SUMMARY
The tumor microenvironment (TME) at the metastatic site consists of multiple components with considerable cellular heterogeneity. To test whether endothelial cells (ECs) associated with lung metastases express a distinct gene expression program that promotes metastatic growth, we isolated CD31+/CD45− cells from lung mammary cancer metastases for RNA sequencing and found CD44 upregulation. Unexpectedly, the CD44+ subset did not comprise authentic ECs nor were they bone-marrow-derived CD45− endothelial progenitor cells. Instead, they were a population of large platelets that are distinct from regular small platelets. These CD44+ large platelets were enriched in lung metastases but not primary mammary tumors and upregulated myeloid cell-regulating chemokines indicative of potential regulation of metastasis via indirect mechanisms. Identification of this cellular player in the TME of metastasis suggests a role for the recently identified lung-resident megakaryocytes (MKs) and offers an unexplored route to discover novel mechanisms and an opportunity for therapeutic interventions.
Graphical Abstract

In Brief
Zheng et al. show a distinct type of large platelets is specifically enriched in lung metastases but not in primary tumors. Identification of this cell type provides insight into the complexity of the metastatic tumor microenvironment. It also helps clarify confusion about the origin of endothelial progenitor cells.
INTRODUCTION
Solid tumors are relatively successfully treated if restricted to the primary site, but current therapeutics are inadequate in metastatic diseases that cause the majority of patient deaths (Steeg, 2016). Anti-angiogenic therapies aim at inhibiting generation of new blood vessels that support tumor growth and have shown some effects in primary cancers, but the benefits for metastatic breast cancer are limited (Potente et al., 2011). Blood vessels are more than passive conduits for delivering nutrients to tumors or for cancer cell dissemination. Instead, they can positively regulate neighboring cells by expression of angiocrine factors (EC-derived paracrine-acting factors) (Butler et al., 2010). However, a comprehensive understanding of what angiocrine factors are at the metastatic site and what biological processes they collectively regulate is fundamentally lacking.
CD31 (also known as PECAM1) is a conventional EC marker that is also expressed to varying degrees in platelets and certain leukocyte subtypes (Lertkiatmongkol et al., 2016). It is also expressed by certain progenitor cells in the bone marrow (BM), including endothelial progenitor cells (EPCs), which are believed to be BM-resident non-hematopoietic progenitors for CD31+ circulating endothelial cells that can be incorporated into the vessel network at the site of angiogenesis (Bertolini et al., 2006; Gao et al., 2008; Patenaude et al., 2010). However, EPCs have remained a controversial concept, as their contribution to vessels in tumors varies from more than 50% to none, and the exact cell of origin of the BM-derived CD31+ cells in tumors has not been fully established (Medina et al., 2017; Patenaude et al., 2010).
Another important component of the TME is the tumor-infiltrating myeloid cells that promote tumor growth and metastasis (Kitamura et al., 2015a; Powell and Huttenlocher, 2016). Since inflammation can prime the endothelium to attract myeloid cells to the site of infection, and inflammation and cancer are intricately linked (Grivennikov et al., 2010), it is thus plausible that disseminated cancer cells in secondary organs may redirect the inflammatory response to recruit metastasis-promoting myeloid cells indirectly via activation of the endothelium.
Based on these data, we initially hypothesized that breast cancer cells disseminated to the lung actively regulate non-sprouting vessels that in turn provide signals to enhance metastatic growth, either directly via activation of cancer cells or indirectly via recruitment and regulation of myeloid cells. Unexpectedly, while attempting to address this hypothesis, we uncovered a previously unrecognized population of large platelets enriched specifically in lung metastases but not in primary tumors.
RESULTS
Gene Expression Profiling of CD31+/CD45− Cells from Lung Metastases Reveals a Distinct CD44+ Population
First, we characterized the vessel phenotypes associated with lung metastases using a spontaneous transgene-induced mouse mammary tumor model of breast cancer (MMTV-PyMT, Figures S1A-S1G) (Guy et al., 1992; Lin et al., 2003) and an experimental metastasis model using tail vein injection of a metastatic mammary cancer cell line, E0771-LG (Kitamura et al., 2015b) (Figures S1H-S1N). In both models, no sprouting angiogenesis was observed in metastatic nodules that were visible to naked eyes and were therefore macro-metastases by definition (Figures S1F, S1G, S1I and S1K). Instead, these intratumor vessels maintain their original anatomical structure found in the alveoli of normal lungs (Figures S1F, S1G, and S1J). This contrasts with the sprouting angiogenesis in primary tumors, as evident by dense microvessels with irregular morphology and sprouting filopodia (Figures S1A-S1C). Not until day 13 in the experimental model, when lesions had taken over much of the space of the lung, did sprouting angiogenesis appear (Figures S1L-S1N). These data suggest that sprouting angiogenesis is not required for the initial and intermediate growth into macro-metastases in the lung that is rich in capillary vessels.
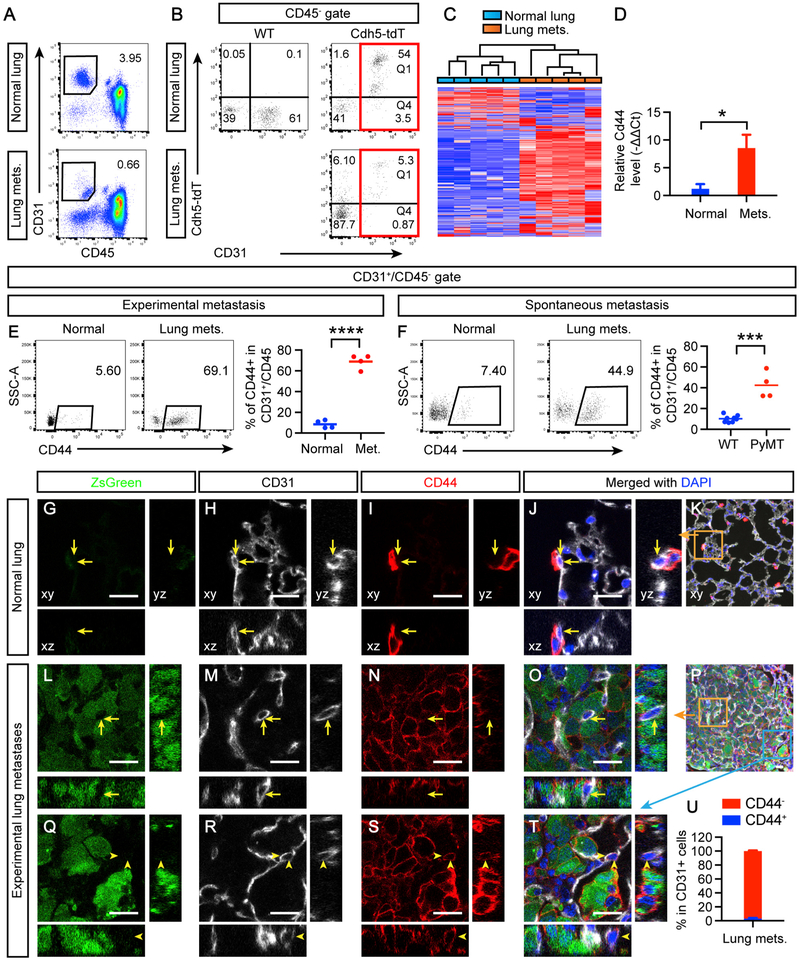
To understand whether non-sprouting vessels in the lung promote metastatic growth via angiocrine factors, we isolated ECs from lung metastases at day 11 post-intravenous (i.v.) injection and compared with those of normal lung vessels using RNA sequencing (RNA-seq) (Table S1). The cells were sorted by fluorescence-activated cell sorting (FACS) based on their expression of the EC marker CD31 and negative expression for the pan-leukocyte marker CD45 (Figure 1A). To further confirm the EC identity, we used double-transgenic mice in which the fluorescent protein tdTomato is expressed upon Cre-mediated recombination in cells that express the EC marker VE-cadherin (Cdh5) (Alva et al., 2006; Madisen et al., 2010). Analysis of lung metastases from these Cdh5-Cre;Rosa26-loxp-stop-loxp-tdTomato mice (Cdh5-tdT thereafter) showed that most of CD31+/CD45− cells were positive for Cdh5-tdT (Figure 1B, Q1/(Q1+Q4) = 86%). Unsupervised hierarchical clustering analysis of the RNA-seq data completely segregated CD31+/CD45− cells from normal lungs and dissected lung metastases into two distinct clusters (Figure 1C), indicating a distinct gene expression program in metastases.
