Abstract
Introduction
In light of the emergence of antibiotic-resistant bacteria, phage (bacteriophage) therapy has been recognized as a potential alternative or addition to antibiotics for use in humans in Western medicine.
Areas covered
This review assessed the scientific literature on phage therapy published between January 1, 2007 and October 21, 2019, with a focus on successes and challenges of this prospective therapeutic.
Expert opinion
Efficacy has been shown in animal models and experimental findings suggest promise for safety of human phagotherapy. Significant challenges remain to be addressed prior to the standardization of phage therapy in the West, including the development of phage resistant bacteria; the pharmacokinetics of phage; and any potential human immune response incited by phagotherapy.
Keywords: Phage, phage resistance, antimicrobial resistance, endolysin, enzybiotics, phage therapy, phagotherapy, antimicrobial therapy
1. Introduction
Less than a century after the discovery of penicillin, the emergence of bacteria no longer susceptible to small-molecule antibiotics is recognized as one of the most significant health threats facing modern medicine. Phages (bacteriophages) are among the strategies being evaluated as a potential replacement or adjunct to antibiotics.
Phages are viruses (with single- or double-stranded DNA or RNA genomes) that exclusively infect bacteria. Individual phages are specific for strains of bacterial species. Like other viruses, phages lack a complete replisome and must therefore assume intracellular infection of a host to propagate. Phages undergo receptor-mediated adsorption to the surface of target bacteria prior to injecting their genetic material into the cytoplasm where bacterial replication machinery is subverted to produce new virions [1]. Phages are released following bacterial lysis, thereafter infecting adjacent bacterial hosts. In therapeutic settings, exclusively virulent phages (i.e., those that do not integrate into bacterial genomes) are generally favored over those that can integrate into bacterial genomes due to the predictable time to lysis, and also because host genomic integration and excision of temperate phages risks mobilization or activation of virulence and/or antibiotic resistance genes, and because infection by temperate phages may prevent subsequent phage infection of their bacterial host [2]. A detailed description of the virulent and lysogenic life cycles of phage is found elsewhere [1].
The earliest accounts of phage, circa 1890s, are as storied as they are disputed, particularly, regarding the individual(s) to whom credit for their discovery should be attributed. The majority of sources contend that in 1896, Ernest Hanbury Hankin, a bacteriologist commissioned by the British Commonwealth, observed in the waters of India’s Ganges and Yamuna rivers an inhibitory phenomenon which thwarted growth of Vibrio cholerae [3], and that in 1898, Russian scientist Nikolay Gamaleya made similar observations in his study of Bacillus subtilis [4]. English bacteriologist Frederick Twort is widely remembered as the next noteworthy individual in the phage narrative; in 1915, he observed bacteriophage in bacterial contaminants in experiments with Vaccinia virus, though he ascribed this to a bacterial co-factor negatively impacting cellular viability [2,3]. While this interpretation was ultimately incorrect, Twort’s research greatly contributed to the field’s understanding of phage biology [2]. Finally, a 1917 publication by Felix d’Herelle at the Pasteur Institute describing an “invisible microbe” [5] leads some sources to ascribe the discovery of phage to him; nevertheless, most sources contend that it was d’Herelle who first developed the notion of using phages therapeutically [6].
Seemingly-successful early human applications of phage in cases of pediatric dysentery [7], cholera [4,8,9], and bubonic plague [6,7] stimulated interest in phage therapy across Europe, the former Soviet Union, and the United States. Failed attempts to reproduce positive findings (possibly secondary to an incomplete biological understanding of phage) however, atrophied scientific interest and even inspired opposition from the Council on Pharmacy and Chemistry of the American Medical Association in 1934 [3,6–9]. The discovery of penicillin, and subsequently other antibiotics, alongside tensions between Eastern and Western political powers, further disincentivized phage use in Western medicine at the time, while they continue to be used in Eastern Europe and the former Soviet Union to this day [6,8]. However, the rise of antibacterial resistance has stimulated renewed Western interest in the therapeutic potential of phage, having already been approved for antibacterial applications in the agriculture and food processing industries [10].
Promising human case reports and case series of Western phagotherapy have been published over the past decade [11–13], including the recovery of a diabetic patient from necrotizing pancreatitis with a multidrug-resistant Acinetobacter baumannii infection [14], bacterial eradication from an aortic graft infected with Pseudomonas aeruginosa [15], treatment of multidrug-resistant P. aeruginosa pneumonia in a cystic fibrosis patient [16], and stabilization of a post-lung transplant Mycobacterium abscessus infection in a cystic fibrosis patient [17]. Further, in vitro and experimental animal data have also demonstrated activity of phage against drug-susceptible and -resistant bacteria (Figures 2 and 3, Table 1). Nevertheless, therapeutic failure of phage in a handful of human studies, including two clinical trials [18,19], suggests that their clinical utility requires careful definition [19–21]. Biological challenges surrounding phagotherapy, which constitute the focus of this literature review, include i) the risk of selecting for phage-resistant bacteria, ii) pharmacodynamic and pharmacokinetic complexities, and iii) human host interactions. Potential solutions to these challenges are also discussed.
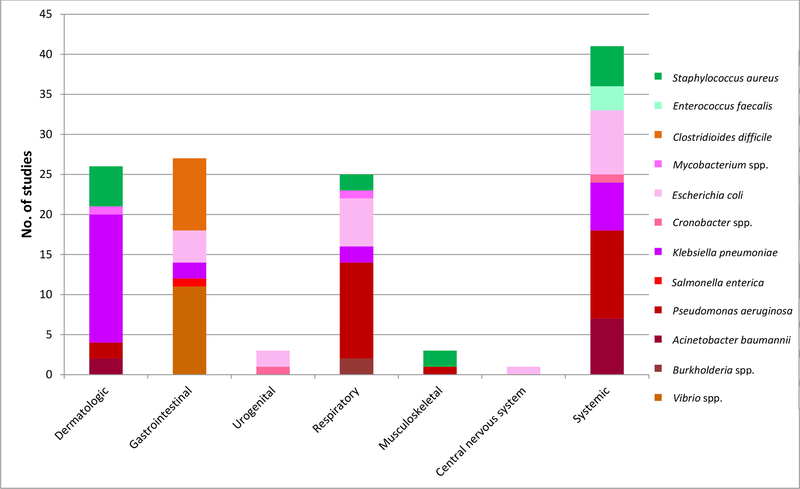
Figure 2.
In vivo phage efficacy studies published between January 1, 2007 and October 21, 2019, by infection and basscterial type.
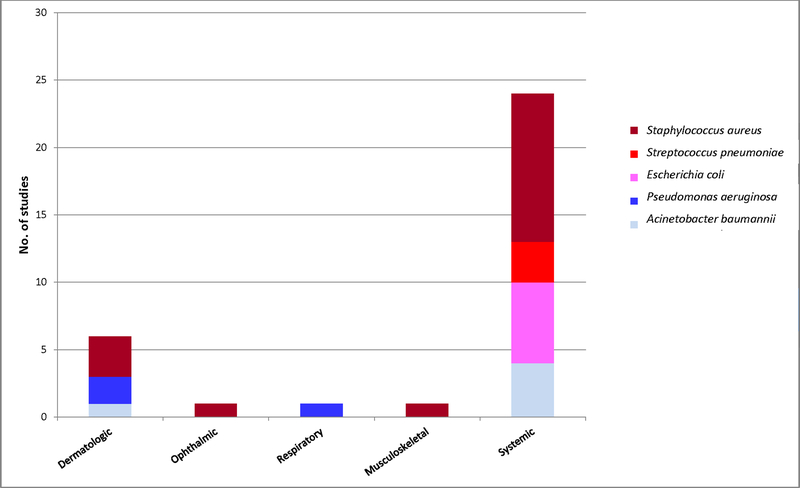
Figure 3.
In vivo lysin efficacy studies published between January 1, 2007 and October 21, 2019, by infection and bacterial type.
Table 1.
Experimental animal and moth larvae model phage efficacy studies published between January 1, 2007 and October 21, 2019.
| Bacterium | Phage | Disease model | Administration (Bacteria [strain(s), if specified]; Phage) | Findings | Reference |
|---|---|---|---|---|---|
| Acinetobacter baumannii | Cocktail: AB-Army1, AB-Navy1, AB-Navy2, AB-Navy3, AB-Navy 4 | Mouse, wound infection | 5×104 cfu AB5075 to wound; 4×109 pfu phage intraperitoneal (IP) and topically (5×109 pfu) 4, 24, and 48 hours post-infection |
Infected, cocktail treated wounds exhibited decreased bacterial abundance at day 5 and wound size at days 9 and 13 compared with AB-Army1-treated and untreated animals | [175] |
| Cocktail: AB-Army1, AB-Navy1, AB-Navy2, AB-Navy3, AB-Navy 4 | Galleria mellonella, systemic infection | Proleg injection (107 cfu AB5075/mL); Proleg injection (107 cfu AB-Army1-primed AB5075/mL) |
95% survival (5 day survival analysis) G. mellonella infected with AB-Army1 primed AB5075 versus 5% survival in wild-type AB5075-infected G. mellonella | [175] | |
| vB_AbaS_D0, vB_AbaP_D2 | Mouse, systemic infection | IP administration (10×LD100, 2×107 cfu AB9/mouse); 100 μL vB_AbaS_D0 (109 pfu/mL), vB_AbaP_D2 (109 pfu/mL), or cocktail (109 pfu/mL) 2 hours post-infection |
0% survival of infected, untreated mice; 50% survival of vB_AbaS_D0-treated mice; 90% survival of vB_AbaP_D2-treated mice; 100% survival of cocktail-treated mice; phage-resistant mutant bacteria sampled from blood 48 hours post-infection demonstrated significant incidence in vB_AbaP_D2-treated mice versus other treatment groups | [24] | |
| vB_AbaM_3090, vB_AbaM_3054 | Mouse, systemic infection | IP administration (6 × 107 cfu FER /100 μL); IP administration of 50mg/kg imipenem, vB_AbaM_3090 and vB_AbaM_3054 alone and in combination (6 ×109 pfu/200 μL) 1 hour post-infection |
At 7 days post-infection, 0% survival of infected, untreated mice, 17% survival of antibiotic-treated mice, and 80–100% survival of monophage and dual phage treated mice, between which differences in survival insignificant | [176] | |
| G. mellonella, systemic infection | Proleg injection (5 ×105 cfu/5μL) [FER]; Proleg injection vB_AbaM_3090 or vB_AbaM_3054 alone or in combination (5 ×107 pfu/10 μL) or imipenem (5 mg/kg) 30 minutes post-infection |
Increased survival among phage-treated and antibiotic-treated groups (83–100% survival at 80 hours post-infection) versus 0% survival of infected, untreated controls | |||
| BΦ-C62 | Mouse (immunocompromised C57BL/6), systemic infection | 1×109 cfu/mL [carbapenem-resistant clinical isolates, n=45] intranasally (IN); Phage IP (MOI 0.1, 1 or 10) 30 minutes post-infection |
0% survival among infected, non-treated mice 3 days post-infection; dose-dependent effects on survival in phage treated mice at 3 days post-infection: 100% at MOI=10, 50% at MOI=1, 16% at MOI=0.1; reduced amounts of bacteria in lungs of phage-treated mice between 6- and 9-fold relative to infected, untreated mice at day 1 post-infection | [177] | |
| Burkholderia pseudomallei | C34 | Mouse, melioidosis | 100 cfu [clinical isolates, n=43] IN; 2×108 pfu 24 hours before or 2 hours after infection |
Phage improved survival to 33% from 0% in controls, with no differences between pre- and post-treated groups, and extended median survival to 13 days in pre-treated and 11 days in post-treated mice compared to 8 days in controls | [178] |
| Burkholderia cenocepacia | BcepIL02 | Mouse, lung infection | 107–108 AU0728 or K56–2 cfu intratracheally; Phage (MOI=100) IN or IP 24 hours post-infection |
Phage treatment (IP) decreased AU0728 bacterial loads in lung but not K-562 bacterial loads under same treatment conditions | [179] |
| Clostridioides difficile | ΦCDHM1 | Hamster, gastroenteritis | 104 cfu/mL spores [CD105HE1] per os (PO) every 8 hours for up to 36 hours; 1×108 pfu/mL individual or mixed phage PO |
Reduction in C. difficile in treated versus control animals with extended survival in treated animals; 2- and 4-phage combination-treated animals exhibited comparable bacterial reductions of 2 and 4 log10 in gut epithelium and lumen, respectively; 4-phage combination prolonged time to death versus infected, untreated animals | [180] |
| ΦCDHM2 | |||||
| ΦCDHM3 | |||||
| ΦCDHM4 | |||||
| ΦCDHM5 | |||||
| ΦCDHS1 | |||||
| ΦCDHS1-ΦCDHS12 | |||||
| ΦCDHS5-ΦCDHS6 | |||||
| Cocktail: ΦCDHM1, ΦCDHM2, ΦCDHM3 ΦCDHM4 ΦCDHM5, ΦCDHS1 | |||||
| Cronobacter sakazakii | vB_CsaM_GAP161 | G. mellonella, systemic infection | 10×105 cfu HPB 3253 (5× LD50) injection; Phage injection, site unspecified (MOI 8) at 1 and 0.5 hours pre-infection, and 0, 1, 2, 4 hours post-infection |
Pre-treatment or simultaneously-treated larvae increased survival versus infected, untreated controls; post-treatment group comparable survival to controls | [164] |
| Cronobacter turicensis | Cocktail: P2, D2 | Mouse, urinary tract infection | Bacterial-seeded (1×1011 cfu 290708/07/mL) transurethral catheter; Immediate IP phage treatment (1011 pfu/mL) |
Renal but not bladder bacterial colonization reduced in phage-treated animals | [181] |
| Escherichia coli | T4 | Mouse, urinary tract infection | 5×109 cfu uropathogenic E. coli (ECU5) transurethrally; Phage IP at various MOI concurrent with bacterial inoculation |
Bacterial inoculum 100% lethal in untreated mice; phage (MOI 60) rescued 100% mice | [182] |
| Unspecified | Mouse, gastroenteritis | 200 μL of 2 ×108 cfu/mL PO; PO administration of ciprofloxacin (160 μL of 0.5 g/mL), phage (200 μL/10 g of 2 ×109 pfu/mL), or a combination 24 hours post-infection, or phage 24 hours pre-infection |
No weight or behavioral changes, or bacterial detection in mice treated with phage 24 hours before or after infection; weight loss and behavioral changes noted in antibiotic or combination-treated animals secondary to dysbiosis, though no bacterial detection observed in these groups | [183] | |
| KEP10 | Mouse, urinary tract infection | 5×109 cfu [uropathogenic E. coli, ECU5] transurethrally; Phage IP at various MOI concurrent with bacterial inoculation |
Bacterial inoculum 100% lethal in infected, untreated mice; phage (MOI 60) rescued 90% mice | [182] | |
| T4 | Rat, gastroenteritis | 105–107 cfu ATCC 11303 PO; 3.6×107 pfu/mL transdermal application |
83% phage-treated versus 0% untreated rats survived | [184] | |
| 536_P1 | Mouse, lung infection | 1×107 or 4×107 cfu 536-lux IN; Phage (MOI 0.3 or 3) IN 2 hours post-infection |
Phage rescued 100% animals from death, compared with 25% survival in infected, non-treated controls; mortality reduction from 80% to 25% with the use of an adapted phage | [185] | |
| 536_P7 | Mouse, lung infection | 1.5×107 cfu PDP302 IN; Phage (MOI 10) IN 2 hours post-infection |
Phage rescued 20% animals from death compared with 12% survival in infected, non-treated controls; phage adaptation increased survival to 75% from 20% | [185] | |
| K1-ind1 | Mouse, systemic infection | 2–3×108 cfu CAB1 or CAB281 IM; 102–108 phage intramuscularly (IM) administered concurrently |
K1 capsule-dependent phages yielded 6 log10 reduction (specimen unspecified) following minimum treatment dose versus K1 capsule-independent phages | [186] | |
| K1-ind2 | |||||
| K1-ind3 | |||||
| K1H | |||||
| K1G | |||||
| K1E | |||||
| K1–5 | |||||
| Cocktail: EcD7, V18, SE40, SI3, CH1, Lm1, ST11 | Mouse, gastroenteritis | 5×107 cfu K12 C600/mL daily for 3 days (route unspecified); 106 pfu/mL PO concomitantly and up to 24 hours post-infection |
Bacteria in stool of untreated mice at 104 cfu/g stool, with no bacteria in phage-treated mice | [187] | |
| Cocktail: CLB_P1, CLB_P2, CLB_P3 | Mouse, gastroenteritis | 55989Str PO (dose unspecified); 3×108–1010 pfu/mL PO for 24 hours days 3 to 4 post-infection |
Bacterial colonization in ileum treated mice reduced by 88% versus controls, although rebound occurred such that median bacterial density was similar across control and treated groups by day 7 post-infection | [188] | |
| EC200PP | Rat, systemic infection | 103–106 cfu S242 (ciprofloxacin-resistant clinical isolate)/mL IP; 108 pfu subcutaneously (SQ) 7 or 24 hours post-infection |
100% rescue and bacterial elimination in blood with 7 hour post-infection treatment; 50% rescue with 24 hour post-infection treatment | [189] | |
| EC200PP | Rat, meningitis | 106–108 cfu S242 (ciprofloxacin-resistant clinical isolate)/mL intrathecally; 108 pfu IP 1 or 7 hours post-infection |
100% untreated meningitis-induced rats died by 36 hours post-infection, treatment with 108 pfu 1 or 7 hours post-infection rescued 100% | [189] | |
| Enterococcus faecalis | ΦEF24C | Mouse, systemic infection | 109–1010 cfu EF14 or VRE2/mL IP (LD100); Range of MOI (100, 10, 1, 0.1, 0.01, 0.001, 0.0001) administered once, IP beginning 20 minutes post-infection |
Phage rescued 100% at MOI 10, 1, 0.1, and 0.01 compared to 0% survival of controls; MOI 0.001 and 0.0001 did not impact survival | [190] |
| SHEF2 | Zebrafish, systemic infection | 3 ×104 OS16 cfu via embryonic microinjection 2 nL (MOI 20) SHEF2 via embryonic microinjection 2 hours post-infection |
84% survival of phage-treated individuals versus 27% survival of infected, non-treated controls at 72 hours post-infection | [191] | |
| EF-P29 | Mouse, systemic infection | 2 ×109 cfu VREF 002 IP (2× LD100); 4×103, 4×104, 4×105, 4×106, 4×107pfu IP 1 hour post-infection |
4×105 and 4×106 pfu phage rescued 100% mice | [192] | |
| Klebsiella pneumonaie | K01 | Mouse, wound infection | 10×106 cfu B5055/mL SQ; Phage SQ or IP (dose unspecified) 30 minutes and 6 hours post-infection |
Bacterial colonization in blood, lung, peritoneum reduced 3+ hours post-infection with IP or SQ phage | [193] |
| Kpn5 | Mouse, burn wound infection | 108 cfu B5055topically (LD100); MOI 1 or 200 topically 4, 12 and 24 hours post-infection |
Survival of low-and high-dose phage-treated animals 0 and 66%, respectively | [194] | |
| vB_KpnP_K1-ULIP33 | G. mellonella, systemic infection | Proleg injection (104 cfu SA12/10 μL); Phage administered via proleg injection (MOI 10) 1 hour pre- or post-infection |
0–30% infected, untreated larvae; 0–30% uninfected, treated larvae; 90% larvae administered phage prophylactically; and 100% larvae treated after bacterial inoculation survived 4 days post-infection | [195] | |
| vB_KpnP_KL106-ULIP47 | Proleg injection (103 cfu 2198/10 μL); Phage administered (vB_KpnP_KL106-ULIP47 and vB_KpnP_KL106-ULIP54 alone and combined) via proleg injection (MOI 10) 1 hour pre- or post-infection |
0–10% infected, untreated larvae; 0–30% uninfected, treated larvae; and 80–100% treated larvae survived 4 days post-infection; no significant difference between survival rates of monophage- versus polyphage-treated larvae | |||
| vB_KpnP_KL106-ULIP54 | |||||
| Cocktail: vB_KpnP_KL06-ULIP47, vB_KpnP_KL1o6-ULIP54 | |||||
| Kpn5 | Mouse, burn wound infection | SQ B5055 (LD100); MOI 1 IP immediately following establishment of wound infection |
Survival 80–100% 72 hours post-infection in each treatment group treated with cocktail and Kpn5 alone achieving similar survival | [196] | |
| Kpn12 | |||||
| Kpn13 | |||||
| Kpn17 | |||||
| Kpn22 | |||||
| Cocktail: Kpn5, Kpn12, Kpn13, Kpn17, Kpn22 | |||||
| Cocktail: KØ1, KØ2, KØ3, KØ4, KØ5 | Mouse, burn wound infection | 105 cfu B5055/mL SQ; MOI 1 IP (liposome- or un-encapsulated phage) 30 minutes post-infection |
Encapsulated phages circulated systemically six times longer than un-encapsulated phages; 100% 7-day survival of animals treated 24 hour post-infection, liposome encapsulated, phage-treated mice compared with 0% 7-day survival of 24 hour post-infection, un-encapsulated phage-treated mice; 100% survival of encapsulated and un-encapsulated treatment groups when administered 30 minutes post-infection | [106] | |
| ΦNK5 | Mouse, liver abscess | 2×108 cfu NK-5 intragastric (IG); 2×105, 2×106, 2×107, 2×108 pfu IP or IG 0.5, 6 or 24 hours post-infection |
IP phage 30 minutes post-infection yielded dose-dependent rescue, with 100% surviving following ≥107 pfu treatment and 30% surviving following 105 pfu treatment; similar dose-dependent effects observed in IG phage 30 minutes post-infection with 100% survival following ≥106 pfu; IP administration supported increased survival at both 6- and 24 h post-infection time points relative to IG dosing | [197] | |
| SS | Mouse, lung infection | 108 cfu B5055 IN; MOI 200 IP administered concomitantly or 6 or 24 hours post-infection compared with phage (1010 pfu/mL) plus amikacin (3.75 mg/25 g) |
No difference in pulmonary bacterial abundance between control and phage-treated mice, or between combination- and phage alone-treated animals | [198] | |
