ABSTRACT
Understanding how events at the molecular and cellular scales contribute to tissue form and function is key to uncovering the mechanisms driving animal development, physiology and disease. Elucidating these mechanisms has been enhanced through the study of model organisms and the use of sophisticated genetic, biochemical and imaging tools. Here, we present an accessible method for non-invasive imaging of Drosophila melanogaster at high resolution using micro-computed tomography (µ-CT). We show how rapid processing of intact animals, at any developmental stage, provides precise quantitative assessment of tissue size and morphology, and permits analysis of inter-organ relationships. We then use µ-CT imaging to study growth defects in the Drosophila brain through the characterization of abnormal spindle (asp) and WD repeat domain 62 (Wdr62), orthologs of the two most commonly mutated genes in human microcephaly patients. Our work demonstrates the power of combining µ-CT with traditional genetic, cellular and developmental biology tools available in model organisms to address novel biological mechanisms that control animal development and disease.
KEY WORDS: Micro-computed tomography, Abnormal spindle, Microcephaly, Drosophila, Human disease modeling, Phenotyping
Summary: Micro-computed tomography in Drosophila is used to visualize all organs in an intact animal at micron resolution, providing a useful tool for whole-animal phenotyping.
INTRODUCTION
Drosophila melanogaster is a foundational model organism that has contributed to our understanding of the fundamental cellular and developmental pathways required to build complex multicellular animals. Furthermore, flies have become an important model for over 230 human diseases, including cancer, cardiovascular diseases, neurological and metabolic disorders, and others that span nearly every organ system (Millburn et al., 2016). About 65% of genes associated with human disease are conserved in flies, and genome engineering (e.g. P-element transformation and CRISPR/Cas9) can be used to manipulate the Drosophila orthologs of these genes, or introduce human transgenes containing exact patient mutations, to improve understanding of disease etiology (Ugur et al., 2016). The powerful genetic tools available in Drosophila, coupled with biochemical, molecular and modern imaging methods, has allowed researchers to uncover the mechanisms underlying normal development, and how failure of these mechanisms can lead to disease.
Visualization of subcellular, cellular and tissue organization in the fly relies heavily on fluorescent-based light microscopy, such as confocal, which has elucidated countless biological mechanisms in the fly. Other imaging techniques, such as electron microscopy and super-resolution microscopy, offer additional advantages with respect to resolution. Together, these imaging modalities allow biological mechanisms to be investigated across wide spatial scales, from molecules to tissues. However, imaging methods that permit whole-animal analysis are grossly underutilized by cell and developmental biologists. This holistic analysis provides significant insight into how animals normally develop and how inter-organ assembly and communication influences disease states. Furthermore, analysis of whole animals can uncover roles for proteins in unexpected tissue types. Thus, there is an opportunity to enhance our understanding of basic developmental mechanisms using imaging technologies that complement commonly used techniques such as light and electron microscopy.
We sought a modality that permits non-invasive imaging of intact animals, which can be easily incorporated into our current genetic, cell and developmental research. We considered several platforms available for medical research, including magnetic resonance imaging (MRI) and optical coherence tomography (OCT), and ultra-microscopy (Holmes, 2009; Jährling et al., 2010; Morton et al., 1990; Null et al., 2008). We focused on micro-computed X-ray tomography (μ-CT), which provides high-resolution and nondestructive imaging of tissues in a commercially available and readily accessible platform. μ-CT relies on attenuation of X-rays to generate image contrast in acquired radiographs, followed by 3D reconstruction algorithms to produce tomograms (Morton et al., 1990). Pre-clinical μ-CT has proven useful for anatomical and developmental assessment of vertebrate models, including mice, rats and zebrafish, which has contributed greatly to our understanding of disease and genetic mechanisms (Dyer et al., 2017; International Mouse Knockout Consortium et al., 2007; Weinhardt et al., 2018; Wong et al., 2012).
μ-CT has also been used for descriptive and comparative analysis of tissue morphology and gross development studies of insects such as bees (Apis and Bombus sp.), blow flies (Calliphora sp.) and hornets (Vespa) (Betz et al., 2007; Metscher, 2009; Smith et al., 2016; Sombke et al., 2015; Wipfler et al., 2016). However, μ-CT is grossly underutilized in Drosophila melanogaster biology; only a handful of studies have used μ-CT (Chen et al., 2017; Fabian et al., 2016; Harrison et al., 2017; Klok et al., 2016; Mattei et al., 2015; Mizutani et al., 2008, 2013), and even fewer studies have used μ-CT alongside traditional molecular, cellular and developmental biology techniques. The ability to use μ-CT as a whole-animal phenotyping tool in this context could significantly enhance our mechanistic understanding of development, anatomy and disease.
Here, we establish μ-CT as a versatile and effective tool to complement traditional imaging platforms for studying organ development and tissue morphogenesis. We highlight how μ-CT provides a detailed atlas of the entire intact animal at micrometer resolution across all developmental stages, allowing for unbiased analysis of mutant animals by investigating all tissues, not simply those predicted to be defective. We provide a labeling and imaging protocol that allows for nondestructive imaging of larval, pupal and adult tissues in their native context. We then demonstrate its use for performing morphometric analysis. Finally, we use μ-CT to phenotype a Drosophila model of autosomal recessive primary microcephaly (MCPH) by investigating mutations in abnormal spindle (asp) and WD repeat domain 62 (Wdr62). Our analysis identified the N-terminal region of Asp as a requirement for the formation of proper adult brain size, and also uncovered previously unreported defects in the gut and visual system in asp mutant animals. These data showcase μ-CT as an accessible component of the model organism research toolkit that can uncover novel developmental mechanisms.
RESULTS
A protocol for micro-computed tomography (μ-CT) of Drosophila melanogaster
Our μ-CT protocol for Drosophila was adapted from methods used to visualize insects (Smith et al., 2016; Sombke et al., 2015; Swart et al., 2016). We prioritized rapid fixation and labeling to best preserve larval, pupal and adult tissues in as close to their native state as possible, although the chemical solvents do cause mild tissue shrinkage (Materials and Methods). μ-CT generates an image through the differential attenuation of X-rays as they pass through a rotating sample towards the detector (Fig. S1A-C). To visualize soft tissue, which do not attenuate X-rays well enough to generate image contrast, elements with high atomic numbers, such as iodine-iodide (I2KI) and phosphotungstic acid (PTA), are used to stain tissue prior to X-ray exposure. We find that the best results are obtained when animals are fixed using Bouin's Solution, washed in an isotonic buffer, then labeled with either iodine or PTA prior to scanning with the animals fully hydrated in water. Iodine is used as a stain that rapidly (1-2 days) labels soft tissue in intact adults. PTA requires longer incubation times (3-5 days) and disruption of the cuticle by poking it with a microdissection needle or removing the mouthparts, but allows for sharper visualization of tissue.
To generate an image, a series of 2D ‘projection’ images are acquired as the sample rotates. Reconstruction software, based on the Feldkamp algorithm, is used to generate stacks of cross-sectional slices with isotropic pixel resolution from the projection images to derive 3D information of the object, which is used for visualization, segmentation and analysis. Scan times per animal range from 20 min to overnight depending on the resolution desired. Spatial resolution in μ-CT depends on many factors, but generally increases with the number of projection images acquired during a scan as the sample rotates. Images suitable for both determining the spatial arrangement of organs and performing morphometric (size and shape) analysis can be derived from ∼300 projections acquired during a 20-30 min ‘fast’ scan (Fig. S2A,A′). Increasing the number of projection images that are collected during a scan (∼1500) leads to better spatial resolution, allowing for more detailed analysis of tissues, such as the individual neuropils of the adult fly brain (Fig. S2B,B′). These ‘slow’ scans require 8-16 h.
Although our scanner is capable of achieving resolutions as high as 5.5 μm under perfect imaging conditions (Morales et al., 2016), our scans average an effective resolution of 10-25 μm for slow scans and 25-50 μm for fast scans (see Materials and Methods). However, our measurements of tissue size are identical between slow and fast scans (Fig. S2C). We therefore use fast scans for all our tissue measurements, and slow scans when evaluating detailed tissue architecture. Using this protocol, we obtain consistent and robust visualization of soft tissue with minimal reagent cost and hands-on time (∼15 min total; Movies 1, 2).
µ-CT visualization of Drosophila larvae
A key advantage of our protocol is that it works for any developmental stage. Drosophila are holometabolous insects, characterized by dramatic changes in morphology as they progress through defined embryonic, larval, pupal and adult stages. Furthermore, many disease phenotypes manifest during periods of rapid cell division and growth, such as those occurring during the larval instars, or during metamorphosis when complex tissue morphogenesis takes place. Light microscopy, enhanced by either chemical clearing methods or larval fillets, has been used to visualize a limited subset of larval tissues (Andlauer and Sigrist, 2012; Parton et al., 2010; Pende et al., 2018), but visualization of all larval organs simultaneously has not been reported.
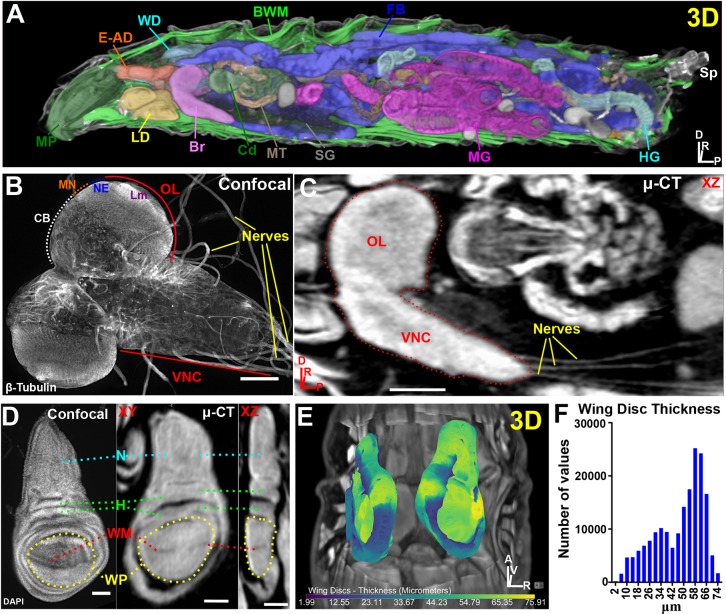
To demonstrate the capabilities of µ-CT for high-resolution whole-animal imaging, we imaged intact wild-type (yw) third instar larvae (Fig. 1). The tomograms produce a fully reconstructed representation of the animal with sufficient resolution and contrast to visualize every internal organ, including those most commonly studied by confocal microscopy such as the imaginal discs, nervous system, gut, and reproductive organs (Fig. 1A, Movie 3). Additionally, tissues much more difficult to dissect are readily identifiable such as the trachea, Malpighian tubules, fat bodies, body wall muscles and the heart (Fig. 1A, Movie 3).
Fig. 1.
