Abstract
Overexpression of NEUROG2 and NEUROG1 (NEUROG2/1) in human embryonic stem cells (hESCs) rapidly produces functional networks of excitatory and inhibitory neurons. To facilitate the use of this efficient inducible human neuron model in neuroscience research, we generated hESCs with doxycycline-inducible NEUROG2/1 via lentivirus and a tdTomato fluorescent reporter knock-in at the MAP2 locus using the CRISPR nuclease Cas9. Upon doxycycline-driven induction of NEUROG2/1, these hESCs differentiate within days into cells that are uniformly MAP2 immunoreactive and tdTomato fluorescent.
1. Resource utility
MAP2 is a widely-used marker for mature neurons. This HUES66 hESC line with doxycycline-inducible NEUROG2 and NEUROG1 and a MAP2-tdTomato knock-in expresses a red fluorescent protein upon neuronal differentiation. This cell line can help identify critical factors in neuronal differentiation or to study how newly-formed neurons integrate into neural networks in vivo.
2. Resource details
We generated a HUES66 human embryonic stem cell (hESC) line with inducible expression of NEUROG2/1 via lentiviral transgenesis and a knock-in MAP2-T2A-tdTomato fluorescent reporter via CRISPR-mediated homologous recombination (HR). As previously described (Busskamp et al., 2014; Lu et al., 2019), doxycycline-inducible expression of NEUROG2/1 was achieved via lentiviral delivery of transgenes for the reverse tetracycline transactivator 3 (rtTA3) linked to the blasticidin resistance gene and tet-inducible expression of the master neuronal transcription factors NEUROG2 and NEUROG1 linked to the puromycin resistance gene.
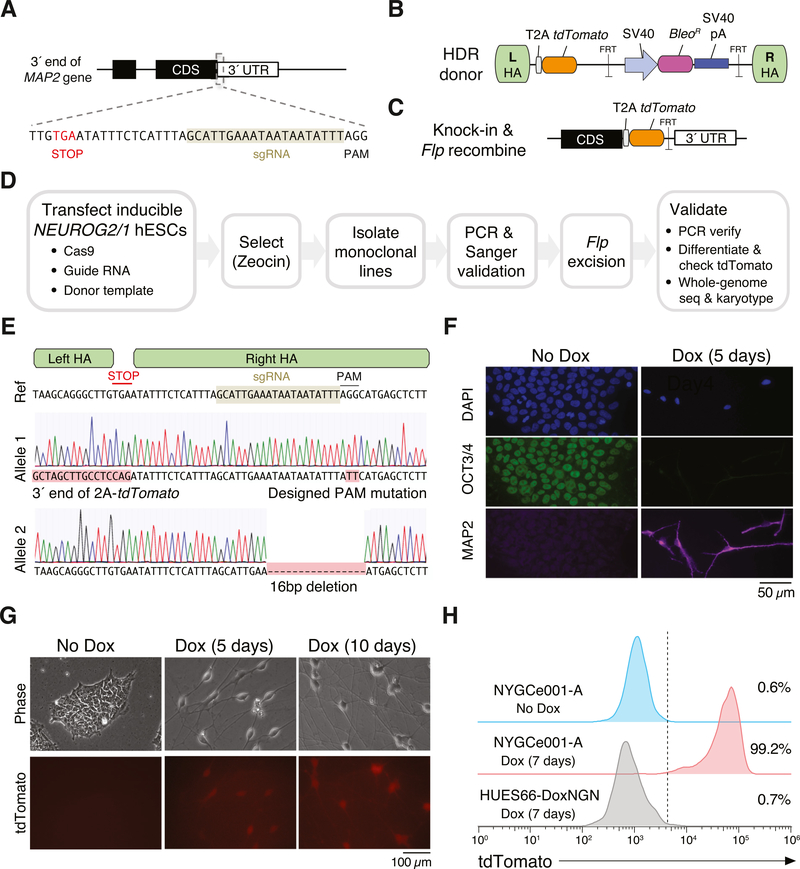
To create the neuron reporter line, we targeted the beginning of the 3′ untranslated region (UTR) of the MAP2 gene by designing a Cas9 single-guide RNA that cuts 31 bp after the MAP2 stop codon (Fig. 1A). We combined the Cas9 plasmid with a donor vector containing ~850 bp homology arms (HA) flanking the MAP2 stop codon (Fig. 1B). The donor vector contains a self-cleaving 2A peptide and tdTomato fluorescent protein that are in frame with the end of MAP2. The donor also contains a zeocin selection cassette flanked by FRT sites to enable selection after successful integration and a small mutation in the right HA at the Cas9 protospacer-adjacent motif (PAM) to prevent Cas9 from cutting either the donor vector or the genome after HR. After integration of the donor, transient expression of Flp recombinase removes the zeocin selection cassette (Fig. 1C).
