Abstract
The bat family Nycteridae contains only the genus Nycteris, which comprises 13 currently recognized species from Africa and the Arabian Peninsula, one species from Madagascar, and two species restricted to Malaysia and Indonesia in South‐East Asia. We investigated genetic variation, clade membership, and phylogenetic relationships in Nycteridae with broad sampling across Africa for most clades. We sequenced mitochondrial cytochrome b (cytb) and four independent nuclear introns (2,166 bp) from 253 individuals. Although our samples did not include all recognized species, we recovered at least 16 deeply divergent monophyletic lineages using independent mitochondrial and multilocus nuclear datasets in both gene tree and species tree analyses. Mean pairwise uncorrected genetic distances among species‐ranked Nycteris clades (17% for cytb and 4% for concatenated introns) suggest high levels of phylogenetic diversity in Nycteridae. We found a large number of designated clades whose members are distributed wholly or partly in East Africa (10 of 16 clades), indicating that Nycteris diversity has been historically underestimated and raising the possibility that additional unsampled and/or undescribed Nycteris species occur in more poorly sampled Central and West Africa. Well‐resolved mitochondrial, concatenated nuclear, and species trees strongly supported African ancestry for SE Asian species. Species tree analyses strongly support two deeply diverged subclades that have not previously been recognized, and these clades may warrant recognition as subgenera. Our analyses also strongly support four traditionally recognized species groups of Nycteris. Mitonuclear discordance regarding geographic population structure in Nycteris thebaica appears to result from male‐biased dispersal in this species. Our analyses, almost wholly based on museum voucher specimens, serve to identify species‐rank clades that can be tested with independent datasets, such as morphology, vocalizations, distributions, and ectoparasites. Our analyses highlight the need for a comprehensive revision of Nycteridae.
Keywords: Africa, biodiversity, Nycteris, species tree, taxonomy
We investigated genetic variation, clade membership, and phylogenetic relationships in the bat family Nycteridae with broad sampling across Africa. At least 16 deeply divergent monophyletic lineages were recovered in both gene tree and species tree analyses. Genetic distances among species‐ranked Nycteris clades (17% for cytb and 4% for concatenated introns) suggest high levels of phylogenetic diversity in Nycteridae. A large number of clades are distributed wholly or partly in East Africa (10/16 clades), suggesting Nycteris diversity has been historically underestimated.
1. INTRODUCTION
The Paleotropical slit‐faced bats, family Nycteridae, all belong to the genus Nycteris with 13 of 16 recognized species found in continental Africa and offshore islands, one species on Madagascar, and two species endemic to South‐East Asia (Mammal Diversity Database, 2019; Simmons, 2005). Members of the Nycteridae are readily recognizable by their nose leaves, which are divided by a deep median furrow running the length of the muzzle, the basis for their common name. They also possess a Y‐shaped terminal caudal vertebra that is unique among mammals. Systematic reviews of the family have not been informed by morphological or molecular phylogenetics, and the most recently named species in the family was described a half‐century ago (N. vinsoni, Dalquest, 1965). To put this taxonomic stasis in context, the number of recognized bat species globally has grown by 26.4% over the last 15 years. In the Paleotropics, this has included a 38% increase in the number of species of Rhinolophidae and a >50% increase in species in the genera Scotophilus and Miniopterus (cf. Simmons, 2005; Mammal Diversity Database, 2019). Here, we use a geographically extensive, multilocus dataset to assay the diversity and infer the evolutionary relationships of Nycteridae in order to establish the foundations for a fuller taxonomic revision.
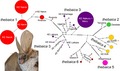
In the first systematic revision of Nycteridae, Andersen (1912) divided then‐known taxa into four species groups: javanica, hispida, aethiopica [now known as macrotis], and thebaica. Later, Aellen (1959) divided the javanica group into two based on tragus and dental characters: javanica (monotypic) and arge, which contained both African and Asian species. Using morphometrics and hyoid morphology, respectively, Van Cakenberghe and De Vree (1993a) and Griffiths (1997) later transferred the Asian member of the arge group, N. tragata, to the javanica group. This five‐group classification has been widely accepted (e.g., Simmons, 2005), but taxonomic membership in these groups has varied, owing to mosaic character variation. For example, the absence of biometrical differences in teeth measurements suggested the conspecificity of N. parisii with N. woodi (Van Cakenberghe & de Vree, 1985), but a subsequent study of bacula strongly supported the validity of both species and suggested their assignment to entirely different species groups (Thomas, Harrison, & Bates, 1994). Although qualitative and mensural characters have been used to characterize and differentiate species, external and skull characters are in conflict with other morphological characters (e.g., Happold, 2013a; Monadjem, Taylor, Cotterill, & Schoeman, 2010; Thomas et al., 1994; Van Cakenberghe & de Vree, 1985, 1993a, 1993b, 1998). Except for Griffiths’ (1997) analysis of the hyoid apparatus, the morphological characters of the species of Nycteridae have not been subjected to explicit phylogenetic analysis. Figure 1 shows the host of names available for Nycteris populations, many of them currently considered synonyms (cf. Simmons, 2005).
Figure 1.