µ-CT reveals all major organs in Drosophila melanogaster larvae. (A) 3D display of a larva optically sliced along the xz axis showing the mouthparts (MP), eye-antennal discs (E-AD), wing discs (WD), body wall muscles (BWM), fat body (FB), spiracles (Sp), hindgut (HG), midgut (MG), salivary glands (SG), Malpighian tubules (MT), cardia (Cd), brain (Br) and leg discs (LD). (B) Confocal image of explanted larval brain labeled with β-tubulin to visualize nerves. Ventral nerve cord (VNC), optic lobe (OL), central brain (CB), medulla neuroblasts (MN), neuroepithelium (NE) and lamina (Lm) neuropil are denoted. (C) µ-CT of larval brain (dotted outline) viewed along the xz axis. Note extension of nerves to target organs from the VNC. (D) Confocal image of DAPI-stained wing disc (left) and a µ-CT-imaged wing disc (right). Notum (N), hinge (H), wing margin (WM) and wing pouch (WP) are labeled; also note epithelial folds in the xz view. (E) 3D representation of wing disc position within the body cavity. Discs are colored by thickness (µm) (see Materials and Methods). (F) Histogram of wing disc thickness values. Axis denotes origin; A, anterior; D, dorsal; P, posterior; R, right; V, ventral. Scale bars: 100 µm (B,C); 50 µm (D). Stained with 0.1 N iodine and scanned in slow mode at an image pixel size of 2 µm.
Beyond identification of the major organs, many tissue details clearly emerge. For example, microtubule-rich nerves extending from the larval ventral nerve cord (VNC) can be imaged by confocal microscopy at high spatial resolution with β-tubulin antibodies, but information about these structures within the context of the intact animal is lost upon dissection (Fig. 1B). In contrast, µ-CT allows nerves extending from the VNC to be traced to their target tissues throughout the body cavity (Fig. 1C). Another example is µ-CT scanning of the major features of wing imaginal discs, routinely studied by confocal microscopy, including the wing margin (dorsal/ventral boundary), epithelial folds, wing pouch, notum and hinge (Fig. 1D). A primary advantage of these µ-CT scans, however, is that they can be used to derive precise morphometric information (Fig. 1E,F), which can then be combined with information gained from light microscopy methods to study comprehensively mechanisms that control tissue shape. Furthermore, the wing disc's relationship to other imaginal disc organs, and to other organs (such as its attachment to the trachea), can be simultaneously assessed in order to understand how spatial organization of tissues within the body cavity influences proper patterning and placement of adult structures (Fig. 1E).
Collectively, these data demonstrate that a simple staining protocol, which requires no dissection and minimal effort/bench time, provides high-quality images from which morphological information can be derived on all organ systems in situ, and allows the detection of defects anywhere in developing larvae.
µ-CT can be used to study one of the most difficult stages of development – pupation
Where µ-CT truly excels is for the observation of metamorphosis during pupation (Fig. 2, Movies 4-6). The rapid morphological changes during pupal development, coupled with the histolysis of larval tissues, makes dissection of intact organs for detailed analysis extremely difficult, sometimes impossible. As a result, many pupal tissues remain highly understudied and therefore our knowledge of the mechanisms driving their transition to adult structures is far from complete. This presents an exciting opportunity for discovery considering that pupation and the pharate adult stage encompass a 5-day window of time, nearly 50% of the pre-adult fly lifespan.
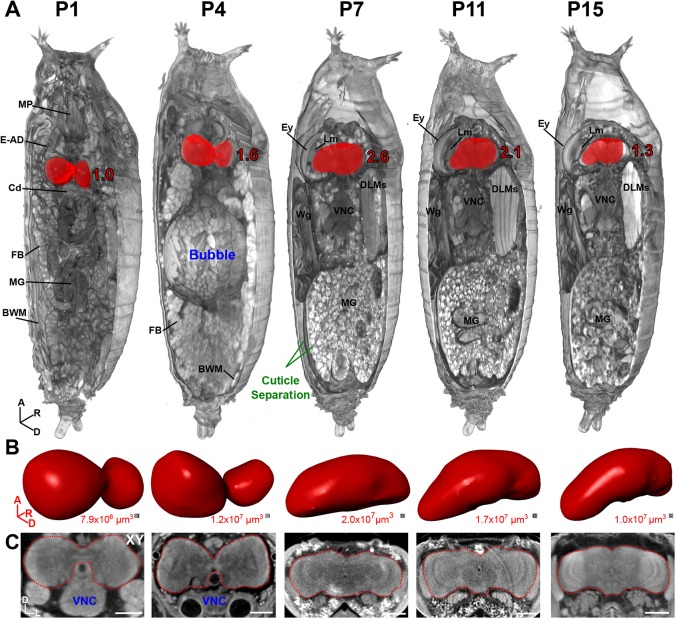
Fig. 2.
Developmental anatomy of Drosophila brains during pupation viewed by µ-CT. (A) 3D representation of pupae at stage P1, early P4, P7, P11 and P15. Part of the pupal cuticle and underlying soft tissue were digitally removed to reveal internal structures (axis denotes origin; A, anterior; D, dorsal; R, right). Brain is highlighted in red; values denote brain size relative to P1. Note the prominent abdominal air bubble at early P4 and cuticle separation between the pupal and adult epidermis at the pharate adult stage P7. BWM, body wall muscles; Cd, cardia; DLMs, dorsal longitudinal muscles; E-AD, eye-antennal discs; Ey, eye; FB, fat body; Lm, lamina; MG, midgut; MP, mouthparts; VNC, ventral nerve cord; Wg, wing. (B) Isolated brains from A, shown in same orientation, rendered as a 3D surface. Volume measurements are given below. (C) 2D xy anterior view of the brains in A. A, anterior; D, dorsal; L, left; R, right. Scale bars: 100 µm. Stained with 0.1 N iodine and scanned in slow mode at an image pixel size of 1.4 µm.
To highlight the capabilities of µ-CT in the study of metamorphosis, we focused on the pupal brain, which undergoes dramatic morphological changes between larval and adult stages. Our µ-CT analysis revealed highly dynamic changes in brain morphology and volume throughout this period, increasing 2.6-fold in size from P1 to P7 (pharate adult) before decreasing to a final volume of 1.3-fold (relative to P1) at P15 prior to eclosion (Fig. 2, Fig. S3A,B, Movies 4, 5). Furthermore, the distinctive morphological features of pupae described by Bainbridge and Bownes, such as the air bubble at P4 and full separation of the pupal and adult cuticle that marks the pharate adult stage, are clearly evident (Fig. 2, Fig. S3A,B) (Bainbridge and Bownes, 1981).
In addition to normal development, µ-CT is advantageous for the analysis of mutations that cause pupal lethality (over 2500 alleles listed on FlyBase). In combination with the morphological characterization of each pupal stage by Bainbridge and Bownes (1981), which are easily visible in the tomograms, one can potentially determine the stage and cause of lethality. Such analysis is difficult using light microscopy because pupal dissection often results in a mass of disintegrating tissue, which makes identification of defects much harder. Tissue-clearing techniques combined with ultramicroscopy can visualize intact pupal stages (Pende et al., 2018), but are suited for tissue-specific analysis rather than a systemic evaluation of all organ systems at once.
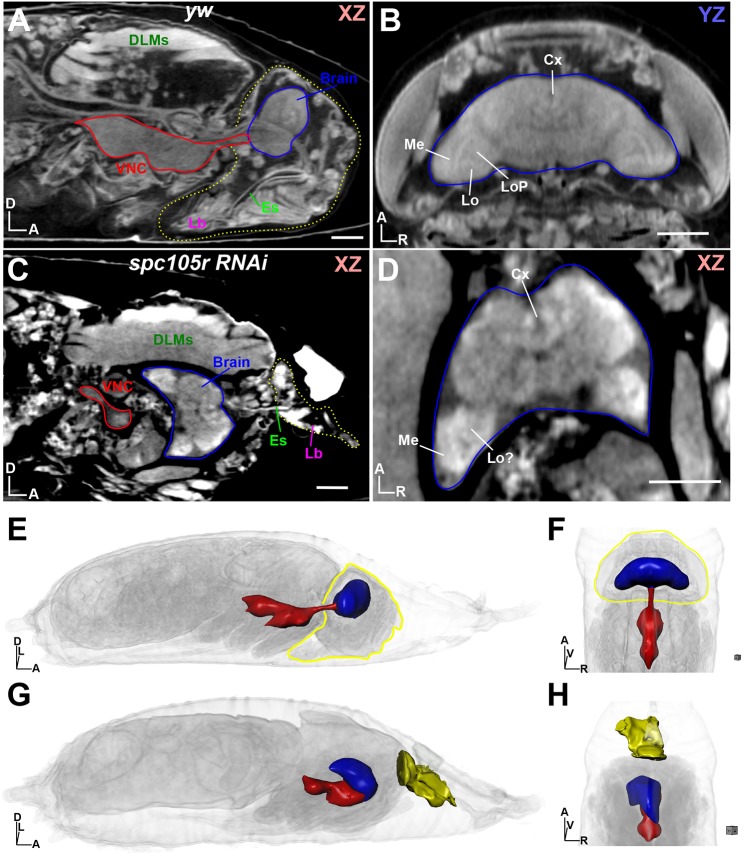
To demonstrate the capabilities of μ-CT in investigating pupal lethal phenotypes, we knocked down Spc105R (ortholog of the human KNL1) with a Gal4 driver used to manipulate the larval brain mushroom body (eyOK107). µ-CT revealed that that pupae die as pharate adults during a variable window spanning P11-P15 (Fig. 3, Movie 6). Moreover, these pupae fail to make proper head structures, instead possessing a remnant structure attached to the thorax that closely resembles the labellum and esophagus, but no antennae, eyes, ocelli or other mouthparts (Fig. 3C). By carefully investigating the µ-CT reconstructions in 2D and 3D, we identified a near-completely developed brain inside the thorax, located ventrally to the dorsal longitudinal muscles (DLMs) and attached to the VNC at an orthogonal angle (Fig. 3C,D,G,H, Movie 6). We were also able to identify specific brain regions (central brain and optic lobe) and individual neuropils, such as the medulla, lobula, lobula plate and central complex, although their morphology was clearly defective compared with P15 yw control brains (Fig. 3A-D, Movie 5). The optic lobes in the Spc105R RNAi flies are deformed toward the posterior, possibly shaped by the confines of the surrounding tissues; we could not readily identify lamina, retina or ocelli structures associated with the ectopically localized brain. Interestingly, our Spc105R RNAi phenotypes (pharate adult lethality, remnant head structure), are similar to those reported for loss of the Pax6 orthologs eyeless (ey) and twin of eyeless (toy) in eye-antennal discs (Wang and Sun, 2012; Zhu et al., 2017). Whether the Pax6 animals also possessed an ectopically developing brain in the thorax was not reported, which may have simply been overlooked owing to technical limitations.
Fig. 3.
Dissection of a ‘pupal lethal’ mutation by µ-CT. (A) Wild-type (yw) pupa at stage P15 shown in xz orientation. The headcase is outlined by a yellow dotted line, with the ventral nerve cord (VNC) and brain outlined in red and blue, respectively. DLMs, dorsal longitudinal muscles; Es, esophagus; Lb, labellum. (B) View of the yw headcase from A, viewing the yz axis from the dorsal perspective. Brain is outlined in blue. Individual brain neuropils are denoted. Cx, central complex; Lo, lobula; LoP, lobula plate; Me, medulla. (C) Analysis of a pupal lethal ‘headless’ fly, resulting from RNAi depletion of spc105r (KNL1) using the eyOK107 Gal4 driver. View shown is xz, with the ‘head remnant’ outlined by a yellow dotted line. Note brain (blue outline) location in the thorax; VNC is outlined in red. (D) View of the mutant brain shown in C; neuropils are highlighted as described in B. (E) 3D view of the yw pupae shown in A; the headcase is outlined in yellow and the brain (blue) and VNC (red) are rendered as 3D surfaces. (F) Alternative view of the yw pupae from E, viewed from the dorsal perspective. (G) 3D view of the spc105r as denoted in E; head remnant containing the labellum is shown as a yellow surface. (H) Alternative view of the spc105r pupae from G, viewed from the dorsal perspective. Body axes are indicated: A, anterior; D, dorsal; L, left; R, right; V, ventral. Scale bars: 100 µm. Stained with 0.1 N iodine and scanned in slow mode at an image pixel size of 1.4 µm.
