Abstract
Adeno-associated viral (AAV) vectors have shown great promise in gene delivery as evidenced by recent FDA approvals. Despite efforts to optimize manufacturing for good manufacturing practice (GMP) productions, few academic laboratories have the resources to assess vector composition. One critical component of vector quality is packaged genome fidelity. Errors in viral genome replication and packaging can result in the incorporation of faulty genomes with mutations, truncations, or rearrangements, compromising vector potency. Thus, sequence validation of packaged genome composition is an important quality control (QC), even in academic settings. We developed Fast-Seq, an end-to-end method for extraction, purification, sequencing, and data analysis of packaged single-stranded AAV (ssAAV) genomes intended for non-GMP preclinical environments. We validated Fast-Seq on ssAAV vectors with three different genome compositions (CAG-GFP, CAG-tdTomato, EF1α-FLuc), three different genome sizes (2.9, 3.6, 4.4 kb), packaged in four different capsid serotypes (AAV1, AAV2, AAV5, and AAV8), and produced using the two most common production methods (Baculovirus-Sf9 and human HEK293), from both common commercial vendors and academic core facilities supplying academic laboratories. We achieved an average genome coverage of >1,400 × and an average inverted terminal repeat coverage of >280 × , despite the many differences in composition of each ssAAV sample. When compared with other ssAAV next-generation sequencing (NGS) methods for GMP settings, Fast-Seq has several unique advantages: Tn5 transposase-based fragmentation rather than sonication, 125 × less input DNA, simpler adapter ligation, compatibility with commonly available inexpensive sequencing instruments, and free open-source data analysis code in a preassembled customizable Docker container designed for novices. Fast-Seq can be completed in 18 h, is more cost-effective than other NGS methods, and is more accurate than Sanger sequencing, which is generally only applied at 1–2 × sequencing depth. Fast-Seq is a rapid, simple, and inexpensive methodology to validate packaged ssAAV genomes in academic settings.
Keywords: ssAAV, next-generation sequencing, transposase, Fast-Seq, method, genome
Introduction
Recombinant adeno-associated virus (rAAV) is the gold standard vector for performing safe viral gene therapy. A naturally replication-incompetent single-stranded DNA virus, rAAV, is widely used for delivering gene therapeutics due to its high transduction efficiency in vivo and non-pathogenicity to humans. rAAV can express payloads stably from unintegrated episomes in nondividing tissues,1 or through integration in actively dividing tissues when appropriately designed homology arms are included in the transfer vector construct.2 As gene therapy grows in popularity, many new academic laboratories have begun preclinical testing of rAAV therapies. However, academic laboratories often lack the time or financial resources to rigorously assess the fidelity of packaged vector genomes. Errors in rAAV genome replication and packaging can result in the incorporation of faulty genomes with truncations, rearrangements, and various mutations.3–6 Such faulty genomes could compromise the functionality of gene therapy vector lots. Sanger sequencing is often the standard method used for sequence verification of rAAV transfer vector plasmids and packaged viral genomes. Although Sanger sequencing does provide high-quality data in a modest turnaround time, the data analysis requires manual evaluation of sequencing reads, rendering this approach very low throughput. In addition, Sanger sequencing depth generally only provides 1–2 × coverage, which results in low confidence single nucleotide polymorphism (SNP) and insertion or deletion (indel) calling. We recommend that academic laboratories sequence both the transfer vector plasmids used for viral production and the resulting packaged genomes to ensure that the genomes packaged faithfully. This is particularly true when the resultant preclinical data are being used to inform planned future clinical trials.
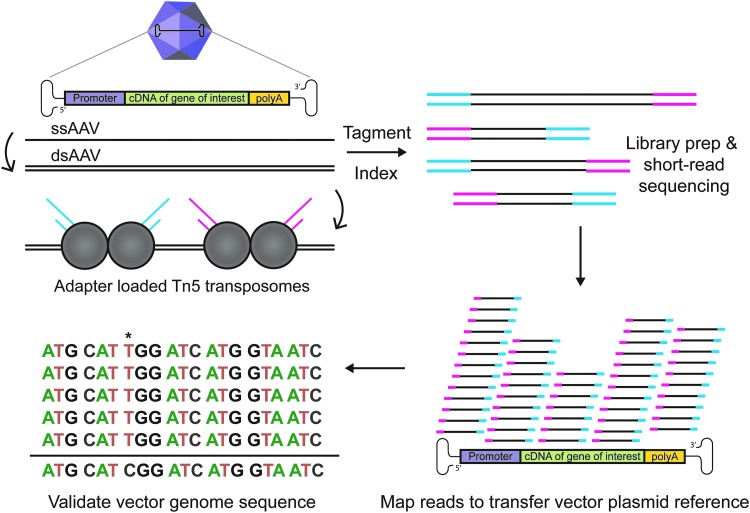
To solve this lack of appropriate methodology, we have adapted existing protocols7–9 to develop Fast-Seq, a new end-to-end method for extraction, purification, sequencing, and data analysis of packaged ssAAV genomes intended for non-GMP academic environments. Other methods exist for self-complementary AAV.3 Fast-Seq was extensively validated on three different ssAAV vector lot compositions (ssAAV-CAG-GFP, ssAAV-CAG-tdTomato, ssAAV-EF1α-FLuc), of different genome sizes (2.9, 3.6, and 4.4 kb), at various dilutions (1E11-1E9 vector genomes input), obtained from two popular vendor types for research-grade vectors for academics (Addgene and a viral vector core facility), and produced by each of the most common manufacturing platforms (transient transfection of human HEK293 and baculoviral infection of Sf9 insect cells). Fifteen independent vector lots were assessed with biological and/or technical replicates by two independent researchers. The Fast-Seq protocol outlines a simple, fast, and inexpensive methodology for sequencing packaged ssAAV genomes through NGS, using Tn5 tagmentation-based libraries (Fig. 1).