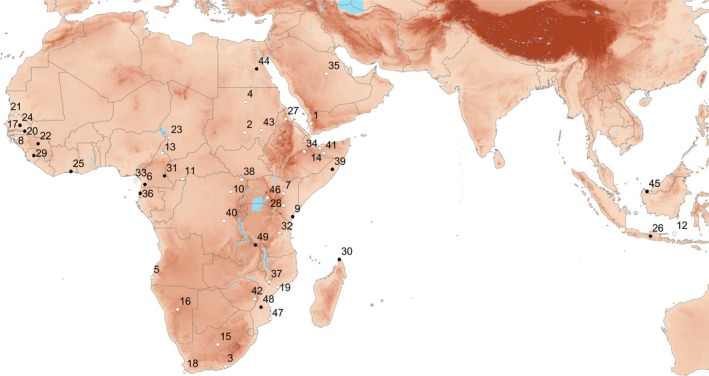
Named taxa of Nycteris, showing type localities for recognized species (filled circles) and subspecies or synonyms (open circles). Number codes are as follows: 1 – adana K. Andersen, 1912; 2 – aethiopica Dobson, 1878; 3 – affinis A. Smith, 1829; 4 –albiventer Wagner, 1840; 5 – angolensis Peters, 1871; 6 – arge Thomas, 1903; 7 – aurantiaca De Beaux, 1923; 8 – aurantiaca Monard, 1939; 9 – aurita K. Andersen, 1912; 10 – avakubia J. A. Allen, 1917; 11 – baikii Gray 1867; 12 – bastiani Bergmans & van Bree, 1986; 13 – benuensis Aellen, 1952; 14 – brockmani K. Andersen, 1912; 15 – capensis A. Smith, 1829; 16 – damarensis Peters, 1871; 17 – daubentonii. Geoffroy, 1813; 18 – discolor Wagner, 1840; 19 – fuliginosa Peters, 1852; 20 – gambiensis K. Andersen, 1912; 21 – geoffroyi Desmarest, 1820; 22 – grandis Peters, 1865; 23 – guineensis Monard, 1939; 24 – hispida Schreber, 1775; 25 – intermedia Aellen, 1959; 26 – javanica. Geoffroy, 1813; 27 – labiata Heuglin, 1861; 28 – luteola Thomas, 1901; 29 – macrotis Dobson, 1876; 30 – madagascariensis G. Grandidier, 1937; 31 – major K. Andersen, 1912; 32 – marica Kershaw, 1923; 33 – martini Fraser, 1843; 34 – media K. Andersen, 1912; 35 – najdiya Nader & Kock, 1982; 36 – nana K. Andersen, 1912; 37 – oriana Kershaw, 1922; 38 – pallida J. A. Allen, 1917; 39 – parisii De Beaux, 1924; 40 – proxima Lonnberg & Gyldenstolpe, 1925; 41 – revoilii Robin, 1881; 42 – sabiensis Roberts, 1946; 43 – senegalensis Hartmann, 1868; 44 – thebaica. Geoffroy, 1818; 45 – tragata K. Andersen, 1912; 46 – tristis G. M. Allen & Lawrence, 1936; 47 – villosa Peters, 1852; 48 – vinsoni Dalquest, 1965; and 49 – woodi K. Andersen, 1914. An additional name, pilosa Gray, 1866 from “Africa,” is not shown
Molecular phylogenetic analyses of the Nycteridae are likewise limited, as they included only a handful of species, each represented by a single sample. Shi and Rabosky (2015) used a concatenated supermatrix and included 7 of 16 Nycteris species in a time‐calibrated analysis of all Chiroptera. They found strong support for the traditional sister relationship between Nycteridae and Emballonuridae (the two families comprising the Emballonuridea of Koopman, 1993). The supermatrix analysis of Amador, Moyers Arévalo, Almeida, Catalano, and Giannini (2018), also based on the same seven Nycteris species, found inconsistent evidence for the endemic Malagasy Myzopodidae joining this group. Nevertheless, both studies recovered Nycteridae as monophyletic and a close relative of Emballonuridae, and both studies recovered the two Asian species, N. tragata and N. javanica, as well‐supported sisters. It should be noted, however, that both studies were based on incomplete supermatrices (71% missing data in Amador et al., 2018 and 83% missing in Shi & Rabosky, 2015). Thus, the diversity and phylogenetic relationships of species in Nycteridae remain largely unresolved and the evolutionary independence of Nycteris lineages has yet to be established.
Bat surveys across Africa over the last two decades have provided substantial new material for the evaluation of phylogenetic relationships and species limits. In addition, recent studies (Demos, Webala, Bartonjo, & Patterson, 2018; Dool et al., 2016; Patterson et al., 2018) have shown that a multilocus intron system based on different chromosomes and enabling independent representation of the nuclear genome offers clear advantages over analyses based only on mitochondrial data. Advantages include better resolution of earlier divergences (e.g., Demos et al., 2019) and improved detection of instances of mitochondrial introgression (e.g., Dool et al., 2016; Hassanin et al., 2018). Here, we address three key aspects of Nycteridae evolution: (a) recognizing monophyletic lineages within Nycteris, focusing on Afrotropical species, and assessing their evolutionary independence using independent nuclear loci under a coalescent framework; (b) evaluating their phylogenetic relationships using both nuclear and mitochondrial data in gene tree, concatenated, and species tree analyses; and (c) assessing the species‐group relationships of Nycteris species that had been classified by morphology alone. This study highlights the need for a comprehensive revision of African Nycteridae. Our analyses and discussion serve to identify species‐rank clades that need to be tested with independent datasets including morphology, vocalizations, distributions, and ectoparasites.