µ-CT as a tool for both morphological and quantitative analysis of Drosophila tissue for phenotyping
The ability to reproduce structures precisely in 3D without any tissue loss or deformation is one key advantage of µ-CT over conventional histology techniques. From a single µ-CT scan, morphometric analysis of tissues can reveal defects in growth, morphogenesis and patterning in an unbiased manner. Once a tissue defect is identified, then traditional light and electron microscopy tools can be used to investigate the tissue at even greater resolution, providing a useful pipeline for evaluating normal development and anatomy as well as relevant human disease phenotypes from the nanometer to the millimeter scale. Here, we present a few examples of adult tissue analysis that can be reliably performed using µ-CT.
Muscle
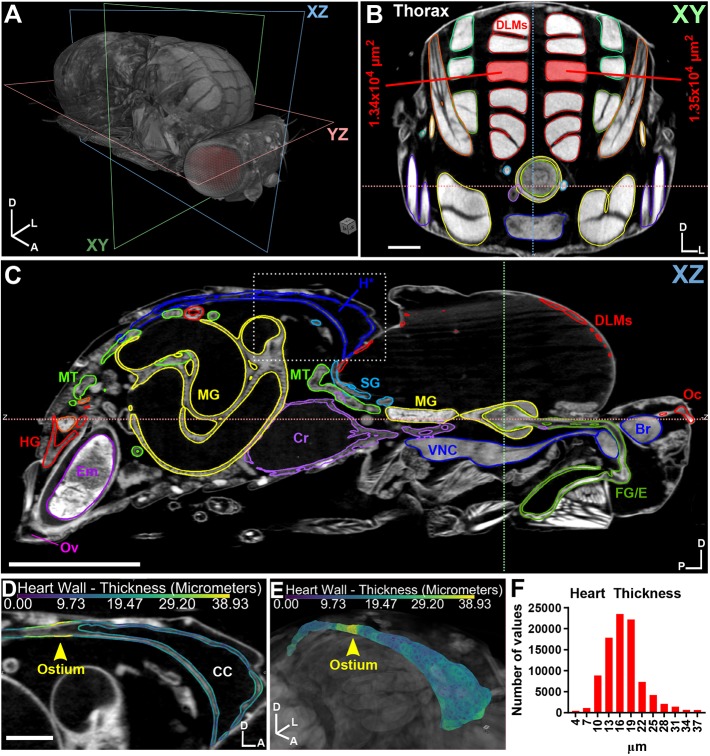
Flies serve as an excellent model for identifying muscle-related defects, similar to those found in human patients with muscle-wasting disorders (Kreipke et al., 2017). Morphometric assessment of 3D muscle volume or 2D cross-sectional area of the DLMs (Fig. 4A,B, Movie 7) can be used to assess phenotypes accurately in a single scan, as opposed to traditional histology techniques, which are labor intensive, prone to tissue deformation artifacts from sectioning and provide only 2D information.
Fig. 4.
µ-CT of adult Drosophila melanogaster highlighting muscle and heart tissue. (A) 3D representation of an adult female fly with the body axis (A, anterior; D, dorsal; L, left) and imaging planes (xy, xz, yz) denoted. (B) 2D xy anterior view of the thorax revealing muscles. A pair of dorsal longitudinal muscles (DLMs) are highlighted and the cross-sectional area is measured. Other muscle groups (red, green, orange, yellow and blue), gut (yellow), salivary glands (aqua blue) and crop (purple) are also shown. (C) xz view; organs are segmented by color. Br, brain; Cr, crop; DLMs, dorsal longitudinal muscles; Em, embryo; FG/E, foregut/esophagus; H, heart; HG, hindgut; MG, midgut; MT, Malpighian tubule; Oc, ocelli; Ov, ovipositor; SG, salivary gland; VNC, ventral nerve cord. (D) Close up of the boxed region in C revealing the heart. Heart wall thickness (µm) is indicated by color code (see Materials and Methods). Position of an ostium is shown (arrowhead). CC, conical chamber. (E) 3D representation of the heart, colored by thickness (µm). (F) Histogram of heart thickness values. Body axes are indicated: A, anterior; D, dorsal; L, left; P, posterior. Scale bars: 100 µm (B,D); 500 µm (C). Stained with 0.1 N iodine and scanned in slow mode at an image pixel size of 1.25 µm.
Heart
The fly heart has become a reliable system in which to study human cardiomyopathies. The adult heart tube consists of only two cell types (cardiomyocytes and pericardial cells), with extracellular matrix (ECM), the ventral longitudinal muscles, and alary muscles providing structural support (Rotstein and Paululat, 2016). We used μ-CT to assess heart morphology and thickness to evaluate both the integrity of the heart wall and the ostia, valve-like structures that direct hemolymph flow in the body cavity (Fig. 4C-F, Movie 8). We found μ-CT to be sensitive enough to easily detect and discriminate between the heart wall (16 μm) and the ostia (35 μm), which presumably appear thicker than the heart wall because of the additional ostial cells that form the valve (Wasserthal, 2007). Thus, μ-CT is an excellent imaging tool for examining extremely delicate tissues such as the heart.
Reproductive system
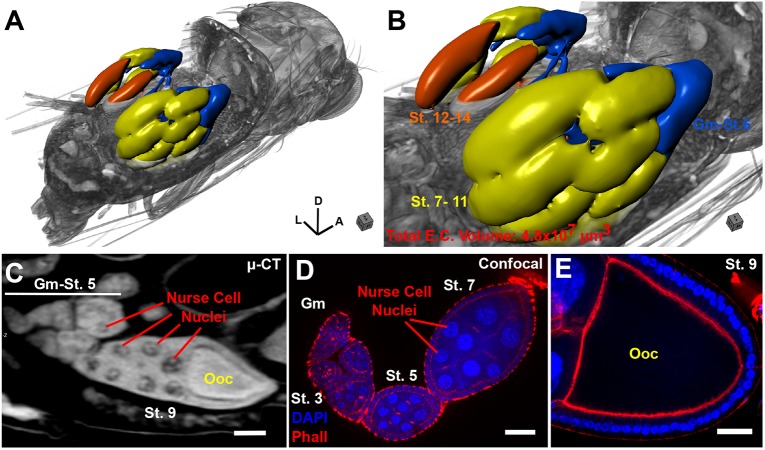
The fly has served as a model for the genetic dissection of sexual reproduction, with almost 2500 genes associated with this term listed on FlyBase. We find that μ-CT can provide useful information for both the male and female reproductive systems (Figs 5, 6, Fig. S10). For example, high-resolution μ-CT scans of an adult female provide a wealth of information about overall ovary organization and individual egg chambers at each stage of development (germarium-stage 14; Fig. 5A-E, Movie 9). We can reliably distinguish nurse cells and their polytene chromosomes, along with clear visualization of the oocyte, similar to confocal microscopy using fluorescent markers (Fig. 5C-E). However, imaging the intact animal using µ-CT allows the entire reproductive system to be studied in its natural spatial arrangement, and its association with other structures, such as the ovipositor (Fig. 4C, OV), is more difficult to study by confocal microscopy. It is noteworthy that the female we present here was withholding eggs, as the embryo seen in the ovipositor has already begun the process of gastrulation, which normally occurs hours after egg laying (Fig. 4C, Movies 1, 2).
Fig. 5.
µ-CT of adult Drosophila melanogaster highlighting the female reproductive system. (A) 3D view of a female fruit fly, with the abdomen digitally removed to reveal the underlying structure of the ovarioles within the ovary. Body axes are denoted: A, anterior; D, dorsal; L, left. (B) Close-up of A; ovarioles are rendered as a surface and colored according to oogenesis stage [blue, germarium (Gm)-Stage (St.) 6); yellow, Stage 7-11; orange, Stage 12-14]. Total egg chamber volume is shown. (C) 2D image of an egg chamber imaged by µ-CT. Stages are denoted along with position of the oocyte (Ooc) and nurse cell nuclei (note visualization of polytene chromosomes). (D) Confocal image of an ovariole stained with DAPI and phalloidin; stages are indicated. (E) Confocal image of an oocyte from a stage 9 egg. Scale bars: 50 µm (C-E). Stained with 0.1 N iodine and scanned in slow mode at an image pixel size of 1.25 µm.
Fig. 6.
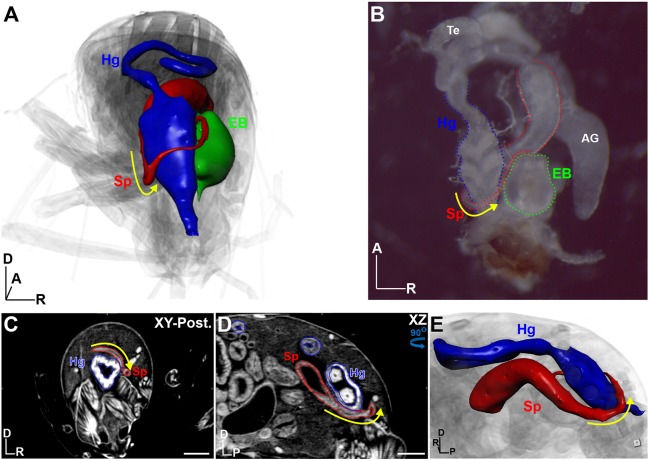
Left-right asymmetry as an example of inter-organ analysis. (A) 3D view from the posterior perspective of a male adult fly. The hindgut (Hg, blue), spermiduct (Sp, red) and ejaculatory bulb (EB, green) are rendered as surfaces. Yellow arrow indicates dextral (left-right) looping of the spermiduct over the hindgut into the ejaculatory bulb, located on the right side of the body axis. (B) Dissected version of the organs outlined in A; testes (Te) and accessory glands (AG) are also labeled. (C) xy view from the posterior of the fly shown in A; the Sp and Hg are outlined. (D) xz view of the male abdomen shown in C. (E) 3D view of the Sp and HG as shown in D, viewed from the left perspective. Yellow arrow denotes dextral looping. Body axes are indicated: A, anterior; D, dorsal; P, posterior; R, right. Scale bars: 100 µm. Flies were stained with 0.1 N iodine and scanned in slow mode at an image pixel size of 1.25 µm.
Inter-organ relationships
The ability to visualize all organs simultaneously in an intact animal permits the study of interactions between organs, a vastly underappreciated area of research that is crucial to understanding the entire phenotypic spectrum of a disease mutation. As an example, left-right asymmetry is a fundamental biological property found throughout metazoans. This is most easily recognized by the human disorder known as situs inversus, in which internal organs are located on the opposite side of the body axis in a mirror-like fashion. In Drosophila, this property is exemplified by the directional looping of the spermiduct over the hindgut in a dextral (clockwise) fashion when viewed from the posterior, which occurs during metamorphosis (Adám et al., 2003) (Fig. 6, Movie 10). μ-CT is perfectly suited to address these types of organ-organ relationships, especially those involving very delicate or interconnected tissues, to probe fundamental questions of development, or to ask questions relating to systemic effects of a diseased tissue.
