Abstract
Particulate matter 2.5 (PM2.5) is a significant risk factor for asthma. A recent study revealed that autophagy was associated with asthma pathogenesis. However, the specific mechanisms underlying PM2.5-induced autophagy in asthma have remained elusive. In the present study, PM2.5-induced autophagy was evaluated in Beas-2B human bronchial epithelial cells and the potential molecular mechanisms were investigated. Using electron microscopy, immunofluorescence staining and immunoblot studies, it was confirmed that PM2.5 induced autophagy in Beas-2B cells as a result of PM2.5-mediated inhibition of the phosphatidylinositol 3-kinase (PI3K)/Akt/mammalian target of rapamycin (mTOR) pathway in Beas-2B cells. LY294002, a PI3K inhibitor, reduced the accumulation of microtubule-associated protein 1 light chain 3 II and attenuated the effect of PM2.5. Phosphorylated (p-)p38, p-extracellular signal-regulated kinase and p-c-Jun N-terminal kinase were dephosphorylated following exposure to PM2.5. The roles of p53, reactive oxygen species scavenger tetramethylthiourea and autophagy inhibitor 3-methyladenine in PM2.5-induced autophagy in Beas-2B cells were also investigated. The results suggested that the PI3K/Akt/mTOR signaling pathway may be a key contributor to PM2.5-induced autophagy in Beas-2B cells. The results of the present study therefore provided an a insight into potential future clinical applications targeting these signaling pathways, for the prevention and/or treatment of PM2.5-induced lung diseases.
Keywords: particulate matter, autophagy, Beas-2B, asthma, phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin
Introduction
Acute and chronic exposure to particulate matter (PM), particularly fine particles with aerodynamic diameters of ≤2.5 µm PM2.5, has been shown to increase the number of hospital admissions for respiratory causes amongst the general population (1). Evidence obtained from environmental and epidemiological studies has revealed a marked association between fine particulate air pollution and multiple health issues, including respiratory illnesses (for example, respiratory track inflammation, asthma, acute bronchitis and lung cancer) as well as cardiovascular disease mortality (2–4).
Increasing evidence supporting a link between traffic-associated air pollution and the incidence of childhood asthma has emerged; however, published estimates highlight the variability observed between populations (5,6). Asthma is a common worldwide respiratory symptom complex, frequently involving airway inflammation, which results in clinically significant physiological airway dysfunction. Asthma pathogenesis is complex, and multiple factors have roles in the development of the disease (7).
Autophagy describes an evolutionarily conserved and tightly regulated lysosomal pathway, responsible for the degradation of macromolecules, including proteins, glycogen, lipids, nucleotides and organelles, via several complex pathways (8). Recently, this process has been demonstrated to be involved in numerous biological processes, including host defense, cell survival and cell death. The detection of autophagosomes in fibroblasts and epithelial cells in tissues from patients with asthma has revealed a potential link between autophagy and asthma pathogenesis (9). Numerous stress signals are able to induce autophagy, including p53, phosphatidylinositol 3-kinase (PI3K)/mammalian target of rapamycin (mTOR) and adenosine monophosphate-activated protein kinase (AMPK) (10,11). The PI3K/Akt/mTOR signaling pathway is required for the regulation of autophagic flux. The mitogen activated protein kinase (MAPK) pathway mediates the transmission of signals between receptors and the nucleus via multiple intermediate proteins, including Ras, Raf, extracellular signal-regulated kinase (ERK) and MAPK/ERK (MEK). Akhtar et al (12) reported that the induction of autophagy via pathways involving ERK1/2, p38 MAPK and Akt enhanced cardiac cell survival following ischemia-reperfusion injury.
A previous study by our group revealed that p53 mediated PM-induced alveolar epithelial cell mitochondria-regulated apoptosis (13). It was also confirmed that the Akt/mTOR and c-Jun N-terminal kinase (JNK) signaling pathways were involved in chrysotile asbestos-induced autophagy in human lung epithelial cells (A549) (14). In the present study, the effect of PM2.5 on the induction of autophagy in vitro and the potential underlying mechanisms were evaluated.
Materials and methods
PM2.5 sampling and composition
The PM2.5 used in the present study was obtained from Zhanjiang, China. The PM2.5 was collected using the QJS-100 multi-level flow particulate matter cutter (Jinzhoulicheng, Jinzhou, China), at a constant aspiration flow rate (100 l/min) over a period of 48 h. The sample containing the PM2.5 fiber was placed in ultrapure water and subjected to ultrasonic oscillations (Xingzhi Biotechnology Co., Ltd., Ningbo, China) for 15 min to elude the particulate matter, vacuum-freeze dried (Xingzhi Biotechnology Co., Ltd.,) for 24 h and formulated into a stock solution by the addition of phosphate-buffered saline (PBS), followed by auto-claving. The PM2.5 suspensions were vortexed and stored at 4°C. PM2.5 filter samples were analyzed according to a previous study (15), the results of which are exhibited in Table I.
Table I.
Concentrations of 15 types of PAHs, transition metals and common metals identified in PM from Zhanjiang, China.
| PM Content | Concentration (pg/µg) |
|---|---|
| PAHs | |
| Napthalene | 23.11 |
| Acenapthylene | 25.00 |
| Fluorene | 42.22 |
| Phenanthrene | 26.89 |
| Anthracene | 20.33 |
| Fluoranthene | 28.00 |
| Perylene | 36.11 |
| 1,2-benza(a)anthracene | 23.33 |
| Chrysene | 27.78 |
| Benzo(b)fluoranthene | 61.56 |
| Benzo(k)fluoranthene | 35.00 |
| Benzo(a)pyrene | 17.22 |
| Ideno(1,2,3-cd)pyrene | 24.44 |
| Dibenzo(ah)anthracene | 30.56 |
| Benzopyrene | 11.33 |
| Total PAHs | 459.56 |
| Metals | Concentration (ng/µg) |
| Na | 12.44 |
| K | 19.44 |
| Mg | 4.67 |
| Ca | 3.89 |
| Fe | 3.00 |
| Cu | <0.1 |
| Cr | 0.11 |
| Co | <0.1 |
| V | <0.1 |
| Ni | 0.72 |
| Cd | <0.1 |
| As | <0.1 |
PAHs, polycyclic aromatic hydrocarbons; PM, particulate matter.