Figure 1. Lung Metastases Induce an Increase of a CD44+ Subset of CD31+/CD45− Cells that Do Not Appear to Be Endothelial Cells.
(A) Lung metastases (mets.) induced by E0771-LG i.v. injection at day 11 were dissected and processed to sort CD31+/CD45− cells and compared with those from the normal lung without tumors.
(B) WT or Cdh5-tdT mice were i.v. injected with E0771-LG cells or untreated, and the lungs were analyzed after 11 days.
(C) The gated cells from (A) were prepared for RNA sequencing. The gene expression data were analyzed using unsupervised hierarchical clustering, with bars indicating individual samples.
(D) Independent qRT-PCR validation of Cd44. Ct, threshold cycle.
(E) Experimental metastases induced by E0771-LG i.v. injection were analyzed for CD44 expression in the CD31+/CD45− population.
(F) Spontaneous lung metastases developed in 14.5-week-old PyMT mice were similarly analyzed.
(G–T) Normal lungs (G and K) or lungs bearing E0771-LG-Luc-ZsGreen (green) metastases (L–T) were sectioned and stained for CD31, CD44, and DAPI for confocal imaging, as presented in xy, xz, and yz views. Yellow arrows indicate CD31+ ECs that do not express CD44, and yellow arrowheads indicate the rare occasions in which CD31+ ECs appear to express CD44. Orange and blue squares in (K) and (P) are zoomed in panels as indicated. Scale bars, 20 μm.
(U) Quantification of CD31+ cells for CD44 positivity. Error bars represent SEM
See also Figures S1-S3.
To understand how potential factors from this CD31+/CD45− population may contribute to metastatic growth indirectly via regulation of myeloid cells, we focused on two of the top ten pathways enriched by the differentially expressed genes related to leukocyte trafficking and function (Table S1, tab “Pathways”). CD44, a transmembrane glycoprotein involved in cell-cell interaction, cell adhesion, and migration (Orian-Rousseau and Ponta, 2015), was identified by the RNA-seq as one of the significantly upregulated genes enriched in these pathways and was also confirmed by qRT-PCR analysis (Table S1, tab ‘Pathways’; Figure 1D). This upregulation was also confirmed at the protein level by FACS in both experimental and spontaneous models (Figures 1E and 1F).
The CD44+ Subset of CD31+/CD45− Cells in Lung Metastases Does Not Appear to Be ECs
To analyze the spatial/anatomic information of CD31+/CD45−/CD44+ cells in relation to metastatic tumor cells, we transduced E0771-LG cells with a lentivirus expressing luciferase-ZsGreen (Luc-ZsG) to visualize tumor cells before i.v. injection (Figures 1G, 1L, and 1Q). As expected in the normal lung, ECs forming pulmonary capillaries were CD44− (Figures 1G-1K). Surprisingly, less than 3% CD31+ ECs appeared to be CD44+ in lung metastases (Figures 1L-1U), in sharp contrast to the 69% observed by flow cytometry (Figure 1E). Similar findings were observed also for the spontaneous metastasis model (Figures 1F and S2). The apparent discrepancy between flow-cytometry-based and histology-based evaluation was not unique to the Cd44 gene but was also for a number of other tested genes, such as Thbd, which showed downregulation in flow cytometry but not in histology (Figures S3A and S3B). Interestingly, most of the CD44+ subset of CD31+/CD45− cells were THBD− (Figure S3A), suggesting that the CD44+ subset in metastases represents a distinct cellular population. Indeed, when backgated to the live-cell gate, the CD44+ appeared to cluster differently from the CD44− subset, with slightly less CD31 and slightly more CD45 staining intensities (Figure S4A).
CD31+/CD45−/CD44+ Cells Associated with Lung Metastases Originate from the CD45+ Hematopoietic Lineage in the BM
Next, we set out to determine whether the distinct CD31intermediate/CD45dim/CD44+ (CD31int/CD45dim/CD44+) population that was also labeled by the Cdh5-tdT reporter (Figure 1B) represents tumor EC heterogeneity (Dudley, 2012) or a yet-unidentified population in the TME. First, we examined two additional parameters commonly used for determining EC identity, VE-cadherin antibody staining, and tomato lectin binding (Baluk and McDonald, 2008; Gao et al., 2008). VE-cadherin antibody staining in flow cytometry may underestimate the true number of positive cells (66.3% of CD31+/CD45−, Figure S4B), possibly due to tissue digestion needed for single-cell preparation. Nevertheless, CD31int/CD45dim/CD44+ cells in lung metastases still expressed a significant level of VE-cadherin, in contrast to the 1.9% of CD45+ cells as a negative control (Figure S4B, Q1/(Q1+Q2)). 92% of CD31int/CD45dim/CD44+ cells in lung metastases were labeled with lectin after i.v. injection (Figure S4C, Q1/(Q1+Q2)). Cdh5-tdT reporter mice also confirmed that most of CD31int/CD45dim/CD44+ cells in lung metastases were positive for the reporter (Figure S4D, Q1/(Q1+Q2)). Thus, flow cytometric analysis with conventional EC markers cannot exclude their EC identity.
We then studied the possible involvement of CD31+ cells that are not ECs. We excluded the possibility of tumor cell contamination due to vasculogenic mimicry, by which tumor cells express CD31 and become part of the endothelium (Seftor et al., 2012), as ZsGreen+ tumor cells were negative for CD31 staining in both histology and flow cytometry (Figures 1L-1T; Figure S4E). They are unlikely to be CD31-expressing monocytic cells either (Kim et al., 2009; Urbich et al., 2003), since, in addition to the relative absence of the pan-leukocyte marker CD45, the myeloid cell marker CD11B expressed by monocytic cells was also negative (Figure S4F).
