Abstract
Background
The information of Oncotype DX applied in Asian breast cancer patients is limited. A recurrence index for distant recurrence (RI-DR) has been developed for early-stage breast cancer (EBC) from tumor samples in Chinese patients. In this study, we compared the prognostic performance of the Oncotype DX (ODx) recurrence score (RS) with the RI-DR for any recurrence risk type.
Materials and methods
One hundred thirty-eight (138) patients with hormone receptor-positive and human epidermal growth factor receptor 2-negative EBC who were previously tested with ODx were included for testing with the RI-DR. The cutoff score to partition the low- and high-risk patients was 26 for RS and 36 for RI-DR. The primary endpoint was recurrence-free survival (RFS).
Results
The concordance between the RI-DR and RS was 83% in N0 patients and 81% in node-positive patients when the RS score cutoff was set at 26. With a median follow-up interval of 36.8 months, the 4-year RFS for the high- and low-risk groups categorized by the RS were 61.9% and 95.0%, respectively (hazard ratio: 10.6, 95.0% confidence interval [CI]: 1.8–62.9). The 4-year RFS in the high- and low-risk groups categorized by the RI-DR were 72.6% and 98.5%, respectively (hazard ratio: 18.9, 95% CI: 1.8–138.8).
Conclusion
This paper illustrated the performance of RI-DR and ODx RS in breast cancer women in Taiwan. There was high concordance between the RI-DR and RS. The RI-DR is not inferior to the RS in predicting RFS in EBC patients. This study will fill the gap between the current and best practice in Chinese patients.
Keywords: breast cancer, distant recurrence, NanoString, gene-expression profiling, prognosis
Utilization of Oncotype DX is uncommon in Asia due to lack of clinical data. We compare a clinical-genomic model, developed from Taiwan, to Oncotype 21-gene panel in Chinese patients.
Introduction
Breast cancer is the most common cancer in women, with around 2.4 million new cases diagnosed in 2015 (1). With improved diagnosis, breast cancer can now be detected at early stages, thus improving survival rates (2). In particular, among node-negative and hormonal receptor-positive (HR+) patients, 80% of those treated with surgery plus tamoxifen have been shown to remain disease-free at 12-year follow-up, according to accumulated information from five randomized trials (3). As a result, overtreatment is common; considering that the risk of recurrence is low, patients may suffer from side effects without receiving the full benefits of adjuvant therapies (4,5). The recent TAILORx trial demonstrated that about 70% of node-negative, HR+, and human epidermal growth factor receptor 2-negative (HER2-) patients can forgo adjuvant chemotherapy (6).
To overcome the imprecision of prognosis based on clinicopathologic factors, genomic tests for breast cancer prognosis, such as the Oncotype DX® (ODx), MammaPrint®, and EndoPredict® assay kits, are frequently utilized, especially in Western countries (7,8). In contrast, these types of multigene panels are not commonly used in Asian countries. One reason for this could be that the majority of such tests were developed based on Caucasian populations. In fact, evidence has shown that ODx may overestimate the risk of recurrence among Japanese populations (9). This may be due to differences in the incidence and lifestyle of breast cancer patients from different ethnic backgrounds. For example, survival rates were shown to differ among Asian, black, and white breast cancer patients (hazard ratios: 0.48, 1.57, and 1, respectively) (10). To fill this gap, an 18-gene classifier (18-GC) based on the gene-expression profiling of Chinese breast cancer patients was developed (11).
In the current study, we combined clinical variables and genetic information to generate a clinical-genomic model: RI-DR, a recurrence index for distant recurrence (based on a genomic model and six clinical variables: age at diagnosis, tumor size, lymph node status, estrogen receptor status, lymphovascular invasion (LVI), and tumor grading). Details of the development of the clinical-genomic model can be found in Supplement 1 (‘From Microarray to NanoString’). By using tissues that had been tested with ODx, in this study, we observed the performance of ODx RS and evaluated the performance of the RI-DR model with that of the ODx assay in the prognosis assessment of a cohort of patients from Taiwan.