| Kpn1 | Mouse, wound infection | 107 cfu B5055/mL topically; MOI 10 topically 6 hours post-infection |
Reduction in bacterial density on days 1–7 following cocktail treatment only | [38] | |
| Kpn2 | |||||
| Kpn3 | |||||
| Kpn4 | |||||
| Kpn5 | |||||
| Cocktail: Kpn1, Kpn2, Kpn3, Kpn4, Kpn5 | |||||
| Kpn5 | Mouse, burn wound infection | 108 cfu B5055 to full thickness burn (LD100); MOI 200 topically as single dose immediately post-infection, compared with silver nitrate or gentamicin daily beginning 24 hours post-infection |
7 day survival 57% and 17% following 0.5% and 0.0005% silver nitrate treatment, respectively; 53% and 13% with 1 g/L and 7 mg/L gentamicin, respectively; 63% with phage; survival in phage-treated versus high dose silver nitrate or gentamicin not different | [199] | |
| NTUH-K2044-K1–1 | Mouse (BALB/cBy1), liver abscess | 3×102 cfu NTUH K2044 IP; 1×108 pfu IP 16 or 24 hours later |
Increase in TNF-α and IL-6 in blood, liver and spleen; survival increased in mice treated with phage 16- or 24 h post-infection compared with controls | [200] | |
| SS | Mouse, lung infection | 108 cfu B5055 IN; IP phage (MOI 200) administration before, after, or concurrent with bacterial inoculation; or combination of IP phage (1010 pfu/mL) plus amikacin (3.75 mg/25 kg) concurrent with bacterial inoculation |
No effect of phage alone or with amikacin | [198] | |
| Mycobacterium tuberculosis | D29 | Mouse, lung infection | Low inoculum (50–100 cfu) or ultra-low inoculum (5–10 cfu) H37Rv inhalation; ≈107 pfu/mouse via inhalation 30 minutes prior to bacterial challenge |
Significant decrease in lung bacterial load at 24 hours and 3 weeks post-infection, but no difference in splenic bacterial load at 3 weeks between low inoculum and untreated groups; while significant decrease was observed at 24 hours post-infection in lungs of treated animals receiving ultra-low inoculum relative to bacterial load versus untreated animals | [201] |
| Mycobacterium ulcerans | D29 | Mouse, Buruli ulcer | 5.5×1010 cfu 1615 isolate footpad injection; 8 ×1010 pfu SQ 33 days post-infection |
Decrease in bacterial abundance in phage-treated animals 68 days post-infection | [202] |
| Pseudomonas aeruginosa | vB_PsaP PAT14 | Rat, foreign body osteomyelitis | Biofilm-coated IV catheter placed into tibial medullary canal; 16 days post-infection, 107 pfu local phage administration for 3 days alone or in combination with IP imipenem (120 mg/kg) and amikacin (25 mg/kg) daily for 14 days |
Antibiotic- and phage-antibiotic-treatment decreased bacterial abundance without affecting biofilm thickness | [203] |
| PAK_P1, PAK_P2, PAK_P3, PAK_P4, PAK_P5, LBL3, LUZ19, PhiKZ | Mouse, lung infection | 1×107 cfu PAK-lumi IN; Phage IM 2 hours post-infection |
Survival: 75–100% (PAK_P1–5), 50% (LBL3), 37% (LUZ19), 15% (PhiKZ); PBS control achieved survival levels “similar” to those of PhiKZ | [204] | |
| Cocktail (CT-PA): Pa193, Pa204, Pa222, Pa223 | Sheep, sinusitis | 2 mL of 108 cfu Aus20 (clinical isolate)/mL frontal sinus inoculation; Nasal rinse twice-daily for 7 days post-infection of 108, 109, or 1010 pfu/mL or saline |
Phage dose-dependent antibacterial effect versus untreated controls, but no differences among groups treated with different phage concentrations | [205] | |
| Cocktail: vB_PaeP_PYO2, vB_PaeP_DEV, vB_PaeM_E215, vB_PaeM_E217 | Zebrafish embryos, systemic infection | 30 cfu PA01 in duct of Cuvier; 2 μL of 5×108 pfu/mL and/or ciprofloxacin (100 mg/mL) 30 minutes or 7 hours post-infection |
Antibiotic and phage treatment alone reduced mortality versus untreated embryos; phage-antibiotic synergy observed; treatment at 30 minutes and 7 hours post-infection exhibited similar effects | [206] | |
| KPP10 | Mouse, systemic infection | 108 cfu/mL D4 PO 1010 pfu PO 1 day before, or 1or 6 days after infection |
Significantly higher survival among 1 day post-infection treated mice than infected, untreated controls, while treatment 1 day pre-infection or 6 days post-infection did not significantly rescue animals versus infected, untreated controls; reduction in fecal shedding of bacteria with phage delivered 1 or 6 days post-infection; reductions in bacterial organ colonization and IL-1β, IL-6, TNF-α inflammatory cytokines 1 day post-infection | [207] | |
| 2.4–300×106 cfu D4 IP; 1010 pfu administered IP 1 day prior, 6 hours after, or concurrently with bacterial inoculation |
Phage administered concurrently with bacterial inoculum (but not 1 day prior or 6 hours post-infection) statistically improved survival rates versus infected, untreated controls | ||||
| PAK_P1 | Mouse, lung infection |
P. aeruginosa PAKlumi 105 or 107 cfu IN; MOI 10 2 hours after bacterial challenge or MOI 100 4 days before bacterial challenge |
Administration 2 hours post-infection rescued 100% of immunocompetent mice; administration 4 days pre-infection rescued 100% of immunocompetent and >90% of lymphocyte-deficient mice; neutropenic mice had 0% survival with phage treatment | [208] | |
| MPK1 | Drosophila melanogaster, systemic infection | 107 cfu PA01/mL injection; 5×107 pfu PO |
Administration of MPK1 and MPK6 to infected D. melanogaster delayed death compared to controls | [209] | |
| MPK6 | Mouse, systemic infection | 2×106 PA01 cfu IP; 106–107 pfu IP or IM 6–12 hours post-infection |
Approximately 1–4 log10 reduction in bacterial loads in lung, spleen and liver 24 hours post-infection wth IP or IM with MPK1 or MPK6 compared with controls | ||
| ΦKZ | G. mellonella, systemic infection | 5×105 cfu PAO1 IP; Phage injection immediately following infection and every 12 hours thereafter |
All phage administrations resulted in prolonged time to death versus infected, untreated controls, with phage cocktail achieving longest mean survival | [210] | |
| Cocktail: 14/1, ΦKZ, PNM, PT7 | 5×105 cfu PAO1 IP; Cocktail injection immediately following infection and every 12 hours thereafter |
||||
| 14/1, PT7, ΦKZ, PNM | 5×105 cfu PAO1 IP; Phage injection sequentially immediately following infection and every 12 hours thereafter |
||||
| PAK-P1 | Mouse, lung infection | 1×107 cfu PAKlumi IN; MOI 0.1, 1 and 10 IN 24 hours before and 2,4, or 6 hours after infection |
Phage resulted in dose-dependent increase in survival, with 100% animals receiving MOI 10 phage surviving to end of 12 day experiment and 80% receiving MOI 1 surviving to same point; treatment delays longer than 2 hours post-infection did not rescue all mice | [211] | |
| CSV-31 | Mouse, systemic infection | 107 cfu YFN-58(clinical isolate) IP (lethal dose); 104, 108, 109 pfu IP 45 minutes after infection |
Phage rescued 100% infected mice when administered as late as 5 hours post-infection at 109 pfu | [212] | |
| Cocktail (composition unspecified) | G. mellonella, systemic infection | 10 or 100 cfu PAO1 hemolymph injection; Phage hemolymph injection 2 hours pre- (MOI 0.1, 1, 10, 100) or post- (MOI 0.1, 1, 10) infection |
Survival improved in pre-treated (90% MOI=100 infected with 10 cfu and 80% MOI=100 infected with 100 cfu) versus post-treated (40% MOI=10 infected with 10 cfu and 20% MOI=10 infected with 100 cfu) | [213] | |
| GNCP | Diabetic mouse, systemic infection | 3×108 cfu/mL IP; 3×1010, 109, 108, 107, 106, and 0 pfu IP ± imipenem IP (30 mg/kg) 20 minutes after infection, or delayed 0, 1, 2, 3, 4, or 6 hours post-infection |
Phage (3×106–3×108 pfu) rescued 90% diabetic and non-diabetic mice from lethal bacteremia versus 20% single-dose imipenem-treated diabetic mice; treatment delays up to 8 hours post-infection rescued fewer diabetic and non-diabetic animals, with 20+ hour treatment delay rescuing 10% non-diabetic mice and 0% diabetic mice | [214] | |
| Cocktail: 1 P. aeruginosa 24, P. aeruginosa 25, P. aeruginosa 7 | Mouse, lung infection | 2.5×106–5×108 cfu/mL IN; 1.2×109 pfu IN simultaneously with infection, 48 hours post-infection or 24 hours pre-infection |
100% mice administered phage simultaneously with bacterial challenge cleared infection, while 5/7 and 6/8 treated mice cleared infection in groups administered phage pre- and post-infection, respectively; all infected, untreated mice exhibited systemic infection | [215] | |
| Cocktail: Pa1, Pa2, Pa11 | Mouse, burn wound infection | 2–3×102 cfu PA01Rif SQ; 3×108 pfu IP, IM, or SQ concurrent with bacterial inoculation |
6% infected, non-treated mice died, while 28%, 22% and 88% infected mice survived when treated with phage IM, SC, or IP, respectively | [216] | |
| Cocktail: ΦBHU49, ΦBHU61, ΦBHU83, ΦBHU89, ΦBHU98, ΦBHU2255, ΦBHU7799, ΦBHU10858, ΦBHU10956, ΦBHU10958, ΦBHU10976 | Mouse, catheter biofilms | SQ biofilm-coated (106 cfu) catheter; Daily 10 μL 107 pfu/mL SQ beginning day of catheter placement for 10 days |
Infected, treated mice exhibited decreased colonization versus untreated animals | [217] | |
| PA10 | Mouse, systemic infection | 104–107 cfu PA01 IP to immunocompetent or 103–105 cfu to neutropenic mice; IP phage (MOI 1, 10, or 100 in immunocompetent l mice; MOI 10 in neutropenic mice) concurrent with bacterial inoculation |
100, 100 and 80% immunocompetent mice treated with MOI 100, 10 or 1 phage, respectively, survived; 0% immunocompromised mice (treated and untreated) survived 48 hours post-infection | [218] | |
| Cocktail: Phagoburn | Rat, endocarditis | 108 cfu CHA; 1010 pfu/mL by bolus or continuous IV administration, IV ciprofloxacin bolus (20 mg/kg), or phage and ciprofloxacin administered by IV bolus 18 h post-infection |
Phage-antibiotic combination resulted in bacterial clearance of 7/11 rats compared with 0/28 rats receiving phage or ciprofloxacin alone | [219] | |
| Salmonella enterica | 1 phage (unspecified) | Mouse, gastroenteritis | 1.5×108 cfu/mL oral gavage; 2×109 pfu/mL at time of infection; 4, 7 and 10 days post-infection; or 4 days pre-infection; ciprofloxacin (0.5 g/mL thrice over 24 hours) evaluated alone and in combination with phage |
S. enterica isolated from stool of untreated mice with all other groups exhibiting absence of S. enterica in stool as early as 7 days post-infection | [220] |
| Staphylococcus aureus | MR-10 | Mouse, wound infection | 105, 106, 107, 108 cfu ATCC 43300/mL hindpaw injection MOI 100 (108 pfu/mL), MOI 100 (108 pfu/mL) + 25 mg/kg PO linezolid or linezolid alone |
Mice receiving phage alone or combination therapy scored similarly in assessment of lesion quality and localized edema with improvement versus untreated, infected controls | [221] |
| Cocktail: 2003, 2002, 3A, K | Rat, lung infection | LD100 (6–8 × 109 cfu) administered intratracheally following 4 hours mechanical ventilation; IV administration of teicoplanin (3 mg/kg), phage cocktail (109 pfu/mL of each of 4 phages), or combination teicoplanin and phage cocktail 2, 12, 48 and 72 hours post-infection |
Each treatment group exhibited similar bacterial loads with significant reduction versus infected, sham-treated controls; non-significant differences in cytokine levels across test groups except for increase in IL-1β levels among infected and non-infected animals treated with phage versus non-phage treated, and increase in IL-6 levels in infected, untreated rats versus uninfected, sham-treated | [222] | |
| P-27/HP | Mouse, systemic infection | 5×108 cfu IP; 107 pfu phage SQ 24 hours post-infection |
Infected, untreated mice 4 log10 bacteria in spleen versus phage-treated mice 2 log10 cfu bacteria in spleen 3 days post-treatment | [223] | |
| Cocktail: (composition unspecified) | Rabbit, wound infection | Wounds inoculated with 100 μL 1.5×108 cfu /mL; 2×108 pfu topical phage immediately before (prevention) or after (treatment) bacterial challenge |
Infected, phage-treated rabbits (both treatment and prevention groups) higher rate of wound healing and lower bacterial wound colonization than infected, untreated animals | [224] | |
| AB-SA01 cocktail | Mouse, lung infection | 3×108 cfu Xen29 IN; 50 μL vehicle or 5×108 pfu IN 2 and 6 hours post-infection or SC vancomycin (110 mg/kg) 2, 6, and 12 hours post-infection |
Phage-treated animals exhibited reductions in bacteria in lung versus untreated animals with comparable amounts to antibiotic-treated animals | [225] | |
| Sb-1 | Rat, foreign body osteomyelitis | Biofilm-coated IV catheter in tibial medullary canal; 16 days post-infection, local phage (107 pfu) for 3 consecutive days alone or with teicoplanin (20 mg/kg) IP daily for 14 days |
Phage-antibiotic-treated group diminished bacterial load compared with control group and other treatment groups; biofilm thickness similar between treatment and control groups | [203] | |
| Stau2 | Mouse, systemic infection | Bacterial challenge (OD600=0.5) S23 IP; MOI 0.1, 1, 10, and 100 IP 0, 30, 60 minutes post-infection with assessment 7 days following infection |
0/10 infected, untreated animals; 10/10 infected, treated receiving phage 0 minutes post-infection (MOI 100); 3/5 infected, treated receiving phage 30 minutes post-infection (MOI 100); 2/5 infected, treated receiving phage 60 minutes post-infection (MOI 100) survived | [226] | |
| SATA-8505 | Mouse, skin and soft tissue infection | 107 cfu USA 300 SQ to immunocompetent or immunosuppressed [chronic granulomatous disease (CGD) model] mice; MOI 1 and 10 IP immediately preceding infection |
MOI 1 reduced lesion size in CGD but not wild-type mice versus respective untreated counterparts with no differences in cfu/lesion among either wild-type or CGD mice; MOI 10 did not influence lesion size among wild-type or CGD mice compared with, untreated counterparts; reduced cfu/lesion among CGD but not wild-type treated mice | [227] | |
| MSa | Mouse, systemic infection | 106–109 cfu A170 SQ, or 5×106 cfu A170; 107–109 pfu SQ concurrent with bacterial inoculation, or 109 pfu IV 10 days post-infection |
Dose-dependent effect of phage with 100% survival in 109 pfu-treated mice versus 40% of 108 pfu-treated, concurrently infected, mice; 100% of 10 day post-infection treated mice exhibit sterility of blood, spleen, kidneys, and heart on day 20 post-infection versus 0% of infected, untreated mice | [228] | |
| MSa | Mouse, local infection | SQ abscess induction with 107 cfu A170; SQ 109 pfu concurrent with bacterial inoculation or 4 days post-infection |
Concurrent phage administration prevented abscess formation in 100% of treated mice; 4 day post-infection phage minimized abscess biomass and bacterial colonization | [228] | |
| S13 | Mouse, systemic infection | 6.4×108 cfu SA27/mL IN; 1010 pfu/mL IP 6 hours after infection |
Phage-treated animals higher survival at day 14 post-infection and decreased bacterial loads in spleen and liver at day 2 post-infection compared with controls | [229] | |
| A5 | Mouse (CBA mice administered busulfan PO and cyclophosphamide IP and syngeneic bone marrow transplant), systemic infection | 1×107 cfu L IV 1×106 pfu IP 4 days post-bone marrow transplant, 30 minutes prior to bacterial challenge |
Phage-treated mice had decreased splenic and hepatic bacterial loads versus controls | [230] | |
| Cocktail: SA-BHU1, SA-BHU2, SA-BHU8, SA-BHU15, SA-BHU21, SA-BHU37, SA-BHU47 | Rabbit, osteomyelitis | 5×106 cfu/mL femur osteomyelitis; 5×1012 pfu/mL beginning 3 weeks or 6 weeks post-infection with 4 doses locally administered 48 hours apart |
Wound swabs culture-negative at conclusion of phage dosing (3 weeks post-infection) in 3 week post-infection treatment group and at conclusion of phage dosing (8 weeks post-infection) in 6 week post-infection treatment group | [231] | |
| Unspecified | Rabbit, wound infection | 106 cfu UAMS-1 or UAMS-929 into wound; 3 days post-infection debridement only, phage only (MOI 1), or phage (MOI 1)-debridement combination every other day for 6–12 days |
Reductions in bacterial counts in debridement-phage combination group only | [232] | |
| Vibrio cholerae | ICP1 | Mouse, gastroenteritis | 5×105 cfu AC53 PO; 106–107pfu PO 3 hours before infection |
Bacterial reductions of at least 2 log10 with phage | [233] |
| ICP2 | |||||
| ICP3 | |||||
| Cocktail: ICP1, ICP2, ICP3 | 5–9×105 cfu or 1×108 cfu AC53 PO; 3×107–3×108 pfu PO 6, 12, or 24 hours before infection |
Reductions in V. cholerae with treatment 6 and 12 hours prior to low-dose challenge and 6, 12, and 24 hours prior to high-dose challenge | |||
| Cocktail: ICP1, ICP2, ICP3 | Rabbit, gastroenteritis | 5×108 cfu AC53 PO; 109 pfu PO 3 or 24 hours before infection |
Reductions in V. cholerae with treatment 3 and 24 hours prior to challenge | ||
| Cocktail: ATCC 51352- B1, -B2, -B3, -B4, -B5 | Rabbit, gastroenteritis | 1×109 cfu ATCC 51352/mL PO; 1×108 pfu/mL (total MOI 0.1) 6 or 12 hours before or 6 or 12 hours after infection |
Phage pre-treatment no effect on fecal shedding of V. cholerae; phage treatment 6 hours post-infection led to reduction in f V. cholerae shedding between 12 and 60 hours post-infection, while phage treatment 12 hours post-infection generated no bacterial reduction over this timespan; infected, untreated controls developed severe diarrhea, pre-treated rabbits developed moderate diarrhea, and post-treated rabbits developed no diarrhea | [234] | |
| Cocktail: ATCC 51352-B1, -B2, -B3, -B4, -B5 | Mouse, gastroenteritis | 1×109 cfu ATCC 51352/mL PO; 1×108 pfu/mL daily PO, ciprofloxacin (40 mg/kg) daily, or reduced osmolarity oral rehydration solution daily starting 24 hours after infection for 3 days |
Ciprofloxacin reduced bacterial counts by more than 2 log10 compared with phage cocktail; both decreased IL-6 and TNF-α levels | [235] | |
| JSF4 | Mouse, gastroenteritis | Human cholera (104–105 cfu) stools - ID50 analysis and competition assay; Phage-containing and phage-free human stool |
Infectious dose (ID50) 10-fold higher in mice inoculated with phage-containing stools versus phage-free stools | [236] | |
| Cocktail: ATCC 51352-B1, -B2, -B3, -B4, -B5 | Rabbit, gastroenteritis | 1×109 cfu MAK 757 intrajejunally; 1×108 pfu concurrently administered intrajejunally with bacterial inoculum |
Administration phage cocktail with bacteria reduced bacterial load, prevented symptom progression and minimized histopathologic findings (intestinal villi morphology, immune cell invasion) | [237] | |
| Phi_1 | Rabbit, gastroenteritis | 5×108 cfu O1 1051 SmR in NaHCO3 Ranitidine (5 mg/kg IP) followed by bacterial challenge and 1×109 pfu PO |
11 of 17 control animals and 0 of 19 experimental animals symptoms by 24 hours post-infection; decrease in intestinal and cecal fluid bacterial load in phage-treated versus control animals | [238] | |
| Vibrio parahaemolyticus | pVp-1 | Mouse, gastroenteritis | 2×106–107 cfu CRS 09–17 (clinical isolate) IP or PO (LD50); 2×108 pfu PO or IP treatment 1 hour post-infection |
56% (IP) and 52% (PO) infected, untreated mice died by 36 hours post-infection, while 8% and 16%, respectively, died in treated groups | [239] |
1.1. Review methods
The search strategy consisted of Embase, Cochrane Central Register of Controlled Trials (CCTR), Cochrane Database of Systematic Reviews (CDSR), Ovid MEDLINE, Ovid MEDLINE Daily, Ovid MEDLINE In-Process, Epub Ahead of Print and Other Non-Indexed Citations database surveys for articles published between January 1, 2007 and October 21, 2019 (Supplemental Table).
