Abstract
The drug-like molecule PF-06446846 (PF846) binds the human ribosome and selectively blocks the translation of a small number of proteins by an unknown mechanism. In structures of PF846-stalled human ribosome nascent chain complexes, PF846 binds in the ribosome exit tunnel in a eukaryotic-specific pocket formed by 28S ribosomal RNA, and alters the path of the nascent polypeptide chain. PF846 arrests the translating ribosome in the rotated state of translocation, in which the peptidyl-tRNA 3’-CCA end is improperly docked in the peptidyl transferase center. Selections of mRNAs from mRNA libraries using translation extracts reveal that PF846 can stall translation elongation, arrest termination, or even enhance translation, depending on nascent chain sequence context. These results illuminate how a small molecule selectively targets translation by the human ribosome, and provides a foundation for developing small molecules that modulate the production of proteins of therapeutic interest.
INTRODUCTION
Most compounds that target the ribosome affect a large number of mRNAs through general inhibition of translation initiation or elongation 1-3. Furthermore, these compounds almost exclusively act as broad-spectrum inhibitors, displaying little sequence specificity 4. Many studies have revealed how these compounds bind the translating ribosome and inhibit its function 5-9. Recently, we described the small molecule PF846 that selectively blocks the translation of individual mRNAs by the human ribosome 10. However, a mechanism to account for the ability of PF846 and related compounds 11,12 to selectively inhibit human protein synthesis remains unknown.
PF846 blocks production of proprotein convertase subtilisin/kexin type 9 (PCSK9)–an important target for regulating plasma low-density lipoprotein cholesterol levels–by interfering with the elongation phase of translation 10; 13. PF846 selectively stalls the ribosome on very few translated protein nascent chains, generally early in their formation and with no clear sequence pattern 10. To gain insight into the mode of action of PF846 in targeting the human ribosome and specific nascent chains, and to identify principles for future drug development, we used single-particle cryogenic electron microscopy (cryo-EM) to determine structures of PF846 stalled ribosome nascent chain (RNC) complexes. We also employed randomized mRNA libraries to identify sequence preferences for PF846-mediated translation stalling.
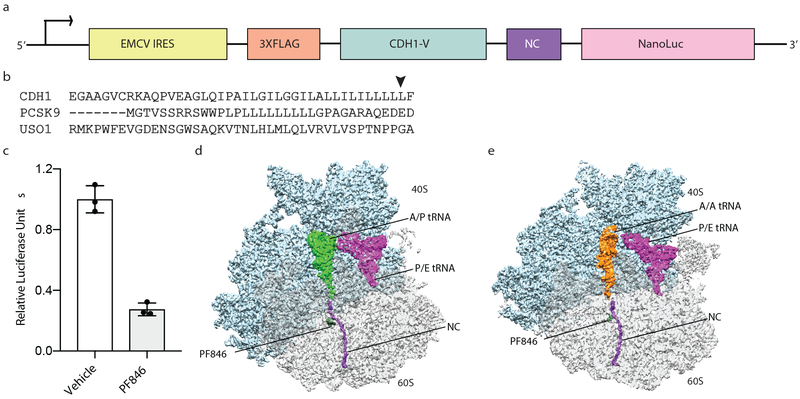
Since the stalling sequences in the few proteins affected by PF846 mostly occur near the N-terminus and would therefore reside in the ribosome exit tunnel 10, we first identified conditions for affinity purification of stable PF846-stalled RNCs from in vitro translation reactions in human cell extracts. The calcium-dependent cell-cell adhesion glycoprotein Cadherin-1 (CDH1) 14 is stalled near its C-terminus by PF846, enabling the extension of the peptide N-terminus beyond the confines of the ribosome exit tunnel 15. Appending an N-terminal affinity tag followed by the CDH1-V domain and the nascent chain sequences targeted by PF846 did not disrupt the ability of PF846 to stall translation and allowed formation and purification of stable PF846-stalled RNCs (Fig. 1a-c, Supplementary Fig. 1). All the following assays with PF846-stalled RNCs used an N-terminal 3X-FLAG tag unless specifically noted.
Fig. 1 ∣. Structural analysis of PF846 stalled RNCs.
a, Schematic representation of the DNA constructs used to prepare PF846 stalled RNCs. b, Protein sequences from CDH1, PCSK9, and USO1 tested for stalling. The arrow indicates the main stalling site predicted from ribosome profiling data 10. c, Luciferase reporter assays using the chimeric PCSK9 nascent chain in the absence (white bar) or presence (grey bar) of PF846 (Data represent mean ± s.d., n = 3 independent experiments). d, Cryo-EM structure of the stalled CDH1-RNC in the rotated state showing A/P tRNA (green), P/E tRNA (magenta), CDH1 nascent chain (purple) and PF846 (dark green) with small and large subunit colored in cyan and grey, respectively. e, Structure of the stalled CDH1-RNC in the rotated state with A/A tRNA (orange), and P/E tRNA (magenta). Source data for panel c are available in Supplementary Data Set 3.
RESULTS
Overall structures of PF846 stalled RNCs
To investigate how PF846 stalls specific nascent chain sequences in the human ribosome, we used cryo-EM to determine structures of PF846-stalled CDH1, PCSK9 and USO1 (USO1 vesicle transport factor) ribosome nascent chain complexes (CDH1-RNC, PCSK9-RNC and USO1-RNC, respectively) (Supplementary Figs. 2, 3, and 4, Tables 1, 2, 3, and 4). In both the CDH1-RNC and PCSK9-RNC samples, particle sorting of the purified RNCs revealed a major population of ribosomes in the rotated state that precedes mRNA and tRNA translocation 16, containing predominantly hybrid A/P site peptidyl-tRNA (i.e. nascent chain-tRNA, or NC-tRNA), but with some rotated-state RNCs containing A/A site NC-tRNA. The USO1-RNC also adopts the rotated state, although the positioning of the A-site tRNA in the A/P or A/A site was less well defined. All three RNCs also included tRNA in the hybrid P/E site and PF846 bound in the ribosome exit tunnel (Fig. 1d, e, Supplementary Fig. 5). The following analyses describe the CDH1-RNCs due to the higher-quality of the CDH1-RNC cryo-EM maps, although we observe similar behavior for the PCSK9-RNC and USO1-RNC samples, as shown in the Supplementary Figures.
Table 1 ∣.
Cryo-EM data collection, refinement and validation statistics of CDH1-RNC
| CDH1-RNC_AP tRNA (EMD-0600, PDB 6OLE.) |
CDH1-RNC_AA tRNA (EMD-0599, PDB 6OLF) |
CDH1-RNC_PP tRNA (EMD-0601, PDB 6OLG) |
|
|---|---|---|---|
| Data collection and processing | |||
| Magnification | 43,478 | 43,478 | 43,478 |
| Voltage (kV) | 300 | 300 | 300 |
| Electron exposure (e−/Å2) | 50 | 50 | 50 |
| Defocus range (μm) | 1.0 to 2.0 | 1.0 to 2.0 | 1.0 to 2.0 |
| Pixel size (Å) | 1.15 | 1.15 | 1.15 |
| Symmetry imposed | C1 | C1 | C1 |
| Initial particle images (no.) | 213,412 | 213,412 | 213,412 |
| Final particle images (no.) | 73,110 | 10,350 | 16,594 |
| Map resolution (Å) | 3.0 | 3.9 | 3.4 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 2.5 to 7.5 | 3.1 to 9 | 2.8 to 8 |
| Refinement | |||
| Initial model used (PDB code) | |||
| 40S subunit | 6ek0 | 6ek0 | 5aj0 |
| 60S subunit | 5aj0 | 5aj0 | 5aj0 |
| tRNA | 3j7r | 3j7r | 5aj0 |
| Model resolution (Å) | 3.0 | 3.9 | 3.4 |
| FSC threshold | 0.5 | 0.5 | 0.5 |
| Model resolution range (Å) | 2.5 to 7.5 | 3.1 to 9 | 2.8 to 8 |
| Map sharpening B factor (Å2) | −71 | −73 | −62 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.007 | 0.019 | 0.009 |
| Bond angles (°) | 1.014 | 1.188 | 1.193 |
| Validation | |||
| MolProbity score | 1.81 | 2.02 | 2.04 |
| Clashscore | 4.89 | 7.02 | 6.89 |
| Poor rotamers (%) | 1.00 | 1.05 | 1.34 |
| Ramachandran plot | |||
| Favored (%) | 89.90 | 87.03 | 89.28 |
| Allowed (%) | 9.76 | 12.63 | 10.08 |
| Disallowed (%) | 0.36 | 0.35 | 0.64 |
Table 2 ∣.