Materials and methods
Patient selection
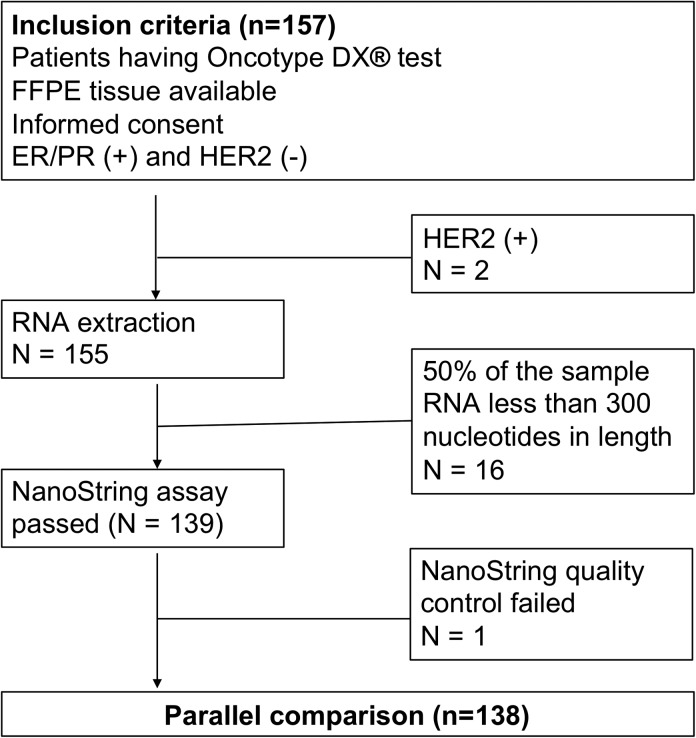
A total of 138 early breast cancer (EBC) patients were included in this head-to-head comparison study, including 42, 15, and 81 patients treated between 2010 and 2016 at McKay Memorial Hospital, Koo Foundation Sun Yat-Sen Cancer Center, and National Taiwan University Hospital, respectively, in Taiwan. The institutional review board of each of the participating medical centers approved the study protocol (IRB numbers: NTUH 201 610 066RINA, KFCC 20 150 327 A, and MMH 17CT040be). Patients that were eligible for this study included those (1) with invasive breast cancer; (2) that had undergone surgery, including mastectomy/breast-conserving surgery (BCS) with sentinel node biopsy/axillary lymph node dissection as first treatment; (3) with ODx testing data available; (4) positive for estrogen or progesterone receptors (HR+) and negative for HER2 receptor (HER2-), as confirmed by immunohistochemistry (IHC) or FISH. Patients with a stage of N3 or M1 were excluded. Figure 1 shows the study CONSORT diagram.
Figure 1.
Consolidated Standards of Reporting Trials (CONSORT) for this study.
18-GC
Development of the 18-GC has been reported previously (11). In summary, it consists of a multifunctional panel of genes associated with cell cycle and proliferation (DDX39, BUB1B, STIL, TPX2, CCNB1), oncogenic processes (BLM, TCF3, PIM1, RCHY1, PTI1), inflammation and immune response (CCR1, NFATC2IP), cell–cell interaction (TRPV6, OBSL1, MMP15), apoptosis (C16ORF7, DTX2), and metabolism (ENSA).
Test procedures using the NanoString nCounter
Each archived FFPE tissue sample was cut into 10-μm-thick sections and stored or transported at 4°C. According to the manufacturer’s instructions for the RNeasy FFPE Kit (Qiagen, Valencia, CA, USA), paraffin was removed, and total RNA extracted from the FFPE tissue sections. The extracted RNA was stored at –80°C until use and quality checked on an Agilent 2100 Bioanalyzer with the Agilent RNA 6000 Nano kit (Agilent Technologies, Santa Clara, CA, USA). To estimate the level of degradation, the RNA integrity number (RIN) was calculated. Custom capturing and reporting probes were designed for five housekeeping genes for normalization. The hybridization procedures were performed following the instructions of the NanoString nCounter system. Briefly, each probe pair consisted of a reporter probe carrying the signal on its 5′ end and a capture probe carrying a biotin on its 3′ end. The color code on each probe was used as its barcode for multiplexing. Data normalization was performed following the manufacturer’s recommended procedures for positive controls and reference genes.
Data adjustment
After normalization with housekeeping genes, a ratio was obtained for each sample by comparing its value against that of a reference sample. To reduce the skew of the distribution, log2 values of the ratios were used to build the model, as follows:
where is the jth gene for the ith sample, and is the jth gene for the fixed reference sample.
Algorithm for recurrence index (RI) calculation
The RI was calculated in two steps: (1) the genomic score was calculated and (2) the final RI was then computed by integration of the clinical score with the genomic score, as follows:
where DGM represents the DR genomic model; CM6 is the clinical score comprising six clinical variables, including age, LVI, ER status, axillary lymph node status, histology grade, and tumor size; βi indicates the weighting of each gene expression level; and Hi indicates the weighting of the clinical or genomic model.
Determination of cutoffs
RI-DR (DGM-CM6)
We transferred our 18-GC platform from the U133 2.0 plus microarray platform using fresh frozen tissues to the NanoString nCounter system using FFPE tumor tissues (details in Supplement 1). For the breakpoint determination, ROC curve analysis was performed with different cutoff points that stratify patients into low- or high-risk of recurrence. Breakpoint score of 36 was selected for a combined greatest sensitivity and specificity. Patients with RI-DR scores ≥ 36 and < 36 were thus defined as being at high and low risk of distant metastasis, respectively.
ODx assay
The ODx assay was performed as part of routine care on sections mailed to Genomic Health, Inc. Redwood City, California, and recurrence score (RS) values were retrieved from medical records. Low, intermediate, and high risk categories were defined as RS values <18, 18–30, and >31, respectively. After the publication of TAILORx (6), cutoffs of <11, 11–25, and ≥26 were used.