2. MATERIALS AND METHODS
2.1. Selection of taxa and sampling
The bats newly sequenced for this study (n = 249) were collected during recent small mammal surveys across sub‐Saharan Africa, with relatively dense sampling in East Africa (see Figure S1 in Supporting Information). Initial assignment of individuals to species for East African specimens was determined using meristic, mensural, and qualitative characters presented in the bat keys of Thorn, Kerbis Peterhans, and Baranga (2009) and Patterson and Webala (2012). Field methods followed mammal collecting guidelines (Sikes, 2016) and were approved under Field Museum of Natural History IACUC #2012‐003. Tissues were taken from euthanized specimens in the course of preparing voucher specimens following IACUC protocols and the respective national collecting permits. Tissues were variously preserved in ethanol, saturated salt solution (EDTA‐DMSO‐NaCl), or liquid nitrogen and stored in liquid nitrogen dewars. Four additional cytochrome b gene (cytb) sequences of Nycteris were downloaded from GenBank. Coleura afra (Emballonuridae) was included as an out‐group. In total, 1–5 genes were analyzed in 253 individuals in this study (see Table S1 in Supporting Information for voucher numbers and locality data and Appendix 1 for GenBank accession numbers). To enable subsequent integrative taxonomic revisions, all but four of the individuals analyzed genetically in this study are accompanied by museum voucher specimens suitable for morphological analysis.
In view of the large number of names (many of which are synonyms; Figure 1) and to avoid contributing to current taxonomic confusion in Nycteris, we utilized a conservative approach in labeling clades. Where a clade's taxonomic identity was ambiguous or unknown, we referred to it simply as a numbered clade. In some cases, even assignment to equivocal groupings was necessary (e.g., hispida/aurita and cf. hispida/aurita). Although used as explicit labels in our study, the validity of these names is provisional. Comprehensive morphological assessments of individual specimens making up these clades included in our analyses will be required in order to verify which, if any, existing names may apply to them.
2.2. Amplification and sequencing
We sequenced one mitochondrial protein‐coding gene cytochrome b (cytb) and the nuclear introns acyl‐CoA oxidase 2 intron 3 (ACOX2), COP9 signalosome subunit 7A intron 4 (COPS7A), rogdi atypical leucine zipper intron 7 (ROGDI), and signal transducer and activator of transcription 5A intron (STAT5A) for specimens of Nycteris and the close emballonurid out‐group Coleura afra. Primers, primer references, and thermocycler conditions are described in Table 1. General methods of DNA extraction, amplification, and sequencing follow Demos et al. (2018) and Patterson et al. (2018). DNA sequences were assembled, aligned, and edited using GENEIOUS PRO v.11.1.5 (Biomatters Ltd.). Alignments were inspected visually and determined to be unambiguous. Several gaps were introduced in the alignments of the four nuclear introns, but their positions were unambiguous. Sequences of cytb were translated to amino acids to confirm the absence of premature stop codons and indels. The cytb alignment was trimmed to 1,121 nucleotides to minimize missing data. Before phylogenetic analyses using mitochondrial data, we reduced the matrix of 253 individuals to the set of unique sequences, resulting in a final matrix of 164 individuals. The matrix used for calculating cytb distances between lineages comprised 250 individuals from the 253 individual alignments. We resolved nuclear DNA to haplotypes with the PHASE program (Stephens, Smith, & Donnelly, 2001) and set the probability threshold to 70%, following Garrick, Sunnucks, and Dyer (2010). PHASE files were formatted and assembled using SeqPhase (Flot, 2010).
Table 1.