Figure 1.
Overview of Fast-Seq, a Tn5-based packaged ssAAV genome sequencing method. Preparations of rAAV undergo ssDNA extraction, second-strand synthesis to generate dsDNA, tagmentation with adapter-loaded Tn5 transposomes, indexing, QC validation, short-read sequencing on Illumina MiSeq, mapping reads to the transfer vector plasmid used to generate the input rAAV, and sequence validation of the packaged genome. An example packaged genome mutation (*) is shown at position 7. dsAAV, double-stranded AAV; dsDNA, double-stranded DNA; rAAV, recombinant AAV; ssAAV, single-stranded adeno-associated virus; Tn5, transposase.
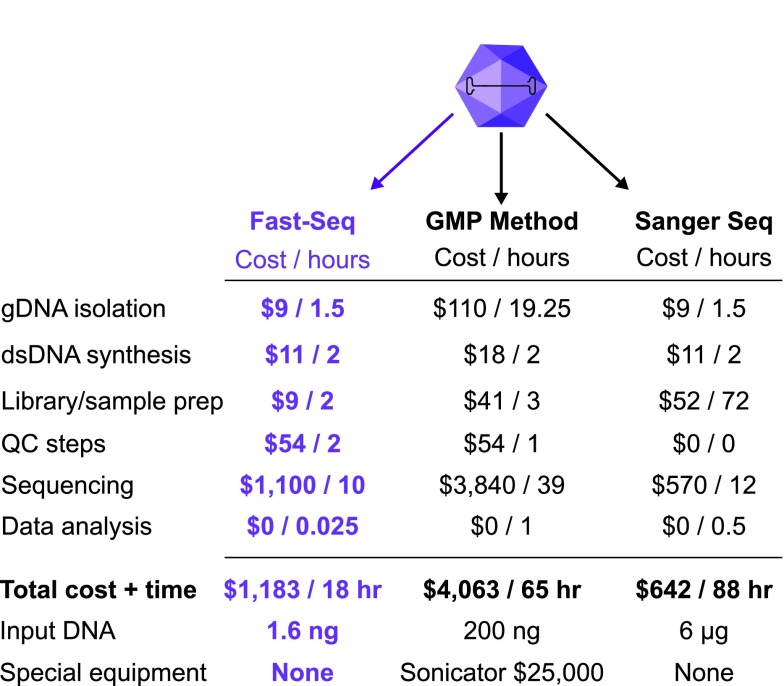
When compared with several other recently published NGS methods for GMP settings,10,11 Fast-Seq has several unique advantages (Fig. 2): fragmentation and simultaneous adapter ligation through Tn5 transposase rather than sonication (no expensive or difficult-to-operate sonicator required), up to 125 × less input DNA, compatibility with inexpensive sequencing instruments such as MiSeq and iSeq, and greater sample retention from fewer magnetic bead purification steps. In addition, we built Fast-Seq to have free open-source code for data analysis available in a prebuilt customizable Docker container accessible on GitHub that requires no prior coding experience. We also provide a freely available detailed laboratory protocol.12 Fast-Seq was optimized for use on the commonly available Illumina MiSeq instrument, but other instruments would also work and should be chosen based on the number of samples and depth required (iSeq, MiniSeq, NextSeq, etc.). Detailed comparative cost breakdowns are given in Supplementary Table S1. Using Fast-Seq, we were able to sequence up to 16 ssAAV samples simultaneously to an average genome coverage of >1,400 × , and an average inverted terminal repeat (ITR) coverage of >280 × , with reads spanning the entire length of the ssAAV genome.
Figure 2.
Comparison of ssAAV packaged genome validation methods. Comparison of the per sample costs and time in hours associated with performing Fast-Seq compared with other existing methods of sequencing packaged ssAAV genomes. gDNA, genomic DNA; QC, quality control; seq, sequencing.
