Abstract
To discover novel genes underlying amyotrophic lateral sclerosis (ALS), we aggregated exomes from 3,864 cases and 7,839 ancestry matched controls. We observed a significant excess of rare protein-truncating variants among ALS cases, which was concentrated in constrained genes. Through gene level analyses, we replicated known ALS genes including SOD1, NEK1, and FUS. We also observed multiple distinct protein-truncating variants in a highly constrained gene, DNAJC7. The signal in DNAJC7 exceeded genome-wide significance and immunoblotting assays showed depletion of DNAJC7 protein in fibroblasts in an ALS patient carrying the p.Arg156Ter variant. DNAJC7 encodes a member of the heat shock protein family (HSP40), which along with HSP70 proteins, facilitate protein homeostasis including folding of newly synthesized polypeptides and clearance of degraded proteins. When these processes are not regulated, misfolding and accumulation of aberrant proteins can occur leading to protein aggregation, a pathological hallmark of neurodegeneration. Our results highlight DNAJC7 as a novel gene for ALS.
Keywords: Amyotrophic lateral sclerosis, protein truncating variants, neurodegeneration, rare variants, DNAJC7
INTRODUCTION
Amyotrophic lateral sclerosis (ALS) is a late-onset neurodegenerative disease characterized primarily by degeneration of motor neurons leading to progressive weakness of limb, bulbar, and respiratory muscles1,2. Genetic variation is an important risk factor for ALS. Given that 5–10% of patients report a positive family history2 and ~10% of sporadic patients carry known familial ALS gene mutations, the distinction between familial and sporadic disease is increasingly blurred3. Until recently, ALS gene discoveries were made through large multigenerational pedigrees in which the gene and the causal variant segregated in an autosomal dominant inheritance pattern with very few cases of autosomal recessive inheritance reported. Collecting sporadic case samples has been valuable for gene discovery in more common disorders such as schizophrenia4, inflammatory bowel disease5, and type 2 diabetes6, and can have profound effects on the success of targeted therapeutic approaches2,7,8. The most recent ALS genetic discoveries using large massively parallel sequencing data yielded several gene discoveries including TBK1, TUBA4A, ANXA11 and NEK1 and KIF5A9–13; in addition to other risk loci in C21orf2, MOBP, and SCFD114.
Herein, we have assembled the largest ALS exome case-control study to date, consisting of 11,703 individuals (3,864 cases and 7,839 controls). We complemented our analysis by leveraging allele frequencies from large external exome sequencing databases in DiscovEHR (>50,000 samples) and a subset of ExAC (>45,000 samples). In our analysis, we observed an excess of rare protein truncating variants in ALS cases, which primarily resided in genes under strong purifying selection and therefore, are less likely to tolerate deleterious mutations (constrained genes). Furthermore, through gene burden testing in which multiple independent variants are harbored in the same gene therefore, implicating that gene in a disease, we confirmed the known association of SOD1, NEK1, and FUS in ALS. Interestingly, we observed multiple, distinct protein-truncating variants in DNAJC7 in our cohort and in an independent, replication cohort. In our analysis, the signal in DNAJC7 exceeded genome-wide significance and immunoblotting showed depletion of DNAJC7 in fibroblasts from an ALS patient carrying the p.Arg156Ter protein truncating variant. DNAJC7 is a highly constrained gene, and encodes a DNAJ molecular chaperone, which facilitates protein maintenance and quality control, such as folding of newly synthesized polypeptides, and clearance of degraded proteins15. Dysregulation of these processes can lead to aberrant protein aggregation, one of the pathological hallmarks of neurodegenerative diseases.
RESULTS
Patient demographics and dataset overview
We processed our initial dataset of 15,722 samples through a rigorous quality control pipeline using Hail, an open-source, scalable framework for exploring and analyzing genomic data https://hail.is/. All samples were screened for the C9orf72 hexanucleotide expansion (G4C2) and positive samples were excluded from our study. We removed samples with poor sequencing quality, high levels of sequence contamination, closely related with one another, ambiguous sex status, or population outliers per PCA (Supplementary Table 1; Supplementary Fig. 1–2). Our final data set consisted of 3,864 cases and 7,839 controls for a total of 11,703 samples. Individuals were of European descent with 7,355 (62.8%) and 4,348 (37.2%) of samples classified as males and females, respectively. Of 3,864 cases, 2,274 (58.9%) and 1,590 (41.1%) samples were classified as males and females, respectively; where 5,081 (64.8%) and 2,758 (35.2%) were classified as males in controls.
Excess of exome-wide rare protein truncating variants
We assessed four models that incorporated different covariates and assessed their stringency and performance by controlling for benign or synonymous variation. Specifically, each model uses firth based logistic regression and incorporates some or all the covariates: 1) sample sex, 2) PC1-PC10, and either 3) the total exome count (summation of synonymous variants, benign missense variants, damaging missense variants, and protein-truncating variants) or 4) benign variation (summation of synonymous and benign missense variants). We show the results from the most conservative model (model 3), which used all the covariates and the total exome count. Under these models, we evaluated four classes of allele frequency thresholds: (1) singletons, which are variants present in a single individual in our dataset (allele count, AC =1); (2) doubletons, which are present in two individuals in our dataset (AC = 2); (3) ultra-rare singletons, which are singletons in our dataset and are absent in DiscovEHR, a large, independent exome dataset (AC = 1, 0 in DiscovEHR); and finally, (4) rare variants, which have an allele frequency of of <0.01% in our dataset (11,703 samples), in ExAC (non-psychiatric studies, >45,000 samples) and in DiscovEHR (>50,000 samples). For a full explanation of these models and allele frequency thresholds, please see the Methods section.
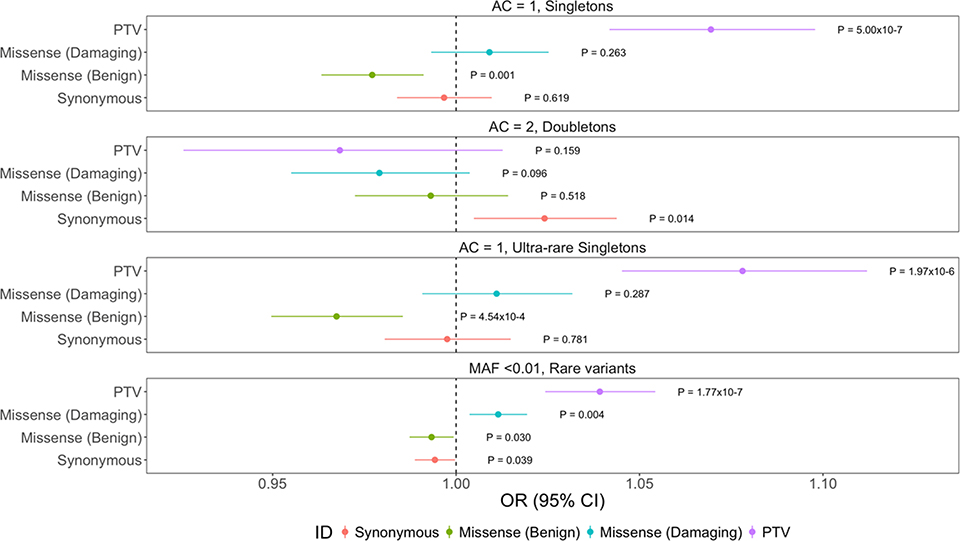
Using model 3, we observed a significant enrichment of singleton protein-truncating variants in ALS cases relative to controls (OR: 1.07, P: 5.00×10–7); ultra-rare singleton PTVs (OR: 1.08, P: 1.97×10–6); and rare PTVs (OR: 1.04, P: 1.77×10–7) (Fig. 1). These values all passed multiple test correction (P<0.0125). The number of doubletons (AC=2) was too low to detect any significant enrichment.
Fig. 1. Exome wide enrichment of protein-truncating variants in ALS cases.
Exome wide analysis of synonymous variants, benign missense variants, damaging missense variants, and protein-truncating variants within singletons, doubletons, ultra-rare singletons, and rare variants. Odds ratios and 95% confidence intervals for each class of variation are depicted by different colors. P-values from firth logistic regression test are also displayed. Multiple test correction P-value: 0.0125. N=3,864 ALS cases; N=7,839 controls. The graph display the mean and standard deviation.
When using model 4 where we restrict to ‘benign variation’ as the final covariate, the protein-truncating variants signal is further enriched among singletons (OR: 1.12, P: <2×10–16); ultra-rare singletons (OR: 1.10, P: 1.53×10–10); and rare variants (OR: 1.04, P: 1.47×10–7). Interestingly, in this analysis, there is a consistent and a significant enrichment of damaging missense variants not observed in the previous analysis: singletons (OR: 1.06, P: <2×10–16); ultra-rare singletons (OR: 1.03, P: 6.33×10–5); and rare variants (OR: 1.01, P: 3.24×10–3).
