Abstract
Increasing evidences have suggested that natural killer (NK) cells in the tumor microenvironment are involved in the regulation of cancer development. However, the potential biological roles and regulatory mechanisms of NK cells in pancreatic cancer (PC) remain unclear. Co-culture system of NK cells with PC cells is used to test the ability of cancer cell proliferation, migration and invasion both in vitro and in vivo. And tail vein intravenous transfer was used to test metastasis in vivo. Meanwhile, extracellular vesicles (EVs) were separated and examined. Furthermore, reporter assay and Biotin-RNA pull down assay were performed to verify the interaction between molecules. NK cells can inhibit the malignant transformation of co-cultured PC cells both in vivo and in vitro, which requires miR-3607-3p. miR-3607-3p is found enriched in the EVs of NK cells and transmitted to PC cells, and low level of miR-3607-3p predicts poor prognosis in PC patients. It can also inhibit proliferation, migration and invasion of PC cells in vitro. Importantly, IL-26 is found to be a direct target of miR-3607-3p in PC cells. miR-3607-3p enriched in EVs derived from NK cells can inhibit the malignant transformation of PC probably through directly targeting of IL-26.
Keywords: natural killer, pancreatic cancer, extracellular vesicles, miR-3607-3p, IL-26
Introduction
Pancreatic cancer (PC) is one of the most lethal human malignancies with poor prognosis (1). Due to the lack of early symptoms and diagnose test, PC is usually found at an incurable advanced stage (2, 3). Presently, the treatment of PC mainly relies on surgical resection and chemotherapy (4). Unfortunately, local deterioration and frequent occurrence of distant metastasis results in poor survival rate of PC patients, with an overall 1-year survival rate at 20% and 5-year survival rate <5% (5). Thus, it is critical to explore the mechanism of malignant transformation of PC, identify novel diagnostic markers and therapeutic targets in PC, which might provide new treatment strategy for PC patients.
Studies have shown that tumor cells can conduct a variety of cross-talks within the tumor microenvironment via chemokines, cytokines, and immune cells to regulate the fate of tumor progression (6, 7). Natural killer (NK) cells, a subset of lymphocytes that play an important role in the rapid innate immune response to viral infection and tumor development, are important components in the tumor microenvironment (8, 9). Increasing studies have shown that NK cells play an important role in inhibiting malignant transformation of tumors, however, the detailed mechanisms are still illusive (10–12). Exosomes are cell-derived vesicles (50–100 nm) that can enclose protein, lipid, mRNA, and microRNA (miRNA) (13–15). Multiple studies have shown that a variety of cells can secrete exosomes that might regulate the development of tumor in the tumor microenvironment including NK cells (16–18).
Here we investigated the function of miR-3607-3p, which was enclosed in NK derived EVs, in pancreatic cancer. Our results suggested that miR-3607-3p enriched in EVs of NK cells, could suppress pancreatic cancer development and malignant transformation by directly targeting IL-26.
Materials and Methods
Patients and Tissue Samples
Forty primary pancreatic cancer (PC) tissues and 40 normal were collected at the First Affiliated Hospital of Wenzhou Medical University from 01/03/2017 to 03/08/2018. All specimens from resection surgery were frozen and stored in liquid nitrogen. This study was performed with the approval of the Ethic and Research Committees of the First Affiliated Hospital of Wenzhou Medical University and these studies was conducted in accordance with the Declaration of Helsinki Principles. Informed written consent was obtained from all subjects.
Human NK Cell Isolation
Human NK cells were isolated as described before using NK cell isolation kit (Miltenyi Biotech, Bergisch Gladbach, Germany) according to the manufacturer's instruction (19). The PBMC were collected according to the protocols approved by the First Affiliated Hospital of Wenzhou Medical University.
Cell Line
Human PC cells (Mia PaCa-2 and PANC-1) were purchased from the American Type Culture Collection (Rockville, USA). The cells were cultured in RPMI 1640 or DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 mg/mL streptomycin at 37°C under 5% CO2 in a humidified chamber.
Transfection
MiR-3607-3p mimics, inhibitor, and negative control were purchased from GenePharma (Shanghai, China). The transfection was performed using Lipofectamine 2000 (Invitrogen, Waltham, MA USA) following the manufacturer's instructions. The knockdown efficiency at the mRNA level was assessed using qRT-PCR assay.
CCK-8 Cell Proliferation Assay
PC cells were seeded in 96-well plates at a density of 5 × 103 cells/well. At the indicated time points, viable cells were examined by cell counting kit-8 (CCK-8) according the manufacturer's instructions.
Migration and Invasion Assay
Cell migration and invasion ability were measured using transwell chambers (24-23ll insert, 8 μm, Millipore, Billerica, MA, USA). The chambers were coated with Matrigel Matrix (Sigma, St. Louis, MO). PC cells were seeded at a density of 5 × 104 cells/well in upper chamber with 200 μL serum free medium. The lower chamber was filled with 600 μL of medium containing 10% FBS. After incubation for 48 h, the number of cells passing through the bottom membrane of the chamber was counted under a microscope.
Colony Formation Assay
PC cells transfected with NC-miRNA or miR-3607-3p were seeded in a 24-well plate at a density of 5 × 104 cells/well and cultured for 24 h. Then, the cells were collected and seeded (1,000–1,500 cells/well) in a dish for 10 days. Surviving colonies were fixed, stained with 5% gentian violet (Sigma) and counted. The experiment was carried out in triplicate wells for three times.
Immunoblotting
Total protein was extracted from the PC cells using RIPA buffer (Invitrogen) according to the manufacturer's protocol. Equivalent amounts of proteins from each sample were subjected to SDS-PAGE electrophoresis and then transferred to a PVDF membrane (Millipore), blocked in 5% fat-free milk for 1 h at room temperature, incubated with specific primary antibodies overnight at 4°C with gentle shaking, and followed by detection with enhanced chemiluminescence system (Pierce, Waltham, MA). Primary antibodies were as follows: CD63, TSG101, IL-26, Actin (Abcam, Cambridge, MA).
