Abstract
BACKGROUND
Habitual alcohol use can be an indicator of alcohol dependence, associated with a wide range of serious health problems.
METHODS
We completed a genomewide association study in 126,936 European-American (EUR) and 17,029 African-American (AFR) subjects in the Veterans Affairs Million Veteran Program (MVP) for a quantitative phenotype based on maximum habitual alcohol consumption (“MaxAlc”).
RESULTS
ADH1B, on chromosome 4, was the lead locus for both populations: for EUR, rs1229984 (p=4.9 × 10−47); for AFR, rs2066702 (p=2.3 × 10−12). In the EUR, we identified three additional genomewide-significant (GWS) MaxAlc loci: on chromosome 17, rs77804065 (p=1.5 × 10−12), at CRHR1 (corticotropin-releasing hormone receptor 1); the protein product of this gene is involved in stress and immune responses; and on chromosomes 8 and 10. EUR and AFR samples were then meta-analyzed; the associated region at CRHR1 increased in significance to 1.02 × 10−13, and we identified two additional genomewide-significant loci, FGF14 (p= 9.86 × 10−9) (chromosome 13), and a locus on chromosome 11. Besides ADH1B, none of the five loci have prior GWS support. Post-GWAS analysis identified genetic correlation to other alcohol-related traits, smoking-related traits, and many others. Replications were observed in UKBiobank data. Genetic correlation between MaxAlc and alcohol dependence was 0.87 (p=4.78 × 10−9). Enrichment for cell types included dopaminergic and GABAergic neurons in midbrain, and pancreatic delta cells.
CONCLUSIONS
The present study supports five novel alcohol use risk loci, with particularly strong statistical support for CRHR1. Additionally, we provide novel insight regarding the biology of harmful alcohol use.
Introduction
The Million Veteran Program (MVP) is a US Department of Veterans Affairs (VA) initiative with a goal of recruiting at least one million VA healthcare beneficiaries, creating a database of genomic and phenotypic information useful for increasing understanding of health and disease (1). The sample is linked both to the VA’s extensive electronic health record (EHR) and to self-report survey information specific to the MVP study. The MVP is particularly valuable for elucidating health problems that are highly prevalent in military veterans, including alcohol use disorder (AUD) and harmful alcohol use.
DSM-IV alcohol dependence (AD), which in DSM-5 is the more severe type of AUD, is moderately heritable; genomewide association studies (GWAS) of AD and habitual alcohol use have been conducted in European (2–8), African (2, 5, 6), and East Asian (6, 9–13) ancestry populations. Most studies of AD diagnosis have been in small samples, but one reported on ~16,000 subjects (2), and the Psychiatric Genomics Consortium has completed a mega-analysis for AD (14). This AD mega-analysis included 14,904 AD cases and 37,944 controls from 28 case-control and family-based studies. Although this study consistently detected AD polygenic architecture, ADH1B risk alleles were the only loci identified, perhaps due to the heterogeneity across the cohorts included (14). Alcohol consumption is the major risk factor for AD and has medical importance per se. For example, alcohol consumption, even in the normal range (“social” drinking) bears a direct relationship to decline in several cognitive measures (15). Studies of large database samples, including the UK Biobank (4), have focused on alcohol consumption and their findings have implications for AD risk. Associations with variants mapped to genes that encode alcohol metabolizing enzymes – generally ADH1B variants in European- and African-ancestry (EUR and AFR) subjects (16, 17) as well as ALDH2*rs671 (18) in Asians – have been observed consistently. Some studies have reported associations at other loci with various alcohol-related traits (2, 4, 8); these reports are comparatively few. One meta-analysis of alcohol drinking (>105,000 EUR individuals) identified associations of daily alcohol intake with KLB, GCKR, and CDH13 (8). The GWAS of alcohol consumption in the UK Biobank sample (4) of >112,000 is the largest to date; this study considered only EUR subjects, and the phenotype was based on average weekly alcohol consumption. Genome-wide significant (GWS) associations were identified at several alcohol dehydrogenase (ADH) loci, in addition to other loci including GCKR, CADM2, and FAM69C.
In the present investigation, we studied the genetic architecture of an alcohol consumption phenotype – maximum habitual (“in a typical month…”) alcohol use, or MaxAlc -- in the MVP sample (19). We used two strategies to increase power for risk variant identification: a large sample size and substantial informativity of the phenotype. We included 143,965 MVP participants, and we used MaxAlc defined as a quantitative phenotype. A different phenotype, “maximum number of drinks consumed in any 24-hour lifetime period” often called MAXDRINKS, has previously been studied (5, 20). The trait definitions differ in that MaxAlc reflects typical habitual (daily) maximum usage, as opposed to the maximum use ever, which might be on a single occasion. Heaviness of habitual alcohol use may be more correlated with risk of AD than MAXDRINKS (21). Accordingly, we expected that our analysis would be informative regarding the mental and physical consequences of excessive alcohol consumption and alcohol dependence.
Methods and Materials
Subject recruitment
Participants were enrollees in the MVP (1) (Table 1). Users of the VHA healthcare system received invitational mailings, encounters with MVP staff while receiving clinical care, or both. Inclusion criteria were ability and willingness to provide informed consent. Research involving MVP in general is approved by the VA Central IRB; the current project was approved by local IRBs in Boston, San Diego, and West Haven.
Table 1.
Demographic characteristics of Million Veteran Program (MVP) European American (EUR) (N=126,936) and African American (AFR) (N=17,029) enrollees with completed Baseline and Lifestyle surveys
| Age (years): | EA (N=126,936) | AA (N=17,029) |
|---|---|---|
| 18–29 | 937 (0.7) | 97 (0.6) |
| 30 – 39 | 2,667 (2.1) | 524 (3.1) |
| 40 – 49 | 6,250 (4.9) | 1,702 (10.0) |
| 50 – 59 | 16,407 (12.9) | 5,207 (30.6) |
| 60 – 69 | 56,805 (44.8) | 6,708 (39.4) |
| 70 – 79 | 28,237 (22.2) | 2,139 (12.6) |
| 80+ | 15,519 (12.2) | 646 (3.8) |
| missing | 114 (0.1) | 6 (0.0) |
| mean (SD) | 66.2 (11.4) | 60.3 (10.6) |
| median | 66 | 61 |
| Sex: | ||
| male | 118,752 (93.6) | 14,981 (88.0) |
| female | 8,070 (6.4) | 2,041 (12.0) |
| missing | 114 (0.1) | 7 (0.0) |
| Ethnicity (self-identified): | ||
| Hispanic | 1,325 (1.0) | 204 (1.2) |
| non-Hispanic | 124,603 (98.2) | 16,603 (97.5) |
| unknown | 894 (0.7) | 216 (1.3) |
| missing | 114 (0.1) | 6 (0.0) |
| Marital status: | ||
| Married | 72873 (57.4) | 6278 (36.9) |
| Divorced | 21294 (16.8) | 3603 (21.2) |
| Civil commitment | 521 (0.4) | 79 (0.5) |
| Never married | 7814 (6.2) | 1812 (10.6) |
| Widowed | 8269 (6.5) | 848 (5.0) |
| Separated | 1713 (1.3) | 941 (5.5) |
| Cohabitating | 3006 (2.4) | 257 (1.5) |
| missing | 11446 (9.0) | 3211 (18.9) |
Two optional surveys were designed to augment data contained in the electronic health record. The MVP Baseline Survey elicits information regarding demographic factors, family pedigree, health status, lifestyle habits, military experience, medical history, family history of specific illnesses, and physical features. The MVP Lifestyle Survey contains questions from validated instruments, in domains selected to provide information on environmental exposures, dietary and other habits, sleep and exercise habits, and sense of well-being. This latter instrument includes the following item: “In a typical month, what is/was the largest number of drinks of alcohol (beer, wine, and/or liquor) you may have had in one day?” The response to this item was used to define the phenotype in the present study, referred to here as MaxAlc. All EUR and AFR subjects who responded to the questionnaire were included. Differences between respondents and non-respondents among MVP participants are shown in Supplementary Table 1.