Primer information for genes amplified in the current study. References indicated by (a) Salicini, Ibáñez, & Juste, 2011; (b) Eick, Jacobs, & Matthee, 2005; (c) Trujillo, Patton, Schlitter, & Bickham, 2009)
| Gene | Primers (5’–3’) | Amplicon length | References | Thermal profile |
|---|---|---|---|---|
| ACOX2 |
ACOX2f CCTSGGCTCDGAGGAGCAGAT ACOX2r GGGCTGTGHAYCACAAACTCCT |
717 bp | a | 3 min at 95°C followed by 10 cycles of 15 s at 95°C, 30 s at 65°C in 1°C decrements from 65°C (64–56°C), and 1 min at 72°C, followed by 36 cycles of 15 s at 95°C, 30 s at 55°C, and 1 min at 72°C, and final 5 min extension at 70°C |
| COPS7A |
COPSf TACAGCATYGGRCGRGACATCCA COPSr TCACYTGCTCCTCRATGCCKGACA |
689 bp | a | Same as ACOX2 above |
| ROGDI |
ROGDIf CTGATGGAYGCYGTGATGCTGCA ROGDIr CACGGTGAGGCASAGCTTGTTGA |
505 bp | a | 3 min at 95°C followed by 10 cycles of 15 s at 95°C, 30 s at 60°C in 1°C decrements from 60°C (59–51°C), and 1 min at 72°C, followed by 36 cycles of 15 s at 95°C, 30 s at 50°C, and 1 min at 72°C, and final 5 min extension at 70°C |
| STAT5A |
STAT5f CTGCTCATCAACAAGCCCGA STAT5r GGCTTCAGGTTCCACAGGTTGC |
530 bp | b | Same as ROGDI above |
| cytb |
LGL−765f GAAAAACCAYCGTTGTWATTCAACT LGL−766r GTTTAATTAGAATYTYAGCTTTGGG |
c | 3 min at 95°C followed by 36 cycles of 45 s at 95°C, 30 s at 50°C, and 2.5 min at 70°C, and final 5 min extension at 70°C |
2.3. Gene trees, networks, species trees, and summary statistics
PartitionFinder 2 (Lanfear, Frandsen, Wright, Senfeld, & Calcott, 2016) on CIPRES Science Gateway v.3.1 (Miller, Pfeiffer, & Schwartz, 2010) was used to determine the appropriate model of sequence evolution using the Bayesian information criterion (BIC) for cytb and the four nuclear introns. Interspecific uncorrected sequence divergences (p‐distances) for cytb were calculated for both positions 1, 2, and 3 and positions 1 and 2 only, and intraspecific distances were calculated using positions 1, 2, and 3 using MEGA X 10.0.5 (Kumar, Stecher, Li, Knyaz, & Tamura, 2018).
Maximum‐likelihood (ML) inference of cytb gene trees and a concatenated alignment using four partitioned nuclear introns were made using the program IQ‐TREE version 1.6.0 (Nguyen, Schmidt, von Haeseler, & Minh, 2015) on the CIPRES portal. Gene tree analyses under a Bayesian inference (BI) framework were carried out in MRBAYES v.3.2.6 (Ronquist et al., 2012) on the CIPRES portal to infer gene trees for cytb and the partitioned alignment of four nuclear introns. Two replicates were run in MrBayes, and nucleotide substitution models were unlinked across partitions for each nuclear locus in the concatenated alignment. Four Markov chains were run for 1 × 107 generations using default heating values and sampled every 1000th generation. Stationarity of the MRBAYES results was assessed in Tracer v1.7 (Rambaut, Drummond, Xie, Baele, & Suchard, 2018). Majority‐rule consensus trees were inferred for each Bayesian analysis. PopART (Leigh & Bryant, 2015) was used to construct a median‐joining network of cytochrome b haplotypes for clades within Nycteris thebaica. Pie charts were used to visualize the relative frequencies and relationships of haplotypes in N. thebaica clades 1–6.
Nycteris taxa were assigned to either species or named clades based on clade support in the analyses of the cytb and nuclear intron datasets. As in Demos et al. (2018), results from gene tree analyses were used to identify populations to be used as “candidate species” for the species tree approach implemented in StarBEAST2 (Ogilvie, Bouckaert, & Drummond, 2017), an extension of BEAST v.2.5.1 (Bouckaert et al., 2014). Species tree analyses were carried out using the four nuclear intron alignments with substitution, clock, and tree models unlinked among loci. The lognormal relaxed‐clock model was applied to each locus using a Yule tree prior and the linear with constant root population size model. Four replicates were carried out, and the analyses were run for 2 × 108 generations with 10% of each run discarded as burn‐in. We used Tracer v.1.7 to assess convergence and stationarity of model parameters based on ESS values and examination of trace files.
Sequence alignments used in this study have been deposited on the Figshare data repository (https://doi.org/10.6084/m9.figshare.8081594.v1). All newly generated sequences are available on GenBank with accession numbers MK837076–MK837603 (see also Appendix 1).
3. RESULTS
3.1. Mitochondrial genetic diversity, gene trees, and haplotype network
Sequences were generated and aligned for cytb (1,121 bp, 99% coverage), ACOX2 (646 bp, 96% coverage), COPS7A (624 bp, 98% coverage), ROGDI (450 bp, 98% coverage), and STAT5A (523 bp, 98% coverage). The concatenated alignment of four introns for 70 individuals was 97.1% complete (mean sequence length 2,166 bp). Models of sequence evolution inferred by PartitionFinder 2 were as follows: cytb, GTR + I+G; ACOX2, TrN + G; COPS7A, TrN + G; ROGDI, TrN + G; and STAT5A, TrN + G. Uncorrected cytb distances for reciprocally monophyletic Nycteris lineages in the 250 sequence cytb alignment ranged from 3.6% to 22.2% for cytb positions 1 + 2 + 3 and 1.0%–8.0% for cytb positions 1 + 2 (Table 2). Within‐lineage variability for cytb positions 1 + 2 + 3 ranged from 0% to 4.9%.
Table 2.