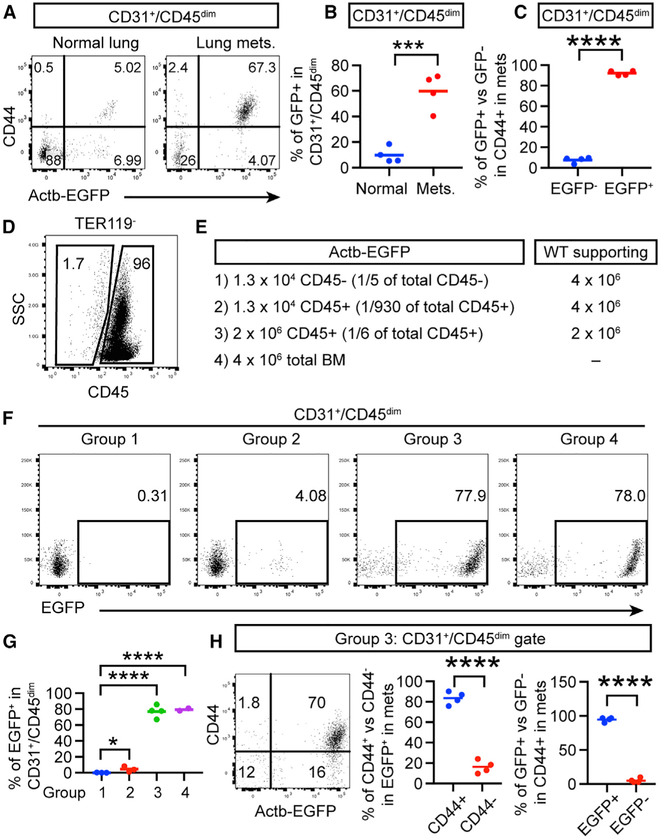
BM-derived EPCs have been suggested as a source for CD31+ cells in tumors (Bertolini et al., 2006; Patenaude et al., 2010). We reasoned that, if the CD44+ subset of CD31+/CD45− cells originates from EPCs in the BM, they would be labeled in a BM transplantation (BMT) experiment using a colored reporter. Thus, we transplanted total BM cells from Actb-EGFP mice (EGFP expression driven by the beta-actin promoter) (Okabe et al., 1997) into wild-type (WT) recipient mice so that BM-derived cells but not pre-existing ECs in lung vessels would be labeled with EGFP. Similar to a previous finding (Gao et al., 2008), the percentage of BM-derived cells in the CD31+/CD45− population was significantly increased in lung metastases compared to that in normal lungs (Figures 2A and 2B). Importantly, the majority of CD44+ cells that increased in the CD31+/CD45− population in lung metastases were EGFP+ (Figure 2C), indicating that BM is the source of these cells. Putative EPCs has been postulated to be CD45− non-hematopoietic progenitors in the BM (Gao et al., 2008; Patenaude et al., 2010). However, formal experimental proof for this hypothesis is lacking. To determine whether EPCs by this definition is the cell of origin of our observed CD31int/CD45dim/CD44+ cells in lung metastases, we FACS-sorted non-hematopoietic cells (CD45−TER119−) and hematopoietic cells not including erythrocytes (CD45+/TER119− from the BM and compared their ability to generate CD31int/CD45dim/EGFP+ cells in lung metastases after BMT (Figures 2D and 2E). The result showed that only progenitors of the hematopoietic lineage (CD45+/TER119− gave rise to CD31+/CD45− cells (of which most were CD44+) in a dose-dependent manner, and, when given at a similar dose, CD45+ engraftment was similarly efficient as total BM cells, in contrast to the undetectable level engrafted by non-hematopoietic CD45− cells (Figures 2F and 2G). In addition, the majority of CD31+/CD45dim/CD44+ cells originated from CD45+ BM progenitors (Figure 2H). Thus, it can be concluded that it is CD45+ hematopoietic progenitor cells in the BM that give rise to metastasis-infiltrating CD31int cells that are CD45dim.
Figure 2. CD31+/CD45−/CD44+ Cells Enriched in Lung Metastases Originate from the CD45+ Hematopoietic Lineage in the BM.
(A–C) Total BM cells from Actb-EGFP donor mice were transplanted into WT recipient mice. After 5 weeks, E0771-LG cells were injected i.v., and the normal lungs without tumor cell injection or lung metastases were analyzed.
(D–G) The CD45−/TER119− and CD45+/TER119− populations were sorted from the total BM cells of Actb-EGFP donor mice and were combined with or without WT supporting cells as indicated for transplantation of WT recipient mice. E0771-LG cells were injected i.v. 5 weeks after transplantation.
(H) Analysis of group 3 in (E) for CD44 and EGFP.
CD31int/CD45dim/CD44+ Cells Accumulated in Lung Metastases Are Large Platelets
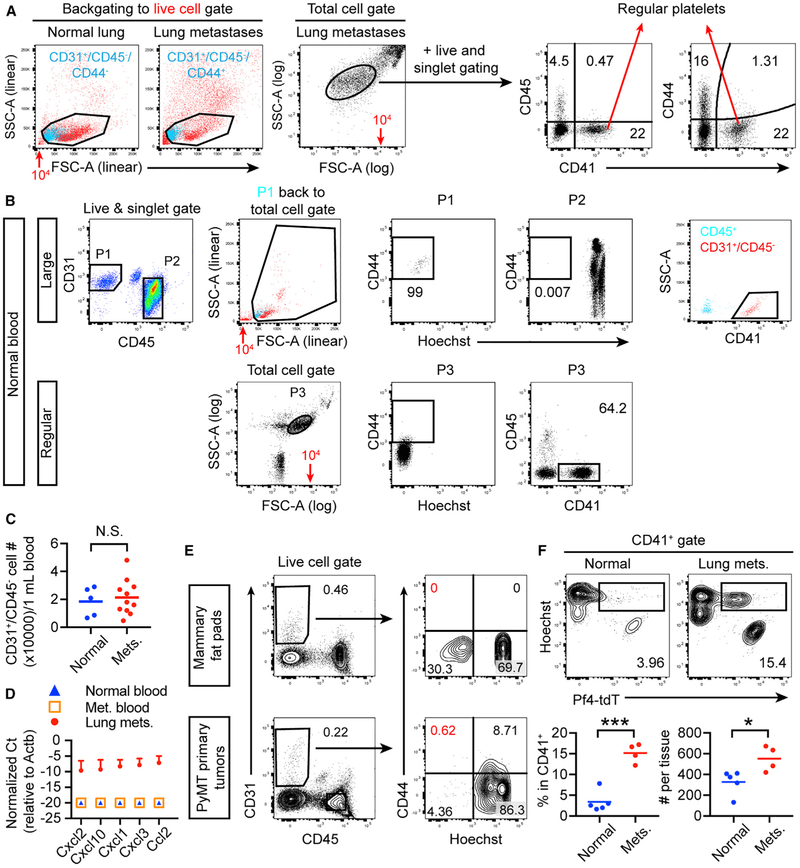
Next, we studied how CD45+ hematopoietic progenitors generated CD31int/CD44+ cells in lung metastases that were CD45dim. We ruled out that they are CD45− erythrocytes in metastases, as they were negative for the lineage marker TER119 (Figure S4G). Platelets also express CD31 and are another subset of hematopoietic cells without CD45 expression (Newman and Newman, 2003). Indeed, we found that CD31int/CD45dim/CD44+ cells in lung metastases were predominantly positive for the platelet marker CD41 (Figures 3A and 3B). Nuclear staining by Hoechst (Hst) confirmed that these cells did not have nuclear DNA (Figures 3C and 3D). Similar results were observed in the spontaneous model (Figure 3E). Next, we FACS-sorted these cells and plated them on a glass slide for fluorescent microscopy. As expected, CD31+/CD45− from the normal lung were positive for CD31 and nuclear staining, while the CD31−/CD45+ leukocytes, despite their nuclear staining by Hst, were CD31− (Figure 3F, left two columns). In comparison, while the CD31int/CD45dim/CD44−/Hst+ cells sorted from metastases appeared to represent authentic ECs due to positive staining for both CD31 and Hst, the CD44+/Hst− subset did not show nuclear staining despite their CD31 staining (Figure 3F, right two column). These cells therefore appeared to be platelets.