2. Biological constraints of phage therapy in humans
2.1. Bacterial resistance to phage
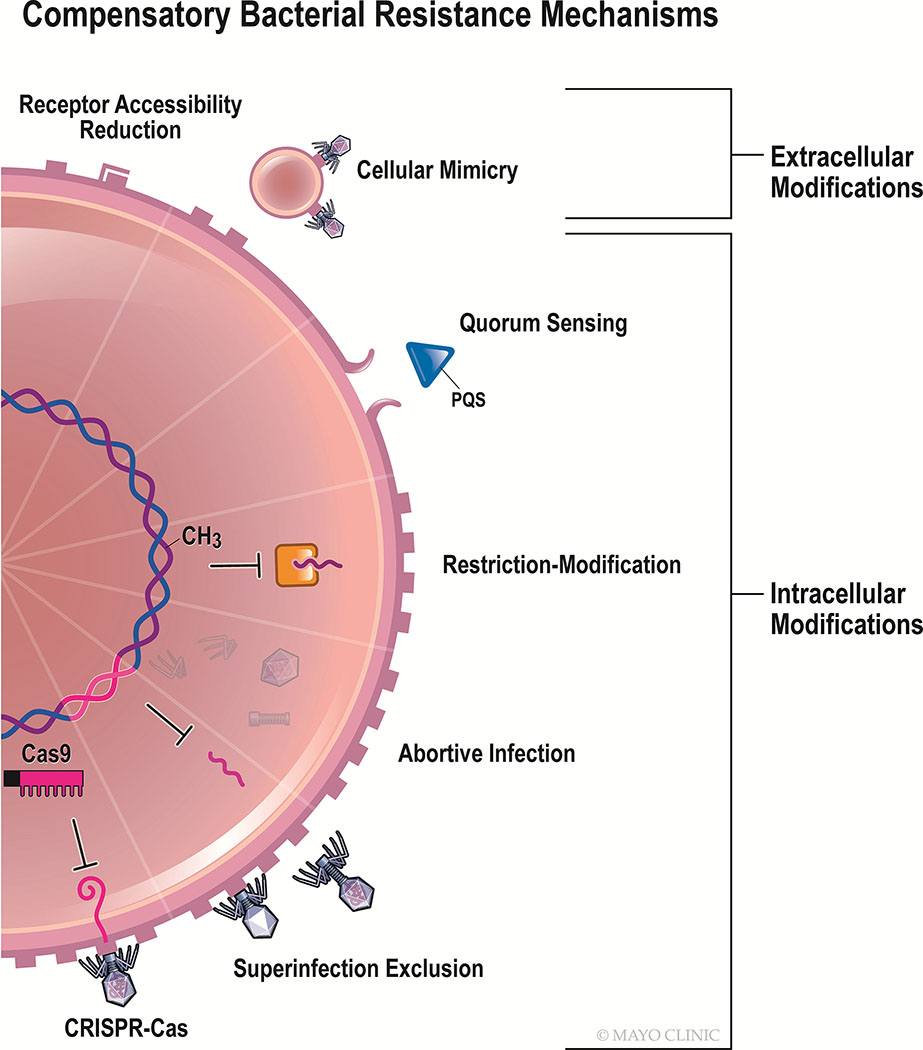
Community dynamics underlying bacterial resistance are complex when the antibacterial agent is biologically dynamic, as occurs in phagotherapy. Unlike small molecule antibiotics, phages are shaped by environmentally-mediated competitive selection in addition to exerting the same upon bacteria. This bidirectional interaction results in co-evolution of bacteria and viruses as each contends for survival. To this end, the former have developed an array of approaches to withstand phage infection that collectively targets every major step of the lytic infection cycle [22] (Figure 1). Phages, in turn, have acquired mechanisms for resisting countermeasures exhibited by bacteria [22,23]. Such an ever-changing environment presents unique challenges in which to assess therapeutic outcomes. It should be noted that the “inevitable” [24] development of phage-resistant bacteria may be biomedically advantageous if the mutation incurred to withstand phage drives a fitness reduction that can be leveraged through secondary antibacterial treatment. For instance, exposure of multidrug-resistant P. aeruginosa clinical isolates to virulent phage OMKO1 selected for the loss of its bacterial receptor, MexAB and MexXY efflux pump-associated outer membrane porin, OprM, ultimately renewing the isolates’ susceptibility to antibiotics [25]. Ho et al. show that lipotechoic acid (LTA) mutations emerge following NPV phage exposure to Enterococcus faecalis, a phenotypic change that can sensitize the organism to daptomycin [26]. Most recently, Sumrall et al. demonstrated in vitro that exposure of Listeria monocytogenes strain 1042 to virulent phage A511 induced loss of glucose and galactose residues on wall techoic acids, a hallmark of the highly pathogenic serotype 4b, to phenotypically resemble the less invasive serotype 4d [27]. Scanlan, Buckling and Hall suggest that bacteria exposed to phage may be more highly evolved as a consequence of increased selective pressure [28]. Such modifications may lead to concomitant changes in bacterial fitness that further impact response to antimicrobial treatment [29]. One extreme example of this phenomenon was demonstrated by Capparelli et al., who generated phage-resistant S. aureus mutants in mice to produce a live-attenuated vaccine [30]. Whereas bacterial fitness attenuation may be associated with phage resistance [31], this is not always the case [32]. Osada et al. found that S. aureus SA003 developed resistance to lytic phage φSA012, with transient fitness reduction, ultimately countered by an elevated growth rate [33]. Similarly, Kashiwagi and Yomo reported that an RNA phage Qβ-resistant Escherichia coli strain incurred mutations in genomic domains governing cell proliferation, resulting in a compensatory increase in doubling time related to evolution of a new pathway enabling sulfur biogenesis [34].
Figure 1.
Compensatory bacterial resistance mechanisms to phage.
The emergence of phage-resistant bacteria must be acknowledged as a certain outcome of human phage therapy. The following are currently-known mechanisms by which bacteria can resist phage.
2.1.1. Cell surface and extracellular modifications
2.1.1.1. Receptor adaptations
Bacterial resistance to phage may be achieved by cell surface modification via receptor downregulation, concealment, or conformational change [22,35,36]. Bacteria may possess sensory mechanisms that detect the preliminary, weak molecular interactions of phage binding to garner infection resistance prior to the irreversible phage binding [37]. Point mutations or epigenetic modifications to bacterial phage receptors [38,39], including lipopolysaccharide (LPS), outer membrane proteins, flagella, pili, and capsule-associated proteins, for example, may result in decreased phage adsorption, and ultimately, infectivity. Le et al. described an observation in P. aeruginosa following exposure to lytic phage whereby 30% of phage-resistant bacteria exhibited genomic deletion of galU, a gene required for biosynthesis of LPS, the particular phage’s receptor [35]. Similarly, Fallico et al. reported biochemical modifications to teichoic and lipoteichoic acids on Lactococcus lactis IL1403 during early phage exposure to contain D-alanine ester, hypothetically resulting in a conformational change to the phage receptors and limiting phage adsorption and infection [40]. Similar examples include O-antigen acetylation of N-acetylmuramic acid in staphylococci [41], and galactosylation of techoic acid in L. lactis subsp. cremoris SK110 and of cell wall techoic acid in Staphylococcus pseudintermedius SP015 and Staphylococcus aureus [39,42].
2.1.1.2. Outer Membrane Vesicles
Outer membrane vesicles (OMVs), nonreplicating structures that are part of a functionally-diverse intercellular transport system, and which bud from Gram-negative bacterial cells, may act as a sink during phage invasion. Phages may bind OMVs as a result of similar surface features to that of their parent cell, thereby reducing the likelihood of cellular infection [22].
2.1.1.3. Quorum Sensing
Bacterial cells may also leverage intercellular communication to limit viral infection within the population. Bru et al. recently reported that in vitro infection of P. aeruginosa UCBPP-PA14 by virulent phage DMS3vir results in decreased swarming motility through a mechanism involving secretion of the quorum sensing molecule PQS thereby spatially segregating uninfected subpopulations; this generalized stress response is also observed when colonies are exposed to aminoglycosides [43]. Induction of PQS also stimulates release of OMVs, potentially compounding the anti-phage effect of quorum sensing [43].
2.1.2. Intracellular Modifications
2.1.2.1. Restriction-Modification (RM) Systems
The most ubiquitous mechanism of defense among bacteria and archaea, RM systems detect and cleave foreign DNA, including phage double-stranded DNA, based on nucleotide methylation of host DNA [44]. (RM systems are considered not to degrade single-stranded DNA or RNA phage.) Harbored by more than 90% of prokaryotes according to some estimates, RM systems play a role in prokaryotic homeostasis analogous to the innate immune system of higher-order organisms [44]. Restriction-modification systems are comprised of two functional subunits – a restriction endonuclease which cleaves (degrades) un-methylated DNA, and a related methyltransferase which methylates host DNA [44]. Such epigenetic modification forms the basis for recognizing and sequestering foreign genomic material [44,45] and is phylogenetically-conserved, with several hypotheses attempting to explain why this might be the case [44]. Korona et al. predict that RM systems stave off global infection of the bacterial community upon phage introduction to allow for the expansion of genetically-diverse bacterial sub-populations[46].
Yet RM systems are notably flawed; inappropriate methylation of foreign DNA may occur with a probability of up to 0.1 per infection allowing certain phages to acquire host-modified nucleic acids, thereby subverting the RM defense barrier [44,47]. If this occurs, all progeny of the methylated phage will maintain this epigenetic mark, enabling viral propagation to continue [44]. Similarly, T-even phages harbor non-canonical base nucleotides that evade recognition and subsequent cleavage by bacterial RM systems [45]. Sneppen et al. used in silico analyses of phage-bacterial systems to conclude that such ‘loopholes’ within RM systems generate diversity among phage and bacteria alike, increasing ecological carrying capacity and portending the emergence of RM-resistant “epigenetic phage variants” [44]. These variants, in turn, promote more balanced population dynamics by reducing the net growth rate of the comparatively-fit RM-harboring bacterial population that remains susceptible to the mutant phage [44].
The defense island system associated with restriction modification (DISARM) is a bacterial and archaeal defense system similar to RM that is also governed by methylated self-recognition, with putative differences between the two systems comprising specific gene networks involved in cleavage of foreign DNA; the mechanism and function of DISARM-mediated methylation is incompletely defined [22,48].
2.1.2.2. Abortive Infection
A lack of consensus exists regarding the scope of the diverse systems of defense comprising abortive infection; some maintain that it necessarily requires host cell lysis, whereas others consider the term to merely require that infectivity of internalized virions be quenched [45,47]. Toxin-antitoxin systems represent a manifestation of abortive infection observed in both bacteria and archaea. An unstable antitoxin molecule binds and neutralizes the effect of its toxin dimer in a virally-uninfected cell, resulting in cell survival, whereas phage infection interferes with antitoxin synthesis such that unbound toxin prevents bacterial translation through RNA-mediated means [49]. Other abortive infection processes include attenuation of bacterial cell membrane potential, or altered gene expression or maturation [47]. A consequence that may be associated with abortive infection is lysis of the cell following deactivation of the internalized phage to prevent infection of adjacent bacteria [45,47,49]. Some bacterial possess phage inhibitory chromosomal islands, which are prokaryotic mobile genetic elements that indirectly minimize the burst size of an active viral infection [50]. While abortive infection appears in Gram-positive and -negative bacteria, it is more prevalent in the former [49].
2.1.2.3. Bacteriophage Exclusion (BREX)
The most recently-identified innate anti-phage defense system, phage exclusion, is a resistance mechanism found in recombinant B. subtilis bearing similarities with RM and abortive infection systems as it enables selective methylation of the host genome to prevent phage propagation; however, it does not subsequently degrade phage [51]. The mechanism underlying phage exclusion is unknown. Such systems may be present in 10% of microbial genomes, according to one estimate [51].
2.1.2.4. Superinfection Exclusion
Superinfection exclusion is a protein-mediated event precluding entry of subsequent, taxonomically-related phage following genomic integration of an earlier temperate phage [32], thereby rendering lysogenized bacteria resistant to secondary infection by certain phages. Superinfection exclusion is a prevalent mechanism in Gram-negative bacteria, occurring less frequently in Gram-positive species.
2.1.2.5. CRISPR/Cas
Clustered regularly interspaced short palindromic repeats (CRISPR), together with CRISPR-associated proteins (Cas), which have revolutionized the field of gene editing, harbor a parallel role in their native bacterial hosts where they are present in 50% of genomes [45,52]. The equivalent of adaptive immunity, CRISPR-Cas systems are comprised of repetitive DNA sequences with exogenously acquired DNA spacers distributed throughout. DNA is inserted into the CRISPR cassette, with Cas1 suspected to play a role [53]. Cells use these spacers as templates to synthesize CRISPR RNAs (crRNA), which may then complex with Cas9 to initiate double strand breaks in the corresponding loci of incoming foreign DNA [53]. Such systems have been shown to maintain viability following recombination: Jakutyte-Giraitiene and Gasiunas recently reported CRISPR3-recombinant B. subtilis sourced from Streptococcus thermophilus. Anti-phage activity between transformed B. subtilis and S. thermophilus was comparable, with both showing high recombination efficiency [52]. CRISPR3 enables recognition of replication gene sequences gp6 and gp58 of lytic phage SPP1 and subsequent destruction of the phage in B. subtilis [52]. The role of this mechanism in the context of phage resistance is incompletely understood. Prokaryotic Argonaute (pAgo) proteins, implicated in other bacterial defense systems, are thought to be involved in a parallel phage immunity scheme utilizing RNA rather than DNA templates to recognize and degrade foreign nucleic acid [22].
3. Phage pharmacokinetics
Pharmacokinetics of phagotherapy are more complex than those of fixed composition small-molecule antibiotics [54,55], more closely resembling therapeutic biologics such as stem cells. While adsorption, distribution, metabolism and excretion of phage can theoretically be computed in silico, ideal human dosing and route(s) of administration are as yet undefined due to potential elimination by the immune system (itself a changing entity), differential access to sites of infection (and oftentimes varying and unknown bacterial abundance at infected sites), inter-individual (human and bacterial) differences, and inter-phage variables, including diverse capacities of different phages to persist and replicate. Unlike antimicrobial chemotherapeutics which are eliminated by human phase I and phase II enzymes, bacteriophages are eliminated by the reticuloendothelial system in the spleen, but their bioavailability; clearance rate; charge and hydrophobicity; binding affinity to plasma proteins such as human serum albumin, lipoproteins, and glycoproteins remain largely unknown.
Pharmacodynamic and pharmacokinetic considerations alike expand in their complexity when phages or lysins are used in combination with other phages or lysins, or other antimicrobial agents altogether. For each, the mechanism of action and removal from the body should be carefully defined prior to determining the dosing regimen in an effort to maximize the antibacterial capacity of such combinations. Some cocktails have shown synergy, while others exhibit additive benefit, and some display antagonism. The order of delivery may impact therapeutic outcome. Kumaran et al. report an experiment in which planktonic and biofilm methicillin-resistant S. aureus was treated with phage SB-1 prior to or at the same time as rifampin, daptomycin, fosfomycin, ciprofloxacin, or vancomycin in vitro with sequential treatment outperforming simultaneous treatment. Similarly, treatment of S. aureus biofilms in vitro with phage SATA-8505 following vancomycin, dicloxacillin, cefazolin, tetracycline, or linezolid abrogated the antimicrobial effect [56].
4. Human interactions
The need to identify the impact of “trans-kingdom interactions” in the context of human phagotherapy is paramount to its widespread clinical usage safely and effectively [57]. Within the triad of phage-treated, human-associated bacteria, interactions between phage and bacteria have been well-characterized, as have those between bacteria and humans. Very few studies have, however, considered the possibility and hypothetical nature of any interactions spanning phage and humans. Given the significant reduction in planktonic and adherent C. difficile cells and significant increase in phage amplification observed in systems containing bacteria, phage, and HT-29 (human colorectal cancer) cells in vitro versus systems lacking HT-29 cells – presumably explained by the close proximity of bacteria and phage that their eukaryotic binding generated – Shan et al. propose that phage-human dynamics be further considered [58].
Perhaps the most obvious eukaryotic niche in which to examine the impact of therapeutic phage is innate and adaptive immune systems. For more than fifty years, phages (particularly, coliphage ΦX174) have upheld a role in clinical practice in the diagnosis of primary and secondary immunodeficiencies and continue to be utilized in this way today [59]. As a neoantigen, ΦX174 is processed only via presentation by T to B cells of the humoral immune system, such that inability to develop cell-mediated immunity against the phage indicates immune deficiency.
Despite the diagnostic role of phage in clinical medicine in addition to their myriad therapeutic application in humans, only a handful of studies have assessed phage safety as a primary endpoint [60]. A 2003 phase I trial in Switzerland demonstrated that oral ingestion of T4 phage resulted in neither entry of phage into the bloodstream nor altered liver enzymes [61]. An Egyptian burn wound trial executed in 1990 in which non-purified lytic phages were applied to wounds was not associated with adverse effects [62]. Even in vulnerable populations, including pediatric and immunocompromised patients, no serious adverse effects have been linked to phage therapy [59,63,64]. Some experts maintain that any safety threat to mankind would have been observed long ago, considering the ubiquitous presence of phages in the natural environment, but this supposition is perhaps an oversimplification [65]. Indeed, the safety of individual phage formulations is not just a function of safety of the phage, but is contingent on preparation methods, which are non-standardized [65]. For example, incomplete purification of phage from its bacterial host could result in inadvertent delivery of a bacterial toxin, such as endotoxin and/or an exotoxin.