Cryo-EM data collection, refinement and validation statistics of PCSK9-RNC
| PCSK9-RNC_AP tRNA (EMD-0596, PDB 6OM0) |
PCSK9-RNC_AA tRNA (EMD-0597, PDB 6OM7) |
PCSK9-RNC_PP tRNA (EMD-0598, PDB 6OLZ) |
|
|---|---|---|---|
| Data collection and processing | |||
| Magnification | 43,478 | 43,478 | 43,478 |
| Voltage (kV) | 300 | 300 | 300 |
| Electron exposure (e−/Å2) | 50 | 50 | 50 |
| Defocus range (μm) | 1.0 to 2.5 | 1.0 to 2.5 | 1.0 to 2.5 |
| Pixel size (Å) | 1.15 | 1.15 | 1.15 |
| Symmetry imposed | C1 | C1 | C1 |
| Initial particle images (no.) | 148,817 | 148,817 | 148,817 |
| Final particle images (no.) | 43,666 | 9,564 | 7,214 |
| Map resolution (Å) | 3.1 | 3.7 | 3.9 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 2.5 to 7.5 | 3 to 9 | 3 to 9 |
| Refinement | |||
| Initial model used (PDB code) | |||
| 40S subunit | 6ek0 | 6ek0 | 5aj0 |
| 60S subunit | 5aj0 | 5aj0 | 5aj0 |
| tRNA | 3j7r | 3j7r | 5aj0 |
| Model resolution (Å) | 3.1 | 3.7 | 3.9 |
| FSC threshold | 0.5 | 0.5 | 0.5 |
| Model resolution range (Å) | 2.5 to 7.5 | 3 to 9 | 3 to 9 |
| Map sharpening B factor (Å2) | −75 | −77 | −80 |
| Model composition | |||
| Nonhydrogen atoms | |||
| Protein residues | |||
| Ligands | |||
| B factors (Å2) | |||
| Protein | |||
| Ligand | |||
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.007 | 0.020 | 0.009 |
| Bond angles (°) | 1.036 | 1.190 | 1.222 |
| Validation | |||
| MolProbity score | |||
| Clashscore | 4.86 | 6.36 | 7.51 |
| Poor rotamers (%) | 1.25 | 1.03 | 0.83 |
| Ramachandran plot | |||
| Favored (%) | 90.02 | 87.57 | 88.18 |
| Allowed (%) | 9.54 | 12.11 | 11.36 |
| Disallowed (%) | 0.45 | 0.32 | 0.45 |
Table 3 ∣.
Cryo-EM data collection of PCSK9s-RNC
| PCSK9s-RNC_AP tRNA (EMD-20134) |
PCSK9s-RNC_PP tRNA (EMD-20135) |
|
|---|---|---|
| Data collection and processing | ||
| Magnification | 40,983 | 40,983 |
| Voltage (kV) | 300 | 300 |
| Electron exposure (e–/Å2) | 50 | 50 |
| Defocus range (μm) | 1.0 to 2.5 | 1.0 to 2.5 |
| Pixel size (Å) | 1.22 | 1.22 |
| Symmetry imposed | C1 | C1 |
| Final particle images (no.) | 8,631 | 8,433 |
| Map resolution (Å) | 4.68 Å | 4.57 Å |
| FSC threshold | 0.143 | 0.143 |
Table 4 ∣.
Cryo-EM data collection, refinement and validation statistics of USO1-RNC
| USO1-RNC_AP tRNA (EMD-0526, PDB 6OLI) |
|
|---|---|
| Data collection and processing | |
| Magnification | 43,478 |
| Voltage (kV) | 300 |
| Electron exposure (e−/Å2) | 55 |
| Defocus range (μm) | 1.5 to 2.5 |
| Pixel size (Å) | 1.15 |
| Symmetry imposed | C1 |
| Initial particle images (no.) | 79,322 |
| Final particle images (no.) | 38,314 |
| Map resolution (Å) | 3.5 |
| FSC threshold | 0.143 |
| Map resolution range (Å) | 2.8 to 8 |
| Refinement | |
| Initial model used (PDB code) | |
| 40S subunit | 6ek0 |
| 60S subunit | 5aj0 |
| tRNA | 3j7r |
| Model resolution (Å) | 3.1 |
| FSC threshold | 0.5 |
| Model resolution range (Å) | 2.5 to 7.5 |
| Map sharpening B factor (Å2) | −91 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.117 |
| Validation | |
| MolProbity score | 1.98 |
| Clashscore | 5.36 |
| Poor rotamers (%) | 1.48 |
| Ramachandran plot | |
| Favored (%) | 89.71 |
| Allowed (%) | 9.95 |
| Disallowed (%) | 0.35 |
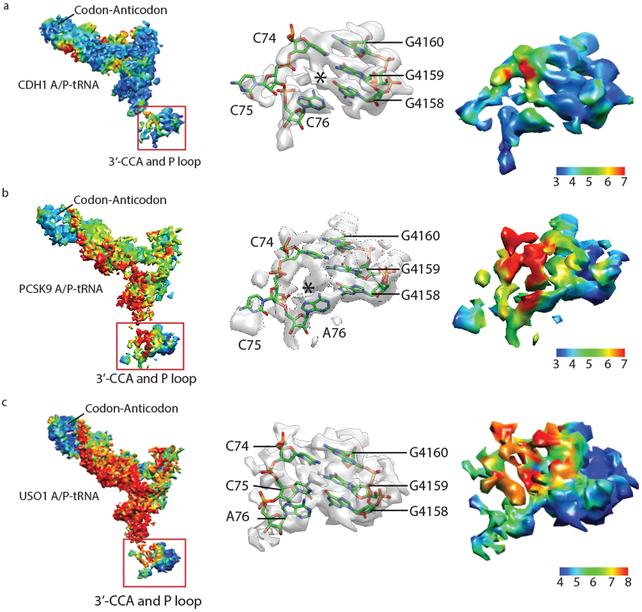
The CDH1-RNC complexes with hybrid A/P site NC-tRNA or with A/A site NC-tRNA yielded cryo-EM maps with an average resolution of 3.0 Å and 3.9 Å, respectively (Fig. 1d, e, Supplementary Fig. 2). The hybrid A/P and P/E NC-tRNAs each represent a mixture of tRNAs, as inferred from the EM map density for nucleotide bases in the codon-anticodon base pairs (Supplementary Fig. 6), consistent with the clustering of stall sites spanning multiple codons observed by ribosome profiling 10. In the CDH1-RNC and PCSK9-RNC samples, we also observed a small population of PF846-stalled RNCs in the non-rotated state bearing a P/P-site NC-tRNA that exists after peptide bond formation (Supplementary Figs. 2, 3, 5). Interestingly, this population appeared in higher abundance when the PCSK9-RNC sample was purified using a short isolation time prior to making cryo-EM grids (Supplementary Fig. 3d-e). These results suggest that the non-rotated RNC is a transient state during PF846 mediated translation stalling. Collectively, our findings suggest that PF846 sequentially stalls RNCs in the rotated state at adjacent codons, whereas the non-rotated state with P/P-site NC-tRNA either participates more readily in translation elongation or can be resolved by ribosome quality control pathways, i.e. those that involve PELO and HBS1L 12.
Eukaryotic-specific small molecule binding pocket
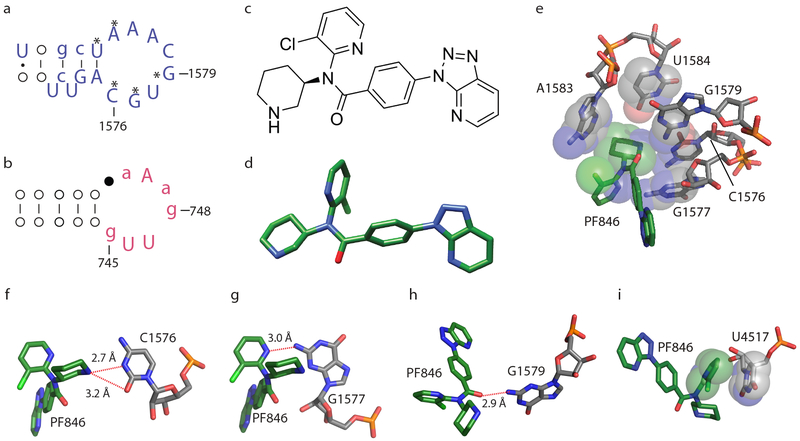
In the stalled RNCs, PF846 occupies a binding pocket in a newly-identified groove of the ribosomal exit tunnel formed by universally conserved 28S rRNA nucleotides 17 (Fig. 2) and a stretch of the stalled nascent chain, best seen in the rotated-state RNCs with A/P NC-tRNAs. PF846 was fit as a rigid body to the density by evaluating its X-ray structures 10 and low energy conformations based on quantum mechanical assessment of accessible torsion angles (Supplementary Fig. 7a-f, Supplementary Table 1), followed by adjustments of the chloropyridine and aryl rings. Consistent with the ability of PF846 to bind to non-translating 80S ribosomes 10, contacts with 28S rRNA make up the bulk of the small molecule interactions with the ribosome (Fig. 2). The piperidine ring extends into the rRNA binding pocket, contacting nucleotides C1576, G1577, G1579, A1583 and U1584 (Fig. 2e, f). The chloropyridine and amide linkage also form hydrogen bonds with highly conserved nucleotides G1577 and G1579 (Fig. 2g, h). Additionally, the interaction is stabilized by U4517, which forms a stacking interaction with the chloro-pyridine ring (Fig. 2i). To explore the structural basis for PF846 action on eukaryotic but not bacterial ribosomes 10, we aligned the small molecule-binding pocket with the corresponding region in the E. coli ribosome (Fig. 2a, b, Supplementary Fig. 7g-k). Phylogenetic analysis revealed key changes to the nucleotide pattern in 23S rRNA in the large ribosomal subunit of bacteria 17 (Fig. 2a, b). Whereas nucleotides C1576, G1577 and G1579 in the human ribosome are highly complementary to the conformation of PF846 (Fig. 2e-h), nucleotides N1-methyl-G745 and ψ746 in bacterial 23S rRNA would clash with the compound (Supplementary Fig. 7i-j). Furthermore, whereas U4517 in the human ribosome stacks on the chloropyridine ring in PF846 (Fig. 2i), the corresponding nucleotide in bacterial 23S rRNA (U2609) would sterically clash with PF846 (Supplementary Fig. 7k). Taken together, these distinctions in nucleotide identity and positions help explain PF846’s specificity for eukaryotic ribosomes.
