Abstract
Mutational profiling has demonstrated utility predicting the likelihood of disease progression in patients with myelofibrosis (MF). However, there is limited data regarding the prognostic utility of genetic profiling in MF patients undergoing allogeneic hematopoietic stem cell transplantation (allo-HCT). We performed high-throughput sequencing of 585 genes on pre-transplant samples from 101 patients with MF who underwent allo-HCT and evaluated the association of mutations and clinical variables with transplantation outcomes
OS at 5 years post-transplant was 52%, and RFS was 51.1 % for this cohort. Non-relapse mortality (NRM) accounted for most deaths. Patient’s age, donor’s age, donor type, and DIPSS at diagnosis did not predict for outcomes. Mutations known to be associated with increased risk of disease progression, such as ASXL1, SRSF2, IDH1/2, EZH2, and TP53, did not impact OS or RFS. The presence of U2AF1 (p=0.007) or DNMT3A (p=0.034) mutations was associated with worse OS. MIPSS-70 score was available on 80 (79%) patients and there were no differences in outcomes between patients with high risk and those with intermediate and low risk scores.
Collectively, these data identify mutational predictors of outcome in MF patients undergoing allo-HCT. These genetic biomarkers in conjunction with clinical variables may have important utility in guiding transplant treatment decision-making.
Keywords: Myelofibrosis, Stem cell transplantation, Molecular mutations
INTRODUCTION
The introduction of the selective inhibitor of Janus kinase (JAK) 1 and 2, ruxolitinib, has significantly improved the outcomes of patients with myelofibrosis (MF) by reducing spleen size and constitutional symptom burden, in addition to improving overall survival (OS)1, 2.However, current JAK inhibitors have limited anti-clonal activity, and partial or complete remissions are not usually observed. Thus, allogeneic hematopoietic stem cell transplantation (allo-HCT) remains the only potential curative treatment for patients with MF. Historical data has indicated highly-variable outcomes for MF patients undergoing allo-HCT and a variety of clinical factors have been identified as potentially impacting transplant outcomes. Data from Rondelli et al demonstrated that in patients who underwent allo-HCT from related or unrelated donors after conditioning with a reduced intensity regimen (RIC), transplant from an unrelated donor, regardless of HLA match status, was associated with worse survival3. At the same time, Kroger et al, in a different prospective study, reported that older age (>55 years) and transplant from HLA mismatched donor were associated with worse survival4. The differing clinical variables contributing to transplant outcomes identified in prior studies highlights the need to develop robust predictive tools to better prognosticate outcomes in patients undergoing allo-HCT, and determine how appropriate allo-HCT may be for a given patient.
The role of genomic alterations in understanding the pathogenesis of myeloproliferative neoplasms (MPNs) has increased tremendously in the past decade. Although activation of the JAK-STAT signaling pathway remains the hallmark of MPN pathogenesis, it has become clear that the presence of additional genomic events, such as mutations in TET25, EZH26 and TP537, alters the biology of disease in preclinical models. Furthermore, retrospective studies have demonstrated that the presence of select mutations may have important prognostic value for MF patients. For example, the presence of ASXL1, EZH2, SRSF2, and IDH1/2 mutations8, as well as a lack of canonical JAK2, MPL or CALR mutations (so-called triple negative status)9 are associated with increased risk of leukemic transformation and poor survival and the presence of any of these mutations is considered high molecular risk disease (HMR). Moreover, the presence of certain genotypes appears to predict less-durable response to JAK inhibitor therapy in MF patients 10, 11,12. Thus, molecular genetic profiling offers important prognostic information in MF patients. Indeed, integration of clinical variables and prognostic mutational data has recently resulted in the development of a novel prognostic tool, the Mutation–Enhanced International Prognostic Score System for transplantation-age patients with primary myelofibrosis (MIPSS-70)13.
To date, limited data is available regarding the ability of genomic alterations to prognosticate outcome in MF patients undergoing allo-HCT14. In order to determine if genomic alterations impact the outcome of MF patients undergoing allo-HCT, as well as to determine how genomic alterations interact with other prognostically important disease and transplant related factors3,15, 16, 17 we undertook comprehensive mutational profiling using a 585-gene panel in a multi-center cohort of MF patients who underwent allo-HCT.
METHODS
Patients
This was a multicenter retrospective analysis including 101 patients diagnosed with primary MF or MF arising from other MPN (ET and PV) and undergoing HCT between 2007 and 2015. The study cohort included patients treated on the myeloproliferative Disorders Research Consortium (MPD-RC) 101 prospective study (n=52) (NCT00572897), and 49 patients with available pre-transplant molecular samples were collected from participating institutions: 19 patients treated at Memorial Sloan Kettering Cancer Center, New-York, NY, 18 patients treated at Princess Margaret Cancer Centre, Toronto, Canada, and 12 patients at Moffitt Cancer Center, Tampa, Florida. Peripheral blood or bone marrow aspirate derived DNA prior to transplant, as well as at the time of relapse for select subjects, was available for sequencing from all patients. All samples were sequenced at one time point retrospectively and therefore molecular data was not available for the treating physicians when transplant decisions took place. Approval for the study was obtained from the Institutional Review Board of Memorial Sloan Kettering Cancer Center, as well as all participating institutions, in accordance with the Declaration of Helsinki.
Sample processing, sequencing and mutation analysis
We performed high-throughput sequencing with a targeted deep sequencing assay of 585 genes (HemePACT) as previously described18. Briefly, tumor tissue (peripheral blood or bone marrow aspirate) was sequenced at an average coverage of 829× (with a standard deviation of 130). The reads were aligned to the human genome (UCSC build hg19) using the Burrows-Wheeler Aligner with maximal exact matches19. We used the Cancer Genome Project pipeline20 and compared the tumor samples to a standard cancer-free germline following the pipeline recommendations. Snpeff 21 was used to annotate variants with functional consequence on genes. We filtered out common population germline variants using the ExAC dataset 22. We only considered variants that were either present in at least two samples or classified as oncogenic or likely oncogenic following criteria published by Papaemmanuil et al.23. The lower limit of detection of the assay employed in this analysis is 0.5% VAF.