Required Materials
Reagent list: name (catalog no., vendor)
Ethanol 200 proof (E7023; Sigma Aldrich)
5 M NaCl solution (AM9759; Thermo Fisher)
HEPES 1 M (15630106; Thermo Fisher)
KOH 8 N (P4494; Millipore Sigma)
0.5 M EDTA solution (15575020; Thermo Fisher)
Dithiothreitol (R0861; Thermo Fisher)
Triton X-100 (85112; Thermo Fisher)
Glycerol (17904; Thermo Fisher)
1 M Tris pH 7.0 (AM9850G; Thermo Fisher)
1 M MgCl2 solution (AM9530G; Thermo Fisher)
NN-dimethylformamide (D4551; Millipore Sigma)
KAPA HiFi PCR kit with dNTPs (NC0142652; Thermo Fisher)
10% SDS (15553027; Thermo Fisher)
i5/i7 index primers (any oligo provider)
DNA LoBind tubes (13-698-791; Thermo Fisher)
Protein LoBind tubes (0030108116; Eppendorf)
SPRIselect (B23317; Beckman Coulter)
Tn5 transposase with Nextera adapters (custom; QB3 Macrolab)
Elution buffer (19086; Qiagen)
DNase endonuclease (DB0715K; Lucigen)
DNase exonuclease (E3101K; Lucigen)
MinElute Virus Spin kit (57704; Qiagen)
dNTP solution mix (N0447S; NEB)
Klenow (M0210L; NEB)
Millex-GP filter unit 0.22 μm (SLGP033RS; EMD)
Random primer mix (S1330S; NEB)
dNTP solution mix (N0447S; NEB)
Ultrapure (10977015; Thermo Fisher)
Qubit dsDNA High Sensitivity Kit (Q33230; Thermo Fisher).
Equipment list
Magnetic tube rack for microcentrifuge tubes
Heat block
Microcentrifuge
Vortexer
Benchtop centrifuge
PCR thermocycler
Qubit fluorometer with provided tubes
Computer with 2GB+ of RAM, with Docker and Python 3.
Optional
Bioanalyzer/tapestation and calibration kit
Sequencer (any model Illumina instrument)
qPCR thermocycler
Illumina Library Quantitation Complete kit (KK4824; Kapa)
MiSeq Nano v2 300-cycle or v3 150-cycle kit (MS-102-2002 or MS-102-3001; Illumina)
HT1 hybridization buffer (FC-131-1024; Illumina).
Reagent Setup
Solutions to prepare in advance and store
(A) Preparation of 50 mL of Tn5 storage buffer
Tn5 storage buffer was prepared by combining all reagents listed, except for the glycerol. The pH of the buffer was then adjusted to 7.2 in a final volume of 20-mL, and sterile filtered with a 0.22-μm filter. 30-mL of sterile glycerol was then added to a final volume of 50-mL. The buffer was stored at −20°C.
| Reagent | [Stock] | [Final] | Volume needed |
|---|---|---|---|
| NaCl | 5 M | 100 mM | 1 mL |
| EDTA | 0.5 M | 0.1 mM | 10 μL |
| HEPES–KOH, pH 7.2 | 1 M | 50 mM | 2.5 mL |
| DTT | 100 mM | 1 mM | 0.5 mL |
| Triton X-100 | 98% | 0.1% | 51 μL |
| Glycerol | 100% | 60% | 30 mL |
| DNase/RNase-free water | — | — | 16 mL |
| Total volume | — | — | 50 mL |
(B) Preparation of 50mL of 5 ñ Tn5 reaction buffer
Tn5 Reaction buffer was prepared by combining the listed reagents, and adjusting the pH to 7.5 in a final volume of 50-mL. The Tn5 reaction buffer was then sterile filtered with a 0.22-μm filter and stored at -20°C.
| Reagent | [Stock] | [Final] | Volume needed |
|---|---|---|---|
| Tris, pH 7.0 | 1 M | 50 mM | 2.5 mL |
| MgCl2 | 1 M | 50 mM | 2.5 mL |
| DNase/RNase-free water | — | — | 45 mL |
| Total volume | — | — | 50 mL |
(C) Preparation of neutralization buffer
Neutralization buffer was used to inactivate the Tn5 enzyme and quench the tagmentation reaction to stop overtagmentation of the AAV libraries.
Neutralization buffer is a 0.1% SDS solution in water. The solution was heated in a 37°C water bath to completely dissolve the SDS and then cooled to room temperature. The neutralization buffer was stored at room temperature.
(D) Qualify each new batch of Nextera adapter-loaded Tn5 enzyme
The dilution of each new batch of adapter-loaded Tn5 enzyme should be empirically determined based on activity. We recommend making a dilution series of loaded enzyme to determine the optimal yield and fragment size. The Tn5 enzyme was diluted in Tn5 storage buffer. For a starting point reference for the dilution series, we have found that adapter-loaded Tn5 at 1 ng/μL in storage buffer works best in our hands; however, working concentration is subject to the activity of the batch. We suggest performing twofold serial dilutions in a range of concentrations both above and below this when qualifying the initial batch of Tn5. Adapter-loaded Tn5 lots can be custom ordered from the UC Berkeley QB3 MacroLab here:
Solutions to be prepared fresh for AAV library preparation
Note: We have provided volumes for one sample. As these are small volumes susceptible to loss and evaporation, we recommend using PCR strip tubes rather than plates, and making master mixes at least 1.5 × the number of samples. For example, if making libraries of 20 samples, master mixes were prepared that are suitable for 1.5 × 20 = 30 samples.