Related theoretical safety concerns include that systemic application of phage may result in emergent toxicity following rapid bacterial lysis and, in the case of Gram-negative bacteria, release of endotoxin in large quantities, although this has not been observed in practice. (In fact, at least one group has observed diminished endotoxin release by phagotherapy versus antibiotic chemotherapy in vitro [66]). Dufour et al. reported on this possibility in an E. coli model in which two lytic coliphages (536_P1 and LM33_P1) or antibiotics were applied in parallel in in vitro systems using E. coli 536 and E. coli LM33 [67]; they found release of extracellular LPS induced by phage to be similar to that observed with amikacin and lower than that observed with cefoxitin or imipenem [66]. Amplification of anti-inflammatory suppressor of cytokine signaling 3 (SOSC3), IL-1 receptor antagonist, and IL-6, as well as reduction in LPS-induced inflammation by NF-κB p65 phosphorylation inhibition are suggestive of the anti-inflammatory properties of phage in some instances [65,68–70], while evidence for the hyperinflammatory potential of therapeutic phage via toll-like receptor 9 and IFN-γ has been demonstrated in animal models to worsen preexisting conditions [71]. However, in one study, although one of two phages utilized in a murine pneumonia model yielded significant increases in IFN-γ, IL-12, monokine induced by IFN-γ (MIG), monocyte chemoattractant protein-1 (MCP-1), and keratinocyte chemoattractant (KC) in the lungs of uninfected mice, there was no statistically significant increase in these acute-phase inflammatory cytokines in infected, phage- or antibiotic-treated mice over the course of a 20–22 hour infection [72]. In this way, modulation of infection-associated inflammation by phage is variable and likely to be phage species-specific.
As viruses, phages are capable of stimulating innate and adaptive immune systems; given their recent detection in the bloodstream via metagenomic analysis as part of the endogenous human phageome, such an interaction is likely and may influence tolerance and/or efficacy of phagotherapy [65]. It is unclear whether phages’ potent bioactivity may be partly attributed to their recruitment of the immune system beyond inherent antibacterial properties [63]. For example, El-Aziz, Elgaml, and Ali observe enhanced complement-mediated antibacterial activity of the innate immune system by virulent phage MMI-Ps1 against P. aeruginosa in a murine model of acute lung infection [73]. On the other hand, Van Belleghem et al. report functional opsonization of phage by binding the surface of invading bacteria [65]; this may result in hyperinflammation or phage neutralization via secondary adaptive immune responses.
In some applications, there is concern that recognition of circulating phages will result in phage elimination, diminishing phage efficacy. Upon systemic introduction, for instance, phage may be intercepted by tissue proteases or the reticuloendothelial system and delivered to the spleen and liver for degradation [65]. Delivery of specific phage in multiple doses, especially systemically, also begets the possibility of neutralizing antibody production against the phage, possibly stimulating phage destruction. Available reports do not agree on the prevalence of phage-neutralizing antibodies and the degree to which they might impact clinical efficacy of phage therapy [3,74]. The extent of antibody production may vary based on routes of phage administration [3,74]. One study administered S. aureus-specific phages 676/Z or A3/R to 122 ill patients twice or thrice daily for 7–91 days and monitored serum anti-phage activity [60]. The investigators found that a cohort of healthy volunteers (n=30) possessed a basal level of anti-phage antibodies similar to that of patients pre-treatment. Serum anti-phage activity increased during phage treatment in a manner dependent upon administration route, with local (which included “gargling, fistula irrigation, irrigation of the abscess cavity, sitz baths, wet compresses, nose drops, ear drops, vaginal irrigation, and inhalations” depending upon infection site [60]) and combined local/oral administration generating greater anti-phage activity than oral or intrarectal administration [75]. Anti-phage activity in patients waned following discontinuation of therapy although it remains a concern whether antibodies demonstrate cross-neutralization of related and unrelated phage species [60].
Finally, a small body of evidence suggests that bacterial lysogeny may directly impact human cells. For instance, one report presented evidence for infection of eukaryotic cells by enterohemorrhagic E. coli (EHEC) temperate phage [76]. The temperate phage studied harbored bacterial genes encoding Shiga toxin which were expressed in adjacent eukaryotic cells, likely via secondary translation pathways or using mitochondrial pathways [77]. This report is consistent with a 1971 report describing translation in mammalian fibroblasts of ß-galactosidase with purported origins from temperate phage [78]. Sweere et al. reported that P. aeruginosa isolated from wounds lysogenized with temperate phage Pf supported chronic bacterial infection compared with non-lysogenized P. aeruginosa, with a proposed mechanism involving phage transcriptome-mediated production of TRIF-dependent type I interferon and reduction of phagocytosis and tumor necrosis factor secretion [79]. The application of a lytic phage treatment in a system containing such lysogenized bacteria might be associated with non-infection via superinfection exclusion or CRISPR, or, as these authors speculate, with exacerbation of bacterial infection by upregulating expression of virulence mechanisms by the bacterium, two clinically-relevant considerations given that temperate phages are present in an estimated 40–50% of bacterial genomes [80].
5. Concessions to limitations of therapeutic phage
5.1. Phage Modification
5.1.1. Adaptation
In 1961, J.F. Vieu described a phenomenon known as phage adaptation, which is described as the repeated passage of a phage in the presence of its target bacterial host or eukaryotic host in order to increase certain therapeutic parameters, such as selecting for long-circulating variants in vivo or evolved receptor binding proteins capable of interacting with evolved bacterial receptors [8,81,82]. Such “directed evolution” has typically been achieved by exposing one phage to one bacterium, and consecutively testing the resulting phage lysate against resistant colonies until susceptibility is observed [83]. However, the Appelmans Protocol is an alternative adaptation technique commonly utilized in the Republic of Georgia, in which a cocktail of phages, rather than a single phage, is tested against a bacterium because the administration of several phages together allows for the possibility of genetic recombination between phages [84].
Though traditionally labor-intensive, the availability of new tools such as multiplex automated genome engineering and phage-assisted continuous evolution has accelerated phage adaptation, subjecting phage to “automatic evolution” and enabling synchrony with adaptation of bacterial hosts [85]. Sybesma et al. found that screening K. pneumoniae and E. coli phages to identify those with an expanded host range improved treatment efficacy; of 38 strains tested, Pyo-phage cocktail susceptibility increased from 66 to 93% [86,87].
Alternatively, phage adaptation may be unnecessary in place of a screening protocol that detected phages which naturally exhibited extended persistence or broad spectra of activity [88].
5.1.1.1. Engineered Phage
Phage researchers have considered optimizing the therapeutic potential of phages via genome engineering [89–91]. Engineering has been employed to increase the antibacterial capacity of phages intended for therapeutic usage. For example, through modifications to the genome of phage M13mp18, the damage incurred following simultaneous application of modified phage and ofloxacin, gentamicin, or ampicillin resulted in augmented killing efficiency by 5-, 3- and 5.5-fold, respectively, compared with bacteria treated with antibiotics alone, though the effect of phage alone was not reported [92,93]. Phages have also been modified to cross eukaryotic membranes in the case of intracellular infection, with some success [94]. Pouillot, Blois and Iris reported a gene editing technology that enabled pausing of the T4 phage replication cycle for insertion of recombinant genes, followed by re-activation of hybrid phage [95]. This technique might be applied for expansion of host range or to encode a bacterial antigen on a phage capsid to augment immune response for ultimate clearance of bacterial pathogens.
The literature describes a diversity of ways in which natural phages have been modified in an attempt to mitigate eukaryotic immune stimulation, which may be a problematic consequence of systemic administration of wild-type phage [87]. A 2005 study conducted in germ-free mice in which animals were treated with lambda phage bearing E158K capsid resulted in extended circulation when administered by intraperitoneal (IP), intravenous (IV) or oral routes of administration[96]. Paul et al. insertionally inactivated the endolysin gene of the S. aureus-specific temperate phage P954 to determine whether compromise to the cell membrane exclusively (i.e., due to holin activity) might lead to bacterial cell death without massive release of bacterial antigen [97], a safety concern of phage therapy. In vitro studies using eight S. aureus strains showed that phages wtP954 and P954Δ cleared over 90% of bacterial cells, without cell lysis (plaque formation) with the latter. In a systemic infection model in neutropenic mice challenged with 5×107 methicillin-resistant S. aureus (MRSA) isolate B911 IP, IP P954Δ phage treatment [200 multiplicity of infection (MOI); defined as the number of virions administered per bacterium] 0- and 2-hours post-infection rescued 100% of mice (n=16) with no adverse events noted, suggesting efficacy of this phage for treatment of bacterial infection in a way that may possibly circumvent lysis-associated safety concerns [97]. (Rescue from lethal bacterial challenge with wtP954 was not reported). Hagens and Bläsi described a similar approach in which modification of phage lambda or phage M13 to exclusively contain a functional holin, resulted in cell death without lysis [98,99], while Bardy et al. developed a holin-deficient T4LyD phage with similar effects [100].
Another widely-recognized exercise in phage modification was described by Lu and Collins in 2009 [92,98]. Following their observations that “directly lethal” antimicrobial approaches resulted in rapid resistance development due to strong selective pressure, the authors overexpressed the nonessential gene φlexA3 for repression of the SOS response as a means of killing bacterial pathogens without incurring phage resistance [92].
Phage engineering was recently employed for the first time in man for the treatment of extrapulmonary tuberculosis in a young lung transplant recipient with early success [17]. Whereas modifications may prompt unique regulations prior to commercial usage, advantages might outweigh drawbacks of phage engineering if antibacterial activity is augmented and/or selection of resistance blunted. Yet Citorik et al. caution against synthetic phage modifications, expressing concern about unintended consequences, including perhaps, attenuated fitness of phage that may abrogate its therapeutic potential [85]. Although the vast number of phages and the ability to generate novel phages through evolution and adaptation renders pan-resistance unlikely [101], caution should be exercised during experimentation to avoid the emergence of bacteria that are resistant to phage and antibiotics, known as “double-resistant variants” [102]. Any such potential consequences of viral genome modification might theoretically be minimized by incorporating synthetic kill switches [103]. It also stands to reason that engineered phages are as susceptible to continuous, directional evolution as wild-type phages, such that the changes they incur may not be observed in any fixed capacity.
5.1.1.2. Encapsulated Delivery
Nanoparticle delivery systems, such as liposome- or polyethylene glycol (PEG)-based platforms, represent another potential strategy to avoid human immune recognition and facilitate phage-bacterial interactions, although phage encapsulation may counterproductively preclude access to its bacterial receptor, resulting in decreased efficacy [93]. Esteban et al. have developed nano-emulsions that complex with phage to quench their negative charge and reduce electrostatic repulsion by bacteria for increased infectivity [104]. Encapsulation of phage may extend circulation in the body up to eight times that of wild-type phage and evade inactivation by neutralizing antibodies [105,106]. In a comparative study, biofilms were preferentially infected by liposome-encapsulated versus wild-type phage in vitro, especially as biofilm age increased; in vivo, liposome-encapsulated phage (KPO1K2) cleared Klebsiella pneumoniae B5055 from the lungs of mice in an experimental model when administered up to 72 hours post-infection, whereas un-encapsulated phage (KPO1K2) cleared infection only when administered no more than 6 hours post-infection [105,107]. In a distinct experimental setup, the same group demonstrated that liposome encapsulation of KPO1K2 (MOI=10) killed 95% of intracellular K. pneumoniae in ex vivo mouse macrophages after 24 hours compared with 21% killed with unencapsulated phage even at increased phage titer [107]. Chadha et al. 2017 similarly considered maintenance in bioactivity of phage when administered intraperitoneally (IP) within liposomes in a mouse K. pneumoniae infected burn model [106]. Concealment of phage from the immune system led to a circulation time of encapsulated phages six times that of un-encapsulated phages. Attenuated phage immunogenicity was observed via reduction of cytokine levels compared to baseline; there was 100% 4-day survival of liposome encapsulated phage-treated mice (n=12) when treatment was delayed by 24 hours post-infection compared with 0% survival of unencapsulated phage-treated mice. Lu and Koeris reported antibody neutralization of PEGylated phage in mice pre-vaccinated with phage, indicating suboptimal effects with repeated exposure to the same phage, even when concealed by PEG [98]. This conclusion is supported and extended by findings of Kim et al. in which prior exposure to wild-type or PEGylated A511 Listeria phage or Felix-O1 Salmonella phage in BALB/c mice resulted in >99% blood clearance of phage within 24 hours following secondary injection, regardless of encapsulation [108].
Hybrid nanoparticle systems have also been employed to optimize phage delivery, as in the case of Chhibber et al.’s use of a transferosome, a synthetic liposome containing phosphatidylcholine, to enhance permeability. Intramuscular (IM) administration of their phage cocktail unbound or transferosome-bound (MOI 10) 30 minutes post-infection with 107 cfu/mL of S. aureus rescued 100% of rats, whereas IM administration 12 hours post-infection rescued 100% of rats treated with transferosome-bound phage and 0% of rats treated with free phage at the study endpoint two weeks later [109].
The effect of encapsulation on phage immunogenicity is more nuanced. In one study, for instance, mice to which K. pneumoniae was intranasally administered were treated with liposome-encapsulated KPO1K2 phage; they exhibited a decrease in proinflammatory IL-1β and TNFα, but an increase in anti-inflammatory IL-10 compared to infected, unencapsulated phage-treated and infected, untreated controls [105]. Other groups utilizing PEG nanoparticles to deliver phage [104,108] collectively report a decreased Th-1 response and decreased levels of inflammatory cytokines IFN-γ and IL-6, in addition to extended circulatory duration [93,100,108].
5.1.1.3. Phage Component-Based Therapies
While whole phages have been the historical focus of phagotherapy, the use of phage component-based molecules as antibacterial agents has been recently investigated. Such approach features simplicity of regulatory execution, and perhaps more limited bacterial resistance [110–113], prevention of genomic integration, rapid antibacterial effect even at low concentrations, and ease of quality control and storage measures relative to conventional phage therapy [114–119]. The dosing regimen of phage components resembles that of small-molecule antibiotics and they also do not replicate in their bacterial hosts as whole phages do, enabling simplified administration [110]. Certain phage components may even directly enhance bacterial sensitivity to conventional antibiotics, such as PA-PP, a serine protease that degrades outer membrane porin protein in P. aeruginosa PAR50 [120]. A potential drawback of employing phage enzymes over conventional phage therapy is the technical difficulty of their acquisition [121].
Over the course of endogenous phage infection of a bacterium, expression of lysins during intracellular biosynthesis provides the means for dissemination of new virions as lysins bind peptidoglycan components from the interior face, perforating the cell wall leading to osmotic rupture. Vázquez and García describe the enzybiotic approach as lysins “repurposed,” due to their extracellular, rather than intracellular, administration to effect the same lethal outcome [115]. Virulent phage enzymes were recently named the most promising alternatives to antibiotics by a pipeline portfolio review in Lancet Infectious Disease [113]. In fact, enzymes secreted over the course of the phage’s life cycle have in recent decades been considered for use as antibacterial agents in place of conventional phage therapy. Two clinically-relevant classes of phage enzymes include depolymerases and endolysins (also known as lysins or peptidoglycan hydrolases [PGHs]). To a lesser extent, the prospect of utilizing “small chemical molecules” that mediate initial phage binding has been envisaged owing to their bacteriostatic effects [113]. Waseh et al. isolated the tailspike protein, P22sTsp, of lytic phage P22, which plays a role in docking P22 at its bacterial receptor. Three doses of P22sTsp were administered to chicks beginning 1 or 18 hours after initial oral challenge with Salmonella enterica serovar Typhimurium and every 24 hours thereafter [122]. P22sTsp caused aggregation of S. enterica serovar Typhimurium concomitant with significant reductions in bacterial density of the liver, spleen and cecum following the final, third dose of P22sTsp among animals initially treated 1 hour post-infection versus sham-treated controls, potentially suggesting an immune-enhancing mechanism underlying phage activity resembling antibody-mediated agglutination. While the precise mechanism of action is unknown, it is speculated that allosteric modulation of O antigen following phage-bacterial complexing impedes the organism’s motility [122].
Endolysins are enzymes produced by double-stranded DNA phages that hydrolyze the cell wall of bacteria. Those targeting Gram-positive bacteria are generally composed of two domains, an enzymatically active domain (EAD) and a cell wall binding domain (CBD) [123]. In contrast, most endolysins targeting Gram-negative bacteria lack a CBD [119], which reflects the presence of the intervening outer membrane; this structure generally limits the efficacy of lysins against these organisms, with exceptional lysins exhibiting amphipathic components capable of traversing the outer membrane [116]. In the same vein, Gram-negative lysins may be synthetically optimized by fusion with outer membrane permeabilizers, as in the case of Artilysin® [113,124,125]. Some lysins harbor two distinct EADs, which may minimize resistance development. As agents that may induce the phenomenon known as “lysis from without” in high concentrations, bacterial entry is not required [126]; their exclusive interaction at the bacterial surface may mitigate resistance development [111]. Their enzymatic activity may promote removal of bacterial structures such as extracellular polymeric substance that may otherwise be unaffected by conventional antimicrobial agents [113]. Lysins are immunogenic due to their protein composition; the dose administered must account for their rapid removal from the circulation [110]. They may also display synergy when combined with antibiotics or one another, or enhance complement-mediated activity of the innate immune system [112,123,127,128] (Table 2).
Table 2.
