Abstract
Myocardial diseases usually appear ischemic. Reperfusion therapy is one of the effective methods that can improve clinical therapeutic efficacy. However, reperfusion results in myocardial injury named I/R injury. Rosuvastatin (RS) is HMG-CoA reductase inhibitor. We investigated the role of RS in the myocardial I/R injury in vitro and its active mechanism. Oxygen-glucose deprivation/reoxygenation (OGD/R) model was applied to investigate I/R in vitro. OGD/R decreased cell viability and increased levels of miR-17-3p and lactate dehydrogenase (LDH) leakage. Besides, RS decreased cleaved caspase-3 level and LDH leakage, promoted the levels of miR-17-3p and LC3II/LC3I, and increased cell viability when H9C2 cell was treated by OGD/R. miR-17-3p inhibitor reduced the H9C2 cell viability and LC3II/LC3I level, whereas miR-17-3p mimics increased H9C2 cell viability and LC3II/LC3I level. RS promoted cell viability and increased LC3II/LC3I level while it lowered LDH leakage, apoptosis rate, and the levels of cleaved caspase-3 and Cyto c. Our study suggested that RS reduced I/R injury in cardiocyte via cleaved caspase-3/Cyto c apoptosis signaling pathway and autophagy. Moreover, the autophagy happens to cardiocyte by upregulating the expression of miR-17-3p.
Keywords: H9C2, ischemia-reperfusion, rosuvastatin, autophagy, miR-17-3p
Introduction
Myocardial tissue in patients suffers from ischemia-reperfusion (I/R) which results in poor clinical outcomes (Prasad et al., 2009). Therefore, effective primary prevention and advances in cardiac treatment are necessary to the improvement of the care of myocardial ischemia patients (Binder et al., 2015). Rosuvastatin (RS), which belongs to statins, is 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase inhibitor used to reduce cholesterol rates. Recent studies indicated that statins had neuroprotective, anti-ischemic, and antioxidant effects (Ajith et al., 2008; Stepien et al., 2005). Besides, particularly, RS upregulated endothelial nitric oxide to protect cells against damage (Laufs et al., 2002).
Oxygen-glucose deprivation/reoxygenation (OGD/R) in cardiocyte is used as a model to study I/R injury in vitro (Li et al., 2018; Zhang et al., 2018). The aim of this study was to investigate whether RS had the effect on H9C2 cell during OGD/R and the mechanism of action.
MicroRNA (miRNA), endogenous short noncoding RNA with a length of 18–25 bp, can inhibit protein expression by combining the target mRNA. miRNAs-mediated gene regulation is related to various mechanism, including proliferation, apoptosis, and differentiation (Shin and Chu, 2014). mir17 microRNA cluster contains miR-17, miR-18, miR-19a, miR-19b, miR-20, miR-25, miR-92, miR-93, miR-106a, and miR-106b (Tanzer and Stadler, 2004). Several studies suggested that miR-17 was associated with cell survival and autophagy process (Comincini et al., 2013; Liao et al., 2017; Yang et al., 2013). Besides, miR-17-3p, processing from the 3′arm of precursor miR-17 can inhibit mitochondrial antioxidant enzymes in cancer cell and cardiac fibroblast senescence in mouse (Du et al., 2015; Xu et al., 2010).
Autophagy is an intracellular self-digesting way to remove long-lived proteins, damaged organelles, and malformed proteins during biosynthesis by lysosomes (Baehrecke, 2005). Chaperone-mediated autophagy, macroautophagy, and microautophagy are three basic types of autophagy. Macroautophagy is responsible for the degradation of cytoplasmic proteins and intracellular organelles, and microautophagy is regarded as a protective mechanism during I/R injury (Ma et al., 2015). Apart from the three basic types of autophagy, studies reported two new types of autophagy named “Rnautophagy” and “DNautophagy,” which proposed that RNA or DNA could be degraded in lysosomes (Fujiwara et al., 2013a, 2013b).
Autophagy is related to normal myocardium and heart disease (Czaja et al., 2013; Li et al., 2015b). Studies on autophagy in myocardium remained controversial. Among them, some studies showed that cardiac autophagy protects against cardiomyocyte apoptosis (He et al., 2013; Ikeda et al., 2015), while others demonstrated that autophagy in cardiomyocyte led to cell injury (Ong and Gustafsson, 2012; Zhang et al., 2012). Whether autophagy protects from or promotes cardiomyocyte and the active mechanism of autophagy need to be studied in the future. Besides, in our study, we also explored the connection between miR-17-3p and autophagy in cardiomyocyte.
Materials and Methods
OGD/R model and cell culture
H9C2 were obtained from ATCC (Manassas, VA). Cells were cultured using free fetal bovine serum (FBS) and no-glucose Dulbecco's modified Eagle's medium (DMEM) (Gibco) with 5% CO2 and 1% O2 at 37°C for 6 hours after the cells having been washed with phosphate-buffered saline (PBS; Gibco) three times. After oxygen-glucose deprivation, the medium was changed with normal medium containing higher-glucose DMEM (Gibco), 10% FBS (Gibco) and 1% 10,000 U/mL penicillin-10,000 μg/mL streptomycin (Gibco) under 5% CO2 and 95% air at 37°C in an incubator (Thermo) for 1 hour. The cells were normally cultured using normal medium with 5% CO2 and 95% air at 37°C in an incubator (Thermo).