Figure 3. CD31int/CD45dim/CD44+ Cells Enriched in Lung Metastases Are CD41+/Hoechst−/Pf4+ Platelets.
(A–D) Lung metastases induced by E0771-LG i.v. injection were analyzed after 11 days for CD41 expression (A and B) or Hoechst (Hst) staining (C and D).
(E) Lung metastases were dissected from 14.5-week-old PyMT mice for similar analyses.
(F) The indicated cell populations from the experimental metastasis model were sorted and fixed on a glass slide for imaging. Arrows indicate CD31+/Hst+ ECs; arrowheads indicate CD31+/Hst− platelets. Scale bars, 25 μm.
(G) WT mice that received Pf4-tdT BMT were either untreated or i.v. injected with E0771-LG. After 11 days, the lungs and lung metastases were analyzed.
(H) Quantification of (G). All error bars represent mean ± SEM.
See also Figure S4.
To further confirm the platelet identity, we used a reporter mouse in which tdTomato expression is controlled by the MK/platelet lineage marker Pf4, Pf4-tdT (Madisen et al., 2010; Tiedt et al., 2007). Whereas mice without tumor cell injection showed little PF4-tdT signal in CD31+/CD45dim cells in the lung, a significant fraction of the CD31+/CD45dim cells in lung metastases highly expressed PF4-tdT (Figures 3G and 3H). Importantly, these PF4-tdT+ cells were predominantly CD44+/CD41+/Hst− (Figure 3H), which reinforces the platelet identity of these cells.
Lung Metastases but Not Primary Tumors Induce Accumulation of Chemokine-Expressing CD31int/CD45dim/CD44+ Large Platelets
To examine whether CD31int/CD45dim/CD44+ platelets exhibit any distinct features compared to regular platelets, we compared their sizes and CD44 expression and found that CD31int/CD45dim/CD44+ were larger and that only these cells express CD44 (Figure 4A). Next, we tested whether CD31int/CD45dim/CD44+ large platelets exist in the blood circulation before they were recruited to lung metastases or even in the blood of normal mice. Indeed, we found CD31int/CD45dim cells in the normal blood as described also by others (Strijbos et al., 2007; Wong et al., 2012) and that these cells were CD44+/CD41+/Hst− (Figure 4B, P1). However, the number of CD31int/CD45dim/CD44+ cells did not increase in the blood of metastasis-bearing mice (Figure 4C), despite their marked increase in lung metastases (Figure 1E), suggesting local production in the lung.
Figure 4. Lung Metastases But Not Primary Tumors Induce Accumulation of Chemokine-Expressing CD31int/CD45dim/CD44+ Large Platelets.
(A and B) Comparison of CD31int/CD45dim/CD44+ large platelets and regular small platelets in the lung (A) and blood (B). Note the same values of 104 (red text and arrows) on different scales of forward scatter (FSC).
(C) Quantification of CD31int/CD45dim/CD44+ large platelets in the blood of normal and metastasis-bearing mice.
(D) CD31int/CD45dim/CD44+ large platelets from the normal blood, the blood of metastasis-bearing mice, and dissected lung metastases were sorted for qRT-PCR analysis.
(E) Primary tumors from 13-week-old MMTV-PyMT mice and normal mammary fat pads from WT littermates were analyzed. The red percentages indicate the essential absence of CD44+/Hoechst− cells.
(F) WT mice that received Pf4-tdT BMT were injected i.v. with E0771-LG, and the lungs and lung metastases were analyzed after 11 days. tdT+/Hst+ cells are quantified as percentage of CD41+ cells and absolute numbers.
See also Figure S5.
Subsequently, we wanted to understand whether CD31int/CD45dim/CD44+ large platelets enriched in lung metastases express chemokines that may regulate recruitment and function of myeloid cells and thus metastatic growth indirectly. qRT-PCR analysis of these cells strikingly revealed that only the CD31int/CD45dim/CD44+ large platelets accumulated in lung metastases highly expressed a number of chemokines identified in the RNA-seq, such as Cxcl2, Cxcl10, Cxcl1, Cxcl3, and Ccl2 data (Table S1, tabs ‘Pathways,’ ‘Agranulocyte Adhesion and Diapedesis,’ and ‘Leukocyte Extravasation Signaling’; Figure 4D). Neither those in the blood of normal mice or metastasis-bearing mice showed detectable levels (Figure 4D). This observation again indicates a local effect in the lung, which prompted us to examine the primary mammary tumors. Notably, very few CD31int/CD45dim/CD44+ large platelets were detected in MMTV-PyMT primary tumors (Figure 4E), in sharp contrast to the lung metastases in this model (Figure 1F). In comparison, regular small platelets did not show such a lung-preferential enrichment, as they were present in a similar level in primary tumors as in normal mammary fat pads (Figure S5). A recent important finding showed that lung megakaryocytes (MKs) in the BM can egress from the BM and reside in the lung and contribute to platelet production (Lefrançais et al., 2017). Consistent with these data, we observed in lung metastases Pf4-tdT+ MKs (Figure 4F) that may represent the source of the chemokine-expressing large platelets enriched only in lung metastases. Interestingly, Pf4-tdT+ MKs in mice that received Pf4-tdT BMT increased in lung metastases (Figure 4F), suggesting that lung metastases may induce MK migration from the BM to the lung or in situ proliferation.
DISCUSSION
We initially set out to test whether cancer cells colonizing the lung induce a distinct expression program in associated ECs. To our surprise, we found in this study a previously unrecognized yet distinct population of CD31int/CD45dim/CD44+ large platelets restricted to lung metastases.
Our unexpected findings have helped to clarify the controversial concept of EPCs. It was postulated but not experimentally proved that EPCs are a population of CD45− non-hematopoietic progenitors in the BM that are able to generate BM-derived CD31+/CD45− cells in tumors, including lung metastases (Gao et al., 2008; Patenaude et al.,2010). However, our lineage tracing experiments that compared CD45+ and CD45− BM cells for this progenitor ability has provided strong evidence that BM-derived CD31+/CD45− cells, at least in lung metastases, originate from CD45+ hematopoietic progenitors. This finding argues against the in vivo existence of a unique population of EPCs located in the BM that are non-hematopoietic. It should be noted that we do not exclude differentiation into ECs from BM-derived hematopoietic progenitor cells (Moschetta et al., 2014; Patenaude et al., 2010) or from vessel-wall-derived progenitors (Ingram et al., 2005; Fang et al., 2012; Wakabayashi et al., 2018). However, this is not the conventional definition of EPCs that emphasizes the BM origin and the non-hematopoietic identity.