Fig. 1.
Generation and characterization of the MAP2 knock-in reporter cell line NYGCe001-A. (A) Schematic of 3′ region of the MAP2 coding sequence with stop codon and Cas9 target site. (B) Schematic of donor plasmid. (C) Schematic of MAP2 locus after homology-directed repair and Flp recombination. (D) Workflow to generate a monoclonal MAP2 reporter cell line. (E) Sanger sequencing of individual MAP2 alleles in NYGCe001-A near the Cas9 target site. (F) Immunocytochemistry for OCT4 and DAPI before and after Dox-induced expression of NEUROG2 and NEUROG1. (G) Endogenous tdTomato fluorescence before and after Dox induction. (H) Flow cytometry of endogenous tdTomato before and after Dox induction in NYGCe001-A and after Dox induction in the parental line (without the MAP2 reporter knock-in).
We transfected HUES66 with inducible expression of NEUROG2/1, a plasmid containing Cas9 nuclease and the sgRNA and a plasmid with the donor template (Fig. 1D). Two days after transfection, zeocin was used to select for cells that underwent successful HR at the MAP2 locus. Zeocin selection was maintained for 5 days to select for cells with stable integration. Using dilution plating, we established monoclonal lines and validated successful HR by PCR amplification with a primer outside the HA and a primer that only binds the tdTomato transgene. After initial validation, we excised the bleomycin resistance cassette by transient transfection of Flp recombinase. We performed a second dilution plating and verified excision using PCR. In total, the HUES66 cells were passaged 14 times over the course of the derivation of the monoclonal reporter line.
The selected monoclonal line (NYGCe001-A) contains the desired in-frame knock-in fragment and a designed GG to TT mutation at the PAM sequence (to prevent re-cutting by Cas9) (Tables 1 and 2). On the other allele, there is a small deletion in a noncoding region near the cut site (3′ UTR) (Fig. 1E). Using low-pass, whole-genome sequencing (WGS) of parental and knock-in lines, we confirmed that NYGCe001-A had no copy number changes at ≤5 Mb resolution. To evaluate the pluripotency and differentiation ability of the selected line, we performed immunocytochemical staining of the pluripotency marker OCT3/4 and the neuron marker MAP2 (Fig. 1F). Prior to differentiation, all NYGCe001-A cells examined were positive for OCT3/4 and negative for MAP2 by immunofluorescence and, using flow cytometry, we found that 99.3% cells were positive for the pluripotency cell surface marker TRA-1–81 by flow cytometry (Supplemental Fig. 1A). Also, using chemical growth factors, we confirmed that NYGCe001-A hESCs could differentiate into all three germ layers via immunofluorescent staining for layer-specific marker genes (Supplemental Fig. 1B).
Table 1.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography | Normal | Fig. 1G |
| Phenotype | Immunocytochemistry | 100% of cells were stained positive for OCT3/4 before Dox induction. 100% of cells were stained positive for MAP2 at day 5 after Dox induction. | Fig. 1F |
| Fluorescent photography | All cells showed tdTomato signal at day 10 after Dox induction. | Fig. 1G | |
| Flow cytometry | 99.2% of NYGCe001-A cells were tdTomato positive at day 7 after Dox induction. | Fig. 1H | |
| Flow cytometry | 99.3% of NYGCe001-A cells were stained positive for TRA-1–81. | Supplemental Fig. 1A | |
| Genotype Identity | Digital karyotype | 46XX : 2.5 – 10 MB resolution | Supplemental Fig. 2 |
| Microsatellite PCR (mPCR) | Not performed | ||
| Identity | STR analysis | 18 sites tested | Submitted in archive with journal |
| Mutation analysis | |||
| Sequencing | Precise insertion in chr2 at desired location; small deletion on the other allele. | Fig. 1E | |
| Mutation analysis | WGS | N/A | |
| Microbiology and virology | Mycoplasma | Mycoplasma testing by luminescence: Negative. | Supplemental Fig. 3 |
| Differentiation potential | NEUROG2/1-overexpression to differentiate neurons | Expression of neuron marker MAP2. | Fig. 1F |
| In vitro differentiation assay | Cells from ectoderm differentiation were stained positive for Otx2; cells from mesoderm differentiation were stained positive for Brachyury; cells from endoderm differentiation were stained positive for SOX17. | Supplemental Fig. 1B |
Table 2.