Reagents and antibodies
Dulbecco's modified Eagle's medium (DMEM), penicillin, streptomycin and fetal bovine serum (FBS) were purchased from Gibco-BRL Life Technologies (Gaithersburg, MD, USA). Rapamycin, dimethyl sulfoxide (DMSO) and trypsin-EDTA were obtained from Sigma-Aldrich (St. Louis, MO, USA). Rapamycin was dissolved in DMSO and the final concentration of DMSO in the culture medium did not exceed 0.2%. The primary antibodies used in western blotting and immunofluorescent analysis are exhibited in Table II. All secondary antibodies were obtained from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). 3-MA, dorsomorphin, pifithrin-α (PFT-α) and tetramethylthiourea (TMTU) were obtained from Sigma-Aldrich. LY294002, U0126, SP600125 and SB203580 were purchased from Calbiochem® (Merck Millipore KGaA, Darmstadt, Germany). pBABE-mCherry-enhanced-green fluorescent protein (eGFP)-microtubule-associated protein 1 light chain 3 (LC3) was purchased from Biovector Science Lab, Inc. (Beijing, China). SDS sample buffer, protease inhibitors, nonfat dry milk, Tris-buffered saline (TBS) and Tween 20 were purchased from Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China). Lipofectamine® 2000 was obtained from Invitrogen Life Technologies (Carlsbad, CA, USA). Glutaraldehyde, parafor-maldehyde, cacodylate buffer, OsO4, ethanol, epoxy resin, lead citrate and uranyl acetate were purchased from Guangzhou Chemical Reagent Factory (Guangzhou, China).
Table II.
Primary antibodies used in the present study.
| Antibody | Source | Clonality | Manufacturer (cat. no.) | Concentration |
|---|---|---|---|---|
| Anti-AKT | Rabbit | Polyclonal | Cell Signaling Technology, Inc. (#9272) | 1:300 (WB) |
| Anti-p-AKT | Rabbit | Monoclonal | Cell Signaling Technology, Inc. (#4058) | 1:300 (WB) |
| Anti-AMPK | Rabbit | Monoclonal | Cell Signaling Technology, Inc. (#5831) | 1:300 (WB) |
| Anti-p-AMPK | Rabbit | Polyclonal | Cell Signaling Technology, Inc. (#2531) | 1:300 (WB) |
| Anti-ERK | Rabbit | Monoclonal | Cell Signaling Technology, Inc. (#4695) | 1:300 (WB) |
| Anti-p-ERK | Rabbit | Monoclonal | Cell Signaling Technology, Inc. (#4377) | 1:300 (WB) |
| Anti-JNK | Rabbit | Monoclonal | Cell Signaling Technology, Inc. (#9258) | 1:300 (WB) |
| Anti-p-JNK | Rabbit | Monoclonal | Cell Signaling Technology, Inc. (#4668) | 1:300 (WB) |
| Anti-p38 | Rabbit | Polyclonal | Cell Signaling Technology, Inc. (#9212) | 1:300 (WB) |
| Anti-p-p38 | Rabbit | Monoclonal | Cell Signaling Technology, Inc. (#4511) | 1:300 (WB) |
| Anti-p53 | Rabbit | Monoclonal | Cell Signaling Technology, Inc. (#2527s) | 1:300 (WB) |
| Anti-p-p53 | Rabbit | Polyclonal | Cell Signaling Technology, Inc. (#9284) | 1:300 (WB) |
| Anti-β-actin | Rabbit | Polyclonal | Santa Cruz Biotechnology, Inc. (sc-1616) | 1:500 (WB) |
| Anti-mTOR | Rabbit | Polyclonal | Cell Signaling Technology, Inc. (#2972) | 1:300 (WB) |
| Anti-p-mTOR | Rabbit | Monoclonal | Cell Signaling Technology, Inc. (#5536) | 1:300 (WB) |
| Anti-LC3 | Rabbit | Polyclonal | Cell Signaling Technology, Inc. (#4108) | 1:300(WB) |
| 1:100(IF) |
WB, western blot; IF, immunofluorescence; p-, phosphorylated; AMPK, adenosine monophosphate-activated protein kinase; ERK, extracellular signal-regulated kinases; JNK, c-Jun N-terminal kinases; mTOR, mammalian target of rapamycin; LC3, microtubule-associated protein 1 light chain 3. Cell Signaling Technology, Inc., Danvers, MA, USA.
Cell culture and treatments
Beas-2B human bronchial epithelial cells were purchased from the American Type Culture Collection (Manassas, VA, USA). Beas-2B cells were cultured in DMEM supplemented with 10% heat-inactivated FBS, penicillin (50 U/ml) and streptomycin (50 U/ml). The cells were incubated at 37°C with humidified 5% CO2. Cells were seeded at a concentration of 1.5×105 cells/well in six-well plates. Following 24 h of culture, cells were treated with PM2.5 and rapamycin at concentrations of 100 µg/ml in 10% FBS supplemented medium for the indicated time-periods. For the analysis of PI3K and MAPK signaling pathway inhibition, Beas-2B cells were pre-treated with inhibitors 3-MA (Sigma-Aldrich; 10 mM), LY294002 (Merck Millipore; 10 nM), U0126 (Merck Millipore; 5 µM), SP600125 (Merck Millipore; 50 µM) and SB203580 (Merck Millipore; 10 µM) for 2 h at 37°C, prior to treatment with 100 µg/ml PM2.5 for 24 h at 37°C in the presence of the inhibitors. The inhibitors were dissolved in DMSO and the final DMSO concentration in the culture medium was ≤0.2%.
Transmission electron microscopy (TEM)
The Beas-2B cells were cultured on plates and treated with PM2.5 for 24 h prior to fixing with 3% glutaraldehyde and 2% paraformaldehyde (PFA) in 0.1 mol/l cacodylate buffer (pH 7.3) for 1 h. Following fixation, the samples were postfixed in 1% OsO4 in identical buffer for 1 h, serially dehydrated with ethanol and embedded in epoxy resin. Subsequently, sections (70 nm) were cut on a Leica Ultra-CUT (Ultra-Microtome; Leica Microsystems GmbH, Wetzlar, Germany) and contrasted with 0.1% lead citrate and 8% uranyl acetate in 50% ethanol. Subsequently, the ultrathin sections were evaluated under a transmission electron microscope (JEM-1400; JEOL Ltd., Tokyo, Japan) operated at 120 kV, and images were captured using a Megaview III CCD camera (Soft Imaging System, Lakewood, CO, USA).
Western blot analysis
Cells were washed twice in PBS and protein extracts were obtained by solubilizing the cells in SDS sample buffer supplemented with protease inhibitors. Soluble proteins were isolated from the untreated or treated Beas-2B cells for western blot analysis as described previously (16). Equal amounts of protein (20 µg) from each sample were separated by electrophoresis on 10% SDS-PAGE, transferred to polyvinylidene difluoride membranes (Merck Millipore KGaA) and blocked with 5% nonfat dry milk in 1X TBS plus 0.1% Tween 20 at room temperature for 1 h. The membranes were subsequently incubated with primary antibodies diluted in 5% nonfat dry milk in 1X TBS plus 0.1% Tween 20 overnight at 4°C. The primary antibody against actin (diluted 1:500) was purchased from Santa Cruz Biotechnology, Inc. (Danvers, MA, USA). Following incubation with primary antibodies (Table II), the membranes were washed and incubated with horseradish peroxidase-conjugated anti-mouse (sc-2025) or anti-rabbit (sc-2027) secondary antibodies for 1 h at room temperature. The bound antibodies were detected using an Enhanced Chemiluminescence Western Blotting system (Amersham Biosciences Corp., Piscataway, NJ, USA).