Clinical performance of and concordance between RI-DR and ODx RS
The clinical performance of the two assays was evaluated by comparing the status predicted by RI-DR or ODx and the actual any breast cancer recurrence status of the corresponding patients. The negative predictive value (NPV), positive predictive value (PPV), sensitivity, and specificity were calculated accordingly. The relapse-free survival probability was plotted against the follow-up time for both low- and high-risk group of patients as determined by RI-DR or ODx assay.
Agreement was analyzed by calculation of Cohen’s kappa coefficient (κ) between RI-DR and ODx scores of each corresponding patients. Score distribution between RI-DR and RS was obtained by plotting score intervals against the percentage of patients in each score interval.
Results
Baseline characteristics of patients
A total of 138 breast cancer patients were included in the study (Table 1), with 70 (51%) of the patients aged > 50 years and 68 (49%) aged ≤ 50 years. Additionally, 80 (58%) patients were at stage N0, 56 (41%) were at N1, and 2 (1%) were at N2. Moreover, 77 (56%) patients had T1 disease, and 61 (44%) had T2–3 disease. The majority (131 or 95%) of the patients exhibited no invasion or focal LVI, and only 7 (5%) patients had prominent LVI. Most patients (128 or 93%) had tumors at grades 1–2, and only 9 (7%) patients had grade 3 tumors. Sixty-one (44%) patients were treated with modified radical mastectomy (MRM), whereas 77 (56%) were treated with BCS. Thirty (22%) and 108 (78%) patients were treated with and without chemotherapy, respectively. All patients were tested with ODx before this study.
Table 1.
Baseline characteristics
| Variables | Patient number (%) |
|---|---|
| Age | |
| Age > 50 | 70 (51%) |
| Age ≤ 50 | 68 (49%) |
| LVI | |
| Absent/focal | 131 (95%) |
| Prominent | 7 (5%) |
| Tumor grade | |
| Grade 1–2 | 128 (93%) |
| Grade 3 | 9 (7%) |
| Tumor category | |
| T1 | 77 (56%) |
| T2–3 | 61 (44%) |
| N category | |
| N0 | 80 (58%) |
| N1 | 56 (41%) |
| N2 | 2 (1%) |
| Ki67 | |
| ≤ 10% | 44 (39%) |
| 11–20% | 31 (28%) |
| > 20% | 37 (33%) |
| Chemotherapy | |
| No | 108 (78%) |
| Yes | 30 (22%) |
| Surgery | |
| Mastectomy | 61 (44%) |
| BCS | 77 (56%) |
BCS: Breast-conserving surgery; LVI: Lymphovascular invasion.
Recurrence index in different patient subgroups
The correlation between the RI-DR and various clinicopathologic factors was analyzed, and RI scores were positively associated with some of the factors (Table 2). For example, the percentage of high-risk patients according to the RI-DR cutoff was higher among those with T2–3 disease than those with T1 disease, among those with tumor grade 3 than those with tumor grade 1, and among tumors with Ki67 expression > 10% than those with Ki67 expression ≤10%. In contrast, no difference in RI-DR was detected between patients ≤ 40 years old and those > 40 years, nor between those with stage N0 and stage N1–2, nor between those positive for both ER and PR and those positive for either ER or PR in this small series.
Table 2.
Correlation between recurrence index (RI) score of the 18-gene classifier panel and clinicopathologic risk factors
| Risk factors | RI-DR low (<36) | RI-DR high (≥36) | P-value |
|---|---|---|---|
| Age | 1 | ||
| ≤ 40 | 14 (13%) | 5 (15%) | |
| > 40 | 90 (87%) | 29 (85%) | |
| T stage | 0.0149 | ||
| T1 | 65 (63%) | 12(35%) | |
| T2–T3 | 39 (38%) | 22(65%) | |
| N stage | 0.8306 | ||
| N0 | 61 (59%) | 19 (56%) | |
| N1–2 | 43 (41%) | 15 (44%) | |
| Tumor grade | 0.0005 | ||
| Grade I–II | 101 (98%) | 27 (79%) | |
| Grade III | 2 (2%) | 7 (21%) | |
| ER/PR status | 1 | ||
| Both (+) | 100 (96%) | 33 (97%) | |
| ER or PR (+) | 4 (4%) | 1 (3%) | |
| Ki67 | 0.0131 | ||
| ≤ 10% | 19 (9%) | 0 (0%) | |
| > 10% | 72 (91%) | 32 (100%) |
ER: estrogen receptor; PR: progesterone receptor; RI-DR: recurrence index for distant recurrence.
Clinical performance of the RI-DR and ODx RS
Among the 138 patients, 104 were classified by the RI-DR as low risk, whereas 34 were classified as high risk (Table 3). When compared against clinical outcomes, one out of the 104 low-risk patients had a recurrence event (NPV = 103/104 = 99%), whereas 5 out of the 34 high-risk patients had a recurrence event (PPV = 5/34 = 15%). Sensitivity was 5/6 (or 83%), and specificity was 103/132 (or 78%).