Uncorrected cytb p‐distances among clades of Nycteris: on and below diagonal based on positions 1, 2, and 3; above diagonal, positions 1 and 2. Clades represented by one individual (N. cf. thebaica 3, N. javanica, N. nana 1) not included
| Taxon | [1] | [2] | [3] | [4] | [5] | [6] | [7] | [8] | [9] | [10] | [11] | [12] | [13] | [14] | [15] | [16] | [17] | [18] | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [1] | arge 1 | 4.3 | 3.7 | 3.8 | 7.6 | 5.3 | 3.7 | 3.6 | 4.2 | 4.1 | 4.9 | 4.0 | 6.5 | 6.6 | 6.4 | 6.4 | 6.5 | 6.6 | 3.1 |
| [2] | arge 2 | 15.9 | 1.1 | 4.5 | 7.3 | 6.3 | 3.7 | 3.4 | 4.5 | 3.6 | 4.6 | 2.6 | 6.4 | 6.7 | 6.5 | 6.7 | 6.7 | 6.5 | 3.6 |
| [3] | cf. hispida/aurita | 16.1 | 17.5 | 0.7 | 8.0 | 6.4 | 3.9 | 4.0 | 5.3 | 4.3 | 5.3 | 4.6 | 7.5 | 7.4 | 7.0 | 7.0 | 7.5 | 7.0 | 4.1 |
| [4] | cf. thebaica 1 | 19.3 | 19.2 | 19.5 | 2.6 | 6.2 | 7.4 | 7.7 | 8.0 | 7.1 | 6.9 | 7.9 | 6.7 | 7.2 | 7.3 | 6.8 | 7.3 | 7.0 | 6.0 |
| [5] | cf. thebaica 2 | 17.0 | 17.8 | 19.0 | 14.9 | 0.1 | 6.0 | 6.3 | 5.8 | 5.5 | 6.3 | 6.1 | 5.9 | 6.1 | 6.0 | 5.7 | 6.0 | 5.8 | 6.0 |
| [6] | grandis | 16.3 | 16.3 | 17.2 | 20.3 | 18.2 | 1.6 | 3.7 | 3.9 | 4.2 | 4.9 | 4.4 | 7.1 | 7.2 | 7.2 | 7.2 | 7.0 | 7.1 | 3.6 |
| [7] | hispida/aurita | 14.5 | 15.4 | 15.0 | 19.7 | 18.0 | 16.1 | 2.5 | 5.0 | 4.5 | 4.8 | 4.0 | 6.9 | 7.4 | 7.2 | 7.0 | 7.5 | 7.5 | 3.2 |
| [8] | macrotis 1 | 17.4 | 18.1 | 18.1 | 20.8 | 19.5 | 17.7 | 17.7 | 2.2 | 3.3 | 4.0 | 4.9 | 6.9 | 7.1 | 6.9 | 7.1 | 6.6 | 6.8 | 3.9 |
| [9] | macrotis 2 | 16.3 | 18.7 | 17.8 | 19.4 | 19.0 | 18.5 | 16.3 | 13.8 | 0.9 | 3.7 | 4.1 | 7.8 | 7.5 | 7.2 | 7.2 | 7.1 | 7.1 | 3.7 |
| [10] | macrotis 3 | 17.6 | 19.0 | 19.2 | 20.1 | 20.0 | 18.7 | 17.8 | 14.3 | 15.0 | 0.4 | 5.0 | 7.4 | 7.8 | 8.0 | 7.7 | 7.6 | 7.7 | 4.4 |
| [11] | nana 2 | 16.1 | 13.2 | 17.1 | 19.3 | 16.8 | 17.0 | 15.3 | 17.8 | 17.4 | 18.0 | 4.9 | 6.9 | 7.1 | 7.0 | 7.0 | 6.9 | 7.0 | 4.4 |
| [12] | thebaica 1 | 18.7 | 18.5 | 19.1 | 18.1 | 17.1 | 19.4 | 19.0 | 19.6 | 22.2 | 20.1 | 18.0 | 0.4 | 2.0 | 1.9 | 2.0 | 2.5 | 1.9 | 6.4 |
| [13] | thebaica 2 | 18.7 | 18.4 | 19.9 | 18.4 | 17.8 | 19.7 | 19.4 | 20.1 | 21.5 | 20.4 | 18.2 | 5.8 | 0.4 | 1.3 | 1.7 | 2.3 | 1.4 | 7.3 |
| [14] | thebaica 3 | 18.9 | 18.6 | 19.5 | 18.2 | 18.0 | 19.9 | 19.6 | 20.4 | 21.8 | 21.1 | 18.5 | 5.0 | 5.0 | 1.6 | 1.2 | 1.6 | 1.0 | 7.1 |
| [15] | thebaica 4 | 18.4 | 18.6 | 19.7 | 17.7 | 17.0 | 19.9 | 19.5 | 19.9 | 21.6 | 20.4 | 17.9 | 5.1 | 4.7 | 3.6 | 1.6 | 2.3 | 1.3 | 6.9 |
| [16] | thebaica 5 | 18.9 | 19.2 | 19.7 | 18.6 | 17.5 | 19.5 | 20.0 | 19.6 | 21.9 | 20.4 | 18.4 | 6.4 | 6.5 | 5.6 | 5.3 | 0.0 | 1.4 | 7.3 |
| [17] | thebaica 6 | 18.2 | 18.1 | 19.5 | 17.4 | 17.0 | 18.8 | 19.4 | 19.8 | 21.4 | 19.7 | 17.4 | 5.4 | 5.2 | 4.7 | 4.2 | 5.4 | 1.7 | 7.3 |
| [18] | tragata | 14.4 | 17.7 | 17.2 | 18.6 | 18.7 | 15.8 | 16.3 | 18.2 | 17.9 | 16.3 | 17.0 | 17.0 | 18.1 | 17.9 | 17.9 | 18.3 | 17.2 | 1.3 |