In our analyses, we use a standard definition of protein-truncating variants as frameshift variants, splice acceptor variants, splice donor variants, or stop gained variants, which are due to insertions or deletions (indels), or single nucleotide variants (SNVs). Given the known elevated error rate in indels we divided all protein-truncating variants as either SNVs or indels and repeated the exome-wide analysis to eliminate any false positive signals. The significant signal is present in both SNVs and indels: SNV singletons (OR: 1.05, P: 2.99×10–3); indel singletons (OR: 1.10, P: 5.75×10–6); SNV ultra-rare singletons (OR: 1.06, P: 4.34×10–3); indel ultra-rare singletons (OR: 1.12, P: 1.96×10–5); and SNV rare variants (OR: 1.03, P: 6.48×10–4); indel rare variants (OR: 1.05, P: 3.30×10–5) (Supplementary Fig. 4). This additional quality control test ensures that the protein-truncating variants signal is driven by both indels and SNVs and is unlikely to be false.
Gene set testing: enrichment of rare variants in constrained genes
To determine whether we could identify the source of the protein-truncating variants enrichment, we assessed multiple different gene sets. We evaluated: (1) constrained genes, which are a set of genes under strong purifying selection; (2) genes known to confer risk to ALS; (3) genes associated with clinically overlapping diseases such as other motor neuron diseases (primary lateral sclerosis, progressive muscular atrophy, progressive bulbar palsy, and spinal muscular atrophy) as well as genes associated with frontotemporal dementia, Parkinson’s disease, Pick’s disease, and Alzheimer’s disease; and finally, (4) genes in which their expression is specific to the brain.
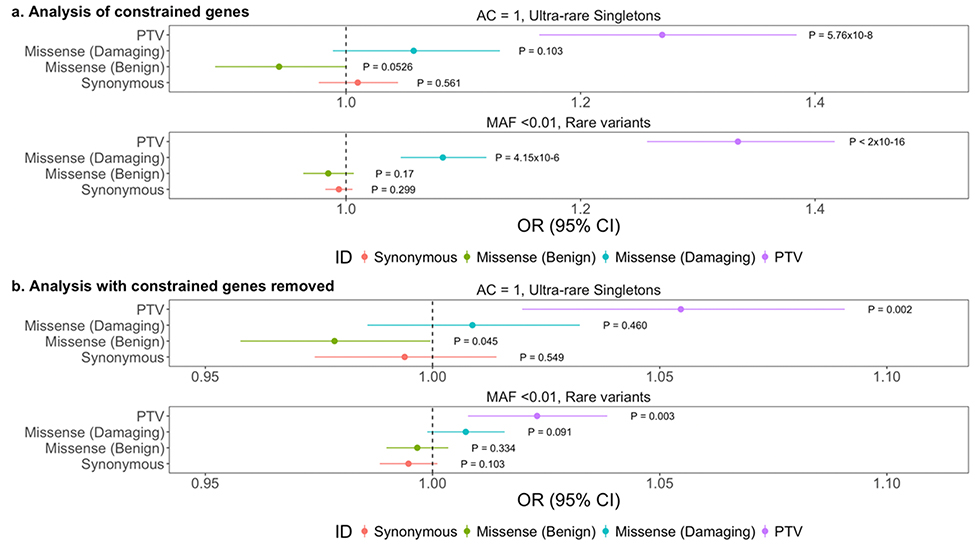
Among constrained genes we observed a significant enrichment of singleton protein-truncating variants (OR: 1.23, P: 7.74×10–7); ultra-rare singletons (OR: 1.27, P: 5.76×10–8), and rare variants (OR: 1.33, P: <2×10–16) (Fig. 2A, Supplementary Fig. 5A). We obtained similar results using model 4 (Supplementary Fig. 5A). To determine whether the entire signal can be explained by constrained genes, we removed them genes and reconducted the analysis. The significant enrichment signal persists however, the effect sizes are attenuated: singleton protein-truncating variants (OR: 1.05, P: 3.30×10–4); ultra-rare singleton protein-truncating variants (OR: 1.05, P: 1.96×10–3); and rare protein-truncating variants (OR: 1.02, P: 2.93×10–3) (Fig. 2B, Supplementary Fig. 5B). This enrichment was also observed in model 4 (Supplementary Fig. 5B).
Fig. 2. Enrichment of protein-truncating variants in constrained genes in ALS cases.
a, Analysis of constrained genes only in synonymous variants, benign missense variants, damaging missense variants, and protein-truncating variants within ultra-rare singletons and rare variants. Odds ratios and 95% confidence intervals for each class of variation are depicted by different colors. P-values from firth logistic regression test are also displayed. Multiple test correction P-value: 0.0125. N=3,864 ALS cases; N=7,839 controls. The graphs display the mean and standard deviation.
b, Exome-wide analysis with constrained genes removed.
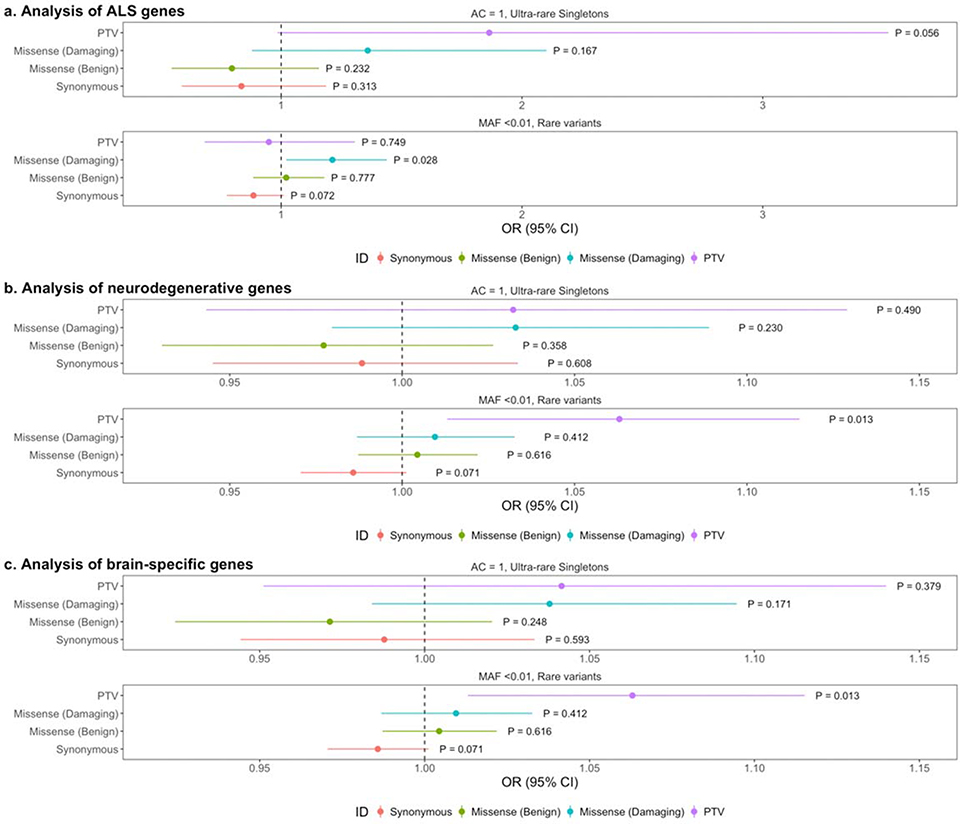
Next, we evaluated the potential effects of known ALS genes. We did not include the ALS genes TBK1, NEK1, KIF5A, C21orf2, MOBP, or SCFD1 as these genes were discovered using datasets that contained a large subset of the same samples and can generate an amplified signal. The known ALS genes had negligible, insignificant effects (Fig. 3A, Supplementary Fig. 6). When including variants from TBK1, NEK1, KIF5A, C21orf2, MOBP, or SCFD1, the negligible signals persist therefore, the initial observation of the exome-wide protein-truncating variant enrichment is not driven by known effects of ALS genes and is likely due to other genomic loci.
Fig. 3. No enrichment of variants in known ALS genes, other related neurodegenerative disease genes, or brain specific genes.
a, Analysis of ALS genes. Synonymous variants, benign missense variants, damaging missense variants, and protein-truncating variants within singletons, doubletons, ultra-rare singletons, and rare variants are shown. Odds ratios and 95% confidence intervals for each class of variation are depicted by different colors. P-values from firth logistic regression test are also displayed. Multiple test correction P-value: 0.0125. N=3,864 ALS cases; N=7,839 controls. The graphs display the mean and standard deviation.
b, Analysis of other neurodegenerative disease genes.
c, Analysis of brain specific genes.
