There was an error in J. Cell Sci. (2019) 132, jcs229500 (doi:10.1242/jcs.229500).
The DMSO lanes in Fig. 2C and D were duplicated in Fig. 2E and F, respectively, which contravenes our image presentation guidelines. The authors supplied full blots for all these panels and prepared a new figure using these.
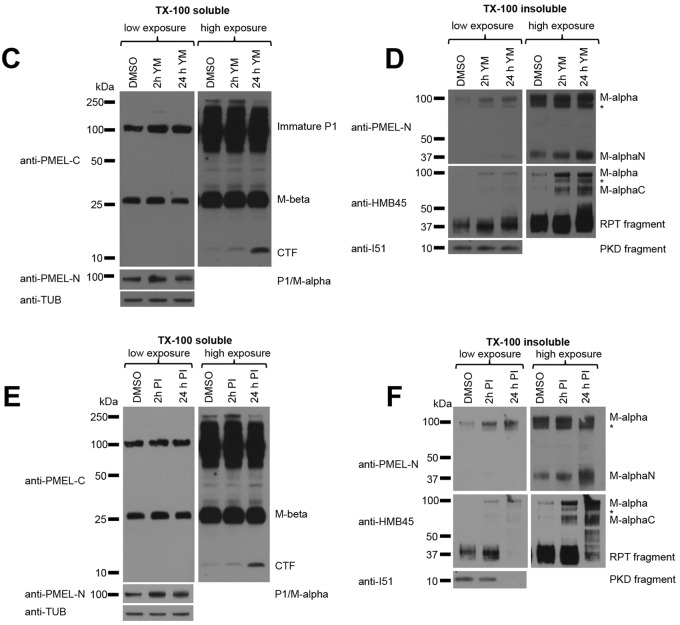
Fig. 2 (original panels).
M-α fragments accumulate upon interference with PIKfyve function or lysosomal protease activity. (C–F) MNT-1 cells were treated for 2 h or 24 h with 1.6 µM YM201636 (C,D) or with a mixture of protease inhibitors (100 µM leupeptin, 10 µM pepstatin A and 10 µM E-64d) (E,F) and Triton X-100-soluble (C,E) and Triton X-100-insoluble (D,F) lysates were analyzed by immunoblotting using antibodies against the PMEL C-terminus (anti-PMEL-C), the PMEL N-terminus (anti-PMEL-N), the PMEL RPT domain (anti-PMEL-HMB45), the PMEL PKD domain (anti-PMEL-I51) and tubulin (anti-TUB), as an equal loading marker. The different PMEL fragments are annotated on the right. Stars indicate M-α fragments derived from another isoform generated by alternative splicing. Right-hand panels show higher exposures.
The corrected and original panels are shown below. Both the online full-text and PDF versions of the article have been updated.
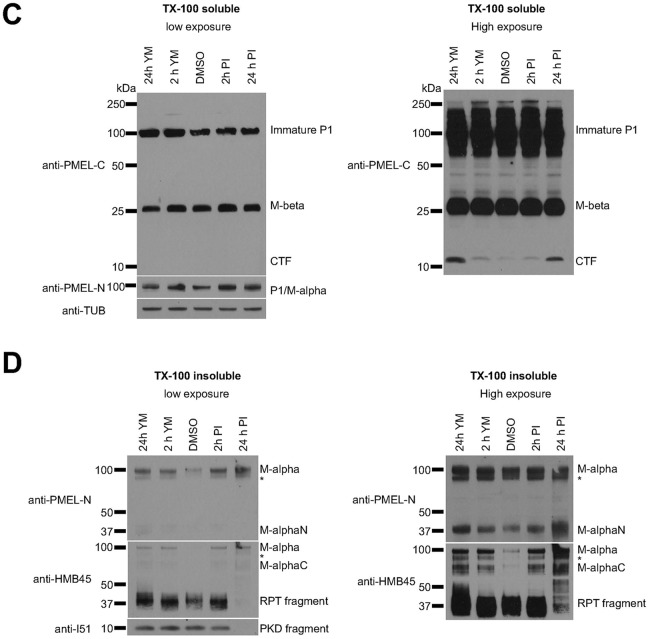
Fig. 2 (corrected panels).
M-α fragments accumulate upon interference with PIKfyve function or lysosomal protease activity. (C,D) MNT-1 cells were treated for 2 h or 24 h with 1.6 µM YM201636 or with a mixture of protease inhibitors (100 µM leupeptin, 10 µM pepstatin A and 10 µM E-64d) and Triton X-100-soluble (C) and Triton X-100-insoluble (D) lysates were analyzed by immunoblotting using antibodies against the PMEL C-terminus (anti-PMEL-C), the PMEL N-terminus (anti-PMEL-N), the PMEL RPT domain (anti-PMEL-HMB45), the PMEL PKD domain (anti-PMEL-I51) and tubulin (anti-TUB), as an equal loading marker. The different PMEL fragments are annotated on the right. Stars indicate M-α fragments derived from another isoform generated by alternative splicing. Right-hand panels show higher exposures.
The authors apologise for this error.