Fig. 2 ∣. Interactions of PF846 with the human ribosome.
a-b, Phylogenetic analysis of the 28S rRNA binding pocket in eukaryotes (a) and bacteria (b). Nucleotides that interact with PF846 are labeled with an asterisk (*). Nucleotides with capital letters (AUCG), are over 98% conserved, nucleotides with small letters (aucg) are 90%-98% conserved. Nucleotides denoted by filled or open circles represent a conservation of 80% to 90%, or less than 80%, respectively. c-d, PF846 chemical and three-dimensional structure, respectively. e, 28S rRNA residues with direct interactions with the piperidine ring of PF846. Surfaces represent the van der Waals radii of the C, N and O atoms. f-h, Interactions of PF846 and nucleotides from 28S rRNA. i, Interaction of the chloro-pyridine ring with U4517, with van der Waals surfaces as in e.
Analysis of stalled nascent chains within the exit tunnel
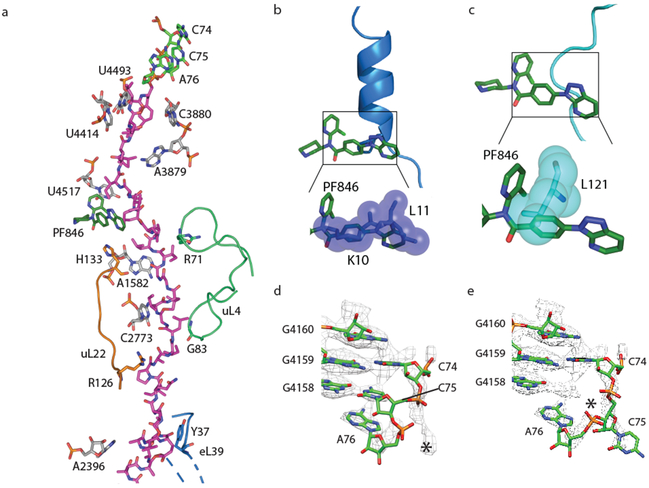
The triazolopyridine ring system of PF846 (Fig. 2c, d) faces the ribosome tunnel and is the only part of the molecule with direct interaction with the stalled nascent chain (Fig. 3a, Supplementary Fig. 8). Notably, the triazolopyridine moiety has incomplete density (Supplementary Fig. 7a-f) that may be caused by the flexibility of this region during nascent chain elongation. Interestingly, the density for the nascent chains in the RNCs with A/P NC-tRNA was mostly well defined in the ribosome exit tunnel with a local resolution of ~4–5 Å, although they are likely comprised of different sequences superimposed on each other corresponding to the cluster of stalled states spanning multiple mRNA codons (Supplementary Fig. 6, Supplementary Fig. 8) 10. All three of the nascent chains (CDH1, PCSK9, USO1) have similar geometry and adopt a predominantly extended conformation in the exit tunnel 18 (Supplementary Fig. 8). The nascent chain spans ~9 residues between the C-terminus of the nascent chains covalently linked to the 3’-CCA end of A/P site tRNA in the peptidyl transferase center (PTC) and the small molecule binding site (Fig. 3a, Supplementary Fig. 8). These residues engage in multiple interactions with nucleotides in 28S rRNA of the 60S subunit, including U4493, U4414, and A3879 (Fig. 3a, Supplementary Fig. 8d, e). Past the compound-binding pocket (i.e. in the N-terminal direction), the nascent chain contacts ribosomal proteins uL4 and uL22, along with A1582 and C2773 in 28S rRNA (Fig. 3a, Supplementary Fig. 8d, e). These contacts distal from the peptidyl transferase center are likely important for the stalling induced by PF846, based on N-terminal deletion analysis of the PCSK9 sequence 10.
Fig. 3 ∣. Nascent chains in the exit tunnel and their effects on the A/P tRNA 3’-CCA end.
a, Molecular model of CDH1 nascent chain (magenta) following the 3’-CCA end of A/P tRNA (green, at the top) in the ribosome exit tunnel with 28S rRNA (grey), uL22 (orange), uL4 (light green) and PF846 (dark green) highlighted. b-c, Superposition of hCMV 20 (blue) and SRP 21 (cyan) stalled nascent chains within the exit tunnel, showing the predicted steric clashes with PF846 (dark green), using van der Waals surfaces. d, Poor positioning of A/P NC-tRNA 3’-CCA end near the ribosomal P-loop. High-resolution structure of the 3’-CCA end of peptidyl-tRNA (PDB: 1vy4) 37 is docked as a rigid body, using nucleotides from 28S rRNA in the PTC as reference. e, Nucleotide C75 in A/P NC-tRNA modeled as flipped to nearby density without pairing with the P-loop (starting model from PDB: 1vy4). The asterisks in (d-e) indicate the positioning of nucleotide C75 in A/P NC-tRNA.
Most reported RNC structures have “kinks” in the nascent chain residing in the ribosome exit tunnel proximal to the PTC 19. The CDH1, PCSK9 and USO1 nascent chains also have a kink in their structures between the PTC active site and PF846-binding pocket (Fig. 3a, Supplementary Fig. 8), but these adopt a different geometry when compared to stalled human cytomegalovirus (hCMV) and signal recognition particle (SRP) nascent chains 20,21 (Fig. 3b, c). In the PF846-stalled RNC structures, the “kinks” assume more acute angles and occur closer to the PTC, to make enough space for the bound small molecule (Fig. 3a, Supplementary Fig. 8), whereas the hCMV and SRP NC positions would occlude PF846 binding (Fig. 3b, c).
Destabilized interaction between the A/P-tRNA and the P-loop in PF846 stalled RNCs
PF846 stalls translation of the ribosome predominantly in the rotated state that precedes mRNA and tRNA translocation to the next codon, presumably impeding the action of eEF2, the GTPase which promotes the translocation reaction 22. In bacterial translation, which is better understood 23,24, translocation proceeds through a series of ribosome rotated states, in which the peptidyl-tRNA body and 3’-CCA end move independently from one another, transiting from an A/A site with 3’-CCA end base pairing with the large subunit rRNA A-loop, to an A/A site in the rotated state in which the 3’-CCA end base pairing with the large subunit rRNA A-loop is retained, and finally to an A/P site with 3’-CCA base pairing with the P-loop 25. At this point, EF-G (the bacterial homologue of eEF2) can catalyze the remaining conformational changes required for complete translocation of the mRNA and tRNAs 23,24. In the PF846-stalled complexes, we observe pre-translocation RNC populations with A/A and A/P NC-tRNAs. However, the A/P-tRNA exhibits incomplete base pairing of the 3’-CCA end with the ribosomal P-loop (Fig. 3d-e, Fig. 4). We propose that PF846 inhibits rapid translocation of specific nascent chains by eEF2, by altering the nascent chain structure within the ribosomal tunnel, causing a kink in the nascent chain closer to the PTC than may normally occur 20,21. This alteration would then prevent the peptidyl-tRNA from binding stably in the hybrid A/P site, which requires the tRNA 3’-CCA end to properly base pair with 28S rRNA nucleotides in the P-loop.
Fig. 4 ∣. Local resolution analysis of A/P tRNAs in PF846-stalled RNCs.
a-c, Local resolution for A/P tRNA in the CDH1-RNC (a), PCSK9-RNC (b) and USO1-RNC (c). Left, global view of tRNA and the P loop in 28S rRNA. Middle and right, zoomed in views of tRNA 3’-CCA and P-loop nucleotides showing cryo-EM density and local resolution, respectively. Resolution scale shown to the right for each local resolution map.
Sequence Determinants of PF846-induced stalling
The few NC sequences stalled by PF846 as previously identified by ribosome profiling do not reveal a simple amino acid “motif” responsible for stalling 10. We therefore used in vitro translation reactions to identify the sequence determinants in the NC required for PF846-induced stalling of RNCs. To assay the sequence-dependence along the NC, we first used a poly-asparagine scan, in which we replaced 3–4 amino acids in the stalling sequence at a time. We used asparagine scanning as opposed to alanine scanning used previously 10 to maintain a rough approximation to the size distribution of the amino acids in the NC, while introducing a hydrophilic side chain. In both the CDH1 and PCSK9 stalling sequences, replacement of leucine- and isoleucine-rich stretches of hydrophobic amino acids predicted to be in the PTC-proximal region of the ribosome exit tunnel abrogated PF846-induced stalling (Supplementary Fig. 9). By comparing to the structural models (Fig. 3a, Supplementary Fig. 8d), the most important sequences for CDH1 stalling span the NC from the PTC to just beyond the PF846 binding site, where multiple interactions with the exit tunnel are observed. For PCSK9, the hydrophobic stretch of amino acids identified to be important for PF846-induced stalling is likely to be positioned just beyond the PF846 binding site in the exit tunnel (Supplementary Fig. 8e).
The Asn-scanning experiments above revealed NC sequences sensitive to mutation, but did not reveal the spectrum of amino acid sequences that can be stalled by PF846. To identify the range of amino acid sequences that can be stalled by PF846 in an unbiased manner, we used an mRNA library-based approach followed by deep sequencing, analogous to ribosome display 26. We randomized four NC amino acids at a time near the PTC and near the PF846 binding site, by introducing (NNK)4 sequences (N, any nucleotide; K, G or U) into mRNAs encoding the CDH1 stalling sequence (Fig. 1a-b, Supplementary Fig. 10a). We used these mRNA libraries in in vitro translation reactions, in the presence or absence of PF846, and then used the 3X-FLAG tag at the N-terminus of the NC to pull down stalled RNCs, which would retain the mRNAs encoding the stalled NC sequences. Next, we used RNase H to eliminate mRNAs translated by elongating ribosomes, i.e. ribosomes in the middle of the luciferase ORF downstream of the stall sequence (Supplementary Fig. 10b). The mRNAs isolated in this manner were then converted into a DNA library and analyzed by deep sequencing.