RNA Isolation and qPCR
Total RNA from PC tissues and cell lines was harvested using the TRIzol reagent (Invitrogen) following the manufacturer's instructions. RNAs were reverse-transcribed into cDNAs using the Prime Script RT reagent Kit (TaKaRa, Dalian, China) according to the manufacturer's protocol. MiR-3607-3p RT primer: GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCATCA; U6 (internal control) RT primer: GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAATA TGGAA. SYBR Taq (TaKaRa) was used to measure the expression of miR-3607-3p and IL-26. The primers used were described below: miR-3607-3p Forward: ATGACTGTAAACGCTTTCTG, Reverse: GTGCAGGGTCCGAGGT;
U6 Forward: TGCGGGTGCTCGCTTCGGCAGC, Reverse: GTGCAGGGTCCGAGGT; IL-26, Forward: AAGCAACGATTCCAGAAGACC; Reverse: AAGTCCTCCACAAAGCGTATTTT; GAPDH, Forward: ACAACTTTGGTATCGTGGAAGG; Reverse: GCCATCACGCCACAGTTTC. Fold changes of miR-3607-3p and IL-26 were calculated by the equation 2−ΔΔCt.
Luciferase Reporter Assays
The dual luciferase reporter assay was performed using Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). Briefly, luciferase reporter vector containing the wild-type (WT) and mutant (MT) miR-3607-3p binding site in the 3′-UTR of IL-26 were constructed transiently transfected into MIA PaCa-2 or PANC-1 cells, together with miR-3607-3p mimics or NC using Lipofectamine 2000. Primers used for cloning WT IL-26 3′UTR, Forward: CTACTCGAGACCAAAGCCAAGTACATT, Reverse: CTAGCGGCCGCGAAGGAAACCCAATTTA; Mutant IL-26 3′UTR, Forward: GCCAAGTACATTGATTCTCCCCT, Reverse: AGGGGAGAATCAATGTACTTGGC. Luciferase activity was measured 48 h later. The renilla luciferase activity were normalized to the firefly luciferase activity in the corresponding well.
Biotin-miRNA Pull-Down Assay
Biotin-miRNA pulldown assay was carried out as described before (20); Briefly, PC cells were washed with cold PBS, harvested by a scraper and treated cell lysis buffer. Supernatant was collected and incubated with biotinylated miR-3607-3p or biotinylated control random RNA for 1 h at 30°C. After incubation, Steptavidin MagneSphere Paramagnetic beads (Promega) was added for 3 h at 4°C. The biotin-miRNA/mRNA complex was eluted and washed for mRNA detection.
Extracellular Vesicle Isolation and Characterization
Extracellular vesicle (EV) isolation and characterization were prepared as described before (21). Purified NK cells were cultured in a serum free medium for 24 h. Cell culture supernatant was centrifuged for 5 min at 500 g and 10 min at 1,500 g to eliminate the cells and debris. EVs were pelleted by ultracentrifuge step at 80, 000 g for 120 min and washed with PBS. EVs were evaluated and characterized by flow cytometry and Scanning Electron Microscopy (SEM). Briefly, isolated EVs were put on a copper grid coated with 0.1% Formvar in chloroform. The grids were stained with 1% (vol/vol) uranyl acetate in ddH2O, and thereafter the EVs were examined immediately by SEM.
In vivo Tumor Formation Assay
Four to six weeks old Male BALB/c nude mice were purchased from SLAC (Shanghai, China). The animal care and experimental protocols were approved by the institutional guidelines of the First Affiliated Hospital of Wenzhou Medical University and by the Animal Care and Use Committee of The First Affiliated Hospital of Wenzhou Medical University. 1 × 107 PC cells were resuspended in 200 μL PBS medium and were subcutaneously injected into the flank of each nude mouse. The tumors were measured weekly and the tumor volume was calculated following the formula length × width2/2. The mice were killed at 6 weeks after inoculation.
Statistical Analysis
All results are shown as mean ± standard deviation (SD) and were analyzed using GraphPad Prism 5 (GraphPad Software, USA) from at least three independent experiments. The differences between groups were analyzed using Two-tail Student's t-test or ANOVA; Chi-squared test was used to analyze the frequency of lung metastases; Pearson correlation analysis was used to analyze the correlation between expressions of two genes; Kaplan–Meier's analysis was used for prognostic analyses. Data were considered statistically significant when p < 0.05.
Results
NK Cells Co-culture Inhibited Tumor Progression of PC Both in vitro and in vivo
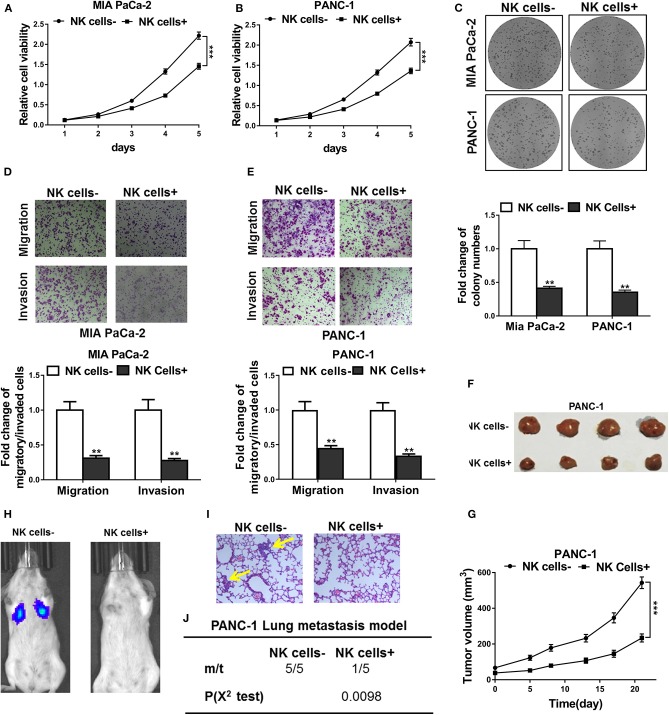
To test the function of NK cells in the tumor microenvironment, we first cultured NK cells together with pancreatic cancer cell line Mia PaCa-2 and PANC-1. CCK-8 assay indicated that the proliferation of Mia PaCa-2 and PANC-1 cells co-cultured with NK cells was significantly suppressed compared with that of cells cultured alone (Figures 1A,B). Colony formation assay, as shown in Figure 1C, showed that cells co-cultured with NK cells had fewer colonies formed. In line with this, after co-culture with NK cells, Mia PaCa-2 and PANC-1 cells had prominent attenuated cell migration and invasive ability (Figures 1D,E). Furthermore, we employed in vivo tumor xenotransplantation mouse model. In situ tumor-bearing experiments showed that NK cell co-inoculation could inhibit the tumor growth ability of PANC-1 cells in vivo (Figures 1F,G). When intravenous transferred in vivo through tail vein, PANC-1 cells co-transferred with NK cells significantly inhibited lung metastasis, as demonstrated by in vivo luminescence imaging and H&E staining of the lung tissues (Figures 1H,I). In summary, NK cell can significantly inhibit the proliferation and metastasis of pancreatic cancer cells in vitro and in vivo.