Nervous system
Studies of the adult fly central nervous system (CNS), consisting of the VNC located in the thorax and the brain located in the headcase, can also be enhanced through μ-CT to understand mechanisms of brain development, homeostasis and neurodegeneration. For example, older adult brains (40 days old) display more vacuoles than young brains. These vacuoles are easily visualized in 2D μ-CT scans and can be rendered in 3D to examine spatial relationships or perform morphometric analysis (Fig. S4). This analysis can be useful for distinguishing between vacuoles and other brain structures. For example, brain vacuoles generally appear ‘spherical’ when rendered in 3D and display higher sphericity (ψ) values (>0.80; a perfect sphere has a value of 1) than other brain structures, such as fiber tracts (<0.80; Fig. S4D). We find the brain vacuoles to be an excellent biomarker for aging phenotypes as well as neurodegenerative disorders, with μ-CT offering advantages over histology techniques, which are more prone to image artifacts (Sunderhaus and Kretzschmar, 2016).
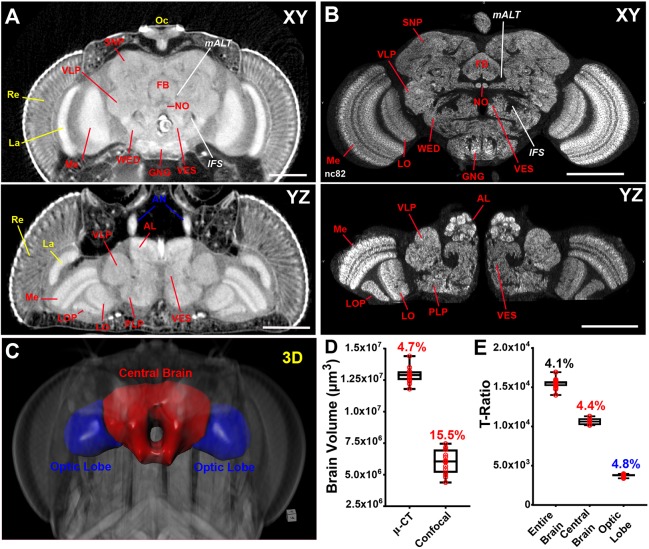
As a final demonstration, we performed an in-depth characterization of the adult nervous system using µ-CT and confocal microscopy (Fig. 7, Movies 11, 12). Confocal microscopy is particularly suitable for high-resolution imaging of brain morphology as it is possible to resolve individual neuropil structures clearly (Ito et al., 2014). Most, but not all neuropils designated from confocal imaging of adult brains can be assigned in our µ-CT scans of PTA-labeled fly heads (Fig. 7A,B, Movie 12). However, µ-CT provides additional information about the organization of the visual circuit by allowing simultaneous observation of the lamina, retina and ocelli as a result of imaging an intact headcase.
Fig. 7.
A comparison of µ-CT and confocal microscopy for quantitative and morphological analysis of the nervous system. (A) Anterior xy (top) and dorsal yz (bottom) view of an adult brain labeled with PTA and scanned in slow mode at an image pixel size of 700 nm. Locations of individual neuropil structures and fiber tracts are noted. (B) xy (top) and yz (bottom) view of an adult brain stained for the synapse marker nc82 (anti-Bruchpilot) and imaged by confocal. AL, antennal lobe; AN, antenna nerve; FB, fan-shaped body; GNG, gnathal ganglia; IFS, inferior fiber system; La, lamina; LO, lobula; LOP, lobula plate; mALT, medial antennal lobe tract; Me, medulla; NO, noduli; Oc, ocelli; PLP, posteriorlateral protocerebrum; Re, retina; SNP, superior neuropils; VES, vest; VLP, ventrolateral protocerebrum; WED, wedge. (C) 3D surface representation of the adult brain, indicating the optic lobes (blue) and central brain (red). (D) Comparison of brain volume measurements between µ-CT and confocal microscopy. Box and whisker plots are shown. Box values represent the 25th, 50th and 75th percentiles. Whiskers represent the minimum/maximum range. Coefficient of variation (CV%) is given. n=15 brains. (E) Measurements of entire brain, central brain and optic lobe volumes normalized to thorax width presented as a T-ratio (raw brain volume/thorax width). All µ-CT measurements were obtained from animals scanned in fast mode (20 min) at an image pixel size of 3 µm. Coefficient of variation (CV%) given. n=5 brains. Scale bars: 100 µm.
Additionally, although both methods provide 3D information about tissues, μ-CT is more precise for morphometric analysis than is confocal microscopy. For example, the average brain volume of 15 animals measured by µ-CT is 12.9×106±6.0×105 µm3 with a coefficient of variation (CV) of 4.7%, compared with a confocal measurement of 6.0×106±9.3×105 µm3 with a CV of 15.5% (Fig. 7C,D). Not only are these µ-CT measurements more precise, but they expose an alarming >50% lower estimate of whole brain volume when measured by confocal imaging. This difference is likely due to solvent effects from chemical clearing of the brains prior to confocal imaging, including the use of ethanol, xylene and DPX (see Materials and Methods; Fig. S5D). These solvents have been reported to have a significant shrinkage effect on tissues, including the fly brain (Nern et al., 2015; Stowell, 1941). Thus, the trade-off should be carefully considered when chemical clearing of tissues is required for light microscopy investigations. Our data also show that any sample preparation method can negatively affect tissue size, including ours for μ-CT (Fig. S5D). It is therefore imperative that these conditions be kept identical for experimental and control samples for accurate phenotyping.
To gain additional precision from µ-CT, measurements can be normalized to thorax width to account for overall body size variation (Fig. S5) (Stillwell et al., 2011). Thorax width is an accurate predictor of body mass in other holometabolous insects such as bumblebees (Hagen and Dupont, 2013; Smith et al., 2016). In Drosophila, thorax width is consistent across different mutant genotypes, is not affected by aging, and accurately reflects the sexual dimorphism in body size, making it a simple and precise measurement (referred to here as the T-ratio) to extract from any tomogram (Fig. 7E, Fig. S5B,C). T-ratio analysis of our dataset revealed a CV of only 4.1%. Thus, we prefer to use T-ratios when performing µ-CT measurements on whole or sub-regions of the brain (Fig. 7E), as well as other tissues.
Taken collectively, these data suggest that performing relative measurements of tissues eliminates sampling error, provides an accurate representation of biological variation in tissue size, and reduces the required sample size, making µ-CT a powerful and reliable tool for size and shape analysis.
µ-CT for modeling human microcephaly in Drosophila
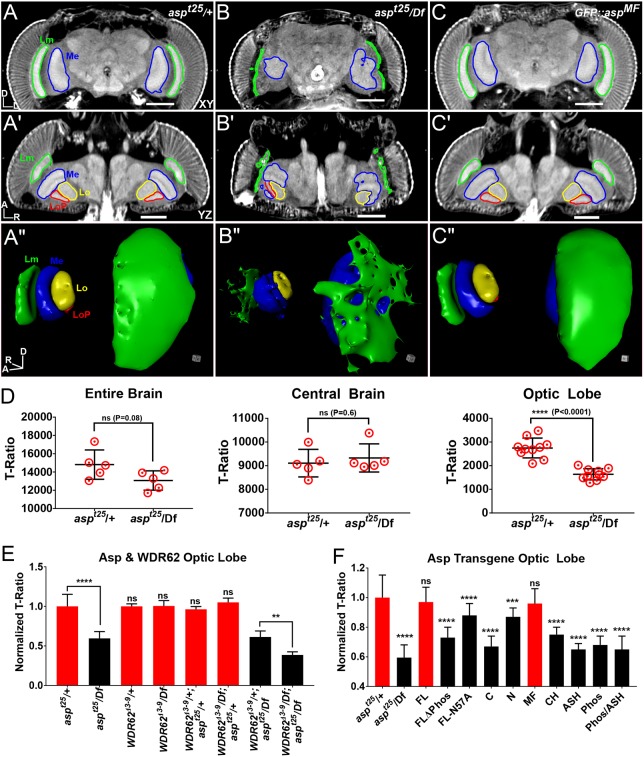
To demonstrate the utility of µ-CT when combined with the genetic power of Drosophila, we applied the technique to investigate brain defects in two models of human microcephaly, which is characterized by reduced brain size, cognitive function, and lifespan (O'Neill et al., 2018). We previously showed that mutations in abnormal spindle (asp), the fly ortholog of abnormal spindle-like microcephaly-associated (ASPM), leads to adult flies displaying a reduction in head and brain size (Schoborg et al., 2015). We used µ-CT to explore heterozygous adult control (aspt25/+; Fig. 8A-A″) and asp mutant (aspt25/Df; Fig. 8B-B″) animals. Morphological examination revealed that optic lobe neuropils (medulla, lobula and lobula plate) in asp mutants were extremely disorganized whereas the central brain neuropils appeared only mildly affected compared with wild-type tissue (Fig. 8A-B″, Fig. S6B), a finding supported by confocal imaging (Fig. S6A).
Fig. 8.
Autosomal recessive primary microcephaly (MCPH) modeled in the adult fly. (A-C″) µ-CT scans of adult heads labeled with PTA and scanned in slow mode at an image pixel size of 1.25 µm from heterozygous control (aspt25/+; A-A″), asp mutant (aspt25/Df; B-B″) and GFP::aspMF transgenic rescue (C-C″) flies. (A-C) Anterior xy view; (A′-C′) dorsal yz view; (A″-C″) 3D rendering of visual system. Medulla (Me), lamina (Lm), lobula (Lo) and lobula plate (LOP) neuropils outlined. (D) T-ratio analysis of asp brain volume for the entire brain, the central brain only, and the optic lobe only. (E) Optic lobe volume of asp and Wdr62 brains expressed as a T-ratio, normalized to Wdr62Δ3-9/+; aspt25/+, which was set to 1.0. (F) Optic lobe volume of each GFP::asp transgene in the aspt25/Df mutant background. All µ-CT measurements were obtained from animals scanned in fast mode (20 min) at an image pixel size of 3 µm. Data are expressed as T-ratios, normalized to the GFP::asp/+; aspt25/+ control, which was set to 1.0. n=5 brains, Welch's t-test. ns, P>0.05; **P≤0.01; ***P≤0.001; ****P≤0.0001. Error bars represent s.d. Significance levels shown were derived from direct pairwise comparison between the control (GFP::asp/+; aspt25/+) and the mutant (GFP::asp/+; aspt25/Df), as shown in Figs S8 and S9. Body axes are indicated: A, anterior; D, dorsal; L, left; R, right. Scale bars: 100 µm.
We also used µ-CT to perform volume analysis on asp− brains. 3D tomograms revealed a ∼12% reduction in entire brain volume in asp− animals. Further analysis of entire brain sub-regions (optic lobes and central brain; Fig. 7C-C″) revealed a ∼30% reduction in optic lobe size (Fig. 8D); central brain volume was not affected. Other asp alleles (aspE3 and aspL1) (Rujano et al., 2013) showed nearly an identical volume decrease in the optic lobe, but also a significant decrease in the central brain (Fig. S6C). This discrepancy could be due to either allele-specific effects or genetic background differences between strains, which can influence the phenotype of a given allele (Chandler et al., 2017). For example, we have observed significant differences in entire brain and sub-region (optic lobe and central brain) size when comparing two independent heterozygous control animals, aspt25/+ and aspt25/TM6B (Fig. S6D). These data suggest that the genetic background should be carefully considered for any μ-CT investigation, because it is sensitive enough to detect small differences in volume for structures (e.g. tissues) that are larger than the effective resolution of the technique (10-50 μm).
asp has previously been shown to interact genetically with another microcephaly gene, Wdr62, to promote proper brain size in vertebrates (Jayaraman et al., 2016). Our analysis found no detectable reduction in the central brain or the optic lobes in Wdr62Δ3-9/Df animals compared with controls (Wdr62Δ3-9/+; Fig. 8E, Fig. S7A,B), suggesting that the smaller brains reported for larval Wdr62 brains do not lead to microcephaly in adults (Ramdas Nair et al., 2016). However, we did detect a genetic interaction between Wdr62 and asp; double mutant (Wdr62Δ3-9/Df; aspt25/Df) optic lobes showed a further ∼25% reduction in volume compared with controls (Fig. 8E) and also a significant decrease in central brain volume (Fig. S7B).