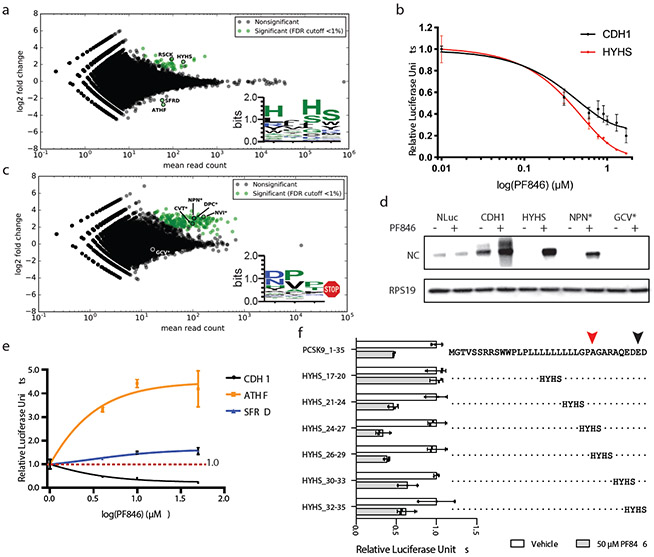
In the mRNA library with randomized sequences near the PTC, we observed strong enrichment for PF846-sensitive stalling sequences strikingly different from the original CDH1 sequence (Fig. 5a, Supplementary Table 2). In particular NC positions −2 and −4 from the predicted stall site are enriched in histidine rather than leucine (Fig. 5a, inset). Other polar and charged amino acids are also enriched in the PF846-sensitive NCs. We validated two of the newly-identified stalling sequences using in vitro luciferase assays. We found PF846 stalled HYHS and RSCK sequences with a similar IC50 compared to LLLL in the original CDH1 sequence (Fig. 5b, Supplementary Fig. 10c), with the HYHS sequence being more potently stalled at high PF846 concentrations (Fig. 5b). Both sequences were also enriched in stalled RNCs, as determined by Western blotting (Fig. 5d, Supplementary Fig. 10d). We tested the HYHS motif in the context of the PCSK9 stalling sequence, and found it does not induce PF846-dependent stalling as seen in the CDH1 sequence background (Fig. 5f). The effects of the HYHS sequence in fact track the poly-asparagine scan of the PCSK9 sequence (Supplementary Fig. 9b), in terms of PF846-dependent stalling. Thus, the HYHS motif on its own is not sufficient to induce PF846-dependent stalling, but requires additional sequences in the NC.
Fig. 5 ∣. Selections of PF846-dependent stalling sequences from randomized mRNA libraries.
a, MA plot (data with M (log ratio) and A (mean average) scales) of sequences enriched for PF846-induced translation elongation stalling, compared to translation reactions in the absence of PF846. Sequences were enriched from the mRNA library of CDH1-derived nascent chains with four amino acid positions randomized near the predicted stall site in the PTC. Log2-fold enrichment of sequences is plotted against total read count, for experiments carried out in duplicate. Green, sequences enriched with an adjusted P-value < 0.01. Inset, consensus sequence logo 38 for enriched sequences. b, In vitro translation assays with luciferase reporters containing the WT CDH1 stalling sequence or with the HYHS motif near the PTC, as a function of PF846 concentration (data represent mean ± s.d., n = 3 independent experiments). c, MA plot of sequences enriched for PF846-induced inhibition of translation termination, compared to translation reactions in the absence of PF846. Sequences were enriched from the mRNA library of CDH1-derived nascent chains with four amino acid positions randomized near the predicted stall site in the PTC, with only sequences containing stop codons shown. Log2-fold enrichment of sequences is plotted against total read count, for experiments carried out in duplicate. Green, sequences enriched with an adjusted P-value < 0.01. Inset, consensus sequence logo for enriched sequences 38. d, Western blot of affinity-purified stalled CDH1-derived nascent chains. Reactions in the absence or presence of 50 μM PF846 are shown. Western blot of ribosomal protein RPS19 for each sample is also shown, as a loading control. Original gels are shown in Supplementary Data Set 1. e, In vitro translation assays of the WT CDH1 stalling sequence, or with the ATHF or SFRD motif near the PTC, in the context of the CDH1 sequence (data represent mean ± s.d., n = 3 independent experiments). f, In vitro translation assays with the PCSK9 nascent chain sequence containing the HYHS motif in different positions in the absence (white bar) or presence (grey bar) of PF846. The stalled PCSK9 sequence is shown in full at the top, and the positions of the HYHS motif are indicated. Luciferase, luciferase-only control sequence. (Data represent mean ± s.d., n = 3 independent experiments). The statistical significance in panels a and c were tested by DESeq2 using the Wald test (two-sided) and adjusted for multiple comparisons by the Benjamini-Hochberg method 63. Source data for panels b, e and f are available in Supplementary Data Set 3.
We also tested two sequences that were depleted from the mRNA library in a PF846-dependent manner (Fig. 5a). Remarkably, in the context of the CDH1 NC, sequences SFRD and ATHF increase overall translation in a PF846-dependent manner (Fig. 5e), with ATHF increasing translation ~4-fold. The results with these sequences suggest that the original sequences identified as stalled by PF846 10 may be inherently poorly translated.
Surprisingly, we observed no enrichment of PF846-stalling dependent sequences in the mRNA library encoding randomized NCs in the ribosome exit tunnel near the PF846 binding site (Supplementary Fig. 10e). This is in striking contrast to the Asn-scanning results, in which mutation of NC sequences near the PF846 binding site disrupted stalling. This is likely due to the fact that Asn-scanning introduced negative determinants of PF846-dependent stalling, whereas enrichment from the mRNA libraries revealed positive determinants of stalling.
Finally, we also identified many sequences with a stop codon at the final position enriched in a PF846-dependent manner in the PTC-proximal mRNA library (Fig. 5c), indicating that PF846 blocked proper translation termination of these NCs. These sequences, roughly 6–7% of the total termination sequences identified, have an entirely different amino acid consensus compared to those predicted to stall translation elongation (Fig. 5a, c, insets). Since PF846 and related compounds had not been observed previously to block translation termination, we tested whether PF846 affects termination at the identified sequences. Consistent with the enrichment observed from the PTC mRNA library, PF846 prevented NC release from ribosomes when sequences NPN, DPC, NVI and CVT–but not GCV–preceded the stop codon (Fig. 5d, Supplementary Fig. 10d). We did not observe an enrichment of eRF1 in the pelleted RNCs (Supplementary Fig. 10d), suggesting that aberrant termination induced by PF846 may prevent eRF1 binding.
Model for PF846 induced translation stalling
The present structures of PF846-stalled CDH1-, PCSK9- and USO1-RNCs provide the first molecular detail for the selective stalling of nascent chains in human ribosomes by a small molecule. PF846 binds a newly identified small molecule-binding pocket in the exit tunnel of the human ribosome and traps select RNCs predominantly in the rotated state (Fig. 6). The ability of PF846 to stall highly specific nascent chain sequences seems to require a distributed set of interactions between the nascent chain and the exit tunnel wall (Fig. 3a, Supplementary Fig. 8d-e, Fig. 5f, Supplementary Fig. 9). PF846-dependent stalling may also be favored on nascent chain sequences that are not translated efficiently, as suggested by the fact that certain amino acid changes in stalled sequences can actually enhance rather that repress translation in a PF846-dependent manner (Fig. 5e). Additionally, we find that the wild-type and RCSK-containing CDH1 NCs are enriched in pelleted ribosomes even in the absence of PF846 (Fig. 5d, Supplementary Fig. 10d), further supporting the notion that these sequences may inherently pause in the ribosome. Finally, the fact that PF846-induced stalling occurs across multiple adjacent codons 10 (Supplementary Fig. 6) also supports a model that these compounds target sequences that are not translated efficiently.
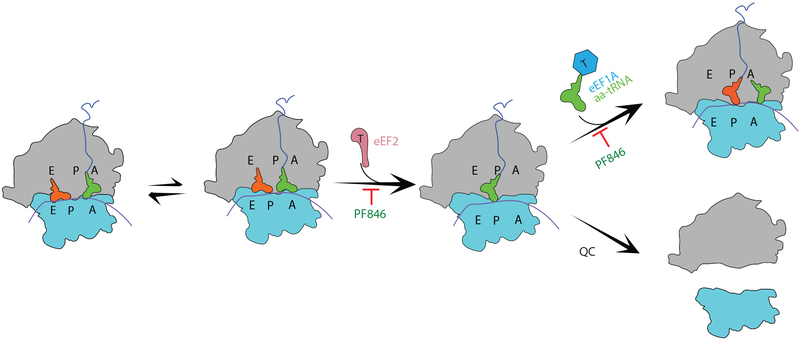
Fig. 6 ∣. Model of the mechanism of PF846-induced stalling.
PF846 binding to the exit tunnel stalls the elongation of the nascent chains in the rotated state, with either A/A NC-tRNA or A/P NC-tRNA. Poor positioning of the 3’-CCA end prevents eEF2-GTP from catalyzing the translocation of hybrid A/P and P/E tRNAs into the P/P tRNA and E/E tRNA sites. PF846 can also stabilize the resulting non-rotated ribosome with an empty A site that may signal quality control (QC) pathways.