Experimental animal and moth larvae model lysin efficacy studies published between January 1, 2007 and October 21, 2019.
| Bacterium | Lysin | Disease Model | Administration (Bacteria; Lysin) | Findings | Reference |
|---|---|---|---|---|---|
| Acinetobacter baumannii | PlyF307 | Mouse, subdermal catheter biofilms | Subdermal catheter with 3 day-old biofilm; Endolysin (250 μL) administered to catheter intraluminally at 24 and 28 hours post-infection; catheters removed 3 hours post-treatment |
2 log reduction in bacterial density on lysin-treated versus lysin-untreated catheters | [124] |
| PlyF307 | Mouse, systemic infection | IP lethal dose (108 cfu); 1 mg PlyF07 IP 2 hours post-infection |
50% lysin-treated versus 10% control mice survived 14 days | [124] | |
| K2 | Galleria mellonella, systemic infection | Proleg injection (106 cfu NIPH 2061/5.5 μL) Proleg injection phage-pretreated (0.25, 0.5, or 3 μg) NIPH 2061 cells or phage (0.25, 0.5, or 3 μg) 30 minutes post-infection |
90% survival of phage-treated larvae at 20 hours and 60% survival at 42 hours post-infection | [240] | |
| Mouse, systemic infection | IP administration 107 cfu following chemically-induced immunosuppression; IP lysin administration 1 hour post-infection |
Pretreated mice survival 53%, 69%, and 88% (0.25, 0.5, or 3 μg/mL, respectively) at 72 hours post-infection. Treated mice survival 15%, 56%, and 70% (0.25, 0.5, or 3 μg/mL, respectively) at 72 hours post-infection | |||
| Ply6A3 | Mouse, systemic infection | IP minimum lethal dose (1×109 cfu AB32/mL); IP administration Ply6A3 (1 mL, 2 mg/mL) or phage PD6A3 (1 mL, 109 pfu/mL); uninfected mice received 14-phage cocktail (1 mL, 109 pfu/mL), Ply6A3 (1 mL, 2 mg/mL), phage PD6A3 (1 mL, 109 pfu/mL), or combined PD6A3 phage and Ply6A3 lysin |
70% survival among Ply6A3-treated mice, 70% survival among combination PD6A3 phage and Ply6A3-treated mice, 60% survival among PD6A3 phage-treated mice, 50% survival among phage cocktail-treated mice, and 0% among infected, shame-treated mice at 7 days post-infection; decrease in WBC counts among all treatment groups versus infected, sham-treated; decrease in WBC counts among lysin Ply6A3 and combination PD6A3 phage and Ply6A3-treated animals versus PD6A3 alone and phage cocktail-treated mice; increase in IL-10 and procalcitonin levels among infected, untreated controls while the same values remained at baseline in uninfected mice, PD6A3-treated mice, and Ply6A3-treated mice | [116] | |
| Escherichia coli | K1E | Mouse, systemic infection | 1–4×108 cfu IM; Capsule depolymerase (0, 2, 5, 20 μg) IM 30 minutes post-infection; Toxicity assessed using IM depolymerase (100 μg) |
20 μg depolymerase (save K1E) rescued most mice from death; toxicity studies revealed no changes in survival (100%), behavior, or body weight of lysin-treated versus lysin-untreated controls | [241] |
| K1F | |||||
| K1H | |||||
| K5 | |||||
| K30gp41 | |||||
| K30gp42 | |||||
| Pseudomonas aeruginosa | PlyPa03 | Mouse, skin infection | Topical application of PA01 (5×106 cfu/mL); Topical treatment 20 hours post-infection with PlyPa03 (200 or 300 μg) or PlyPa91 (100 μg) |
Dose-dependent reduction in bacterial load (>2 log in PlyPa03-treated group and 1 log in PlyPa91-treated group) | [125] |
| PlyPa91 | |||||
| PlyPa91 | Mouse, lung infection | Two sequential intranasal administrations of PA01 (50 μL 108 cfu/mL each); Two doses (two intranasal or one intranasal and one intratracheal; each 50 μL [1.8 mg/mL]) of PlyPa91 administered 3 or 6 hours post-infection |
10-day survival rates 0% infected, untreated controls; 20% infected mice treated with two intranasal phage doses; 70% infected mice treated with one intratracheal and one intranasal dose | [125] | |
| Staphylococcus aureus | Ply187 | Mouse, endophthalmitis | 500 cfu intravitreally; Endolysin intravitreally 6 and 12 hours post-infection |
Improvement in 24 hour post-infection clinical score of lysin-treated eyes at 6 and 12 hours post-treatment versus controls; bacterial abundance reduced 24 hours post-treatment in 6 hour post-infection treated mice (3.6×103 cfu/eye) and 12 hours post-infection treated mice (2.9×104 cfu/eye) versus untreated mice (1.0×105 cfu/eye) | [242] |
| Exebacase | Mouse, systemic infection | 106–109 cfu IP; Lysin IP (0.25–5.0 μg/mL) 2 hours post-infection with or without vancomycin or daptomycin |
Lysin associated with 2 log10 reduction in bacteria in blood; increased efficacy with combination of lysin and vancomycin or daptomycin; fourfold maximal increase in lysin MIC | [128] | |
| P-27/HP | Mouse, systemic infection | 5×108 cfu IP; Lysin (250 μg SQ) 24 hours later |
Infected, untreated mice 4 log10 bacteria in spleen versus endolysin-treated mice 2 log10 cfu bacteria in spleen 4 days post-treatment | [243] | |
| MR-10 | Mouse, burn wound infection | 105–108 cfu/mL SQ; Lysin (50 μg SQ) and/or minocycline (50–100 mg/kg PO) 3 hours post-infection |
Infected, untreated mice 100% mortality (7 days); animals treated with either agent alone 35% mortality by day 5; animals treated with high-dose minocycline or combination therapy no mortality (7 days) | [244] | |
| MR-10 | Mouse, burn wound infection | 107–109 cfu topical; Lysin (50 μg SQ) and/or minocycline (50–100 mg/kg PO) 3 hours post-infection |
Combination therapy-treated and high-dose minocycline-treated animals nearly total wound closure 12 days post-treatment versus infected, untreated control animals (28%), low-dose minocycline treated (43%), and lysin only-treated (49%) animals | [244] | |
| 80α | Mouse, systemic infection | 4×107 cfu IP; Endolysins (200 μg) IP 30 minutes post-infection |
Six of the eight PGH compounds administered intraperitoneally (200 μg) 30 minutes after microbial challenge exhibited similar activity to vancomycin, with the six compounds and vancomycin all effecting 100% survival | [131] | |
| Phi11 | |||||
| LysK | |||||
| P68 | |||||
| 2638A | |||||
| Twort | |||||
| PhiSH2 | |||||
| WMY | |||||
| Exebacase | Rat, osteomyelitis | Intratibial injection of 10 μL arachidonic acid (50 μg/mL) and 50 μL S. aureus (107 cfu/mL); IP administration of daptomycin (60 mg/kg twice daily doses for 4 days), IV Exebacase (single dose of 40 mg/kg), or daptomycin and Exebacase combination therapy, all beginning one week post-infection |
Reduction in bacterial loads in all treatment groups compared with infected, untreated controls 4 days post-treatment; reduction in bacterial loads among combination treated animals versus animals treated with Exebacase or antibiotic alone | [127] | |
| S25–3lys-his | Mouse, impetigo | 2 ×108 cfu administered topically to inner pinna; Topical administration of 2 μL (1 mg/mL) lysin immediately post-infection |
Lysin-treated animals had decreased bacterial densities, pustule surface area (but not pustule quantity), and epidermal invasion and increased species richness of pinna epidermal microbiome versus infected, untreated pinna | [245] | |
| Streptococcus pneumoniae | PL3 chimeric N-acetylmuramoyl-l-alanine amidase | Zebrafish embryos, systemic infection | Embryos cultured with 108 cfu/mL D39; Single dose of PL3 (15 or 20 μg) post-infection (timing unspecified) |
0% mortality with single dose 20 μg PL3 versus 40% in untreated, infected embryos | [129] |
| ClyJ | Mouse, systemic infection | 2.68 × 107 cfu IP (LD100); IP ClyJ (0.3 or 0.4 mg) or penicillin G (0.25 mg) 1 hour post-infection |
0, 90, 100 and 40%, survival among untreated, low- and high-dose phage- and antibiotic-treated animals, respectively, at 10 days post-treatment | [246] | |
| Cpl-711 | Zebrafish, systemic infection | 10μL 106 cfu/mL strain 48 IP; 1 hour post-infection, subtherapeutic doses of Cpl-711 (10 μL 0.5 μg/mL) or PL3 (10 μL 0.15 μg/mL) administered IP, alone or in combination (10 μL 0.125 μg/mL Cpl-711, 10μL 0.08 μg/mL PL3) |
72 hours post-infection, 28% survival of infected, sham-treated animals; 44% survival of Cpl-711-only treated animals; 50% survival of PL3-only treated animals; 78% survival of Cpl-711 and PL3 combination-treated animals; 100% survival of uninfected, lysin-treated animals | [115] | |
| PL3 |
Recombinant lysin systems have also been developed, typically capitalizing on protein engineering technology to modify the lysin gene product with the purpose of increasing its spectrum of activity (spanning genera, in some instances [125]), or increasing its circulation time in order to reach the infected site [113,115]. To better address Streptococcus pneumoniae infections, Blazquez et al. synthesized a chimeric lysin comprised of natural phage lysins Pal and LytA, both of which are N-acetylmuramoyl-L-alanine-amidases with bactericidal activity against S. pneumoniae, with the latter representing the major pneumococcal autolysin [129]. The recombination of the Pal catalytic domain with the consolidated CBD of LytA formed the PL3 chimeric endolysin, resulting in a reduction of all 10 tested choline-containing Gram-positive bacterial strains including S. pneumoniae and other streptococci in vitro; Pal and LytA were not tested alone, though other studies have demonstrated comparatively reduced potency and efficacy. Application of PL3 in a zebrafish (Danio rerio) embryo S. pneumoniae D39 infection model reduced mortality to 0%, from 40% in non-treated, infected embryos [129]. Daniel et al. developed a chimeric lysin, ClyS, in which the N-terminal domain of the Twort phage was fused with the C-terminal cell wall targeting domain of φNM3, which lacks Sh3b domain binding peptidoglycan peptide bridges, to which bacteria may easily become resistant [112]. Others have considered cleaving the C-terminal to increase lysis, as the binding component precludes the catalytic subunit from functioning [130]. In one of the largest comparative studies of recombinant lysins, Schmelcher et al. reported that six of the nine PGH compounds administered IP 30 minutes after IP challenge of BALB/c mice with MRSA NRS382 exhibited similar activity to vancomycin [131]. The experimental success of recombinant lysin technology has piqued the interest of the pharmaceutical industry. Zhang et al. demonstrated activity of phage IME-EF1 endolysin against enterococci in vitro; this phage was active against four of 20 strains tested, whereas treatment with its endolysin killed 11 strains [132]. Subcutaneous implantation of a catheter colonized with A. baumannii and subsequently treated with PlyF307 endolysin resulted in a population reduction of 2 logs by 28 hours later, with the same endolysin able to clear bacterial EPS in vitro [124]. Lastly, Hathaway and colleagues loaded either a combination of endolysin CHAPK and lysostaphin into poly(N-isopropylacrylamide) (PNIPAM)-based vesicles creating a thermoresponsive polymer-based approach which allowed for the ejection of antimicrobial cargo following phase transition induced by inflammation-associated heat release for topical administration to staphylococcal infections [133,134]. In vitro testing demonstrated a 0.5 fold reduction in bacterial density at 32°C in PNIPAM-encapsulated versus unencapsulated cocktail, but a fourfold reduction in cell density at 37°C in PNIPAM-encapsulated versus unencapsulated cocktail [133].
Table 2 summarizes experimental animal and moth larval studies involving lysins. Examples of endolysin products under development include Tonabacase (Intron Biotechnology, Inc.), a S. aureus-specific endolysin currently in a phase II clinical trial for IV administration in bacteremia [135], and Exebacase (previously CF-301, ContraFect) which in 2018 completed a phase II clinical trial of IV administration for treatment of S. aureus bacteremia and endocarditis [136,137]. Results of the trial demonstrated that combination therapy of Exebacase and standard of care antibiotics generated a significant increase in response rate versus antibiotics alone in patients with MRSA bacteremia or endocarditis [137].
5.2. Pharmacologic Optimization
5.2.1. Phage Cocktails
The application of phage ‘cocktails’ comprised of phages bearing divergent mechanisms of action may increase the formulation’s spectrum of activity and decrease the potential for development of resistance [138]. Experts have advised that cocktails be comprised of between two and 10 phages [24,139]. The in vitro study of a cocktail of phage DRA88 and phage K containing distinct host spectra showed activity against 74% of 95 S. aureus isolates tested in the planktonic state, with individual applications being active against only 60 and 64%, respectively, as measured by spot plating, a modification of the double overlay plaque assay [140]. In vitro population reduction of phage cocktail-treated S. aureus was observed for all three isolates tested (15981, MRSA 252, H325), with a range of MOIs achieving similar effects, although lower MOIs required an extended time period to do so [140]. Nonetheless, phage formulation via cocktails is not a foolproof antibacterial strategy; if each phage in a cocktail is not maintained at a sufficient dose, or if constituent phages do not target the same bacteria, resistance may occur as readily as it would with single phages [141]. The majority of clinical trials evaluating phage therapy past and present have been administered as cocktails rather than individual viruses (Table 3).
Table 3.
Phage and phage lysin therapy in human clinical trials published between January 2007 and March 2019.
| Trial Start | Trial Registration Number | Ref. | Phage/Lysin Formulation (Investigator/Sponsor) | Bacteria Targeted | Treatment Indication | Phase | Outcome |
|---|---|---|---|---|---|---|---|
| 2006 | [21] | WPP-201 cocktail (Southwest Regional Wound Care Center) | Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus | Infected venous leg ulcers | I | No adverse effects observed | |
| 2009 | [61] | T4 coliphage cocktail: AB2, 4, 6, 11, 46, 50, 55, JS34, 37, 98, D1.4 (Nestlé) | E. coli | Dysentery | I/II | No adverse effects or benefits noted; study prematurely discontinued | |
| 2009 | EudraCT 2004–001691–39 | [247] | Biophage-PA (Biocontrol Ltd) | P. aeruginosa | Otitis externa | I/II | No adverse effects noted; at 42 days post-treatment, phage-treated cohort exhibited reduced median bacterial abundance compared to placebo group |
| 2015 | [21] | PhagoBurn (Pherecydes Pharma) | P. aeruginosa, E. coli | Infected burn wounds | I/II | No adverse effects noted; time to “sustained bacterial reduction” longer in phage treatment arm; study prematurely discontinued | |
| 2017 | N/A | SAL200 (Tonabacase) (iNtRON Biotech) | S. aureus | S. aureus bacteremia | II | Ongoing | |
| 2017 | [137] | Exebacase (ContraFect Corp) | S. aureus | Bacteremia, including endocarditis (single intravenous dose added to standard of care antibiotics) | II | Well tolerated; higher clinical response rate compared to standard of care antibiotics alone in methicillin-resistant but not methicillin-susceptible S. aureus subgroups | |
| 2017 | N/A | Pyophage (Tzulakidze National Center of Urology) | Enterococcus species, E. coli, Proteus mirabilis, staphylococci, streptococci, P. aeruginosa | Urinary tract infections | II/III | Results pending (personal communication, Thomas Kessler) | |
| 2019 | N/A | EcoActive (Intralytix, Inc.) | Adherent invasive E. coli (AIEC) | Exacerbation of inflammation in Crohn disease secondary to AIEC | I/II | Recruitment ongoing | |
| 2019 | N/A | PhagoPied (Pherecydes Pharma) | S. aureus | Diabetic wounds | I/II | Not yet recruiting |
Table reproduced from [174]
5.2.2. Combination Therapy
Phages have also been combined with other antimicrobial agents, including antibiotics, with considerable success. One objective of combination therapy is to reduce the mutant selection window, or the range in drug concentration spanning the minimum inhibitory concentration and the mutant prevention concentration, to minimize resistance-selecting population expansion in the presence of the therapeutic [142]. Combination therapy using phage may be pursued with natural substances and/or antibiotics. First described in 2007, phage antibiotic synergy (PAS) is a phenomenon wherein the antibacterial effect observed following the combination of phage with sub-inhibitory concentrations of antibiotic enhances phage activity [143,144]. Johnson and Garcia evaluated hydrogel-embedded MR-5 phage, linezolid, or a combination thereof for their abilities to inhibit MRSA biofilm formation in vitro. Combination therapy resulted in significant reductions in bacterial colonization versus no treatment or phage or linezolid alone, and was associated with lower mutation rates compared with those observed in the monotherapy groups [145]. The binding of phage to surface receptors that contribute to bacterial pathogenicity can diminish bacterial virulence and/or lead to selection against receptor expression among progeny, resulting in comparatively treatable infection, as discussed above [143]. Combination therapy was shown to limit the acquisition of φSan23 phage resistance to ~33% of a S. enterica subsp. enterica serovar Enteritidis population, while treatment with phage alone resulted in phage resistance in ~90% of the population [36]. Higher drug concentrations combined with phages do not always support greater antibacterial effects since phages require a certain bacterial density (replication threshold) before phage replication can occur [143]. Recent work from Kim et al. revealed the highly-anticipated mechanism underlying some forms of PAS to be cellular expansion mediated by DNA damage response pathways in the presence of antibiotic or other environmental stressors, including, but not limited to, reactive oxygen species [146]. Phages in combination with antibiotics may also, however, elicit additive, antagonistic, or facilitative effects (the last being an effect in which combination therapy generates greater activity than either agent alone but less activity than the sum of the effects of the individual agents) beside synergistic effects [143].
PAS is observed in biofilms as well as planktonic cultures, as demonstrated by Akturk et al. in the treatment of mono- and dual-species biofilms with in vitro phage, antibiotics, or combination therapy administered simultaneously or staggered [147]. They report that treatment of 48 hour monospecies biofilms with phage for 6 hours, MIC or 8 x MIC concentrations of gentamicin, ciprofloxacin, and meropenem for 24 hours, combination phage and MIC or 8 x MIC concentrations of gentamicin, ciprofloxacin, or meropenem for 24 hours, or combination phage (for 6 hours) and MIC or 8 x MIC concentrations of gentamicin, ciprofloxacin, or meropenem for 18 hours thereafter led to statistical reductions in P. aeruginosa biofilm-associated bacteria, while significant reductions were only seen in S. aureus biofilms treated with phage and ciprofloxacin or gentamicin at MIC and 8 x MIC concentrations. Overall, combination treatment of phage for six hours followed by 18 hours gentamicin (MIC or 8 x MIC) or ciprofloxacin (8 x MIC) supported the greatest reduction in P. aeruginosa monospecies biofilm-associated bacterial density.
In 48 hour dual-species biofilms, simultaneous combination therapy did not yield synergistic activity. Sequential combination therapy of EPA1 followed 6 hours later by gentamicin (MIC) yielded anti-biofilm synergy against P. aeruginosa constituents while S. aureus density was not significantly altered.
Although phage antibiotic synergy classically implicates lytic phages, recent data proposes that a similar phenomenon may occur by temperate phages. Population modelling suggests that possession of a prophage may render its P. aeruginosa host more sensitive to antibiotics, even if the host is antibiotic-resistant, as antibiotic exposure may place stress on the bacterium driving induction of temperate phage and resulting in cell lysis [148].
Overall, combination therapies offer a potentially effective, though complex, solution to phage resistance.
5.2.3. Selective Administration
Adjusting therapeutic phage formulations has been shown to increase biological stability of phage as well as increase the dose that reaches the site(s) of infection. In a cohort of pediatric patients with pulmonary infection, Leung et al. evaluated the impact on phage activity in spray-dried versus spray freeze-dried formulations. Superior results were found with the latter, which is able to accommodate larger viral loads, although some damage was observed to phage through the delivery process, dependent to an extent on the phage utilized [149]. The addition of L-leucine and trehalose excipients increased stability and bioactivity of phage solutions targeting P. aeruginosa, increasing the mouth-throat fraction to 58%, and preserving properties exhibited by phage prior to drying [150]. Results were corroborated in a similar study subjecting P. aeruginosa and Burkholdheria cenocepacia phages to spray-drying [151]. In a similar manner, antacids may be used to neutralize gastric acidity, allowing phage transit following oral administration [152]. Likewise, dosing schedules may be designed to deliver higher phage titers at fewer time points to minimize development of antibodies and/or avoid consequences of antigen-antibody binding [102].
Phage therapy may not be ideal for every infection. Administration into various body fluids may functionally reduce viral titers via biochemical interactions. For instance, serum, but not albumin or fibrinogen [153], inhibits phage-bacterial relations, and, as mentioned above, phages administered orally may be inactivated by the acidity of the gastric compartment [154]. Further, not all bacteria are equally susceptible to infection by lytic phage, at least those discovered by extant detection methods, suggesting the need for improved “phage hunting” methods [155]. Indeed, for some microorganisms, such as C. difficile, only a small number of lytic phages have been identified. Nevertheless, efforts to identify additional phages or lysing targeting C. difficile are underway [50,156–159].
6. Conclusion
While experimental findings suggest promise for the safety and, in some cases, efficacy of phage therapy, many questions remain [160]. Improved in vivo models and human studies are needed to better understand selection of phage resistance, and to inform pharmacokinetics and pharmacodynamics of phage and its interactions with human tissues, in the context of human phage therapy [161]. As with other aspects of this highly individualized treatment regimen, the precise effects of phage administration upon the prokaryotic and eukaryotic milieu to which it is introduced is likely to be phage species-dependent [88,162,163].
7. Expert opinion
Although multiple factors led to the discontinuation of therapeutic phage use in Western medicine until recently, the impending antimicrobial resistance crisis justifies its reconsideration. Moreover, the authors contend that the role of phage therapy may not be limited to cases of dire resistance, but that phages may be considered much earlier in a patient’s therapeutic course, perhaps even as first-line therapy, allowing for the preservation of conventional antibiotics. This would represent a major paradigm-shift in the treatment of infections with the potential for a significant impact on reducing antibiotic usage. Phage therapy therefore should be seen as not only a reaction to antibiotic resistance, but more significantly, a way to address a root cause of development of resistance.
The authors encourage well-documented, well-controlled clinical trials of phage (and/or phage products) to more fully establish the disease(s) against which they may be beneficial, how best to select appropriate phages (and/or phage products), how best to dose and administer them, their potential interaction with the immune system and its impact on therapeutic outcome, and the selection of phage (and/or phage product) resistance.