Immunofluorescent microscopy of cells exhibiting LC3-positive vesicles
For indirect immunofluorescent staining, cells were fixed with 4% PFA for 20 min, incubated with blocking buffer (comprised of 3% bovine serum albumin and 0.01% saponin) for 45 min. In order to analyze the autophagic flux, Beas-2B cells were transfected with mCherry-eGFP-LC3-expressing plasmids. The cells were pooled and seeded in chamber slides at a density of 2×104. The level of autophagic flux was determined via examination of the punctate pattern of eGFP and mCherry expression by counting the puncta per cell. Fluorescent images were captured with a confocal microscope (TCS SP5 II; Leica Microsystems GmbH) and analyzed using Cell M software (TCS SP5 II).
Flow cytometric analysis of apoptosis
Apoptosis analysis was performed using flow cytometry with propidium iodide (PI) staining. Cells were seeded at a density of 5×105 cells/well in six-well plates and following 24 h of culture, were treated with 100 µg/ml PM2.5 for a further 24 h at 37°C. Following PM2.5 exposure, cells were harvested and subjected to centrifugation at 400 × g for 5 min at 4°C. The cells were fixed by incubation with cold 75% ethanol for 24 h and subsequently stained with PI solution, which was comprised of 45 mg/ml PI, 10 mg/ml RNase A and 0.1% Triton X-100. Following incubation at 4°C for 1 h in the dark, the fluorescence-activated cells were evaluated using a FACScan flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Statistical analysis
Values are presented as the mean ± standard deviation. Statistical differences among experimental groups were evaluated using one-way analysis of variance with repeated measures. P<0.05 was considered to indicate a statistically significant difference between values. The statistical analysis software package SPSS 11.0 for Windows (SPSS Inc., Chicago, IL, USA) was used to conduct the statistical analyses.
Results
PM2.5 induces the expression of morphological and biochemical markers of autophagy in Beas-2B cells
Treatment of human Beas-2B cells with PM2.5, induced the expression of morphological and biochemical markers of autophagy. Initially, the effects of PM2.5 were analyzed by incubating Beas-2B cells with three concentrations of PM2.5 (25, 50 and 100 µg/ml), or with PBS as the negative control for 24 h (Fig. 1A). Exposure to PM2.5 for 24 h resulted in an up to two-fold, dose-dependent increase in LC3I to LC3II processing, with an apparent maximum at 50 µg/ml. Exposure of Beas-2B cells to 100 µg/ml PM2.5 for 12, 24 or 48 h induced a time-dependent increase in LC3I to LC3II processing, which commenced at 12 h and continued to increase up until 24 h of exposure. Whether there was efficient autophagic flux upon exposure to PM2.5 was subsequently examined. Beas-2B cells were transfected with the tandem-tagged, fluorescent reporter plasmid mCherry-eGFP-LC3II (Fig. 1B). An increase in autophagic flux is indicated by enhanced expression of yellow and red puncta within the cells, whereas blocking of autophagic flux is indicated by an increase in yellow puncta, without an accompanying increase in the number of red puncta in the cells (17). PM2.5 treatment of Beas-2B cells revealed significant accumulation of yellow and red foci, indicative of an enhanced population of immature autophagosomes. These data suggested that PM2.5 induced autophagy and enhanced autophagic flux in Beas-2B cells.
Figure 1.
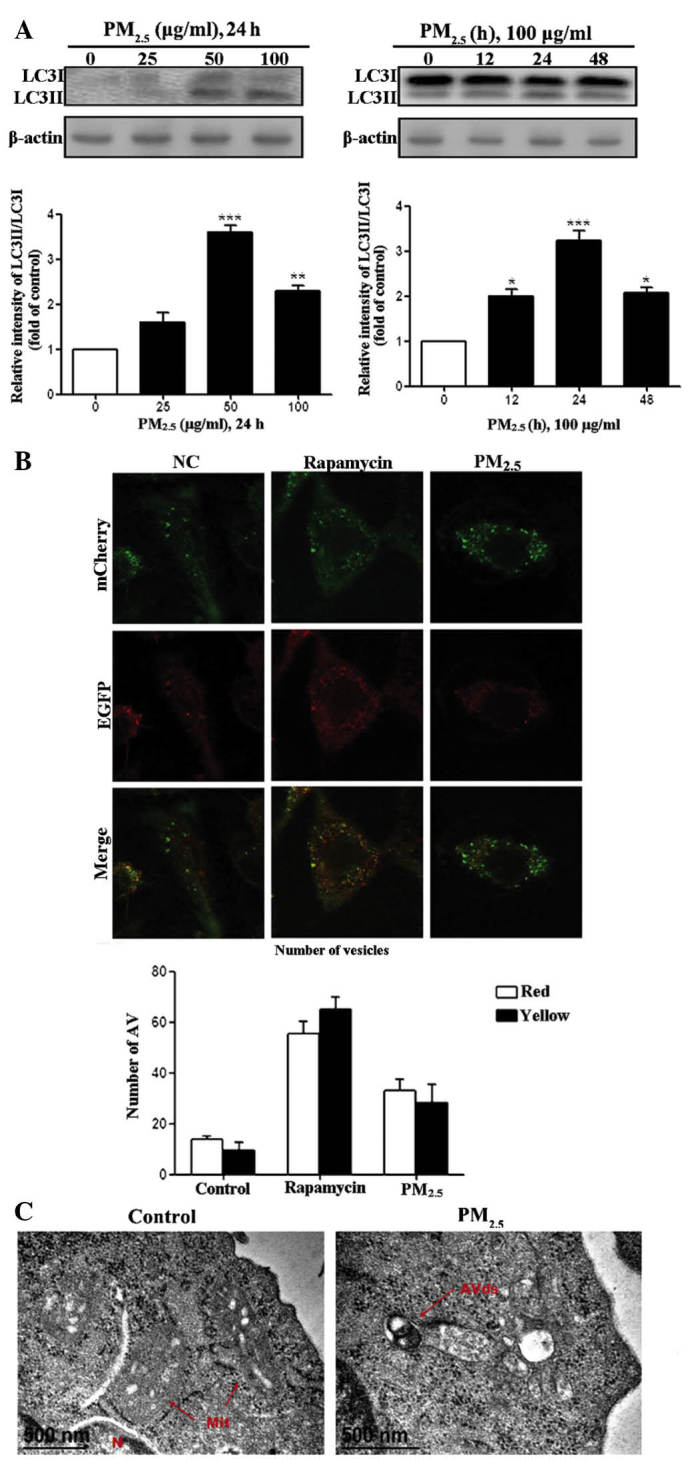
Autophagy is induced in Beas-2B cells following exposure to PM2.5. (A) Beas-2B human bronchial epithelial cells were treated with the indicated concentrations of PM2.5 for 24 h or with 100 µg/ml PM2.5 for the indicated time-periods. Cell lysates were subjected to immunoblot analysis for detection of LC3 levels and β-actin was used as loading control. Quantification of the results are presented as the amount of LC3II normalized against LC3I. (B) Beas-2B cells were transfected with mCherry-eGFP-LC3 and treated with 100 µg/ml PM2.5 for 24 h. Beas-2B cells were treated with phosphate-buffered saline and rapamycin as negative and positive controls, respectively. Cells were examined by fluorescent microscopy, and representative cells were selected and photographed. (C) PM2.5 induced ultrastructural features of autophagy. Beas-2B cells were treated with 100 µg/ml PM2.5 for 24 h and processed for electron microscopy. Note the double membrane structure of the autophagic vacuoles. Degrading autophagic vacuoles (AVds) are indicated. Scale bar, 500 nm. Values are expressed as the mean ± standard deviation of three independent experiments. *P<0.05, **P<0.01 and ***P<0.001 vs. control. All above experiments were repeated three times. PM2.5, particle matter 2.5; LC3, microtubule-associated protein 1 light chain 3; NC, normal control; N, nucleus; Mit, mitochondria; eGFP, enhanced green fluorescent protein; AV, autophagic vesicles.