Phenotype distribution is shown in Supplementary Figure 1.
Genotyping and Microarray
Genotyping was accomplished via a 723,305-SNP Affymetrix Axiom biobank array, customized for the MVP (1, 22). Additional information is provided in Supplementary Methods.
GWAS Analyses
We performed single variant tests using RVTEST(23) software, including the first 10 principal components, age, and sex as covariates in the linear regression association analyses, separately for EUR and AFR. The significance threshold was p=5×10−8.
Post-GWAS analyses
To investigate shared genetic and molecular mechanisms, we tested genetic overlap (i.e., shared risk alleles) of MaxAlc with a wide range of phenotypes. Genetic correlations were calculated using the LD score regression method (https://github.com/bulik/ldsc) (24). LDSC results regarding 232 traits were extracted from the data available at LD Hub v1.4.0 (http://ldsc.broadinstitute.org/ldhub/)(25). Genetic correlations for an additional 1,547 traits were calculated using the GWAS summary association results available at https://sites.google.com/broadinstitute.org/ukbbgwasresults; these GWAS used data from ~337,000 unrelated British individuals from the UK Biobank (26).
To explore further the functional role of the GWS variants identified, we conducted an expression quantitative trait locus (eQTL) analysis using GTEx V7 data (30). FDR correction, MAGMA (28), FUMA (31), and eQTL analyses are described further in Supplementary Methods.
Results
We identified an unusual instance of Hardy-Weinberg disequilibrium (HWD). ADH1B rs1229984, the most consistently associated alcohol risk variant in European populations (16), was initially excluded from analysis because it deviated from Hardy-Weinberg equilibrium expectations (HWEE) (p=1.46e-43). This variant is functional (32), presents very strong allele frequency differences among human populations (33), and has undergone selection in Asian and European populations (17, 34), although there is an open debate about the presence of convergent evolution in Europeans (35). Since ADH1B rs1229984 is the most relevant locus associated with alcohol drinking behaviors that has a very well established causative mechanism (17), we investigated the cause for HWD further to avoid unnecessary exclusion of this variant, which would have highlighted the association of other variants in the same region due to the LD without reflecting the real causal mechanism. This is described in Supplementary Results.
Primary GWAS Analysis
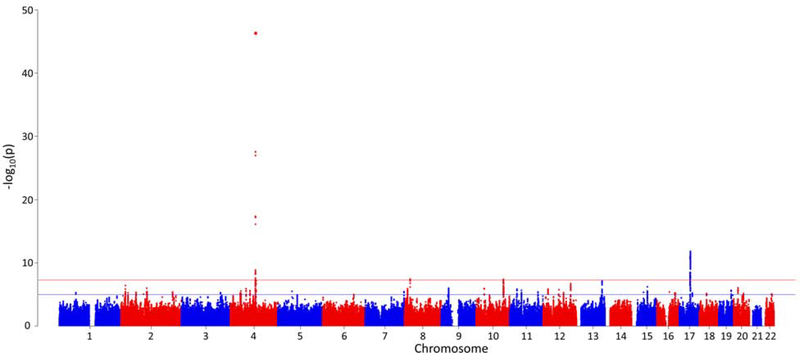
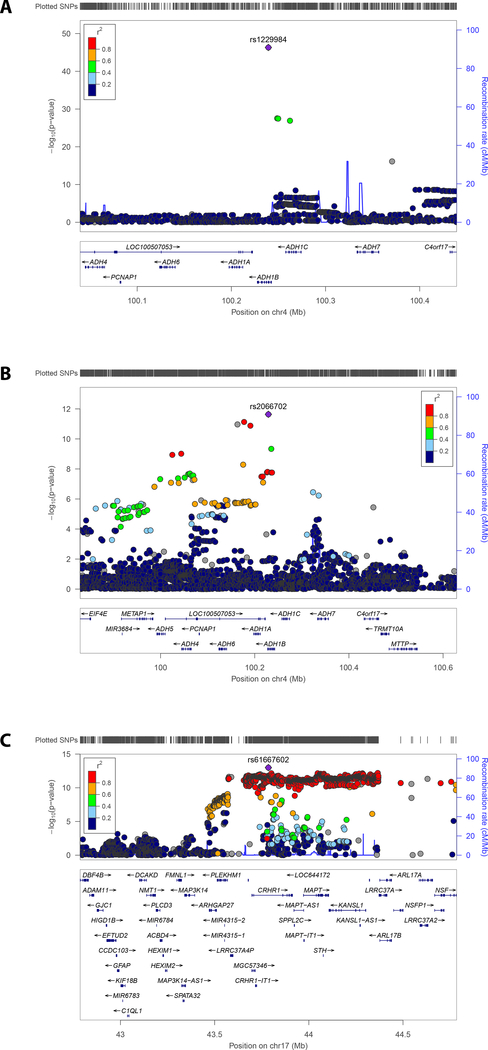
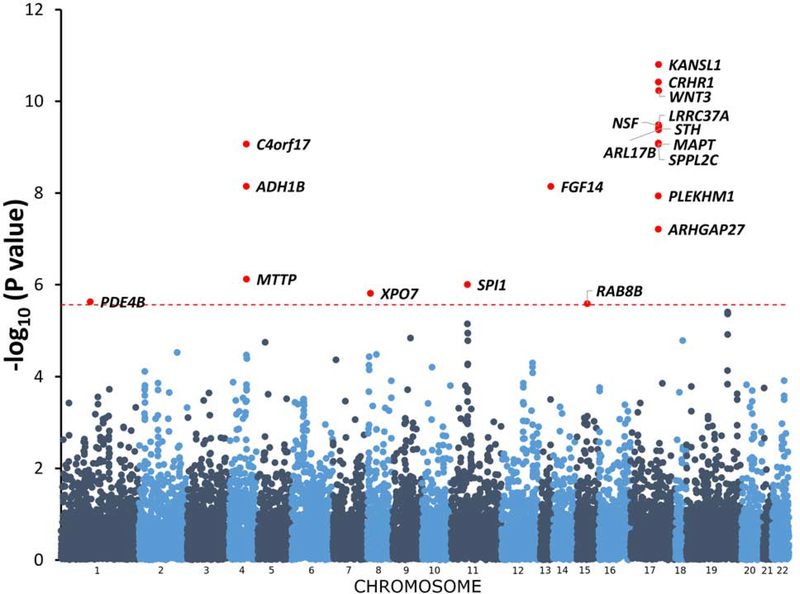
We observed 7.8% SNP-based heritability (p = 1.01×10−40) calculated on the basis of the summary association data in “G1” EUR via LD score regression (LDSC). As with other large-scale GWAS(25), an inflated lambdagc value was observed in the summary association data (λgc=1.16; Supplementary Figure 2). The LDSC intercept was 1.011 (SE = 0.0091), however, demonstrating that this inflation was due to polygenicity and not to population stratification, phenotype distribution, or other confounders. (25). In the smaller AFR sample (n=17,029), no effect of polygenicity was observed in the summary association data (λgc=1.01; Supplementary Figure 3). Four independent GWS regions were identified in “G1” EUR (Figure 1). The lead region was on chromosome 4, lead SNP rs1229984 (p=4.9×10−47; Figure 2a); gene ADH1B, (beta subunit, class I alcohol dehydrogenase). GWS SNPs mapped to numerous loci in the region, so we performed conditional analysis for these loci using GCTA with EUR summary statistics and 1000G data as reference LD. This analysis confirmed that there are only four independent signals, i.e. no associated region reflected more than one independent signal. The other three associated regions map to chromosome 17, lead SNP rs77804065 (p=1.5×10−12; Supplementary Figure 4a), at CRHR1, corticotropin-releasing hormone receptor 1, with the protein product of this gene involved in stress and immune responses (numerous additional GWS SNPs were found in the chromosome 17 region, including variants that map to KANSL1, KAT8 Regulatory NSL Complex Subunit 1); chromosome 8, lead SNP rs7821592 (p=3.6×10−08; Supplementary Figure 4b), closest gene XPO7, exportin 7, the protein product of which mediates nuclear export of proteins; and chromosome 10, lead SNP rs1577857 (p=4.2×10−08; Supplementary Figure 4c), at LOC105378478, which has unknown function (closest gene, RNU6–53P).