Platelets are released from MKs with an intermediate stage of proplatelets (Patel et al., 2005). The CD31int/CD45dim/CD44+ large platelets identified in our study may represent this poorly understood intermediate form. Recently Lefrancais et al. showed that MKs commonly found in the BM can migrate to and reside in the lung, where they contribute up to 50% of the total platelet production (Lefrançais et al., 2017). These lung-resident MKs are likely the source of the CD31int/CD45dim/CD44+ large platelets that are enriched and activated only in lung metastases. Future work such as intravital imaging is required to provide direct evidence of this assertion and to obtain the spatial-temporal information of the multiple cell types in metastases. Nevertheless, our findings have extended the significance of the lung-resident population of MKs by showing that they represent a distinct player that releases chemokine-expressing large platelets to metastatic cancer cells in the lung.
The exact function of CD31int/CD45dim/CD44+ large platelets requires further study. They may use similar mechanisms as regular small platelets to promote cancer cell extravasation and metastatic growth (Haemmerle et al., 2018), or they may also have distinct roles. For example, only the CD31int/CD45dim large platelets but not regular small ones expressed CD44. Since CD44 binds to HA, and HA deposition by tumor and stromal cells is enhanced upon their interaction (Kimata et al., 1983; Knudson et al., 1984), CD31int/CD45dim/CD44+ large platelets may use this adhesion mechanism to accumulate in metastases. Elucidation of the specific activities of this population will require advances that allow perturbation of only these atypical platelets but not the regular ones. Unfortunately, unique tools to study this cell population do not exist. Despite the limitations, our observation that lung metastases induced upregulation of chemokines in CD31int/CD45dim/CD44+ large platelets has shed important mechanistic insights, since many of these chemokines have been shown in a variety of studies to regulate the recruitment and activity of monocytes, macrophages, and neutrophils to promote extravasation, seeding, and colonization of disseminated tumor cells (Coffelt et al., 2016; Kitamura et al., 2015a). Additional future work is needed to determine the amount and functional effects of these chemokines as compared to those from other stromal cells in the TME including regular platelets.
Identification of this platelet population has advanced our understanding of the heterogeneity and complexion of the TME in metastases. Our study suggests that metastatic cancer cells may trigger organ-specific responses. Elucidation of these mechanisms may offer opportunities for design of novel therapeutics.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jeffrey Pollard (jeffrey.pollard@einstein.yu.edu). The stable cell line generated in this study will be made available on request, but we may require a payment and/or a completed Materials Transfer Agreement if there is potential for commercial application.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All procedures involving mice were conducted in accordance with National Institutes of Health regulations concerning the use and care of experimental animals and were approved by the Albert Einstein College of Medicine Animal Use Committee. MMTV-PyMT mice were provided by W.J. Muller (McMaster University, Hamilton, Ontario, Canada) (Guy et al., 1992) and bred in house in the FVB background. Cdh5-Cre (Stock number 006137), Rosa26-LSL-tdTomato (Stock number 007914), Actb-EGFP (Stock number 006567), Pf4-Cre (Stock number 008535) and C57BL6/J WT mice were purchased from the Jackson Laboratory. Only female mice were used in breast cancer models.
Cell lines
E0771-LG (Kitamura et al., 2015b) and 293T cells were cultured in DMEM supplemented with 10% v/v fetal bovine serum and penicillin/streptomycin.
METHOD DETAILS
Lung metastasis assays
Spontaneous metastases in the lung developed in MMTV-PyMT mice were collected when mice were 14-15 weeks old. For experimental metastasis assays, 1 × 106 E0771-LG were injected intravenously through the tail vein of syngenic C57BL6/J female mice (6-8 wk old unless otherwise specified). For in vivo analysis of lectin binding, 50μg biotinylated tomato-lectin (Vector Lab, B-1175) was injected into the retro-orbital sinus and was analyzed 10 min after injection by staining with streptavidin-conjugated BUV395 (BD Biosciences 564176).
Viral production and transduction of tumor cells
293T cells were transfected with pHIV-Luc-ZsGreen (gift from Dr. Bryan Welm, Addgene #39196), pMD2.G (gift from Dr. Didier Trono, Addgene #12259) and pxPAX2 (gift from Dr. Didier Trono, Addgene #12260) at 4:3:1 in μg using Lipofectamien 2000 (Invitrogen, #11668-019) according to the manufacturer’s manual. Medium was replaced 4-6 hours (h) after transfection with DMEM containing 2% v/v FBS. Viral supernatants were collected 48 and 72 h after transfection, pooled, cleared with a 0.2 μm filter and used for transduction. Target cells were seeded at about 50% confluency, incubated with the viral supernatant and 10 μg/mL polybrene (Santa Cruz, sc-134220) and centrifuged at 1000 G for 30 min at room temperature (RT). The virus was removed and the cells were then cultured for 3 days before FACS sorting for ZsGreen+ cells.
Flow cytometry and cell sorting
Lungs were perfused with PBS through the right ventricle, dissected and minced. Lung metastases (< 1 mm in diameter) were dissected using a dissection microscope and pooled from 2-4 mice. The tissues were then digested with an enzyme mix of Liberase DL (Sigma-Aldrich 5466202001, 0.52 U/mL), TL (Sigma-Aldrich 5401020001, 0.26 U/mL) and DNase I (Sigma-Aldrich DN25, 150 μg/mL) diluted in basal DMEM medium with rotation for 30 min at 37°C and filtered (70-μm membrane). For cells sorted for qRT-PCR, transcription inhibitors alpha-amanitin (Sigma-Aldrich A2263, 5 μg/mL) and actinomycin D (Sigma-Aldrich A1410, 1 μg/mL) were also added in the digestion buffer. Mouse peripheral blood was collected from the major abdominal vessels (inferior vena cava) with a syringe containing 150 μL ACD buffer [85 mM trisodium citrate, 71 mM citric acid, and 111 mM dextrose (pH 4.5)] (Gil-Bernabé et al., 2012). Red blood cells were removed by incubation with the RBC Lysis Buffer (Biolegend, 420301) for 5 min on ice (once for tissues and twice for blood). Before antibody labeling, cells were blocked with anti-mouse CD16/CD32 antibody (BD Biosciences 553141) for 10 min on ice. Flow cytometry was performed with a LSRII cytometer (BD Biosciences) and the data were analyzed using Flowjo software (TreeStar). FACSAria II (BD Biosciences) and Moflo Astrios (Beckman Coulter) were used for cell sorting.