(A) 1.6 × Tris-N,N-dimethylformamide buffer
If this will be a regular assay, we recommend premixing and aliquoting the 5 × Tn5 reaction buffer and water and storing as aliquots. Importantly, N,N-dimethylformamide (DMF) should be added fresh, right at the time of the assay.
| Reagent | 1 sample (μL) |
|---|---|
| 5 × Tn5 reaction buffer | 0.64 |
| DNase/RNase-free water | 1.2 |
| 100% DMF* | 0.16 |
| Total volume | 2 |
Caution: DMF is a volatile carcinogen and should be handled with appropriate safety precautions in a chemical fume hood. When handling DMF, wear appropriate PPE, including a lab coat, goggles, closed toe shoes and double gloves. Dispose of any pipette tips that contact DMF in a labeled solid waste container. Clean pipettes after use.
PPE, personal protective equipment.
(B) Tagmentation reaction mixture
| Reagent | 1 sample (μL) |
|---|---|
| 1.6 × Tris-DMF buffer | 2 |
| Tn5-enzyme (adapter loaded) | 0.4 |
| Total volume | 2.4 |
(C) Index PCR master mix
This can be prepared while the AAV samples are tagmenting.
| Reagent | 1 sample (μL) |
|---|---|
| 5 × KAPA HiFi buffer | 1.6 |
| dNTP mix | 0.24 |
| DNase/RNase-free water | 0.4 |
| KAPA HiFi polymerase | 0.16 |
| Total volume | 2.4 |
Method Steps
Isolating pure ssDNA AAV genomes from the vector lot
(1) We recommend extracting total gDNA from 1E11 full ssAAV particles. However, this protocol has been successfully validated for inputs as low as 1E9 full ssAAV particles. We suggest including a known prevalidated control vector with replicates for every run. To meet the minimum input DNA requirement for NGS, we recommend preparing at least eight samples (this can be composed of biological/technical replicates or dilutions of the sample and controls) for sequencing.
(2) To remove unincorporated DNA, ssAAV samples were incubated with 40-U exonuclease and 10-U endonuclease for 30-min at 37°C in the provided 10 × buffer, supplemented with 1 mmol/L of the provided ATP in a final volume of 200-μL in a protein low-binding microcentrifuge tube.
| Reagent | [Stock] | [Final] | Volume needed (μL) |
|---|---|---|---|
| 10 × buffer | 10 × | 1 × | 20 |
| Endonuclease | 1 U/μL | 10 U | 10 |
| Exonuclease | 10 U/μL | 40 U | 4 |
| ATP | 1 mM | 25 mM | 8 |
| ssAAV 1E11 full particles | — | 1E11 | X |
| DNase/RNase-free water | — | — | X |
| Total volume | — | — | 200 |
(3) The samples were incubated at 65°C for 10-min to stop the nuclease digestion.
*Critical step*—The entire 200 μL reaction mixture from Step 3 should be used as the input for the kit for Step 4.
(4) The ssAAV gDNA extraction was continued using a Qiagen MinElute Virus Spin Kit following the manufacturer's protocol. Additional reagents and equipment required for the kit: 100% ethanol, 1.5 mL microcentrifuge tubes, heat block, microcentrifuge, and vortexer. The final gDNA was eluted into 28-μL, into a 1.5-mL DNA low-binding microcentrifuge tube. The extracted gDNA was stored short-term (<24-hr) at 4°C wrapped in parafilm, or long-term (>24-hr) at -20°C wrapped in parafilm.
*Pause point*
Second-strand DNA synthesis
(5) The gDNA from Step 4 was incubated at 95°C for 5-min and then quenced on ice.
(6) Second-strand synthesis was performed by combining the listed reagents in a PCR tube in a final volume of 50-μL. Note: The 10 × NEB2 buffer listed is the buffer provided with the DNA polymerase I (Klenow) reagent.
| Reagent | [Stock] | [Final] | Volume needed (μL) |
|---|---|---|---|
| Random primer mix | 60 μM | 6 μM | 5 |
| dNTPs | 10 mm each | 2 mM | 10 |
| Klenow | 5 U/μL | 10 U | 2 |
| 10 × NEB2 buffer | 10 × | 1 × | 5 |
| gDNA from Step 5 | — | — | 28 |
| Total volume | — | — | 50 |
(7) Randomly primed DNA synthesis was performed in a thermocycler using a ramp of 0.1°C/second until 37°C, followed by a 1 hour incubation at 37°C.
(8) dsDNA AAV genomes were stored short-term (<24-hr) on ice, or long-term (<24-hr) at -20°C wrapped in parafilm.
*Pause point*
AAV tagmentation with Tn5
For one reaction (multiply volumes to scale for more reactions):
(9) dsDNA AAV genomes were diluted to 1-ng/μL and verified with a Qubit fluorometer, using Qubit thin-walled assay tubes and the Qubit dsDNA High Sensitivity Assay Kit. We do not recommend using a NanoDrop to measure DNA concentration here, as these readings can be inaccurate given the low concentrations.