Although ALS is traditionally considered to be a disease of upper and lower motor neurons, more than 50% of ALS patients exhibit neuropsychological and cognitive deficits, with up to 30% of ALS patients meeting some diagnostic criteria for frontotemporal dementia, and some patients may also exhibit Parkinsonism or Parkinsonism-dementia1,16–20. We tabulated a list of genes associated with other motor neuron diseases such as primary lateral sclerosis, progressive muscular atrophy, progressive bulbar palsy, and spinal muscular atrophy. We also included genes associated with frontotemporal dementia, Parkinson’s disease, Pick’s disease, and Alzheimer’s disease (Supplementary Table 5). We did not observe a significant enrichment of variants in any class of variation, suggesting that the initial observation of protein-truncating variant enrichment is unlikely to be explained by only these genes (Fig. 3B, Supplementary Fig. 7).
Finally, we tested whether there is a signal in brain specific genes as ALS is a neurodegenerative disease with the predominant symptoms affecting the central nervous system. We extracted a list of genes with specific brain expression generated using GTEx and performed the same burden analysis across classes of variation. We did not observe any significant differences in protein-truncating variants or damaging missense variation in any allele frequency threshold (Fig. 3C, Supplementary Fig. 8).
Single gene burden analysis replicates previous ALS associations
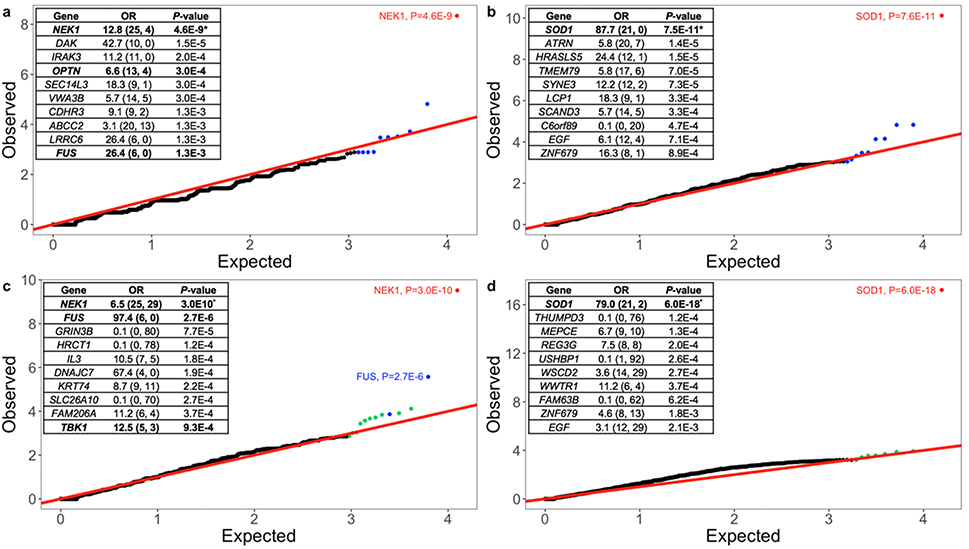
To determine whether a single gene is enriched for variation in ALS cases (ALS-associated) or depleted in ALS cases (ALS-protective), we evaluated ultra-rare (AC=1, absent in DiscovEHR) and rare (MAF <0.001% in our dataset, DiscovEHR, and ExAC) protein-truncating variants and damaging missense variants. Within the ultra-rare variant category, no individual gene passed exome-wide significance. However, the top genes were known ALS genes: (1) NEK1 (PTVs, OR: 12.21, P: 7.32×10–5); (2) OPTN (PTVs, OR: 20.33, P: 1.2×10–4); and (3) SOD1 (dmis, OR: 46.91, P: 5.03×10–6) (Supplementary Fig. 9). Within rare protein-truncating variants, only NEK1 (OR: 12.8, P: 4.59×10–9), passed exome-wide significance; the next top 9 most significant genes, which include FUS, a known ALS gene (OR: 26.4, P: 1.29×10–3), are displayed in Table 1, Fig. 4A. Similarly, within damaging missense variants, SOD1 (OR: 87.7, P: 7.5×10–11) was the only gene to pass exome-wide significance; the top 9 most significant genes are displayed in Table 1, Fig. 4B. In Supplementary Tables 2 and 3, we tabulate the results of the single gene burden analysis for the proposed ALS genes based on the literature, as well as their odds ratio and P-values.
Table 1.
Top hits in protein-truncating variants model in initial (3,864 cases and 7,839 controls), and secondary datasets (3,864 cases and 28,910 controls).
| Gene | Initial OR | Initial P-value | Secondary OR | Secondary P-value |
|---|---|---|---|---|
| Protein truncating variants model | ||||
| NEK1 | 12.8 (25, 4) | 4.6×10–9* | 6.5 (25, 29) | 3.0×10–10# |
| DAK | 42.7 (10, 0) | 1.5×10–5 | 5.8 (10, 13) | 1.4×10–4 |
| IRAK3 | 11.2 (11, 0) | 2.0×10–4 | 2.1 (11, 40) | 0.05 |
| OPTN | 6.6 (13, 4) | 3.0×10–4 | 2.6 (13, 38) | 6.9×10–3 |
| SEC14L3 | 18.3 (9, 1) | 3.0×10–4 | 1.4 (9, 48) | 0.31 |
| VWA3B | 5.7 (14, 5) | 3.0×10–4 | 3.0 (14, 50) | 0.02 |
| CDHR3 | 9.1 (9, 2) | 1.3×10–3 | 1.4 (9, 70) | 0.9 |
| ABCC2 | 3.1 (20, 13) | 1.3×10–3 | 1.8 (20, 84) | 0.03 |
| LRRC6 | 26.4 (6, 0) | 1.3×10–3 | 1.2 (6, 38) | 0.64 |
| FUS | 26.4 (6, 0) | 1.3×10–3 | 97.4 (6, 0) | 2.7×10–6# |
| GRIN3B | 0.4 (0, 10) | 0.04 | 0.05 (0, 80) | 7.7×10–5# |
| HRCT1 | 0.2 (0, 15) | 4.1×10–3 | 0.05 (0, 78) | 1.2×10–4# |
| IL3 | 14.2 (7, 1) | 2.4×10–3 | 10.5 (7, 5) | 1.8×10–4# |
| DNAJC7 | 18.3 (4, 0) | 0.01 | 67.4 (4, 0) | 1.9×10–4# |
| KRT74 | 3.0 (9, 6) | 0.05 | 8.7 (9, 11) | 2.2×10–4# |
| SLC26A10 | 0.1 (0, 9) | 0.03 | 0.1 (0, 70) | 2.7×10–4# |
| FAM206A | 4.1 (6, 3) | 0.07 | 11.2 (6, 4) | 3.7×10–4# |
| TBK1 | 22.3 (5, 0) | 3.9×10–3 | 12.5 (5, 3) | 9.3×10–4# |
| KLHDC4 | 0.1 (0, 14) | 7.4 ×10–3 | 0.1 (0, 61) | 9.8×10–4# |
| DUOXA2 | 2.4 (7, 6) | 0.14 | 5.8 (7, 9) | 1.4×10–3# |
| Damaging missense variants model | ||||
| SOD1 | 87.7 (21, 0) | 7.5×10–11* | 79.0 (21, 2) | 6.0×10–18# |
| ATRN | 5.8 (20, 7) | 1.4×10–5 | 2.0 (20, 74) | 9.2×10–3 |
| HRASLS5 | 24.4 (12, 1) | 1.5×10–5 | 1.6 (12, 56) | 0.13 |
| TMEM79 | 5.8 (17, 6) | 7.0×10–5 | 1.8 (17, 70) | 0.03 |
| SYNE3 | 12.2 (12, 2) | 7.3×10–5 | 1.1 (12, 79) | 0.62 |
| LCP1 | 18.3 (9, 1) | 3.3×10–4 | 3.7 (9, 18) | 2.8×10–3 |
| SCAND3 | 5.7 (14, 5) | 3.3×10–4 | 1.8 (14, 58) | 0.06 |
| C6orf89 | 0.05 (0, 20) | 4.7×10–4 | 0.05 (0, 47) | 5.2×10–3 |
| EGF | 6.1 (12, 4) | 7.1×10–4 | 3.1 (12, 29) | 2.1×10–3 |
| ZNF679 | 16.3 (8, 1) | 8.9×10–4 | 4.6 (8, 13) | 1.8×10–3 |
| THUMPD3 | 0.07 (0, 15) | 4.1×10–3 | 0.08 (0, 76) | 1.2×10–4# |
| MEPCE | 9.1 (9, 2) | 1.3×10–3 | 6.7 (9, 10) | 1.3×10–4# |
| REG3G | 5.4 (8, 3) | 8.2×10–3 | 7.5 (8, 8) | 2.0×10–4# |
| USHBP1 | 0.09 (1, 22) | 1.6×10–3 | 0.08 (1, 92) | 2.6×10–4# |
| WSCD2 | 3.6 (14, 8) | 4.8×10–3 | 3.6 (14, 29) | 2.7×10–4# |
| WWTR1 | 26.4 (6, 0) | 1.3×10–3 | 11.2 (6, 4) | 3.7×10–4# |
| FAM63B | 0.08 (0, 12) | 0.01 | 0.06 (0, 62) | 6.2×10–4# |
Passed exome-wide significance (P-value <2.5×10–6) in first analysis (3,864 cases and 7,839 controls.