Gating of single cells using FSC-A/H and SSC-A/W and exclusion of dead cells with DAPI, Zombie Yellow (Biolegend 423103) or Zombie Green (Biolegend 423111) staining were performed routinely during analysis. For Hoechst staining, cells were incubated with 5 μg/mL Hoechst 33342 (BD Biosciences, 561908) for 10 min at 37°C before antibody staining. For visualization of sorted cells on a glass slide for microscopy, the sorted cells were pelleted, resuspended in 10 μL PBS, pipetted to a glass slide enclosed by a hydrophobic pen (Vector Laboratories, H-4000) and air-dried in a laminar hood. The cells were then fixed with 4% w/v paraformaldehyde (PFA) for 10 min at room temperature and mounted with VECTASHIELD Mounting Medium (Vector Laboratories, H-1000). For quantification of cell numbers, CountBrignt abosolute counting beads (ThermoFisher Scientific, Cat# C36950) were used according to the manual.
BMT
Total BM cells were extracted from femurs, tibiae and the spine by grinding in a mortar. For BMT involving cell sorting, an additional step was performed to purify low-density mononuclear cells by using density gradient centrifugation with Histopaque 1083 (Sigma-Aldrich 1083-1) before antibody staining. Female recipient mice of 4-6 weeks old were irradiated with 10 Gy gamma rays (split into 2 doses with a 4-h interval) the day before iv injection of 5-10 × 106 BM cells unless otherwise specified. Transplanted mice were used for experiments after 4-5 weeks.
RNA isolation, qRT-PCR and RNA sequencing
Cells were sorted directly into the Extraction Buffer of the Picopure RNA Isolation kit (Arcturus KIT0202). RNA sequencing of mouse total RNA was performed at Beijing Genomic Institute, using the Ovation® RNA-Seq System V2 kit for library construction, pair-end 100 bp and Hiseq 4000, which generated 60-80M reads per sample. The reads were aligned to the mouse reference genome (GRCm38/mm10) using Tophat (v2.0.13) (Kim et al., 2013). Uniquely mapped reads were counted for each gene using htseq-count in the HTSeq package (v0.6.1) with gene models from UCSC RefGene (Anders et al., 2015). FPKM values were generated using cufflinks v2.2.1 (Trapnell et al., 2013). Lowly expressed genes (mean FPKM values < 1 in both groups under comparison) were excluded from differential expression analysis. DESeq2 was used to identify differentially expressed genes (Love et al., 2014), of which fulfilling the following criteria were used for enrichment of Canonical Pathways in the Ingenuity Pathway Analysis (QIAGEN): log2 (fold change) > 1.5; false discovery rate (FDR) < 0.03; for upregulated genes, mean FPKMs of lung metastases > 20; for downregulated genes, mean FPKMs of normal lungs > 20.
For qRT-PCR, the isolated RNAs were reverse transcribed and amplified using QuantiTect® Whole Transcriptome (QIAGEN 207043) before qPCR. Gene expression was normalized to beta-actin. Relative expression is calculated using the formula –ΔΔCt, where Ct stands for threshold cycles. Undetectable expressions were assigned a relative Ct value of −20 with respect to the Ct of beta-actin. The following Taqman gene expression assays (ThermoFisher Scientific) were used: Actb (Mm02619580_g1), Ccl2 (Mm00441242_m1), Cd44 (Mm01277161_m1), Cxcl1 (Mm04207460_m1), Cxcl2 (Mm00436450_m1), Cxcl3 (Mm01701838_m1), and Cxcl10 (Mm00445235_m1).
Immunofluorescent staining and microscopy
Mouse lungs were processed for frozen sectioning using a method modified from a previous study (Favre et al., 2003). Briefly, the lung were perfused with 1% w/v PFA through the right ventricle, and then 2% agarose diluted in PBS were infused into the lung via the trachea to expand alveoli. After dissection with the associated trachea, the lungs were kept in the inflated form in a histology cassette for further immersion fixation in 4% w/v PFA for 1 h at +4°C, and were then washed with PBS and incubated with 25% w/v sucrose before frozen embedding in the OCT Compound (Fisher HealthCare 4585). Twenty-μm frozen sections were cut, air-dried for 1 h, permeabilized and blocked using Donkey Immunomix (PBS containing 5% v/v normal donkey serum, 0.2% w/v bovine serum albumin, 0.3% Triton-X and 0.05% w/v sodium azide) for 1 h, stained overnight at +4°C with primary antibodies, washed and then stained with secondary antibodies for 1-1.5 h at RT. Sections were then mounted with VECTASHILD Mounting Medium (Vector Laboratories, #H-1200). Samples were imaged using the Leica SP8 confocal microscope (Leica). For confocal images, 3D projections and orthogonal views were digitally reconstructed from Z stacks using ImageJ. Brightness and contrast of the images were adjusted uniformly to entire images using Adobe Photoshop where appropriate. Quantification of the percentage of CD44+ cells out of CD31+ ECs was performed on confocal images. A cell was identified as double positive only when they colocalized to the same cell from all xy, xz and yz views. A total of approximately 50 CD31+ cells per mouse were analyzed.
QUANTIFICATION AND STATISTICAL ANALYSIS
The majority of experiments were repeated two or more times as indicated in the figure legends. A few experiments were performed once, with N ≥ 3 mice per group. Student’s t test (2 groups) or one-way ANOVA (> 2 groups) were used to compare means. Two-way ANOVA was used when data from multiple experiments were pooled. When variances are unequal among different groups as determined by F test, logarithmic transformation to base 10 is used to compute the statistics. Data are presented as mean ± standard error of mean (SEM). Holm-Sidak test was used as a post hoc analysis when more than two groups were compared. Statistical analyses were carried out with Graphpad Prism (version 7), and the significance is indicated in the figures as follows: p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), p < 0.0001 (****), and not significant (N.S.). For RNA-seq analysis, multiple hypothesis testing was adjusted using the Benjamini and Hochberg false-discovery-rate method.
Sample sizes for quantifications are as follows. Figures 1D and 1E: N = 4 mice for normal lungs and 8 mice pooled into 4 samples for metastases. Figure 1F: N = 4 samples pooled from 4 independent experiments, each pooled from 5-7 mice. N = 8 mice for WT in total. Figures 2B and 2C: N = 4 mice for normal lungs and 8 mice pooled into 4 samples for lung metastases. Figure 2H: N = 2-4 mice per group. Figures 3A and 3B, N = 6 mice for normal, 12 mice pooled into 5 samples for metastases, and 2 independent experiments are pooled. Figures 4C and 4D: N = 4 mice for normal lungs and 8 mice pooled into 4 samples for metastases. Figure 3E: N = 4 samples from 4 independent experiments, each pooled from 5-7 mice. Figure 3H: N = 7 mice pooled from 2 independent experiments. Figure 4C: N = 5-11 samples pooled from 10-22 mice from 3 independent experiments in total. Figure 4D: N = 2-6 pooled from 8-21 mice from 3 independent experiments in total. Figure 4E: N = 4-5 mice in total from two independent experiments. Figure 4F: N = 4-5 mice. All bar graphs represent means ± SEM. Lines in scatterplots indicate means.