(10) 2.4-μL of Tagmentation Reaction Mix was added per PCR tube.
*Critical step*—Ensure that each new batch of adapter-loaded Tn5 enzyme is qualified before using.
(11) 1.6-μL of DNA solution was added per PCR tube.
(12) PCR tubes were spun down briefly in a benchtop centrifuge to collect reagents at the bottom of the tube.
(13) Tagmentation of samples was performed in a thermocycler with the following conditions: 1 cycle of 55°C for 10 min; 1 cycle of 72°C for 3 min; 1 cycle of 10°C kept at an infinite hold.
Neutralization
(14) 0.8-μL of Neutralization Buffer was added per PCR tube.
(15) The PCR tubes were spun down in a benchtop centrifuge at room temperature for 5-min at 1,500-G.
Index PCR
(16) 2.4-μL of Index PCR Master Mix was added per PCR tube.
(17) 1.6-μL of premixed Illumina Nextera i5/i7 indexing primers previously diluted to 5-μM were added per PCR tube.
Index 1 (i7):
CAAGCAGAAGACGGCATACGAGATNNNNNNNNNNNNGTCTCGTGGGCTCGG
Index 2 (i5):
AATGATACGGCGACCACCGAGATCTACACNNNNNNNNNNNNTCGTCGGCAGCGTC
bold = P5/P7 adapter sequence
italics = unique 12 bp barcode index
underline = primer for mosaic ends added during tagmentation.
Additional guidance for designing adapters can be found at: (https://support.illumina.com/content/dam/illumina-support/documents/documentation chemistry_documentation/experiment-design/illumina-adapter-sequences-1000000002694-09.pdf).
(18) PCR tubes were spun down briefly in a benchtop centrifuge to collect reagents at the bottom of the tube.
(19) Index PCR was performed in a thermocycler with the following conditions: 1 cycle of 72°C for 3 min; 1 cycle of 95°C for 30 sec; 12 cycles of (98°C for 10 sec; 67°C for 30 sec; 72°C for 1 min); 1 cycle of 10°C for an infinite hold.
*Pause point*
Pooling and purification of AAV libraries
(20) 8-μL of every PCR tube was pooled into a 1.5-mL DNA low-binding microcentrifuge tube.
(21) To remove adapter dimers and undesired fragment sizes from the pooled libraries, two Solid Phase Reversible Immobilization (SPRI) bead purifications were performed:
(a) Purification 1: 0.6 × volume of sample, followed with 2 × wash with 80% ethanol and elution into 100-μL Elution Buffer.
(b) Purification 2: 0.7 × volume of sample, followed with 2 × wash with 80% ethanol and elution into 30-μL Elution Buffer.
(22) The final DNA concentration was quantified with a Qubit, using Qubit thin-walled assay tubes and the Qubit dsDNA High Sensitivity Assay Kit. Note: the final DNA concentration at this step must be at least 2-nM to sequence the sample on an Illumina MiSeq.
(23) Option A: If the samples will be QC validated and sequenced by a core facility or external contract research organization, submit the samples at this stage. Skip Steps 24–26 and resume at Step 27 for ‘Data Analysis’ once raw sequencing data has been received from the sequencing provider. Option B: If the samples will be QC validated and sequenced internally, continue to the next step.
Option B: If we are planning on doing the library QC and sequencing, the next step is continued.
*Pause point*
QC libraries and load sequencer
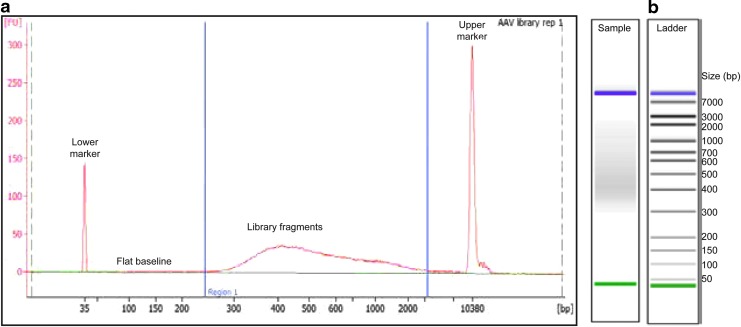
(24) Pooled and purified libraries were analyzed on a bioanalyzer for fragment size verification, using the high sensitivity DNA kit (Fig. 3a). Traces should be free from adapter-dimers, which are generally 100–150 bp. The absolute shape and bp size of the peaks in the traces are less important than having a large smooth proportion of inserts in the 200–800 bp range. Inserts in this size range from AAV genomes are optimal for cluster formation on Illumina flow cells. If the fragment size range is not between 200–800 bp, we recommend reviewing the AAV tagmentation step to determine if the library is over- or underfragmented (Fig. 3b).
Figure 3.
Example of a clean bioanalyzer run of fragmented DNA after successful Fast-Seq library preparation. (a) The fragmented library trace shown was generated from pooling eight AAV library samples following the Fast-Seq protocol, and which generated successful sequencing results. Ideally, the electropherogram will have a flat baseline (indicated) preceding the sample peak and a gradual curve centered on the library product. bp, base pair; FU, arbitrary fluorescence intensity signal. (b) Example of a good ladder from a high-sensitivity bioanalyzer run, showing adequate separation of the control size bands.