Figure 1.
Manhattan plot
Figure 2. Regional Manhattan Plots:
A. Regional Manhattan plot, chromosome 4 ADH genes, EUR
B. Regional Manhattan plot, chromosome 4 ADH genes, AFR
C. Regional Manhattan plot, meta-analysis of EUR and AFR, chromosome 17 (CRHR1) region
The MVP includes mostly male subjects (93.6%). Although sex was included as a covariate, males and females differ in their prevalence of and genetic liability to AUDs (36, 37), so we evaluated whether inclusion of females affected the results substantively by repeating the analysis excluding females. No major differences were observed between GWAS of both-sexes and male-only samples (Supplementary Table 2).
In the AFRs, one GWS region was identified, lead SNP ADH1B*rs2066702 (2.29×10−12; Figure 2b). Conditional analysis, with AFR LD reference, confirmed that this reflects a single peak.
When EUR and AFR results were meta-analyzed (n=143,965 subjects total), we identified two additional GWS loci, uncharacterized LOC105376602 (p= 4.63×10−08) on chromosome 11, and FGF14 (p= 9.86×10−09) on chromosome 13 (Supplementary Figure 5b and 5e). In addition, the associated region at CRHR1 increased in statistical significance to p=1.02×10−13. Comparing EUR results (Supplementary Figure 4a) with EUR-AFR meta-analysis (Figure 2c), we observed different lead variants on chromosome 17, but they both indicated CRHR1 as a credible gene responsible for the association observed. Results are summarized in Table 2 and more extensively in Supplementary Table 3.
Table 2.
Most significant SNPs
| EA | ||||||||||
| uniqID | rsID | chr | pos | P-value | start | end | nSNPs | nGWASSNPs | LeadSNPs | Gene |
| 4:100239319:C:T | rs1229984 | 4 | 100239319 | 4.91E-47 | 100019089 | 100638613 | 111 | 107 | rs1229984 | ADH1B |
| 8:21811530:C:G | rs7821592 | 8 | 21811530 | 3.63E-08 | 21777476 | 21869727 | 43 | 30 | rs7821592 | XPO7 |
| 10:110572259:G:T | rs1577857 | 10 | 110572259 | 4.15E-08 | 110462973 | 110635222 | 116 | 90 | rs1577857 | RNU6–53P |
| 17:43810896:C:T | rs77804065 | 17 | 43810896 | 1.54E-12 | 43463493 | 44865603 | 3414 | 2979 | rs77804065;rs199447 | CRHR1 |
| AA | ||||||||||
| uniqID | rsID | chr | pos | P-value | start | end | nSNPs | nGWASSNPs | LeadSNPs | Gene |
| 4:100229017:A:G | rs2066702 | 4 | 100229017 | 2.29E-12 | 99986965 | 100263535 | 54 | 51 | rs2066702 | ADH1B |
| META | ||||||||||
| uniqID | rsID | chr | pos | P-value | start | end | nSNPs | nGWASSNPs | LeadSNPs | Gene |
| 4:100239319:C:T | rs1229984 | 4 | 100239319 | 1.07E-49 | 100019089 | 100638613 | 110 | 106 | rs1229984 | ADH1B |
| 8:21827162:C:T | rs2291317 | 8 | 21827162 | 2.48E-08 | 21777476 | 21869727 | 43 | 30 | rs2291317 | XPO7 |
| 10:110572259:G:T | rs1577857 | 10 | 110572259 | 4.15E-08 | 110462973 | 110635222 | 116 | 90 | rs1577857 | RNU6–53P |
| 11:28648185:C:T | rs7931459 | 11 | 28648185 | 4.63E-08 | 28591168 | 28704399 | 172 | 84 | rs7931459 | LOC105376602 |
| 13:102868108:C:T | rs1360983 | 13 | 102868108 | 9.86E-09 | 102860527 | 102911712 | 51 | 44 | rs1360983 | FGF14 |
| 17:43785349:C:T | rs61667602 | 17 | 43785349 | 1.02E-13 | 43463493 | 44865603 | 3423 | 2976 | rs61667602;rs1378358 | CRHR1 |
To verify our results in an independent sample, we used summary association data from the AD GWAS conducted by the Psychiatric Genomics Consortium (PGC) (38). Although to date this is the largest AD GWAS, its effective sample size (39) is much smaller than the one used in our analysis (PGC = 31,819; MVP = 143,965) so there is low statistical power to replicate our findings. Nevertheless, considering our six GWS results in trans-ancestry meta-analysis, we observed genome-wide significant replication of the chromosome 4 ADH1B*rs1229984 association (p=2.18×10−11), a nominal replication of chromosome 10 rs1577857 (p=2.44×10−3) and direction replication (i.e., the loci showed the same effect direction in both MaxAlc and AD) for all loci (Supplementary Table 4). We estimate that the probability to observe a direction replication of all six MVP-identified loci in PGC AD GWAS by chance is 1.7% (Supplementary Figure 6). Leveraging the polygenic architecture of the complex traits investigated, in the EUR sample, MaxAlc in MVP showed rg=0.87 with AD in the PGC cohort (p=4.78×10−9) by LDSC. For additional replication, we investigated UK Biobank data regarding nine traits related to alcohol use (Supplementary Table 5). To identify the phenotypes most closely related to MaxAlc, we performed a genetic-correlation analysis and observed the strongest correlation with “Amount of alcohol drunk on a typical drinking day” (rg=0.81, p=5.83×10−40). Significant correlations were also observed with the other traits, including “Frequency of consuming six or more units of alcohol” (rg=0.70, p=2.72×10−30), “Ever been injured or injured someone else through drinking alcohol” (rg=0.84, p=8.56×10−5), and “Ever had known person concerned about, or recommend reduction of, alcohol consumption” (rg=0.64, p=3.79×10−14). Considering the most strongly genetically correlated alcohol-use trait (i.e., “Amount of alcohol drunk on a typical drinking day”), we observed replications (Supplementary Table 4) for chromosome 4 rs1229984 (p=3.77×10−32), chromosome 10 rs1577857 (p=0.027), and chromosome 17 rs77804065 (p=2.67×10−6) and rs61667602 (p=1.25×0−6).
We evaluated possible association of genes identified as associated in previous investigations of alcohol consumption phenotypes: GCKR, CADM2, FAM69C, KLB, and CDH13. No GWS results were observed, but suggestive results were observed in EUR at two of these loci, GCKR (min p=5.78×10−6) and KLB (min p=5.54×10−6), and nominally significant signals were observed in the remaining genomic regions (Regional Manhattan Plots for all five of these are in Supplementary Figure 7). This could be attributable to the polygenic architecture of complex traits, where loci have very small effect sizes, and a much larger sample size will be needed to replicate these loci at a genome-wide significance level; or to the difference between MaxAlc and AD, which has a high correlation with PGC AD (see above) and the consumption phenotypes wherein these other markers were identified.