Figure 1.
NK cells co-culture inhibited tumor progression of pancreatic cancer both in vitro and in vivo. (A,B) CCK-8 assay showed cell viability of Mia PaCa-2 and PANC-1 cells co-cultured with (NK cells+) or without (NK cells-) natural killer cells. (C) Colony formation assay showed cell proliferation of Mia PaCa-2 and PANC-1 cells co-cultured with (NK cells+) or without (NK cells-) natural killer cells. (D,E) Transwell assays showed cell migratory and invasive ability of Mia PaCa-2 and PANC-1 cells co-cultured with (NK cells+) or without (NK cells-) natural killer cells. (F,G) PANC-1 cells were implanted into the flank of mice (n = 4 each group), without (NK cell–) or with co-injection of natural killer cells (NK cell+), respectively, followed by growth curve evaluation on the indicated day after injection. (H–J) Representative in vivo images showed tumor colonization in the lungs of mice (n = 5 each group) following tail vein injection of PANC-1 cells, without (NK cell–) or with co-injection of natural killer cells (NK cell+), respectively, H&E staining of lung sections of mice (metastatic nodules were indicated by yellow arrow, 200×) and incidence of lung metastasis in mice following tail vein injection of the respective PANC-1 cells. The data represent the mean ± SD from three independent experiments. **P < 0.01; ***P < 0.001, two-way ANOVA for (A,B,G), χ2 test for j, Student's t-test for others.
MiR-3607-3p Is Enriched in EVs of NK Cells
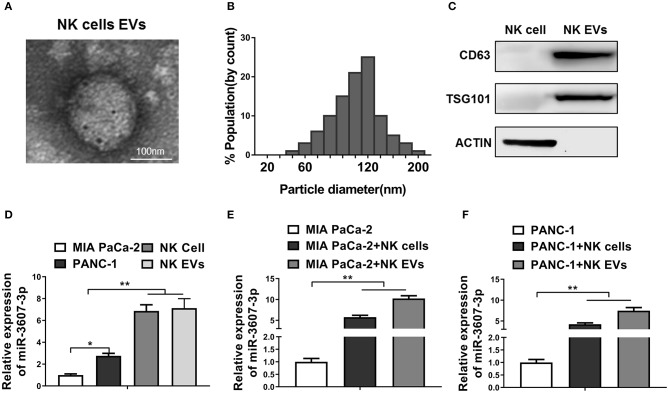
To explore the underlying mechanisms by which NK cells inhibit malignant transformation of pancreatic cancer cells, we speculate that NK cells might act on pancreatic cancer cells through EVs. EVs were isolated and identified from the culture medium of NK cells (Figures 2A–C). In addition, the level of miR-3607-3p in NK cells and its EVs was significantly higher than that in Mia PaCa-2 and PANC-1 (Figure 2D). Furthermore, when co-cultured Mia PaCa-2 and PANC-1 with NK cells or NK cells EVs, we found that the levels of miR-3607-3p were significantly up-regulated in co-cultured Mia PaCa-2 and PANC-1 cells (Figures 2E,F). In summary, the results suggest that NK cells might transmit miR-3607-3p to pancreatic cancer cells via secreted EVs.
Figure 2.
MiR-3607-3p was enriched in EVs of NK cells and can be transferred to pancreatic cancer cells. (A) Transmission electron microscopy showed the representative image of NK cell-derived EVs (scale bar, 100 nm). (B) The particle diameter (nm) of the purified EVs was showed in histogram. (C) Exosomal markers (CD63, TSG101) were detected in NK cellular protein and corresponding EVs by western blotting. (D) qRT-PCR detection of miR-3607-3p in Mia PaCa-2, PANC-1, NK cells, and NK EVs. (E,F) qRT-PCR detection of miR-3607-3p in Mia PaCa-2 or PANC-1 cultured alone or co-cultured with NK cells or NK EVs. The data represent the mean ± SD from three independent experiments. *P < 0.05; **P < 0.01. One-way ANOVA analysis.