Morphological indices of autophagy were also evaluated in PM2.5-treated Beas-2B cells. Exposure to PM2.5 for 24 h resulted in an increase in the formation of immature and degradative autophagic vesicles in Beas-2B cells, as detected by electron microscopy (Fig. 1C). Quantification of electron micrographs revealed an approximately three-fold increase in the number of autophagy-positive cells in PM2.5-exposed Beas-2B cells compared with that of PM2.5-untreated cells. These data further supported the hypothesis that PM2.5 induces autophagy in Beas-2B cells.
The AMPK signaling pathway is activated, but not required, for PM2.5-induced autophagy
Previous studies have demonstrated that AMPK functions as an upstream kinase for mTOR, and that AMPK activation downregulates mTOR signaling (18). It was therefore hypothesized that AMPK activation may provide the upstream kinase involved in autophagy activation. In order to investigate the effect of the AMPK signaling pathway, the expression and phosphorylation of AMPK (phosphorylation sites at Thr-172) was examined in Beas-2B cells. As shown in Fig. 2, PM2.5 enhanced the phosphorylation of AMPK following 24 h of incubation. Immunoblot analysis revealed that this enhanced phosphorylation did not occur in tandem with an increase in total AMPK expression (Fig. 2A). Dorsomorphin, a specific AMPK inhibitor, was used to block AMPK phosphorylation. Western blot analysis demonstrated that dorsomorphin effectively blocked AMPK activation in Beas-2B cells; however, PM2.5-induced autophagy was not significantly decreased by dorsomorphin treatment as revealed by LC3II expression analysis (Fig. 2). These results indicated that AMPK is activated by PM2.5 exposure for 24 h; however, this activation is not required for PM2.5-induced autophagy.
Figure 2.
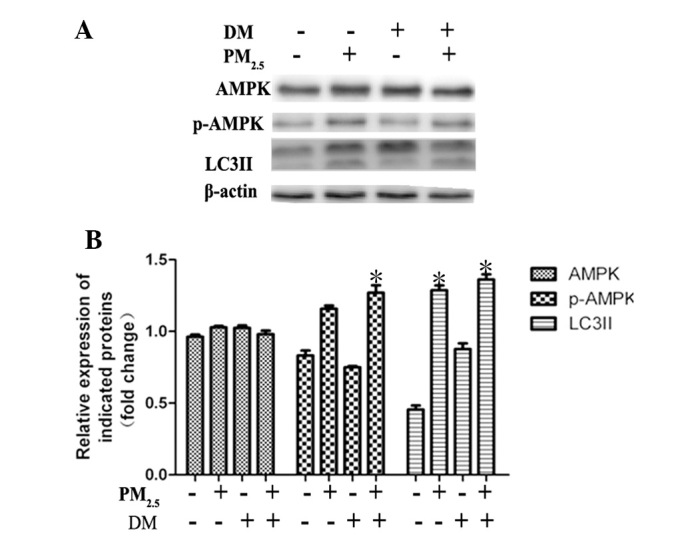
The AMPK signaling pathway positively regulates PM2.5-mediated autophagy in Beas-2B cells. (A) Western blot analysis of AMPK, p-AMPK and LC3II expression. (B) Quantification relative to β-actin expression. Values are expressed as the mean ± standard deviation of ≥3 separate experiments. Cells were treated with 100 µg/ml PM2.5 for 24 h, following 2 h pre-treatment with the AMPK inhibitor DM (40 µm/l). Dimethyl sulfoxide was tested as a control. PM2.5 enhanced AMPK phosphorylation in Beas-2B cells; however, blocking AMPK activation did not significantly influence PM2.5-mediated autophagy. *P<0.05, vs. control. DM, dorsomorphin; PM2.5, particle matter 2.5; AMPK, adenosine monophosphate-activated protein kinase; p-, phosphorylated; LC3II, microtubule-associated protein 1 light chain 3 II.
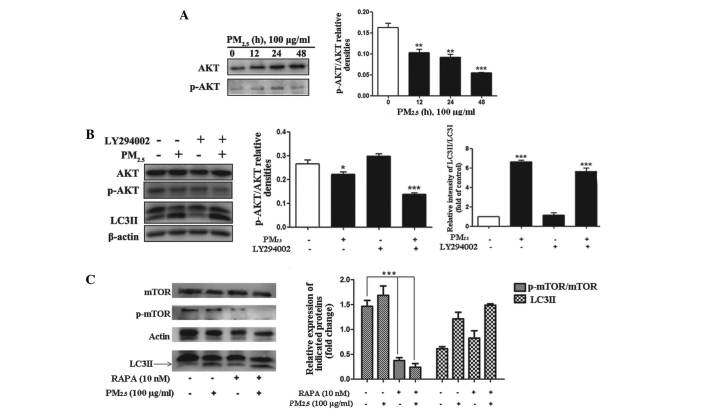
The Akt/mTOR signaling pathway is involved in PM2.5-induced autophagy
The PI3K/Akt/Raptor-mTOR (mTORC1) signaling pathway is a well characterized signaling cascade, which is involved in the regulation of autophagy (19,20). The Akt/mTOR signaling pathway in particular has a significant role in the regulation of autophagy (21,22). For these reasons, whether PM2.5 regulated autophagy via activation of Akt/mTOR signaling was evaluated. The phosphorylation levels of Akt and mTOR were examined by western blotting. Significant dephosphorylation of Akt and mTOR were detected following PM2.5 treatment for 12, 24 and 48 h, (Fig. 3A–C; n=3) with the peak effect occurring following 48 h of exposure. These results indicated that PM2.5-induced Akt activation may be one of the signaling pathways, which contribute to the progression of autophagy.