OR direction is maintained in secondary analysis (3,864 cases and 28,910 controls) and P-value is lower. Bolded genes have been previously reported in ALS. The results displayed are from a burden analysis using Fisher’s exact test as well as SKAT, with previously defined covariates (sample sex, PC1-PC10, and total exome count). Exome-wide correction for multiple testing was set at (P<2.5×10–6), which was the 5% type-I error rate multiplied by the number of genes tested.
Fig. 4. Quantile-quantile plot of discovery results for rare variants.
a, Rare protein-truncating variants in ALS dataset. X and Y axis represent the negative logarithm P-value. N=3,864 ALS cases; N=7,839 controls. The top 10 genes with their P-values are displayed. Genes in red and blue pass or approach exome-wide significance, respectively. The results displayed are from a burden analysis using Fisher’s exact test as well as SKAT, with previously defined covariates (sample sex, PC1-PC10, and total exome count). Exome-wide correction for multiple testing was set at (P<2.5×10–6), which was the 5% type-I error rate multiplied by the number of genes tested.
b, Rare damaging missense variants in ALS dataset.
c, Rare protein-truncating variants in ALS cases with an additional 21,071 non-Finnish European controls for a total of 28,910 controls. Genes in blue were the most significant genes in the discovery analysis. Genes in green were the most significant genes in the secondary analysis.
d, Rare damaging missense variants in ALS cases and 28,910 controls. The top 10 genes with their P-values are displayed.
To determine if we can reproduce the initial signals observed, we included an additional 21,071 controls from ExAC that are of European descent (non-Finnish) and were not a part of any psychiatric or brain related studies, to eliminate any sample overlap. We performed the same burden analyses using 3,864 cases and 28,910 controls (7,839 controls within our dataset and 21,071 additional controls). In Tables 1 and 2, we display the most significant genes that were identified in the initial discovery and tabulate their OR and P-values for both the initial discovery cohort (3,864 cases and 7,839 controls) and the secondary analysis (3,864 cases and 28,910 controls). Within protein-truncating variants, NEK1 is still the only gene that exceeds exome-wide significance (OR: 6.5, P: 3.03×10–10) (Fig. 4C). Of the next 9 most significant genes in the initial analysis, the only signal that was strengthened was in FUS (OR: 97.4, P: 2.68×10–6). This finding suggests that the other genes may not be true positives or will need further evidence to support their association with ALS. Interestingly, the signal in OPTN, a proposed ALS associated gene, decreased (OR: 6.6, P: 3.0×10–3 to OR: 2.6, P: 6.9×10–3) however, this may be explained in part by the observation that OPTN protein-truncating variants tend to manifest as a recessive form of ALS, which may not be detected in our burden model. With the additional controls, multiple genes had similar ORs as the discovery analysis, with their respective P-values approaching significance (P-values ranging from 7.7×10–5-1.4×10–3). Most notably, the signal in TBK1, a proposed ALS gene based on Cirulli et al. strengthened: (initial analysis; OR: 22.3, P: 3.9×10–3; secondary analysis: OR: 12.5, P: 9.35×10–4). Within damaging missense variants, SOD1 is still the only gene that exceeds exome-wide significance (OR: 79.0, P: 6.0×10–18); however, the next 9 most significant genes no longer approach statistical significance. Similarly, when integrating additional controls, multiple genes approach significance (P-values ranging from 1.2×10–4-6.2×10–4) (Fig. 4D).
Table 2.
Protein-truncating variants and ‘damaging’ missense variants in DNAJC7.
| Variant type | Variant location | cDNA change | Protein change | Cases (n=5,095) | Controls (n=28,910) | gnomAD (non-neuro) AF | CADD | MPC |
|---|---|---|---|---|---|---|---|---|
| Stop gain | 17:g.40152569C>A | c.97G>T | p.E33X | 1 | 0 | 0 | 39 | |
| Stop gain | 17:g.40148376G>A | c.358C>T | p.Q120X | 1 | 0 | 0 | 37 | |
| Stop gain | 17:g.40146902G>A | c.466C>T | p.R156X | 2 | 0 | 0 | 41 | |
| Frameshift | 17:g. 40142393delA | c.488delT | p. F163fs | 1 | 0 | 0 | ||
| Stop gain | 17:g.40141529G>A | c.646C>T | p.R216X | 2 | 0 | 0 | 40 | |
| Essential splice site | 17:g. 40135656T>C | c.1011–2A>G | 1 | 0 | 0 | 26.3 | ||
| Missense | 17:g.40169413C>G | c.22G>C | p.D8H | 1 | 0 | 1.985×10–5 | 25 | 0.78 |
| Missense | 17:g.40149189G>A | c.235C>T | p.R79W | 0 | 1 | 1.204×10–5 | 35 | 1.58 |
| Missense | 17:g. 40141544C>T | c. 631G>A | p. D211N | 1 | 0 | 0 | 26.4 | 0.94 |
| Missense | 17:g.40134023G>A | c.1234C>T | p.R412W | 1 | 0 | 4.029×10–6 | 34 | 1.66 |
| Missense | 17:g.40133984C>T | c.1273G>A | p.E425K | 2 | 0 | 0 | 35 | 1.69 |
AF, allele frequency; empty cell denotes inappliscable information.
Loss of function variants in DNAJC7 in ALS patients
DNAJC7, which is a highly constrained gene (pLI = 0.99) had 4 protein-truncating variants carriers in cases (3,864) and 0 in controls (7,839) in the discovery analysis (OR: 18.3, P: 0.01); and 0 protein-truncating variants in total controls (28,910) (OR: 96.1, P: 1.9×10–4). While DNAJC7 did not initially exceed genome-wide significance, its high constraint score and role in neurodegeneration as a member of the heat shock protein 40 (HSP40) family, encouraged us to evaluate additional datasets to determine its loss of function mutation frequency.
We surveyed data from the UK Motor Neurone Disease Association (n=1,135) and The Agnes Ginges Center for Human Neurogenetics at the Hadassah-Hebrew University Medical Center in Israel (n=96). We observed an additional 4 carriers for a total of 6 distinct protein-truncating variants in 8 individuals with ALS (cases: 5,095; controls: 28,910; OR: 96.6, P: 2.5×10–7) (Table 2). These DNAJC7 variants are extremely rare or completely absent from large population datasets such as gnomAD (Table 2). The DNAJC7 p.Phe163fs variant was observed in the Israeli cohort. As gnomAD does not currently provide variant frequency on individuals of Middle Eastern ethnicity, we screened an additional 3,244 controls from a mixture of Middle Eastern ethnicities for the p.Phe163fs variant and did not observe any carriers further demonstrating its rarity in the general population and an ancestry matched population. In addition, we also observed 15 rare missense variants in DNAJC7, of which 4 are predicted to exert a damaging effect in 5 ALS cases and 1 in control (Table 2).