DISCUSSION
The specific sequence determinants required for PF846-induced stalling of translation elongation extend roughly one-third of the way through the exit tunnel, beginning at the PTC and extending past the binding pocket for PF846 (Supplementary Fig. 8). Notably, sequences close to the PTC exert a more dominant effect on stalling potency, as revealed from the mRNA libraries (Fig. 5, Supplementary Fig. 10). However, sequences close to the PTC are not sufficient for stalling, as revealed in the Asn scans (Supplementary Fig. 9), and in experiments with the HYHS sequence embedded in the PCSK9 context (Fig. 5f). In the case of endogenous PCSK9, the NC likely needs to span the length of the exit tunnel, as N-terminal truncations of PCSK9 reporters prevented PF846-dependent stalling 10. The distributed nature of PF846-induced stalling is reminiscent of those observed in naturally-occurring stalling sequences in bacteria 19,27,28 and in eukaryotes 29. By contrast, the more general stalling induced by antibiotics bound to the bacterial ribosome exit tunnel may only require localized rearrangements between the drug binding site and the peptidyl transferase center, as observed for macrolide antibiotics 30,31.
Redirection of the nascent chain by PF846 seems to prevent stable base pairing of the NC-tRNA 3’-CCA end with the ribosomal P-loop, likely making these RNCs poor substrates for the GTPase eEF2 responsible for mRNA and tRNA translocation. Consistent with this model for stalling, mutations in the 3’-CCA end of A/P tRNA or P-loop of the bacterial ribosome that disrupt base pairing slow or prevent translocation 32. Furthermore, the SecM stalling peptide in bacteria is also capable of trapping the ribosome in the rotated state with hybrid A/P NC-tRNA, in which the 3’-CCA end of the tRNA is not properly base paired with the P-loop 27. Proper 3’-CCA end positioning has also been shown to be important for the peptidyl transferase reaction. Peptide bond formation is disrupted by certain amino acid sequences such as proline stretches that prevent pairing of the 3’-CCA end with the P-loop in ribosomal RNA. These sequences require dedicated translation elongation factors, EF-P in bacteria and eIF5A in eukaryotes, to rescue stalled ribosome by stabilizing the CCA end of the P-site tRNA in the conformation that favors peptide bond formation 33. Notably, although PF846 does not stall elongating ribosomes on NCs with proline sequences enriched near the PTC (Fig. 5a, inset), PF846 blocks termination when proline-rich sequences occur immediately adjacent to a stop codon positioned at the C-terminus of the CDH1 stalling sequence (Fig. 5c, inset). The broader sequence determinants for PF846-induced inhibition of termination remain to be elucidated, but may be distributed over a longer stretch of the NC, as seen for the hCMV stalling peptide, which ends in the sequence IPP but requires specific sequences in the NC that are span at least one-third the length of the exit tunnel 20,34. The mechanism of PF846-induced inhibition of translation termination differs from that of the hCMV peptide, which traps termination factor eRF1 on the ribosome in an inactive conformation 20,34. By contrast, the PF846-induced block of termination seems to have no observable effect on eRF1 binding (Supplementary Fig. 10d). This may be due to the PF846-stalled termination complex occupying the rotated state, thereby preventing eRF1 from accessing the tRNA A site on the small subunit, and/or preventing the 3’-CCA end of P-site tRNA from properly docking in the PTC 35.
Stalling of RNCs in the rotated state may serve as a critical determinant of PF846 selectivity by evading cellular quality control pathways that recognize aberrantly stalled ribosomes and initiate recycling 36. For example, Pelota (PELO, Dom34 in yeast) and its cofactors are involved in ribosome-associated quality control pathways that recognize stalled ribosome with an empty A site, to which Pelota binds and promotes ribosome subunit dissociation 36. Consistent with the hypothesis that quality control pathways resolve PF846-stalled RNCs, ribosome profiling experiments did not reveal a buildup of stalled RNCs in cells treated with PF846 10. Furthermore, PELO and HBS1L are genetically linked to cellular toxicity of PF846-related compounds administered in high doses 12. The experimental methods presented here provide a means for probing the sequence specificity of PF846-like compounds, and should enable the discovery of other compounds that target the translating human ribosome to inhibit translation elongation, termination, or even enhance translation for therapeutic effect.
Methods
DNA constructs and in vitro transcription
The DNA fragments needed to assemble the constructs encoding the affinity tagged nascent chains (EMCV IRES, CDH1 sequence encoding amino acids 586–750 and PCSK9 sequence encoding amino acids 1–35; Fig. 1a, Supplementary Table 3) were amplified from previous plasmids encoding full length PCSK9 and CDH1 10. For the CDH1 nascent chain used for structural studies, we fused the EMCV IRES, affinity tags, CDH1 sequence encoding amino acids 586–750 and a NanoLuc luciferase reporter gene together by overlap PCR (Supplementary Table 3). For the PCSK9 nascent chain, CDH1 amino acids 716–750 were replaced with PCSK9 amino acids 1–35, leaving the CDH1-V domain intact (Supplementary Table 3). The USO1 construct was made by replacing the sequence encoding CDH1 amino acids 709–750 with the USO1 nascent chain sequence spanning that found previously to cause PF846-induced stalling (Supplementary Table 3)10. The NanoLuc control without stalling sequence was obtained by fusing the EMCV IRES and the NanoLuc open reading frame using overlap PCR (Supplementary Table 3). All PCR products were purified via spin columns (Qiagen) before their use in in vitro transcription reactions.
In vitro transcription reactions were performed using PCR products generated with primers encoding a flanking T7 RNA polymerase promoter and a poly-A tail. Reactions were set up with 20 mM Tris-HCl pH 7.5, 35 mM MgCl2, 2 mM spermidine, 10 mM DTT, 1 U/mL pyrophosphatase (Sigma), 7.5 mM each NTP, 0.2 U/L SUPERaseIn RNase Inhibitor (ThermoFisher), 0.1 mg/mL T7 RNA polymerase and 40 ng/μL DNA. After 3 hours incubation at 37 °C, 0.1 U/μL DNase I (Promega) was added to the reactions, and incubated at 37 °C for 30 min to remove the template DNA. RNA was precipitated for 2–3 hours at −20 °C after adding 1/2 volume of 7.5 M LiCl/50 mM EDTA, and the resulting pellet was washed with cold 70% ethanol and dissolved with RNase free water. The mRNA was further purified by Zymo RNA Clean and Concentrator (Zymo research) before use in in vitro translation reactions.
In vitro translation reactions
The HeLa cell extract was made as described previously 10. Briefly, a frozen HeLa cell pellet was thawed and suspended with an equal volume of lysis buffer (20 mM HEPES pH 7.5, 10 mM potassium acetate, 1.8 mM magnesium acetate, and 1 mM DTT). After incubation on ice for 20 min, the cells were lysed with a Dounce homogenizer 150 times, followed by centrifugation twice at 1,200 x g for 5 min. The supernatant after centrifugation was aliquoted to avoid freeze-thaw cycles, and flash-frozen in liquid nitrogen. For a 10 μL reaction, 5 μL HeLa cell extract was used in a buffer containing final concentrations of 20 mM Hepes pH 7.4, 120 mM KOAc, 2.5 mM Mg(OAc)2, 1 mM ATP/GTP, 2 mM creatine phosphate (Roche), 10 ng/μL creatine kinase (Roche), 0.21 mM spermidine, 0.6 mM putrescine, 2 mM TCEP (tris(2-carboxyethyl)phosphine), 10 μM amino acids mixture (Promega), 1 U/μL murine RNase inhibitor (NEB), 200 ng mRNA, and 0.1 mM PF846 in 1% DMSO, or 1% DMSO as control. Translation reactions were incubated for 23 min at 30 °C, after which luciferase activity was monitored with a Microplate Luminometer (Veritas). To account for batch-to-batch variability, all assays presented in a given figure were carried out using the same in vitro translation extract and same preparation of mRNAs. Although the IC50 value of PF846-mediated inhibition was consistent across batches, we did observe some variability in the absolute extent of inhibition at high PF846 concentrations. Before preparation of PF846-dependent stalled RNCs, all chimeric mRNAs were tested in in vitro translation reactions for PF846-induced stalling, using reactions plus or minus 50 μM PF846.
Purification of stalled RNCs
1.5 mL in vitro translation reactions with 0.2 mM PF846 were incubated at 30 °C for 23 min and then centrifuged at 11,400 r.p.m. for 5 min. The supernatant was incubated with 50 μL (packed volume) of anti-FLAG M2 agarose beads (Sigma) for 30 min at room temperature with gentle mixing. To avoid the binding of nonspecific ribosomes and other proteins, the beads were washed at room temperature three times with 500 μL RNC buffer (20 mM Hepes pH 7.4, 300 mM potassium acetate, 5 mM magnesium acetate, 1 mM DTT, 0.2 mM PF846), then three times with 500 μL RNC buffer plus 0.1% TritonX-100, followed by three times with 300 μL RNC buffer plus 0.5% TritonX-100, and finally washed twice with 300 μL RNC buffer. The PF846 stalled RNCs, bound to the FLAG beads by the N-terminal 3xFLAG tag, were eluted twice at room temperature for 20 min each time, with 30 μL 0.2 mg/mL 3X-FLAG peptide (Sigma) in RNC buffer. The eluted fractions were combined and loaded onto a 50% sucrose cushion prepared with cushion buffer (25 mM HEPES-KOH pH 7.5, 120 mM KOAc and 2.5 mM Mg(OAc)2, 1 M sucrose, 1 mM DTT, 0.2 mM PF846), and centrifuged for 1 hour in an MLA rotor (Beckman Coulter) with 100,000 r.p.m. (~603,000 g) at 4 °C. The pellet was suspended in ice-cold RNC buffer and was immediately used for cryo-EM grid preparation. The concentration of purified RNC was determined using a NanoDrop Microvolume Spectrophotometer and calculated using 1 A260 unit corresponding to 20 pmol of ribosome.
RNaseA digestion and Western blot
RNCs released from the anti-FLAG beads and pelleted through the sucrose cushion were resuspended in 20 μL to an A260 O.D. of around 1 and then incubated with or without 100 μg/mL DNase free Rnase A (ThermoFisher) and 50 mM EDTA at 37 °C for 30 min followed by Western blot of the FLAG-tagged peptides. For the Western blots, Monoclonal ANTI-FLAG® M2-Peroxidase (HRP) antibody (SIGMA, cat. number: A8592) at 1:10,000 dilution was used.