Statistical analysis
The overall survival (OS) and relapse-free survival (RFS), in which relapse and death were defined as event of interest, for the whole cohort were estimated using Kaplan-Meier method. To investigate the association between clinical characteristics (10 demographic and clinical variables pre-specified) and gene mutations (22 individual genes with gene mutation frequency greater than 1%, and 4 groups of gene variables including triple negative MPN, presence of HMR mutations, MPN driver mutation groups, and presence of 3 or more somatic mutations) and OS as well as RFS outcomes, Univariate Cox regression was used to estimate the hazard ratio for each potential predictor. All the potential predictors (total of 36 variables) were put in the multiple Cox regression model and forward selection using 0.1 as the significant level was used to choose the final variables in the multivariable model. Non-relapse mortality (NRM) and relapse cumulative incidence were estimated using Fine and Gray’s method 24 in the presence of competing risk (i.e., relapse as competing risk for NRM and death without relapse as competing risk for relapse). A proportional hazards model for the sub distribution of NRM was used to estimate the hazard ratio for each potential predictor for NRM. SAS 9.3 was used to analyze the data.
RESULTS
Genomic analysis of pre-transplant MF cohort
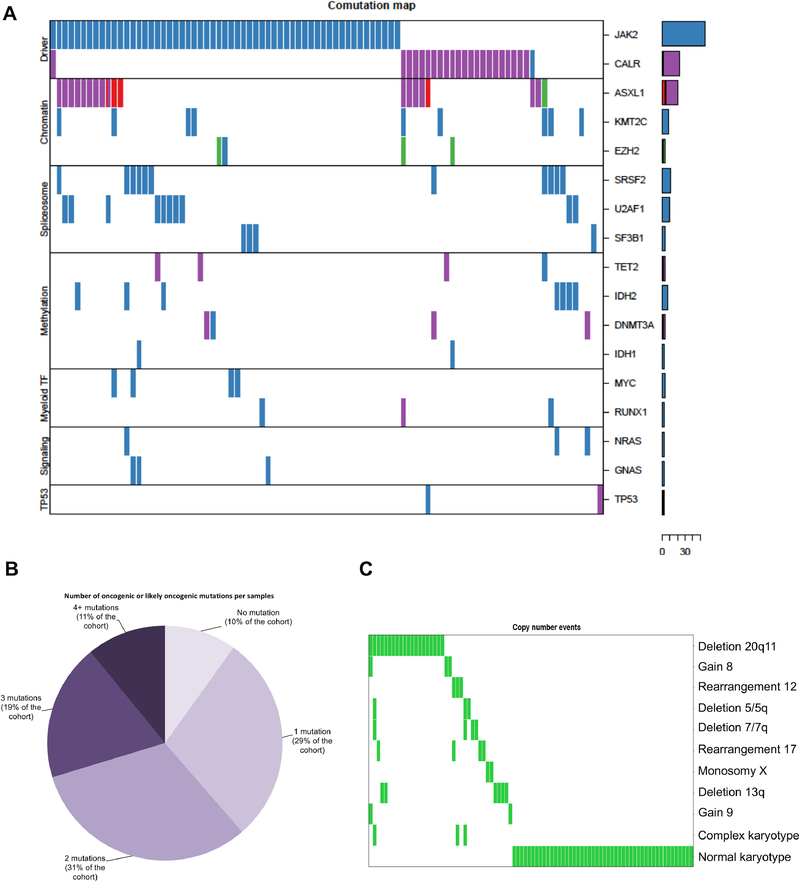
High-throughput sequencing using a panel of 585 cancer-related genes was carried out on peripheral blood or bone marrow samples obtained prior to allo-HCT as described above (Figure 1A; Supplemental Table 1). The majority of patients had activating JAK2 mutations (56.4%). Mutations in chromatin modifiers (ASXL1 18% and EZH2 4%) as well splicing factors (SRSF2 12%, U2AF1 10% and SF3B1 4%) were the most frequently observed class of non-JAK-STAT mutations in this cohort. Less-frequent mutations were identified in genes involved in DNA methylation regulation, such as IDH2 and TET2 (8% each) and DNMT3A (5%). Notably, we identified recurrent mutations in KMT2C in 11% of patients.
Figure 1: Summary of mutations and cytogenetic abnormalities detected in 101 patients with myelofibrosis.
Figure 1A: Spectrum and frequency of mutations. Mutations are grouped according to mechanism. Figure 1B: Number of mutations per sample Figure 1C: Summary of Cytogenetic data, which was available for 86 patients.
As mentioned above, the presence of mutations in ASXL1, SRSF2, IDH1/2, and EZH2 have been previously associated with increased risk of leukemic transformation, and TP53 mutations are enriched in post-MPN AML8. Collectively, these high-molecular risk mutations (HMR) occurred in 36.6% of patients in this cohort. Lack of an identifiable JAK-STAT driver mutation (triple negative status; TN) was identified in 22 patients (21.8%) of the cohort. 51 (50.5%) patients in this cohort had either HMR risk status, TN status, or both. Thus, this cohort of patients was highly enriched for high-risk genomic alterations. Further, prior data has indicated that increasing numbers of mutations per patient are associated with increased risk of leukemic transformation and impaired survival 25. 62 patients (61.4%) in this cohort had more than 1 mutation, inclusive of the driver mutation (Figure 1B).
Cytogenetic data were available for 86 patients (85%) in this cohort. Unfavorable cytogenetics were found in 24.7% of patient and 31.3% of evaluable patients did not have any cytogenetic abnormalities (Figure 1C). The most common cytogenetic abnormality was del20q, identified in 18.6% of patients. Complex karyotype was identified in 3 patients.