MiR-3607-3p Is Down-Regulated in PC and Decreased miR-3607-3p Level Predicts Poor Prognosis in PC Patients
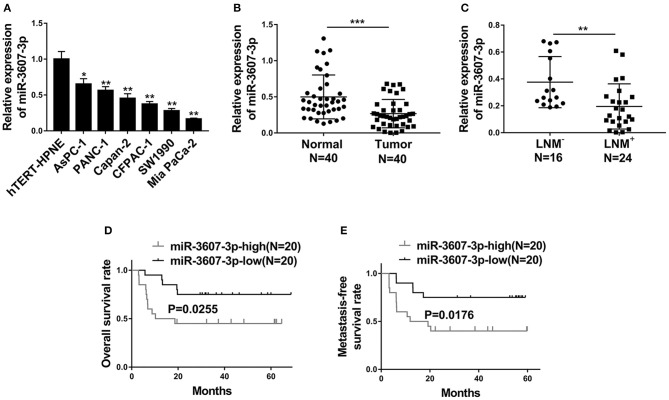
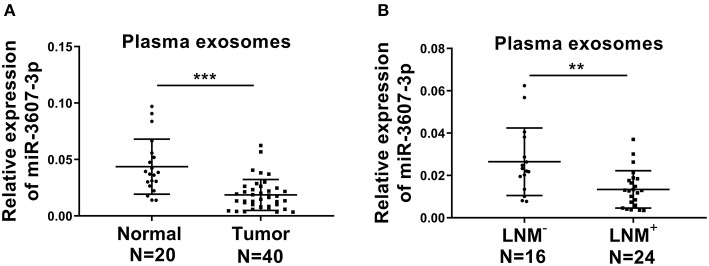
Next, miRNA-3607-3p level was analyzed in different PC cell lines (AsPC-1, PANC-1, Capan-2, CFPAC-1, SW1990 and Mia PaCa-2) and a normal human pancreatic ductal cell control (hTERT-HPNE). As shown in Figure 3A, the level of miR-3607-3p was significantly lower in PC cell lines compared with that in control cell line. Consistently, the level of miR-3607-3p was significantly lower in PC tissues compared with that in normal tissues (Figure 3B). In addition, PC patients with lymph node metastasis (LNM+) further decreased the expression of miR-3607-3p compared to that in lymph node metastasis free (LNM−) patients (Figure 3C). Kaplan–Meier's analysis of the correlation between miR-3607-3p expression and the metastasis-free survival of PC patients indicated that low levels of miR-3607-3p were characterized by worse overall survival and metastasis-free survival rate (Figures 3D,E). We also revealed the similar expression pattern of miR-3607-3p in plasma exosomes in PC patients and plasma exosomal miR-3607-3p expression in PC patients with LNM+ was significantly lower than that in LNM− PC patients (Figures 4A,B). Our results indicate that miR-3607-3p acts as a tumor suppressor in PC and is likely to be involved in tumor metastasis.
Figure 3.
MiR-3607-3p was down-regulated in PC and decreased miR-3607-3p level predicted poor prognosis. (A) qRT-PCR analysis of the expression of miR-3607-3p in six pancreatic cancer cell lines (AsPC-1, PANC-1, Capan-2, CFPAC-1, SW1990, and Mia PaCa-2) and a normal human pancreatic ductal cell line (hTERT-HPNE). (B) qRT-PCR analysis of miR-3607-3p expression in 40 PC tissues and 40 normal human pancreas tissues. (C) qRT-PCR analysis of miR-3607-3p expression in 24 PC tissues from lymph node metastasis (LNM+) patients compared to 16 PC tissues from lymph node metastasis free (LNM−) patients (D) Kaplan–Meier's analysis of the correlation between miR-3607-3p expression and the overall survival of PC patients. (E) Kaplan–Meier's analysis of the correlation between miR-3607-3p expression and the metastasis-free survival of PC patients. *P < 0.05; **P < 0.01; ***P < 0.001. One-way ANOVA for a, Student's t-test for (B,C).
Figure 4.
The expression levels of plasma exosomal miR-3607-3p in PC patients. (A) qRT-PCR analysis of plasma exosomal miR-3607-3p expression from 40 PC patients (Tumor) and 20 normal donors (Normal). (B) qRT-PCR analysis of plasma exosomal miR-3607-3p expression from 24 lymph node metastasis (LNM+) PC patients and 16 lymph node metastasis free (LNM−) PC patients. **P < 0.01; ***P < 0.001. Student's t-test.
MiR-3607-3p Suppresses Proliferation, Migration, and Invasion of PC Cells
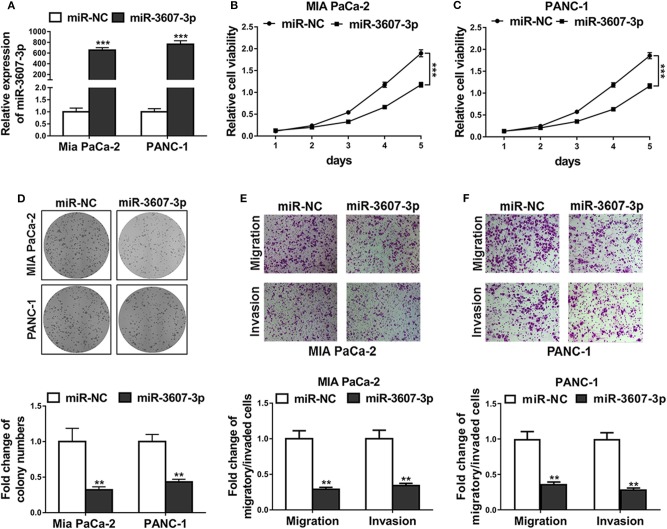
To investigate the function of miR-3607-3p in pancreatic cancer cells, miR-3607-3p mimics was transfected to overexpress miR-3607-3p in PC cell line Mia PaCa-2 and PANC-1. As shown in Figures 5A–D, CCK-8 assay and colony formation assays demonstrated that overexpression of miR-3607-3p inhibited the proliferation of Mia PaCa-2 and PANC-1 in vitro; Transwell assay results showed that overexpression of miR-3607-3p inhibited the migration and invasion of pancreatic cancer cells in vitro (Figures 5E, 4F). Interestingly, NK cells extracellular vesicles (NK EVs) treatment was found to be able to inhibit cell viability (Figures S1A,B), proliferation (Figure S1C), migration (Figures S1D–F) and inhibited IL-26 production (Figure S1G) in both Mia PaCa-2 and PANC-1 cells. In conclusion, miR-3607-3p inhibits the malignant transformation of pancreatic cancer cells.