Transfection
miR-17-3p mimics and miR-17-3p inhibitor were purchased from Biomics (Nantong, China). miR-17-3p mimics, miR-17-3p inhibitor, and lipofectamine were diluted by free-FBS higher DMEM. Mimics or inhibitor was mixed with lipofectamine solution. After the cells were washed using PBS, the solution was added. The cells were then cultured normally for 24 hours.
Cell viability assay
H9C2 viability was measured using MTT assay. The cells were seeded into 96-well plate (Corning) at 4 × 103 cells/hole and incubated for 24 hours. Then, cells were treated with drugs, or cells were exposed to OGD/R. MTT (Invitrogen) solution (0.5 mg/mL) was dissolved in PBS (Gibco). Liquid in hole was removed. Two hundred microliters MTT solution was added to hole, and the sample were incubated in an incubator (Thermo) at 37°C for 2–3 hours. Finally, 100 μL DMSO (Invitrogen) was added to lyse the cells after MTT solution had been removed. The samples were put in a dark room at 37°C for 15 minutes. The absorbance was measured at 490 nm using a microplate reader (Thermo).
Apoptosis
Cell apoptosis was assessed using dead cell apoptosis kit with Annexin V-FITC/PI (Invitrogen), and the operation was conducted following manufacturer's instructions. The cells were planted in a 75 mm dish at 1 × 106 cells/dish for 24 hours. The cells were then treated with drugs or OGD/R. The cells were collected and resuspended in 100 μL cell solution at a concentration of 1 × 106 cells/mL. Next, 5 μL FITC Annexin V and 5 μL PI working solution were added to cell solution to incubate for 15 minutes in dark room at room temperature. FITC and PI were measured by a flow cytometer (Thermo Scientific) at an excitation wavelength of 488 nm and emission wavelength of 530 nm.
Colorimetric assays
Lactate dehydrogenase (LDH) has been reported to assess the apoptotic cell or cell injury (Cao et al., 2016; Lobner, 2000). In brief, the cells were seeded at 96-well plates (Corning) at 4 × 103 cells/hole and incubated for 24 hours. Then, the cells were treated with drugs or OGD/R. Supernatant in hole was collected in EP tube (Corning). The amount of LDH in supernatant was detected using LDH Activity Assay Kit (Thermo Scientific). The determination operation was performed following the product instruction. The absorbance was read by a microplate reader (Thermo Scientific) at the wavelength of 490 nm.
Western bolt
After the cells being treated with drugs or OGD/R, protein was extracted using a cell lysis buffer (Invitrogen) at 4°C 12,000 rad/min for 15 minutes. The concentration of protein was determined by BCA Protein Assay Kit (Thermo Scientific). Equal contents of protein were separated by SDS-PAGE, and protein was transferred to PVDF membranes (Invitrogen), which were blocked in 5% bovine serum albumin (BSA; Thermo Scientific) in TBST buffer (containing Tween-20) for 2 hours at room temperature. Primary antibody was dissolved in TBST, and the ratio of primary antibody to TBST was determined according to the product instructions. Primary antibody solution was used to incubate the membranes overnight at 4°C.
Primary antibody contained cleaved caspase-3 antibody (#9661; Cell Signaling Technology), LC3B antibody (ab48394; Abcam), Cyto c antibody (ab133504; Abcam), and GAPDH antibody (ab9485; Abcam). Secondary antibody conjugated HRP (ab6721; Abcam) was used to incubate the membranes for 2 hours at room temperature. ECL reagent (Thermo Scientific) was added to membranes in a dark room at room temperature. The fluorescence was analyzed using ImageJ software.
Quantitative reverse transcription-polymerase chain reaction
Total RNA was extracted from the cells treated with drugs or OGD/R using TRIzol kit (Invitrogen). The cDNA was synthesized from total RNA (0.5 μg) at 48°C for 30 minutes and at 95°C for 10 minutes. The cDNA (0.7 μg) was subjected to polymerase chain reaction (PCR) by 45 cycles at 94°C for 45 seconds, 56°C for 30 seconds, and 72°C for 30 seconds. The primers and RNA were obtained from TianGen (Beijing, China).
These primers used in quantitative reverse transcription-PCR were as follows: miR-17-3p primer (forward: 5′-TGCGTTGACGTCACTCCCG-3′, reverse: 5′-GTGCAGGGTCCGAGGT-3′) and U6 primer (forward: 5′-CTCGCTTCGGCAGCACA-3′, reverse: 5′-ACGCTTCACGAATTTGCGT-3′). The relative expression level was analyzed using 2−ΔΔCt method.
Statistical analysis
Data were shown as the mean ± SD, and all experiments were repeated at least three times. Significant differences between the two groups were analyzed by ANOVA with Tukey's test. Statistical significance can be accepted when p < 0.05. The images were analyzed by ImageJ system.