Analysis of co-occurrence of mutational events and karyotype did not reveal a statistically significant association between any individual mutations and cytogenetic abnormalities (data not shown).
Impact of clinical, genetic and treatment factors on transplant outcomes
The median age of the cohort was 59 years (range 30–73.4 years). 56 patients (55.5%) had DIPSS risk score of Intermediate-2 or higher. MIPSS-70 score was available on 80 (79%) of patients: 3 patients with low risk, 29 with intermediate risk and 48 with high risk scores. 69 patients had splenomegaly present at the time of transplant (68.3%), and 11 patients (10.9%) had undergone prior splenectomy. The donor utilized in 98 out of the 101 patients was matched (46 related, 52 unrelated). Most patients in this cohort received RIC regimens (Table 1).
Table 1:
Disease and transplant characteristics of evaluated patients
| (N=101) | |
|---|---|
| Age at transplant | |
| Median, Range | 59 (30.0–73.4) |
| < 50 | 13 (12.9%) |
| 50 – 65 | 75 (74.3%) |
| > 65 | 13 (12.9%) |
| Gender: Male | 60 (59.4%) |
| Diagnosis | |
| PMF | 62 (61.4%) |
| Post ET MF | 20 (19.8%) |
| Post PV MF | 18 (17.8%) |
| MPN-U | 1 (1.0%) |
| DIPSS | |
| Low Risk | 9 (8.9%) |
| Int-1 | 36 (35.6%) |
| Int-2 | 41 (40.6%) |
| High Risk | 15 (14.9%) |
| MIPSS-70 | |
| Missing | 21 |
| High Risk | 48 (60.0%) |
| Intermediate Risk | 29 (36.3%) |
| Low Risk | 3 (3.8%) |
| Cytogenetics | |
| Missing | 15 |
| Favorable | 61 (70.9%) |
| Unfavorable | 25 (29.1%) |
| 3 or more somatic mutations | |
| Yes | 30 (29.7%) |
|
HMR: Presence of one of the mutations ASXL1/SRSF2/IDH1/2/EZH2/T P53 |
|
| Yes | 37 (36.6%) |
| MPN Triple Negative (no for JAK2,MPL and CALR) | |
| Yes | 22 (21.8%) |
| Spleen status | |
| Splenectomy | 11 (10.9%) |
| Splenomegaly | 69 (68.3%) |
| No splenomegaly | 21 (20.8%) |
| Time from Diagnosis to Transplant (years) | |
| Median, range | 1.9 (0.1–28.4) |
| Donor | |
| MRD | 46 (45.5%) |
| MUD | 52 (51.5%) |
| Mismatch | 3 (3.0%) |
| Donor age | |
| Missing | 17 |
| Median, Range | 45.5 (18.0–73.0) |
| Conditioning Regimen | |
| MAC | 18 (17.8%) |
| RIC | 83 (82.2%) |
The median follow-up for this cohort was 972 days (2.6 years, 95% CI: 770 to 1124).
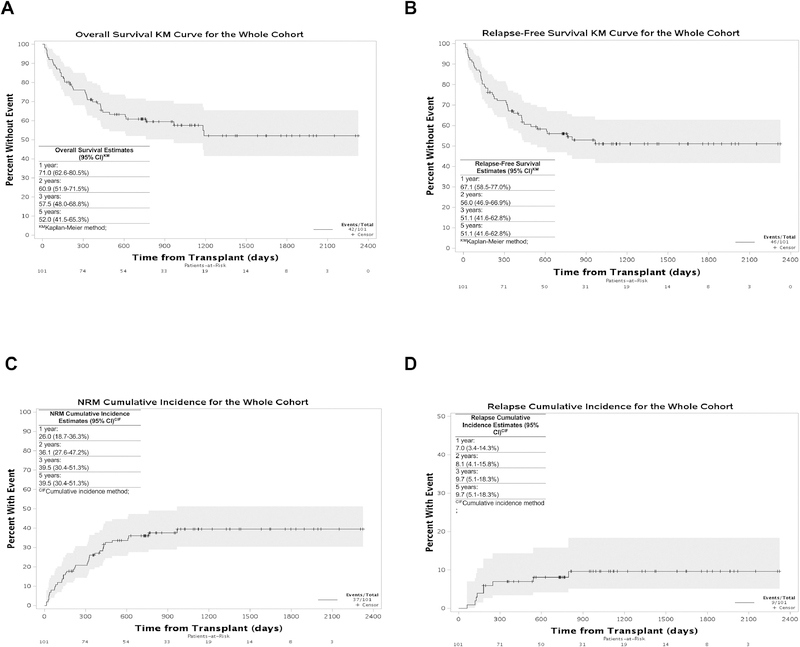
The OS for the cohort was 57.5% (48.0–68.8%) at 3 years and 52.0% (41.5–65.3%) at 5 years post-transplant (Figure 2A); the RFS was 51.1% (41.6–62.8%) at 3 and 5 years post-transplant (Figure 2B). Notably, non-relapse mortality (NRM) accounted for the majority of deaths in the cohort; the cumulative incidence (CI) of NRM was 25.9% (18.6–36.2%) at 1 year post-transplant and 39.0% (30.1–50.7%) at 3 and 5 years post-transplant and the CI of relapse was 7.0% (3.4–14.3%) and 9.7 (5.1–18.3%), respectively (Figure 2C and 2D respectively). The most common cause of death in this cohort (28.5%) was attributed to GVHD.
Figure 2: Kaplan-Meier curves for the whole cohort.