DATA AND CODE AVAILABILITY
The accession number for RNA sequencing data deposited in NCBI Gene Expression Omnibus is GEO: GSE123520.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE Antibodies |
SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD11B, PE-Cy7, clone M1/70 | BioLegend | Cat# 101215, RRID:AB_312798 |
| CD11B, PE, clone M1/70 | BD Biosciences | Cat# 561689, RRID:AB_10893803 |
| CD31, clone MAB1398Z | Millipore | Cat# MAB1398Z, RRID:AB_94207 |
| CD31, clone MEC13.3 | BD Biosciences | Cat# 550274, RRID:AB_393571 |
| CD31, BV421, clone 390 | BD Biosciences | Cat# 563356, RRID:AB_2738154 |
| CD31, AF647, clone 390 | BD Biosciences | Cat# 563608, RRID:AB_2738313 |
| CD44, clone IM7 | BD Biosciences | Cat# 553131, RRID:AB_394646 |
| CD41, BUV395, clone MWReg30 | BD Biosciences | Cat# 565980, RRID:AB_2739432 |
| CD41, PE, clone MWReg30 | BD Biosciences | Cat# 561850, RRID:AB_10896980 |
| CD44, FITC, clone IM7 | BioLegend | Cat# 103021, RRID:AB_493684 |
| CD44, PE-Cy7, clone IM7 | BioLegend | Cat# 103030, RRID:AB_830787 |
| CD45, PerCP-Cy5.5, clone 30-F11 | BD Biosciences | Cat# 550994, RRID:AB_394003 |
| CD45, APC, clone 30-F11 | BD Biosciences | Cat# 559864, RRID:AB_398672 |
| Donkey anti-rat, AF488 | ThermoFisher | Cat# A-21208, RRID:AB_2535794 |
| Donkey anti-rat, AF594 | ThermoFisher | Cat# A-21209, RRID:AB_2535795 |
| Goat anti-hamster, AF647 | Jackson ImmunoResearch | 127-605-160, RRID:AB_2339001 |
| Goat anti-hamster, AF488 | Jackson ImmunoResearch | Cat# 127-545-160, RRID:AB_2338997 |
| Streptavidin BUV395 | BD Biosciences | Cat# 564176 |
| TER119, PE-Cy7, clone TER-119 | BD Biosciences | Cat# 557853, RRID:AB_396898 |
| THBD, PE, clone LS17-9 | BD Biosciences | Cat# 566338, RRID:AB_2739695 |
| THBD | R&D | Cat# AF3894, RRID: AB_664141 |
| VE-cadherin, APC, clone 11D4.1 | BD Biosciences | Cat# 562242 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Liberase DL | Sigma-Aldrich | 5466202001 |
| Liberase TL | Sigma-Aldrich | 5401020001 |
| Hoechst 33342 | BD Biosciences | 561908 |
| Histopaque 1083 | Sigma-Aldrich | 1083-1 |
| Deposited Data | ||
| RNA sequencing | This paper | GEO: GSE123520 |
| Experimental Models: Cell Lines | ||
| E0771-LG | Jeffrey Pollard lab (Kitamura et al., 2015b) | N/A |
| E0771-LG-luciferase-ZsGreen | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Actb-EGFP | Jackson Laboratory | 006567 |
| C57/Bl/6J | Jackson Laboratory | C57/Bl/6J |
| Cdh5-Cre | Jackson Laboratory | 006137 |
| MMTV-PyMT | William Muller Lab (Guy et al., 1992) | |
| Pf4-Cre | Jackson Laboratory | 008535 |
| Rosa26-loxp-stop-loxp-tdTomato | Jackson Laboratory | 007914 |
| Oligonucleotides | ||
| Actb | ThermoFisher | Mm02619580_g1 |
| Ccl2 | ThermoFisher | Mm00441242_m1 |
| Cd44 | ThermoFisher | Mm01277161_m1 |
| Cxcl1 | ThermoFisher | Mm04207460_m1 |
| Cxcl10 | ThermoFisher | Mm00445235_m1 |
| Cxcl2 | ThermoFisher | Mm00436450_m1 |
| Cxcl3 | ThermoFisher | Mm01701838_m1 |
| Recombinant DNA | ||
| pHIV-Luc-ZsGreen | Bryan Welm Lab | Addgene 39196 |
| pMD2.G | Didier Trono | Addgene 12259 |
| psPAX2 | Didier Trono | Addgene 12260 |
| Software and Algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Prism 7.0 | GraphPad Software | https://www.graphpad.com/ |
| FlowJo | FlowJo | https://www.flowjo.com/ |
| Ingenuity Pathway Analysis | Ingenuity System Inc | https://www.ingenuity.com/ |
| Tophat v2.0.13 | Kim et al., 2013 | https://ccb.jhu.edu/software/tophat/index.shtml |
| HTSeq v0.6.1 | Anders et al., 2015 | https://htseq.readthedocs.io/en/release_0.10.0/overview.html |
| DESeq2 v1.6.3 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Cufflinks v2.2.1 | Trapnell et al., 2013 | http://cole-trapnell-lab.github.io/cufflinks/manual/ |
Highlights.
Atypical large platelets are specifically enriched in lung metastases
They are produced by lung-resident megakaryocytes and highly express chemokines
They share markers with endothelial cells but have a distinct bone marrow origin
ACKNOWLEDGMENTS
We thank Drs. Shentong Fang, Antonio Di Cristofano, and Donald McDonald for valuable discussion. We acknowledge support from the Flow Cytometry Core Facility (partially supported by NCI P30CA013330) and the Analytical Imaging Facility (funded by NCI P30CA013330). The Leica SP8 confocal microscope was funded by the NIH 1S10OD023591-01. This work was supported by NIH grant P30CA0133330.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.10.016.