Figure 5.
MiR-3607-3p suppressed proliferation, migration and invasion of pancreatic cancer cells. (A) Mia PaCa-2 and PANC-1 cells were transfected with miR-3607-3p mimics (miR-3607-3p) or mimics negative control (miR-NC), miR-3607-3p expression levels were quantified by qRT-PCR analysis. (B,C) CCK-8 assay showed cell viability of Mia PaCa-2 and PANC-1 cells transfected with miR-3607-3p mimics (miR-3607-3p) or mimics negative control (miR-NC). (D) Colony formation assay showed cell proliferation of Mia PaCa-2 and PANC-1 cells transfected with miR-3607-3p mimics (miR-3607-3p) or mimics negative control (miR-NC). (E,F) Transwell assays showed cell migratory and invasive ability of Mia PaCa-2 and PANC-1 cells transfected with miR-3607-3p mimics (miR-3607-3p) or mimics negative control (miR-NC). The data represent the mean ± SD from three independent experiments. **P < 0.01; ***P < 0.001. Two-way ANOVA for b and c, Student's t-test for others.
IL-26 Is a Direct Target of miR-3607-3p in PC Cells
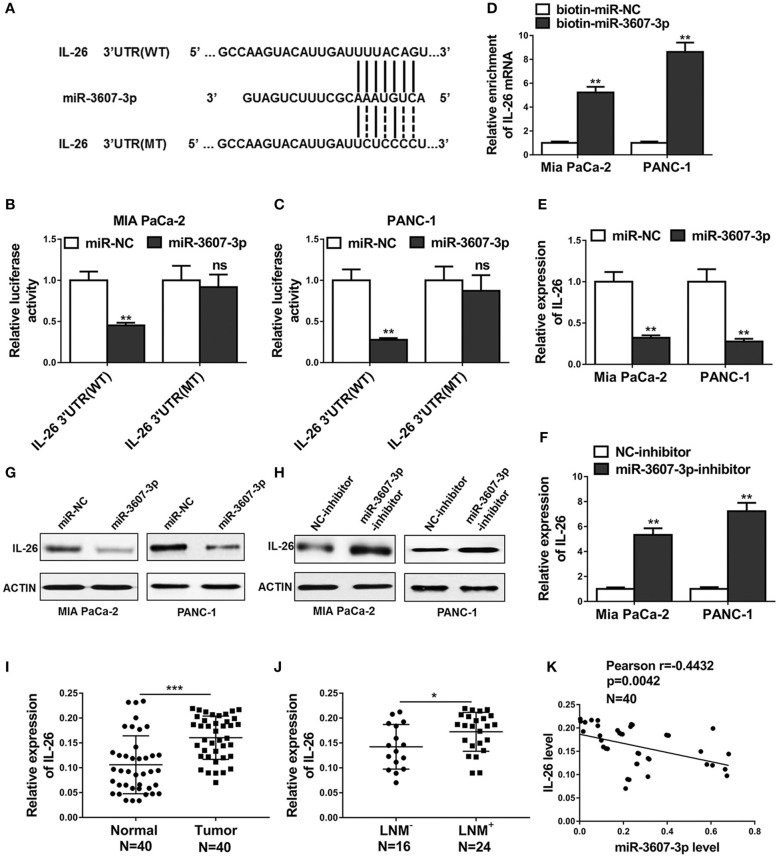
To explores the mechanism how miR-3607-3p regulates malignant transformation of PC, we performed bioinformatics analysis and Targetscan (http://www.targetscan.org/vert_71/) was used to predict potential downstream targets of miR-3607-3p. IL-26 was predicted as a potential miR-3607-3p target binding to the IL-26 3′UTR (Figure 6A). Luciferase reporter vector containing IL-26 3' UTR (WT) or IL-26 mutated 3' UTR (MT) was cotransfected with miRNA control or miR-3607-3p mimics into MIA PaCa-2 and PANC-1 cells. The dual luciferase reporter assay demonstrated that miR-3607-3p bound to the WT IL-26 3′UTR to inhibit the luciferase activity, but not to the mutant IL-26 3′UTR (Figures 5C, 6B). Furthermore, using the biotin-labeled RNA pulldown technique, we found that miR-3607-3p directly interacted with IL-26 mRNA (Figure 5D). We confirmed that miR-3607-3p inhibited IL-26 expression at both mRNA and protein levels while miR-3607-3p inhibitor enhanced IL-26 mRNA and protein levels in MIA PaCa-2 and PANC-1 cells (Figures 6E–H). Finally, we examined the mRNA levels of IL-26 in 40 pancreatic cancer tissues and 40 normal tissues. IL-26 was highly expressed in pancreatic cancer tissues compared with that in normal tissue (Figure 6I). Consistently, IL-26 mRNA level was significantly higher in tissues from LNM+ PC patients that that in tissues from LNM− PC patients (Figure 6J). Correlation analysis showed that there was a significant negative correlation between the expression levels of miR-3607-3p and IL-26 in pancreatic cancer tissues. In summary, IL-26 is a direct target of miR-3607-3p in pancreatic cancer cells.
Figure 6.
IL-26 was a direct target of miR-3607-3p in pancreatic cancer cells. (A) Schematic diagram of miR-3607-3p binding sites in the IL-26 3′UTR. Sequences were compared between the mature miR-3607-3p and wild-type [IL-26 3′UTR (WT)] or mutant [IL-26 3′UTR (MT)] putative target sites in the 3′UTR of IL-26. (B,C) Luciferase reporter assay was performed in Mia PaCa-2 and PANC-1 cells co-transfected with plasmid containing IL-26 3′UTR(WT) or IL-26 3′UTR(MT) and miR-3607-3p or miR-NC. (D) Detection of IL-26 mRNAs in biotinylated miRNA/target mRNA complex by real-time RT-PCR. The relative levels of IL-26 mRNA in the complex pulled down by using biotinylated miR-3607-3p was compared to that of the complex pulled down by using the biotinylated control random RNA. (E–H) The relative expression levels of IL-26 in Mia PaCa-2 and PANC-1 cells transfected with indicated microRNA mimics and microRNA inhibitors or their respective negative controls were detected by qRT-PCR and western blot. (I) qRT-PCR analysis of IL-26 expression in 40 PC tissues and 40 normal human pancreas tissues. (J) qRT-PCR analysis of IL-26 expression in 24 PC tissues from lymph node metastasis (LNM+) patients compared to 16 PC tissues from lymph node metastasis free (LNM−) patients. (K) Correlation analysis between IL-26 and miR-3607-3p in 40 PC tumor samples. *P < 0.05; **P < 0.01; ***P < 0.001. Student's t-test.