Figure 3.
Role of the PI3K/AKT/mTOR signaling pathways in PM2.5-induced autophagy. Cells were treated with PM2.5 (0, 12, 24 or 48 h) and the expression levels of the indicated proteins were analyzed by immunoblotting. (A) Phosphorylation status of AKT and protein expression levels of AKT were examined by western blot assay in PM2.5-treated Beas-2B cells. (B) Cells were treated with 100 µg/ml PM2.5 for 24 h following 2 h pretreatment with the inhibitor, LY294002. The phosphorylation status of AKT and protein expression level of AKT were analyzed by western blotting in PM2.5-treated Beas-2B cells. (C) Cells were treated with 100 µg/ml PM2.5 for 24 h, following 2 h pre-treatment with rapamycin. The phosphorylation status of mTOR and protein expression levels of mTOR were analyzed by western blotting in PM2.5-treated Beas-2B cells. Values are expressed as the mean ± standard deviation of three independent experiments. *P<0.05, **P<0.01 and ***P<0.001 vs. control. PI3K, phosphatidylinositol 3-kinase; mTOR, mammalian target of rapamycin; PM2.5, particle matter 2.5; p-, phosphorylated; RAPA, rapamycin; LC3II, microtubule-associated protein 1 light chain 3 II.
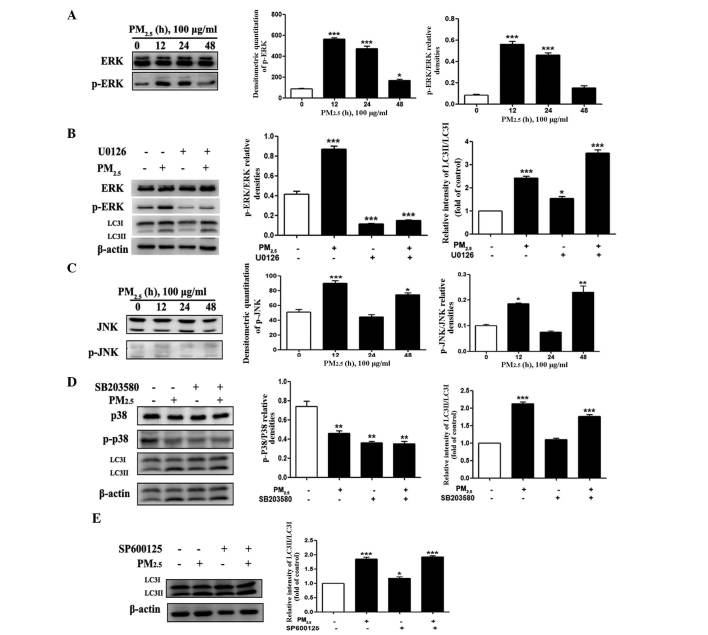
The ERK1/2, but not the JNK or p38 MAPK, pathway is involved in PM2.5-induced autophagy in Beas-2B cells
MAPKs, including p38, JNK, and ERK have key roles in the mediation of autophagy involved in cell death or survival (23). To explore whether MAPK signaling pathways were involved in PM2.5-induced autophagy, the phosphorylation of ERK1/2, JNK and p38 MAPK with LC3 expression and the accumulation of its active form (LC3II) were evaluated following exposure of Beas-2B cells to 100 µg/ml PM2.5 for 12, 24 and 48 h. As shown in Fig. 4A, PM2.5 rapidly and markedly increased ERK phosphorylation following 12 and 24 h of exposure, though little effect was observed following 48 h of exposure. To evaluate the involvement of ERK activation in the modulation of PM2.5-induced autophagy, cells were pre-treated with ERK inhibitor U0126 and the levels of ERK and p-ERK were examined by western blot analysis following PM2.5 exposure. As shown in Fig. 4B, the phosphorylation of ERK was markedly decreased; however the expression of LC3II was not significantly altered following U0126 administration with PM2.5 compared with PM2.5 exposure only.
Figure 4.
Role of MAPK signaling pathways in PM2.5-induced autophagy (A and C) Western blot assays were used to examine the total and phosphorylated protein levels of ERK, JNK and β-actin. (B) Western blot assays were used to examine the expression of LC3II/LC3I, ERK and p-ERK when ERK inhibitor U0126 (5 µM) was added for 1 h before PM2.5 treatment. (D) Western blot assays were used to examine the expression of LC3II/LC3I, p38 and p- p38 when p38 inhibitor SB203580 (10 µM) was added for 1 h before PM2.5 treatment. (E) Western blot assays were used to examine the expression of LC3II/LC3I when JNK inhibitor SP600125 (50 µM) was added for 1 h before PM2.5 treatment. Values are expressed as the mean ± standard deviation of three independent experiments. *P<0.05, **P<0.01 and ***P<0.001 vs. control. MAPK, mitogen activated protein kinase; PM2.5, particle matter 2.5; ERK, extracellular signal-regulated kinase; JNK, c-Jun N-terminal kinase; LC3, microtubule-associated protein 1 light chain 3; p-, phosphorylated.
JNK and p38-MAPK protein expression levels were also evaluated by western blotting to determine whether they were involved in PM2.5-induced autophagy. It was demonstrated that the phosphorylation of JNK was increased 12 h after PM2.5 treatment (Fig. 4C). To confirm the underlying mechanism of the JNK and p38 MAPK signaling pathway involvement in PM2.5-induced autophagy, JNK inhibitor (SP600125) and p38 inhibitor (SB203580) were applied, following 24 h exposure of Beas-2B cells to 100 µg/ml PM2.5. With regard to p38 (Fig. 4D), pre-treatment with SB203580 decreased the phosphorylation level of p38 and elevated LC3II expression. As shown in Fig. 4E, an increase in the conversion of LC3I to LC3II was still observed after pretreatment with a JNK inhibitor. These data suggested that with continuous exposure of Beas-2B cells to PM2.5, p-ERK negatively regulated PM2.5-induced autophagy; whereas, p-JNK and p38 (of the MAPK signaling pathway) did not have a significant effect on PM2.5-induced autophagy in Beas-2B cells.