(A) Overall survival, (B) Relapse-Free survival, (C) Non-relapse mortality and (D) Cumulative incidence of relapse
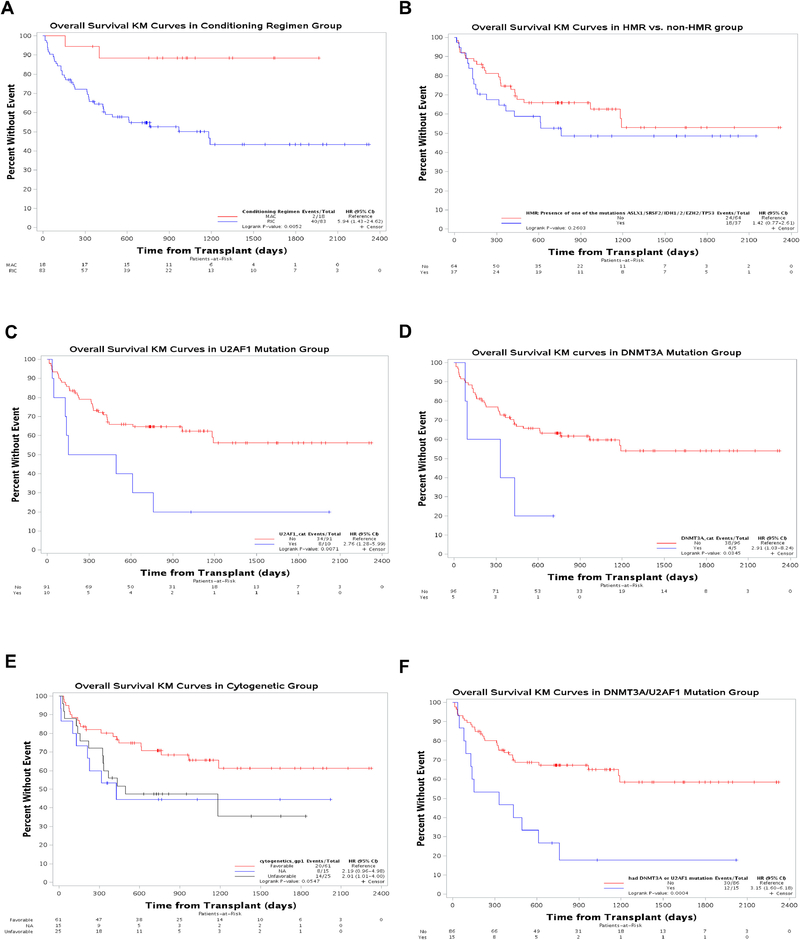
We examined the impact of patient-related characteristics such as age, gender, disease risk (by DIPSS) as well as transplant-related characteristics such as conditioning, type of donor and donor age on overall outcomes for patients in this cohort. Patients who received a RIC regimen had a worse OS compared to those who had a MAC regimen (HR 5.94, 95% CI 1.43–24.62, p=0.005) (Table 2, Figure 3A) in a univariate analysis. Comparison of the MAC and RIC groups demonstrated that the 2 groups were similar with no significant statistical differences between the groups with regard to patient’s age, gender, cytogenetics risk group, DIPSS, time from diagnosis to transplant, number of mutations and presence of high risk mutations, but with a higher proportion of mismatched donors in the MAC group (16.7% vs 0%, p=0.008) (Supplemental Table 2). The majority of the patients who had a MAC regimen received a T cell depleted transplant (ex vivo CD34+ selected allo-graft26) (13/18, 72.2%). In this analysis, patients age had no impact on outcomes, nor did the source of the graft (related vs unrelated), contrasting with what has been previously reported3,4 (Table 2).
Table 2:
Univariate analysis of clinical characteristics and mutations analysis for overall survival
| Effect | Level | HR (95% CI) | P value |
|---|---|---|---|
| Age at transplant | 50 – 65 vs. < 50 | 1.10 (0.43,2.84) | 0.9635 |
| > 65 vs. < 50 | 1.01 (0.31,3.31) | ||
| Gender | F vs. M | 0.95 (0.51,1.76) | 0.8613 |
|
Cytogenetic risk |
unfavorable vs. favorable | 2.01 (1.01,4.00) | 0.0547 |
| NA vs. favorable | 2.19 (0.96,4.98) | ||
| DIPSS | High Risk vs. Low Risk | 1.33 (0.40,4.42) | 0.4426 |
| Int-1 vs. Low Risk | 1.24 (0.42,3.68) | ||
| Int-2 vs. Low Risk | 0.73 (0.24,2.24) | ||
| Spleen status | Splenectomy vs. No splenomegaly |
1.95 (0.67,5.63) | 0.4527 |
| Splenomegaly vs. No splenomegaly |
1.43 (0.65,3.14) | ||
| Conditioning intensity | RIC vs. MAC | 5.94 (1.43,24.62) | 0.0052 |
|
Time from diagnosis to transplant |
>2 years vs. <= 2 years | 1.07 (0.58,1.95) | 0.8363 |
|
Primary vs seconday MF |
Other dx vs. PMF | 0.75 (0.40,1.43) | 0.3816 |
| Donor | Unrelated vs. Related | 1.59 (0.85,2.96) | 0.1436 |
| Donor age | >=50 vs. <50 | 0.91 (0.46,1.80) | 0.2808 |
| NA vs. <50 | 0.46 (0.18,1.22) | ||
| Mutations | At least one positive vs. triple negative |
1.22 (0.56,2.64) | 0.6145 |
| HMR presence | Yes vs. No | 1.36 (0.73,2.56) | 0.3334 |
|
3 or more somatic mutations |
Yes vs. No | 1.22 (0.64,2.31) | 0.5467 |
| JAK2 | Yes vs. No | 1.34 (0.71,2.53) | 0.3572 |
| CALR | Yes vs. No | 0.72 (0.32,1.63) | 0.4328 |
| ASXL1 | Yes vs. No | 1.39 (0.67,2.92) | 0.3755 |
| SRSF2 | Yes vs. No | 0.95 (0.37,2.42) | 0.9174 |
| KMT2C | Yes vs. No | 0.78 (0.28,2.19) | 0.6342 |
| U2AF1 | Yes vs. No | 2.76 (1.28,5.99) | 0.0071 |
| TET2 | Yes vs. No | 1.60 (0.63,4.08) | 0.317 |
| IDH2 | Yes vs. No | 2.23 (0.94,5.29) | 0.0626 |
| DNMT3A_cat | Yes vs. No | 2.91 (1.03,8.24) | 0.0345 |
| MIPSS-70 | High Risk vs. Intermediate/low risk |
1.25 (0.62,2.52) | 0.5372 |
Figure 3: Kaplan-Meier curves for overall survival (OS) by conditioning intensity, mutations and cytogenetic abnormalities.