(25) While library concentration can be estimated with a Qubit, we highly recommend using qPCR instead as a gold standard method to determine the library concentration. We recommend following the detailed Illumina protocol here:
(26) Following QC validation, libraries were loaded for sequencing according to the manufacture's protocol. For our purposes, we characterized this assay on a MiSeq using a MiSeq Nano V2 300 cycle kit, or a MiSeq Nano V3 150 cycle kit, with 2 × 75-bp paired-end sequencing. Additional information on MiSeq specifications and expected outputs and run times can be found here:
https://www.illumina.com/systems/sequencing-platforms/miseq/specifications.html
Data Analysis
Requirements:
Computer with 2GB RAM to be dedicated to Java heap, ideally 8GB+
Docker
Python 3
GitHub account
(27) We have developed a free open-source computational workflow for analysis of NGS data intended for novices with no prior coding experience. This pipeline uses a simple provided Python script to launch a prebuilt Docker container that contains all the needed sequence processing tools. You can input custom ssAAV transfer vector sequences for alignment of NGS reads. Go to the GitHub repository and find instructions on how to run the preassembled Docker container:
(https://github.com/paulk-lab/FastSeq-pipeline).
*Critical step* Please read the GitHub README file before beginning for important instructions.
Summary
NGS data in fastq format will first be filtered using Trimmomatic13 to remove adapter sequences, low-quality reads (PHRED score <20, or length <50 bp), and unpaired reads. Trimmed reads will then be aligned to the rAAV transfer vector plasmid reference sequence with BWA v0.7.17,14 using the mem algorithm. Alignments will be saved as BAM files, which will then be used to generate Variant Call Format (VCF) files using GATK Haplotype Caller algorithm.15 SNPs and indels identified in VCF files will be filtered using BCFtools filter algorithm,16 with a 10 × depth threshold and a 0.7 allele fraction requirement. A consensus sequence will be generated using BCFtools consensus algorithm. Alignment and fragment distribution statistics will be obtained with Picard tools.17 The code can be modified as needed without the need for incorporation into a new Docker container. The code and tooling are licensed under the Creative Commons with Attribution license.
This code generates an output folder in the data directory, containing sample-specific folders and a compiled .csv file containing NGS statistics for all samples. Each sample-specific folder contains all the files necessary for obtaining alignments and statistics. Of note, please look at the following important files:
BAM alignment file
Inferred consensus sequence in FASTA format
PDF depiction of the library preparation fragment distribution
Raw and filtered VCF file, containing all called SNPs and indels.
Reported statistics include mean coverage, standard deviation of coverage, number of SNPs/indels, mean fragment size, and other relevant metrics. Resultant BAM files can be aligned to the AAV transfer vector plasmid reference sequence and viewed in a genome browser, such as Integrative Genomics Viewer (IGV), which can be downloaded here:
(https://software.broadinstitute.org/software/igv/download).
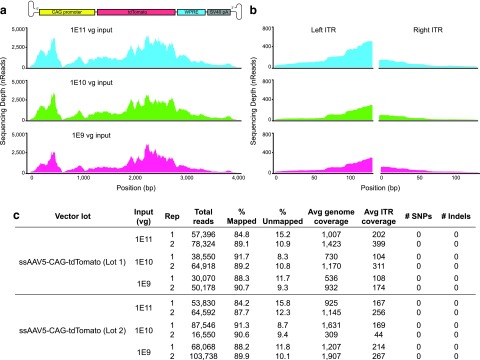
Plots of sequencing depth and coverage can be generated in R using ggplot (Fig. 4), with instructions at:
Figure 4.
Dilution series validates input requirements for sequencing ssAAV genomes and ITRs with Fast-Seq. (a) NGS sequence validation of recombinant packaged ssAAV genomes depicting the expected sequencing depth and coverage from a standard MiSeq run after generating libraries with Fast-Seq. As shown, the coverage distribution will vary along the length of the genome due to bias in Tn5 fragmentation. (b) Zoom-in on the depth and coverage of the corresponding ITRs from (a). (c) Table highlighting the relevant metrics from the dilution series from (a) and (b). Avg, average; rep, replicate; Vg, vector genomes.
Results
For the initial Fast-Seq validation, we extracted, purified, and sequenced genomes from a set of ssAAV-CAG-tdTomato (3.6 kb) vector lots over a dilution series ranging from 1E9 to 1E11 input vector genomes (Fig. 4). This study was completed in biological and technical duplicate by two independent researchers. We found no significant differences in read number, read mapping percentage, or sequencing depth across the input genome dilution series (Fig. 4c, Supplementary Table S2). Owing to known biases in Tn5 transposase insertion,18,19 read depth across the genome is nonuniform, but sufficient for high-confidence base calling. With the exception of a low coverage point in the known difficult GC-rich CAG promoter,20 all samples sequenced to a significant depth, regardless of input dilutions tested. Coverage spanned the entirety of the genome, including both ITRs (Fig. 4b). It is important to note that other common methods of DNA fragmentation like sonication also introduce biases.21 Given that the preference for sequences is not binary, data should not be accepted or rejected based on the presence or absence of such preferred sequences. The focus needs to be on ensuring that there are sufficient reads per AAV sample to ensure that any low coverage region still achieves sufficient depth for high-confidence base calling. Typically this threshold is 15 × 22,23 and Fast-Seq meets these criteria for sequencing AAV genomes.