Phenome-wide Genetic Correlations
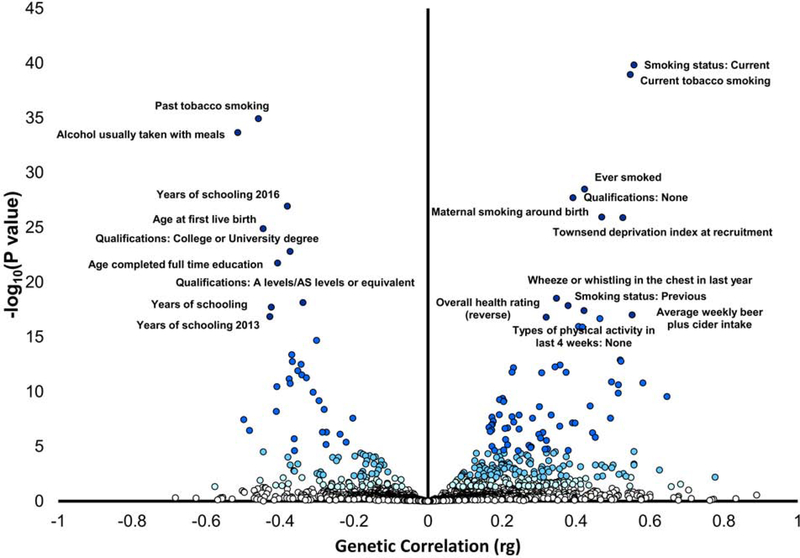
LDSC revealed significant genetic correlations (FDR q<0.05) with 238 of nearly 1800 traits (Figure 3; Supplementary Table 6). The most significant observed correlations (Supplementary Table 3 shows all results at FDR q <0.05) were with respect to smoking and alcohol-drinking traits, where the top correlations were with current smoking status (positive correlation, rg= 0.55, p=1.30×10−39), the degree of past smoking (past tobacco smoking; negative correlation, rg= −.46, p=5.49×10−36) and “healthy” alcohol-drinking behaviors (e.g., alcohol usually taken with meals; negative correlation, rg= −.50, p= 5.44×10−34). Among the other highly significant correlations, several were related to level of education (e.g. years of schooling, rg= −.37 p=1.53×10−25) and socio-economic status (Townsend deprivation index, rg= 0.53, p=3.69×10−27). Numerous correlations were also found with measures of physical activity (e.g., no physical activity in the last four weeks, rg=0.41, p=3.06×10−17). Other noteworthy correlations included mood swings (rg=0.20, p=1.05×10−5) and risk taking (rg=.20, p=2.74×10−5). Considering psychiatric traits, we observed significant genetic correlations with depressive symptoms (rg = 0.22, p=4×10−4), schizophrenia (rg = 0.13, p=0×10−4), and attention deficit hyperactivity disorder (rg = 0.32, p = 0.023).
Figure 3.
Phenome-wide genetic-correlation analysis. Blue shades corresponds to significance strength, from white, non-significant (p > 0.05), to very light blue (p < 0.05), light blue (FDR q < 0.05), to blue (Bonferroni correction p < 2.81 × 10−40), and dark blue (top-20 results). Phenotype labels are included for the top 20 results.
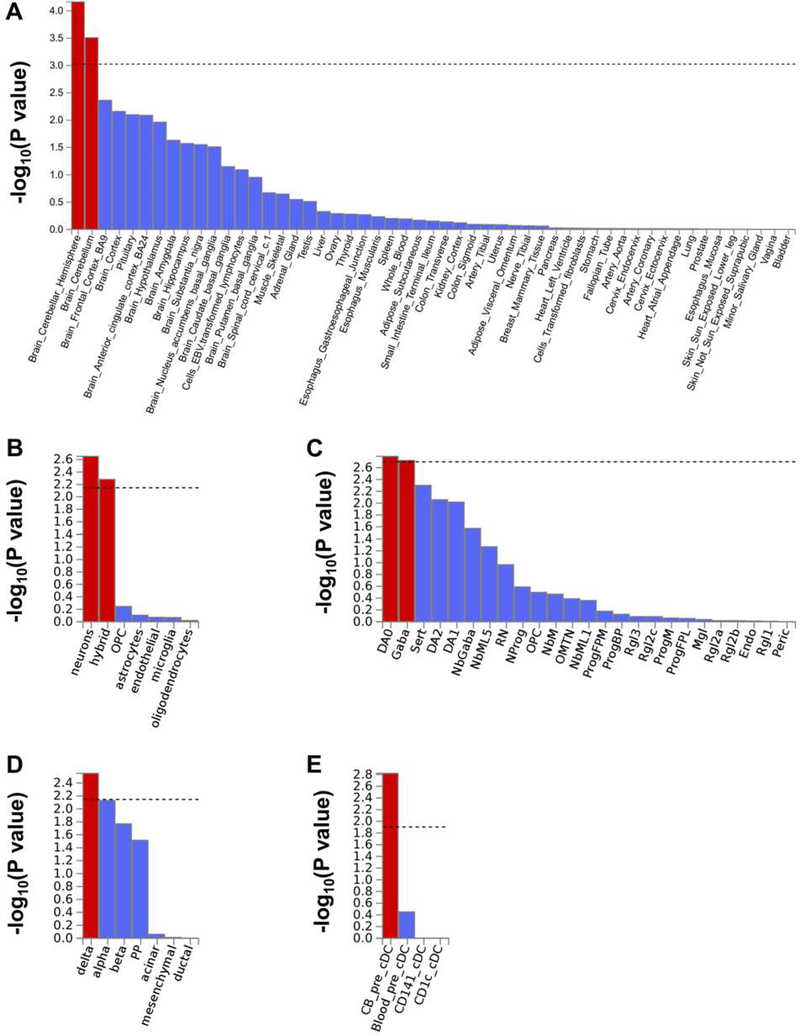
Gene-based association and Tissue and Cell-type Enrichment analysis
Gene-based association analysis and tissue and cell type enrichment results are shown in Figures 1 and 5 and described in Supplementary Methods.
Figure 5.
A. Statistical significance of the enrichments for tissue-specific gene expression. Detailed results are reported in Supplementary Table 4.
B. Statistical significances for cell types in human cortex from adult samples. “Hybrid” refers to a mixture of oligodendrocyte progenitor cells (OPC), oligodendrocytes, and neurons. Detailed results are reported in Supplementary Table 5.
C. Statistical significances for cell types in human midbrain. Detailed results and acronym legends are reported in Supplementary Table 6.
D. Statistical significances for cell types in human pancreas. Detailed results are reported in Supplementary Table 7.
E. Statistical significances for cell types in conventional dendritic cells (cDC). Detailed results are reported in Supplementary Table 8.
eQTL analysis
After applying a FDR 5% correction for the variants, genes, and tissues tested, we observed 212 significant eQTLs out of 2,855 tests conducted with respect to the GWS loci observed in the trans-ancestry meta-analysis. Considering the top CNS tissue for each eQTL surviving multiple testing correction (Table 3), we observed 37 significant results. Thirty-four relate to rs61667602 (chromosome 17), associated with the expression of multiple genes, where the strongest significance was mostly observed in the cerebellum transcriptomic profile (22/34). Additionally, we identified significant eQTLs with respect to rs1360983 on chromosome 13 (FGF14-AS2, top CNS tissue: spinal cord) and rs2291317 on chromosome 8 (BIN3, top CNS tissue: nucleus accumbens; FAM160B2, top CNS tissue: substantia nigra). Consistent with the strong linkage disequilibrium with the loci identified in the trans-ancestry analysis, similar eQTL results were observed with respect to the variants identified in the EUR analysis.