Finally, to understand better the domains of Asp required for its ability to promote brain size, we imaged asp mutants expressing various truncated N-terminal constructs tagged with GFP (Fig. S7C-F). This revealed that the first 573 amino acids of Asp were sufficient to rescue both brain morphology and size (Fig. 8A-C″,F, Fig. S7C-F, Fig. S9A, Movie 15); we term this fragment AspMF (‘minimal fragment’). Shorter truncations, including AspASH (ASH domain) and AspPhos (predicted CDK1 phosphorylation region; Saunders et al., 1997), could not rescue optic lobe size even when co-expressed in the same animal (AspASH/AspPhos) (Fig. 8F). To test the necessity of the ASH domain or the phosphorylation region, we generated transgenic animals expressing full-length Asp carrying either a mutation of a highly conserved asparagine residue (Ponting, 2006) (AspFL-N57A), or a deletion of the phosphorylation region (AspFLΔPhos). Both failed to restore optic lobe size to the level of the AspFL control (Fig. 8F, Fig. S8B,C), demonstrating that the ASH domain and the phosphorylation region are necessary, but not sufficient, for Asp's function in specifying brain size. Although the precise functions of these two regions remain unknown, an equivalent deletion of the AspMF region in the human ASPM gene has been identified in an individual with MCPH (Nicholas et al., 2009), further highlighting conserved mechanisms of brain development.
A tool for unbiased phenotyping in Drosophila
As a final highlight of the capabilities of μ-CT, we used our asp dataset to perform phenotyping analysis to identify additional tissue defects and provide a more complete description of asp function. In addition to the well-characterized small brain phenotype (Fig. 8, Figs S6-S8; Rujano et al., 2013; Schoborg et al., 2015), our analysis revealed severe defects in the visual circuit. The size and morphology of the lamina and the retina were severely compromised, and the ocelli were reduced in size, extremely disorganized or completely absent in asp mutant animals. This suggests that Asp is essential for proper development of the entire visual circuit and that the reduction in head size of asp mutant animals results from multiple tissue defects, rather than just a small brain per se (Figs 8A,B and 9, Movies 13, 14).
Fig. 9.
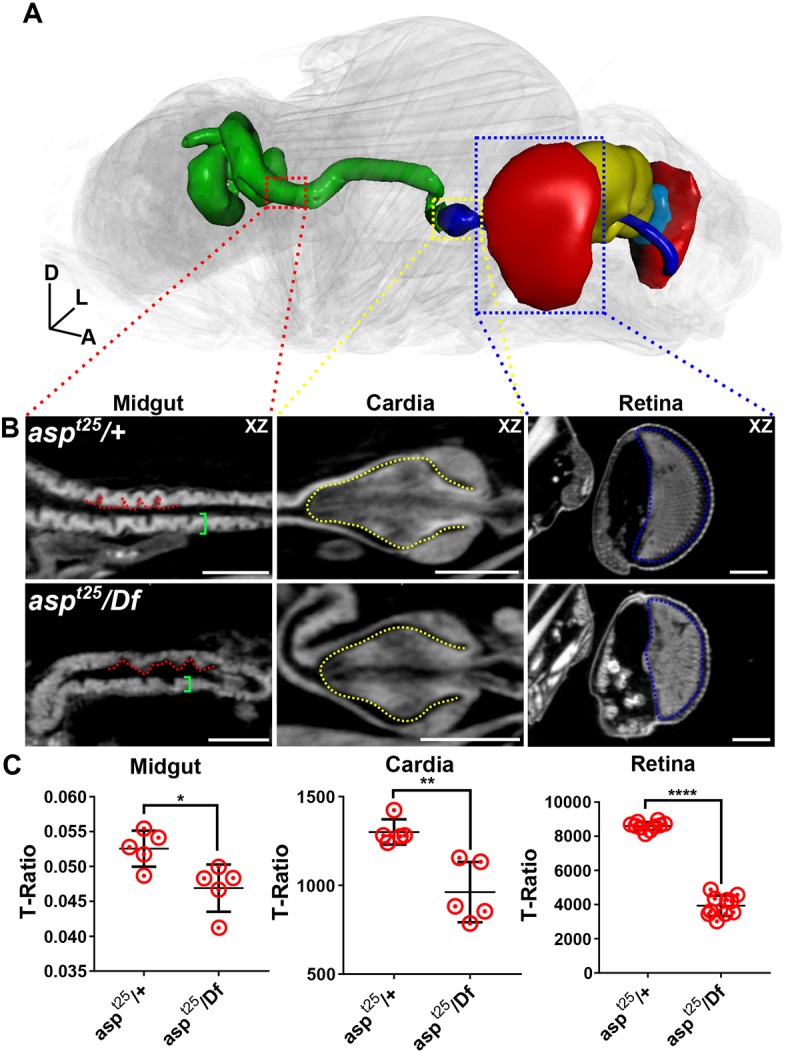
Phenotyping asp mutant animals with μ-CT. Morphometric analysis of additional tissues in heterozygous control (aspt25/+) and asp mutant (aspt25/Df) animals scanned in slow mode at an image pixel size of 1.25 µm. (A) 3D representation of an asp mutant with affected tissues shown: midgut (green); cardia and foregut (blue), retina (red), brain (yellow), lamina (aqua blue). (B) 2D xy views of the midgut, cardia and retina. Red dotted line outlines villi-like structures in the gut epithelium; green brackets highlight wall thickness. Yellow dotted line outlines the cardia. Blue dotted line outlines the retina. (C) T-ratio analysis of midgut epithelial thickness, cardia volume and retina volume. Error bars represent s.d. Body axes are indicated: A, anterior; D, dorsal; L, left. Scale bars: 100 µm. n=5, Welch's t-test. *P≤0.05; **P≤0.01; ****P≤0.0001.
We also found novel defects in the gut of aspt25 animals. The cardia, which functions as a sphincter to regulate the passage of food from the foregut to the midgut, was significantly reduced in volume. The epithelium of the midgut was thinner in asp mutants and the morphology of the villi-like structures was also altered (Fig. 9B,C). Egg chambers within the ovary were also defective, consisting of only a few early-stage oocytes and likely explaining the sterility defects observed in asp mutant females (Fig. S10A,B). These tissue defects are not pleiotropic, as we did not find defects in the heart, flight muscles, hindgut or other tissues (Fig. S10C-E).
Together, these data suggest that Asp has tissue-specific functions, and demonstrates how whole-animal phenotyping with μ-CT can direct new investigations at the cellular and molecular level (e.g. the retina or gut) to achieve a complete understanding of gene function.
DISCUSSION
This work demonstrates the advantages of combining non-traditional imaging methods (µ-CT) with traditional genetic, cell and developmental biology tools utilized by the model organism community to answer novel questions regarding anatomy, development and disease effectively. We have focused on Drosophila melanogaster, a well-characterized model system that, surprisingly, has few imaging tools available for phenotyping whole animals. Studies in other model organisms, such as mice, rats and zebrafish, have also demonstrated the powerful morphometric analyses that can be performed with μ-CT when combined with genetic mutant studies and the wealth of knowledge that can be added to developmental atlases (Dyer et al., 2017; Weinhardt et al., 2018; Wong et al., 2012). μ-CT has also played a key role in the work of the Mouse International Phenotyping Consortium, whose efforts have provided an invaluable resource for understanding development and disease mechanisms (International Mouse Knockout Consortium et al., 2007).
Our work presented here suggests that the ability of μ-CT to image intact flies at any developmental stage and provide precise 3D morphometric analysis, all within a commercially available platform (costing ∼400,000 USD), makes it suitable for similar phenotyping efforts in the fly. Analysis of our asp mutant provides an excellent example of de novo phenotype identification in tissues other than the brain. Also, our finding that only a small portion of Asp's N-terminus is needed for proper brain size highlights the novel questions that can be addressed by μ-CT when combined with traditional genetic tools in the fly.
We envisage μ-CT data having a broad impact on the Drosophila community as morphometric and anatomical assessment of organs throughout post-embryonic developmental stages provides a wealth of anatomical data, serving as a modern and complementary atlas to Volker Hartenstein's Atlas of Drosophila Development (Hartenstein, 1993). Additionally, our analysis of the brain could serve as a companion to atlases of the fly brain, such as Virtual Fly Brain (www.virtualflybrain.org). We are currently working to implement an online repository of our data that can be viewed by other researchers in the community and serve to host future data. Automating the segmentation and analysis (the most time-consuming part) of μ-CT datasets using machine-learning approaches, will be the most significant hurdle to overcome for this technology to be used in a high-throughput fashion for large-scale fly phenotyping.
X-ray-based techniques have additional advantages when it comes to whole-animal imaging. For example, μ-CT and synchrotron X-ray imaging have been used to visualize living insect physiology in real time (Poinapen et al., 2017; Westneat et al., 2003), providing knowledge of physiological mechanisms that simply cannot be determined from dead flies. Synchrotron X-ray sources have also recently been used for whole-animal phenotyping of fixed zebrafish (Ding et al., 2019), providing superior temporal and spatial resolution capabilities over benchtop scanners but requiring collaboration with a Department of Energy National Laboratory (e.g. Argonne) for access to beamlines. Furthermore, genetic labeling tools have also been developed that allow for tissue-specific visualization by electron microscopy and X-ray microscopy, such as miniSOG or APEX2 (Bushong et al., 2015; Martell et al., 2017; Ng et al., 2016). Recently, a technique that combined genetic labeling in chemically cleared flies was reported that allows high-resolution ultramicroscopy of tissues (Pende et al., 2018). This could provide an alternative imaging workflow, whereby μ-CT is first used to identify defects de novo from a systemic assessment, followed by ultramicroscopy to visualize specific defects at high resolution. When combined with traditional cellular and molecular analysis, this workflow has the potential to provide a near-complete understanding of a given biological system.
MATERIALS AND METHODS
µ-CT
This protocol was adapted from previous work (Smith et al., 2016; Sombke et al., 2015; Swart et al., 2016) and allows for visualization of larval, pupal and adult stages in Drosophila.
Adult labeling
Between five and 50 adult flies were anesthetized using CO2, and transferred to a 1.5 ml Eppendorf tube containing 1 ml of 0.5% Triton X-100 in PBS (0.5% PBST). Tubes were capped, tapped gently on the bench top and incubated for 5 min. This step removes wax on the fly cuticle and, once complete, most of the flies will sink to the bottom of the tube. Flies were then fixed in 1 ml Bouin's solution (5% acetic acid, 9% formaldehyde, 0.9% picric acid; Sigma-Aldrich) for 16-24 h, washed 3×30 min in 1 ml of µ-CT Wash Buffer (0.1 M Na2HPO4, 1.8% sucrose) and stained with 1 ml of a 0.1 N solution of I2KI (Lugol's solution) or 0.5% PTA solution, diluted in water. I2KI penetrates quicker and can label tissue in 1-2 days without mechanical disruption of adult cuticle, whereas PTA requires five or more days plus disruption of the cuticle. This is due to the higher molecular weight and hydrodynamic radius of PTA versus I2KI. However, the differential uptake of PTA by tissue yields better contrast ratios that result in excellent visualization of tissue substructure for finer morphological and anatomical detail. To achieve sufficient brain staining with PTA, the mouthparts were carefully removed with a pair of forceps. Flies were then washed in two changes of ultrapure water and stored at room temperature in ultrapure water until scanned. Samples can be stored indefinitely this way, but we recommend scanning within a month for optimal morphological and quantitative analysis. At no point should flies be kept at 4°C, as this causes the formation of air bubbles, particularly in the headcase, when flies return to room temperature. Although the chemical solvents used in this protocol cause mild tissue shrinkage, the use of ethanol should also be avoided because it causes tissue dehydration, thus significantly altering size and quantitative measurements. It is also recommended that all animal genotypes, including relevant controls, are processed and scanned in the same experiment to ensure the highest confidence when comparing datasets. Finally, this protocol does not require rotation/nutation and can be done simply by incubating flies on the benchtop.
