microRNA processing by Lin28. Science 320, 97-100. 10.1126/science.1154040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan S. R., Powers J. T., Einhorn W., Hoshida Y., Ng T. L., Toffanin S., O'Sullivan M., Lu J., Phillips L. A., Lockhart V. L. et al. (2009). Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 41, 843-848. 10.1038/ng.392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Ren D., Guo W., Huang S., Wang Z., Li Q., Du H., Song L. and Peng X. (2016). N-cadherin promotes epithelial-mesenchymal transition and cancer stem cell-like traits via ErbB signaling in prostate cancer cells. Int. J. Oncol. 48, 595-606. 10.3892/ijo.2015.3270 [DOI] [PubMed] [Google Scholar]
- Wang L., Huang D., Huang C., Yin Y., Vali K., Zhang M. and Tang Y. (2017). Enhanced human somatic cell reprogramming efficiency by fusion of the MYC transactivation domain and OCT4. Stem Cell Res. 25, 88-97. 10.1016/j.scr.2017.10.014 [DOI] [PubMed] [Google Scholar]
- Worringer K. A., Rand T. A., Hayashi Y., Sami S., Takahashi K., Tanabe K., Narita M., Srivastava D. and Yamanaka S. (2014). The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell 14, 40-52. 10.1016/j.stem.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Zhang M., Li W., Zhu X., Bao X., Qin B., Hutchins A. P. and Esteban M. A. (2016). Transcriptional control of somatic cell reprogramming. Trends Cell Biol. 26, 272-288. 10.1016/j.tcb.2015.12.003 [DOI] [PubMed] [Google Scholar]
- Yan D., Wiesmann M., Rohan M., Chan V., Jefferson A. B., Guo L., Sakamoto D., Caothien R. H., Fuller J. H., Reinhard C. et al. (2001). Elevated expression of axin2 and hnkd mRNA provides evidence that Wnt/beta-catenin signaling is activated in human colon tumors. Proc. Natl. Acad. Sci. USA 98, 14973-14978. 10.1073/pnas.261574498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Yao H., Zhu P., Zhang X., Pan Q., Gong C., Huang Y., Hu X., Su F., Lieberman J. et al. (2007a). let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131, 1109-1123. 10.1016/j.cell.2007.10.054 [DOI] [PubMed] [Google Scholar]
- Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R. et al. (2007b). Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917-1920. 10.1126/science.1151526 [DOI] [PubMed] [Google Scholar]
- Zhang P., Chang W.-H., Fong B., Gao F., Liu C., Al Alam D., Bellusci S. and Lu W. (2014). Regulation of induced pluripotent stem (iPS) cell induction by Wnt/beta-catenin signaling. J. Biol. Chem. 289, 9221-9232. 10.1074/jbc.M113.542845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ratanasirintrawoot S., Chandrasekaran S., Wu Z., Ficarro S. B., Yu C., Ross C. A., Cacchiarelli D., Xia Q., Seligson M. et al. (2016). LIN28 regulates stem cell metabolism and conversion to primed pluripotency. Cell Stem Cell 19, 66-80. 10.1016/j.stem.2016.05.009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.