Table 3.
Oncotype DX recurrence score (RS) and recurrence index for distant recurrence (RI-DR) with clinical outcomes
| Risk group | Patient # | Any recurrences | |
|---|---|---|---|
| RI-DR | Low (<36) | 104 | 1 (1%) |
| High (≥36) | 34 | 5 (15%) | |
| Oncotype DX RS | Low (26) | 121 | 3 (2%) |
| High (≥26) | 17 | 3 (18%) | |
| RI-DR (>50yo) | Low (<36) | 54 | 0 |
| High (≥36) | 16 | 2 (12.5%) | |
| Oncotype DX RS (>50yo) | Low (<26) | 63 | 1 (1.6%) |
| High (≥26) | 7 | 1 (14.3%) | |
| Total | 70 | 2 (2.9%) | |
| RI-DR (≤50yo) | Low (<36) | 50 | 1 (2%) |
| High (≥36) | 18 | 3 (16.7%) | |
| Oncotype DX RS (≤50yo) | Low (<26) | 58 | 2 (3.4%) |
| High (≧26) | 10 | 2 (20%) | |
| Oncotype DX RS (≤50yo) | Low (<16) | 29 | 0 |
| High (≧16) | 39 | 4 (10.3%) | |
| Total | 68 | 4 (5.9%) |
In contrast, 121 of the 138 patients were classified by the ODx RS as low risk (cutoff < 26), whereas 17 were classified as non-low risk (Table 3). When compared against clinical outcomes, three of the 121 low-risk patients had a recurrence event (NPV = 118/121 = 98%), whereas three of the 17 non-low-risk patients had a recurrence event (PPV = 3/17 = 18%). Sensitivity was 3/6 (or 50%), and specificity was 118/132 (or 89%).
The observation was similar if patients were divided into age > 50 years and age ≤ 50 years. The risk classification by ODx RS and RI-DR was comparable, however, the RS cutoff of 16 performed better than the cutoff of 26 for women with age ≤ 50 years (Table 3).
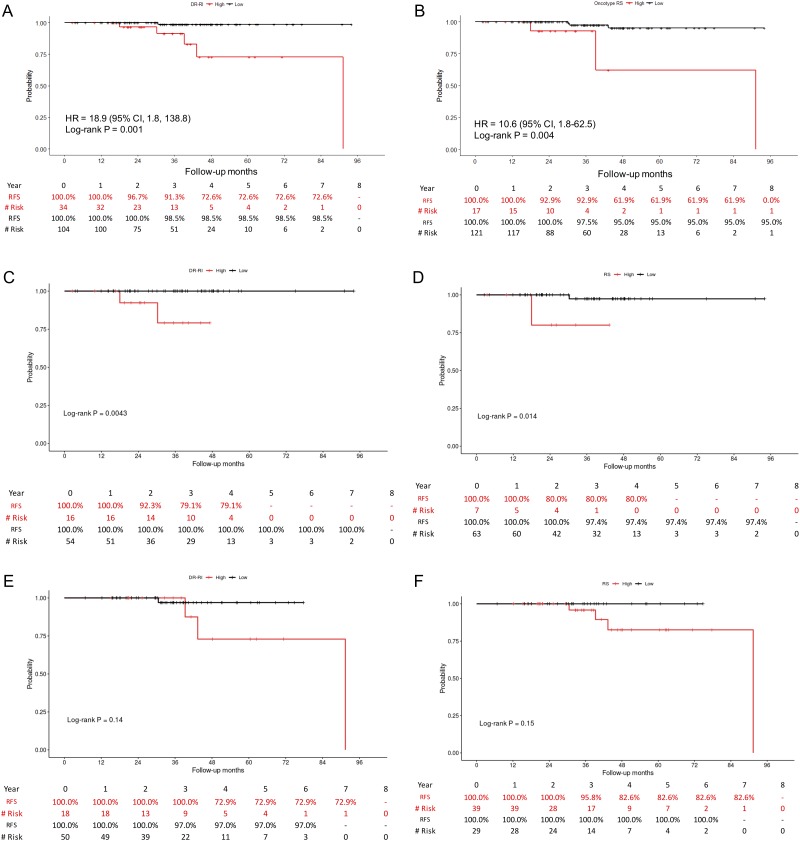
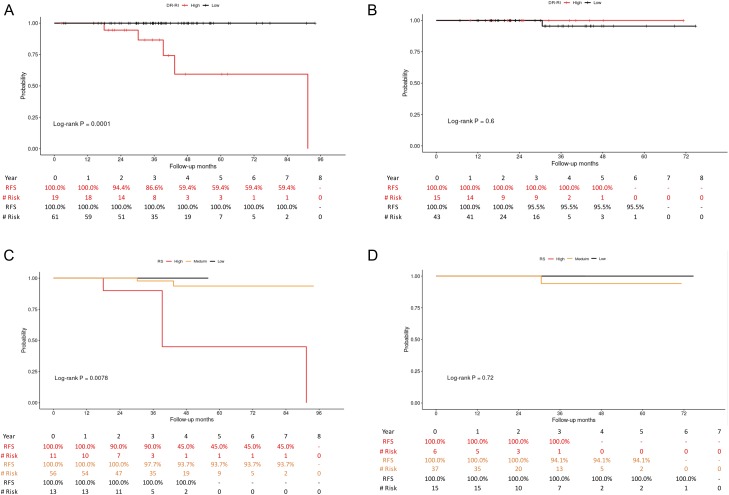
With a median follow-up interval of 36.8 months (ranging from 2.5 to 94.1 months), the survival probabilities of the low- and high-risk patients were plotted against the disease-free survival time (Fig. 2A-B). The results showed that patients classified as having low risk of recurrence by the RI-DR had a recurrence-free survival (RFS) rate of 98.5% at 4 years. In contrast, for those classified as having a high risk of recurrence, the 4-year RFS rate declined to 72.6% (P = 0.001, hazard ratio: 18.9, 95% confidence interval [CI]: 1.8–138.3). Similar trends were noticed by plotting the ODx RS: the 4-year RFS was 95.0% in the low-risk group and 61.9% in the high-risk group (P = 0.004, hazard ratio: 10.6, 95% CI: 