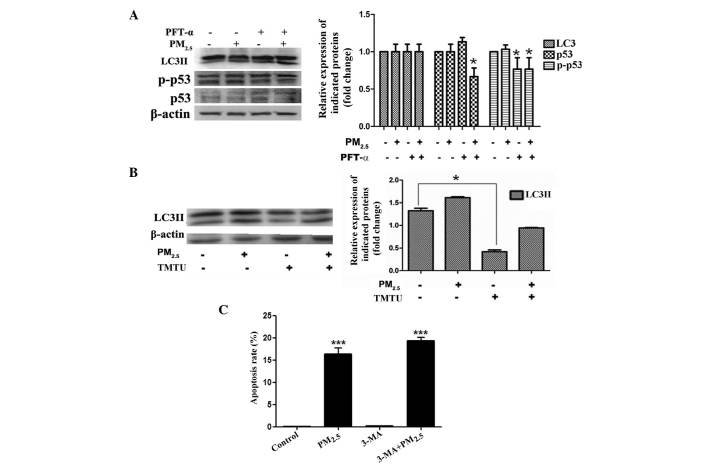
Expression of p53 in PM2.5-induced autophagy
To observe whether PM2.5 enhanced the expression and phosphorylation of p53, immunoblot analysis of Beas-2B cells following exposure to PM2.5 was performed. The total protein expression and phosphorylation of p53 were not significantly altered following PM2.5 exposure (Fig. 5A). To assess the function of p53, p53 activity was blocked with the p53 inhibitor, PFT-α, in Beas-2B cells treated for 24 h with or without PM2.5. Western blot analysis revealed that LC3 protein expression was not markedly altered by PM2.5 exposure with or without PFT-α (Fig. 5A). These data indicated that PM2.5-induced autophagy was p53-independant.
Figure 5.
PM2.5-mediated autophagy is p53-independant. (A) Beas-2B cells were treated with PM2.5 in the absence or presence of PFT-α for 24 h. LC3 and p-p53/p53 protein expression levels were evaluated by western blot analysis (n=3). β-actin was used as an internal control. (B) Beas-2B cells were treated with PM2.5 in the absence or presence of TMTU for 24 h. LC3 protein expression levels were determined by western blot analysis (n=3). β-actin was used as an internal control. (C) Propidium iodide staining for PM2.5-induced apoptosis was measured using a flow cytometer, with or without pretreatment of 3-MA (10 mM) and PM2.5 (100 µg/ml). Values are expressed as the mean ± standard deviation of three independent experiments. *P<0.05 and ***P<0.001 vs. control. PM2.5, particle matter 2.5; PFT-α, pifithrin-α; LC3, microtubule-associated protein 1 light chain 3; TMTU, tetramethylthiourea; 3-MA, 3-methyladenine.
PM2.5-induced Beas-2B cell autophagy is dependent on reactive oxygen species (ROS) production
In order to determine whether ROS generation was involved in the PM2.5-induced autophagy signaling pathway in Beas-2B cells, LC3II expression was evaluated by western blot analysis in non-treated cells and in cells pre-incubated with or without TMTU for 1 h, followed by treatment with 100 µg/ml PM2.5 for 24 h. The activation of LC3II by PM2.5 was diminished by pre-treatment with TMTU (Fig. 5B). These data suggested that PM2.5-induced intracellular autophagy in Beas-2B cells was dependent on ROS.
Suppression of autophagy enhances cytoxicity of PM2.5-induced apoptosis in Beas-2B cells
In order to investigate whether PM2.5-induced autophagy was involved in apoptosis, PI staining for apoptosis analysis was performed. Beas-2B cells were analyzed following treatment with or without autophagy inhibitor 3-MA for 1 h, followed by 100 µg/ml PM2.5 exposure for 24 h. As shown in Fig. 5C, the apoptotic rate of Beas-2B cells in the PM2.5-treated group was elevated compared with that of the non-treated group. However, pre-treatment with 3-MA induced an increase in the level of apoptosis (Fig. 5C). These results suggested that PM2.5-induced autophagy had a pro-survival function in Beas-2B cells.
Discussion
Asthma is a chronic inflammatory disease that influences >300 million individuals worldwide, in which the majority of cases are characterized by an allergic response, (24). A previous study provided evidence demonstrating an association between air pollution and the development of asthma (25).
Multiple studies, utilizing genetic and histological approaches, have also indicated that autophagy is associated with asthma pathogenesis (9,26–28). The present study aimed to investigate the molecular mechanisms underlying PM2.5-induced autophagy at a cellular level.
In the current study, multiple experimental techniques were used to evaluate PM2.5-induced autophagy in Beas-2B cells, including fluorescent microscopy, western blot analysis and TEM, which revealed the formation of characteristic autophagosomes following 24 h of treatment with PM2.5. It is well known that the Akt-mTOR signaling pathway is a significant negative regulator of autophagy, and in the present study, western blot analyses revealed that PM2.5-induced autophagy in Beas-2B cells was mediated by dephosphorylation of Akt/mTOR signaling. Conversely, PM2.5 induced activation of nutrient sensor AMPK; however, the expression of LC3II was not reduced upon inhibition of AMPK phosphorylation by DM. These results suggested that the AMPK pathway was not involved in PM2.5-mediated induction of autophagy in Beas-2B cells.
In the present study, PM2.5 was demonstrated to induce the expression and activation of autophagic protein LC3II, and to stimulate autophagosome formation, two characteristics of autophagic pathway initiation. Prolonged exposure to PM2.5 resulted in time- and dose-dependent changes in the extent of LC3I to LC3II conversion. In addition, MAPKs are upstream regulators of mTOR that mediate responses to various extracellular stimuli (29). All four categories of MAPK (ERK, p38, JNK/stress-activated protein kinases and big MAPK) have previously been reported to regulate autophagy (30–32). Enhanced phosphorylation of the ERK and JNK signaling pathways was also observed following Beas-2B cell exposure to PM2.5. Notably, inhibition of ERK activity or expression did not abrogate PM2.5-induced autophagy in Beas-2B cells. Furthermore, although the ERK inhibitor effectively blocked the increase in p-ERK associated with PM2.5 exposure, the expression of LC3II remained unchanged by this inhibition, indicating that the MEK-ERK1/2 pathway was not involved in autophagy induction by PM2.5.
Whether the JNK and p38 MAPK signaling pathways were involved in PM2.5-induced autophagy in Beas-2B cells was also examined. The phosphorylation of JNK and p38 MAPK was evaluated, but neither was found to be significantly involved in PM2.5-induced autophagy in Beas-2B cells, indicating a lack of MAPK signaling significance in this process.