Table 3.
Significant eQTLs observed with respect to the GWS variants identified in trans-ancestry meta-analysis considering 13 CNS tissues. TSS: transcription start site.
| rsID | Gene | TSS distance | slope | se | P value | FDR Q | Top CNS Tissue |
|---|---|---|---|---|---|---|---|
| rs1360983 | FGF14-AS2 | −178872 | −0.361 | 0.078 | 2.00E-05 | 1.72E-04 | Spinal cord (cervical c-1) |
| rs61667602 | LRRC37A2 | −803528 | 1.169 | 0.063 | 2.29E-36 | 4.69E-34 | Cerebellum |
| LRRC37A4P | 157648 | −1.096 | 0.061 | 4.20E-34 | 7.89E-32 | Nucleus accumbens (basal ganglia) | |
| AC005829.1 | −559054 | 1.132 | 0.068 | 1.67E-32 | 2.18E-30 | Cerebellum | |
| KANSL1-AS1 | −485593 | 1.202 | 0.072 | 3.39E-32 | 4.05E-30 | Cortex | |
| AC005829.2 | −552623 | 1.219 | 0.076 | 2.65E-31 | 2.45E-29 | Cerebellum | |
| PLEKHM1 | 217234 | −0.951 | 0.062 | 7.95E-30 | 5.43E-28 | Cerebellum | |
| ARL17A | −871739 | 1.130 | 0.076 | 5.96E-29 | 3.80E-27 | Cerebellum | |
| MAPK8IP1P2 | 105643 | 1.093 | 0.083 | 5.77E-25 | 2.30E-23 | Cerebellum | |
| MAPK8IP1P1 | −535623 | 1.157 | 0.085 | 1.25E-24 | 4.78E-23 | Cerebellar Hemisphere | |
| DND1P1 | 122112 | 1.166 | 0.096 | 3.14E-22 | 9.89E-21 | Cortex | |
| LINC02210 | 87655 | 0.690 | 0.059 | 3.02E-21 | 8.66E-20 | Cortex | |
| SPPL2C | −136907 | 0.734 | 0.067 | 8.67E-20 | 2.18E-18 | Cerebellum | |
| LRRC37A | −584750 | 1.004 | 0.093 | 3.42E-19 | 7.91E-18 | Cerebellum | |
| AC091132.1 | 204723 | −0.922 | 0.098 | 4.67E-16 | 7.83E-15 | Cerebellum | |
| FAM215B | −854812 | 0.844 | 0.090 | 6.89E-16 | 1.14E-14 | Cerebellum | |
| FMNL1 | 485759 | −0.672 | 0.074 | 2.54E-15 | 3.87E-14 | Cerebellum | |
| AC091132.3 | 176406 | 0.757 | 0.120 | 6.44E-09 | 6.76E-08 | Cerebellar Hemisphere | |
| ARHGAP27 | 273562 | 0.459 | 0.078 | 4.94E-08 | 4.93E-07 | Nucleus accumbens (basal ganglia) | |
| AC091132.2 | 255139 | −0.573 | 0.101 | 1.03E-07 | 1.00E-06 | Cerebellum | |
| AC008105.3 | 486194 | −0.469 | 0.086 | 3.15E-07 | 2.99E-06 | Cerebellum | |
| MAPT-AS1 | −187617 | 0.508 | 0.096 | 6.23E-07 | 5.72E-06 | Cerebellum | |
| MAPT | −186399 | −0.316 | 0.065 | 3.62E-06 | 3.22E-05 | Cerebellum | |
| CRHR1 | 86082 | −0.399 | 0.098 | 1.01E-04 | 8.31E-04 | Putamen (basal ganglia) | |
| RPS26P8 | 99440 | 0.698 | 0.169 | 1.11E-04 | 9.12E-04 | Spinal cord (cervical c-1) | |
| KANSL1 | −517384 | 0.355 | 0.090 | 1.42E-04 | 0.001 | Cerebellum | |
| NMT1 | 656319 | −0.211 | 0.057 | 3.44E-04 | 0.003 | Cerebellum | |
| NSF | −882686 | −0.158 | 0.043 | 3.93E-04 | 0.003 | Cerebellum | |
| AC008105.1 | 466248 | −0.247 | 0.072 | 7.87E-04 | 0.006 | Cerebellum | |
| AC091132.4 | 162179 | −0.387 | 0.118 | 0.001 | 0.010 | Cerebellum | |
| AC015936.1 | 760069 | 0.575 | 0.173 | 0.002 | 0.011 | Spinal cord (cervical c-1) | |
| CR936218.1 | −327330 | −0.393 | 0.122 | 0.002 | 0.013 | Putamen (basal ganglia) | |
| ACBD4 | 575382 | −0.175 | 0.057 | 0.003 | 0.020 | Cerebellum | |
| PLCD3 | 574628 | 0.189 | 0.067 | 0.006 | 0.041 | Hypothalamus | |
| ARL17B | −653781 | 0.351 | 0.127 | 0.007 | 0.043 | Cerebellum | |
| rs2291317 | BIN3 | −699499 | 0.212 | 0.074 | 0.005 | 0.034 | Nucleus accumbens (basal ganglia) |
| FAM160B2 | −119533 | 0.251 | 0.089 | 0.007 | 0.043 | Substantia nigra |
Discussion
We report here findings from a GWAS of maximum habitual alcohol use from the US MVP sample, in EUR and AFR. In EUR, we observed 7.8% SNP-based heritability that is consistent with other large-GWAS of alcohol-related traits which also range from 5% to 10% (14). These SNP-based heritability estimates account for about 15–25% of the heritability reported by twin studies (40). The phenotype tested (i.e., “In a typical month, what is/was the largest number of drinks of alcohol (beer, wine, and/or liquor) you may have had in one day?”) has genetic overlap with both alcohol consumption (UK Biobank: “Amount of alcohol drunk on a typical drinking day” and “Frequency of consuming six or more units of alcohol”) and with alcohol misuse (PGC: DSM-IV Alcohol Dependence; UK Biobank: “Ever been injured or injured someone else through drinking alcohol” and “Ever had known person concerned about, or recommend reduction of, alcohol consumption”).
Our findings provide strong support for association in the chromosome 4 ADH region, for ADH1B*rs1229984, as has been reported multiple times previously (here with p=4.9×10−47), spanning a lengthy chromosomal region (Figure 2a). A different signal at the same locus, rs2066702 (2.29×10−12; Figure 2b), was the only GWS result in AFR. We also report three additional regions in EUR, with prior varying, but never GWS, support: a region on chromosome 17 including CRHR1*rs77804065 (p=1.5×10−12; Figure 2c); chromosome 8, lead SNP rs7821592 (3.6×10−8), closest to XPO7; and at chromosome 10, lead SNP rs1577857 (4.2×10−08), LOC105378478, which has unknown function. The trans-population meta-analysis added two additional novel GWS regions, FGF14 (p=9.86×10−09) and LOC105376602 (p=4.63×10−08)(19).
Lead SNP ADH1B*rs1229984 is a long-established risk locus from the pre-GWAS era that has been strongly confirmed by GWAS (2, 6). To identify rs1229984 as the lead variant, we needed to address a data-cleaning dilemma, as this variant was initially excluded from analysis on HWE criteria. Knowing the importance of the variant, we investigated the situation further, and discovered two subpopulations within the EUR, one with higher rs1229984 MAF (that clusters with Ashkenazi Jews(41)), and another much larger subpopulation with lower MAF. HWE criteria for this key variant were met within both of these individual subpopulations. In the initial quality control investigation, the violation of HWE expectations was, we conclude, attributable to this demonstrable violation of the random mating assumption (and not to a problem with data quality). We recommend that studies that may have excluded ADH1B*rs1229984 on HWE grounds examine this same issue.