We then sought to further validate Fast-Seq on ssAAV vectors with different genome compositions (CAG-GFP, EF1α-FLuc), different genome sizes (2.9–4.4 kb), different capsid serotypes (AAV1, AAV2, AAV5, and AAV8), and different production methods (Baculovirus-Sf9 and human HEK293) from both common commercial vendors and academic core facilities supplying academic laboratories. These data are summarized in Supplementary Table S2 and Supplementary Fig. S1. We achieved significant coverage across all vector genomes, despite the many differences in the composition of each AAV lot. In total, we sequenced ssAAV genomes from 15 different vector lots with numerous replicates to thoroughly validate Fast-Seq.
Timeline
-
Isolating pure ssDNA AAV genomes
Steps 1–4: 1.5 h
-
Second-strand DNA synthesis
Steps 5–8: 2 h
*Prepare upcoming solutions during incubation
-
AAV tagmentation with Tn5
Steps 9–13: 30 min
-
Neutralization
Steps 14–15: 5 min
-
Index PCR
Steps 16–19: 1 h
-
Pooling and cleaning AAV libraries
Steps 20–23: 20 min
-
QC libraries
Steps 24–25: 2 h
-
Sequencing (optional)
Steps 25: 10 h
-
Data analysis
Steps 27–30: 1.5 min per sample (assuming Docker is run locally)
Total time: 5.5–17.5 h (depending on whether we run the library QC and sequencing).
Troubleshooting
Final DNA concentration at Step 22 is <2 nM
Remeasure DNA concentration from Step 9 with a Qubit to ensure that concentration is indeed 1 ng/μL. If the concentration is 1ng/μL, consider increasing the input AAV gDNA at Step 9 to 2-5ng/μL or increasing the number of PCR cycles at Step 19 to 15 cycles. Another alternative is to increase the number of samples to be sequenced (we recommend a minimum of eight samples/replicates). If concentration at Step 9 is <1 ng/μL, even without diluting, increase the amount of input AAV. Additional samples can be added to the library by completing the protocol a second time with more samples (be sure to use unique indices), pooling the two libraries together and performing a final 0.7 × SPRI purification, eluting into 30-μL.
Fragment size at Step 25 is far <200 bp
The DNA is overfragmented. Possible causes are: a) input DNA to Step 9 is low quality. QC the starting material to ensure high quality dsDNA by absorbance; b) input DNA to Step 9 is too low. Quantitate DNA from Step 9 again with a Qubit; c) the ratio of DNA to Tn5 is too low. Remeasure DNA concentration from Step 9 again with a Qubit to ensure that concentration is indeed 1-ng/μL. If the concentration is 1-ng/μL, consider increasing the DNA input at Step 9 to 2-5ng/μL or increasing the number of samples to be sequenced. If the issue persists, we recommend requalifying your adapter-loaded Tn5 enzyme.
Fragment size at Step 25 is >800 bp
The DNA is underfragmented. Possible causes are: a) too much DNA was input to Step 9. Quantitate starting DNA from Step 9 again with a Qubit; b) the ratio of DNA to Tn5 is too high. Remeasure the DNA concentration at Step 9 to ensure the concentration is indeed 1-ng/μL. If the concentration is 1-ng/μL, consider increasing the tagmentation time at Step 13 from 10 minutes at 55°C to 20 minutes, or decreasing the number of samples to be sequenced; c) the Tn5 transposase enzyme is inhibited. Requalify the adapter-loaded Tn5 enzyme or check the DNA purification kit (if the suggested Qiagen MinElute Virus Spin kit was not used) for inhibitors.
No library peak at Step 25
This library should not be sequenced. Possible causes are: a) loss of sample during SPRI purification; b) low quality or insufficient input DNA to Step 9; c) loss of sample due to evaporation during steps involving the thermocycler; d) Tn5 transposase enzyme has expired and/or gone through multiple freeze thaws; e) a sharp peak is observed at 100–150 bp which represents adapter dimer that will dominate the run if sequenced. If adapter-dimers are present in the bioanalyzer trace, consult the following SPRI manual for purification strategies:
https://www.beckman.com/reagents/genomic/cleanup-and-size-selection/size-selection
How to qualify each new batch of Nextera adapter-loaded Tn5 enzyme
A dilution series of Nextera adapter-loaded Tn5 enzyme in Tn5 Storage Buffer was performed with template DNA. We have found that 1-ng/μL of Tn5 enzyme is optimal for Fast-Seq. We recommend using this concentration as a middle starting position and testing Tn5 enzyme concentrations above and below this when qualifying batches of Tn5. To assess the efficacy of each dilution following fragmentation, perform a bioanalyzer run on all dilution samples and determine which concentrations of Tn5 yields fragments between 200–800 bp. Choose the lowest concentration that reproducibly works. Additional help with troubleshooting common library preparation problems can be found here:
Supplementary Material
Authors' Contributions
L.M. conceived the idea for the study. L.M., A.P.M., and N.K.P. designed the experiments. L.M., O.S., E.T., S.H., W.T., R.L., N.P.T., and N.K.P. generated reagents, protocols, performed experiments, and analyzed data. L.M. and N.P.T. wrote the code. N.P.T. built the Docker container and GitHub repository. L.M. and N.K.P. wrote the article. L.M. and N.K.P. generated the figures. All authors reviewed, edited, and approved the article.