MiR-3607-3p Was Required for NK Cell-Induced Inhibition of PC Progression
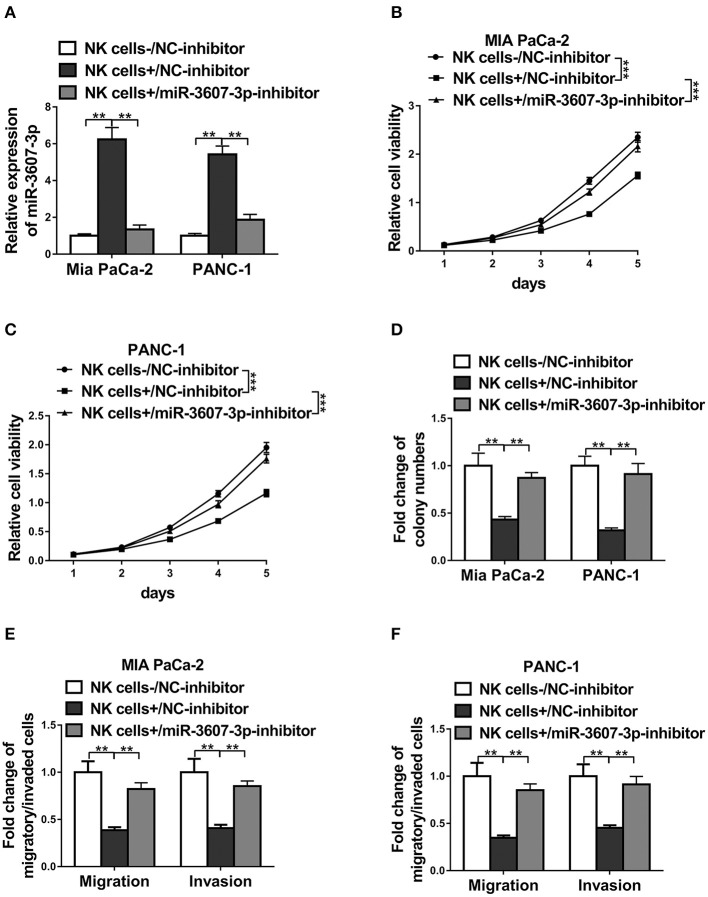
To test whether the inhibition of malignant transformation of PC by NK cells depends on miR-3607-3p, we cultured Mia PaCa-2 or PANC-1 cells together with NK cells transfected NC-inhibitor or miR-3607-3p-inhibitor, respectively. The miR-3607-3p levels were confirmed by qPCR (Figure 7A). The results showed that Mia PaCa-2 and PANC-1 cells co-cultured with NK cells transfected with miR-3607-3p-inhibitor had similar levels of cell proliferation, colony formation and cell migration/invasion compared with that of cells co-cultured with only NC-inhibitor (Figures 7B–F). Inhibition of cell proliferation, migration and invasion of Mia PaCa-2 or PANC-1 cells co-cultured with NK cells transfected with NC-inhibitor was significantly restored in cells co-cultured with NK cells transfected with miR-3607-3p-inhibitor (Figures 7B–F). Thus, NK cells inhibited malignant transformation of pancreatic cancer cells at least partially depending on miR-3607-3p.
Figure 7.
MiR-3607-3p was required for NK cell-induced inhibition of pancreatic cancer progression. (A) qRT-PCR detection of miR-3607-3p in Mia PaCa-2 or PANC-1 co-cultured with only NC-inhibitor (NK cells-/C-inhibitor), NC-inhibitor transfected NK cells (NK cells+/NC-inhibitor) or miR-3607-3p-inhibitor transfected NK cells (NK cells+/miR-3607-3p-inhibitor). (B,C) CCK-8 assay showed cell viability of Mia PaCa-2 and PANC-1 cells co-cultured with only NC-inhibitor (NK cells-/NC-inhibitor), NC-inhibitor transfected NK cells (NK cells+/NC-inhibitor) or miR-3607-3p-inhibitor transfected NK cells (NK cells+/miR-3607-3p-inhibitor). (D) Colony formation assay showed cell proliferation of Mia PaCa-2 and PANC-1 cells co-cultured with only NC-inhibitor (NK cells-/NC-inhibitor), NC-inhibitor transfected NK cells (NK cells+/NC-inhibitor) or miR-3607-3p-inhibitor transfected NK cells (NK cells+/miR-3607-3p-inhibitor). (E,F) Transwell assays showed cell migratory and invasive ability of Mia PaCa-2 and PANC-1 cells co-cultured with only NC-inhibitor (NK cells-/NC-inhibitor), NC-inhibitor transfected NK cells (NK cells+/NC-inhibitor) or miR-3607-3p-inhibitor transfected NK cells (NK cells+/miR-3607-3p-inhibitor). The data represent the mean ± SD from three independent experiments. **P < 0.01; ***P < 0.001. Two-way ANOVA for (B,C), one-way ANOVA for others.