Results
OGD/R decreases H9C2 cell viability and increases miR-17-3p level in H9C2 cell
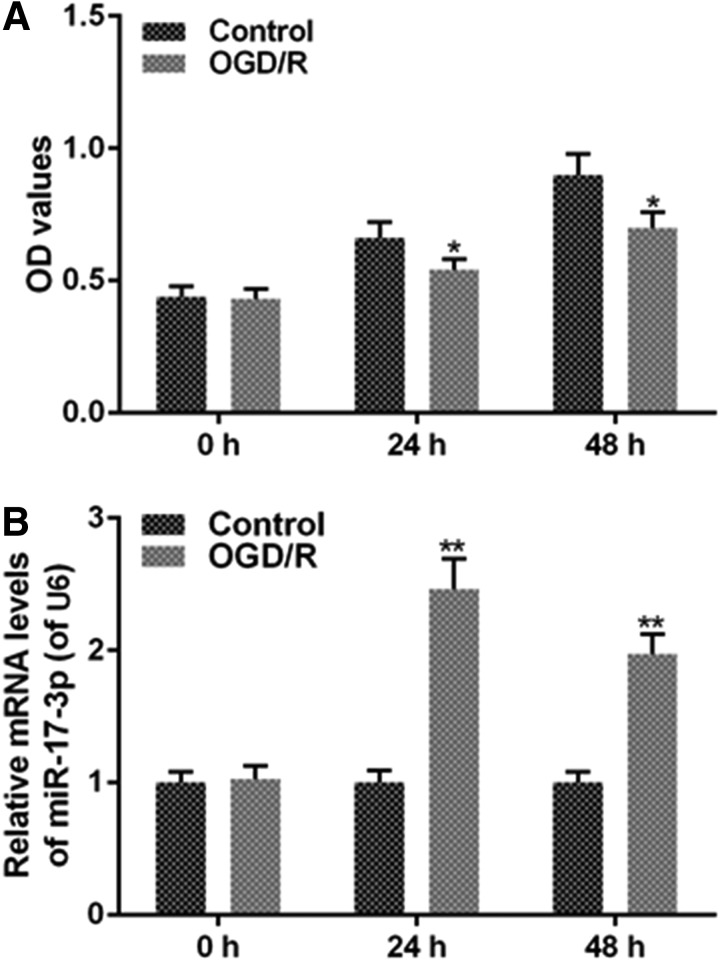
H9C2 cells were treated with OGD/R and then normally cultured. OGD/R decreased OD values (Fig. 1A) and increased miR-17-3p level (Fig. 1B), compared with the control group. Hence, H9C2 cell had a lower viability and a higher expression of miR-17-3p when the cells were in OGD/R.
FIG. 1.
OGD/R decreases H9C2 cell viability and increases the expression of miR-17-3p. OGD/R was performed by means of free FBS and no-glucose Dulbecco's modified Eagle's medium (DMEM) in 5% CO2, 1% O2, and 37°C for 6 hours, then higher-glucose DMEM, 10% FBS and 1% penicillin-streptomycin in 5% CO2, 95% air, and 37°C for 1 hour. H9C2 cells were seeded in 96-well plate at 4 × 103 cells/hole for 24 hours. Then, H9C2 cells were treated with OGD/R for 24 hours or 48 hours. (A) Cell viability was detected by MTT assay. (B) The expression level of miR-17-3p was analyzed via qRT-PCR assays. Values were presented by mean ± SD, and the relationship between the two groups was analyzed by ANOVA with Tukey's test (* vs. Control group, *p < 0.05, **p < 0.01). FBS, fetal bovine serum; OGD/R, oxygen-glucose deprivation/reoxygenation; qRT-PCR, quantitative reverse transcription-polymerase chain reaction.
Effects of RS on OGD/R injury and the levels of miR-17-3p and LC3II/LC3I in H9C2 cell
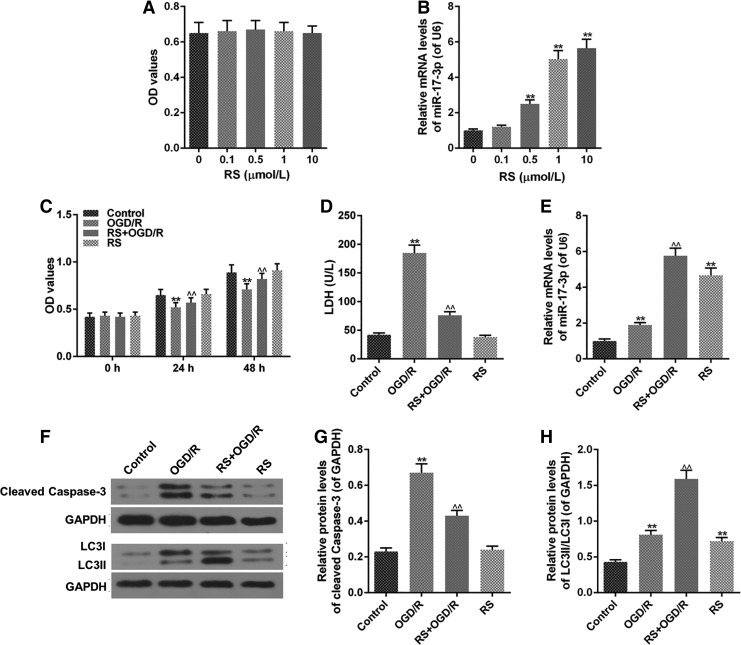
OD values did not have significant variation when the cells were treated with RS, showing that 0–10 μM RS did not exert effects on H9C2 cell viability (Fig. 2A). Besides, H9C2 cells treated with RS had a higher miR-17-3p level, and the miR-17-3p level increased as the concentration of RS increased (Fig. 2B). RS + OGD/R group had higher OD values, compared to OGD/R group, indicating that RS decreased the cell death caused by OGD/R (Fig. 2C). OGD/R resulted in a higher LDH leakage, which was decreased by RS, however, RS had no significant effects on LDH levels in normal cells (Fig. 2D).
FIG. 2.