OS (A) compared between myeloablative conditioning (MAC) and reduced intensity conditioning (RIC), (B) Presence or absence of high molecular risk (HMR) mutations (C) Presence or absence of U2AF1 mutations (D) Presence or absence of DNMT3A mutations (E) Favorable and unfavorable cytogenetic abnormalities and (F) The combined effect of conditioning intensity and presence or absence of U2AF1 and DNMT3A mutations.
We next sought to determine the impact of molecular and cytogenetic parameters on survival by univariate analysis. The total number of mutations per patient was not associated with increased mortality risk (HR for mortality with 3 or more mutations compared to less was 1.22, 95% CI 0.64–2.31, p=0.546), indicating that allo-HCT may be able to overcome the poor prognostic impact of multiple mutations in patients with MPNs. Furthermore, the presence of HMR mutations did not impact survival of patients in this cohort (HR for mortality with HMR mutation compared to none was 1.42, 95% CI 0.77–2.61, p=0.2603, Figure 3B). Analysis of the impact of individual mutations revealed that the presence of U2AF1 or DNMT3A was associated with worse OS (U2AF1: hazard ratio for death 2.76; 95% confidence interval, 1.28 to 5.99, p=0.007, DNM3TA: HR 2.91; 95% CI, 1.03 – 8.24, p=0.034, Figure 3C and 3D respectively). Notably, three out of four cases of mortality due to graft failure occurred in patients with U2AF1 mutations. As well, the presences of U2AF1, DNMT3A, or IDH2 mutations were associated with increased risk of NRM (Supplemental Table 3). Recent data has indicated that the variant allele fraction (VAF) of mutant genes such as TP53 27 in myelodysplastic syndrome may impact the clinical outcomes of patients. Assessment of the impact of VAF of the most common mutations in this cohort (JAK2, CALR, ASXL1), stratified by median VAF, did not demonstrate an impact on survival or relapse.
Analysis of the impact of cytogenetics categorization (as defined in DIPSS-plus scoring system) 28 demonstrated that patients with unfavorable cytogenetic abnormalities had worse OS, with a trend towards significance, with HR 2.01 (95% CI 1.01–4.00, p=0.05) (Figure 3E)
MIPSS-70 score was available in 79% of the patients in this cohort. Patients with intermediate and high-risk score comprised the majority of the cohort (96%). Notably, there were no differences in transplant outcomes when comparing those with high risk and intermediate risk (Table 2).
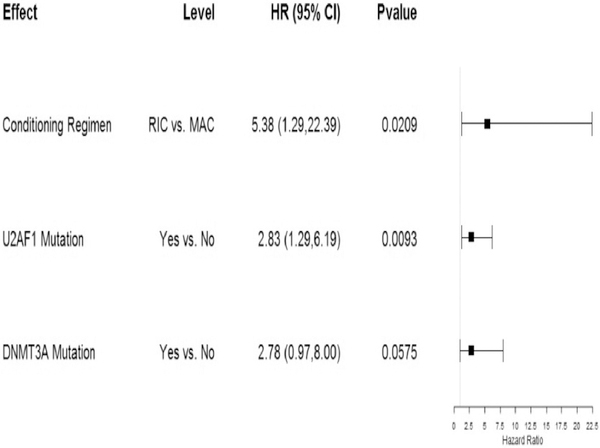
In multivariate analysis, both RIC (HR 5.38, 95% CI 1.29–22.39, p=0.02) and the presence of U2AF1 mutations (HR 2.83, 95% CI 1.29–6.19, p=0.009) remained negatively associated with OS. DNMT3A was also associated with worse survival, although this association did not reach statistical significance (HR 2.78, 95% CI 2.78, 0.97–8.0, p=0.057) (Figure 4).
Figure 4: Multivariate analysis for overall survival shown by forest plot.
With regard to relapse-free survival (RFS), analysis of clinical factors, molecular mutations and cytogenetics demonstrated similar patterns to those seen with OS. Univariate analysis demonstrated that RIC was associated with worse RFS compared to MAC (HR 2.96, 95% CI 1.06–8.26, p=0.03) and presence of U2AF1 mutations (HR 2.37, 95% CI 1.10–5.08, p=0.026) and DNMT3A mutations (HR 4.02 95% CI 1.56–10.35, p=0.0018) were associated with worse RFS (Supplemental Table 4, Supplemental Figure 1). In multivariate analysis, only U2AF1 and DNMT3A retained significant association with reduced RFS, though there was a strong trend for RIC regimen as well (p=0.0598) (Supplemental Figure 2).
Genomic analysis of post-transplant relapse samples
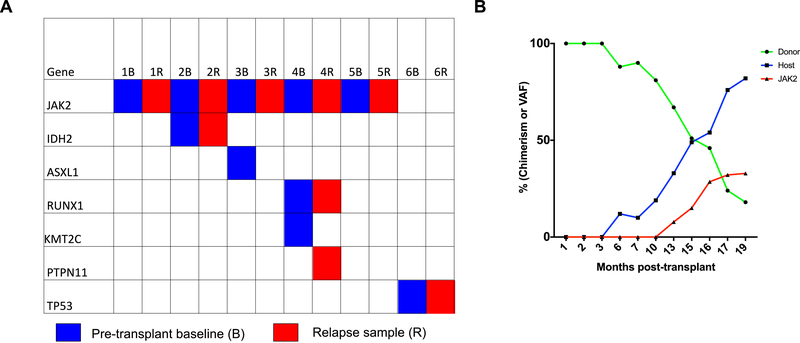
Among patients who relapsed following allo-HCT, 6 patients had samples pre-transplant and at time of relapse on which analysis of paired samples was carried out. This analysis demonstrated that the relapsed sample in many cases contained the same clonal architecture as the pre-transplant samples (Figure 5A). In only one case (number 4) was a new mutation detected at time of relapse. Analysis of chimerism over time following transplant demonstrates that in many cases, loss of donor engraftment is detected prior to detection of the JAK2V617F allele (Figure 5B; Supplemental Table 5).