Disclaimer
The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the funding bodies, respective universities, or organizations.
Author Disclosure
No corporate funding was used for this study. No authors have stock and/or equity in companies with technology related to data in this study. No authors have paid board positions, or accepted travel money, or had paid speaking engagements for companies with technology related to this study. No authors have commercial positions or affiliations related to data in this study. No authors are employed by an organization with a financial stake in data presented in this study. No authors have patents related to the content of this study. All authors, therefore, declare no real or perceived conflicts of interest for the data presented in this study.
Funding Information
This research was primarily supported by funding from the Chan Zuckerberg Biohub, as well as grants to N.K.P. from the NIH (K01-DK107607, U01-HL145795), an American Society of Gene & Cell Therapy Career Development Award, and the Sandler Family Foundation.
Supplementary Material
References
- 1. Nakai H, Yant SR, Storm TA, et al. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. J Virol 2001;75:6969–6976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alexander IE, Russell DW, Spence AM, et al. Effects of gamma irradiation on the transduction of dividing and nondividing cells in brain and muscle of rats by adeno-associated virus vectors. Hum Gene Ther 1996;7:841–850 [DOI] [PubMed] [Google Scholar]
- 3. Tai PWL, Xie J, Fong K, et al. Adeno-associated virus genome population sequencing achieves full vector genome resolution and reveals human-vector chimeras. Mol Ther Methods Clin Dev 2018;9:130–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu Z, Yang H, Colosi P. Effect of genome size on AAV vector packaging. Mol Ther 2010;18:80–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dong JY, Fan PD, Frizzell RA. Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Hum Gene Ther 1996;7:2101–2112 [DOI] [PubMed] [Google Scholar]
- 6. Dong B, Nakai H, Xiao W. Characterization of genome integrity for oversized recombinant AAV vector. Mol Ther 2010;18:87–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tabula Muris Consortium. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 2018;562:367–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hennig BP, Velten L, Racke I, et al. Large-scale low-cost NGS library preparation using a robust Tn5 purification and tagmentation protocol. G3 2018;8:79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Picelli S, Björklund AK, Reinius B, et al. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res 2014;24:2033–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lecomte E, Tournaire B, Cogné B, et al. Advanced characterization of DNA molecules in rAAV vector preparations by single-stranded virus next-generation sequencing. Mol Ther Nucleic Acids 2015;4:e260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Penaud-Budloo M, Lecomte E, Guy-Duché A, et al. Accurate identification and quantification of DNA species by next-generation sequencing in adeno-associated viral vectors produced in insect cells. Hum Gene Ther Methods 2017;28:148–162 [DOI] [PubMed] [Google Scholar]
- 12. Maynard L, Smith O, Tilmans N, et al. Fast-Seq, a universal method for rapid and inexpensive genomic validation of rAAV vectors in preclinical settings v1 (protocols.io.utzewp6) n.d. DOI:10.17504/protocols.io.utzewp6 https://www.protocols.io/view/fast-seq-a-universal-method-for-rapid-and-inexpens-utzewp6
- 13. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011;27:2987–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Picard Tools. By Broad Institute n.d. https://broadinstitute.github.io/picard (last accessed November13, 2018)
- 18. Ason B, Reznikoff WS. DNA sequence bias during Tn5 transposition. J Mol Biol 2004;335:1213–1225 [DOI] [PubMed] [Google Scholar]
- 19. Green B, Bouchier C, Fairhead C, et al. Insertion site preference of Mu, Tn5, and Tn7 transposons. Mobile DNA 2012;3:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kondratov O, Marsic D, Crosson SM, et al. Direct head-to-head evaluation of recombinant adeno-associated viral vectors manufactured in human versus insect cells. Mol Ther 2017;25:2661–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Poptsova M, Il'icheva I, Nechipurenko D, et al. Non-random DNA fragmentation in next-generation sequencing. Sci Reports 2014;4:4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu L, Yavas G, Hong H, et al. Direct comparison of performance of single nucleotide variant calling in human genome with alignment-based and assembly-based approaches. Sci Reports 2017;7:10963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reiber N, Zapatka M, Lasitschka B, et al. Coverage bias and sensitivity of variant calling for four whole-genome sequencing technologies. PLoS One 2013;8:e66621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.