RS reduces OGD/R injury in H9C2 cell. H9C2 cells were seeded in 96-well plate at 4 × 103 cells/hole. After 24 hours, the cells were treated with different concentrations of RS for 24 hours. RS + OGD/R group was performed as follows: H9C2 cells were incubated by 1 μM RS for 3 hours. Then, the cells were performed by OGD/R and normal circumstance for 21 hours after the cells were washed by PBS. (A, C) MTT assay detected cell viability. (B, E) miR-17-3p level was evaluated by qRT-PCR assays. (D) LDH leakage was assessed by colorimetric assays. (F-H) The levels of LC3II/LC3I and cleaved caspase-3 were measured by western blot. Values were presented by mean ± SD, and the relationship between two groups was analyzed by ANOVA with Tukey's test (* vs. Control group, ^ vs. OGD/R group; **^^p < 0.01). RS, rosuvastatin; LDH, lactate dehydrogenase.
In addition, RS increased the expression of miR-17-3p in H9C2 cell treated with OGD/R (Fig. 2E). OGD/R group had the highest expression of cleaved caspase-3, compared to other groups, whereas RS had a lower level of cleaved caspase-3; however, RS decreased cleaved caspase-3 level in H9C2 cell treated with OGD/R and had no significant effects on cleaved caspase-3 expression in normal cells (Fig. 2F, G). Besides, RS increased protein levels of LC3II/LC3I in OGD/R group, and meanwhile OGD/R and RS also increased ratio of LC3II/LC3I, compared with control group (Fig. 2F, H).
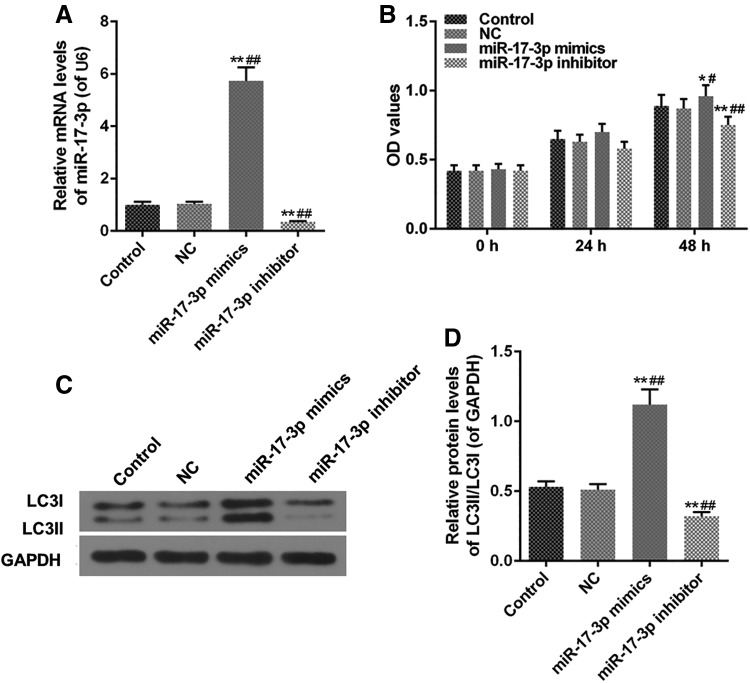
Effects of miR-17-3p mimics and miR-17-3p inhibitor on cell viability and LC3II/LC3I levels in H9C2 cell
miR-17-3p mimics upregulated the level of miR-17-3p, however, miR-17-3p inhibitor weakened the expression of miR-17-3p (Fig. 3A). Besides, miR-17-3p inhibitor decreased OD values and miR-17-3p mimics (Fig. 3B). The levels of LC3II/LC3I in miR-17-3p mimics group had been dynamically increased by miR-17-3p mimics, and LC3II/LC3I levels were decreased in miR-17-3p inhibitor group by miR-17-3p inhibitor, compared with control group (Fig. 3C, D).
FIG. 3.
Effects of miR-17-3p mimics and miR-17-3p inhibitor on H9C2 cell. H9C2 cells were treated with miR-17-3p mimics and miR-17-3p inhibitor for 24 hours. (A) The level of miR-17-3p was measured by qRT-PCR assays. (B) Cell viability was evaluated using MTT assays. (C, D) The protein level of LC3II/LC3I was analyzed via western blot. Values were presented by mean ± SD, and the relationship between the two groups was analyzed by ANOVA with Tukey's test (* vs. Control group, # vs. NC group; *^p < 0.05, **##p < 0.01).