1.8–62.5). Survival probabilities for ages ≤50 and >50 were plotted separately in Fig. 2C-D (RI-DR) and Fig. 2E-F (RS). Figures of survival plots for node-negative and node-positive patients categorized by RI-DR and RS are provided in Fig. 3. RI-DR could differentiate low- and high-risk N0 patients (P = 0.0001); however, whereas RS < 11 and 11-25 had similar outcome, RS ≥ 26 had the poorest prognosis (P = 0.0078).
Figure 2.
Relapse-free survival rates of (A) low-risk (scores <36) and high-risk (scores ≥36) patients determined by RI-DR; (B) low-risk (scores <26) and high-risk (scores ≥26) patients determined by RS; (C) Age > 50: low-risk (scores <36) and high-risk (scores ≥36) by RI-DR; (D) Age > 50, low-risk (scores <26) and high-risk (scores ≥26) by RS; (E) Age ≤50: low-risk (scores <36) and high-risk (scores ≥36) by RI-DR; (F) Age ≤50, low-risk (scores <16) and high-risk (scores ≥16) by RS.
Figure 3.
Relapse-free survival rates of low- (scores <36) and high-risk (scores ≥36) patients determined by RI-DR: (A) node negative, (B) node positive. Relapse-free survival rates of low- (scores <11), medium- (scores 11-25) and high-risk (scores ≥26) patients determined by RS: (C) node negative, (D) node positive.
Concordance between RI-DR and ODx RS
There were two cutoffs used for the ODx RS. First, cutoffs of < 18 and > 30 were used to define the low- and high-risk groups, respectively. Using the revised cutoffs, RS values of < 11 and > 25 were used to define the low- and high-risk groups, respectively.
Using the initial RS cutoffs of 18 and 30, the agreement between RI-DR and RS in the low- and high-risk groups was 90% (74/82). The RS intermediate group (score 18–30) was classified as low-risk by the RI-DR in 63% of cases (Table 4).
Table 4.
Concordance between recurrence index for distant recurrence (RI-DR) and Oncotype DX recurrence score (RS)
| All patients | RI-DR | ||||
|---|---|---|---|---|---|
| Cutoff 18–30 | Low (<36) | High (≥36) | Total | Relapse (%) | |
| Oncotype DX RS | Low (<18) | 67 (92%) | 6 (8%) | 73 | 1 (1.4%) |
| Middle (18–30) | 35 (63%) | 21 (37%) | 56 | 4 (7.1%) | |
| High (≥31) | 2 (22%) | 7 (78%) | 9 | 1 (11.1%) | |
| Cutoff 11–25 | |||||
| Oncotype DX RS | Low (≤10) | 26 (93%) | 2 (7%) | 28 | 0 |
| Middle (11–25) | 74 (80%) | 19 (20%) | 93 | 3 (3.2%) | |