Larval/pupal labeling
Larval and pupal samples were heated in 1 ml of 0.5% PBST to 100°C for 20 s in a heat block, followed by cooling at room temperature for 5 min. This kills the animals immediately so that they do not progress to later developmental time points. Bouin's solution was then added as described for adults. After 2 h, the Bouin's solution was discarded and the cuticle of each animal disrupted by poking with a microdissection needle near the anterior and posterior ends, avoiding areas of soft tissue. Fresh Bouin's solution was added and flies were incubated for another 16-22 h. The 2-h fixation stiffens the larvae or pupae just enough to prevent hemolymph from leaking out of the animals upon poking with the needle, which can alter morphology. Disrupting the cuticle allows for easy passage of the Bouin's solution to the interior of the animal and the additional incubation time ensures even fixation of all tissues. Animals were then washed 3×30 min in µ-CT Wash Buffer. Iodine was then added and the procedure carried out as described for adults.
Sample mounting
There is no defined way to mount samples for scanning and this may require resourcefulness on the investigator's part. The biggest priority is that the sample does not move or dry out during imaging. We used a custom-made brass holder with an indention at the tip that perfectly fits a small piece of standard plastic capillary. A P10 pipette tip was then wedged into the capillary tube and filled with water. Parafilm was used to wrap connection points to prevent water leaking into the scanner. Using forceps, flies were then transferred into the pipette tip and a dulled 20-gauge needle was used to gently push the fly downward until it became wedged along the wall of the tip and was unable to move. A piece of Parafilm was used to cover the opening of the pipette tip to prevent evaporation during overnight scans. It is important that the capillary tube and pipette tip are as closely aligned along the long axis of the brass sample holder as possible to avoid wobble during the 360° sample rotation, resulting in the highest quality reconstructions (the alignment in Fig. S1C is slightly off axis).
Scanning
Samples were scanned using a SkyScan 1172 desktop scanner controlled by Skyscan software (Bruker) operating on a Dell workstation computer [Intel Xeon X5690 processor at 3.47 GHz (12 CPUs), 50 GB RAM and an NVIDIA Quadro 5000 (4 GB available graphics memory) GPU]. X-Ray source voltage and current settings: 40 kV, 110 μA, 4 W of power. A Hamamatsu 10 Mp camera with a 11.54 μm pixel size coupled to a scintillator was used to collect X-rays converted to photons. Medium camera settings at an image pixel size of 2.95 μm were used for fast scans (∼20 min), which consists of about 300 projection images. Small camera settings at an image pixel size of 0.7 μm-2 μm and utilizing 360° of sample rotation (∼1500 projection images) were used for the slow overnight scans. Random movement, which is an algorithm that can correct for any mechanical issues (e.g. jitteriness) during sample rotation, was set to 10. Frame averaging, which averages a defined number of projection images to increase signal to noise, ranged from four to eight.
Reconstruction
Tomograms were generated using NRecon software (Bruker MicroCT, v1.7.0.4). Reconstruction is sensitive to the acquired image data including image brightness, contrast and optical bench geometry. Reconstruction parameters may vary by sample preparation and acquisition conditions; therefore, in most cases the process is iterative in setting misalignment compensation values, ring artifact reduction settings and beam-hardening compensation values to create the highest quality reconstructions. NRecon allows for individual fine-tuning of each of these components using user-defined parameter steps. For our scans, we utilized a built-in shift correction that uses reference scans to compensate for any sample movement during scanning (i.e. thermal fluctuations or slowly varying sample movements) and any remaining misalignment was manually fine-tuned using the misalignment compensation function (Salmon et al., 2009). Ring artifact correction ranged from 10 to 20, with beam hardening set to 0%.
Image resolution
With the X-ray source settings used in this paper (40 kV, 4 W), our scanner has a measured resolution (based on the spot size of the X-ray beam) of 5.5 μm under perfect imaging conditions (Morales et al., 2016). However, because many extrinsic factors determine the final spatial resolution of μ-CT tomograms, including stain type, sample movement/deformation, scanner vibration, reconstruction parameters, etc., scans taken with identical imaging parameters will often differ in their actual spatial resolution. As a result, we avoid the use of the term ‘spatial resolution’ in this manuscript and instead use image pixel size as determined by the camera setting (slow or fast mode) to describe each dataset. Nonetheless, comparisons of brain structures of known size (e.g. individual neuropils and fiber tracts) from confocal microscopy, plus the ability to resolve non-neuronal cells in the headcase with μ-CT (see Fig. 7 and Movie 12), suggest that we routinely achieve a ‘practical resolution’ of a single cell (10-25 μm), but not subcellular resolution (<10 μm).
µ-CT image analysis and statistics
Dragonfly [v3.6, Object Research Systems (ORS), 2018; software available at http://www.theobjects.com/dragonfly] was operated on a Dell workstation [Intel Xeon CPU E5-2623 at 3.00 GHz (16 CPUs), 32 GB RAM, NVIDIA Quadro M2000 GPU (20 GB available graphics memory)]. Segmentation of structures into individual regions of interest (ROIs) was performed manually using thresholded images with the 3D paintbrush function. To eliminate human bias during segmentation, a single threshold value was selected that encompassed the entire structure of interest in all images that were to be compared. This threshold value was then applied to all images and used for segmentation of tissues into ROIs. ROIs were then converted to meshes for quantitative analysis and visualization. The built-in 3D movie maker was used to generate all movies.
Thickness values
Thickness values for the wing disc and heart (Figs 1E,F and 3D-F) were calculated based on the diameter (µm) of a hypothetical sphere that fits between two boundary points within the tissue, which defines the local thickness at that point (the adult heart wall contains over 90,000 of these points). A LUT colored by thickness (µm) was then used to map the diameter of each sphere within the tissue to provide a visual representation of tissue thickness. The thickness values at each point were also exported as individual values and represented as histograms. Fiji (ImageJ v1.52a, National Institutes of Health) was used for all confocal images (Schindelin et al., 2012). Measurements of brain volume from confocal images was achieved by thresholding brains labeled with DAPI and anti-nc82 (also known as Brp; see below for antibody details) and creating a binary mask of the entire z-stack, which was then measured using the 3D ImageJ suite (Ollion et al., 2013).
Sphericity
To calculate how closely an object matches the shape of a sphere (e.g. brain vacuoles), the following equation was used: ψ=π1/3(6Vp)2/3/Ap, where Vp is the volume of the particle and Ap is the surface area. A value of 1 represents a perfect sphere.
T-ratio
We used thorax width to normalize all brain measurements to account for any body size variation between samples. Thorax width was measured by drawing a line across the widest point of the thorax cuticle when viewed from the yz imaging plane (Fig. S5). To calculate the T-ratio, raw brain volume was divided by thorax width for each animal. Normalized T-ratios were derived by setting the T-ratio from the relevant control genotype to 1 and normalizing the relevant mutant genotypes accordingly. Statistical analysis and graph generation were performed with GraphPad Prism 7 software. An unpaired, two-tailed Welch's t-test (α=0.05) was used for pair-wise brain size comparisons. All brain size measurements in Figs 7D,E and 8, and Figs S6-S9 were derived from animals scanned in fast mode (20 min) at an image pixel size of 3 μm.
Immunostaining, antibodies and microscopy
Adult brain
To ensure optimal imaging by laser-scanning confocal microscopy, we used a xylene-based chemical clearing protocol followed by mounting tissue in DPX (distyrene, plasticizer and xylene) (Nern et al., 2015). DPX has a refractive index of 1.52 n20/D, which is identical to the refractive index of the immersion oil used for confocal microscopy in this study (Carl Ziess Immersol 518F; 1.518 n20/D). This eliminates light-scattering effects at deeper z-depth and provides excellent image quality throughout the entire volume of the adult brain (∼120 μm thick). First, adult brains were carefully dissected in SF900 cell culture media and transferred to tubes containing 1.2% paraformaldehyde (PFA) diluted in SF900. Tubes were transferred to a nutator at 4°C for 24 h. Brains were washed 3×10 min in 0.5% PBST then blocked with 5% normal goat serum (NGS) in 0.5% PBST for 2 h at room temperature. Primary antibodies were diluted in NGS/PBST and brains were incubated for 48 h at 4°C. Following a 3×10 min wash in PBST, secondary antibodies were added for an additional 48 h. Brains were washed 3×10 min in PBST, then post-fixed in 4% PFA/SF900 for 4 h at room temperature followed by a 3×10 min wash in PBS. Brains were then transferred with forceps to a coverslip coated with poly-L-lysine (PLL), dipped in an H2O bath and then dehydrated in an ethanol series (30%, 50%, 75%, 95%, 100%, 100%, 100%; 10 min each). Coverslips were then soaked in a xylene bath, 3×5 min each in a fume hood. DPX (350 μl) was then added dropwise to the coverslip, which was placed face down on a microscope slide using two separate coverslips as spacers to prevent compression of the brains. Slides were imaged by laser-scanning confocal microscopy 3 days later to allow the DPX to fully cure. For more information, see the Janelia FlyLight Project (https://www.janelia.org/project-team/flylight/protocols). This protocol, while providing superior image quality, does result in tissue shrinkage as noted by us (Fig. 7, Fig. S5D) and others (Nern et al., 2015).
Other tissues
Larval wing discs were fixed in 4% PFA for 20 min at room temperature, washed in 0.5% PBST and incubated with 1× DAPI for 5 min. Larval brains and ovaries were fixed in 9% PFA for 30 min at room temperature, washed 3×10 min in 0.5% PBST and incubated with primary and secondary antibodies overnight. Tissues were then mounted in Vectashield for imaging.
Antibodies
For adult brain, anti-Bruchpilot [nc82, Brp, Developmental Studies Hybridoma Bank (DHSB)] or anti-Cadherin (DN-Ex #8; DHSB) were used at 1:20 (nc82) or 1:30 (DN-Ex #8). For other tissues, β-Tubulin antibody (E7, DHSB) was used at 1:50. Phalloidin was used at 1:500. Secondary antibodies conjugated to Alexa Fluor 488, 568 and 647 (A32723, A-11031, A32728, Life Technologies) were used at 1:500.
Microscopy
Laser-scanning confocal imaging of adult brains was performed on a Zeiss LSM 880 with Airyscan and imaged using a 63×/1.4 NA plan Apo objective, controlled by Zen Black software. Fixed and live spinning-disc confocal imaging of larval wing discs, brains and adult ovaries were performed using a Nikon Eclipse Ti inverted microscope with a 10×/0.30 NA plan Fluor or 40×/1.30 NA plan Fluor objective, a CSU-22 spinning disc module (Yokogawa), an ORCA-Flash4.0 CMOS camera (Hamamatsu) or interline-transfer cooled charge-coupled device camera (CoolSNAP HQ2; Photometrics) and 405 nm, 491 nm, 562 nm and 647 nm solid-state illumination lasers (VisiTech International). The microscope was controlled by and images acquired using MetaMorph (v7.7.10, Molecular Devices).