As with other newly emerging individualized therapies, phage therapy also has the potential to challenge the current pathways by which drugs are regulated. Individualized gene and cell therapies call for a re-evaluation of current regulatory processes, and bring to light new ethical and safety questions.
7.1. Novel technologies beget novel potential
Although the current ‘re-discovery’ of therapeutic phages exemplifies an unusual narrative in scientific discovery, the present is an ideal time for the revitalization of phage research [63] given recent technological advancements that afford new experimental possibilities. For instance, the past decade has witnessed the refinement of animal models through which to investigate preclinical utility, including use of the greater wax moth (Galleria mellonella) as a small, cost-effective, IACUC-exempt organism that is easy to maintain and possesses an immune system analogous to that of humans [164]. The sequencing revolution, the advent of -omics technologies, the emergence of the multidisciplinary field of systems biology, as well as the breadth of molecular [165–167] and visualization techniques [168–170] all poise the field to address unanswered questions asked a century ago which, in many ways, precluded the possibility of evidence-based clinical application [98,165]. For example, genome sequencing of candidate phages can now provide information regarding their ability to lysogenize bacteria and therefore to mobilize or enhance production of virulence factors [171]. Genome engineering technologies have afforded the expression of phage recombinases in bacteria that indicate prior phage exposure under laboratory conditions by way of altered gene expression [85]. Computational prediction models using inter-species genomics data through the use of machine learning is a promising method by which phage-bacteria pairs can be identified rapidly and on a large scale [172].
Systems biology has also lent contributions which have included the development of tools that accelerate phage adaptation to maintain relevance as a bacterial predator. Insights to be gleaned from this approach include determination of phage predation rates amd phage behavior in diverse, multicellular communities [85]. While laboratory experiments are needed to confirm the veracity of in silico predictions, modeling represents a powerful tool for exploring ecological parameters governing phage-bacterial interactions in a low-risk environment prior to in vitro and in vivo experimentation [36,142].
7.2. The need for more, better preclinical and clinical studies
We recognize that the greatest impediment to standardized phagotherapy is well-controlled clinical trials. The lack of randomized controlled trial data and the complexity of regulatory restrictions required has driven much of the recent human data generation to be from compassionate use cases. However, case reports and series often include the concomitant use of other antimicrobial agents given the primary clinical objective of positive patient outcomes [11]. This kind of approach can make it difficult to draw conclusions as to the effects of phage therapy alone. Additional insight is critical for the determination of how best to select appropriate phages or phage enzymes, as well as how to dose and administer them; pharmacokinetic and pharmacodynamic properties must also be better defined. Perhaps, randomized controlled clinical trials performed in well-defined infectious diseases (even if not caused by drug-resistant bacteria) should be considered. As an unconventional approach, bioethicists Anomaly and Savulescu suggest recruiting healthy individuals to a representative population sample for infectious diseases studies in which participants are challenged with a pathogen and subsequently administered an experimental treatment regimen, and compensating them fairly for the potentially high risks of involvement [173].
If shown to be efficacious, a goal of present and future research vis-à-vis phage therapy will be to develop a pipeline through which phage(s) targeting individual patients’ bacterial isolates can be identified and administered as primary or adjunctive therapy, or to identify appropriate phage cocktails. Some countries have begun this process already: The Queen Astrid Military Hospital in Brussels, Belgium has published standardized guidelines for phage treatment in the setting of osteomyelitis, while Germany’s Phage 4 Cure initiative aims to develop Good Manufacturing Practice (GMP)-grade processes for the assimilation of phage therapy into the regulatory confines of Western medicine expected to meet “international quality standards” [12]. Nevertheless, several challenges remain, including the creation of clinical laboratory standards for phage testing and appropriate regulation for such a potentially individualized anti-infective approach [174].
Supplementary Material
Article highlights.
Growing antibiotic resistance, in addition to promising historical and current phage studies, has renewed interest in human phagotherapy
To expedite consideration of phages as antibacterial agents, certain biological challenges associated with their use should be addressed, including acquired bacterial resistance to phage infection; the pharmacologic complexity of phage relative to small-molecule antibiotics; and interactions between the human immune system and phage
Challenges may be allayed through the use of phage cocktails, phage modifications, encapsulation, and/or deployment of phage products
Acknowledgements
The authors thank Larry J. Prokop, MLS, for his careful search and compilation of source material from several medical databases and support throughout the writing and revision process, and to Kerryl Greenwood-Quaintance, MS, proofreading the manuscript.
Funding
R Patel is supported by UM1 AI104681, R01 AR056647, R01 AI091594, R01 AI134770 and R21 AI125870. K M Caflisch is supported by T32 AR056950.
Declaration of interest
R Patel has research grant support from ContraFect, a company which has financial interest in the subject matter discussed in this manuscript. In addition, she reports other relationships not directly relevant to the subject matter discussed in this manuscript, including research grant support from CD Diagnostics, Merck, Hutchison Biofilm Medical Solutions, Accelerate Diagnostics, ContraFect, TenNor Therapeutics Limited and Shionogi, serving a consultant to Curetis, Specific Technologies, Next Gen Diagnostics, PathoQuest, Selux Diagnostics and Qvella (all monies are paid to Mayo Clinic), and receiving travel reimbursement from ASM and IDSA, an editor’s stipend from IDSA, and honoraria from the NBME, Up-to-Date and the Infectious Diseases Board Review Course. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Footnotes
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
- 1.Guttman B, Raya R, Kutter E. Basic phage biology In: Kutter E, Sulakvelidze A, editors. Bacteriophages: Biology and applications: CRC Press; 2004. [Google Scholar]
- 2.Summers W History of phage research and phage therapy In: Waldor M, DI F,SL A, editors. Phages: their role in bacterial pathogenesis and biotechnology: ASM Press; 2005. p. 3–17. [Google Scholar]
- 3.Wittebole X, De Roock S, Opal SM. A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens [Review]. Virulence. 2014;5(1):209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chanishvili N Bacteriophages as therapeutic and prophylactic means: summary of the Soviet and post Soviet experiences [Review]. Curr Drug Deliv. 2016. 01 May;13(3):309–323. [DOI] [PubMed] [Google Scholar]
- 5.On an invisible microbe antagonistic to dysentery bacilli. Note by d’Herelle MF, presented by Roux M Comptes Rendus Academie des Sciences 1917; 165:373–5. Bacteriophage. 2011 2011/01/01;1(1):3–5. [Google Scholar]
- 6.Fruciano E, Bourne S. Phage as an antimicrobial agent: d’Herelle’s heretical theories and their role in the decline of phage prophylaxis in the West [Conference Paper]. Can J Infect Dis Med. 2007. Jan-Feb;18(1):19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Summers WC. Bacteriophage research: early history In: Kutter E, Sulakvelidze A, editors. Bacteriophages: biology and applications. 1 ed: CRC Press; 2004. p. 5–27. [Google Scholar]
- 8.Abedon ST, Kuhl SJ, Blasdel BG, et al. Phage treatment of human infections [Review]. Bacteriophage. 2011. Mar-Apr;1(2):66–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Summers WC. History of phage research and phage therapy In: Waldor MK, Friedman DI, Adhya SL, editors. Phages: their role in bacterial pathogenesis and biotechnology. Washington, D.C.: ASM Press; 2005. p. 3–17. [Google Scholar]
- 10.Vivas R, Barbosa AAT, Dolabela SS, et al. Multidrug-resistant bacteria and alternative methods to control them: an overview. Microb Drug Resist. 2019. Jul-Aug;25(6):890–908. [DOI] [PubMed] [Google Scholar]
- 11.Tkhilaishvili T, Winkler T, Muller M, et al. Bacteriophages as adjuvant to antibiotics for the treatment of periprosthetic joint infection caused by multidrug-resistant Pseudomonas aeruginosa [Journal Article]. Antimicrob Agents Chemother. 2019. September 16;16:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Onsea J, Soentjens P, Djebara S, et al. Bacteriophage application for difficult-to-treat musculoskeletal infections: development of a standardized multidisciplinary treatment protocol. Viruses. 2019;11(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maddocks S, Petrovic Fabijan A, Ho J, et al. Bacteriophage therapy of ventilator-associated pneumonia and empyema caused by Pseudomonas aeruginosa. Am J Respir Crit Care Med. 2019;200(9):1179–1181. [DOI] [PubMed] [Google Scholar]
- 14.Schooley RT, Biswas B, Gill JJ, et al. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob Agents Chemother. 2017;61(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan BK, Turner PE, Narayan D, et al. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol Med Public Health. 2018;2018(1):60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Law N, Logan C, Yung G, et al. Successful adjunctive use of bacteriophage therapy for treatment of multidrug-resistant Pseudomonas aeruginosa infection in a cystic fibrosis patient. Infection. 2019. May 17. [DOI] [PubMed] [Google Scholar]
- 17.Dedrick RM, Guerrero-Bustamante CA, Garlena RA, et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat Med. 2019. 05;25(5):730–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jault P, Leclerc T, Jennes S, et al. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind phase 1/2 trial. Lancet Infect Dis. 2019;19(1):35–45. [DOI] [PubMed] [Google Scholar]
- 19.Sarker SA, Sultana S, Reuteler G, et al. Oral phage therapy of acute bacterial diarrhea with two coliphage preparations: a randomized trial in children from Bangladesh. EBioMedicine. 2016. 01 February;4:124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jennes S, Merabishvili M, Soentjens P, et al. Use of bacteriophages in the treatment of colistin-only-sensitive Pseudomonas aeruginosa septicaemia in a patient with acute kidney injury-a case report. Crit Care. 2017;21(1):129–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhoads DD, Wolcott RD, Kuskowski MA, et al. Bacteriophage therapy of venous leg ulcers in humans: results of a phase I safety trial. J Wound Care. 2009. June;18(6):237–8, 240–3. [DOI] [PubMed] [Google Scholar]
- 22.Azam AH, Tanji Y. Bacteriophage-host arm race: an update on the mechanism of phage resistance in bacteria and revenge of the phage with the perspective for phage therapy. Appl Microbiol Biotechnol. 2019. March;103(5):2121–2131. [DOI] [PubMed] [Google Scholar]
- 23.Taylor VL, Fitzpatrick AD, Islam Z, et al. The diverse impacts of phage morons on bacterial fitness and virulence. Adv Virus Res. 2019;103:1–31. [DOI] [PubMed] [Google Scholar]
- 24.Yuan Y, Wang L, Li X, et al. Efficacy of a phage cocktail in controlling phage resistance development in multidrug resistant Acinetobacter baumannii. Virus Res. 2019. 15 October;272(197734). [DOI] [PubMed] [Google Scholar]
- 25.Chan BK, Sistrom M, Wertz JE, et al. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci Rep. 2016. 05/26/online;6:26717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho K, Huo W, Pas S, et al. Loss-of-function mutations in epaR confer resistance to ϕNPV1 infection in Enterococcus faecalis OG1RF. Antimicrob Agents Chemother. 2018;62(10):e00758–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sumrall ET, Shen Y, Keller AP, et al. Phage resistance at the cost of virulence: Listeria monocytogenes serovar 4b requires galactosylated teichoic acids for InlB-mediated invasion. PLoS Pathog 2019. 01 October;15(10):e1008032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scanlan PD, Buckling A, Hall AR. Experimental evolution and bacterial resistance: (co)evolutionary costs and trade-offs as opportunities in phage therapy research. Bacteriophage. 2015;5(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chatterjee A, Johnson CN, Luong P, et al. Bacteriophage resistance alters antibiotic-mediated intestinal expansion of enterococci. Infect Immun. 2019;87 (6)(e00085–19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Capparelli R, Nocerino N, Lanzetta R, et al. Bacteriophage-resistant Staphylococcus aureus mutant confers broad immunity against staphylococcal infection in mice. PLoS One. 2010;5(e11720). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kvachadze L, Balarjishvili N, Meskhi T, et al. Evaluation of lytic activity of staphylococcal bacteriophage Sb-1 against freshly isolated clinical pathogens. Microb Biotechnol. 2011. September;4(5):643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Obeng N, Pratama AA, Elsas JDV. The significance of mutualistic phages for bacterial ecology and evolution [Review]. Trends Microbiol. 2016. 01 June;24(6):440–449. [DOI] [PubMed] [Google Scholar]
- 33.Osada K, Takeuchi I, Miyanaga K, et al. Coevolution between Staphylococcus aureus isolated from mastitic milk and its lytic bacteriophage ΦSA012 in batch co-culture with serial transfer. Biochem Eng J. 2017;126:16–23. [Google Scholar]
- 34.Kashiwagi A, Yomo T. Ongoing phenotypic and genomic changes in experimental coevolution of RNA bacteriophage Qβ and Escherichia coli. PLoS Genet. 2011;7(8):e1002188–e1002188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le S, Yao X, Lu S, et al. Chromosomal DNA deletion confers phage resistance to Pseudomonas aeruginosa. Sci Rep-UK. 2014;4:4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holguin AV, Cardenas P, Prada-Penaranda C, et al. Host resistance, genomics and population dynamics in a Salmonella Enteritidis and phage system. Viruses. 2019. February 22;11(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Debarbieux L Bacterial sensing of bacteriophages in communities: the search for the Rosetta stone [Review]. Curr Opin Microbiol. 2014. August;20:125–130. [DOI] [PubMed] [Google Scholar]
- 38.Chadha P, Katare OP, Chhibber S. In vivo efficacy of single phage versus phage cocktail in resolving burn wound infection in BALB/c mice [Journal Article]. Microb Pathog. 2016. October;99:68–77. [DOI] [PubMed] [Google Scholar]
- 39.Azam AH, Kadoi K, Miyanaga K, et al. Analysis host-recognition mechanism of staphylococcal kayvirus SA039 reveals a novel strategy that protects Staphylococcus aureus against infection by Staphylococcus pseudintermedius Siphoviridae phages. Appl Microbiol Biotechnol. 2019. 16 August;103(16):6809–6823. [DOI] [PubMed] [Google Scholar]
- 40.Fallico V, Ross RP, Fitzgerald GF, et al. Genetic response to bacteriophage infection in Lactococcus lactis reveals a four-strand approach involving induction of membrane stress proteins, D-alanylation of the cell wall, maintenance of proton motive force, and energy conservation. J Virol. 2011. November;85(22):12032–12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bera A, Biswas R, Herbert S, et al. The presence of peptidoglycan O-acetyltransferase in various staphylococcal species correlates with lysozyme resistance and pathogenicity. Infect Immun. 2006. August;74(8):4598–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sijtsma L, Wouters JT, Hellingwerf KJ. Isolation and characterization of lipoteichoic acid, a cell envelope component involved in preventing phage adsorption, from Lactococcus lactis subsp. cremoris SK110. J Bacteriol. 1990. December;172(12):7126–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bru JL, Rawson B, Trinh C, et al. PQS produced by the Pseudomonas aeruginosa stress response repels swarms away from bacteriophage and antibiotics. J Bacteriol. 2019;26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sneppen K, Semsey S, Seshasayee ASN, et al. Restriction modification systems as engines of diversity. Front Microbiol. 2015;6 June(528). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stern A, Sorek R. The phage-host arms race: shaping the evolution of microbes [Review]. BioEssays. 2011. January;33(1):43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Korona R, Levin BR. Phage-mediated selection and the evolution and maintenance of restriction-modification. Evolution. 1993. April;47(2):556–575. [DOI] [PubMed] [Google Scholar]
- 47.Seed KD. Battling phages: how bacteria defend against viral attack. PLoS Pathog. 2015. June;11(6):e1004847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ofir G, Melamed S, Sberro H, et al. DISARM is a widespread bacterial defence system with broad anti-phage activities. Nat Microbiol. 2018. January;3(1):90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fineran PC, Blower TR, Foulds IJ, et al. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. P Natl Acad Sci USA. 2009. 20 January;106(3):894–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dalmasso M, Hill C, Ross RP. Exploiting gut bacteriophages for human health [Review]. Trends Microbiol. 2014. July;22(7):399–405. [DOI] [PubMed] [Google Scholar]
- 51.Goldfarb T, Sberro H, Weinstock E, et al. BREX is a novel phage resistance system widespread in microbial genomes. EMBO J. 2015;34(2):169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jakutyte-Giraitiene L, Gasiunas G. Design of a CRISPR-Cas system to increase resistance of Bacillus subtilis to bacteriophage SPP1. J Ind Microbiol Biot. 2016. 01 August;43(8):1183–1188. [DOI] [PubMed] [Google Scholar]
- 53.Makarova KS, Wolf YI, van der Oost J, et al. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol Direct. 2009. August 25;4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chan BK, Abedon ST. Phage therapy pharmacology: phage cocktails. Adv Appl Microbiol. 2012;78:1–23. [DOI] [PubMed] [Google Scholar]
- 55.Abedon ST, Thomas-Abedon C. Phage therapy pharmacology [Review]. Curr Pharm Biotechnol. 2010. January;11(1):28–47. [DOI] [PubMed] [Google Scholar]
- 56.Kumaran D, Taha M, Yi Q, et al. Does treatment order matter? Investigating the ability of bacteriophage to augment antibiotic activity against Staphylococcus aureus biofilms. Front Microbiol. 2018;9:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Santiago-Rodriguez TM, Hollister EB. Human virome and disease: high-throughput sequencing for virus discovery, identification of phage-bacteria dysbiosis and development of therapeutic approaches with emphasis on the human gut. Viruses. 2019. July;11 (7)(656). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shan J, Ramachandran A, Thanki AM, et al. Bacteriophages are more virulent to bacteria with human cells than they are in bacterial culture; insights from HT-29 cells. Sci Rep. 2018. 23 March;8(1):5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith LL, Buckley R, Lugar P. Diagnostic immunization with bacteriophage ΦX 174 in patients with common variable immunodeficiency/hypogammaglobulinemia. Front Immunol. 2014;5:410–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lusiak-Szelachowska M, Zaczek M, Weber-Dabrowska B, et al. Phage neutralization by sera of patients receiving phage therapy. Viral Immunol. 2014;27(6):295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vandenheuvel D, Lavigne R, Brussow H. Bacteriophage therapy: advances in formulation strategies and human clinical trials [Review]. Ann Rev Virol. 2015. 09 November;2:599–618. [DOI] [PubMed] [Google Scholar]
- 62.Abul-Hassan HS E-TkMB, Gomaa R. Bacteriophage therapy of Pseudomonas burn wound sepsis. Ann Burns Fire Disasters. 1990;3(4). [Google Scholar]
- 63.Borysowski J, Gorski A. Is phage therapy acceptable in the immunocompromised host? [Short Survey]. Int J Infect Dis. 2008. September;12(5):466–471. [DOI] [PubMed] [Google Scholar]
- 64.Fortuna W, Miedzybrodzki R, Weber-Dabrowska B, et al. Bacteriophage therapy in children: facts and prospects [Review]. Med Sci Monit. 2008. August;14(8):RA126–RA132. [PubMed] [Google Scholar]