This variant has many orders of magnitude greater support for association than the next-best-supported independent region on chromosome 17, lead SNP rs77804065, which maps to CRHR1, observed p=1.5×10−12 in EUR. CHRH1 variants were previously implicated in candidate gene studies of alcohol use phenotypes(42, 43) and in an animal study regarding sensitivity to relapse into alcohol seeking induced by environmental stress (44). This GWS association signal maps to a well-known 900 kb inversion region (45) containing numerous other genes, some of which could also be considered MaxAlc candidate loci. The inversion is much less common in Africans (45, 46) consistent with the complex evolutionary history at this locus (47), so meta-analysis between EUR and AFR could potentially narrow the associated region greatly, if there is association information in that population as well, even if non-significant taken only in AFR. Indeed, the transpopulation meta-analysis showed that statistical significance increased by over an order of magnitude (to 1.02×10−13) with improved evidence for localization of the lead SNP at CHRH1 (Figure 2c). A similar phenomenon has been observed in narrowing associated regions for schizophrenia when meta-analyzing EUR and Asian GWAS results (48). Gene-based analyses and the replication in the UK Biobank provided additional evidence supporting CRHR1 as a risk locus.
On chromosome 8, rs7821592, the implicated locus is XPO7. Although this locus was identified as being of interest in a prior sparse “pooled GWAS” study of AD (49), and was identified in a study of AD comorbid with bipolar disorder (50), it has never previously been identified for these traits at anything approaching GWS. Finally, in EUR, rs1577857 (LOC105378478) on chromosome 10 has apparently not been reported previously. Although this variant is located in a non-coding RNA gene not previously associated with any human phenotype, the association in the MVP cohort was also replicated in PGC and UK Biobank cohorts. Additionally, the regulatory functional significance of this locus is supported by the fact that the variant is in a DNase I hypersensitivity site detected in twelve different cell types(51).
In AFR, we identified a single region led by ADH1B rs2066702 which, like rs1229984 in EUR, is well replicated (2).
The trans-population meta-analysis added two novel GWS loci, i.e., six in the meta-analysis, vs. only four in the European-only analysis (and one, overlapping with an EUR-associated region, GWS in AFR taken individually, albeit with a different SNP). These were FGF14 - Fibroblast Growth Factor 14 – at p=9.86×10−09: a gene implicated in inherited cerebellar ataxias (52), among other traits, which regulates KCNQ2/3 potassium channels (53); and an uncharacterized RNA gene locus, LOC105376602 (at p=4.63×10−08). FGF14 is particularly relevant because KLB, a locus previously identified as associated to alcohol consumption (4, 6, 8, 54) and replicated in MVP (min p=5.54×10−6), is a receptor that acts as a targeting signal for several FGF genes (55), suggesting the strong possibility of wider involvement of the FGF family in predisposition to alcohol consumption.
Thus, although our AFR sample was too small for novel locus identification when taken individually, trans-population meta-analysis was very valuable because of the differences in local LD (allowing improved CRHR1 region mapping) and additional association information for risk regions apparently in common between these populations.
The phenome-wide genetic-correlation analyses identified correlations with numerous traits including tobacco smoking behaviors, socioeconomic status, physical activity, reproductive behaviors, fat mass, personality traits, and, to a lesser extent, certain psychiatric disorders. Similar findings have been reported previously, even with small numbers of markers or with ADH1B*rs1229984 taken individually (56). These genetic relationships of MaxAlc are consistent with the pervasive role of alcohol use and abuse on human morbidity and mortality (57). Gene-based analysis, besides supporting CRHR1 as noted above, supported other genes associated in SNP-based analysis such as XPO7 and FGF14, as well as, for example, KANSL1, which maps to the same inversion region as CRHR1, and PDE4B (Phosphodiesterase 4B), previously implicated in other neuropsychiatric disorders. Tissue and human cell-type expression enrichments were noted for cerebellar hemisphere and cerebellum; dopaminergic and GABAergic neurons in human midbrain; and delta cells in pancreas. The cerebellar enrichment is particularly relevant with respect to the known effects of alcohol on this brain region: ethanol is the most common injurious agent to Purkinje cells (58, 59). In this context, inter-individual variability in the genetic regulation of cerebellum may be linked to the ability to drink large amounts of alcohol. Additionally, alcohol affects the type-A γ-aminobutyric acid (GABAA) receptor, which mediates autocrine signaling mechanisms in pancreatic cells (60). Individuals with high resistance to the effects of ethanol on this system may be able to drink larger amounts of alcohol; subjects at risk for alcohol dependence tend to have lower levels of response to measures including body sway (61) which is presumably at least in part cerebellar in origin. In a mouse model, it was demonstrated that genetically-influenced differences in cerebellar alcohol response affect alcohol consumption (62). eQTL analysis provide further evidence for functional effects of risk loci, particularly those mapped to chromosome 17, in central nervous system, particularly cerebellum.
In summary, we mapped a) four risk loci for MaxAlc in EUR, of which only one (ADH1B) was previously known; b) one in AFR, which was previously known (a different marker in ADH1B than in EUR); and c) an additional two loci, both novel, in the transpopulation meta-analysis. MaxAlc is a clinically meaningful trait that differs from, but is genetically correlated with, DSM diagnosis of AUD. It is unclear to what extent the novel findings are due to the phenotype definition, or to the size and other characteristics of the clinical sample. MaxAlc, relating not merely to habitual alcohol use but to maximal habitual use, is more strongly related to the pathological range of alcohol use than some other measures such as the Alcohol Use Disorders Identification Test - Consumption (AUDIT-C) or MAXDRINKS (63). The negative correlation with “healthy” alcohol use behaviors, such as “alcohol usually taken with meals,” supports this interpretation.
Although our study is based on a large sample, we are still underpowered to conduct additional analyses to dissect the differences in the polygenic architecture of excessive drinking behaviors between sexes and age classes. MaxAlc, although a valid and useful phenotype, has previously been used only rarely. The high correlation with AUD per se may encourage more use in future studies, in the context of the results we report here. Additionally, the MVP uses an array that, while adequate for studies of EUR, is sparse for AFR and accordingly leaves much of the genome unstudied (64). This is the case because AFR are a genetically older population than EUR and have lower linkage disequilibrium genomewide; hence each SNP tends to query a shorter genomic region. For studies including large AFR populations, a more informative array would, ideally, be employed.
Finally, this study demonstrates the tremendous utility of the MVP sample for locus discovery. The large sample and informative set of surveys (combined with electronic health record data, which were not used here) will permit powerful and virtually unprecedented association studies of a vast array of traits and diseases. Furthermore, the inclusion of a sizeable sample of individuals of African descent contributes to additional locus identification opportunities.
Supplementary Material
Figure 4.
Manhattan plot, gene-based association results
Acknowledgments
This research is based on data from the Million Veteran Program (MVP), Office of Research and Development, Veterans Health Administration, and was supported by MVP and the VA Cooperative Studies Program (CSP) study #575B. We also appreciate the availability of summary statistic data from the Psychiatric Genomics Consortium (PGC) Substance Use Disorders (SUD) working group. The PGC-SUD is supported by funds from NIDA and NIMH to MH109532 and, previously, had analyst support from NIAAA to U01AA008401 (COGA). PGC-SUD gratefully acknowledges its contributing studies and the participants in those studies without whom this effort would not be possible.