The ML phylogeny for Nycteridae based on cytb shows division of the family into four deeply diverged subclades (labeled as clades 1A, 1B, 2A, and 2B in Figure 2a). The topology of the maximum clade credibility tree is substantially similar in topology to the maximum‐likelihood tree presented here. The monophyly of all named clades was strongly supported with the exception of Nycteris thebaica clade 6. Relationships among clades were generally well supported with the exception of the position of (a) the relationships of the geographically delimited clades within N. thebaica, (b) N. cf. thebaica clade 3, and (c) the relationship of N. arge clade 1 and N. tragata + N. javanica. Two nodes had equivocal support (bootstrap (BS) ≥70%, posterior probability (PP) <0.95): the node uniting N. thebaica clades 1–6 and N. cf. thebaica clades 1 + 2 and the node uniting N. arge clade 2 and N. nana clade 1. Several clades with broad geographic sampling showed relatively high levels of within‐clade genetic variation (i.e., N. hispida/aurita, N. grandis, and N. macrotis clade 1). For those clades with limited geographic sampling, we recovered high levels of divergence among populations in N. cf. thebaica 1 and N. nana clade 2. Both ML and BI analyses strongly supported N. arge clade 1 (Central African Republic [CAR], Democratic Republic of Congo [DRC], Gabon, Uganda) + N. tragata (Malaysia) + N. javanica (Borneo) as nested well within the other African Nycteris clades. The ML and BI trees support multiple deeply divergent clades separated by >10% cytb distances. The number of deeply diverged clades that include individuals from East Africa (Kenya, Tanzania, and Uganda) is high: 10 of 16 clades in the trees include individuals from this region.
Figure 2.
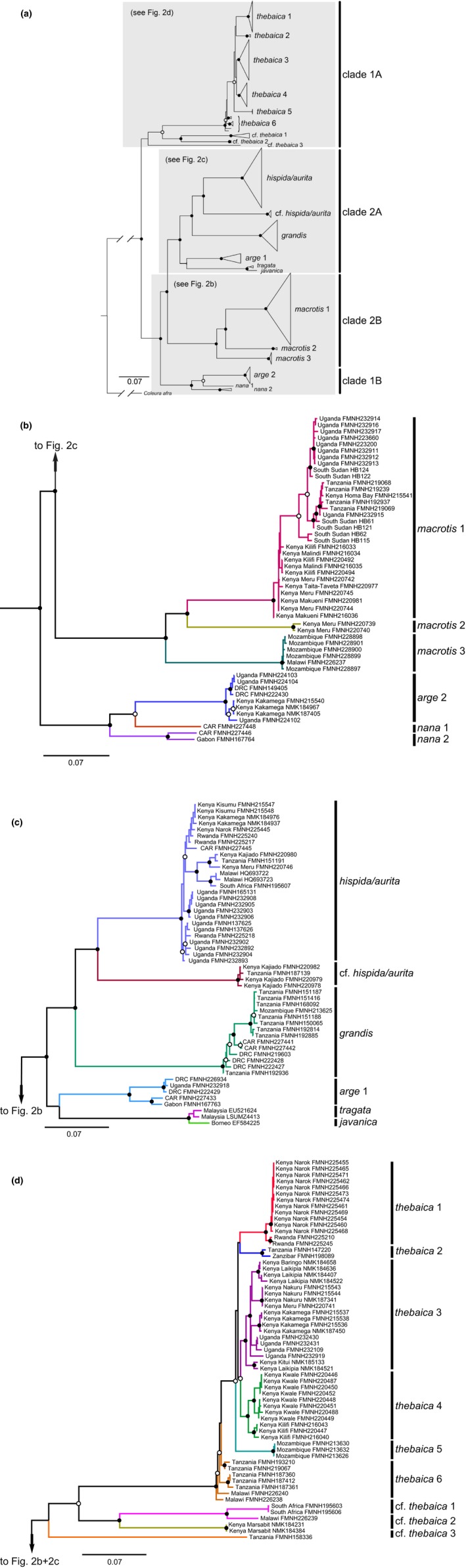
(a) Maximum‐likelihood phylogeny of 163 Nycteris specimens based on cytochrome b. The phylogeny was inferred in IQ‐TREE and its topology closely resembled the phylogeny calculated in MrBayes under a Bayesian framework. Filled circles on nodes denote bootstrap values (BS) ≥70% and Bayesian posterior probabilities (PP) ≥0.95, open circles outlined in black indicate BS ≥ 70% and PP < 0.95, and unmarked nodes indicate BS < 70% and PP < 0.95. Support values for most minor clades are not shown. Species names assigned on basis of preliminary field identifications or examination of museum specimens. (b–d) enlarged sections of the complete cytb tree showing individual relationships. Specimen localities include counties for densely sampled Kenya. CAR refers to Central African Republic and DRC to Democratic Republic of the Congo. Museum acronyms are defined in Appendix 1
The median‐joining network of cytb haplotype diversity for the six allopatric populations within N. thebaica showed no shared alleles among clades (Figure 3). The haplotype network revealed the existence of six well‐differentiated clades (minimum separation of clades was 19 substitutions), although N. thebaica clade 4 (coastal Kenya) clusters ambiguously between N. thebaica clade 5 (Mozambique) and N. thebaica clade 2 (Tanzania and Zanzibar).