- 65.Van Belleghem JD, Dabrowska K, Vaneechoutte M, et al. Interactions between bacteriophage, bacteria, and the mammalian immune system. Viruses. 2018. December 25;11(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dufour N, Delattre R, Ricard JD, et al. The lysis of pathogenic Escherichia coli by bacteriophages releases less endotoxin than by β-lactams. Clin Infect Dis. 2017. June 1;64(11):1582–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dufour N, Clermont O, La Combe B, et al. Bacteriophage LM33_P1, a fast-acting weapon against the pandemic ST131-O25b: H4 Escherichia coli clonal complex. J Antimicrob Chemother. 2016. 01 November;71(11):3072–3080. [DOI] [PubMed] [Google Scholar]
- 68.Van Belleghem JD, Clement F, Merabishvili M, et al. Pro- and anti-inflammatory responses of peripheral blood mononuclear cells induced by Staphylococcus aureus and Pseudomonas aeruginosa phages. Sci Rep. 2017. August 14;7(1):8004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang L, Hou X, Sun L, et al. Corrigendum: Staphylococcus aureus bacteriophage suppresses LPS-induced inflammation in MAC-T bovine mammary epithelial cells. Front Microbiol. 2018;9:2511–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang L, Hou X, Sun L, et al. Staphylococcus aureus bacteriophage suppresses LPS-induced inflammation in MAC-T bovine mammary epithelial cells. Front Microbiol. 2018;9:1614–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gogokhia L, Buhrke K, Bell R, et al. Expansion of bacteriophages is linked to aggravated intestinal inflammation and colitis. Cell Host Microbe. 2019. February 13;25(2):285–299.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dufour N, Delattre R, Chevallereau A, et al. Phage therapy of pneumonia is not associated with an overstimulation of the inflammatory response compared to antibiotic treatment in mice. Antimicrob Agents Chemother. 2019;63 (8)(e00379–19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Abd El-Aziz AM, Elgaml A, Ali YM. Bacteriophage therapy increases complement-mediated lysis of bacteria and enhances bacterial clearance after acute lung infection with multidrug-resistant Pseudomonas aeruginosa. J Infect Dis. 2019;219(9):1439–1447. [DOI] [PubMed] [Google Scholar]
- 74.Qadir MI. Phage therapy: a modern tool to control bacterial infections [Review]. Pak J Pharm Sci. 2015. 01 January;28(1):265–270. [PubMed] [Google Scholar]
- 75.Lusiak-Szelachowska M, Zaczek M, Weber-Dabrowska B, et al. Antiphage activity of sera during phage therapy in relation to its outcome. Future Microbiol. 2017. February;12(2):109–117. [DOI] [PubMed] [Google Scholar]
- 76.Bentancor LV, Bilen MF, Mejias MP, et al. Functional capacity of Shiga-toxin promoter sequences in eukaryotic cells. PLoS One. 2013;8(2):e57128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bentancor LV, Mejias MP, Pinto A, et al. Promoter sequence of Shiga toxin 2 (Stx2) is recognized in vivo, leading to production of biologically active Stx2. MBio. 2013. October 1;4(5):e00501–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lengeling A, Mahajan A, Gally DL. Bacteriophages as pathogens and immune modulators? mBio. 2013;4 (6)(e00868–13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sweere JM, Van Belleghem JD, Ishak H, et al. Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science. 2019;363(6434):eaat9691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Howard-Varona C, Hargreaves KR, Abedon ST, et al. Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J. 2017;11(7):1511–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Merril CR, Biswas B, Carlton R, et al. Long-circulating bacteriophage as antibacterial agents. Proc Natl Acad Sci USA. 1996;93(8):3188–3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vieu J Interet des bacteriophages dans le traitement de staphylococcies. Vie Med. 1961;42:823–829. [PubMed] [Google Scholar]
- 83.Botka T, Pantucek R, Maslanova I, et al. Lytic and genomic properties of spontaneous host-range Kayvirus mutants prove their suitability for upgrading phage therapeutics against staphylococci. Sci. 2019. 02 April;9(1):5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Burrowes BH, Molineux IJ, Fralick JA. Directed in vitro evolution of therapeutic bacteriophages: the Appelmans Protocol. Viruses. 2019;11(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Citorik RJ, Mimee M, Lu TK. Bacteriophage-based synthetic biology for the study of infectious diseases [Review]. Curr Opin Microbiol. 2014. June;19(1):59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sybesma W, Zbinden R, Chanishvili N, et al. Bacteriophages as potential treatment for urinary tract infections. Front Microbiol. 2016. 11 April;7 Apr(465). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Goodridge LD. Designing phage therapeutics [Review]. Curr Pharm Biotechnol. 2010. January;11(1):15–27. [DOI] [PubMed] [Google Scholar]
- 88.Serwer P, Wright ET, Lee JC. High murine blood persistence of phage T3 and suggested strategy for phage therapy. BMC Res Notes. 2019;12(1):560–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Diez-Martinez R, De Paz HD, Garcia-Fernandez E, et al. A novel chimeric phage lysin with high in vitro and in vivo bactericidal activity against Streptococcus pneumoniae. J Antimicrob Chemother. 2014. 12 November;70(6):1763–1773. [DOI] [PubMed] [Google Scholar]
- 90.Weber W, Fussenegger M. Emerging biomedical applications of synthetic biology [Review]. Nat Rev Genet. 2012. January;13(1):21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Principi N, Silvestri E, Esposito S. Advantages and limitations of bacteriophages for the treatment of bacterial infections [Review]. Front Pharmacol. 2019;10 (May)(513). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lu TK, Collins JJ. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. P Natl Acad Sci USA. 2009. 24 March;106(12):4629–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Viertel TM, Ritter K, Horz HP. Viruses versus bacteria-novel approaches to phage therapy as a tool against multidrug-resistant pathogens. J Antimicrob Chemother. 2014. September;69(9):2326–2336. [DOI] [PubMed] [Google Scholar]
- 94.Puiu M, Julius C. Bacteriophage gene products as potential antimicrobials against tuberculosis [Review]. Biochem Soc Trans. 2019;47(3):847–860. [DOI] [PubMed] [Google Scholar]
- 95.Pouillot F, Blois H, Iris F. Genetically engineered virulent phage banks in the detection and control of emergent pathogenic bacteria. Biosecur Bioterror. 2010. 01 June;8(2):155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vitiello CL, Merril CR, Adhya S. An amino acid substitution in a capsid protein enhances phage survival in mouse circulatory system more than a 1000-fold. Virus Res. 2005. December;114(1–2):101–3. [DOI] [PubMed] [Google Scholar]
- 97.Paul V, Sundarrajan S, Rajagopalan S, et al. Lysis-deficient phages as novel therapeutic agents for controlling bacterial infection. BMC Microbiol. 2011;11(195). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lu TK, Koeris MS. The next generation of bacteriophage therapy [Review]. Curr Opin Microbiol. 2011. October;14(5):524–531. [DOI] [PubMed] [Google Scholar]
- 99.Hagens S, Blasi U. Genetically modified filamentous phage as bactericidal agents: a pilot study. Lett Appl Microbiol. 2003;37(4):318–23. [DOI] [PubMed] [Google Scholar]
- 100.Bardy P, Pantucek R, Benesik M, et al. Genetically modified bacteriophages in applied microbiology [Review]. J Appl Microbiol. 2016. 01 September;121(3):618–633. [DOI] [PubMed] [Google Scholar]
- 101.Donlan RM. Preventing biofilms of clinically relevant organisms using bacteriophage. Trends Microbiol. 2009. February;17(2):66–72. [DOI] [PubMed] [Google Scholar]
- 102.Gorski A, Miedzybrodzki R, Weber-Dabrowska B, et al. Phage therapy: combating infections with potential for evolving from merely a treatment for complications to targeting diseases [Review]. Front Microbiol. 2016. 26 September;7(1515). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Planson AG, Carbonell P, Grigoras I, et al. A retrosynthetic biology approach to therapeutics: from conception to delivery [Review]. Curr Opin Biotechnol. 2012. December;23(6):948–956. [DOI] [PubMed] [Google Scholar]
- 104.Esteban PP, Jenkins ATA, Arnot TC. Elucidation of the mechanisms of action of bacteriophage K/nano-emulsion formulations against S. aureus via measurement of particle size and zeta potential. Colloids Surf B Biointerfaces. 2016. March 01;139:87–94. [DOI] [PubMed] [Google Scholar]
- 105.Singla S, Harjai K, Katare OP, et al. Bacteriophage-loaded nanostructured lipid carrier: improved pharmacokinetics mediates effective resolution of Klebsiella pneumoniae-induced lobar pneumonia. J Infect Dis. 2015. 15 July;212(2):325–334. [DOI] [PubMed] [Google Scholar]
- 106.Chadha P, Katare OP, Chhibber S. Liposome loaded phage cocktail: enhanced therapeutic potential in resolving Klebsiella pneumoniae mediated burn wound infections [Journal Article]. Burns. 2017. November;43(7):1532–1543. [DOI] [PubMed] [Google Scholar]
- 107.Singla S, Harjai K, Katare OP, et al. Encapsulation of bacteriophage in liposome accentuates its entry in to macrophage and shields it from neutralizing antibodies. PLoS One. 2016. April;11(e0153777). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kim KP, Cha JD, Jang EH, et al. PEGylation of bacteriophages increases blood circulation time and reduces T-helper type 1 immune response. Microb Biotechnol. 2008. May;1(3):247–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chhibber S, Shukla A, Kaur S. Transfersomal phage cocktail is an effective treatment against methicillin-resistant Staphylococcus aureus-mediated skin and soft tissue infections. Antimicrob Agents Chemother. 2017;61(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pastagia M, Euler C, Chahales P, et al. A novel chimeric lysin shows superiority to mupirocin for skin decolonization of methicillin-resistant and -sensitive Staphylococcus aureus strains. Antimicrob Agents Chemother. 2011. February;55(2):738–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ji W, Huang Q, Sun L, et al. A novel endolysin disrupts Streptococcus suis with high efficiency. FEMS Microbiol Lett. 2015. 31 October;362(fnv205). [DOI] [PubMed] [Google Scholar]
- 112.Daniel A, Euler C, Collin M, et al. Synergism between a novel chimeric lysin and oxacillin protects against infection by methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2010. April;54(4):1603–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Abdelkader K, Gerstmans H, Saafan A, et al. The preclinical and clinical progress of bacteriophages and their lytic enzymes: the parts are easier than the whole. Viruses. 2019. January 24;11(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Benesik M, Novacek J, Janda L, et al. Role of SH3b binding domain in a natural deletion mutant of Kayvirus endolysin LysF1 with a broad range of lytic activity [In Press]. Virus Genes. 2017. 29 August:1–10. [DOI] [PubMed] [Google Scholar]
- 115.Vazquez R, Garcia P. Synergy between two chimeric lysins to kill Streptococcus pneumoniae. Front Microbiol. 2019;10 (June)(1251). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wu M, Hu K, Xie Y, et al. A novel phage PD-6A3, and its endolysin Ply6A3, with extended lytic activity against Acinetobacter baumannii. Front Microbiol. 2019;10 (January)(3302). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schuch R, Khan BK, Raz A, et al. Bacteriophage lysin CF-301, a potent antistaphylococcal biofilm agent. Antimicrob Agents Chemother. 2017. July;61(e02666–16). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhou Y, Zhang H, Bao H, et al. The lytic activity of recombinant phage lysin LysKΔamidase against staphylococcal strains associated with bovine and human infections in the Jiangsu province of China. Res Vet Sci. 2017. 01 April;111:113–119. [DOI] [PubMed] [Google Scholar]
- 119.Schmelcher M, Donovan DM, Loessner MJ. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012;7(10):1147–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Al-Wrafy F, Brzozowska E, Gorska S, et al. Identification and characterization of phage protein and its activity against two strains of multidrug-resistant Pseudomonas aeruginosa. Sci Rep. 2019. 17 September;9(1):13487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen W, Hwang YY, Gleaton JW, et al. Optimization of a peptide extraction and LC-MS protocol for quantitative analysis of antimicrobial peptides. Future Sci OA. 2019;5 (1)(FSO348). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Waseh S, Hanifi-Moghaddam P, Coleman R, et al. Orally administered P22 phage tailspike protein reduces Salmonella colonization in chickens: prospects of a novel therapy against bacterial infections. PLoS One. 2010;5(e13904). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Latka A, Maciejewska B, Majkowska-Skrobek G, et al. Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process [Review]. Appl Microbiol Biotechnol. 2017. 01 April;101(8):3103–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lood R, Winer BY, Pelzek AJ, et al. Novel phage lysin capable of killing the multidrug-resistant Gram-negative bacterium Acinetobacter baumannii in a mouse bacteremia model. Antimicrob Agents Chemother. 2015. 01 April;59(4):1983–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Raz A, Serrano A, Hernandez A, et al. Isolation of phage lysins that effectively kill Pseudomonas aeruginosa in mouse models of lung and skin infection. Antimicrob Agents Chemother. 2019;63 (7)(e00024–19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Abedon ST. Lysis from without. Bacteriophage. 2011. Jan-Feb;1(1):46–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Karau MJ, Schmidt-Malan SM, Yan Q, et al. Exebacase in addition to daptomycin is more active than daptomycin or Exebacase alone in methicillin-resistant Staphylococcus aureus osteomyelitis in rats. Antimicrob Agents Chemother. 2019:AAC.01235–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schuch R, Lee HM, Schneider BC, et al. Combination therapy with lysin CF-301 and antibiotic is superior to antibiotic alone for treating methicillin-resistant Staphylococcus aureus-induced murine bacteremia. J Infect Dis. 2014;209(9):1469–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Blazquez B, Fresco-Taboada A, Iglesias-Bexiga M, et al. PL3 amidase, a tailor-made lysin constructed by domain shuffling with potent killing activity against pneumococci and related species. Front Microbiol. 2016. 28 July;7 Jul(1156). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shang Nelson D. Contributions of net charge on the PlyC endolysin CHAP domain. Antibiotics. 2019. 05/28;8:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schmelcher M, Shen Y, Nelson DC, et al. Evolutionarily distinct bacteriophage endolysins featuring conserved peptidoglycan cleavage sites protect mice from MRSA infection. J Antimicrob Chemother. 2014. 20 October;70(5):1453–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang W, Mi Z, Yin X, et al. Characterization of Enterococcus faecalis phage IME-EF1 and its endolysin. PLoS One. 2013. 13 November;8(e80435). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hathaway H, Ajuebor J, Stephens L, et al. Thermally triggered release of the bacteriophage endolysin CHAPK and the bacteriocin lysostaphin for the control of methicillin resistant Staphylococcus aureus (MRSA) [Journal Article]. J Control Release. 2017. January 10;245:108–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hathaway H, Alves DR, Bean J, et al. Poly(N-isopropylacrylamide-co-allylamine) (PNIPAM-co-ALA) nanospheres for the thermally triggered release of bacteriophage K. Eur J Pharm Biopharm. 2015. October;96:437–41. [DOI] [PubMed] [Google Scholar]
- 135.ClinicalTrials.gov. Phase IIa clinical study of N-Rephasin® SAL200 2017. [updated 8 Aug 2018]. Available from: https://clinicaltrials.gov/ct2/show/NCT03089697?term=SAL-200&rank=3
- 136.ClinicalTrials.gov. Safety, efficacy and pharmacokinetics of CF-301 vs. placebo in addition to antibacterial therapy for treatment of S. aureus bacteremia 2018. Available from: https://clinicaltrials.gov/ct2/show/NCT03163446?term=CF-301&rank=1
- 137.Fowler J, Vance G., Das A, Lipka J, et al. Exebacase (lysin CF-301) improved clinical responder rates in methicillin resistant Staphylococcus aureus (MRSA) bacteremia including endocarditis compared to standard of care antibiotics (SOC) alone in a first-in-patient phase II study [30 April 2019]. Available from: https://d1io3yog0oux5.cloudfront.net/_53ec10a3e327f1e291fe5c77b7504a7a/contrafect/db/226/1306/pdf/ECCMID+ORAL+Ph2_FINAL.pdf [Google Scholar]
- 138.Worthington RJ, Melander C. Combination approaches to combat multidrug-resistant bacteria. Trends Biotechnol. 2013;31(3):177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chan BK, Abedon ST, Loc-Carrillo C. Phage cocktails and the future of phage therapy [Review]. Future Microbiol. 2013. June;8(6):769–783. [DOI] [PubMed] [Google Scholar]
- 140.Alves DR, Gaudion A, Bean JE, et al. Combined use of bacteriophage K and a novel bacteriophage to reduce Staphylococcus aureus biofilm formation. Appl Environ Microb. 2014. November;80(21):6694–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nilsson AS. Phage therapy--constraints and possibilities. Ups J Med Sci. 2014;119(2):192–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hill JA, Cowen LE. Using combination therapy to thwart drug resistance. Future Microbiol. 2015;10(11):1719–26. [DOI] [PubMed] [Google Scholar]
- 143.Tagliaferri TL, Jansen M, Horz H-P. Fighting pathogenic bacteria on two fronts: phages and antibiotics as combined strategy. Front Cell Infect Microbiol. 2019;9:22–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Comeau AM, Tetart F, Trojet SN, et al. Phage-antibiotic synergy (PAS): beta-lactam and quinolone antibiotics stimulate virulent phage growth. PLoS One. 2007. August 29;2(8):e799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kaur S, Harjai K, Chhibber S. Bacteriophage mediated killing of Staphylococcus aureus in vitro on orthopaedic K wires in presence of linezolid prevents implant colonization. PLoS ONE. 2014;9(3):e90411–e90411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kim M, Jo Y, Hwang YJ, et al. Phage-antibiotic synergy via delayed lysis. Appl Environ Microbiol. 2018. November 15;84(22). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Akturk E, Oliveira H, Santos SB, et al. Synergistic action of phage and antibiotics: parameters to enhance the killing efficacy against mono and dual-species biofilms. Antibiotics (Basel). 2019. July 25;8(3):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Clifton SM, Kim T, Chandrashekhar JH, et al. Lying in wait: modeling the control of bacterial infections via antibiotic-induced proviruses. mSystems. 2019;4 (5)(e00221–19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Leung SSY, Parumasivam T, Gao FG, et al. Production of inhalation phage powders using spray freeze drying and spray drying techniques for treatment of respiratory infections. Pharm Res. 2016. 01 June;33(6):1486–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chang RY, Wong J, Mathai A, et al. Production of highly stable spray dried phage formulations for treatment of Pseudomonas aeruginosa lung infection. Eur J Pharm Biopharm. 2017. December;121:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Matinkhoo S, Lynch KH, Dennis JJ, et al. Spray-dried respirable powders containing bacteriophages for the treatment of pulmonary infections. J Pharm Sci. 2011. December;100(12):5197–5205. [DOI] [PubMed] [Google Scholar]
- 152.Huh H, Wong S, St Jean J, et al. Bacteriophage interactions with mammalian tissue: therapeutic applications. Adv Drug Deliv Rev. 2019. January 17. [DOI] [PubMed] [Google Scholar]
- 153.Dixon DV, Hosseinidoust Z, Tufenkji N. Effects of environmental and clinical interferents on the host capture efficiency of immobilized bacteriophages. Langmuir. 2014. March 25;30(11):3184–90. [DOI] [PubMed] [Google Scholar]
- 154.Miedzybrodzki R, Klak M, Jonczyk-Matysiak E, et al. Means to facilitate the overcoming of gastric juice barrier by a therapeutic staphylococcal bacteriophage A5/80. Front Microbiol. 2017;8:467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Garretto A, Miller-Ensminger T, Wolfe AJ, et al. Bacteriophages of the lower urinary tract [Review]. Nat Rev Urol. 2019. 01 July;16(7):422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Abedon ST. Ecology of anti-biofilm agents II: bacteriophage exploitation and biocontrol of biofilm bacteria [Review]. Pharmaceuticals. 2015. 09 September;8(3):559–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hargreaves KR, Clokie MRJ. Clostridium difficile phages: still difficult? Front Microbiol. 2014;5 April(184). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rashid SJ, Barylski J, Hargreaves KR, et al. Two novel myoviruses from the north of Iraq reveal insights into Clostridium difficile phage diversity and biology. Viruses. 2016. 16 November;8(310). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yang J, Yang H. Non-antibiotic therapy for Clostridioides difficile infection: a review [Review]. Crit Rev Clin Lab Sci. 2019. [DOI] [PubMed] [Google Scholar]