Discussion
Mounting evidences have shown that exosomal miRNA are involved in cancer biology and clinical applications (14, 22). By using in vitro cell co-culture and in vivo tumor-bearing experiments, we described a previously un-identified miR-3607-3p which is enriched in EVs derived from NK cells, suppressing PC development and malignant transformation. Furthermore, we showed that PC patients had low level of miR-3607-3p and decreased miR-3607-3p level could predicte poor prognosis. Most importantly, IL-26 was demonstrated as a direct target candidate of miR-3607-3p. Overall, these results help us to further understand the function of miR-3607-3p in PC and provide a potential novel diagnostic and therapeutic target for PC.
NK cells display critical antitumor properties as they respond rapidly to metastatic and malignancies (23). In PC, NK cell activation and function are compromised due to the extensive crosstalk between NK cell and tumor microenvironment, and different strategies are employed to immunomodulate NK cells in anti-tumor therapies (24). Studies have shown that exosomes derived from NK cells exert anti-tumor effect in different cancers, such as melanoma and neuroblastoma (18, 21). Here we showed that co-cultured NK cells inhibited tumor progression of PC both in vitro and in vivo (Figure 1). However, more effects are needed to distinguish the human NK cells from the endogenous mouse NK cells and to verify the transplanted human NK cells were in the metastasized region of the lung. To understand how NK cell co-culture affects PC progression, we investigated the EVs derived from NK cell. mRNA and microRNAs have been identified in EVs, which could be transmitted to neighboring cells and function in recipient cells (25). Potential target microRNAs were screened and miR-3607-3p was found to be enriched in NK secreted EVs (Figure 2).
miR-3607-3p, a recent defined novel miRNA, has been demonstrated to suppress non-small cell lung cancer development (26). However, other studies have suggested that miR-3607 might promote cancer development and metastasis (27, 28). To explore the function of miR-3607-3p in PC, we examined the levels of miR-3607-3p in PC patients. Our results indicate that miR-3607-3p is down-regulated in PC and decreased miR-3607-3p level predicts poor prognosis in PC patients (Figure 3). Furthermore, the function of miR-3607-3p was confirmed in vitro as it could suppress PC cell proliferation, migration and invasion (Figure 4). Using bioinformatics analysis, we predicted IL-26 as a potential miR-3607-3p target and confirmed that using luciferase reporter assay and biotin-miRNA pull-down assay (Figures 5A–D). Consistently, IL-26 expression was much higher in LNM+ PC patients and there was a significant negative correlation between the expression levels of miR-3607-3p and IL-26 in pancreatic cancer tissues (Figures 5I–K).
IL-26 is a member of the IL-10 cytokine family and often co-expressed with IL-22 by activated T cells, especially Th17 cells (29). However, the function of IL-26 in human tumor remains largely unknown. Wei You et.al, reported that IL-26 expression was enhanced in human gastric cancer cells and enhanced IL-26 facilitated gastric cancer cell proliferation and survival by regulating STAT1/STAT3 signaling (30). Here we also found increased IL-26 levels in PC patients compared with that in control group. Intriguingly, IL-26 level was further enhanced in LNM+ PC patient compared with that in LNM− PC patient (Figures 5I,J), suggesting a role in PC metastasis. However, the downstream signaling of IL-26 in PC is unclear, and whether IL-26 could be used a diagnose marker in PC still need further studies.
Conclusions
In conclusion, our findings suggest that miR-3607-3p enriched in EVs derived from NK cells inhibits the malignant transformation of PC probably through direct targeting of IL-26, implicating that targeting tumor microenvironment and related molecular cross-talk could be a new strategy of PC therapy.
Data Availability Statement
The datasets analyzed in this article are not publicly available. Requests to access the datasets should be directed to MZ, qianazi@yeah.net.
Ethics Statement
The animal study was reviewed and approved by Ethic and Research Committees of the First Affiliated Hospital of Wenzhou Medical University.
Author Contributions
HS, KS, KQ, HK, JZ, SD, WY, TD, and QH: conception and design. HS, KS, KQ, and HK: collection and assembly of data. HS, and MZ: manuscript writing. HS, KS, KQ, HK, JZ, SD, WY, TD, QH, and MZ: final approval of manuscript.
Conflict of Interest
QH was employed by company Singlera Genomics Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Glossary
Abbreviations
- NK
natural killer
- PC
pancreatic cancer
- miRNA
microRNA.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.02819/full#supplementary-material
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. (2011) 61:69–90. 10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- 2.Edwards BK, Brown ML, Wingo PA, Howe HL, Ward E, Ries LA, et al. Annual report to the nation on the status of cancer, 1975-2002, featuring population-based trends in cancer treatment. J Natl Cancer Inst. (2005) 97:1407–27. 10.1093/jnci/dji289 [DOI] [PubMed] [Google Scholar]
- 3.Arslan AA, Helzlsouer KJ, Kooperberg C, Shu XO, Steplowski E, Bueno-de-Mesquita HB, et al. Anthropometric measures, body mass index, and pancreatic cancer: a pooled analysis from the Pancreatic Cancer Cohort Consortium (PanScan). Arch Intern Med. (2010) 170:791–802. 10.1001/archinternmed.2010.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fogel EL, Shahda S, Sandrasegaran K, DeWitt J, Easler JJ, Agarwal DM, et al. A Multidisciplinary approach to pancreas cancer in 2016: a review. Am J Gastroenterol. (2017) 112:537–54. 10.1038/ajg.2016.610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tuveson DA, Neoptolemos JP. Understanding metastasis in pancreatic cancer: a call for new clinical approaches. Cell. (2012) 148:21–3. 10.1016/j.cell.2011.12.021 [DOI] [PubMed] [Google Scholar]