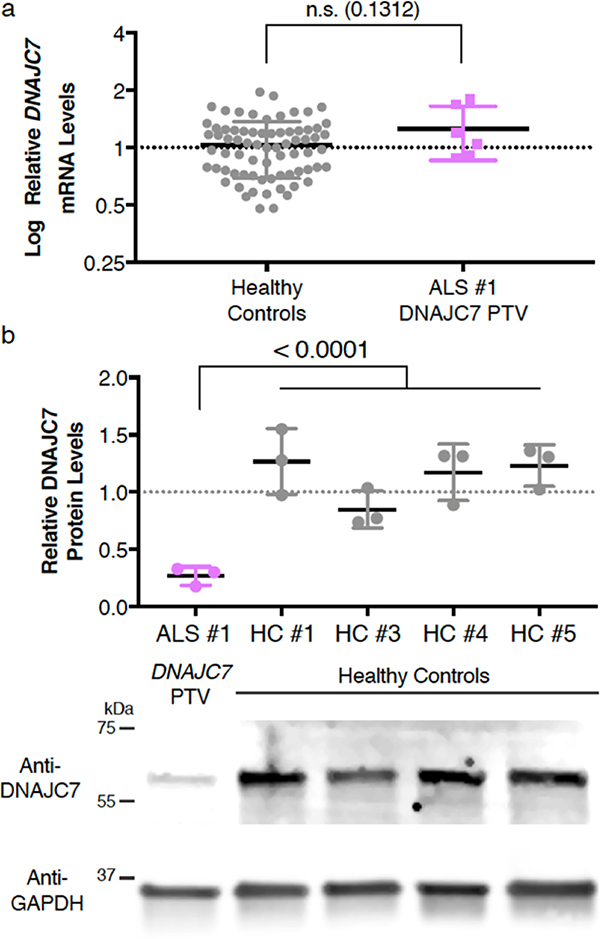
We next proceeded to ask if any of the protein-truncating variants in DNAJC7 can affect its mRNA or protein levels. Accordingly, we collected total RNA from human fibroblasts derived from healthy controls and a patient with a DNAJC7 protein-truncating variant p.Arg156Ter and performed qRT-PCR with two different sets of primer pairs to investigate DNAJC7 transcript levels (Supplementary Fig. 10A and B). These data indicate that DNAJC7 mRNA abundance is not significantly altered in fibroblasts harboring a DNAJC7 protein-truncating variant (Fig. 5A). We next carried out immunoblot assays on protein lysates from fibroblasts and determined that DNAJC7 protein levels were significantly reduced in the ALS patient fibroblasts (Fig. 5B). Although this protein-truncating variant could potentially yield a 17.5 kDa protein, no evidence for such a product was detected (Supplementary Fig. 10C). Together, our findings indicate the protein-truncating variants we identified in DNAJC7 leads to decreased protein levels of this heat shock protein co-chaperone.
Fig. 5. Effects of DNAJC7 protein-truncating variant p.Arg156Ter on transcript and protein levels.
a, qRT-PCR analysis of DNAJC7 transcripts in human fibroblasts from healthy controls or a patient harboring a DNAJC7 protein-truncating variant p.Arg156Ter. Data were normalized to GAPDH and displayed as mean of 3 technical replicates with s.d. from two independent experiments with n=12 control and 1 patient lines (unpaired t test, two-sided, P<0.05). P-value is displayed, 0.1312.
b, Immunoblot analysis for DNAJC7 protein levels in human fibroblast lysates. Protein levels were normalized to GAPDH and displayed relative to the average levels in healthy controls. Data are displayed as mean with s.d. of n=3 technical replicates (unpaired t test, two-sided, P< 0.05). P-value is displayed, <0.0001. The blot image was cropped to make this figure, for the full scan of the blot, please see Supplementary Fig. 10.
DISCUSSION
Herein, we have assembled the exomes of 3,864 ALS cases and 7,839 controls and observed an exome-wide enrichment of protein-truncating variants, which typically result in protein loss-of-function. The abundance of protein-truncating variants in ALS cases seems to be primarily driven by constrained genes, which are under strong purifying selection. When removing constrained genes, the initial exome-wide enrichment of protein-truncating variants remains; however, the effect sizes are much smaller, suggesting that while constraint genes may explain much of protein-truncating variant enrichment, there may be minor residual effects elsewhere in the genome. Accordingly, we examined the effects of ALS associated genes and did not observe any significant enrichment. Importantly, a subset of cases was pre-screened for known pathogenic variants in a select number of known ALS genes and positive cases were eliminated prior to assembling the dataset, which attenuated the effect size estimates and significance for genes in this gene set.
Acknowledging the phenotypic variability of ALS, we also evaluated the effects of genes implicated in other motor neuron diseases such as primary lateral sclerosis, progressive muscular atrophy, progressive bulbar palsy, and spinal muscular atrophy; as well as genes associated with frontotemporal dementia, Parkinson’s disease, Pick’s disease, and Alzheimer’s disease. We did not observe a significant enrichment in any class of variation, suggesting that the initial observation of excess protein-truncating variants do not reside in these genes. Lastly, the genes implicated in the development of ALS are not specifically expressed in motor neurons, nor are they brain specific, despite the specific degree of degeneration of upper and lower motor neurons. Nevertheless, we tested whether the signal in protein-truncating variants is concentrated in brain specific genes, a much larger gene set than ALS genes only. We did not observe any significant enrichment within brain specific genes.
The single gene burden analysis identified the most significant genes as SOD1, NEK1, and FUS, which are known ALS genes. No other individual gene passed exome-wide significance within our dataset (3,864 cases and 7,839 controls) and the additional controls in the secondary analysis (3,864 cases and 28,910 controls). Notably, in the secondary analysis, multiple genes with consistent OR and lower P-values than the initial analysis, surfaced. Within protein-truncating variants, these include: GRIN3B, HRCT1, IL3, and DNAJC7. Interestingly, protein-truncating variants in GRIN3B and HRCT1 may offer protection against ALS: OR: 0.05, P: 7.7×10–5; OR: 0.05, P: 1.2×10–4, respectively; while protein-truncating variants in IL3 and DNAJC7 may confer risk: OR: 10.5, P: 1.8×10–4; OR: 67.4, P: 1.9×10–4).
In this analysis, DNAJC7 had 4 protein-truncating variant carriers in 3,864 cases and 0 in 7,839 and 28,910 controls additionally, when integrating data from the UK Motor Neurone Disease Association, we observed an additional 4 protein-truncating variant carriers for a total of 6 distinct protein-truncating variants in 8 individuals (initial analysis P: 0.01; secondary analysis P: 1.9×10–4; replication analysis P: 2.5×10–7). According to the HPA RNA-seq normal tissues project21 and the Genotype-Tissue Expression (GTEx) project22, DNAJC7 is ubiquitously expressed with elevated expression in the brain. DNAJC7 encodes a molecular chaperone, DnaJ heat shock protein family (HSP40) member C7, and like all DNAJ proteins, contains an approximately 70 amino acid J-domain, which is critical for binding to HSP70 proteins23. There are approximately 50 DNAJ proteins, which are also classified as HSP40 proteins, that facilitate protein maintenance and quality control, such as folding of newly synthesized polypeptides, and clearance of degraded proteins15,24,25. Specifically, DNAJs act as co-chaperones for HSP70 proteins by regulating ATPase activity, aid in polypeptide binding, and prevention of premature polypeptide folding25,26.
Aberrant protein aggregation due to accumulation of misfolded proteins, is one of the pathological hallmarks of neurodegenerative diseases like Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, prion disease, and ALS27–32. HSP proteins have a conserved and central role in protein function by aiding in their folding and stabilization, and the clearance of misfolded proteins, ultimately diminishing protein aggregates and the associated pathologies. However, genetic aberrations or cellular stress such as exposure to environmental toxins, fluctuations in temperature, chemical stress, cell injury, or aging, can influence the dynamics of the protein quality control network allowing misfolded proteins to go undetected thereby triggering neurotoxicity33,34. Furthermore, abnormal expression of HSP70 and DNAJ genes leads to the formation of protein aggregates in models of Alzheimer’s disease35, Parkinson’s disease36,37, Huntington’s disease35,38, prion disease39,40, and ALS41–43. In light of these studies, elevated HSP expression is thought to be beneficial in preventing or in halting neurodegenerative disease progression44. For example, overexpression of DNAJB6b and DNAJB8 suppressed toxic protein aggregation45; while overexpression of HSP70 in neuroglioma cells decreased the formation of alpha-synuclein fibrils46. Within ALS models, overexpression of HSPB8 promoted clearance of mutant SOD147; double transgenic mice overexpressing HSP27 and mutated SOD1 exhibited increased survival of spinal motor neurons than mice overexpressing a SOD1 mutation only, however, the neuroprotective effects were not sustained in later stages of the disease48. Finally, DNAJB2, which when mutated can cause autosomal recessive spinal muscular atrophy, was overexpressed in mice motor neurons also expressing a SOD1 mutation (p.Gly93Ala), and led to reduced mutant SOD1 aggregation and improved motor neuron survival49. In Supplementary Table 4, we tabulated additional HSP genes that have been reported to harbor pathogenic or likely pathogenic mutations in patients with neurodegenerative diseases.
In summary, we observed a significant exome-wide enrichment of protein-truncating variants, which seem to primarily reside in constrained genes. Through gene burden tests, we confirmed the known association of ALS genes SOD1, NEK1, and FUS, and also observed multiple protein-truncating variants in ALS cases in a highly constrained, HSP40 gene, DNAJC7. Our replication of protein-truncating variants in DNAJC7 in an independent ALS cohort as well as functional validation highlights loss of DNAJC7 as a novel genetic risk factor for ALS.
METHODS
Study overview
The familial ALS (FALS) and the ALS Genetics (ALSGENS) consortia were assembled to aggregate the existing ALS sequencing data in the community to improve the power to discover novel genetic risk factors for ALS. Herein, we describe our approach of assembling the largest ALS exome case-control study to date.