Asp transgene expression levels
We attempted to measure Asp transgene levels using standard western blotting techniques with anti-GFP antibodies; however, this approach consistently failed despite numerous attempts to optimize lysis and running conditions (Rujano et al., 2013; Schoborg et al., 2015). Instead, we directly measured GFP fluorescence from dissected larval brains that were imaged live to determine expression levels. At least ten third instar larvae per genotype were washed in a dish of 1× PBS. The brains were dissected in room temperature Schneider's media with 1× Antibiotic-Antimycotic (15240062, Thermo Fisher Scientific) and kept in media until imaging. Brains were placed into a 25-50 µl droplet of media on the gas-permeable membrane surface of a 50 mm Lumox dish (Sarstedt). Four drops of Halocarbon oil 700 (Sigma-Aldrich) were placed on membrane surface to make the corners of a square corresponding to the size of a 22 × 22 mm, 1.5 mm thickness microscope coverslip (Fisher Scientific). The coverslip was gently placed on top of the sample, and excess media was wicked away using a Kimwipe to flatten and secure the brains. Brains were imaged live on a Nikon Eclipse Ti Spinning Disc confocal microscope with a 10×/0.30 NA objective and a 491 nm laser with 2000 ms exposure and 100% laser power. For each data point, a single image was taken of the entire isolated CNS including the brain and VNC. At least two independent experiments were performed for each genotype. All data analysis was performed using ImageJ. Mean pixel intensity (MPI) was calculated for (1) an entire outline of each asp transgenic animal CNS (MPICNS), (2) an area away from the sample for background (MPIBkg), and (3) average MPI for wild-type (yw) CNS (MPIyw). The final fluorescence measurement, MPIfinal, was calculated by MPICNS−MPIBkg−MPIyw. All data were normalized to Asp full-length GFP.
Solvents effects on brain size
Ten adult brains from yw female flies (3-5 days old) were dissected in SF900 and imaged under the following solvent conditions. (1) PBS: Freshly dissected brains were transferred to a glass-bottom dish (MatTek) coated with PLL and imaged in 1× PBS. (2) Vectashield: Brains were fixed in 1.2% PFA/SF900 overnight at 4°C, washed 3×10 min in 0.5% PBST, then 3×5 min in PBS. Brains were then transferred to a PLL dish and imaged in Vectashield. (3) Ethanol/xylene/DPX: Brains were treated as described for Vectashield, but transferred to a coverslip coated with PLL then dehydrated in an ethanol series (30%, 50%, 75%, 95%, 100%, 100%, 100%, 10 min each). The coverslip was then soaked in xylene (3×5 min each) and 350 μl of DPX was added dropwise and mounted on a microscope slide between two coverslips that functioned as spacers to prevent the brains from being compressed. Brains were imaged 3 days after curing. Brightfield microscopy using a Nikon Eclipse Ti inverted microscope with a 10×/0.30 NA plan Fluor objective and a CoolSNAP HQ2 camera was used to acquire images of brains. Brain size was measured by manually outlining each brain as a ROI using Fiji, then deriving the area (pixels2) of each ROI.
Fly stocks and husbandry
All stocks and crosses were maintained on standard cornmeal-agar media at room temperature (20-22°C). Fly stocks used were as follows: control (yw); aspt25/TM6B, Hu, Tb (Schoborg et al., 2015); w1118; Df(3R)BSC519/TM6C, Sb1 cu1 (asp deficiency, Bloomington Stock #25023); w; e11, aspE3/TM3, Ser, Act-GFP& aspL1/TM6B, Hu, Tb (Gonzalez et al., 1998); Wdr62Δ3-9/CyO, GFP (Cabernard Lab; Ramdas Nair et al., 2016); w1118; Df(2L)Exel8005/CyO (Wdr62 deficiency, Bloomington Drosophila Stock Center, #7779); OK107-GAL4 (w*; P{w[+mW.hs]=GawB}OK107 eyOK107/In(4)ciD, ciD panciD svspa-pol; Bloomington Drosophila Stock Center, #854); Spc105r RNAi (y1 sc* v1; P{TRiP.HMS01752}attP2/TM3, Sb1; Bloomington Drosophila Stock Center, #38534). All asp transgenic animals were produced in yw genetic background (BestGene). asp transgene genotype combinations were used as follows: control (yw; GFP-asp*/+; aspt25/+); mutants (yw; GFP-asp*/+; aspt25/aspDf).
Pupal staging
Pupae (yw) were staged according to Bainbridge and Bownes (1981), whose morphological descriptions were accomplished with a ‘dissecting microscope with lateral illumination close to the stage, interposing the tip of a pair of watch maker's forceps between the lamp and the puparium to cast a shadow over any feature which may be obscured by surface reflection but which would show up with light scattered inside the animal’. Remarkably, nearly all these features described over 30 years ago were easily visualized in our µ-CT scans, allowing for unambiguous identification of the pupal stage.
Supplementary Material
Acknowledgements
We thank Carey Fagerstrom for molecular cloning of Asp transgenes; Brian Galletta for critical reading of the manuscript and help with asp larval brain dissections and fluorescent measurements; Ryan O'Neil for critical reading of the manuscript and providing the spc105RNAi animals; Karen Plevock Haase for providing aged flies; Rachel Ng for assisting with µ-CT scans; Danielle Donahue and Brenda Klaunberg of the NIH Mouse Imaging Facility for µ-CT training, advice and discussion; and Mike Marsh from Object Research Systems for Dragonfly technical support.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: T.A.S.; Methodology: T.A.S., H.D.M.; Software: T.A.S., H.D.M.; Validation: T.A.S.; Formal analysis: T.A.S., S.L.S., L.N.S.; Investigation: T.A.S., S.L.S., L.N.S.; Resources: T.A.S., H.D.M.; Data curation: T.A.S., S.L.S., L.N.S.; Writing - original draft: T.A.S.; Writing - review & editing: T.A.S., N.M.R.; Visualization: T.A.S.; Supervision: T.A.S., N.M.R.; Project administration: N.M.R.; Funding acquisition: N.M.R.
Funding
This work is supported by the Division of Intramural Research at the National Institutes of Health/National Heart, Lung, and Blood Institute (1ZIAHL006126 to N.M.R. and 1K22HL137902 to T.A.S.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.176685.supplemental
References
- Adám G., Perrimon N. and Noselli S. (2003). The retinoic-like juvenile hormone controls the looping of left-right asymmetric organs in Drosophila. Development 130, 2397-2406. 10.1242/dev.00460 [DOI] [PubMed] [Google Scholar]
- Andlauer T. F. M. and Sigrist S. J. (2012). In vivo imaging of the Drosophila larval neuromuscular junction. Cold Spring Harb. Protoc. 2012, 481-489. 10.1101/pdb.prot068593 [DOI] [PubMed] [Google Scholar]
- Bainbridge S. P. and Bownes M. (1981). Staging the metamorphosis of Drosophila melanogaster. J. Embryol. Exp. Morphol. 66, 57-80. [PubMed] [Google Scholar]
- Betz O., Wegst U., Weide D., Heethoff M., Helfen L., Lee W.-K. and Cloetens P. (2007). Imaging applications of synchrotron X-ray phase-contrast microtomography in biological morphology and biomaterials science. I. General aspects of the technique and its advantages in the analysis of millimetre-sized arthropod structure. J. Microsc. 227, 51-71. 10.1111/j.1365-2818.2007.01785.x [DOI] [PubMed] [Google Scholar]
- Bushong E. A., Johnson D. D., Kim K.-Y., Terada M., Hatori M., Peltier S. T., Panda S., Merkle A. and Ellisman M. H. (2015). X-ray microscopy as an approach to increasing accuracy and efficiency of serial block-face imaging for correlated light and electron microscopy of biological specimens. Microsc. Microanal. 21, 231-238. 10.1017/S1431927614013579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler C. H., Chari S., Kowalski A., Choi L., Tack D., DeNieu M., Pitchers W., Sonnenschein A., Marvin L., Hummel K. et al. (2017). How well do you know your mutation? Complex effects of genetic background on expressivity, complementation, and ordering of allelic effects. PLoS Genet. 13, e1007075 10.1371/journal.pgen.1007075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.-C., Chen H.-Y., Liao P.-C., Wang S.-J., Tsai M.-Y., Chen Y.-H. and Lin W.-Y. (2017). Toward a new insight of calcium oxalate stones in Drosophila by micro-computerized tomography. Urolithiasis 46, 149-155. 10.1007/s00240-017-0967-0 [DOI] [PubMed] [Google Scholar]
- Ding Y., Vanselow D. J., Yakovlev M. A., Katz S. R., Lin A. Y., Clark D. P., Vargas P., Xin X., Copper J. E., Canfield V. A. et al. (2019). Computational 3D histological phenotyping of whole zebrafish by X-ray histotomography. eLife 8, e44898 10.7554/eLife.44898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer E. L., Gray Roncal W., Prasad J. A., Fernandes H. L., Gürsoy D., De Andrade V., Fezzaa K., Xiao X., Vogelstein J. T., Jacobsen C. et al. (2017). Quantifying mesoscale neuroanatomy using X-ray microtomography. Eneuro 4, ENEURO.0195-17.2017 10.1523/ENEURO.0195-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian B., Schneeberg K. and Beutel R. G. (2016). Comparative thoracic anatomy of the wild type and wingless (wg1cn1) mutant of Drosophila melanogaster (Diptera). Arthropod. Struct. Dev. 45, 611-636. 10.1016/j.asd.2016.10.007 [DOI] [PubMed] [Google Scholar]
- Gonzalez C., Sunkel C. E. and Glover D. M. (1998). Interactions between mgr, asp, and polo: asp function modulated by polo and needed to maintain the poles of monopolar and bipolar spindles. Chromosoma 107, 452-460. 10.1007/s004120050329 [DOI] [PubMed] [Google Scholar]
- Hagen M. and Dupont Y. L. (2013). Inter-tegular span and head width as estimators of fresh and dry body mass in bumblebees (Bombus spp.). Insectes Soc. 60, 251-257. 10.1007/s00040-013-0290-x [DOI] [Google Scholar]
- Harrison J. F., Waters J. S., Biddulph T. A., Kovacevic A., Klok C. J. and Socha J. J. (2017). Developmental plasticity and stability in the tracheal networks supplying Drosophila flight muscle in response to rearing oxygen level. J. Insect Physiol. 106, 189-198. 10.1016/j.jinsphys.2017.09.006 [DOI] [PubMed] [Google Scholar]
- Hartenstein, V (1993). Atlas of Drosophila Development. Cold Spring Harbor Laboratory Press. [Google Scholar]
- Holmes J. (2009). In vivo real-time optical coherence tomography imaging of Drosophila for cardiovascular research. Nat. Methods 6, 782 10.1038/nmeth.f.270 [DOI] [Google Scholar]
- International Mouse Knockout Consortium, Collins F. S., Rossant J. and Wurst W. (2007). A mouse for all reasons. Cell 128, 9-13. 10.1016/j.cell.2006.12.018 [DOI] [PubMed] [Google Scholar]
- Ito K., Shinomiya K., Ito M., Armstrong J. D., Boyan G., Hartenstein V., Harzsch S., Heisenberg M., Homberg U., Jenett A. et al. (2014). A systematic nomenclature for the insect brain. Neuron 81, 755-765. 10.1016/j.neuron.2013.12.017 [DOI] [PubMed] [Google Scholar]