REFERENCES
- Alva JA, Zovein AC, Monvoisin A, Murphy T, Salazar A, Harvey NL, Carmeliet P, and Iruela-Arispe ML (2006). VE-Cadherin-Cre-recombinase transgenic mouse: a tool for lineage analysis and gene deletion in endothelial cells. Dev. Dyn 235, 759–767. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W (2015). HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluk P, and McDonald DM (2008). Markers for microscopic imaging of lymphangiogenesis and angiogenesis. Ann. N Y Acad. Sci 1131, 1–12. [DOI] [PubMed] [Google Scholar]
- Bertolini F, Shaked Y, Mancuso P, and Kerbel RS (2006). The multifaceted circulating endothelial cell in cancer: towards marker and target identification. Nat. Rev. Cancer 6, 835–845. [DOI] [PubMed] [Google Scholar]
- Butler JM, Nolan DJ, Vertes EL, Varnum-Finney B, Kobayashi H, Hooper AT, Seandel M, Shido K, White IA, Kobayashi M, et al. (2010). Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell Stem Cell 6, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffelt SB, Wellenstein MD, and de Visser KE (2016). Neutrophils in cancer: neutral no more. Nat. Rev. Cancer 16, 431–446. [DOI] [PubMed] [Google Scholar]
- Dudley AC (2012). Tumor endothelial cells. Cold Spring Harb. Perspect. Med 2, a006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Wei J, Pentinmikko N, Leinonen H, and Salven P (2012). Generation of functional blood vessels from a single c-kit+ adult vascular endothelial stem cell. PLoS Biol. 10, e1001407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre CJ, Mancuso M, Maas K, McLean JW, Baluk P, and McDonald DM (2003). Expression of genes involved in vascular development and angiogenesis in endothelial cells of adult lung. Am. J. Physiol. Heart Circ. Physiol 285, H1917–H1938. [DOI] [PubMed] [Google Scholar]
- Gao D, Nolan DJ, Mellick AS, Bambino K, McDonnell K, and Mittal V (2008). Endothelial progenitor cells control the angiogenic switch in mouse lung metastasis. Science 319, 195–198. [DOI] [PubMed] [Google Scholar]
- Gil-Bernabé AM, Ferjancic S, Tlalka M, Zhao L, Allen PD, Im JH, Watson K, Hill SA, Amirkhosravi A, Francis JL, et al. (2012). Recruitment of monocytes/macrophages by tissue factor-mediated coagulation is essential for metastatic cell survival and premetastatic niche establishment in mice. Blood 119,3164–3175. [DOI] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, and Karin M (2010). Immunity, inflammation, and cancer. Cell 140, 883–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CT, Cardiff RD, and Muller WJ (1992). Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell. Biol 12, 954–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haemmerle M, Stone RL, Menter DG, Afshar-Kharghan V, and Sood AK (2018). The Platelet Lifeline to Cancer: Challenges and Opportunities. Cancer Cell 33, 965–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram DA, Mead LE, Moore DB, Woodard W, Fenoglio A, and Yoder MC (2005). Vessel wall-derived endothelial cells rapidly proliferate because they contain a complete hierarchy of endothelial progenitor cells. Blood 105, 2783–2786. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Kim J-S, Papadopoulos J, Wook Kim S, Maya M, Zhang F, He J, Fan D, Langley R, and Fidler IJ (2009). Circulating monocytes expressing CD31: implications for acute and chronic angiogenesis. Am. J. Pathol 174, 1972–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata K, Honma Y, Okayama M, Oguri K, Hozumi M, and Suzuki S (1983). Increased synthesis of hyaluronic acid by mouse mammary carcinoma cell variants with high metastatic potential. Cancer Res. 43, 1347–1354. [PubMed] [Google Scholar]
- Kitamura T, Qian B-Z, and Pollard JW (2015a). Immune cell promotion of metastasis. Nat. Rev. Immunol 15, 73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T, Qian B-Z, Soong D, Cassetta L, Noy R, Sugano G, Kato Y, Li J, and Pollard JW (2015b). CCL2-induced chemokine cascade promotes breast cancer metastasis by enhancing retention of metastasis-associated macrophages. J. Exp. Med. 212, 1043–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson W, Biswas C, and Toole BP (1984). Interactions between human tumor cells and fibroblasts stimulate hyaluronate synthesis. Proc. Natl. Acad. Sci. USA 81, 6767–6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrançais E, Ortiz-Muñoz G, Caudrillier A, Mallavia B, Liu F, Sayah DM, Thornton EE, Headley MB, David T, Coughlin SR, et al. (2017). The lung is a site of platelet biogenesis and a reservoir for haematopoietic progenitors. Nature 544, 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lertkiatmongkol P, Liao D, Mei H, Hu Y, and Newman PJ (2016). Endothelial functions of platelet/endothelial cell adhesion molecule-1 (CD31). Curr. Opin. Hematol 23, 253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, and Pollard JW (2003). Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol 163,2113–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15,550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina RJ, Barber CL, Sabatier F, Dignat-George F, Melero-Martin JM, Khosrotehrani K, Ohneda O, Randi AM, Chan JKY, Yamaguchi T, et al. (2017). Endothelial Progenitors: A Consensus Statement on Nomenclature. Stem Cells Transl. Med 6, 1316–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschetta M, Mishima Y, Sahin I, Manier S, Glavey S, Vacca A, Roccaro AM, and Ghobrial IM (2014). Role of endothelial progenitor cells in cancer progression. Biochim. Biophys. Acta 1846, 26–39. [DOI] [PubMed] [Google Scholar]
- Newman PJ, and Newman DK (2003). Signal transduction pathways mediated by PECAM-1: new roles for an old molecule in platelet and vascular cell biology. Arterioscler. Thromb. Vasc. Biol 23, 953–964. [DOI] [PubMed] [Google Scholar]
- Okabe M, Ikawa M, Kominami K, Nakanishi T, and Nishimune Y (1997). ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 407, 313–319. [DOI] [PubMed] [Google Scholar]
- Orian-Rousseau V, and Ponta H (2015). Perspectives of CD44 targeting therapies. Arch. Toxicol 89, 3–14. [DOI] [PubMed] [Google Scholar]
- Patel SR, Hartwig JH, and Italiano JE Jr. (2005). The biogenesis of platelets from megakaryocyte proplatelets. J. Clin. Invest 115, 3348–3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patenaude A, Parker J, and Karsan A (2010). Involvement of endothelial progenitor cells in tumor vascularization. Microvasc. Res 79, 217–223. [DOI] [PubMed] [Google Scholar]
- Potente M, Gerhardt H, and Carmeliet P (2011). Basic and therapeutic aspects of angiogenesis. Cell 146, 873–887. [DOI] [PubMed] [Google Scholar]
- Powell DR, and Huttenlocher A (2016). Neutrophils in the Tumor Microenvironment. Trends Immunol. 37, 41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seftor REB, Hess AR, Seftor EA, Kirschmann DA, Hardy KM, Margaryan NV, and Hendrix MJC (2012). Tumor cell vasculogenic mimicry: from controversy to therapeutic promise. Am. J. Pathol 181, 1115–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeg PS (2016). Targeting metastasis. Nat. Rev. Cancer 16, 201–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strijbos MH, Kraan J, den Bakker MA, Lambrecht BN, Sleijfer S, and Gratama JW (2007). Cells meeting our immunophenotypic criteria of endothelial cells are large platelets. Cytometry B Clin. Cytom 72, 86–93. [DOI] [PubMed] [Google Scholar]
- Tiedt R, Schomber T, Hao-Shen H, and Skoda RC (2007). Pf4-Cre transgenic mice allow the generation of lineage-restricted gene knockouts for studying megakaryocyte and platelet function in vivo. Blood 109, 1503–1506. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, and Pachter L (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol 31, 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbich C, Heeschen C, Aicher A, Dernbach E, Zeiher AM, and Dimmeler S (2003). Relevance of monocytic features for neovascularization capacity of circulating endothelial progenitor cells. Circulation 108, 2511–2516. [DOI] [PubMed] [Google Scholar]
- Wakabayashi T, Naito H, Suehiro JI, Lin Y, Kawaji H, Iba T, Kouno T, Ishikawa-Kato S, Furuno M, Takara K, et al. (2018). CD157 Marks Tissue-Resident Endothelial Stem Cells with Homeostatic and Regenerative Properties. Cell Stem Cell 22, 384–397. [DOI] [PubMed] [Google Scholar]
- Wong CKE, Namdarian B, Chua J, Chin X, Speirs R, Nguyen T, Fankhauser M, Pedersen J, Costello AJ, Corcoran NM, and Hovens CM (2012). Levels of a subpopulation of platelets, but not circulating endothelial cells, predict early treatment failure in prostate cancer patients after prostatectomy. Br. J. Cancer 107, 1564–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for RNA sequencing data deposited in NCBI Gene Expression Omnibus is GEO: GSE123520.