| High (≥26) | 4 (24%) | 13 (76%) | 17 | 3 (17.6%) | |
| Total | 104 (75%) | 34 (25%) | 138 | ||
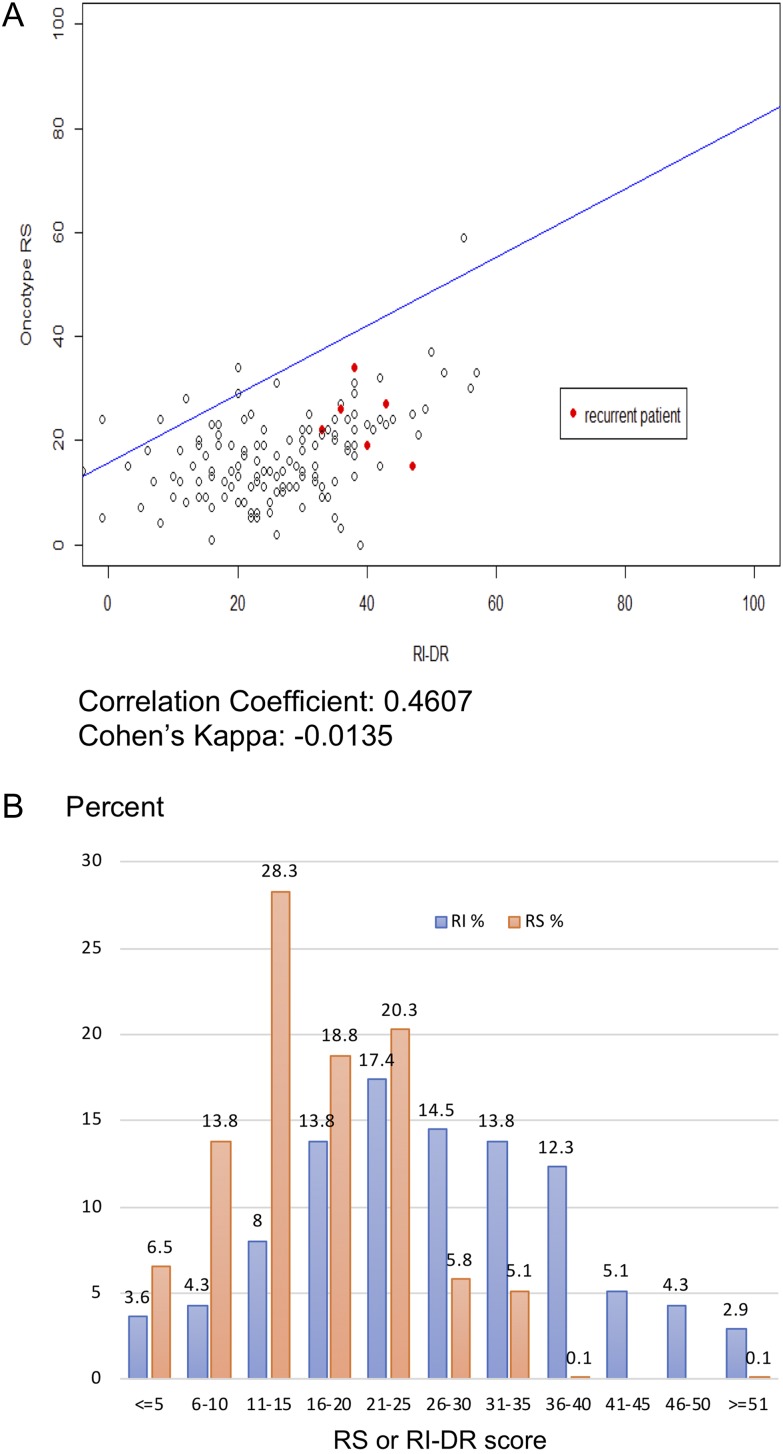
Using the new cutoffs of 11 and 25, 26 out of the 28 (93%) RS low-risk patients were also classified as low-risk by the RI-DR (Table 4). In contrast, 13 (76%) of the 17 RS high-risk patients were defined as high-risk group by the RI-DR. Patients with RS scores of 11–25 were categorized as low-risk by the RI-DR in 80% of cases (74/93). The total agreement using the revised cutoffs (<26 as low risk and ≥ 26 as high risk) was thus 113/138 (82%), with a Cohen’s k of 0.41, indicating moderate agreement (Fig. 4A).
Figure 4.
(A) Correlation between RI-DR and RS. The x-axis provides the RI-DR, and the y-axis shows the RS. Red dots represent recurrent patients. (B) Score distribution between RI-DR and RS. The x-axis provides the RS and RI-DR score intervals, and the y-axis shows the percentage of patients in each score interval.
Both RS score cutoffs (18 and 30 or 11 and 25) showed that the high-risk group was at high risk of recurrence, with recurrence rates of 11.1% and 17.6%, respectively (Table 4). The relapse rate in the intermediate-risk group defined by scores of 11–25 was more reasonable (3.2%) than that in the group defined by scores of 18–30 (7.1%).
The RI-DR scores were distributed normally; the median score was 26, and the lower and upper quantile scores were 19 and 35, respectively. The median score of ODx RS score was 17, while the lower and upper quantile were 11 and 22, respectively; only 2 patients had scores > 35 in this series (Fig. 4B).
Recurrent cases
Only six out of the 138 patients experienced recurrence (five LRR and one distant metastasis). Compared with the RI-DR, the ODx RS failed to predict the outcomes of cases 1 and 4. Case 4, who had a T3 tumor and was younger than 50 years of age, would be considered clinically high-risk and be responsible for her local recurrence. According to the subgroup analysis of the TAILORx trial (6), women aged ≤ 50 years with RS > 16 would be classified as at high risk of distant recurrence. Both RS and RI-DR failed to predict the relapse of case 2; one possible explanation for this could be the lack of adjuvant radiotherapy after BCS.