Reagents details.
| Antibodies used for immunocytochemistry/flow-citometry | |||
|---|---|---|---|
| Antibody | Dilution | Company Cat # and RRID | |
| Pluripotency Marker | Mouse anti-OCT3/4 | 1:500 | Santa Cruz Biotech Cat# sc-5279,RRID: AB_628051 |
| Mouse anti-TRA-1–81 (Podocalyxin), PE | 1:20 | ThermoFisher Cat# 12–8883–80 RRID: AB_891608 | |
| Differentiation Marker | Guinea pig anti-MAP2 | 1:1000 | Synaptic Systems Cat # 188 004,RRID: AB_2138181 |
| Goat anti-SOX17 | 1:500 | R&D Systems Cat# AF1924, RRID: AB_355060 | |
| Goat anti-Otx2 | 1:500 | R&D Systems Cat# AF1979, RRID: AB_2157172 | |
| Goat anti-Brachyury | 1:500 | R&D Systems Cat# BAF2085 RRID: AB_2303014 | |
| Secondary antibodies | Donkey anti-guinea pig Alexa 647 | 1:1000 | Jackson ImmunoResearch Cat# 706–605–148, RRID: AB_2340476 |
| Goat anti-mouse Alexa 488 | 1:1000 | ThermoFisher Scientific Cat# A32723, RRID: AB_2633275 | |
| Donkey anti-goat Northern Lights 557 | 1:200 | R&D Systems Cat# NL001, RRID: AB_663766 | |
| Primers | |||
| Target | Forward/Reverse primer (5′–3′) | ||
| Targeted mutation analysis/sequencing | MAP2 exon 15, tdTomato knock-in | aggtcccagattgtagacctgg/ cgtccatgccgtacaggaa | |
| Targeted mutation analysis/sequencing | tdTomato knock-in, MAP2 3′ UTR | tcggaatcgttttccgggac/ gccttcaggcacatgaaaggt | |
After 5 days of doxycycline-induced (NEUROG2/1-driven) neural differentiation, all cells examined stained negative for OCT3/4 and positive for MAP2 by immunofluorescence. Similarly, the endogenous tdTomato fluorescence signal for the selected line was negative prior to doxycycline induction, but, after doxycycline treatment, tdTomato expression was initially visible at day 5 and intensified at day 10 (Fig. 1G). Flow cytometry at day 7 post-doxycycline induction indicated that 99.2% of cells were tdTomato positive (Fig. 1H).
3. Materials and methods
3.1. Cell culture
HUES66 human embryonic stem cells (hESCs) were maintained according to the Enhanced Culture Platform, which is less prone to spontaneous differentiation and aids in culturing hESCs as single cells for selection after genome engineering (Lu et al., 2019; Peters et al., 2013; Schinzel et al., 2011). Briefly, hESCs were maintained in Essential 8 media (ThermoFisher) supplemented with 100 μg/mL Normocin (InvivoGen) and cultured in standard tissue culture dishes coated with Geltrex (ThermoFisher) at 37 °C in 5% CO2. For passaging, the cells were dissociated using Accutase (STEMCELL). When subculturing or growing at very low density, we added 10 μM Rho kinase inhibitor Y-27632 (MilliporeSigma) to the culture medium to increase survival.
3.2. Lentiviral transduction
Lentiviral delivery was used to introduce constitutive expression of reverse tetracycline transactivator (rtTA3) driven by the human elongation factor-1a promoter (hEF1a) and inducible expression of NEUROG2/1 driven by a tetracycline response element promoter into hESCs. Lentivirus was produced as previously described (Sanjana et al., 2014). The two viral constructs were packaged separately and then pooled at the time of transduction.
3.3. Transfection
We cloned a single guide RNA (sgRNA) targeting the 3′ end of the MAP2 gene into lentiCRISPRv2 (Addgene 52961), a plasmid that contains both Cas9 and a sgRNA cassette, as previously described (Sanjana et al., 2014). We transfected hESCs using Lipofectamine 3000 (ThermoFisher) with this CRISPR construct and a plasmid donor template T2A-tdTomato-FRT-SV40 promoter-Zeo-SV40pA-FRT flanked by 837 bp left and 876 bp right homology arms. We plated 0.4 million cells per well into a 12-well dish 6 h before transfection. A total of 2 μg plasmid was transfected into the plated cells with 2 μl of Lipofectamine 3000. Culture media was fully refreshed 12 h after transfection. After successful homology-directed integration of the donor, we removed the zeocin selection cassette using recombinase plasmid pCAG-Flpe (Addgene 13787).
3.4. In vitro differentiation assay
NYGCe001-A cells were differentiated to ectoderm, mesoderm, and endoderm using the media supplements and growth factors in the Human Pluripotent Stem Cell Functional Identification Kit (R&D Systems SC027B). To validate lineage commitment, differentiated cells were stained with anti-Otx2 (ectoderm), anti-Brachyury (mesoderm), and anti-SOX17 (endoderm).