Figure 5: Mutations analysis of cases of disease relapse post-transplant.
(A) Sequencing analysis of 6 paired pre-transplant and post-transplant relapse cases. (B) Trend over time of chimerism and recurrence of JAK2V617F mutation post-transplant.
DISCUSSION
Molecular genetic and cytogenetic analyses have been merged with analysis of clinical parameters to develop tools for prognostication in MF. Furthermore, molecular profiling has identified ruxolitinib-treated patients with decreased time-to-treatment failure 10, thus allowing for prediction of patients at risk of poor response to ruxolitinib. By contrast, few predictive models exist for MF patients being considered for allo-HCT, thus complicating treatment decisions for physicians and patients, particularly given the risks of allo-HCT. We have therefore sought to extend the impact of mutational profiling as a prognostic tool to patients undergoing allo-HCT.
In multivariate analysis, mutations previously associated with worse outcome in MF patients, such as ASXL1, EZH2, SRSF2, IDH1/2, and TP53 mutations were not found to affect OS or RFS in MF patients undergoing transplant. Further, the number of mutations per patient did not impact OS or RFS. These findings suggest that allo-HCT can overcome the poor prognosis associated with these mutations. It may further imply that patients with HMR or those who are likely to have short-duration of benefit on ruxolitinib should be referred for earlier allo-HCT evaluation. We identified U2AF1 mutations as a risk factor for decreased OS, and U2AF1 and DNMT3A mutations were both associated with impaired RFS. Mutations in U2AF1 have been reported in about 10–15% of patients with MF and have been shown to strongly correlate with the degree of anemia 29,30 and also with worse OS compared to patients with unmutated U2AF1. Interestingly, in our cohort, 4 cases of mortality were secondary to graft failure, 3 of which had mutated U2AF1. We were unable to identify other factors related to disease, donor or transplant that placed these patients at higher risk for graft failure relative to the rest of the cohort. All 4 patients received reduced intensity conditioning and were transplanted from a matched unrelated donor. Two patients had intermediate-1 disease and 2 had intermediate-2 disease by DIPSS.arrow microenvironment. DNMT3A mutations appear to mediate anthracycline-based chemotherapy resistance in AML and DNMT3A R882 in particular predicts for minimal residual disease in AML31. Thus, it is possible that the presence of DNMT3A mutations renders MPN hematopoietic stem cells relatively resistant to effects of conditioning. The biological impact of U2AF1 and DNMT3a mutations may thus alter the likelihood of transplant success. Further genomic and biological studies are required to validate these observations.
The mutational profile of our cohort was similar to prior reports of MF patients across the literature. However, we did identify mutations in KMT2C. KMT2C mutations have been described in a variety of solid tumors 32, 33, 34, 35 and were recently described by Durham et al. in classical and variant hairy cell leukemia 36. Chang et al also recently reported KMT2C mutations in a group of patients with TN MPN37. The biological contribution of KMT2C mutations to MPN pathogenesis remains to be determined.
Most cases of mortality in this cohort were not related to relapse, and indeed the incidence of relapse was surprisingly low despite the fact that 55% of the patients in this cohort had advanced disease (Intermediate-II and high-risk disease), and many patients had HMR mutations. By contrast, data from MDS and AML literature indicates that certain mutations predict for very poor prognosis post allo-HCT, mostly due to disease relapse 38, 39. As well, our findings are in contrast to data recently published by Kroger et al14 that demonstrated ASXL1 mutations are associated with higher relapse risk. In our cohort, among 19 patients who had mutated ASXL1, nine patients died without relapse at a median of 4 months post-transplant. Differences between the cohorts, and the resulting differences in RFS and NRM, may account for differences in the observed impact of mutations on outcomes. Thus, larger cohorts of patients will be required to validate observations from these studies.
Recent analysis by Wolschke 40 et al examined the impact of minimal residual disease by molecular studies, post allo-HCT in patients with MF. This study demonstrated that patients who had persistent evidence of disease at the molecular level at days 100 or 180 post allo-HCT had a significantly higher relapse incidence compared to patients without molecular residual disease (62% vs 10%). In congruence with this observation, we detected the same mutational profile pre-transplant and post-transplant in most cases in patients suffering relapse, without evidence of clonal evolution. In two patients, loss of clones containing ASXL1 and KMT2C mutations were noted, suggesting some degree of selective pressure by allo-HCT on different subclones. These observations suggest that mutational analysis may play an important adjunctive role (together with chimerism analysis) in minimal residual disease monitoring. Further studies of depth of molecular response are required to define clinically meaningful molecular minimal residual disease.
The majority of patients in this retrospective analysis were conditioned with a reduced intensity regimen, and a myeloablative regimen was used mostly in the context of T cell depleted (TCD) transplants (Ex-vivo positive selection of CD34+ stem cells by the CliniMACS CD34 Reagent System 26). Use of MAC was associated with better OS in this analysis, which could not be accounted for by the patient’s baseline characteristics. Historically, patients with MF who underwent allo-HCT with a MAC regimen had a high incidence of NRM and therefore patients with MF are mostly offered a RIC. It is important to note that the MAC regimen used with TCD transplants was chemotherapy based and did not include total body irradiation (TBI). This may explain the better outcomes compared to those historically reported with MAC regimens16, 41. It is also possible that with a TCD transplant, the lack of need for calcineurin inhibitors for GVHD prophylaxis as well as lower GVHD incidence in these patients accounted for the better outcomes compared to what historically has been reported with MAC in patients with MF. These finding are important in the context of the MAC vs RIC study42 in patients with MDS and AML where the MAC regimen was superior in patients with AML. We also recognize the limitations associated with interpreting this when using a small cohort of patients and we believe that further prospective studies to address intensity of conditioning regimen in patients with MF is important.