Sample acquisition
Blood samples were collected from subjects following appropriate and informed consent in accordance with the Research Ethics Board at each respective recruiting site within the CReATe, FALS, and ALSGENS consortia. All samples known to be carriers of the C9orf72 hexanucleotide expansion (G4C2) were excluded from the study. Additionally, prior to exome sequencing, a subset of the samples (approximately 2,000) were genotyped and screened for known variants in known ALS genes, SOD1, FUS, and TARDBP; and were only included in our study if they were found to be negative for the variants tested.
Exome sequencing data for control and a subset of case samples were downloaded from dbGAP and were not enriched for (but not specifically screened for) ALS or other neurodegenerative disorders. Control samples were matched to case samples with respect to similar capture kits and coverage levels. The age of control samples was not provided for all samples but in general, controls were older than typical age of onset of ALS. The data are available under the following accession codes: MIGen Exome Sequencing: Ottawa Heart (phs000806.v1.p1); MIGen Exome Sequencing: Leicester UK Heart Study (phs001000.v1.p1); Swedish Schizophrenia Population-Based Case-control Exome Sequencing (phs000473.v2.p2); Genome-Wide Association Study of Amyotrophic Lateral Sclerosis (phs000101.v5.p1).
No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications9. Randomization of experimental groups was not applicable to this study. The experimental conditions are determined by each individual’s genetics, which are fixed at conception. This reflects a randomization of the alleles inherited from each individual’s parents (i.e.mendelian randomization), but it does not involve randomization of experimental parameters. Blinding was not relevant to the study as this study was composed of cases and controls. Therefore, the analyst needed to know the case-control status of every participant.
Whole exome sequencing
15,722 DNA samples were sequenced at the Broad Institute, Guy’s Hospital, McGill University, Stanford University, HudsonAlpha, and University of Massachusetts, Worcester. Samples weresequenced using the exome Agilent All Exon (37MB, 50MB, or 65MB), Nimblegen SeqCap EZ V2.0 or 3.0 Exome Enrichment kit, Illumina GAIIx, HiSeq 2000, or HiSeq 2500 sequencers according to standard protocols.
All samples were joint called together and were aligned to the consensus human genome sequence build GRCh37/hg19; and BAM files were processed using BWA Picard. Genotype calling was performed using the Genome Analysis Toolkit’s (GATK) HaplotypeCaller and was performed at the Broad Institute as previously described50,51.
Hail software and quality control
Code availability: we used Hail, an open-source, scalable framework for exploring and analyzing genomic data https://hail.is/ to process the data. All quality control steps were performed using Hail 0.1 (Supplementary Table 1).
(1). Sample QC and Variant QC
Samples with high proportion of chimeric reads (>5%) and high contamination (>5%) were excluded. Samples with poor call rates (<90%), mean depth <10x, or mean genotype-quality <65 were also eliminated from further analysis.
For variant QC, we restricted to GENCODE coding regions, independent of capture interval, where both Agilent and Illumina exomes surpass 10x mean coverage. We restricted to ‘PASS’ variants in GATK’s Variant Quality Score Recalibration (VQSR) filter. Individual genotypes were filtered (set to missing) if they did not meet the following criteria: 1) genotype depth (g.DP) 10 or greater 2) Allele balance >=0.2 in heterozygous sites or <= 0.8 for homozygous reference and homozygous alternate variants 3) Genotype quality (GQ)> 20. Finally, we selected variants with call rate >90% and Hardy-Weinberg equilibrium test P-value >1×10–6. For quality control analysis, see Supplementary Table 1 and Supplementary Fig. 1.
(2). Sex imputation
We used the X chromosome inbreeding coefficient to impute sample sex. Samples with an X chromosome inbreeding coefficient >0.8 were classified as males and samples with an X chromosome inbreeding coefficient <0.4 were classified as females. Samples within <0.8 and >0.4 were classified as having ambiguous sex status, and therefore were excluded from the dataset (Supplementary Table 1).
(3). Principal component analysis
Principal component analysis (PCA) was performed using Hail. We used a subset of high confidence SNPs in the exome capture region to calculate the principal components. We used only ancestry-matched cases and controls as indicated by overlapping population structure. Furthermore, we used 1000 Genomes samples to determine the general ethnicity of the ALS dataset. The majority of the samples in the ALS dataset were reported to be of European descent and this was confirmed by PCA with 1000 Genomes samples (Supplementary Fig. 2, Supplementary Table 1).
(4). Relatedness check
We included only unrelated individuals (IBD proportion < 0.2) (Supplementary Table 1).
(5). Variant annotation
We annotated protein-coding variants into four classes: (1) synonymous; (2) benign missense; (3) damaging missense; and (4) protein-truncating variants (PTV). Using VEP annotations (Version 85)52, we classified synonymous variants as: “synonymous_variant”, “stop_retained_variant”, and “incomplete_terminal_codon_variant”. Missense variants were classified as: “inframe_deletion”, “inframe_insertion”, “missense_variant”, “stop_lost”, “start_lost”, and “protein_altering_variant”. Furthermore, benign missense variants were predicted as “tolerated” and “benign” by PolyPhen-2 and SIFT, respectively; whereas damaging missense variants were predicted as “probably damaging” and “deleterious”. Finally, protein-truncating variants were classified as: “frameshift_variant”, “splice_acceptor_variant”, “splice_donor_variant”, and “stop_gained”.
(6). Allele frequency categorization
Allele frequencies were estimated within our case-control sample, and from two external exome sequence databases, DiscovEHR and ExAC53. DiscovEHR is a publicly available database with >50,000 exomes of participants who may have some health conditions however, they do not have ALS. ExAC is a mixture of healthy controls and complex disease patients, and we restricted to the non-psychiatric subset of ExAC for allele frequency estimation. Of note, many of our controls are present in the ExAC database, so we restricted to the DiscovEHR cohort to determine ultra-rare singletons. We did not use gnomAD for this analysis as our cases and our controls have been deposited into this resource.
We classified variant allele frequency using the following criteria: (1) singletons, which are variants present in a single individual in our dataset (allele count, AC =1); (2) doubletons, which are present in two individuals in our dataset (AC = 2); (3) ultra-rare singletons, which are singletons in our dataset and are absent in DiscovEHR (AC = 1, 0 in DiscovEHR); and finally, (4) rare variants, which have a MAF of <0.01% in our dataset (11,703 samples), in ExAC (non-psychiatric studies, >45,000 samples) and in DiscovEHR (>50,000 samples).
Multivariate models used for analysis
To determine whether an enrichment of a specific class of variation was present in ALS cases versus controls, we ran multiple Firth logistic regression models. The Firth penalization is used in the likelihood model due to the low counts in many tests, and helps to minimize the type I error rate when multiple covariates are included in the model54. Model 1 predicted ALS case-control status solely from variant count; Model 2 incorporated multiple covariates: (1) sample sex, (2) sample population structure from the first 10 principal components; Model 3 incorporated all covariates used the second model along with (3) sample total exome count, which is the exome-wide count of variants in the specific frequency class tested. Finally, Model 4 is similar to Model 3, but instead uses the “benign variant” count as a covariate, which is the exome-wide count of synonymous variants and benign missense variants only, rather than total exome count. Model 3, which we considered to be the most conservative model to represent the dataset, was used as the preferred model for our analysis (Supplementary Fig. 3).
Exome-wide burden
The four Firth logistic regression models above were used to predict case-control status from exome-wide counts of synonymous, missense, and protein-truncating variants. Given that sequencing errors are more prevalent when calling insertions or deletions (indels)55,56, we divided variants within the protein-truncating variants category as either 1) SNV-based protein-truncating variants or 2) indel-based protein-truncating variants, due to single nucleotide variants (SNVs) or indels, respectively. This ensures that any enrichment observed in protein-truncating variants is not solely from indel-based protein-truncating variants.
Gene sets
(1). Constrained genes (pLI genes: 3,488, constrained missense genes: 1,730)
We evaluated whether variation in loss of function intolerant (pLI) genes are associated with ALS using the same approach as described in the exome-wide approach however, we extracted only high pLI genes from the exome. We obtained the genic pLI intolerance metrics from Lek et al., 2016 available online: (ftp://ftp.broadinstitute.org/pub/ExAC_release/release0.3/functional_gene_constraint/). For protein-truncating variants, we used genes with a probability of loss-of-function intolerant (pLI) >0.9. We also evaluated missense constrained genes generated by Samocha et al., 201457. For missense variants, we used genes with a z-score of >3.09.
(2). ALS associated genes (38 genes)
We also examined exome-wide burden with known ALS genes removed. The list of ALS genes are as follows: TARDBP, DCTN1, ALS2, CHMP2B, ARHGEF28, MATR3, SQSTM1, FIG4, HNRNPA2B1, C9orf72, SIGMAR1, VCP, SETX, OPTN, PRPH, HNRNPA1, DAO, ATXN2, ANG, FUS, PFN1, CENPV, TAF15, GRN, MAPT, PNPLA6, UNC13A, VAPB, SOD1, NEFH, ARPP21, and UBQLN2. We did not remove TBK1, NEK1, KIF5A, C21orf2, MOBP, or SCFD1 as these genes were discovered using datasets that contained a large subset of the same samples. We also performed an analysis with all proposed ALS genes.