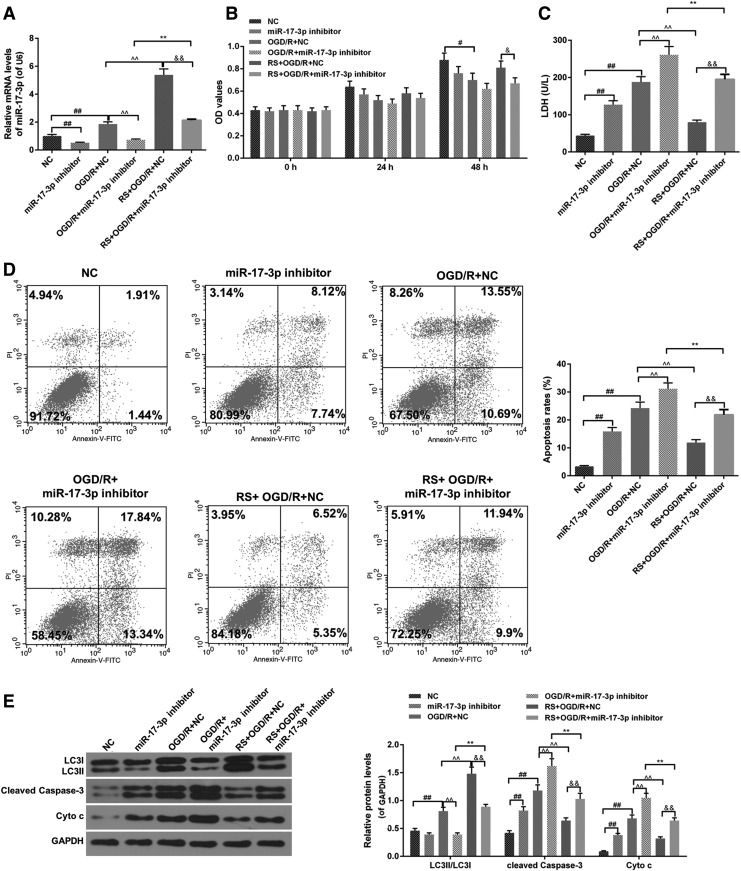
Effects of miR-17-3p inhibitor combined RS on OGD/R injury and the levels of miR-17-3p, LC3II/LC3I, cleaved caspase-3, and Cyto c in H9C2 cell
miR-17-3p inhibitor decreased miR-17-3p level of H9C2 cell in OGD/R, and RS enhanced miR-17-3p level of H9C2 cell in the presence of OGD/R in combination with miR-17-3p inhibitor (Fig. 4A). Moreover, miR-17-3p inhibitor attenuated cell viability in OGD/R group, whereas RS augmented OD values in OGD/R + miR-17-3p inhibitor group (Fig. 4B). miR-17-3p inhibitor promoted LDH leakage in H9C2 cell treated with OGD/R, however, RS reduced the amount of LDH leakage in H9C2 cell treated with OGD/R in combination with miR-17-3p inhibitor (Fig. 4C). As for apoptosis, miR-17-3p inhibitor and OGD/R increased H9C2 cell apoptosis, and RS reduced apoptosis rate in H9C2 cell treated with OGD/R or OGD/R in combination with miR-17-3p inhibitor (Fig. 4D).
FIG. 4.
Effects of miR-17-3p inhibitor combined RS on OGD/R injury. NC group: NC transfected H9C2 cell for 24 hours; miR-17-3p inhibitor group: miR-17-3p inhibitor transfected H9C2 cells for 24 hours; OGD/R + NC group: NC transfected H9C2 cell for 24 hours, meanwhile, H9C2 cells were put in OGD/R model and normal culture conditions; OGD/R + miR-17-3p inhibitor group: miR-17-3p inhibitor transfected H9C2 cells for 24 hours, meanwhile, H9C2 cells were placed in OGD/R model and normal culture conditions; RS + OGD/R + NC group: RS and NC treated H9C2 cell for 3 hours. Then, H9C2 cells-treated NC were placed in OGD/R model and normal culture conditions for 21 hours; RS + OGD/R + miR-17-3p inhibitor group: RS and miR-17-3p inhibitor treated H9C2 cell for 3 hours. Then, H9C2 cells-transfected miR-17-3p inhibitor were placed in OGD/R model and normal culture conditions for 21 hours; (A) The expression of miR-17-3p was analyzed using qRT-PCR assays. (B) Cell viability was measured with MTT assays. (C) LDH leakage was detected by colorimetric assays. (D) Cell apoptosis was evaluated using flow cytometer with dead cell apoptosis kit. (E) The levels of LC3II/LC3I, cleaved caspase-3, and Cyto c were detected by western blot. Values were presented by mean ± SD, and the relationship between the two groups was analyzed by ANOVA with Tukey's test (# vs. NC group, ^ vs. OGD/R + NC group, & vs. RS + OGD/R + NC group; #&p < 0.05, **##&&p < 0.01).
Furthermore, miR-17-3p inhibitor and OGD/R increased the protein levels of cleaved caspase-3 and Cyto c in H9C2 cell, and RS decreased the levels of cleaved caspase-3 protein and Cyto c in H9C2 cell treated with OGD/R or OGD/R in combination with miR-17-3p inhibitor (Fig. 4E). Besides, RS increased the ratio of LC3II to LC3I in miR-17-3p inhibitor group and miR-17-3p inhibitor + OGD/R group (Fig. 4E).
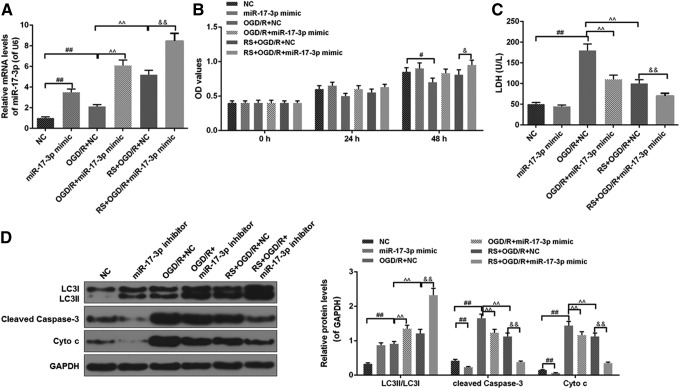
Effects of miR-17-3p mimic combined RS on OGD/R injury and the levels of miR-17-3p, LC3II/LC3I, cleaved caspase-3, and Cyto c in H9C2 cell
The results showed that miR-17-3p mimic significantly increased the level of miR-17-3p (Fig. 5A). The cell viability of the OGD/R+ miR-17-3p mimic group was higher than that of the OGD/R+NC group, and the cell viability ancestors in the RS+ OGD/R+ miR-17-3p mimic group were higher than that in OGD/R+ miR-17-3p mimic group or RS+ OGD/R +NC group (Fig. 5B). The LDH levels of the RS+ OGD/R+ miR-17-3p mimic group were significantly lower than those in the OGD/R+ miR-17-3p mimic group and the RS+ OGD/R+NC group (Fig. 5C). Furthermore, miR-17-3p mimic reduced apoptosis rate in H9C2 cell treated with OGD/R or OGD/R in combination with RS (Fig. 5D). In addition, miR-17-3p mimic further promoted LC3II/LC3I levels in cells treated with OGD/R in the presence of RS (Fig. 5D) and inhibited cleaved caspase-3 and Cyto c expressions (Fig. 5E).