Our data establishes that genomic alterations have predictive value with regard to allo-HCT, and are likely useful in guiding transplant treatment decision-making in MF patients. It also suggests that mutations that are associated with poor prognosis and progression to AML do not predict for post-transplant outcomes. Moreover, these observations raise new questions about how genomic alterations may impact transplant outcomes in MF and whether interventions to eliminate the mutated clone, particularly in patient with mutated U2AF1, will impact transplant outcomes (notably, clinical trials of inhibitors targeting splicing factors are currently underway; NCT02841540). onsidering the rarity of MF and the relatively small numbers of allo-HCT performed for this disease we strongly believe that further analysis with larger cohorts is needed to confirm the findings of this analysis. Last, prospective studies are needed to assess the optimal conditioning regimen in patients with MF.
Supplementary Material
Highlights.
In this retrospective analysis high-risk molecular mutations were found not to affect outcomes of patients with myelofibrosis (MF) undergoing Allo-HCT.
The presence of U2AF1 mutations was associated with worsened overall survival and relapse-free survival in patients undergoing allo-HCT for MF.
Further studies with larger cohorts are needed to further assess the role of molecular mutations in the field of MF and allo-HCT.
Acknowledgements:
This study was supported by the National Institutes of Health; National Cancer Institute grant 1 P01 CA108671-01A2 (principal investigator: R.H.); Cancer Center Support Grant/Core Grant to Memorial Sloan Kettering Cancer Center (P30 CA008748); NCI 1K08CA188529-01 (R.K.R); grant # UL1 TR001866 from the National Center for Advancing Translational Sciences (NCATS, National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) program
Disclosure of Conflicts of Interest:
C.M. received honoraria from Novartis
A.K. received honoraria from Celgene and has consultancy agreement with Janssen.
J.O.M serves on the clinical trials steering committee of Celgene, Incyte, Roche.
R.M. received honoraria from Novartis and research support from Incyte, CTI, Genentech, Celgene
R.L.L. is on the supervisory board of Qiagen and is a scientific advisor to Loxo, Imago, C4 Therapeutics and Isoplexis. He receives research support from and consulted for Celgene and Roche, research support from Prelude Therapeutics, and has consulted for Novartis and Gilead. He has received honoraria from Lilly and Amgen for invited lectures.
R.H. serves on the advisory Board Novartis and La Jolla Pharmaceuticals
R.K.R has received consulting fees from Incyte corporation, Celgene corporation, Agios Pharmaceuticals, Apexx oncology, and Jazz Pharmaceuticals, and has received research funding from Constellation pharmaceuticals, Incyte corporation, and Stemline Therapeutics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Verstovsek S, Mesa RA, Gotlib J, et al. A double-blind, placebo-controlled trial of ruxolitinib for myelofibrosis. N Engl J Med. 2012;366(9):799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vannucchi AM, Kantarjian HM, Kiladjian JJ, et al. A pooled analysis of overall survival in COMFORT-I and COMFORT-II, 2 randomized phase III trials of ruxolitinib for the treatment of myelofibrosis. Haematologica. 2015;100(9):1139–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rondelli D, Goldberg JD, Isola L, et al. MPD-RC 101 prospective study of reduced-intensity allogeneic hematopoietic stem cell transplantation in patients with myelofibrosis. Blood. 2014;124(7):1183–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kroger N, Holler E, Kobbe G, et al. Allogeneic stem cell transplantation after reduced-intensity conditioning in patients with myelofibrosis: a prospective, multicenter study of the Chronic Leukemia Working Party of the European Group for Blood and Marrow Transplantation. Blood. 2009;114(26):5264–5270. [DOI] [PubMed] [Google Scholar]
- 5.Chen E, Schneider RK, Breyfogle LJ, et al. Distinct effects of concomitant Jak2V617F expression and Tet2 loss in mice promote disease progression in myeloproliferative neoplasms. Blood. 2015;125(2):327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimizu T, Kubovcakova L, Nienhold R, et al. Loss of Ezh2 synergizes with JAK2-V617F in initiating myeloproliferative neoplasms and promoting myelofibrosis. J Exp Med. 2016;213(8):1479–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rampal R, Ahn J, Abdel-Wahab O, et al. Genomic and functional analysis of leukemic transformation of myeloproliferative neoplasms. Proc Natl Acad Sci U S A. 2014;111(50):E5401–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vannucchi AM, Lasho TL, Guglielmelli P, et al. Mutations and prognosis in primary myelofibrosis. Leukemia. 2013;27(9):1861–1869. [DOI] [PubMed] [Google Scholar]
- 9.Tefferi A, Lasho TL, Finke CM, et al. CALR vs JAK2 vs MPL-mutated or triple-negative myelofibrosis: clinical, cytogenetic and molecular comparisons. Leukemia. 2014;28(7):1472–1477. [DOI] [PubMed] [Google Scholar]
- 10.Patel KP, Newberry KJ, Luthra R, et al. Correlation of mutation profile and response in patients with myelofibrosis treated with ruxolitinib. Blood. 2015;126(6):790–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pardanani A, Abdelrahman RA, Finke C, et al. Genetic determinants of response and survival in momelotinib-treated patients with myelofibrosis. Leukemia. 2015;29(3):741–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spiegel JY, McNamara C, Kennedy JA, et al. Impact of genomic alterations on outcomes in myelofibrosis patients undergoing JAK1/2 inhibitor therapy. Blood Adv. 2017;1(20):1729–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guglielmelli P, Lasho TL, Rotunno G, et al. MIPSS70: Mutation-Enhanced International Prognostic Score System for Transplantation-Age Patients With Primary Myelofibrosis. J Clin Oncol. 2018;36(4):310–318. [DOI] [PubMed] [Google Scholar]