(3). Neurodegenerative disease genes (120 genes)
We investigated whether genes associated with other neurodegenerative phenotypes showed enrichment in ALS cases. We included the following motor neuron diseases: primary lateral sclerosis, progressive muscular atrophy, progressive bulbar palsy, and spinal muscular atrophy. We also used genes associated with Parkinson’s disease, frontotemporal dementia, Pick’s disease, and Alzheimer’s disease as patients with ALS can also present with frontotemporal dementia, cognitive impairment, or Parkinsonism (Supplementary Table 5).
(4). Brain expressed genes (2,650 genes)
We evaluated whether genes expressed specifically in the brain were enriched for variation in our dataset. For this analysis, we used brain specific genes generated by Ganna et al., 2016.
Single gene burden analysis
(1). ALS dataset (3,864 cases and 7,839 controls)
To determine whether a single gene is enriched or depleted for rare protein-coding variation in ALS cases, we performed a burden analysis using Fisher’s exact test as well as SKAT, with previously defined covariates (sample sex, PC1-PC10, and total exome count). Exome-wide correction for multiple testing was set at (P<2.5×10–6), which was the 5% type-I error rate multiplied by the number of genes tested. We performed four different tests in ALS cases and controls: (1) ultra-rare protein-truncating variants (AC=1 and absent in DiscovEHR); (2) ultra-rare damaging missense variants (AC=1 and absent in DiscovEHR); (3) rare protein-truncating variants (MAF <0.001% in the dataset, DiscovEHR, and ExAC); and (4) rare damaging missense variants (MAF <0.001% in the dataset, DiscovEHR, and ExAC).
(2). ALS dataset and additional controls (3,864 cases and 28,910 controls)
We also included an additional 21,071 samples from ExAC that are of European descent (non-Finnish) and were not a part of any psychiatric or brain related studies, to eliminate any sample overlap. Furthermore, to mitigate against false discoveries, in addition to passing our QC filters, we ensured each variant also passed gnomAD (123,136 exomes and 15,496 genomes) QC filters. We included variants that were either a singleton (AC=1) in gnomAD or completely absent to ensure we minimize the inclusion of an excess of variants that passed gnomAD QC, that were rare (MAF <0.001%), yet were still observed in a very high number of individuals and were likely, false positive variants. The additional 21,071 samples allowed us to perform a secondary analysis of the genes that approached statistical significance (P<2.5×10–6) and determine whether their OR and P-values are maintained and exceed statistical significance, respectively. Additionally, we also used the 21,071 controls to increase statistical power to detect any gene discoveries not detected in the original dataset. Importantly, we did not perform a joint PCA on the 21,071 non-Finnish European controls and our dataset, therefore, we are unable to completely match the ancestry of our dataset.
Cell acquisition culture and authentication
The fibroblasts used in this study were previously approved by the institutional review boards (IRBs) of Harvard University, Massachusetts General Hospital, and Columbia University. Specific point mutations were confirmed by PCR amplification followed by Sanger sequencing. Weekly, cultures were checked for mycoplasma contamination using the MycoAlert kit (Lonza) with no cell lines used in this study testing positive. The use of these cells at Harvard was further approved and determined not to constitute Human Subjects Research by the Committee on the Use of Human Subjects in Research at Harvard University. Human fibroblasts were grown with DMEM (Invitrogen) supplemented with 15% fetal bovine serum (VWR), 10 mM MEM Non-essential amino acid (Millipore), and B-mercaptoethanol 55 μM (Invitrogen), and cultured on tissue culture dishes maintained in 5% CO2 incubators at 37°C. Fibroblasts were passaged after reaching confluency using trypsin (Invitrogen).
Immunoblot assays
For analysis of DNAJC7 protein expression levels, fibroblasts were lysed in RIPA buffer (150mM Sodium Chloride; 1% Triton X-100; 0.5% sodium deoxycholate; 0.1% SDS; 50 mM Tris pH 8.0) containing protease and phosphatase inhibitors (Roche) for 20 min on ice, and centrifuged at high speed to remove insoluble components. 500 μL of RIPA buffer per well of a 6-well plate were routinely used, which yielded ~20μg of total protein as determined by BCA (Thermo Scientific). For immunoblot assays, 1 μg of total protein was separated by SDS-PAGE (BioRad), transferred to PDVF membranes (BioRad) and probed with antibodies against DNAJC7 (1:1000, Abcam, Clone EPR13349) and GAPDH (1:1000, Millipore, Clone 6C5). LI-COR software (Image Studios) was used to quantitate protein band signal, and GAPDH levels were used to normalized each sample. Data are from three technical replicates with n=12 control and 1 patient lines. To analyze the results from this experiment, we used an unpaired t test, two-sided with a statistical threshold of P<0.05.
RNA preparation and qRT-PCR
Total RNA was isolated from fibroblasts using Trizol (Invitrogen) according to manufacturer’s instructions. 500 μL of Trizol were added per well of the 6-well cultures. A total of 300–1000ng of total RNA was then used to synthesize cDNA by reverse transcription according to the iSCRIPT kit (Bio-rad). Quantitative RT-PCR (qRT-PCR) was then performed using SYBR green (Bio-Rad) and the iCycler system (Bio-rad). Quantitative levels for all genes assayed were normalized using GAPDH expression. For comparison between control and patient lines, normalized expression was displayed relative to the average of pooled data points from the healthy controls. The primer sequences (forward, reverse) are for GAPDH (AATGGTGAAGGTCGGTGTG, GTGGAGTCATACTGGAACATGTAG), DNAJC7 Exons 4–6 (CAGTGAGGTTGGATGACAGTT, ACTCTTGTTGTGCCTGAGC), DNAJC7 Exons 13–14 (TACTATCCTCTCTGATCCCAAGA, CCTTGTTCTCCAGCTGAGAG). Data are from three technical replicates with n=12 control and 1 patient lines. To analyze the results from this experiment, we used an unpaired t test, two-sided with a statistical threshold of P<0.05.
Data presentation and statistical analysis
In the figure elements, points and lines represent the median and standard deviation, respectively. The plots display the minimum to maximum. Data distribution was assumed to be normal but this was not formally tested. For the exome-wide and gene specific test, we build four models that use firth logistic regression, please refer to ‘Multivariate models used for analysis’ in the Materials and Methods section. Multiple test correction P-value < 0.0125 was considered significant. For gene specific analyses, a multiple test correction P-value <2.5×10–6 was considered significant. For the immunoblot and qPCR assays, the statistical analyses were performed using a two-tail unpaired Student’s t-test, with a P value of *P<0.05 considered as significant using Prism 7 (Graph Pad).
Reporting Summary
Further information on research design is available in the Nature Research Life Sciences Reporting Summary linked to this article.
Data availability
The sequencing data discussed in this publication were obtained through dbGaP and are available under the following accession codes: MIGen Exome Sequencing: Ottawa Heart (phs000806.v1.p1); MIGen Exome Sequencing: Leicester UK Heart Study (phs001000.v1.p1); Swedish Schizophrenia Population-Based Case-control Exome Sequencing (phs000473.v2.p2); Genome-Wide Association Study of Amyotrophic Lateral Sclerosis (phs000101.v5.p1).
Code availability
Code used to conduct the analysis is provided online.
Supplementary Material
ACKNOWLEDGEMENTS
We thank and acknowledge the consent and cooperation of all study participants. Many thanks to F. Cerrato for helping us assemble the dataset and providing general project management; and to T. Poterba, J. Bloom, D. King, and C. Seed for their assistance in Hail. Data used in this research were in part obtained from the UK MND Collections for MND Research, funded by the MND Association and the Wellcome Trust. We would like to thank people with MND and their families for their participation in this project. The project is supported through the following funding organisations under the aegis of JPND - www.jpnd.eu (United Kingdom, Medical Research Council (MR/L501529/1; MR/R024804/1) and Economic and Social Research Council (ES/L008238/1)) and through the Motor Neurone Disease Association. This study represents independent research part funded by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. Samples used in this research were in part obtained from the UK National DNA Bank for MND Research, funded by the MND Association and the Wellcome Trust. We acknowledge sample management undertaken by Biobanking Solutions funded by the Medical Research Council at the Centre for Integrated Genomic Medical Research, University of Manchester. The CReATe consortium (U54NS092091) is part of Rare Diseases Clinical Research Network (RDCRN), an initiative of the Office of Rare Diseases Research (ORDR), NCATS. This consortium is funded through collaboration between NCATS, and the NINDS. Additional support is provided by the ALS Association (17-LGCA-331). S.M.K. Farhan is supported by the ALS Canada Tim E. Noël Postdoctoral Fellowship. J.R. Klim was supported by the Project ALS Tom Kirchhoff Family Postdoctoral Fellowship and acknowledges K. Mamia and L.T. Kane for their work banking fibroblasts.