In the present study, it was revealed that Akt/mTOR had a negatively regulatory role in PM2.5-induced autophagy in Beas-2B cells. However, this autophagy was independent of JNK, concurrent with previous findings (33–35), indicating that a different MAPK response to PM2.5 is dependent on cell-type specification.
Urich et al (36) examined PM2.5-induced apoptosis in the alveolar epithelium, and identified that it required transcriptional activation of p53 and its phosphorylation; however, the results of the present study did not indicate that p53 was activated by PM2.5 in Beas-2B cells. The autophagy induced by PM2.5 exposure was also demonstrated to be independent of p53. p53 has previously been shown to serve a dual role in the control of autophagy (37,38). In addition, it was demonstrated that ROS scavenger TMTU was able to diminish the expression of LC3II protein induced by PM2.5, indicating that ROS were involved in PM2.5-induced autophagy. Previous studies have demonstrated that apoptosis is implicated in asthma pathogenesis (39,40) and, in the context of PM2.5 exposure, it was demonstrated that pre-treatment of Beas-2B cells with 3-MA enhanced PM2.5-inducible apoptosis. These results indicated that the autophagy induced by PM2.5 had a pro-survival function in Beas-2B cells. Together, the results of the present study indicated that multiple autophagy-associated signaling pathways are activated following PM2.5 exposure, and that alterations in the Akt-mTOR inhibition pathway likely have major roles in the induction of autophagy.
The role of autophagy in the regulation of Beas-2B cell survival and apoptosis is complex. Further studies are required in order to characterize the specific autophagic signaling pathways activated by exposure to PM2.5 (for example, macroautophagy, microautophagy and chaperone-mediated autophagy) in isolated primary mouse bronchial epithelial cells. Although the evaluation of PM2.5-induced autophagy in Beas-2B cells in vitro is not necessarily indicative of the conditions in vivo, particularly in the context of human diseases, it was hypothesized that PM2.5-induced autophagy may be a pro-survival response to PM2.5 exposure. Additional studies are required in order to elucidate the molecular mechanisms underlying PM2.5-induced autophagy and apoptosis in the context of health and disease.
In conclusion, the results of the present study indicated that PM2.5 exposure stimulated autophagy in human bronchial epithelial cells, identified by ultrastructural and biochemical features of autophagy. PM2.5-induced autophagy was also demonstrated to be associated with altered signaling via the Akt/mTOR pathway, but was independent of p53. Furthermore, PM2.5-induced autophagy was regulated in part by ROS-associated mechanisms. However, the specific molecular mechanisms underlying the in vivo significance of these results remain to be elucidated. The results of the present study provide a mechanistic basis for the development of clinical applications targeting these signaling pathways for the prevention and/or treatment of PM2.5-induced lung disease.
Acknowledgments
The present study was supported by the Science and Technology Innovation Fund of Guangdong Medical College (no. STIF201109), the Natural Science Foundation of China project (no. NSFC81172615), Guangdong Natural Science Foundation (no. S20122010008299) and the Science and Technology Planning Project of Guangdong Province (no. 2012B031800223).
Abbreviations
- mTOR
mammalian target of rapamycin
- LC3
microtubule-associated protein 1 light chain 3
- AMPK
adenosine monophosphate-activated protein kinase
- p38
p38 mitogen-activated protein kinase
- JNK
c-Jun N-terminal kinase
- ERK
extracellular signal-regulated kinase
- MAPK
mitogen-activated protein kinases
- 3-MA
3-methyladenine
- PM2.5
particulate matter 2.5
References
- 1.Zanobetti A, Franklin M, Koutrakis P, Schwartz J. Fine particulate air pollution and its components in association with cause-specific emergency admissions. Environ Health. 2009;8:58. doi: 10.1186/1476-069X-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delfino RJ, Sioutas C, Malik S. Potential role of ultrafine particles in associations between airborne particle mass and cardiovascular health. Environ Health Perspect. 2005;113:934–946. doi: 10.1289/ehp.7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dockery DW, Pope CA, III, Xu X, et al. An association between air pollution and mortality in six U.S. cities. N Engl J Med. 1993;329:1753–1759. doi: 10.1056/NEJM199312093292401. [DOI] [PubMed] [Google Scholar]
- 4.Trejo Bittar HE, Yousem SA, Wenzel SE. Pathobiology of severe asthma. Annu Rev Pathol. 2015;10:511–545. doi: 10.1146/annurev-pathol-012414-040343. [DOI] [PubMed] [Google Scholar]
- 5.Mehta M, Chen LC, Gordon T, Rom W, Tang MS. Particulate matter inhibits DNA repair and enhances mutagenesis. Mutat Res. 2008;657:116–121. doi: 10.1016/j.mrgentox.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nordling E, Berglind N, Melén E, et al. Traffic-related air pollution and childhood respiratory symptoms, function and allergies. Epidemiology. 2008;19:401–408. doi: 10.1097/EDE.0b013e31816a1ce3. [DOI] [PubMed] [Google Scholar]
- 7.Anderson HR, Favarato G, Atkinson RW. Long-term exposure to air pollution and the incidence of asthma: meta-analysis of cohort studies. Air Qual Atmos Health. 2013;6:47–56. doi: 10.1007/s11869-011-0144-5. [DOI] [Google Scholar]