Footnotes
Disclosures
Drs. Gelernter and Kranzler are named as co-inventors on PCT patent application #15/878,640 entitled: “Genotype-guided dosing of opioid agonists,” filed January 24, 2018.
Dr. Stein has in the past three years been a consultant for Actelion, Aptinyx, Bionomics, Dart Neuroscience, Healthcare Management Technologies, Janssen, Jazz Pharmaceuticals, Neurocrine Biosciences, Oxeia Biopharmaceuticals, and Pfizer. Dr. Stein owns founders shares and stock options in Resilience Therapeutics and has stock options in Oxeia Biopharmaceticals.
Dr. Kranzler has been an advisory board member, consultant, or CME speaker for Alkermes, Indivior and Lundbeck. He is also a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which was supported in the last three years by AbbVie, Alkermes, Ethypharm, Indivior, Lilly, Lundbeck, Otsuka, Pfizer, Arbor, and Amygdala Neurosciences.
All other authors report no biomedical financial interests or potential conflicts of interest.
Data Sharing
Summary statistics for all GWAS analyses are freely available. The dbGaP accession assigned to the Million Veteran Program is phs001672.v1.p. https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001672.v1.p1
Additionally, investigators who wish to gain access to the individual-level data may contact Dr Gelernter or Dr Stein; access to these data will be available in one of our laboratories on a collaborative basis. MVP is presently working towards developing ways to make individual-level coded data more broadly accessible as allowed by the consent and consistent with the MVP data access policies and procedures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gaziano JM, Concato J, Brophy M, Fiore L, Pyarajan S, Breeling J, et al. (2016): Million Veteran Program: A mega-biobank to study genetic influences on health and disease. Journal of clinical epidemiology. 70:214–223. [DOI] [PubMed] [Google Scholar]
- 2.Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, et al. (2014): Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Molecular psychiatry. 19:41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mbarek H, Milaneschi Y, Fedko IO, Hottenga JJ, de Moor MH, Jansen R, et al. (2015): The genetics of alcohol dependence: Twin and SNP-based heritability, and genome-wide association study based on AUDIT scores. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 168:739–748. [DOI] [PubMed] [Google Scholar]
- 4.Clarke TK, Adams MJ, Davies G, Howard DM, Hall LS, Padmanabhan S, et al. (2017): Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N=112 117). Molecular psychiatry. 22:1376–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu K, Kranzler HR, Sherva R, Sartor CE, Almasy L, Koesterer R, et al. (2015): Genomewide Association Study for Maximum Number of Alcoholic Drinks in European Americans and African Americans. Alcoholism, clinical and experimental research. 39:1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jorgenson E, Thai KK, Hoffmann TJ, Sakoda LC, Kvale MN, Banda Y, et al. (2017): Genetic contributors to variation in alcohol consumption vary by race/ethnicity in a large multi-ethnic genome-wide association study. Molecular psychiatry. 22:1359–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, et al. (2011): Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proceedings of the National Academy of Sciences of the United States of America. 108:7119–7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schumann G, Liu C, O’Reilly P, Gao H, Song P, Xu B, et al. (2016): KLB is associated with alcohol drinking, and its gene product beta-Klotho is necessary for FGF21 regulation of alcohol preference. Proceedings of the National Academy of Sciences of the United States of America. 113:14372–14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quillen EE, Chen XD, Almasy L, Yang F, He H, Li X, et al. (2014): ALDH2 is associated to alcohol dependence and is the major genetic determinant of “daily maximum drinks” in a GWAS study of an isolated rural Chinese sample. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 165B:103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang X, Lu X, Wang L, Chen S, Li J, Cao J, et al. (2013): Common variants at 12q24 are associated with drinking behavior in Han Chinese. The American journal of clinical nutrition. 97:545–551. [DOI] [PubMed] [Google Scholar]
- 11.Baik I, Cho NH, Kim SH, Han BG, Shin C (2011): Genome-wide association studies identify genetic loci related to alcohol consumption in Korean men. The American journal of clinical nutrition. 93:809–816. [DOI] [PubMed] [Google Scholar]
- 12.Park BL, Kim JW, Cheong HS, Kim LH, Lee BC, Seo CH, et al. (2013): Extended genetic effects of ADH cluster genes on the risk of alcohol dependence: from GWAS to replication. Human genetics. 132:657–668. [DOI] [PubMed] [Google Scholar]
- 13.Gelernter J, Zhou H, Nunez YZ, Mutirangura A, Malison RT, Kalayasiri R (2018): Genomewide association study of alcohol dependence and related traits in a Thai population. Alcoholism, clinical and experimental research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walters RK, Polimanti R, Johnson EC, McClintick JN, Adams MJ, Adkins AE, et al. (2018): Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat Neurosci. 21:1656–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Topiwala A, Allan CL, Valkanova V, Zsoldos E, Filippini N, Sexton C, et al. (2017): Moderate alcohol consumption as risk factor for adverse brain outcomes and cognitive decline: longitudinal cohort study. BMJ (Clinical research ed). 357:j2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li D, Zhao H, Gelernter J (2011): Strong association of the alcohol dehydrogenase 1B gene (ADH1B) with alcohol dependence and alcohol-induced medical diseases. Biological psychiatry. 70:504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polimanti R, Gelernter J (2018): ADH1B: From alcoholism, natural selection, and cancer to the human phenome. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 177:113–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li D, Zhao H, Gelernter J (2012): Strong protective effect of the aldehyde dehydrogenase gene (ALDH2) 504lys (*2) allele against alcoholism and alcohol-induced medical diseases in Asians. Human genetics. 131:725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Concato J SN, Lu Q, Hu Y, Li B, Chen Q, Aslan M, Radhakrishnan K, Cheung KH, Li Y, Pietrzak R, Rajeevan N, Sayward F, Cho K, Harrington K, Honerlaw J, Pyarajan S, Quaden R, Gaziano JM, Zhao H, Stein MB, and Gelernter J on behalf of the VA Million Veteran Program (2017): Genetic associations of maximum regular alcohol intake in the Million Veteran Program. Presented at the Annual Meeting of The American Society of Human Genetics. Abstract/Program #275. [Google Scholar]
- 20.Saccone NL, Kwon JM, Corbett J, Goate A, Rochberg N, Edenberg HJ, et al. (2000): A genome screen of maximum number of drinks as an alcoholism phenotype. American journal of medical genetics. 96:632–637. [DOI] [PubMed] [Google Scholar]
- 21.Grant JD, Agrawal A, Bucholz KK, Madden PAF, Pergadia ML, Nelson EC, et al. (2009): Alcohol Consumption Indices of Genetic Risk for Alcohol Dependence. Biological psychiatry. 66:795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klarin D, Damrauer SM, Cho K, Sun YV, Teslovich TM, Honerlaw J, et al. (2018): Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nature genetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhan X, Hu Y, Li B, Abecasis GR, Liu DJ (2016): RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics (Oxford, England). 32:1423–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, et al. (2015): An atlas of genetic correlations across human diseases and traits. Nature genetics. 47:1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J (2015): LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. 47:291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. (2017): Genome-wide genetic data on ~500,000 UK Biobank participants. bioRxiv. [Google Scholar]
- 27.Benjamini YHY (1995): Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B 57:289–300. [Google Scholar]