3.5. Immunocytochemistry
Cells were grown on Geltrex-coated coverslips and fixed with 4% paraformaldehyde and 4% sucrose in PBS at room temperature for 5 min followed by washing 3 times with PBS. The fixed cells were then permeabilized with 0.1% Triton X-100 in PBS for 5 min, washed 3 times with PBS, and incubated in 3% bovine serum albumin in PBS for 1 h to block nonspecific binding. The cells were incubated overnight with the primary antibodies at 4 °C overnight followed by incubation with the secondary antibodies at room temperature for 1 h. The images were collected using an Axio Observer Z1 microscope (Zeiss).
3.6. Flow cytometry
The cells were dissociated into a single cell suspension using Accutase and passed through a 35 μm cell strainer (BD Falcon). Flow cytometry data was acquired on a Sony SH800 and analyzed using FlowJo v10.5 (BD).
3.7. Karyotype analysis
Karyotyping was done using the digital karyotyping service from FIND Genomics. Briefly, DNA was extracted using the GeneJET DNA Purification Kit (ThermoFisher), followed by nanopore library preparation using the Rapid Barcoding Kit (Oxford Nanopore). The gDNA library was sequencing using a MinION nanopore sequencer (Oxford Nanopore). The collected DNA data was analysed using the FIND Cell software (FIND Genomics) and analyzed to confirm cell population identity and digital karyotype with a structural variant detection resolution of 5Mb or greater. Digital karyotyping were done on 3 separate genomic DNA samples.
3.8. Mycoplasma detection
We verified the absence of mycoplasma in the culture medium using a luminescence-based assay, MycoAlert PLUS (Lonza).
3.9. Short tandem repeat analysis
STR analysis was performed by ATCC using a PowerPlex 18D kit (Promega) and the Prism 3500xl Genetic Analyzer (Applied Biosystems). STR profiles were analyzed using GeneMapper ID-X v1.2 (Applied Biosystems).
Supplementary Material
Resource table:
| Unique stem cell line identifier | NYGCe001-A |
|---|---|
| Alternative name(s) of stem cell line | HUES66-DoxNGN-MAP2-tdTomato |
| Institution | New York Genome Center |
| Contact information of distributor | Neville Sanjana (neville@sanjanalab.org) |
| Type of cell line | hESC |
| Origin | Human |
| Additional origin info | Sex: Female |
| Cell Source | Harvard Stem Cell Institute |
| Clonality | Monoclonal |
| Method of reprogramming | N/A |
| Genetic Modification | Yes |
| Type of Modification | Lentiviral transgenic for Dox-inducible Neurogenin2/1 and CRISPR-Cas9 knock-in of tdTomato at MAP2 locus |
| Associated disease | N/A |
| Gene/locus | Chr2/MAP2 |
| Method of modification | Lentiviral transgenic; CRISPR-Cas9 homology-mediated repair following by Flp recombinase |
| Name of transgene or resistance | Transgenes: NEUROG2, NEUROG1, rtTA3, tdTomato; Resistance markers: blasticidin, puromycin |
| Inducible/constitutive system | TET-ON (rtTA3) for NEUROG2–2A-NEUROG1–2A-PuroR |
| Date archived/stock date | N/A |
| Cell line repository/bank | N/A |
| Ethical approval | New York Genome Center IBC (ISCRO003-Y2-M0); HUES66 line from Harvard Stem Cell Institute |
Acknowledgments
We thank the entire Sanjana laboratory for support and advice. We also thank G. Garipler and E. Mazzoni for the homology donor vector and D. Peters and C. Cowan for guidance with stem cell culture. N.E.S. is supported by NYU and NYGC startup funds, NIH/NHGRI (R00HG008171, DP2HG010099), NIH/NCI (R01CA218668), DARPA (D18AP00053), the Sidney Kimmel Foundation, and the Brain and Behavior Foundation.
Footnotes
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.scr.2019.101643.
References
- Busskamp V, Lewis NE, Guye P, Ng AHM, Shipman SL, Byrne SM, Sanjana NE, Murn J, Li Y, Li S, et al. , 2014. Rapid neurogenesis through transcriptional activation in human stem cells. Mol. Syst. Biol 10, 760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Shi X, Allen A, Baez-Nieto D, Nikish A, Sanjana NE, Pan JQ, 2019. Overexpression of NEUROG2 and NEUROG1 in human embryonic stem cells produces a network of excitatory and inhibitory neurons. FASEB J. 33, 5287–5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters DT, Cowan CA, Musunuru K, 2013. Genome editing in human pluripotent stem cells. Stembook. Harvard Stem Cell Institute, CambridgeMA. [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F, 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinzel RT, Ahfeldt T, Lau FH, Lee Y-K, Cowley A, Shen T, Peters D, Lum DH, Cowan CA, 2011. Efficient culturing and genetic manipulation of human pluripotent stem cells. PLoS One 6, e27495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.