FIG. 5.
Effects of miR-17-3p mimic combined RS on OGD/R injury. NC group: NC transfected H9C2 cells for 24 hours; miR-17-3p mimic group: miR-17-3p mimic transfected H9C2 cells for 24 hours; OGD/R + NC group: NC transfected H9C2 cells for 24 hours, meanwhile, H9C2 cells were put in OGD/R model and normal culture conditions; OGD/R + miR-17-3p mimic group: miR-17-3p mimic transfected H9C2 cells for 24 hours, meanwhile, H9C2 cells were placed in OGD/R model and normal culture conditions; RS + OGD/R + NC group: RS and NC treated H9C2 cells for 3 hours. Then, H9C2 cells-treated NC were placed in OGD/R model and normal culture conditions for 21 hours; RS + OGD/R + miR-17-3p mimic group: RS and miR-17-3p mimic treated H9C2 cells for 3 hours. Then H9C2 cells-transfected miR-17-3p mimic were placed in OGD/R model and normal culture conditions for 21 hours; (A) The expression of miR-17-3p was analyzed using qRT-PCR assays. (B) Cell viability was measured with MTT assays. (C) LDH leakage was detected by colorimetric assays. (D) The levels of LC3II/LC3I, cleaved caspase-3, and Cyto c were detected by western blot. Values were presented by mean ± SD, and the relationship between two groups was analyzed by ANOVA with Tukey's test (# vs. NC group, ^ vs. OGD/R + NC group, & vs. RS + OGD/R + NC group; #&p < 0.05, **##&&p < 0.01).
Discussion
Ischemia leads to myocardial infarction, and reperfusion therapy is an effective method to enhance clinical outcome, whereas reperfusion contributes to myocardial cell death (Bainey and Armstrong, 2014), therefore, I/R injury occurs in cardiac therapy in clinical (Herr et al., 2015). Apoptosis in cardiomyocyte is well known to be an important mechanism of I/R injury (Freude et al., 2000; Gottlieb et al., 1994). In our study, we demonstrated that OGD/R induced cell apoptosis and decreased cell viability (Figs. 1 and 4). Besides, RS increased H9C2 cell viability and decreased H9C2 cell apoptosis (Fig. 2).
LDH is one of major metabolic enzymes in the heart and can be released into serum when cardiac damage (Zhang et al., 2013) takes place. LDH leakage was regarded as an indicator to the evaluation of the degree of cardiac myocytes injury (Amani et al., 2013). Light-chain 3 (LC3) recruitment to phagosomes is required for phagosome maturation (Romao and Munz, 2014). Membrane association of LC3 family proteins is achieved by the formation of a covalent bond to phosphatidylethanolamine (PE). ATG7 acts as the LC3 activating enzyme, while ATG3 acts as the LC3 conjugating enzyme that transfers LC3 to PE to form LC3–PE/LC3-II (Dancourt and Melia, 2014). The lipidated protein then drives autophagy (Nath et al., 2014).
Therefore, lipidation of LC3 leads to the increase of the ratio of LC3-II/LC3-I, which is usually used to be a marker for autophagy in various tissues (Fritzen et al., 2016). OGD/R increased H9C2 cell injury, and RS attenuated cell apoptosis induced by OGD/R and increased cell viability and autophagy in H9C2 cell treated with OGD/R (Fig. 4).
Cleaved caspase-3 and cytochrome c (Cyto c) are related to cell apoptosis signaling pathway, and upregulated cleaved caspase-3 and Cyto c can increase cell apoptosis (Chang et al., 2014; Martinez-Fabregas et al., 2014). Recent studies suggested that autophagy may protect against I/R injury, and it increases cell viability via autophagy pathway (Li et al., 2015a; Wang et al., 2014). RS attenuated OGD/R-induced apoptosis by improving autophagy.
To investigate target gene affected by autophagy, we determined the expression of miR-17-3p and autophagy during OGD/R, miR-17-3p mimics, miR-17-3p inhibitor, and RS. We found that higher expression of miR-17-3p promoted autophagy, and RS improved the expression of miR-17-3p during OGD/R, which was in accordance with the improvement of autophagy. Besides, RS also promoted expression of miR-17-3p during H9C2 cell treated with OGD/R and miR-17-3p inhibitor; meanwhile, RS improved autophagy. The results in our study showed that RS promoted autophagy in H9C2 cell by upregulating the expression of miR-17-3p.