- 14.Kroger N, Panagiota V, Badbaran A, et al. Impact of Molecular Genetics on Outcome in Myelofibrosis Patients after Allogeneic Stem Cell Transplantation. Biol Blood Marrow Transplant. 2017;23(7):1095–1101. [DOI] [PubMed] [Google Scholar]
- 15.Scott BL, Gooley TA, Sorror ML, et al. The Dynamic International Prognostic Scoring System for myelofibrosis predicts outcomes after hematopoietic cell transplantation. Blood. 2012;119(11):2657–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abelsson J, Merup M, Birgegard G, et al. The outcome of allo-HSCT for 92 patients with myelofibrosis in the Nordic countries. Bone Marrow Transplant. 2012;47(3):380–386. [DOI] [PubMed] [Google Scholar]
- 17.Gupta V, Malone AK, Hari PN, et al. Reduced-intensity hematopoietic cell transplantation for patients with primary myelofibrosis: a cohort analysis from the center for international blood and marrow transplant research. Biol Blood Marrow Transplant. 2014;20(1):89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kucine N, Viny AD, Rampal R, et al. Genetic analysis of five children with essential thrombocytosis identified mutations in cancer-associated genes with roles in transcriptional regulation. Haematologica. 2016;101(6):e237–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.H L. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 2013. [Google Scholar]
- 20.Igarashi M, Osuga J, Uozaki H, et al. The critical role of neutral cholesterol ester hydrolase 1 in cholesterol removal from human macrophages. Circ Res. 2010;107(11):1387–1395. [DOI] [PubMed] [Google Scholar]
- 21.Cingolani P, Platts A, Wang le L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374(23):2209–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. Journal of the American Statistical Association. 1999;94(446):496–509. [Google Scholar]
- 25.Lundberg P, Karow A, Nienhold R, et al. Clonal evolution and clinical correlates of somatic mutations in myeloproliferative neoplasms. Blood. 2014;123(14):2220–2228. [DOI] [PubMed] [Google Scholar]
- 26.Keever-Taylor CA, Devine SM, Soiffer RJ, et al. Characteristics of CliniMACS(R) System CD34-enriched T cell-depleted grafts in a multicenter trial for acute myeloid leukemia-Blood and Marrow Transplant Clinical Trials Network (BMT CTN) protocol 0303. Biol Blood Marrow Transplant. 2012;18(5):690–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sallman DA, Komrokji R, Vaupel C, et al. Impact of TP53 mutation variant allele frequency on phenotype and outcomes in myelodysplastic syndromes. Leukemia. 2016;30(3):666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gangat N, Caramazza D, Vaidya R, et al. DIPSS plus: a refined Dynamic International Prognostic Scoring System for primary myelofibrosis that incorporates prognostic information from karyotype, platelet count, and transfusion status. J Clin Oncol. 2011;29(4):392–397. [DOI] [PubMed] [Google Scholar]
- 29.Barraco D, Elala YC, Lasho TL, et al. Molecular correlates of anemia in primary myelofibrosis: a significant and independent association with U2AF1 mutations. Blood Cancer J. 2016;6:e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tefferi A, Lasho TL, Finke CM, et al. Targeted deep sequencing in primary myelofibrosis. Blood Adv. 2016;1(2):105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guryanova OA, Shank K, Spitzer B, et al. DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling. Nat Med. 2016;22(12):1488–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anjanappa M, Hao Y, Simpson ER, et al. A system for detecting high impact-low frequency mutations in primary tumors and metastases. Oncogene. 2018;37(2):185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiappetta C, Mancini M, Lessi F, et al. Whole-exome analysis in osteosarcoma to identify a personalized therapy. Oncotarget. 2017;8(46):80416–80428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Sanz P, Trivino JC, Mota A, et al. Chromatin remodelling and DNA repair genes are frequently mutated in endometrioid endometrial carcinoma. Int J Cancer. 2017;140(7):1551–1563. [DOI] [PubMed] [Google Scholar]
- 35.Song Y, Li L, Ou Y, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509(7498):91–95. [DOI] [PubMed] [Google Scholar]
- 36.Durham BH, Getta B, Dietrich S, et al. Genomic analysis of hairy cell leukemia identifies novel recurrent genetic alterations. Blood. 2017;130(14):1644–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang YC, Lin HC, Chiang YH, et al. Targeted next-generation sequencing identified novel mutations in triple-negative myeloproliferative neoplasms. Med Oncol. 2017;34(5):83. [DOI] [PubMed] [Google Scholar]
- 38.Lindsley RC, Saber W, Mar BG, et al. Prognostic Mutations in Myelodysplastic Syndrome after Stem-Cell Transplantation. N Engl J Med. 2017;376(6):536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bejar R, Stevenson KE, Caughey B, et al. Somatic mutations predict poor outcome in patients with myelodysplastic syndrome after hematopoietic stem-cell transplantation. J Clin Oncol. 2014;32(25):2691–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolschke C, Badbaran A, Zabelina T, et al. Impact of molecular residual disease post allografting in myelofibrosis patients. Bone Marrow Transplant. 2017;52(11):1526–1529. [DOI] [PubMed] [Google Scholar]
- 41.Nivison-Smith I, Dodds AJ, Butler J, et al. Allogeneic hematopoietic cell transplantation for chronic myelofibrosis in Australia and New Zealand: older recipients receiving myeloablative conditioning at increased mortality risk. Biol Blood Marrow Transplant. 2012;18(2):302–308. [DOI] [PubMed] [Google Scholar]
- 42.Scott BL, Pasquini MC, Logan BR, et al. Myeloablative Versus Reduced-Intensity Hematopoietic Cell Transplantation for Acute Myeloid Leukemia and Myelodysplastic Syndromes. J Clin Oncol. 2017;35(11):1154–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.