- Jährling N., Becker K., Schönbauer C., Schnorrer F. and Dodt H.-U. (2010). Three-dimensional reconstruction and segmentation of intact Drosophila by ultramicroscopy. Front. Syst. Neurosci. 4, 1 10.3389/neuro.06.001.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman D., Kodani A., Gonzalez D. M., Mancias J. D., Mochida G. H., Vagnoni C., Johnson J., Krogan N., Harper J. W., Reiter J. F. et al. (2016). Microcephaly proteins wdr62 and aspm define a mother centriole complex regulating centriole biogenesis, apical complex, and cell fate. Neuron 92, 813-828. 10.1016/j.neuron.2016.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klok C. J., Kaiser A., Socha J. J., Lee W.-K. and Harrison J. F. (2016). Multigenerational effects of rearing atmospheric oxygen level on the tracheal dimensions and diffusing capacities of pupal and adult Drosophila melanogaster. Adv. Exp. Med. Biol. 903, 285-300. 10.1007/978-1-4899-7678-9_20 [DOI] [PubMed] [Google Scholar]
- Kreipke R. E., Kwon Y. V., Shcherbata H. R. and Ruohola-Baker H. (2017). Drosophila melanogaster as a model of muscle degeneration disorders. Curr. Top. Dev. Biol. 121, 83-109. 10.1016/bs.ctdb.2016.07.003 [DOI] [PubMed] [Google Scholar]
- Martell J. D., Deerinck T. J., Lam S. S., Ellisman M. H. and Ting A. Y. (2017). Electron microscopy using the genetically encoded APEX2 tag in cultured mammalian cells. Nat. Protoc. 12, 1792-1816. 10.1038/nprot.2017.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattei A. L., Riccio M. L., Avila F. W. and Wolfner M. F. (2015). Integrated 3D view of postmating responses by the Drosophila melanogaster female reproductive tract, obtained by micro-computed tomography scanning. Proc. Natl. Acad. Sci. USA 112, 8475-8480. 10.1073/pnas.1505797112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metscher B. D. (2009). MicroCT for comparative morphology: simple staining methods allow high-contrast 3D imaging of diverse non-mineralized animal tissues. BMC Physiol. 9, 11 10.1186/1472-6793-9-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millburn G. H., Crosby M. A., Gramates L. S., Tweedie S. and FlyBase Consortium (2016). FlyBase portals to human disease research using Drosophila models. Dis. Model. Mech. 9, 245-252. 10.1242/dmm.023317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani R., Takeuchi A., Akamatsu G., Uesugi K. and Suzuki Y. (2008). Element-specific microtomographic imaging of Drosophila brain stained with high-Z probes. J. Synchrotron Radiat. 15, 374-377. 10.1107/S0909049508003725 [DOI] [PubMed] [Google Scholar]
- Mizutani R., Saiga R., Takeuchi A., Uesugi K. and Suzuki Y. (2013). Three-dimensional network of Drosophila brain hemisphere. J. Struct. Biol. 184, 271-279. 10.1016/j.jsb.2013.08.012 [DOI] [PubMed] [Google Scholar]
- Morales A. G., Stempinski E. S., Xiao X., Patel A., Panna A., Olivier K. N., McShane P. J., Robinson C., George A. J., Donahue D. R. et al. (2016). Micro-CT scouting for transmission electron microscopy of human tissue specimens. J. Microsc. 263, 113-117. 10.1111/jmi.12385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton E. J., Webb S., Bateman J. E., Clarke L. J. and Shelton C. G. (1990). Three-dimensional x-ray microtomography for medical and biological applications. Phys. Med. Biol. 35, 805-820. 10.1088/0031-9155/35/7/001 [DOI] [PubMed] [Google Scholar]
- Nern A., Pfeiffer B. D. and Rubin G. M. (2015). Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc. Natl. Acad. Sci. USA 112, E2967-E2976. 10.1073/pnas.1506763112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng J., Browning A., Lechner L., Terada M., Howard G. and Jefferis G. S. X. E. (2016). Genetically targeted 3D visualisation of Drosophila neurons under Electron Microscopy and X-Ray Microscopy using miniSOG. Sci. Rep. 6, 38863 10.1038/srep38863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas A. K., Swanson E. A., Cox J. J., Karbani G., Malik S., Springell K., Hampshire D., Ahmed M., Bond J., Di Benedetto D. et al. (2009). The molecular landscape of ASPM mutations in primary microcephaly. J. Med. Genet. 46, 249-253. 10.1136/jmg.2008.062380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Null B., Liu C. W., Hedehus M., Conolly S. and Davis R. W. (2008). High-resolution, in vivo magnetic resonance imaging of Drosophila at 18.8 Tesla. PLoS ONE 3, e2817 10.1371/journal.pone.0002817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollion J., Cochennec J., Loll F., Escudé C. and Boudier T. (2013). TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics 29, 1840-1841. 10.1093/bioinformatics/btt276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill R. S., Schoborg T. A. and Rusan N. M. (2018). Same but different: pleiotropy in centrosome-related microcephaly. Mol. Biol. Cell 29, 241-246. 10.1091/mbc.E17-03-0192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton R. M., Vallés A. M., Dobbie I. M. and Davis I. (2010). Drosophila larval fillet preparation and imaging of neurons. Cold Spring Harb. Protoc. 2010, pdb.prot5405 10.1101/pdb.prot5405 [DOI] [PubMed] [Google Scholar]
- Pende M., Becker K., Wanis M., Saghafi S., Kaur R., Hahn C., Pende N., Foroughipour M., Hummel T. and Dodt H.-U. (2018). High-resolution ultramicroscopy of the developing and adult nervous system in optically cleared Drosophila melanogaster. Nat. Commun. 9, 4731 10.1038/s41467-018-07192-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinapen D., Konopka J. K., Umoh J. U., Norley C. J. D., McNeil J. N. and Holdsworth D. W. (2017). Micro-CT imaging of live insects using carbon dioxide gas-induced hypoxia as anesthetic with minimal impact on certain subsequent life history traits. BMC Zool. 2, 9 10.1186/s40850-017-0018-x [DOI] [Google Scholar]
- Ponting C. P. (2006). A novel domain suggests a ciliary function for ASPM, a brain size determining gene. Bioinformatics 22, 1031-1035. 10.1093/bioinformatics/btl022 [DOI] [PubMed] [Google Scholar]
- Ramdas Nair A., Singh P., Salvador Garcia D., Rodriguez-Crespo D., Egger B. and Cabernard C. (2016). The microcephaly-associated protein Wdr62/CG7337 is required to maintain centrosome asymmetry in Drosophila neuroblasts. Cell Rep. 14, 1100-1113. 10.1016/j.celrep.2015.12.097 [DOI] [PubMed] [Google Scholar]
- Rotstein B. and Paululat A. (2016). On the morphology of the Drosophila heart. J. Cardiovasc. Dev. Dis. 3, 15 10.3390/jcdd3020015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujano M. A., Sanchez-Pulido L., Pennetier C., le Dez G. and Basto R. (2013). The microcephaly protein Asp regulates neuroepithelium morphogenesis by controlling the spatial distribution of myosin II. Nat. Cell Biol. 15, 1294-1306. 10.1038/ncb2858 [DOI] [PubMed] [Google Scholar]
- Salmon P. L., Liu X. and Sasov A. (2009). A post-scan method for correcting artefacts of slow geometry changes during micro-tomographic scans. J. Xray. Sci. Technol. 17, 161-174. [DOI] [PubMed] [Google Scholar]
- Saunders R. D. C., Avides M. C., Howard T., Gonzalez C. and Glover D. M. (1997). The Drosophila gene abnormal spindle encodes a novel microtubule-associated protein that associates with the polar regions of the mitotic spindle. J. Cell Biol. 137, 881-890. 10.1083/jcb.137.4.881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoborg T., Zajac A. L., Fagerstrom C. J., Guillen R. X. and Rusan N. M. (2015). An Asp-CaM complex is required for centrosome-pole cohesion and centrosome inheritance in neural stem cells. J. Cell Biol. 211, 987-998. 10.1083/jcb.201509054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. B., Bernhardt G., Raine N. E., Abel R. L., Sykes D., Ahmed F., Pedroso I. and Gill R. J. (2016). Exploring miniature insect brains using micro-CT scanning techniques. Sci. Rep. 6, 21768 10.1038/srep21768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sombke A., Lipke E., Michalik P., Uhl G. and Harzsch S. (2015). Potential and limitations of X-Ray micro-computed tomography in arthropod neuroanatomy: a methodological and comparative survey. J. Comp. Neurol. 523, 1281-1295. 10.1002/cne.23741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillwell R. C., Dworkin I., Shingleton A. W. and Frankino W. A. (2011). Experimental manipulation of body size to estimate morphological scaling relationships in Drosophila. J. Vis. Exp., e3162 10.3791/3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell R. E. (1941). Effect on tissue volume of various methods of fixation, dehydration, and embedding. Stain Technol. 16, 67-83. 10.3109/10520294109106200 [DOI] [Google Scholar]
- Sunderhaus E. R. and Kretzschmar D. (2016). Mass histology to quantify neurodegeneration in Drosophila. J. Vis. Exp., e54809 10.3791/54809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart P., Wicklein M., Sykes D., Ahmed F. and Krapp H. G. (2016). A quantitative comparison of micro-CT preparations in Dipteran flies. Sci. Rep. 6, 39380 10.1038/srep39380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugur B., Chen K. and Bellen H. J. (2016). Drosophila tools and assays for the study of human diseases. Dis. Model. Mech. 9, 235-244. 10.1242/dmm.023762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.-W. and Sun Y. H. (2012). Segregation of eye and antenna fates maintained by mutual antagonism in Drosophila. Development 139, 3413-3421. 10.1242/dev.078857 [DOI] [PubMed] [Google Scholar]
- Wasserthal L. T. (2007). Drosophila flies combine periodic heartbeat reversal with a circulation in the anterior body mediated by a newly discovered anterior pair of ostial valves and “venous” channels. J. Exp. Biol. 210, 3707-3719. 10.1242/jeb.007864 [DOI] [PubMed] [Google Scholar]
- Weinhardt V., Shkarin R., Wernet T., Wittbrodt J., Baumbach T. and Loosli F. (2018). Quantitative morphometric analysis of adult teleost fish by X-ray computed tomography. Sci. Rep. 8, 16531 10.1038/s41598-018-34848-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westneat M. W., Betz O., Blob R. W., Fezzaa K., Cooper W. J. and Lee W.-K. (2003). Tracheal respiration in insects visualized with synchrotron x-ray imaging. Science 299, 558-560. 10.1126/science.1078008 [DOI] [PubMed] [Google Scholar]
- Wipfler B., Pohl H., Yavorskaya M. I. and Beutel R. G. (2016). A review of methods for analysing insect structures - the role of morphology in the age of phylogenomics. Curr. Opin. Insect Sci. 18, 60-68. 10.1016/j.cois.2016.09.004 [DOI] [PubMed] [Google Scholar]
- Wong M. D., Dorr A. E., Walls J. R., Lerch J. P. and Henkelman R. M. (2012). A novel 3D mouse embryo atlas based on micro-CT. Development 139, 3248-3256. 10.1242/dev.082016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Palliyil S., Ran C. and Kumar J. P. (2017). Drosophila Pax6 promotes development of the entire eye-antennal disc, thereby ensuring proper adult head formation. Proc. Natl. Acad. Sci. USA 114, 5846-5853. 10.1073/pnas.1610614114 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.