Discussion
The current 18-gene-based clinical-genomic model (RI-DR) identified 75% (104/138) of HR-positive and HER2-negative EBC patients to be at low-risk; these patients had an excellent 4-year RFS of 98.5%. According to recurrence and outcome patterns, more than 60% of recurrences happen within 4 years after surgery (12); thus, recurrence would be too rare among these patients for them to obtain any benefit from adjuvant chemotherapy. Breast cancer treatments not only carry the risk of individual side effects but also have become a substantial public health burden (1). Considering the decreased morbidity and mortality rates in EBC patients, numerous healthcare providers have begun to make efforts to find new methods and biomarkers for tailoring treatments to patients and improving their quality of life, such as avoiding axillary lymph node dissection and forgoing adjuvant chemotherapy in selected patients (13,14). The current study offers another option for identifying low-risk patients after surgery.
It has also been shown that clinical manifestations of breast cancer in Asian patients may differ from those in Caucasian counterparts. Assays developed based on Western populations may thus not be suitable for predicting recurrence in Asian breast cancer patients. For example, Toi et al. showed that the risk of recurrence in some Japanese patients was overestimated, as those classified as having intermediate risk experienced LRR at a rate of only 2.5% and no distant recurrence (9). In contrast to this, we have found that patients from Taiwan with an RS of 18–30 had a high rate of recurrence (7.1%) with a median follow-up only 36.8 months (Table 4). Of the four patients with LRR, three would be at high-risk of distant metastasis, as two had chest wall recurrence after mastectomy and one had supraclavicular lymph node recurrence after BCS (Table 5). Asian patients were previously found to be less likely to be classified as high risk than non-Hispanic whites (15). It may thus prove critical to test Asian patients with more ODx assays or a more suitable multi-gene prognostic assay, e.g., one that was developed based on Asian genomic data.
Table 5.
Recurrent case study: treatments and failure patterns
| Case | Age | Surgery | TNM | R/T | C/T | Relapse site | ODx RS26 | RI-DR36 |
|---|---|---|---|---|---|---|---|---|
| 1 | 56–60 | BCS | T1N0 | Yes | No | Breast | 15 | 47 |
| 2 | 46–50 | BCS | T2N1 | No | No | Breast/axilla | 22 | 33 |
| 3 | 41–45 | MRM | T1N0 | No | Yes | Local | 26 | 36 |
| 4 | 41–45 | MRM | T3N0 | No | No | Local | 19 | 40 |
| 5 | 36–40 | BCS | T2N0 | Yes | No | Supraclavicular lymph node | 27 | 43 |
| 6 | 51–55 | MRM | T2N0 | No | Yes | Distant | 34 | 38 |
BCS: Breast-conserving surgery; MRM: Modified radical mastectomy.
The 18-gene-based clinical-genomic model (RI-DR) that was developed by our group decades ago has been described previously and was demonstrated to be an independent prognostic factor in Asian patients with luminal-type breast cancer (16). FFPE sections can be used for analysis with the multiplex probe-based NanoString system for RI-DR testing, and the validation work had already been completed (Supplement 1). In the current study, among the 104 patients classified by the RI-DR as low risk, only one had a recurrence event, resulting in an NPV of 99%. In contrast, five of the 34 high-risk patients had a recurrence event, resulting in a PPV of 15%. In comparison, even though the ODx assay also had an NPV of 98%, three patients classified as low risk had recurrence. The RI-DR and ODx RS exhibited moderate agreement (Cohen’s k = 0.41), and the total agreement was 82% if the high-risk cutoff of ≥26 was used. It is noteworthy that while correctly identifying non-recurrent patients (i.e., high NPV) is critical when evaluating a prognostic test, identifying recurrent patients (i.e., high sensitivity) is even more important. The results indicate that the RI-DR, which has both a relatively high NPV and a relatively high sensitivity, could be a useful tool for identifying low-risk breast cancer patients, especially those with Asian genetic backgrounds, such that they may forego adjuvant chemotherapy.
There are some limitations to the current study. First, we designed the study to compare the performance of the two panels using two different RS cutoffs because the updated results from the TAILORx study (6) were published during our study period. Although this is likely to result in an inevitable bias, we still achieved moderate agreement with the RS score in all patients, reflecting the reliable performance of RI-DR at the very least. Second, luminal breast cancer has a relatively good prognosis and may therefore result in more late relapses of >5 years after primary surgery in comparison to other subtypes of EBC (12). The present study has a relatively short follow-up interval, and therefore the results should be interpreted with caution; however, the RFS benefit from adjuvant chemotherapy usually is observed within the first 5 years after surgery (3). Finally, the RI-DR predictions performed well with regard to the low-risk patients (n = 103), with a 4-year RFS of 98.5%, which poses an interesting question regarding whether patients deemed at low-risk by our panel could forgo adjuvant chemotherapy.