Figure 3.
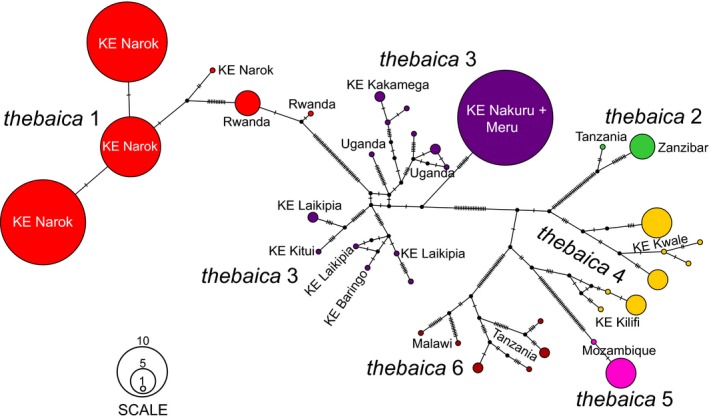
PopART network median‐joining analysis of cytochrome b haplotypes for 127 individuals representing Nycteris thebaica clades 1 to 6. Colored circles represent different sampled haplotypes, and black circles represent inferred missing or unsampled states. Hatch marks each denote a mutational step between haplotypes. CAR refers to Central African Republic, DRC to Democratic Republic of the Congo, and KE to Kenya
3.2. Concatenated nuclear gene trees
The ML gene tree inferred from the concatenated nuclear genes ACOX2, COPS7A, ROGDI, and STAT5A (70 individuals; matrix > 97% complete) is shown in Figure 4. This tree was similar to the BI tree with strong support for 22 of 25 major nodes. All of the named clades are strongly supported as monophyletic. Unlike the cytb gene trees, the position of N. arge clade 2 + N. nana clade 1 + N. nana clade 2 is ambiguous, while N. cf. thebaica clade 3 is strongly supported as part of the N. thebaica group. Nycteris tragata from SE Asia is strongly supported as nested within African Nycteris clades but is not sister to N. arge clade 1 as in the cytb gene trees. The most striking difference between the concatenated nuclear trees and the mitochondrial gene trees is the absence of support for genetic structure among the numbered lineages of N. thebaica. None of the clades named as N. thebaica 1–6 are supported as monophyletic, and relationships among individuals are poorly supported.
Figure 4.
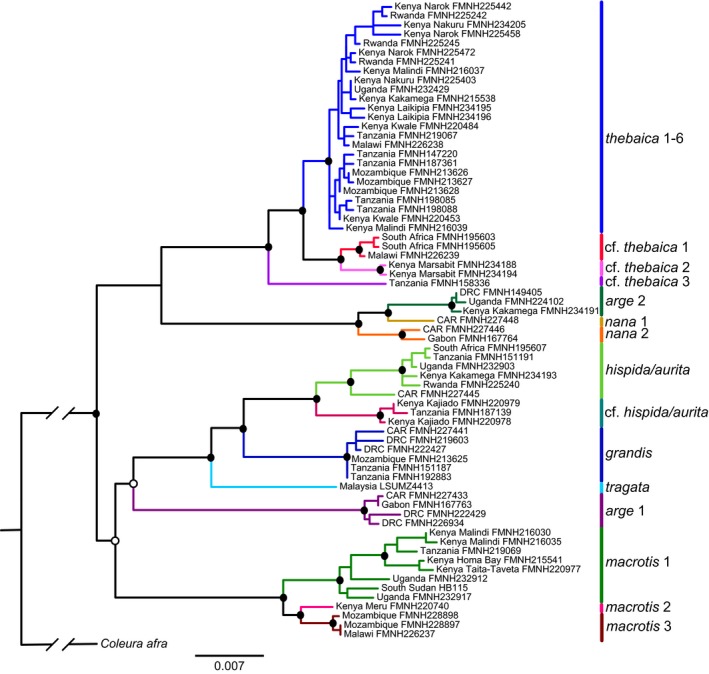
Concatenated Bayesian phylogeny of four independent nuclear introns of Nycteris. Filled circles at nodes denote ML bootstrap values (BS) ≥70% and Bayesian posterior probabilities (PP) ≥0.95, open circles outlined in black indicate BS ≥ 70% and PP < 0.95, and unmarked nodes indicate BS < 70% and PP < 0.95. Support values for most minor clades are not shown. Specimen localities include counties for Kenya. CAR refers to Central African Republic and DRC to Democratic Republic of the Congo. Museum acronyms are defined in Appendix 1
3.3. Species trees
Samples from parameter values of the four StarBEAST analyses had ESS values >200, with the exception of the five tree‐height parameters which all had values >100. We discarded the first 10% of each run, leaving 18,000 species trees in the posterior distributions that were then merged using LogCombiner. The topology of the maximum clade credibility tree (Figure 5) was identical across all four replicates. Species tree analysis using StarBEAST resulted in a topology that is strongly supported, with 12 of 13 nodes having PP ≥ 0.95. As in the concatenated nuclear gene trees, but unlike the cytb gene trees, Nycteris cf. thebaica 3 is strongly supported as sister to the other N. thebaica clades. There is strong support for the node uniting N. arge 2 + N. nana 1 + N. nana 2 with the N. thebaica clades, resolving a relationship that was poorly supported in all of the gene tree analyses. Most relationships among N. thebaica clades 1–6 are poorly supported and minimally diverged, consistent with the assignment of individuals from all six clades to N. thebaica (Supporting Information Figure S1). N. arge 1 is weakly supported as sister to the strongly supported grouping N. hispida/aurita + N. cf. hispida/aurita + N. grandis + N. tragata. Nycteris tragata, the only Asian species tested, is well supported within the African clades.