Cryo-sample preparation and Data collection
Approximately 3 μL of freshly made RNC at a concentration of 40 to 60 nM were incubated for 1 min on plasma-cleaned 300-mesh holey carbon grids (C-flat R2/2), on which a home-made continuous carbon film was pre-deposited. Grids were blotted for 4 seconds with 100% humidity at 4 °C and plunge-frozen in liquid ethane using an FEI Vitrobot. Automated data collection was performed on a Titan Krios electron microscope (FEI) equipped with a K2 Summit direct detector and GIF Quantum filter (Gatan) at 300 kV (Tables 1-4). The total exposure time was 9 seconds, with a total dose of 50 electrons per Å2 (frame dose 1.3 electrons per Å2) 39.
Data Processing
Frames were aligned using MotionCor2 with FtBin 2 and dose weighting 40. The final two frames were not used for image alignment to avoid excessive beam damage. We used Gautomatch (http://www.mrc-lmb.cam.ac.uk/kzhang/) for automatic particle picking. Power spectra, defocus values and astigmatism were determined with Gctf software with per-particle CTF estimation 41. All the subsequent data processing was performed in RELION 2.0 42 and RELION 3.0 43 unless specifically noted. To speed up computing, we binned the particles by 8 times during particle extraction. Reference free 2D classification was performed to remove particles with ice or other contaminants, followed by 3D classification to select RNC particles. The initial 3D classification roughly separated the ribosome particles from the non-ribosome particles without yielding structurally distinct classes. All of the final reconstructions used dose-weighted images.
For the CDH1-RNC data set, we divided the whole data set into three subsets and carried out particle sorting as shown in Supplementary Fig. 2a. After initial 3D classification and refinement, an additional round of 3D classification was performed with a local angular search that separated CDH1-RNCs in rotated states from a class with apo-80S ribosomes or ribosomes with weak density for tRNA (Supplementary Fig. 2a). A total number of 84,437 particles representing the CDH1-RNC in the rotated state were combined and subjected to 3D refinement, which generate a map of 4.0 Å without the use of a mask. Signal subtraction was conducted to improve the density of the nascent peptide chain 44. Starting with the aligned 84,437 particles, a region with CDH1 nascent chain, PF846 and A/P NC-tRNA was extracted and classified by applying a soft mask generated in RELION. Focused 3D classification separated the rotated state into two major classes, one with A/P NC-tRNA and P/E tRNA (84% of particles), and the other with A/A NC-tRNA and P/E tRNA (15% of the particles). The more-abundant class with A/P NC-tRNA had good electron density for the CDH1 nascent chain, which was used for model building (Supplementary Fig. 2a). The overall resolutions of the CDH1-RNC’s with A/P NC-tRNA and A/A NC-tRNA were 3.0 Å and 3.9 Å, respectively, using the gold-standard Fourier shell correlation (FSC=0.143) criterion (Supplementary Fig. 2b, Table 1).
To identify other CDH1-RNC classes, we combined the remaining classes after sorting the RNC with hybrid tRNAs (Supplementary Fig. 2a) for an additional 3D refinement and classification with a local angular search, from which three major classes were observed: from left to right, 80S ribosome without tRNA bound, CDH1-RNC with P/P tRNA, and the large (60S) subunit. Another round of signal subtraction for the CDH1-RNC with P/P tRNA was conducted based on the P/P tRNA density, resulting in ~61% particles which were chosen for the final reconstruction of the non-rotated state RNC.
Two data sets for the PCSK9-RNC were collected on the same Titan Krios microscope within a week of each other. After overall refinement, an additional round of 3D classification was performed with a local angular research, which separated RNCs in the rotated state (Supplementary Fig. 3a). We combined a total of 53,247 particles with hybrid state tRNAs from both data sets for another round of 3D auto refinement. The post-processing step implemented in RELION 3.0 was used to generate the final maps using the FSC=0.143 criterion indicating an average resolution of 3.0 Å (Supplementary Fig. 3b, Table 2) 45,46. Signal subtraction for PCSK9-RNC with either hybrid tRNAs or P/P tRNA was conducted in the same way as described for the CDH1-RNC complexes 44 (Supplementary Fig. 3). Data for the USO1-RNC were collected in a single session and processed similarly to the PCSK-RNC sample, except signal subtraction yielded only a single tRNA population, with A/P NC-tRNA (Supplementary Fig. 4, Table 4).
For the data set of stalled PCSK9-RNC prepared using a short time interval, the selected particles after RELION 2D classification were processed using CryoSPARC 47 for further 3D classification and homogeneous refinement, which generated two maps representing different states of the ribosome at an average resolution of 4.6 Å and 4.8 Å, respectively, according to the “gold standard” FSC=0.143 (Supplementary Fig. 3d, e, Table 3) 45,48.
Local resolution estimation was carried out using the program Resmap 49.
Model Building and Refinement
Initial rigid body fitting of the human 40S ribosomal subunit structure (PDB: 6ek0) 50 and 60S subunit structure (PDB: 5aj0) 51 was done for all the rotated RNCs (CHD1-RNCs and PCSK9-RNCs with either A/A NC-tRNA and A/P NC-tRNA, and USO1-RNCs with A/P NC-tRNA), using UCSF Chimera 52. We then docked the ribosomal proteins and RNAs of the 40S and 60S subunits separately into the EM map, with parts of the model manually adjusted to the EM map using COOT 53. For rotated-state RNCs, helix H69 in 28S rRNA was modeled based on the rotated state structure in PDB entry 4u4r 7, followed by manual rebuilding in COOT. All the tRNAs (A/A, A/P, P/E tRNA) for the RNCs in the rotated state were docked into the EM maps using reported tRNA structures (PDB: 3j7r) 54, followed by manual rebuilding in COOT 53. The anticodon nucleotides from 34 to 36 in the tRNA were modeled as one sequence (CCG). RNCs with P/P site tRNA were initially modeled using rigid-body docking of the human ribosome structure (PDB: 5aj0) 51 and then corrected based on the EM density in COOT. The nascent chains for the CDH1-RNC and PCSK9-RNC were manually built into the density in one sequence register (CDH1_NC sequence: CRKAQPVEAGLQIPAILGILGGILALLILILLLLLF and PCSK9_NC sequence: WWPLPLLLLLLLLLGPAGARAQED) in COOT. The USO1-RNC NC was modeled as poly-alanine, due to the lack of recognizable density features for side chains. All RNC structures were refined in PHENIX using phenix.real_space_refine with secondary structure restraints imposed 55. Model refinement and validation statistics are provided in Tables 1-4.
To model the interaction of E. coli 23S rRNA and PF846, the high-resolution model (PDB: 4ybb) 56 was docked into the map by rigid-body docking. For the analysis of the pairing between the A/P-tRNA 3’-CCA end and 28S rRNA, we docked the high resolution tRNA (PDB: 1vy4) 37 as a rigid body, using nucleotides in the ribosomal P-loop as reference. Subsequent modeling of C75 used manual rebuilding in Coot followed by real-space refinement, enforcing minimal changes to the positions of C74 and A76.
Modeling of codon-anticodon base pairs
Correlations between modeled nucleotides and observed cryo-EM density at 3.0 Å resolution are dominated by the phosphate-ribose backbone and the common position of the nucleobase in pyrimidines and five-membered ring in purines. Thus, it is difficult to distinguish purines from pyrimidines at this resolution using only real-space correlations. We therefore used helix h24 in human 18S rRNA (nts 1037–1078) to assess the ability of real-space difference density maps to distinguish purines and pyrimidines in base pairs, and guanosines from adenosines in base pairs, in ~3.0 Å resolution cryo-EM maps. We generated map fragments from the CDH1-RNC cryo-EM map with A/P NC-tRNA and made mutations in nucleotides using COOT 53, followed by real-space refinement of models in Phenix (phenix.real_space_refine) 55, with secondary structure restraints generated by phenix.secondary_structure_restraints and verified manually. We then generated real-space difference maps in Chimera 52, as follows. We set the minimum threshold of the observed cryo-EM map to zero (vop threshold minimum 0.), and used the model coordinates to generate a calculated map at 3.0 Å resolution (molmap). Then, we generated (observed – calculated) difference maps by minimizing the root-mean-square of the differences (vop subtract minRMS true). From these maps, we found that negative difference density was diagnostic for positions that are incorrectly modeled as purine, due to negative density on the N2, C2, N1, C6, and O6/N6 positions of the modeled purine ring. Notably, we also observed positive difference density in these maps consistent with the position of a purine incorrectly modeled as a pyrimidine, but not reproducibly. This is possibly due to the distribution in density in the map calculated from model coordinates with ADP values grouped at the nucleotide level, currently the only setting available in phenix.real_space_refine 55.
We observed that, in the CDH1-RNC and PCSK9-RNC reconstructions at ~3.0 Å resolution, the cryo-EM density between the nucleobases for base pairs in 18S rRNA was clearly separated, and could be easily assigned to purines versus pyrimidines visually. However, the density between the nucleobases in the tRNA anticodon-mRNA codon base pairs was fused. Furthermore, the density shapes did not clearly align with either a pyrimidine or purine base, as shown in Supplementary Fig. 6.