Conclusion
In conclusion, the current study shows that 18-GC, when integrated with clinicopathologic models, can accurately identify early-stage luminal-like breast cancer patients who are at low risk of recurrence. Nevertheless, further follow-up is needed to demonstrate the effectiveness of the 18-GC panel in predicting recurrence over a longer period. Future study with a larger cohort size is also warranted.
Supplementary Material
Funding
This work was supported by 1) Amwise Diagnostics Pte. Ltd., Singapore; and 2) National Taiwan University Hospital.
Clinical Trials
ClinicalTrials.gov Identification No.: NCT03772197. Retrieved from https://clinicaltrials.gov/ct2/show/NCT03772197
Conflict of interest statement
None declared.
Author Contributions
Conception/Design: SHC, CSH
Provision of study material or patients: PSY, YHL, CFC, YCC, BLY, CSH
Collection and/or assembly of data: KHS
Data analysis and interpretation: KHS, SHC, CSH
Manuscript writing: JL, SHC
Final approval of manuscript: All authors
Disclosure of Potential Conflicts of Interest
SHC, co-founder of Amwise Diagnositics
KHS and JL, employees of Amwise Diagnostics
Ethics Statements
This study was carried out in accordance with the recommendations of International Compilation of Human Research Standards with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki. The protocol was approved by the institutional review board of each of the participating medical centers (IRB numbers: NTUH 201610066RINA, KFCC 20150327A, and MMH 17CT040be).
References
- 1. Global Burden of Disease Cancer C. Fitzmaurice C, Allen C, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol 2017;3:524–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tabar L, Fagerberg G, Duffy S, et al. Update of the Swedish two-county program of mammographic screening for breast cancer. Radiol Clin North Am 1992;30:187–210. [PubMed] [Google Scholar]
- 3. Dignam JJ, Dukic V, Anderson SJ, et al. Hazard of recurrence and adjuvant treatment effects over time in lymph node-negative breast cancer. Breast Cancer Res Treat 2009;116:595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Esserman LJ, Thompson IM, Reid BJJ. Overdiagnosis and overtreatment in cancer: an opportunity for improvement. JAMA 2013;310:797–8. [DOI] [PubMed] [Google Scholar]
- 5. Cheng SH-C, Yu B-L, Horng C-F, et al. Long-term survival and stage I breast cancer subtypes. J Cancer Res Pract 2016;3:1–8. [Google Scholar]
- 6. Sparano JA, Gray RJ, Makower DF, et al. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N Engl J Med 2018;379:111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Naoi Y. Noguchi SJBc: Multi-gene classifiers for prediction of recurrence in breast cancer patients. Breast Cancer 2016;23:12–8. [DOI] [PubMed] [Google Scholar]
- 8. Harris LN, Ismaila N, McShane LM, et al. Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early-stage invasive breast cancer: American society of clinical oncology clinical practice guideline. J Clin Oncol 2016;34:1134–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Toi M, Iwata H, Yamanaka T, et al. Clinical significance of the 21‐gene signature (Oncotype DX) in hormone receptor‐positive early stage primary breast cancer in the Japanese population. Cancer 2010;116:3112–8. [DOI] [PubMed] [Google Scholar]
- 10. Iqbal J, Ginsburg O, Rochon PA, et al. Differences in breast cancer stage at diagnosis and cancer-specific survival by race and ethnicity in the United States. JAMA 2015;313:165–73. [DOI] [PubMed] [Google Scholar]
- 11. Cheng SH, Horng CF, Huang TT, et al. An eighteen-gene classifier predicts locoregional recurrence in post-mastectomy breast cancer patients. EBioMedicine 2016;5:74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cossetti RJ, Tyldesley SK, Speers CH, et al. Comparison of breast cancer recurrence and outcome patterns between patients treated from 1986 to 1992 and from 2004 to 2008. J Clin Oncol 2015;33:65–73. [DOI] [PubMed] [Google Scholar]
- 13. Giuliano AE, Ballman KV, McCall L, et al. Effect of axillary dissection vs no axillary dissection on 10-year overall survival among women with invasive breast cancer and sentinel node metastasis: the ACOSOG Z0011 (Alliance) Randomized Clinical Trial. JAMA 2017;318:918–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harbeck N, Sotlar K, Wuerstlein R, et al. Molecular and protein markers for clinical decision making in breast cancer: today and tomorrow. Cancer Treat Rev 2014;40:434–44. [DOI] [PubMed] [Google Scholar]
- 15. Kizy S, Huang JL, Marmor S, et al. Distribution of 21-gene recurrence scores among breast cancer histologic subtypes. Arch Pathol Lab Med 2018;142:735–41. [DOI] [PubMed] [Google Scholar]
- 16. Cheng SH, Huang TT, Cheng YH, et al. Validation of the 18-gene classifier as a prognostic biomarker of distant metastasis in breast cancer. PLoS One 2017;12:e0184372. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.