- 8.Zeng Y, Yang X, Wang J, Fan J, Kong Q, Yu X. Aristolochic acid I induced autophagy extenuates cell apoptosis via ERK 1/2 pathway in renal tubular epithelial cells. PloS One. 2012;7:e30312. doi: 10.1371/journal.pone.0030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poon AH, Chouiali F, Tse SM, et al. Genetic and histologic evidence for autophagy in asthma pathogenesis. J Allergy Clin Immunol. 2012;129:569–571. doi: 10.1016/j.jaci.2011.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maiuri MC, Galluzzi L, Morselli E, Kepp O, Malik SA, Kroemer G. Autophagy regulation by p53. Curr Opin Cell Biol. 2010;22:181–185. doi: 10.1016/j.ceb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akhtar S, Yousif MH, Chandrasekhar B, Benter IF. Activation of EGFR/ERBB2 via pathways involving ERK1/2, P38 MAPK, AKT and FOXO enhances recovery of diabetic hearts from ischemia-reperfusion injury. PloS One. 2012;7:e39066. doi: 10.1371/journal.pone.0039066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soberanes S, Panduri V, Mutlu GM, Ghio A, Bundinger GR, Kamp DW. p53 mediates particulate matter-induced alveolar epithelial cell mitochondria-regulated apoptosis. Am J Respir Crit Care Med. 2006;174:1229–1238. doi: 10.1164/rccm.200602-203OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin Z, Liu T, et al. AKT/mTOR and c-Jun N-terminal kinase signaling pathways are required for chrysotile asbestos-induced autophagy. Free Radic Biol Med. 2014;72:296–307. doi: 10.1016/j.freeradbiomed.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang L, Liu G, Lin Z, et al. Pro-inflammatory response and oxidative stress induced by specific components in ambient particulate matter in human bronchial epithelial cells. Environ Toxicol. 2014 Dec 23; doi: 10.1002/tox.22102. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 16.Wiltfang J, Smirnov A, Schnierstein B, et al. Improved electrophoretic separation and immunoblotting of beta-amyloid (A beta) peptides 1–40, 1–42, and 1–43. Electrophoresis. 1997;18:527–532. doi: 10.1002/elps.1150180332. [DOI] [PubMed] [Google Scholar]
- 17.Zhou C, Zhong W, Zhou J, et al. Monitoring autophagic flux by an improved tandem fluorescent-tagged LC3 (mTagRFP-mWasabi-LC3) reveals that high-dose rapamycin impairs autophagic flux in cancer cells. Autophagy. 2012;8:1215–1226. doi: 10.4161/auto.20284. [DOI] [PubMed] [Google Scholar]
- 18.Bolster DR, Crozier SJ, Kimball SR, Jefferson LS. AMP-activated protein kinase suppresses protein synthesis in rat skeletal muscle through down-regulated mammalian target of rapamycin (mTOR) signaling. J Biol Chem. 2002;277:23977–23980. doi: 10.1074/jbc.C200171200. [DOI] [PubMed] [Google Scholar]
- 19.Corcelle EA, Puustinen P, Jäättelä M. Apoptosis and autophagy: Targeting autophagy signalling in cancer cells – ‘trick or treats’? FEBS J. 2009;276:6084–6096. doi: 10.1111/j.1742-4658.2009.07332.x. [DOI] [PubMed] [Google Scholar]
- 20.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Codogno P, Meijer AJ. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2005;12(Suppl 2):1509–1518. doi: 10.1038/sj.cdd.4401751. [DOI] [PubMed] [Google Scholar]
- 22.Roy B, Pattanaik AK, Das J, et al. Role of PI3K/Akt/mTOR and MEK/ERK pathway in Concanavalin A induced autophagy in HeLa cells. Chem Biol Interact. 2014;210:96–102. doi: 10.1016/j.cbi.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Hamamura K, Goldring MB, Yokota H. Involvement of p38 MAPK in regulation of MMP13 mRNA in chondrocytes in response to surviving stress to endoplasmic reticulum. Arch Oral Biol. 2009;54:279–286. doi: 10.1016/j.archoralbio.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dougherty R, Fahy JV. Acute exacerbations of asthma: epidemiology, biology and the exacerbation-prone phenotype. Clin Exp Allergy. 2009;39:193–202. doi: 10.1111/j.1365-2222.2008.03157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delfino RJ, Wu J, Tjoa T, Gullesserian SK, Nickerson B, Gillen DL. Asthma morbidity and ambient air pollution: effect modification by residential traffic-related air pollution. Epidemiology. 2014;25:48–57. doi: 10.1097/EDE.0000000000000016. [DOI] [PubMed] [Google Scholar]
- 26.Jyothula SS, Eissa NT. Autophagy and role in asthma. Curr Opin Pulm Med. 2013;19:30–35. doi: 10.1097/MCP.0b013e32835b1150. [DOI] [PubMed] [Google Scholar]
- 27.Martin LJ, Gupta J, Jyothula SS, et al. Functional variant in the autophagy-related 5 gene promotor is associated with childhood asthma. PLoS One. 2012;7:e33454. doi: 10.1371/journal.pone.0033454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poon A, Eidelman D, Laprise C, Hamid Q. ATG5, autophagy and lung function in asthma. Autophagy. 2012;8:694–695. doi: 10.4161/auto.19315. [DOI] [PubMed] [Google Scholar]
- 29.Albert L, Karsy M, Murali R, Jhanwar-Uniyal M. Inhibition of mTOR activates the MAPK pathway in glioblastoma multiforme. Cancer Genomics Proteomics. 2009;6:255–261. [PubMed] [Google Scholar]
- 30.Hazzalin CA, Mahadevan LC. MAPK-regulated transcription: a continuously variable gene switch? Nat Rev Mol Cell Biol. 2002;3:30–40. doi: 10.1038/nrm715. [DOI] [PubMed] [Google Scholar]
- 31.Webber JL, Tooze SA. Coordinated regulation of autophagy by p38alpha MAPK through mAtg9 and p38IP. EMBO J. 2010;29:27–40. doi: 10.1038/emboj.2009.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de la Cruz-Morcillo MA, Valero ML, Callejas-Valera JL, et al. P38MAPK is a major determinant of the balance between apoptosis and autophagy triggered by 5-fluorouracil: implication in resistance. Oncogene. 2012;31:1073–1085. doi: 10.1038/onc.2011.321. [DOI] [PubMed] [Google Scholar]
- 33.Brigelius-Flohé R. Commentary: oxidative stress reconsidered. Genes Nutr. 2009;4:161–163. doi: 10.1007/s12263-009-0131-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng Y, Qiu F, Tashiro S, Onodera S, Ikejima T. ERK and JNK mediate TNFalpha-induced p53 activation in apoptotic and autophagic L929 cell death. Biochem Biophys Res Commun. 2008;376:483–488. doi: 10.1016/j.bbrc.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 35.Kabesch M, Adcock IM. Epigenetics in asthma and COPD. Biochimie. 2012;94:2231–2241. doi: 10.1016/j.biochi.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 36.Urich D, Soberanes S, Burgess Z, et al. Proapoptotic Noxa is required for particulate matter-induced cell death and lung inflammation. FASEB J. 2009;23:2055–2064. doi: 10.1096/fj.08-114546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCarthy N. Autophagy: Directed development. Nat Rev Cancer. 2014;14:74–75. doi: 10.1038/nrc3673. [DOI] [PubMed] [Google Scholar]
- 38.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim MN, Lee KE, Hong JY, et al. Involvement of the MAPK and PI3K pathways in chitinase 3-like 1-regulated hyperoxia-induced airway epithelial cell death. Biochem Biophys Res Commun. 2012;421:790–796. doi: 10.1016/j.bbrc.2012.04.085. [DOI] [PubMed] [Google Scholar]
- 40.Takada E, Furuhata M, Nakae S, Ichijo H, Sudo K, Mizuguchi J. Requirement of apoptosis-inducing kinase 1 for the induction of bronchial asthma following stimulation with ovalbumin. Int Arch Allergy Immunol. 2013;162:104–114. doi: 10.1159/000353240. [DOI] [PubMed] [Google Scholar]