Quantum mechanical calculations of PF846 conformations
All torsional assessments and geometry optimizations of PF846 were performed by quantum mechanical calculations using TeraChem 57,58 using a DFT-(b3lyp) level of theory and a 6–31gs basis set. Dihedral angle energy profiles from model systems derived from truncation of the small molecule X-ray structure of PF846 were used to create low energy poses for fitting to the density. All poses were geometry optimized before fitting. Supplementary Table 1 shows the calculated energies for the poses in Supplementary Fig. 7a-f. After rigid-body docking of PF846 pose 4, the torsion angles between the chloropyridine and amide linkage, and between the aryl ring and amide linkage were manually adjusted by ~15° each to better fit the map density.
DNA libraries with randomized codons in the CDH1 stalling sequence
DNA libraries encoding the open reading frame containing the CDH1 stalling sequence in Fig. 1b, with stretches of 4 random codons encoded by NNK sequences (N=any nucleotide, K=G or T) to introduce random mutations in different sites of the CDH1 nascent chain within the ribosome exit tunnel, were generated by PCR amplification. PCR products were purified using spin columns (Qiagen, catalog number: 28506), then treated with Dpn1 nuclease at 37 °C for 1 hour to remove template DNA. A double-stranded DNA fragment encoding a T7 RNA polymerase promoter was then ligated to the 5’ end of the treated DNA, along with a double-stranded DNA encoding a poly-A tail to the 3’ end, using T4 DNA ligase and T4 Polynucleotide Kinase at 16 °C overnight. The ligation reaction was followed by PCR amplification using primers flanking the T7 promoter and poly-A tail to generate the final DNA library. The oligonucleotides used to prepare the DNA libraries are presented in Supplementary Table 3.
Selection of ribosome nascent chain complexes stalled by PF846 using mRNAs encoding random libraries
The mRNAs encoding the NNK libraries were prepared by T7 RNA polymerase transcription of the above NNK DNA libraries, followed by mRNA purification as described above. The mRNA libraries were incubated in a 1.0 mL in vitro translation reaction with 50 μM PF-06446846 at 30 °C for 28 min. Stalled RNCs were then purified using anti-FLAG peptide beads as described above, with the following modifications. Stalled ribosome nascent-chain complexes (RNCs) were bound to the beads in the presence of 1 μM DNA oligonucleotide 5’-TCTCCTCCGAAGAAA-3’ (targeting DNA) and 2 μL RNase H (5 units/μL, NEB) were added to the reaction. The targeting DNA was designed to base pair with the CDH1 mRNA codons protected in the mRNA binding channel of stalled RNCs, but that would be exposed in free mRNAs, or in mRNAs bound to elongating ribosome complexes. RNCs were then eluted from the anti-FLAG peptide beads using the competitor FLAG peptide, and pelleted through a sucrose cushion as described above. After the sucrose cushion pelleting step, the pellet was resuspended in 40 μL RNase H buffer (20 mM Tris 7.5, 150 mM KCl, 2.5 mM MgCl2, 2mM DTT) and treated with 1 μL (5 units) RNase H and 1 μM of the targeting DNA oligonucleotide at 37 °C for 40 min.
Total RNA was then extracted from the purified and RNase H treated RNCs using TRIzol LS (Thermo Fisher Scientific, cat. number: 10296010). First strand cDNAs were synthesized using a primer specific to a nucleotide sequence in the open reading frame of Fig. 1a located 3’ of the stalling sequence, using Superscript II (Invitrogen, cat. number: 18064014), according to the manufacturer’s protocol. The cDNA was then used in a 70 μL PCR reaction with Q5 DNA polymerase with optimized cycles of 10 s at 98 °C, 30 s at 66 °C and 15 s at 72 °C, followed by a two step purification with SPRIselect beads (Beckman Coulter, cat. number: B23317), first by adding 42 μL beads (0.6X amount of the PCR reaction) and then applying the flowthrough to another 56 μL beads (0.8X amount of the PCR reaction). The resulting bound DNA was eluted in 15 μL molecular biology grade water. The DNA libraries were sequenced using an Illumina HiSeq 4000. The oligonucleotides used to prepare the sequencing libraries are presented in Supplementary Table 3.
Analysis of the Illumina sequencing libraries
UMIs were extracted from raw reads using umi_tools 59 and sequences were aligned to an index of all 12-mer DNA sequences using seqkit 60 and bowtie2 61 (parameters --N 0, --no-1mm-upfront, -norc --L 24). No mismatches were allowed in the 6 bp sequences flanking the library bases to ensure each read was a correctly generated library member. Alignments were sorted and indexed with samtools 62 and deduplicated with umi_tools using the ‘unique’ method. Deduplicated reads were tabulated by translated library sequence, replicate, and condition (PF846 treated or untreated) in python, and PF846 sensitivity was analyzed using DESeq2 63. PF846-sensitive or -resistant library members were called with a false discovery rate cutoff of 1%. The complete list of sequences recovered from sequencing the mRNA libraries is included in Supplementary Data Set 2 and sequencing data for those statistically enriched or depleted is included in Supplementary Table 2.
Validation of PF846-dependent stalling sequences and inhibition of termination
To validate the sequences identified in the mRNA selections are bona-fide targets of PF846-dependent stalling, we cloned reporter mRNAs with representative examples into the CDH1-chimeric mRNA construct described above and shown in Fig. 1a. We replaced codons for amino acids L726-L729 in the CDH1 sequence with codons for the following sequences: HYHS and RSCK for sequences stalled by PF846 during elongation, and NPN*, CVT*, DPC*, and NVI* for sequences that lead to PF846-dependent inhibition of termination (*, stop codon). We also used GCV*, which was not enriched in the experiment, as a negative control for PF846-dependent inhibition of translation termination. Codons used for each sequence are listed in Supplementary Table 3. As an additional negative control, we cloned nanoluciferase with an N-terminal 3X-FLAG tag.
To test whether the sequences HYHS and RSCK, and the ones with stop codons, lead to PF846-dependent stalling in the context of the CDH1 sequence, we synthesized mRNAs as described above and subjected them to in vitro translation reactions, also as described above, with the following modification. We carried out the in vitro translation reactions for nascent chains with HYHS and RSCK sequences using different concentration of PF846, in order to determine the IC50 values for PF846 for each sequence, in comparison to that for the WT CDH1 sequence. To assess the amount of NCs that remain associated with ribosomes after the reactions, we also carried out in vitro translation reactions with or without 50 μM PF846, followed by pelleting ribosomes through a sucrose cushion and Western blotting with an anti-FLAG antibody (Monoclonal ANTI-FLAG® M2-Peroxidase (HRP) antibody (SIGMA), cat. number: A8592) and RPS19 antibody (Bethyl Laboratories, cat. number: A304–002A ) as a loading control. We also probed for the presence of eRF1 using Western blotting after the ribosome pelleting step (anti-eRF1 antibody from Cell Signaling Technology, cat. number: 13916). Membrane imaging was carried out using iBright CL1000 (Thermo Fisher Scientific) and images were automatically adjusted using the “auto contrast” function.
To test whether the HYHS sequence can also promote stalling in the context of the PCSK9 sequence, we replaced regions of the PCSK9 sequences with codons for HYHS in different registers, prepared mRNAs and carried out translation reactions as described above, in the presence or absence of 50 μM PF846.
To test whether the SFRD or ATHF sequences near the PTC found to be depleted from the mRNA library affect stalling in the CDH1 context, we replaced codons for amino acids L726-L729 in the CDH1 sequence with codons for SFRD or ATHF, prepared mRNAs and carried out translation reactions as described above, in the absence or presence of different concentrations of PF846 as indicated.
Figure preparation
Figures were generated with UCSF Chimera 52, PyMOL (Schrödinger) and ChemDraw (PerkinElmer), and WebLogo 38.
Data availability
The cryo-EM maps have been deposited with the Electron Microscopy Data Bank under the accession codes EMD-0599 (CDH1-RNC with A/A and P/E tRNA), EMD-0600 (CDH1-RNC with A/P and P/E tRNA), EMD-0601 (CDH1-RNC with P/P tRNA), EMD-0597 (PCSK9-RNC with A/A and P/E tRNA), EMD-0596 (PCSK9-RNC with A/P and P/E tRNA), EMD-0598 (PCSK9-RNC with P/P tRNA), EMD-20134 (PCSK9-RNC in rotated state with sample prepared with a short sample incubation time), EMD-20135 (PCSK9-RNC in non-rotated state with sample prepared with a short sample incubation time) and EMD-0526 (USO1-RNC with A/P and P/E tRNA). Atomic Coordinates have been deposited in the Protein Data Bank with accession codes 6OLF (CDH1-RNC with A/A and P/E tRNA), 6OLE (CDH1-RNC with A/P and P/E tRNA), 6OLG (CDH1-RNC with P/P tRNA), 6OM7 (PCSK9-RNC with A/A and P/E tRNA), 6OM0 (PCSK9-RNC with A/P and P/E tRNA), 6OLZ (PCSK9-RNC with P/P tRNA), and 6OLI (USO1-RNC with A/P and P/E tRNA). Sequencing data is available at the Sequence Read Archive (SRA) under accession (PRJNA533782). Analysis of all reads from the sequencing data for the PTC library and PF library is available in Supplementary Data Set 2. Source data for Figs. 1c, 5b, 5e, 5f, Supplementary Figs. 1a, 9, and 10c are available with in Supplementary Data Set 3. All other data that support the findings of this study are available from the corresponding author upon reasonable request. The uncropped images for the main text are available in Supplementary Data Set 1.
Supplementary Material
Acknowledgments:
We thank Dan Toso (Bay Area Cryo-EM consortium) and Paul Tobias for help with microscope operation and data collection; Vignesh Kasinath, Junjie Liu, Kelly Nguyen, Basil Greber, Fuguo Jiang and Alexander Myasnikov for discussions about EM data processing; Nadège Liaud, Amy Lee, Meihua Tu, Nathanael Lintner, and Nicholas Ingolia for helpful suggestions. This work was funded by the Pfizer Emerging Science Fund, and by the NIH (R01-GM065050).