- 6.Soysal SD, Tzankov A, Muenst SE. Role of the tumor microenvironment in breast cancer. Pathobiology. (2015) 82:142–52. 10.1159/000430499 [DOI] [PubMed] [Google Scholar]
- 7.Yaacoub K, Pedeux R, Tarte K, Guillaudeux T. Role of the tumor microenvironment in regulating apoptosis and cancer progression. Cancer Lett. (2016) 378:150–9. 10.1016/j.canlet.2016.05.012 [DOI] [PubMed] [Google Scholar]
- 8.Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, et al. Innate or adaptive immunity? The example of natural killer cells. Science. (2011) 331:44–9. 10.1126/science.1198687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jewett A, Kos J, Fong Y, Ko MW, Safaei T, Perisic Nanut M, et al. NK cells shape pancreatic and oral tumor microenvironments; role in inhibition of tumor growth and metastasis. Semin Cancer Biol. (2018) 53:178–88. 10.1016/j.semcancer.2018.08.001 [DOI] [PubMed] [Google Scholar]
- 10.Yan Y, Steinherz P, Klingemann HG, Dennig D, Childs BH, McGuirk J, et al. Antileukemia activity of a natural killer cell line against human leukemias. Clin Cancer Res. (1998) 4:2859–68. [PubMed] [Google Scholar]
- 11.Ljunggren HG, Malmberg KJ. Prospects for the use of NK cells in immunotherapy of human cancer. Nat Rev Immunol. (2007) 7:329–39. 10.1038/nri2073 [DOI] [PubMed] [Google Scholar]
- 12.Cheng M, Zhang J, Jiang W, Chen Y, Tian Z. Natural killer cell lines in tumor immunotherapy. Front Med. (2012) 6:56–66. 10.1007/s11684-012-0177-7 [DOI] [PubMed] [Google Scholar]
- 13.Thery C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep. (2011) 3:15. 10.3410/B3-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Penfornis P, Vallabhaneni KC, Whitt J, Pochampally R. Extracellular vesicles as carriers of microRNA, proteins and lipids in tumor microenvironment. Int J Cancer. (2016) 138:14–21. 10.1002/ijc.29417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tkach M, Thery C. Communication by extracellular vesicles: where we are and where we need to go. Cell. (2016) 164:1226–32. 10.1016/j.cell.2016.01.043 [DOI] [PubMed] [Google Scholar]
- 16.Whiteside TL. Tumor-derived exosomes and their role in cancer progression. Adv Clin Chem. (2016) 74:103–41. 10.1016/bs.acc.2015.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng P, Chen L, Yuan X, Luo Q, Liu Y, Xie G, et al. Exosomal transfer of tumor-associated macrophage-derived miR-21 confers cisplatin resistance in gastric cancer cells. J Exp Clin Cancer Res. (2017) 36:53. 10.1186/s13046-017-0528-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu L, Kalimuthu S, Gangadaran P, Oh JM, Lee HW, Baek SH, et al. Exosomes derived from natural killer cells exert therapeutic effect in melanoma. Theranostics. (2017) 7:2732–45. 10.7150/thno.18752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang LL, Zhang LF, Shi YB. miR-24 inhibited the killing effect of natural killer cells to colorectal cancer cells by downregulating Paxillin. Biomed Pharmacother. (2018) 101:257–63. 10.1016/j.biopha.2018.02.024 [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto K, Ito S, Hanafusa H, Shimizu K, Ouchida M. Uncovering direct targets of MiR-19a involved in lung cancer progression. PLoS ONE. (2015) 10:e0137887. 10.1371/journal.pone.0137887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shoae-Hassani A, Hamidieh AA, Behfar M, Mohseni R, Mortazavi-Tabatabaei SA, Asgharzadeh S. NK cell-derived exosomes from NK cells previously exposed to neuroblastoma cells augment the antitumor activity of cytokine-activated NK cells. J Immunother. (2017) 40:265–76. 10.1097/CJI.0000000000000179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang G, et al. Effect of exosomal miRNA on cancer biology and clinical applications. Mol Cancer. (2018) 17:147. 10.1186/s12943-018-0897-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholson SE, Keating N, Belz GT. Natural killer cells and anti-tumor immunity. Mol Immunol. (2019) 110:40–7. 10.1016/j.molimm.2017.12.002 [DOI] [PubMed] [Google Scholar]
- 24.Van Audenaerde JRM, Roeyen G, Darcy PK, Kershaw MH, Peeters M, Smits ELJ. Natural killer cells and their therapeutic role in pancreatic cancer: a systematic review. Pharmacol Ther. (2018) 189:31–44. 10.1016/j.pharmthera.2018.04.003 [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, Li S, Li L, Li M, Guo C, Yao J, et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. (2015) 13:17–24. 10.1016/j.gpb.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao P, Wang H, Yu J, Zhang J, Yang Z, Liu M, et al. miR-3607-3p suppresses non-small cell lung cancer (NSCLC) by targeting TGFBR1 and CCNE2. PLoS Genet. (2018) 14:e1007790. 10.1371/journal.pgen.1007790 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Lin Y, Gu Q, Sun Z, Sheng B, Qi C, Liu B, et al. Upregulation of miR-3607 promotes lung adenocarcinoma proliferation by suppressing APC expression. Biomed Pharmacother. (2017) 95:497–503. 10.1016/j.biopha.2017.08.052 [DOI] [PubMed] [Google Scholar]
- 28.Wu J, Jiang Z, Chen C, Hu Q, Fu Z, Chen J, et al. CircIRAK3 sponges miR-3607 to facilitate breast cancer metastasis. Cancer Lett. (2018) 430:179–92. 10.1016/j.canlet.2018.05.033 [DOI] [PubMed] [Google Scholar]
- 29.Donnelly RP, Sheikh F, Dickensheets H, Savan R, Young HA, Walter MR. Interleukin-26: an IL-10-related cytokine produced by Th17 cells. Cytokine Growth Factor Rev. (2010) 21:393–401. 10.1016/j.cytogfr.2010.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.You W, Tang Q, Zhang C, Wu J, Gu C, Wu Z, et al. IL-26 promotes the proliferation and survival of human gastric cancer cells by regulating the balance of STAT1 and STAT3 activation. PLoS ONE. (2013) 8:e63588. 10.1371/journal.pone.0063588 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed in this article are not publicly available. Requests to access the datasets should be directed to MZ, qianazi@yeah.net.