- 28.de Leeuw CA, Mooij JM, Heskes T, Posthuma D (2015): MAGMA: generalized gene-set analysis of GWAS data. PLoS computational biology. 11:e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe K, Taskesen E, van Bochoven A, Posthuma D (2017): FUMA: Functional mapping and annotation of genetic associations. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Battle A, Brown CD, Engelhardt BE, Montgomery SB (2017): Genetic effects on gene expression across human tissues. Nature. 550:204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe K, Taskesen E, van Bochoven A, Posthuma D (2017): Functional mapping and annotation of genetic associations with FUMA. Nature communications. 8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edenberg HJ (2007): The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol research & health : the journal of the National Institute on Alcohol Abuse and Alcoholism. 30:5–13. [PMC free article] [PubMed] [Google Scholar]
- 33.Polimanti R, Yang C, Zhao H, Gelernter J (2015): Dissecting ancestry genomic background in substance dependence genome-wide association studies. Pharmacogenomics. 16:1487–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galinsky KJ, Bhatia G, Loh PR, Georgiev S, Mukherjee S, Patterson NJ, et al. (2016): Fast Principal-Component Analysis Reveals Convergent Evolution of ADH1B in Europe and East Asia. American journal of human genetics. 98:456–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen Q-K, Sulaiman X, Yao Y-G, Peng M-S, Zhang Y-P (2016): Was ADH1B under Selection in European Populations? American journal of human genetics. 99:1217–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kendler KS, Edwards AC, Gardner CO (2015): Sex differences in the pathways to symptoms of alcohol use disorder: a study of opposite-sex twin pairs. Alcoholism, clinical and experimental research. 39:998–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hasin DS, Grant BF (2015): The National Epidemiologic Survey on Alcohol and Related Conditions (NESARC) Waves 1 and 2: review and summary of findings. Social psychiatry and psychiatric epidemiology. 50:1609–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walters RK, Adams MJ, Adkins AE, Aliev F, Bacanu S-A, Batzler A, et al. (2018): Trans-ancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willer CJ, Li Y, Abecasis GR (2010): METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics (Oxford, England). 26:2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verhulst B, Neale MC, Kendler KS (2015): The heritability of alcohol use disorders: a meta-analysis of twin and adoption studies. Psychological medicine. 45:1061–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lencz T, Guha S, Liu C, Rosenfeld J, Mukherjee S, DeRosse P, et al. (2013): Genome-wide association study implicates NDST3 in schizophrenia and bipolar disorder. Nature communications. 4:2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Treutlein J, Kissling C, Frank J, Wiemann S, Dong L, Depner M, et al. (2006): Genetic association of the human corticotropin releasing hormone receptor 1 (CRHR1) with binge drinking and alcohol intake patterns in two independent samples. Molecular psychiatry. 11:594–602. [DOI] [PubMed] [Google Scholar]
- 43.Ray LA, Sehl M, Bujarski S, Hutchison K, Blaine S, Enoch MA (2013): The CRHR1 gene, trauma exposure, and alcoholism risk: a test of G × E effects. Genes, brain, and behavior. 12:361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hansson AC, Cippitelli A, Sommer WH, Fedeli A, Bjork K, Soverchia L, et al. (2006): Variation at the rat Crhr1 locus and sensitivity to relapse into alcohol seeking induced by environmental stress. Proceedings of the National Academy of Sciences of the United States of America. 103:15236–15241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stefansson H, Helgason A, Thorleifsson G, Steinthorsdottir V, Masson G, Barnard J, et al. (2005): A common inversion under selection in Europeans. Nature genetics. 37:129. [DOI] [PubMed] [Google Scholar]
- 46.Cáceres A, Sindi SS, Raphael BJ, Cáceres M, González JR (2012): Identification of polymorphic inversions from genotypes. BMC bioinformatics. 13:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alves JM, Lima AC, Pais IA, Amir N, Celestino R, Piras G, et al. (2015): Reassessing the Evolutionary History of the 17q21 Inversion Polymorphism. Genome biology and evolution. 7:3239–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lam M, Chen C-Y, Li Z, Martin A, Bryois J, Ma X, et al. (2018): Comparative genetic architectures of schizophrenia in East Asian and European populations. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson C, Drgon T, Liu QR, Walther D, Edenberg H, Rice J, et al. (2006): Pooled association genome scanning for alcohol dependence using 104,268 SNPs: validation and use to identify alcoholism vulnerability loci in unrelated individuals from the collaborative study on the genetics of alcoholism. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 141b:844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lydall GJ, Bass NJ, McQuillin A, Lawrence J, Anjorin A, Kandaswamy R, et al. (2011): Confirmation of prior evidence of genetic susceptibility to alcoholism in a genome-wide association study of comorbid alcoholism and bipolar disorder. Psychiatric genetics. 21:294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sheffield NC, Thurman RE, Song L, Safi A, Stamatoyannopoulos JA, Lenhard B, et al. (2013): Patterns of regulatory activity across diverse human cell types predict tissue identity, transcription factor binding, and long-range interactions. Genome research. 23:777–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dalski A, Atici J, Kreuz FR, Hellenbroich Y, Schwinger E, Zuhlke C (2005): Mutation analysis in the fibroblast growth factor 14 gene: frameshift mutation and polymorphisms in patients with inherited ataxias. European journal of human genetics : EJHG. 13:118–120. [DOI] [PubMed] [Google Scholar]
- 53.Pablo JL, Pitt GS (2017): FGF14 is a regulator of KCNQ2/3 channels. Proceedings of the National Academy of Sciences of the United States of America. 114:154–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanchez-Roige S, Palmer AA, Fontanillas P, Elson S, Team aR, Consortium tSUDWGotPG, et al. (2019): Genome-wide association study meta-analysis of the Alcohol Use Disorder Identification Test (AUDIT) in two population-based cohorts (N=141,958). The American journal of psychiatry. 176:107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Agrawal A, Parlee S, Perez-Tilve D, Li P, Pan J, Mroz PA, et al. (2018): Molecular elements in FGF19 and FGF21 defining KLB/FGFR activity and specificity. Molecular metabolism. 13:45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Polimanti R, Kranzler HR, Gelernter J (2016): Phenome-Wide Association Study for Alcohol and Nicotine Risk Alleles in 26394 Women. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 41:2688–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kunzmann AT, Coleman HG, Huang WY, Berndt SI (2018): The association of lifetime alcohol use with mortality and cancer risk in older adults: A cohort study. PLoS Med. 15:e1002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koeppen AH (2018): The neuropathology of the adult cerebellum. Handbook of clinical neurology. 154:129–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de la Monte SM, Kril JJ (2014): Human alcohol-related neuropathology. Acta neuropathologica. 127:71–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang S, Luo Y, Feng A, Li T, Yang X, Nofech-Mozes R, et al. (2014): Ethanol induced impairment of glucose metabolism involves alterations of GABAergic signaling in pancreatic beta-cells. Toxicology. 326:44–52. [DOI] [PubMed] [Google Scholar]
- 61.Schuckit MA (1988): Reactions to alcohol in sons of alcoholics and controls. Alcoholism, clinical and experimental research. 12:465–470. [DOI] [PubMed] [Google Scholar]
- 62.Kaplan JS, Nipper MA, Richardson BD, Jensen J, Helms M, Finn DA, et al. (2016): Pharmacologically Counteracting a Phenotypic Difference in Cerebellar GABAA Receptor Response to Alcohol Prevents Excessive Alcohol Consumption in a High Alcohol-Consuming Rodent Genotype. The Journal of neuroscience : the official journal of the Society for Neuroscience. 36:9019–9025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanchez-Roige S, Fontanillas P, Elson SL, andMe Research T, Gray JC, de Wit H, et al. (2017): Genome-wide association study of alcohol use disorder identification test (AUDIT) scores in 20 328 research participants of European ancestry. Addiction biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nelson SC, Doheny KF, Pugh EW, Romm JM, Ling H, Laurie CA, et al. (2013): Imputation-based genomic coverage assessments of current human genotyping arrays. G3 (Bethesda, Md). 3:1795–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.