Overall, our findings revealed that RS improved cardiocyte viability or reduced cell death via cleaved caspase-3/Cyto c apoptosis signaling pathway. We also found that upregulated miR-17-3p can promote autophagy in cardiocyte, and autophagy promoted cardiocyte viability. The relationship among RS, autophagy, and miR-17-3p has been investigated in this study, and it is clear that they play important roles in I/R injury. Our data suggest that RS reduced myocardial I/R injury by inhibiting autophagy caused by upregulation of miR-17-3p.
Author Disclosure Statement
The authors declare they have no financial conflicts of interest.
Funding Information
No funding was received for this work.
References
- Ajith T.A., Riji T., and Anu V. (2008). In vitro anti-oxidant and DNA protective effects of the novel 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitor rosuvastatin. Clin. Exp. Pharmacol. Physiol. 35, 625–629 [DOI] [PubMed] [Google Scholar]
- Amani M., Jeddi S., Ahmadiasl N., Usefzade N., and Zaman J. (2013). Effect of HEMADO on level of CK-MB and LDH enzymes after ischemia/reperfusion injury in isolated rat heart. BioImpacts 3, 101–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baehrecke E.H. (2005). Autophagy: Dual roles in life and death? Nat. Rev. Mol. Cell Biol. 6, 505–510 [DOI] [PubMed] [Google Scholar]
- Bainey K.R., and Armstrong P.W. (2014). Clinical perspectives on reperfusion injury in acute myocardial infarction. Am. Heart J. 167, 637–645 [DOI] [PubMed] [Google Scholar]
- Binder A., Ali A., Chawla R., Aziz H.A., Abbate A., and Jovin I.S. (2015). Myocardial protection from ischemia-reperfusion injury post coronary revascularization. Expert Rev. Cardiovasc. Ther. 13, 1045–1057 [DOI] [PubMed] [Google Scholar]
- Cao Y., Zhang L., Sun S., Yi Z., Jiang X., and Jia D. (2016). Neuroprotective effects of syringic acid against OGD/R-induced injury in cultured hippocampal neuronal cells. Int. J. Mol. Med. 38, 567–573 [DOI] [PubMed] [Google Scholar]
- Chang Z., Xing J., and Yu X. (2014). Curcumin induces osteosarcoma MG63 cells apoptosis via ROS/Cyto-C/Caspase-3 pathway. Tumour Biol. 35, 753–758 [DOI] [PubMed] [Google Scholar]
- Comincini S., Allavena G., Palumbo S., Morini M., Durando F., Angeletti F., Pirtoli L., and Miracco C. (2013). microRNA-17 regulates the expression of ATG7 and modulates the autophagy process, improving the sensitivity to temozolomide and low-dose ionizing radiation treatments in human glioblastoma cells. Cancer Biol. Ther. 14, 574–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaja M.J., Ding W.X., Donohue T.M., Jr, Friedman S.L., Kim J.S., Komatsu M., Lemasters J.J., Lemoine A., Lin J.D., Ou J.H. and others. (2013). Functions of autophagy in normal and diseased liver. Autophagy 9, 1131–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancourt J., and Melia T.J. (2014). Lipidation of the autophagy proteins LC3 and GABARAP is a membrane-curvature dependent process. Autophagy 10, 1470–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W.W., Li X., Li T., Li H., Khorshidi A., Liu F., and Yang B.B. (2015). The microRNA miR-17-3p inhibits mouse cardiac fibroblast senescence by targeting Par4. J. Cell Sci. 128, 293–304 [DOI] [PubMed] [Google Scholar]
- Freude B., Masters T.N., Robicsek F., Fokin A., Kostin S., Zimmermann R., Ullmann C., Lorenz-Meyer S., and Schaper J. (2000). Apoptosis is initiated by myocardial ischemia and executed during reperfusion. J. Mol. Cell. Cardiol. 32, 197–208 [DOI] [PubMed] [Google Scholar]
- Fritzen A.M., Frosig C., Jeppesen J., Jensen T.E., Lundsgaard A.M., Serup A.K., Schjerling P., Proud C.G., Richter E.A., and Kiens B. (2016). Role of AMPK in regulation of LC3 lipidation as a marker of autophagy in skeletal muscle. Cell. Signal. 28, 663–674 [DOI] [PubMed] [Google Scholar]
- Fujiwara Y., Furuta A., Kikuchi H., Aizawa S., Hatanaka Y., Konya C., Uchida K., Yoshimura A., Tamai Y., Wada K. and others. (2013a). Discovery of a novel type of autophagy targeting RNA. Autophagy 9, 403–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y., Kikuchi H., Aizawa S., Furuta A., Hatanaka Y., Konya C., Uchida K., Wada K., and Kabuta T. (2013b). Direct uptake and degradation of DNA by lysosomes. Autophagy 9, 1167–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb R.A., Burleson K.O., Kloner R.A., Babior B.M., and Engler R.L. (1994). Reperfusion injury induces apoptosis in rabbit cardiomyocytes. J. Clin. Invest. 94, 1621–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Zhu H., Li H., Zou M.H., and Xie Z. (2013). Dissociation of Bcl-2-Beclin1 complex by activated AMPK enhances cardiac autophagy and protects against cardiomyocyte apoptosis in diabetes. Diabetes 62, 1270–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr D.J., Aune S.E., and Menick D.R. (2015). Induction and assessment of ischemia-reperfusion injury in Langendorff-perfused rat hearts. J. Vis. Exp. 101, e52908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y., Shirakabe A., Maejima Y., Zhai P., Sciarretta S., Toli J., Nomura M., Mihara K., Egashira K., Ohishi M. and others. (2015). Endogenous Drp1 mediates mitochondrial autophagy and protects the heart against energy stress. Circ. Res. 116, 264–278 [DOI] [PubMed] [Google Scholar]