Footnotes
Conflict of interest: The authors declare competing interests. RGD is employed by Pfizer, Inc. KFM and SL were employed by Pfizer, Inc. The authors have filed a patent application related to this work.
References
- 1.Lin J, Zhou D, Steitz TA, Polikanov YS & Gagnon MG Ribosome-Targeting Antibiotics: Modes of Action, Mechanisms of Resistance, and Implications for Drug Design. Annu. Rev. Biochem 87, 451–478 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rinehart KL Jr et al. Didemnins: antiviral and antitumor depsipeptides from a caribbean tunicate. Science 212, 933–935 (1981). [DOI] [PubMed] [Google Scholar]
- 3.Myasnikov AG et al. Structure-function insights reveal the human ribosome as a cancer target for antibiotics. Nat. Commun 7, 12856 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Almutairi MM et al. Co-produced natural ketolides methymycin and pikromycin inhibit bacterial growth by preventing synthesis of a limited number of proteins. Nucleic Acids Res. 45, 9573–9582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kannan K & Mankin AS Macrolide antibiotics in the ribosome exit tunnel: species-specific binding and action. Ann. N. Y. Acad. Sci 1241, 33–47 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Wilson DN Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol 12, 35–48 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Garreau de Loubresse N et al. Structural basis for the inhibition of the eukaryotic ribosome. Nature 513, 517–522 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Marks J et al. Context-specific inhibition of translation by ribosomal antibiotics targeting the peptidyl transferase center. Proc. Natl. Acad. Sci. U. S. A 113, 12150–12155 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanchard SC, Cooperman BS & Wilson DN Probing translation with small-molecule inhibitors. Chem. Biol 17, 633–645 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lintner NG et al. Selective stalling of human translation through small-molecule engagement of the ribosome nascent chain. PLoS Biol. 15, e2001882 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Londregan AT et al. Small Molecule Proprotein Convertase Subtilisin/Kexin Type 9 (PCSK9) Inhibitors: Hit to Lead Optimization of Systemic Agents. J. Med. Chem 61, 5704–5718 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Liaud N et al. Cellular response to small molecules that selectively stall protein synthesis by the ribosome. PLOS Genetics 15, e1008057 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petersen DN et al. A Small-Molecule Anti-secretagogue of PCSK9 Targets the 80S Ribosome to Inhibit PCSK9 Protein Translation. Cell Chem Biol 23, 1362–1371 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Hirano S, Nose A, Hatta K, Kawakami A & Takeichi M Calcium-dependent cell-cell adhesion molecules (cadherins): subclass specificities and possible involvement of actin bundles. J. Cell Biol 105, 2501–2510 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhushan S et al. alpha-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat. Struct. Mol. Biol 17, 313–317 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Noller HF, Lancaster L, Mohan S & Zhou J Ribosome structural dynamics in translocation: yet another functional role for ribosomal RNA. Q. Rev. Biophys 50, e12 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Cannone JJ et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3, 2 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson DN & Beckmann R The ribosomal tunnel as a functional environment for nascent polypeptide folding and translational stalling. Curr. Opin. Struct. Biol 21, 274–282 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Su T et al. The force-sensing peptide VemP employs extreme compaction and secondary structure formation to induce ribosomal stalling. Elife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matheisl S, Berninghausen O, Becker T & Beckmann R Structure of a human translation termination complex. Nucleic Acids Res. 43, 8615–8626 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voorhees RM & Hegde RS Structures of the scanning and engaged states of the mammalian SRP-ribosome complex. Elife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dever TE & Green R The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb. Perspect. Biol 4, a013706 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wasserman MR, Alejo JL, Altman RB & Blanchard SC Multiperspective smFRET reveals rate-determining late intermediates of ribosomal translocation. Nat. Struct. Mol. Biol 23, 333–341 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belardinelli R et al. Choreography of molecular movements during ribosome progression along mRNA. Nat. Struct. Mol. Biol 23, 342–348 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Altman RB & Blanchard SC Insights into the molecular determinants of EF-G catalyzed translocation. RNA 17, 2189–2200 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li R, Kang G, Hu M & Huang H Ribosome Display: A Potent Display Technology used for Selecting and Evolving Specific Binders with Desired Properties. Mol. Biotechnol (2018). doi: 10.1007/s12033-018-0133-0 [DOI] [PubMed] [Google Scholar]
- 27.Zhang J et al. Mechanisms of ribosome stalling by SecM at multiple elongation steps. Elife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai A, Kornberg G, Johansson M, Chen J & Puglisi JD The dynamics of SecM-induced translational stalling. Cell Rep. 7, 1521–1533 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei J, Wu C & Sachs MS The arginine attenuator peptide interferes with the ribosome peptidyl transferase center. Mol. Cell. Biol 32, 2396–2406 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sothiselvam S et al. Macrolide antibiotics allosterically predispose the ribosome for translation arrest. Proc. Natl. Acad. Sci. U. S. A 111, 9804–9809 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kannan K et al. The general mode of translation inhibition by macrolide antibiotics. Proc. Natl. Acad. Sci. U. S. A 111, 15958–15963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dorner S, Brunelle JL, Sharma D & Green R The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat. Struct. Mol. Biol 13, 234–241 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huter P et al. Structural Basis for Polyproline-Mediated Ribosome Stalling and Rescue by the Translation Elongation Factor EF-P. Mol. Cell 68, 515–527.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Alderete JP, Jarrahian S & Geballe AP Translational effects of mutations and polymorphisms in a repressive upstream open reading frame of the human cytomegalovirus UL4 gene. J. Virol 73, 8330–8337 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown A, Shao S, Murray J, Hegde RS & Ramakrishnan V Structural basis for stop codon recognition in eukaryotes. Nature 524, 493–496 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joazeiro CAP Ribosomal Stalling During Translation: Providing Substrates for Ribosome-Associated Protein Quality Control. Annu. Rev. Cell Dev. Biol 33, 343–368 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Polikanov YS, Steitz TA & Innis CA A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat. Struct. Mol. Biol 21, 787–793 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crooks GE WebLogo: A Sequence Logo Generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grant T & Grigorieff N Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. Elife 4, e06980 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang K Gctf: Real-time CTF determination and correction. J. Struct. Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheres SHW RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zivanov J et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bai X-C, Rajendra E, Yang G, Shi Y & Scheres SH W. Sampling the conformational space of the catalytic subunit of human γ-secretase. Elife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosenthal PB & Henderson R Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol 333, 721–745 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Chen S et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 48.van Heel M & Schatz M Fourier shell correlation threshold criteria. J. Struct. Biol 151, 250–262 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Kucukelbir A, Sigworth FJ & Tagare HD Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Natchiar SK, Myasnikov AG, Kratzat H, Hazemann I & Klaholz BP Visualization of chemical modifications in the human 80S ribosome structure. Nature 551, 472–477 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Behrmann E et al. Structural snapshots of actively translating human ribosomes. Cell 161, 845–857 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voorhees RM, Fernández IS, Scheres SHW & Hegde RS Structure of the mammalian ribosome-Sec61 complex to 3.4 Å resolution. Cell 157, 1632–1643 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Afonine PV et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Noeske J et al. High-resolution structure of the Escherichia coli ribosome. Nat. Struct. Mol. Biol 22, 336–341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ufimtsev IS & Martinez TJ Quantum Chemistry on Graphical Processing Units. 3. Analytical Energy Gradients, Geometry Optimization, and First Principles Molecular Dynamics. J. Chem. Theory Comput 5, 2619–2628 (2009). [DOI] [PubMed] [Google Scholar]
- 58.Titov AV, Ufimtsev IS, Luehr N & Martinez TJ Generating Efficient Quantum Chemistry Codes for Novel Architectures. J. Chem. Theory Comput 9, 213–221 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Smith T, Heger A & Sudbery I UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 27, 491–499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shen W, Le S, Li Y & Hu F SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS One 11, e0163962 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM maps have been deposited with the Electron Microscopy Data Bank under the accession codes EMD-0599 (CDH1-RNC with A/A and P/E tRNA), EMD-0600 (CDH1-RNC with A/P and P/E tRNA), EMD-0601 (CDH1-RNC with P/P tRNA), EMD-0597 (PCSK9-RNC with A/A and P/E tRNA), EMD-0596 (PCSK9-RNC with A/P and P/E tRNA), EMD-0598 (PCSK9-RNC with P/P tRNA), EMD-20134 (PCSK9-RNC in rotated state with sample prepared with a short sample incubation time), EMD-20135 (PCSK9-RNC in non-rotated state with sample prepared with a short sample incubation time) and EMD-0526 (USO1-RNC with A/P and P/E tRNA). Atomic Coordinates have been deposited in the Protein Data Bank with accession codes 6OLF (CDH1-RNC with A/A and P/E tRNA), 6OLE (CDH1-RNC with A/P and P/E tRNA), 6OLG (CDH1-RNC with P/P tRNA), 6OM7 (PCSK9-RNC with A/A and P/E tRNA), 6OM0 (PCSK9-RNC with A/P and P/E tRNA), 6OLZ (PCSK9-RNC with P/P tRNA), and 6OLI (USO1-RNC with A/P and P/E tRNA). Sequencing data is available at the Sequence Read Archive (SRA) under accession (PRJNA533782). Analysis of all reads from the sequencing data for the PTC library and PF library is available in Supplementary Data Set 2. Source data for Figs. 1c, 5b, 5e, 5f, Supplementary Figs. 1a, 9, and 10c are available with in Supplementary Data Set 3. All other data that support the findings of this study are available from the corresponding author upon reasonable request. The uncropped images for the main text are available in Supplementary Data Set 1.