- 160.Bassetti M, Poulakou G, Ruppe E, et al. Antimicrobial resistance in the next 30 years, humankind, bugs and drugs: a visionary approach [Review]. Intens Care Med. 2017. 01 October;43(10):1464–1475. [DOI] [PubMed] [Google Scholar]
- 161.Bull JJ, Gill JJ. The habits of highly effective phages: population dynamics as a framework for identifying therapeutic phages. Front Microbiol. 2014;5(618). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Akanda ZZ, Taha M, Abdelbary H. Current review-The rise of bacteriophage as a unique therapeutic platform in treating peri-prosthetic joint infections. Journal of orthopaedic research : official publication of the Orthopaedic Research Society. 2018. April;36(4):1051–1060. [DOI] [PubMed] [Google Scholar]
- 163.Naghizadeh M, Karimi Torshizi MA, Rahimi S, et al. Effect of serum anti-phage activity on colibacillosis control by repeated phage therapy in broilers. Vet Microbiol. 2019. July;234:61–71. [DOI] [PubMed] [Google Scholar]
- 164.Abbasifar R, Kropinski AM, Sabour PM, et al. Efficiency of bacteriophage therapy against Cronobacter sakazakii in Galleria mellonella (greater wax moth) larvae. Arch Virol. 2014. September;159(9):2253–2261. [DOI] [PubMed] [Google Scholar]
- 165.Eyer L, Pantucek R, Ruzickova V, et al. New perspectives of the phage therapy [Review]. [Nove perspektivy fagove terapie.]. Klin Mikrobiol Infekc Lek. 2007;13(6):231–235. [PubMed] [Google Scholar]
- 166.Eyer L, Pantucek R, Zdrahal Z, et al. Structural protein analysis of the polyvalent staphylococcal bacteriophage 812. Proteomics. 2007. January;7(1):64–72. [DOI] [PubMed] [Google Scholar]
- 167.Maszewska A, Wojcik E, Ciurzynska A, et al. Differentiation of polyvalent bacteriophages specific to uropathogenic Proteus mirabilis strains based on the host range pattern and RFLP. Acta Biochim Pol. 2016;63(2):303–310. [DOI] [PubMed] [Google Scholar]
- 168.Zhang X, Guo H, Jin L, et al. A new topology of the HK97-like fold revealed in Bordetella bacteriophage by cryoEM at 3.5 A resolution. eLife. 2013. 17 December;2013(e01299). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Schofield DA, Molineux IJ, Westwater C. ‘Bioluminescent’ reporter phage for the detection of Category A bacterial pathogens. JoVE. 2011. (53):e2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Dubrovin EV, Popova AV, Kraevskiy SV, et al. Atomic force microscopy analysis of the Acinetobacter baumannii bacteriophage AP22 lytic cycle. PLoS One. 2012. 11 October;7(e47348). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.McCallin S, Alam Sarker S, Barretto C, et al. Safety analysis of a Russian phage cocktail: from metagenomic analysis to oral application in healthy human subjects. Virology. 2013. 01 September;443(2):187–196. [DOI] [PubMed] [Google Scholar]
- 172.Leite DMC, Brochet X, Resch G, et al. Computational prediction of inter-species relationships through omics data analysis and machine learning. BMC Bioinformatics. 2018. November 20;19(Suppl 14):420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Anomaly J, Savulescu J. Compensation for cures: Why we should pay a premium for participation in ‘challenge studies’. Bioethics. 2019. 01 September;33(7):792–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Caflisch KM, Patel R. Implications of bacteriophage- and bacteriophage component-based therapies for the clinical microbiology laboratory. J Clin Microbiol. 2019:JCM.00229–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Regeimbal JM, Jacobs AC, Corey BW, et al. Personalized therapeutic cocktail of wild environmental phages rescues mice from Acinetobacter baumannii wound infections. Antimicrob Agents Chemother. 2016. October;60(10):5806–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Leshkasheli L, Kutateladze M, Balarjishvili N, et al. Efficacy of newly isolated and highly potent macteriophages in a mouse model of XDRAB bacteremia. J Glob Antimicrob Resist. 2019;14. [DOI] [PubMed] [Google Scholar]
- 177.Jeon J, Ryu CM, Lee JY, et al. In vivo application of bacteriophage as a potential therapeutic agent to control OXA-66-like carbapenemase-producing Acinetobacter baumannii strains belonging to sequence type 357. Appl Environ Microbiol. 2016. July 15;82(14):4200–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Guang-Han O, Leang-Chung C, Vellasamy KM, et al. Experimental phage therapy for Burkholderia pseudomallei infection. PLoS One. 2016. July;11(e0158213). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Carmody LA, Gill JJ, Summer EJ, et al. Efficacy of bacteriophage therapy in a model of Burkholderia cenocepacia pulmonary infection. J Infect Dis. 2010. 15 January;201(2):264–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Nale JY, Spencer J, Hargreaves KR, et al. Bacteriophage combinations significantly reduce Clostridium difficile growth in vitro and proliferation in vivo. Antimicrob Agents Chemother. 2016. February;60(2):968–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Tothova L, Celec P, Babickova J, et al. Phage therapy of Cronobacter-induced urinary tract infection in mice. Med Sci Monit. 2011;17(7):BR173–BR178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Nishikawa H, Yasuda M, Uchiyama J, et al. T-even-related bacteriophages as candidates for treatment of Escherichia coli urinary tract infections. Arch Virol. 2008. March;153(3):507–515. [DOI] [PubMed] [Google Scholar]
- 183.Vahedi A, Soltan Dallal MM, Douraghi M, et al. Isolation and identification of specific bacteriophage against enteropathogenic Escherichia coli (EPEC) and in vitro and in vivo characterization of bacteriophage. FEMS Microbiol Lett. 2018;365(16). [DOI] [PubMed] [Google Scholar]
- 184.Rastogi V, Yadav P, Verma A, et al. Ex vivo and in vivo evaluation of microemulsion based transdermal delivery of E. coli specific T4 bacteriophage: a rationale approach to treat bacterial infection. Eur J Pharm Sci. 2017. 30 September;107:168–182. [DOI] [PubMed] [Google Scholar]
- 185.Dufour N, Debarbieux L, Fromentin M, et al. Treatment of highly virulent extraintestinal pathogenic Escherichia coli pneumonia with bacteriophages. Crit Care Med. 2015. 20 June;43(6):e190–e198. [DOI] [PubMed] [Google Scholar]
- 186.Bull JJ, Otto G, Molineux IJ. In vivo growth rates are poorly correlated with phage therapy success in a mouse infection model. Antimicrob Agents Chemother. 2012. February;56(2):949–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Aleshkin AV, Rubalskii EO, Volozhantsev NV, et al. A small-scale experiment of using phage-based probiotic dietary supplement for prevention of E. coli traveler’s diarrhea. Bacteriophage. 2015;5(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Maura D, Galtier M, Le Bouguenec C, et al. Virulent bacteriophages can target O104:H4 enteroaggregative Escherichia coli in the mouse intestine. Antimicrob Agents Chemother. 2012. December;56(12):6235–6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pouillot F, Chomton M, Blois H, et al. Efficacy of bacteriophage therapy in experimental sepsis and meningitis caused by a clone O25b: H4-ST131 Escherichia coli strain producing CTX-M-15. Antimicrob Agents Chemother. 2012. July;56(7):3568–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Uchiyama J, Rashel M, Takemura I, et al. In silico and in vivo evaluation of bacteriophage phiEF24C, a candidate for treatment of Enterococcus faecalis infections. Appl Environ Microbiol. 2008. July;74(13):4149–4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Al-Zubidi M, Widziolek M, Court EK, et al. Identification of novel bacteriophages with therapeutic potential targeting Enterococcus faecalis. Infect Immun. 2019;26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Cheng M, Liang J, Zhang Y, et al. The bacteriophage EF-P29 efficiently protects against lethal vancomycin-resistant Enterococcus faecalis and alleviates gut microbiota imbalance in a murine bacteremia model. Front Microbiol. 2017. 09 May;8:837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Malik R, Chhibber S. Protection with bacteriophage KO1 against fatal Klebsiella pneumoniae-induced burn wound infection in mice [Journal Article]. J Microbiol Immunol Infect. 2009. April;42(2):134–40. [PubMed] [Google Scholar]
- 194.Kumari S, Harjai K, Chhibber S. Topical treatment of Klebsiella pneumoniae B5055 induced burn wound infection in mice using natural products. J Infect Dev Ctries. 2010. June 30;4(6):367–77. [PubMed] [Google Scholar]
- 195.Thiry D, Passet V, Danis-Wlodarczyk K, et al. New bacteriophages against emerging lineages ST23 and ST258 of Klebsiella pneumoniae and efficacy assessment in Galleria mellonella larvae. Viruses. 2019. May;11 (5)(411). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kumari S, Harjai K, Chhibber S. Efficacy of bacteriophage treatment in murine burn wound infection induced by Klebsiella pneumoniae [Journal Article]. J Microbiol Biotechnol. 2009. June;19(6):622–8. [DOI] [PubMed] [Google Scholar]
- 197.Hung C-H, Kuo C-F, Wang C-H, et al. Experimental phage therapy in treating Klebsiella pneumoniae-mediated liver abscesses and bacteremia in mice. Antimicrob Agents Chemother. 2011. April;55(4):1358–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Chhibber S, Kaur S, Kumari S. Therapeutic potential of bacteriophage in treating Klebsiella pneumoniae B5055-mediated lobar pneumonia in mice. J Med Microbiol. 2008. December;57(12):1508–1513. [DOI] [PubMed] [Google Scholar]
- 199.Kumari S, Harjai K, Chhibber S. Bacteriophage versus antimicrobial agents for the treatment of murine burn wound infection caused by Klebsiella pneumoniae B5055. J Med Microbiol. 2011. February;60(Pt 2):205–10. [DOI] [PubMed] [Google Scholar]
- 200.Lin TL, Hsieh PF, Huang YT, et al. Isolation of a bacteriophage and its depolymerase specific for K1 capsule of Klebsiella pneumoniae: implication in typing and treatment. J Infect Dis. 2014;210(11):1734–1744. [DOI] [PubMed] [Google Scholar]
- 201.Carrigy NB, Larsen SE, Reese V, et al. Prophylaxis of Mycobacterium tuberculosis H37Rv infection in a preclinical mouse model via inhalation of nebulized bacteriophage D29. Antimicrob Agents Chemother. 2019;16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Trigo G, Martins TG, Fraga AG, et al. Phage therapy is effective against infection by Mycobacterium ulcerans in a murine footpad model. PLoS Negl Trop Dis. 2013. April;7 (4)(e2183). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yilmaz C, Colak M, Yilmaz BC, et al. Bacteriophage therapy in implant-related infections: an experimental study [Journal Article]. J Bone Joint Surg Am. 2013. January 16;95(2):117–25. [DOI] [PubMed] [Google Scholar]
- 204.Henry M, Lavigne R, Debarbieux L. Predicting in vivo efficacy of therapeutic bacteriophages used to treat pulmonary infections. Antimicrob Agents Chemother. 2013. December;57(12):5961–5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Fong SA, Drilling AJ, Ooi ML, et al. Safety and efficacy of a bacteriophage cocktail in an in vivo model of Pseudomonas aeruginosa sinusitis. Transl Res. 2019. April;206:41–56. [DOI] [PubMed] [Google Scholar]
- 206.Cafora M, Deflorian G, Forti F, et al. Phage therapy against Pseudomonas aeruginosa infections in a cystic fibrosis zebrafish model. Sci Rep. 2019 2019/02/06;9(1):1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Watanabe R, Matsumoto T, Sano G, et al. Efficacy of bacteriophage therapy against gut-derived sepsis caused by Pseudomonas aeruginosa in mice. Antimicrob Agents Chemother. 2007. February;51(2):446–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Roach DR, Leung CY, Henry M, et al. Synergy between the host immune system and bacteriophage is essential for successful phage therapy against an acute respiratory pathogen. Cell Host Microbe. 2017. 12 July;22(1):38–47.e4. [DOI] [PubMed] [Google Scholar]
- 209.Heo YJ, Lee YR, Jung HH, et al. Antibacterial efficacy of phages against Pseudomonas aeruginosa infections in mice and Drosophila melanogaster. Antimicrob Agents Chemother. 2009. June;53(6):2469–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hall AR, De Vos D, Friman VP, et al. Effects of sequential and simultaneous applications of bacteriophages on populations of Pseudomonas aeruginosa in vitro and in wax moth larvae. Appl Environ Microbiol. 2012. August;78(16):5646–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Debarbieux L, Leduc D, Maura D, et al. Bacteriophages can treat and prevent Pseudomonas aeruginosa lung infections. J Infect Dis. 2010. 01 April;201(7):1096–1104. [DOI] [PubMed] [Google Scholar]
- 212.Vinodkumar C, Kalsurmath S, Neelagund Y. Utility of lytic bacteriophage in the treatment of multidrug-resistant Pseudomonas aeruginosa septicemia in mice. Indian J Pathol Micr. 2008. 01 July;51(3):360–366. [DOI] [PubMed] [Google Scholar]
- 213.Beeton ML, Alves DR, Enright MC, et al. Assessing phage therapy against Pseudomonas aeruginosa using a Galleria mellonella infection model. Int J Antimicrob Agents. 2015. 20 July;46(2):196–200. [DOI] [PubMed] [Google Scholar]
- 214.Shivshetty N, Hosamani R, Ahmed L, et al. Experimental protection of diabetic mice against lethal P. aeruginosa infection by bacteriophage. Biomed Res Int. 2014;2014(793242). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Pabary R, Singh C, Morales S, et al. Antipseudomonal bacteriophage reduces infective burden and inflammatory response in murine lung. Antimicrob Agents Chemother. 2016. February;60(2):744–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.McVay CS, Velasquez M, Fralick JA. Phage therapy of Pseudomonas aeruginosa infection in a mouse burn wound model. Antimicrob Agents Chemother. 2007. June;51(6):1934–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Basu S, Agarwal M, Kumar Bhartiya S, et al. An in vivo wound model utilizing bacteriophage therapy of Pseudomonas aeruginosa biofilms [Journal Article]. Ostomy Wound Manage. 2015. August;61(8):16–23. [PubMed] [Google Scholar]
- 218.Tiwari BR, Kim S, Rahman M, et al. Antibacterial efficacy of lytic Pseudomonas bacteriophage in normal and neutropenic mice models. J Microbiol. 2011. December;49(6):994–999. [DOI] [PubMed] [Google Scholar]
- 219.Oechslin F, Piccardi P, Mancini S, et al. Synergistic interaction between phage therapy and antibiotics clears Pseudomonas aeruginosa infection in endocarditis and reduces virulence. J Infect Dis. 2017;215(5):703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Nikkhahi F, Dallal MMS, Alimohammadi M, et al. Phage therapy: assessment of the efficacy of a bacteriophage isolated in the treatment of salmonellosis induced by Salmonella enteritidis in mice. Gastroenterol Hepatol Bed Bench. 2017;10(2):125–130. [PMC free article] [PubMed] [Google Scholar]
- 221.Chhibber S, Kaur T, Sandeep K. Co-therapy using lytic bacteriophage and linezolid: effective treatment in eliminating methicillin resistant Staphylococcus aureus (MRSA) from diabetic foot infections [Journal Article]. PLoS One. 2013;8(2):e56022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Prazak J, Iten M, Cameron DR, et al. Bacteriophages improve outcome in experimental Staphylococcus aureus ventilator associated pneumonia. Am J Respir Crit Care Med. 2019;01. [DOI] [PubMed] [Google Scholar]
- 223.Gupta R, Prasad Y. Efficacy of polyvalent bacteriophage P-27/HP to control multidrug resistant Staphylococcus aureus associated with human infections. Curr Microbiol. 2011. January;62(1):255–260. [DOI] [PubMed] [Google Scholar]
- 224.Rasool MH, Yousaf R, Siddique AB, et al. Isolation, characterization, and antibacterial activity of bacteriophages against methicillin-resistant Staphylococcus aureus in Pakistan. Jundishapur J Microbiol. 2016. October;9(e36135). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lehman SM, Mearns G, Rankin D, et al. Design and preclinical development of a phage product for the treatment of antibiotic-resistant Staphylococcus aureus infections. Viruses. 2019. January 21;11(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Hsieh SE, Lo HH, Chen ST, et al. Wide host range and strong lytic activity of Staphylococcus aureus lytic phage Stau2. Appl Environ Microb. 2011. February;77(3):756–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Pincus NB, Reckhow JD, Saleem D, et al. Strain specific phage treatment for Staphylococcus aureus infection is influenced by host immunity and site of infection. PLoS One. 2015. 24 April;10(e0124280). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Capparelli R, Parlato M, Borriello G, et al. Experimental phage therapy against Staphylococcus aureus in mice. Antimicrob Agents Chemother. 2007. August;51(8):2765–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Takemura-Uchiyama I, Uchiyama J, Osanai M, et al. Experimental phage therapy against lethal lung-derived septicemia caused by Staphylococcus aureus in mice. Microbes Infect. 2014. June;16(6):512–517. [DOI] [PubMed] [Google Scholar]
- 230.Zimecki M, Artym J, Kocidba M, et al. Prophylactic effect of bacteriophages on mice subjected to chemotherapy-induced immunosuppression and bone marrow transplant upon infection with Staphylococcus aureus. Med Microbiol Immunol. 2010. May;199(2):71–79. [DOI] [PubMed] [Google Scholar]
- 231.Kishor C, Mishra RR, Saraf SK, et al. Phage therapy of staphylococcal chronic osteomyelitis in experimental animal model. Indian J Med Res. 2016. January;143:87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Seth AK, Geringer MR, Nguyen KT, et al. Bacteriophage therapy for Staphylococcus aureus biofilm-infected wounds: a new approach to chronic wound care. Plast Reconstr Surg. 2013. February;131(2):225–34. [DOI] [PubMed] [Google Scholar]
- 233.Yen M, Cairns LS, Camilli A. A cocktail of three virulent bacteriophages prevents Vibrio cholerae infection in animal models. Nat Commun. 2017. 01 February;8(14187). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Jaiswal A, Koley H, Ghosh A, et al. Efficacy of cocktail phage therapy in treating Vibrio cholerae infection in rabbit model. Microbes Infect. 2013. February;15(2):152–156. [DOI] [PubMed] [Google Scholar]
- 235.Jaiswal A, Koley H, Mitra S, et al. Comparative analysis of different oral approaches to treat Vibrio cholerae infection in adult mice. Int J Med Microbiol. 2014. May;304(3–4):422–430. [DOI] [PubMed] [Google Scholar]
- 236.Shamim Hasan Zahid M, Nashir Udden SM, Faruque ASG, et al. Effect of phage on the infectivity of Vibrio cholerae and emergence of genetic variants. Infect Immun. 2008. November;76(11):5266–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Bhowmick TS, Koley H, Das M, et al. Pathogenic potential of vibriophages against an experimental infection with Vibrio cholerae O1 in the RITARD model. Int J Antimicrob Ag. 2009. June;33(6):569–573. [DOI] [PubMed] [Google Scholar]
- 238.Bhandare S, Colom J, Baig A, et al. Reviving phage therapy for the treatment of cholera. J Infect Dis. 2019. February 15;219(5):786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Jun JW, Shin TH, Kim JH, et al. Bacteriophage therapy of a Vibrio parahaemolyticus infection caused by a multiple-antibiotic-resistant O3:K6 pandemic clinical strain. J Infect Dis. 2014. 01 July;210(1):72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Oliveira H, Mendes A, Fraga AG, et al. K2 capsule depolymerase is highly stable, is refractory to resistance, and protects larvae and mice from Acinetobacter baumannii sepsis. Appl Environ Microbiol. 2019;85(17). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Lin H, Paff ML, Molineux IJ, et al. Therapeutic application of phage capsule depolymerases against K1, K5, and K30 capsulated E. coli in mice. Front Microbiol. 2017. 16 November;8(2257). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Singh PK, Donovan DM, Kumar A. Intravitreal injection of the chimeric phage endolysin Ply187 protects mice from Staphylococcus aureus endophthalmitis. Antimicrob Agents Chemother. 2014. August;58(8):4621–4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Gupta R, Prasad Y. P-27/HP endolysin as antibacterial agent for antibiotic resistant Staphylococcus aureus of human infections. Curr Microbiol. 2011. July;63(1):39–45. [DOI] [PubMed] [Google Scholar]
- 244.Chopra S, Harjai K, Chhibber S. Potential of combination therapy of endolysin MR-10 and minocycline in treating MRSA induced systemic and localized burn wound infections in mice. Int J Med Microbiol. 2016. 01 December;306(8):707–716. [DOI] [PubMed] [Google Scholar]
- 245.Imanishi I, Uchiyama J, Tsukui T, et al. Therapeutic potential of an endolysin derived from kayvirus S25–3 for staphylococcal impetigo. Viruses. 2019. September;11 (9)(769). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Yang H, Gong Y, Zhang H, et al. ClyJ Is a novel pneumococcal chimeric lysin with a cysteine- and histidine-dependent amidohydrolase/peptidase catalytic domain. Antimicrob Agents Chemother. 2019. April;63(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Wright A, Hawkins CH, Anggard EE, et al. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; A preliminary report of efficacy. Clin Otolaryngol. 2009. August;34(4):349–357. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.