- Laufs U., Gertz K., Dirnagl U., Bohm M., Nickenig G., and Endres M. (2002). Rosuvastatin, a new HMG-CoA reductase inhibitor, upregulates endothelial nitric oxide synthase and protects from ischemic stroke in mice. Brain Res. 942, 23–30 [DOI] [PubMed] [Google Scholar]
- Li H., Zou T., Meng S., Peng Y.Z., and Yang J.F. (2018). p21 protects cardiomyocytes against ischemia-reperfusion injury by inhibiting oxidative stress. Mol. Med. Rep. 17, 4665–4671 [DOI] [PubMed] [Google Scholar]
- Li L., Li L., Zhang Z., and Jiang Z. (2015a). Hypoxia promotes bone marrow-derived mesenchymal stem cell proliferation through apelin/APJ/autophagy pathway. Acta Biochim. Biophys. Sin. 47, 362–367 [DOI] [PubMed] [Google Scholar]
- Li Z., Wang J., and Yang X. (2015b). Functions of autophagy in pathological cardiac hypertrophy. Int. J. Biol. Sci. 11, 672–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X.H., Xiang Y., Yu C.X., Li J.P., Li H., Nie Q., Hu P., Zhou J., and Zhang T.C. (2017). STAT3 is required for MiR-17-5p-mediated sensitization to chemotherapy-induced apoptosis in breast cancer cells. Oncotarget 8, 15763–15774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobner D. (2000). Comparison of the LDH and MTT assays for quantifying cell death: Validity for neuronal apoptosis? J. Neurosci. Methods 96, 147–152 [DOI] [PubMed] [Google Scholar]
- Ma S., Wang Y., Chen Y., and Cao F. (2015). The role of the autophagy in myocardial ischemia/reperfusion injury. Biochim. Biophys. Acta 1852, 271–276 [DOI] [PubMed] [Google Scholar]
- Martinez-Fabregas J., Diaz-Moreno I., Gonzalez-Arzola K., Janocha S., Navarro J.A., Hervas M., Bernhardt R., Velazquez-Campoy A., Diaz-Quintana A., and De la Rosa M.A. (2014). Structural and functional analysis of novel human cytochrome C targets in apoptosis. Mol. Cell. Proteomics 13, 1439–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath S., Dancourt J., Shteyn V., Puente G., Fong W.M., Nag S., Bewersdorf J., Yamamoto A., Antonny B., and Melia T.J. (2014). Lipidation of the LC3/GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3. Nat. Cell Biol. 16, 415–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S.B., and Gustafsson A.B. (2012). New roles for mitochondria in cell death in the reperfused myocardium. Cardiovasc. Res. 94, 190–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A., Stone G.W., Holmes D.R., and Gersh B. (2009). Reperfusion injury, microvascular dysfunction, and cardioprotection: The “dark side” of reperfusion. Circulation 120, 2105–2112 [DOI] [PubMed] [Google Scholar]
- Romao S., and Munz C. (2014). LC3-associated phagocytosis. Autophagy 10, 526–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin V.Y., and Chu K.M. (2014). MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J. Gastroenterol. 20, 10432–10439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepien K., Tomaszewski M., and Czuczwar S.J. (2005). Neuroprotective properties of statins. Pharmacol. Rep. 57, 561–569 [PubMed] [Google Scholar]
- Tanzer A., and Stadler P.F. (2004). Molecular evolution of a microRNA cluster. J. Mol. Biol. 339, 327–335 [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Lin J.H., Muharram A., and Liu W.G. (2014). Beclin-1-mediated autophagy protects spinal cord neurons against mechanical injury-induced apoptosis. Apoptosis 19, 933–945 [DOI] [PubMed] [Google Scholar]
- Xu Y., Fang F., Zhang J., Josson S., St Clair W.H., and St Clair D.K. (2010). miR-17* suppresses tumorigenicity of prostate cancer by inhibiting mitochondrial antioxidant enzymes. PLoS One 5, e14356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Du W.W., Li H., Liu F., Khorshidi A., Rutnam Z.J., and Yang B.B. (2013). Both mature miR-17-5p and passenger strand miR-17-3p target TIMP3 and induce prostate tumor growth and invasion. Nucleic Acids Res. 41, 9688–9704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Shao M., He X., Wang B., Li Y., and Guo X. (2018). Overexpression of microRNA-146 protects against oxygen-glucose deprivation/recovery-induced cardiomyocyte apoptosis by inhibiting the NF-kappaB/TNF-alpha signaling pathway. Mol. Med. Rep. 17, 1913–1918 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yan H., Lv S.G., Wang L., Liang G.P., Wan Q.X., and Peng X. (2013). Effects of glycyl-glutamine dipeptide supplementation on myocardial damage and cardiac function in rats after severe burn injury. Int. J. Clin. Exp. Pathol. 6, 821–830 [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.L., Fan Y., and Liu M.L. (2012). Ginsenoside Rg1 inhibits autophagy in H9c2 cardiomyocytes exposed to hypoxia/reoxygenation. Mol. Cell. Biochem. 365, 243–250 [DOI] [PubMed] [Google Scholar]