Footnotes
COMPETING INTERESTS
MN participation is supported by a consulting contract between Data Tecnica International and the National Institute on Aging, NIH, Bethesda, MD, USA, as a possible conflict of interest. MN also consults for Lysosomal Therapeutics Inc, the Michael J. Fox Foundation and Vivid Genomics among others. The other authors declare no competing interests.
REFERENCES
- 1.Strong MJ et al. Amyotrophic lateral sclerosis - frontotemporal spectrum disorder (ALS-FTSD): Revised diagnostic criteria. Amyotroph Lateral Scler Frontotemporal Degener 18, 153–174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Chalabi A, van den Berg LH & Veldink J Gene discovery in amyotrophic lateral sclerosis: implications for clinical management. Nat Rev Neurol 13, 96–104 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Al-Chalabi A Perspective: Don’t keep it in the family. Nature 550, S112 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Singh T et al. Rare loss-of-function variants in SETD1A are associated with schizophrenia and developmental disorders. Nat Neurosci 19, 571–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohanan V et al. C1orf106 is a colitis risk gene that regulates stability of epithelial adherens junctions. Science 359, 1161–1166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manning A et al. A Low-Frequency Inactivating AKT2 Variant Enriched in the Finnish Population Is Associated With Fasting Insulin Levels and Type 2 Diabetes Risk. Diabetes 66, 2019–2032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamburg MA & Collins FS The path to personalized medicine. N Engl J Med 363, 301–4 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Nelson MR et al. The support of human genetic evidence for approved drug indications. Nat Genet 47, 856–60 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Cirulli ET et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science 347, 1436–41 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith BN et al. Exome-wide rare variant analysis identifies TUBA4A mutations associated with familial ALS. Neuron 84, 324–31 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith BN et al. Mutations in the vesicular trafficking protein annexin A11 are associated with amyotrophic lateral sclerosis. Sci Transl Med 9(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kenna KP et al. NEK1 variants confer susceptibility to amyotrophic lateral sclerosis. Nat Genet 48, 1037–42 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicolas A et al. Genome-wide Analyses Identify KIF5A as a Novel ALS Gene. Neuron 97, 1268–1283 e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Rheenen W et al. Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat Genet 48, 1043–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lackie RE et al. The Hsp70/Hsp90 Chaperone Machinery in Neurodegenerative Diseases. Front Neurosci 11, 254 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swinnen B & Robberecht W The phenotypic variability of amyotrophic lateral sclerosis. Nat Rev Neurol 10, 661–70 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Farhan SM et al. The Ontario Neurodegenerative Disease Research Initiative (ONDRI). Can J Neurol Sci 44, 196–202 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Farhan SMK, Gendron TF, Petrucelli L, Hegele RA & Strong MJ OPTN p.Met468Arg and ATXN2 intermediate length polyQ extension in families with C9orf72 mediated amyotrophic lateral sclerosis and frontotemporal dementia. Am J Med Genet B Neuropsychiatr Genet 177, 75–85 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Aarsland D, Zaccai J & Brayne C A systematic review of prevalence studies of dementia in Parkinson’s disease. Mov Disord 20, 1255–63 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Hely MA, Reid WG, Adena MA, Halliday GM & Morris JG The Sydney multicenter study of Parkinson’s disease: the inevitability of dementia at 20 years. Mov Disord 23, 837–44 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Fagerberg L et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics 13, 397–406 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Consortium GT The Genotype-Tissue Expression (GTEx) project. Nat Genet 45, 580–5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang J et al. Structural basis of J cochaperone binding and regulation of Hsp70. Mol Cell 28, 422–33 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kampinga HH & Craig EA The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol 11, 579–92 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayer MP & Bukau B Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci 62, 670–84 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clerico EM, Tilitsky JM, Meng W & Gierasch LM How hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. J Mol Biol 427, 1575–88 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uddin MS et al. Autophagy and Alzheimer’s Disease: From Molecular Mechanisms to Therapeutic Implications. Front Aging Neurosci 10, 04 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irwin DJ, Lee VM & Trojanowski JQ Parkinson’s disease dementia: convergence of alpha-synuclein, tau and amyloid-beta pathologies. Nat Rev Neurosci 14, 626–36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imarisio S et al. Huntington’s disease: from pathology and genetics to potential therapies. Biochem J 412, 191–209 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Brundin P, Melki R & Kopito R Prion-like transmission of protein aggregates in neurodegenerative diseases. Nat Rev Mol Cell Biol 11, 301–7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ross CA & Poirier MA Protein aggregation and neurodegenerative disease. Nat Med 10 Suppl, S10–7 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Winklhofer KF, Tatzelt J & Haass C The two faces of protein misfolding: gain- and loss-of-function in neurodegenerative diseases. EMBO J 27, 336–49 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR & Morimoto RI Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 311, 1471–4 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Voisine C, Pedersen JS & Morimoto RI Chaperone networks: tipping the balance in protein folding diseases. Neurobiol Dis 40, 12–20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brehme M et al. A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease. Cell Rep 9, 1135–50 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roodveldt C et al. Chaperone proteostasis in Parkinson’s disease: stabilization of the Hsp70/alpha-synuclein complex by Hip. EMBO J 28, 3758–70 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auluck PK, Chan HY, Trojanowski JQ, Lee VM & Bonini NM Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science 295, 865–8 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Wacker JL et al. Loss of Hsp70 exacerbates pathogenesis but not levels of fibrillar aggregates in a mouse model of Huntington’s disease. J Neurosci 29, 9104–14 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kovacs GG et al. Prominent stress response of Purkinje cells in Creutzfeldt-Jakob disease. Neurobiol Dis 8, 881–9 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Jones G, Song Y, Chung S & Masison DC Propagation of Saccharomyces cerevisiae [PSI+] prion is impaired by factors that regulate Hsp70 substrate binding. Mol Cell Biol 24, 3928–37 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen HJ et al. The heat shock response plays an important role in TDP-43 clearance: evidence for dysfunction in amyotrophic lateral sclerosis. Brain 139, 1417–32 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Udan-Johns M et al. Prion-like nuclear aggregation of TDP-43 during heat shock is regulated by HSP40/70 chaperones. Hum Mol Genet 23, 157–70 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang YJ et al. Phosphorylation regulates proteasomal-mediated degradation and solubility of TAR DNA binding protein-43 C-terminal fragments. Mol Neurodegener 5, 33 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benatar M et al. Randomized, double-blind, placebo-controlled trial of arimoclomol in rapidly progressive SOD1 ALS. Neurology 90, e565–e574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hageman J et al. A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation. Mol Cell 37, 355–69 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Outeiro TF et al. Formation of toxic oligomeric alpha-synuclein species in living cells. PLoS One 3, e1867 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crippa V et al. The small heat shock protein B8 (HspB8) promotes autophagic removal of misfolded proteins involved in amyotrophic lateral sclerosis (ALS). Hum Mol Genet 19, 3440–56 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Sharp PS et al. Protective effects of heat shock protein 27 in a model of ALS occur in the early stages of disease progression. Neurobiol Dis 30, 42–55 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Novoselov SS et al. Molecular chaperone mediated late-stage neuroprotection in the SOD1(G93A) mouse model of amyotrophic lateral sclerosis. PLoS One 8, e73944 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 50.Ganna A et al. Ultra-rare disruptive and damaging mutations influence educational attainment in the general population. Nat Neurosci 19, 1563–1565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ganna A et al. Quantifying the Impact of Rare and Ultra-rare Coding Variation across the Phenotypic Spectrum. Am J Hum Genet 102, 1204–1211 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McLaren W et al. The Ensembl Variant Effect Predictor. Genome Biol 17, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lek M et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–91 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X Firth logistic regression for rare variant association tests. Front Genet 5, 187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lam HY et al. Performance comparison of whole-genome sequencing platforms. Nat Biotechnol 30, 78–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Rawe J et al. Low concordance of multiple variant-calling pipelines: practical implications for exome and genome sequencing. Genome Med 5, 28 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Samocha KE et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet 46, 944–50 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data discussed in this publication were obtained through dbGaP and are available under the following accession codes: MIGen Exome Sequencing: Ottawa Heart (phs000806.v1.p1); MIGen Exome Sequencing: Leicester UK Heart Study (phs001000.v1.p1); Swedish Schizophrenia Population-Based Case-control Exome Sequencing (phs000473.v2.p2); Genome-Wide Association Study of Amyotrophic Lateral Sclerosis (phs000101.v5.p1).