Figure 5.
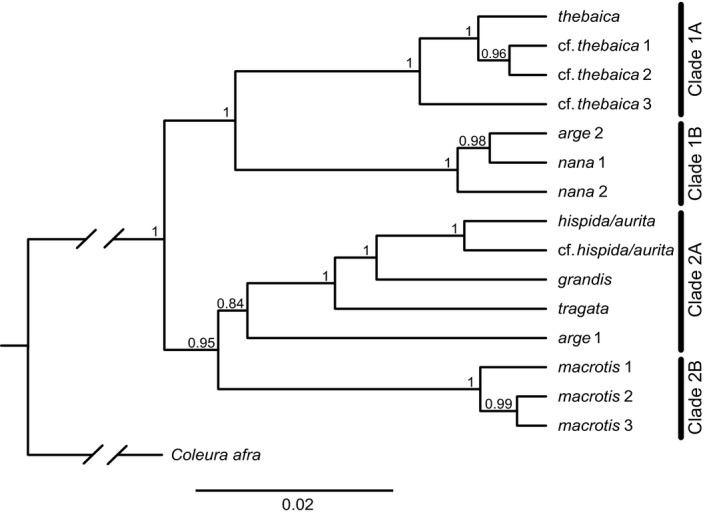
Species tree for Nycteris inferred using four nuclear loci in StarBEAST. Nodes are labeled with posterior probabilities
4. DISCUSSION
4.1. Multiple deeply diverged lineages
The monogeneric Nycteridae has been estimated to have diverged from Emballonuridae 51–53 Mya (Amador et al., 2018; Shi & Rabosky, 2015), and the most recent common ancestor age for the family has been placed variously at 18 mya (Shi & Rabosky, 2015) to 33.9 mya (Amador et al., 2018); Nycteridae ranks as a relatively ancient lineage among Chiroptera. Ours is the most taxonomically and geographically comprehensive phylogenetic study of Nycteridae to date. We recovered multiple instances of deep lineage divergence at both the inter‐ and intra‐clade levels. Mean pairwise uncorrected genetic distances among species‐ranked Nycteris clades for cytb were 0.17. In comparison, and in equivalent systematic surveys, overall cytb distances in Scotophilus (0.10; Demos et al., 2018) and Rhinolophus (0.10; Demos et al., 2019) were less than that of Nycteris. Overall mean genetic distances for concatenated intron datasets showed parallel variation: The mean distance of Nycteris was 0.04, Rhinolophus was 0.02, and Scotophilus was 0.01. As elaborated below, two deeply diverged multispecies clades are apparent in all of the phylogenetic analyses that we executed.
One of the most striking contrasts between the cytb gene tree (Figure 2d) and both the concatenated nuclear tree and species tree (Figure 4 and Supporting Information Figure S2) is the pattern of fine‐scale geographic structure for N. thebaica apparent only in the mitochondrial tree: There is strong support for monophyly of 5 of 6 labeled N. thebaica clades. Population‐level sampling recovered well‐supported and geographically restricted clades in (1) Kenya + Rwanda, (2) Tanzania. (3) Kenya + Uganda, (4) Kenya, and (5) Mozambique (Figure 3). The most divergent of these clades, N. thebaica clade 5 from Mozambique, is >5% cytb diverged from sister N. thebaica clades (Figure 2a, d). However, little population structure is present in either the concatenated nuclear analyses (Figure 4) or in the alternate species tree analysis where individuals were assigned to “species” based on clade membership in the mitochondrial tree (Supporting Information Figure S2). Although incomplete lineage sorting may be expected to play a role in mitonuclear discordance at this phylogenetic level, we note that other haplogroups did not exhibit such discordance at similar levels of divergence (e.g., N. arge 1 with subclades in West‐Central vs. East‐Central Africa, and N. tragata + N. javanica). This raises the possibility that the pattern results from sex‐biased dispersal within the N. thebaica species group. Monadjem (2005) longitudinal study of N. thebaica survivorship in Swaziland offers robust evidence for female philopatry and male‐biased dispersal. Of 39 females he banded as adults, nearly a quarter were living in the same culverts 4.5 years later, whereas only one of the 29 banded males was recaptured. Although other Nycteris dispersal studies are lacking, his observations are compatible with the strongly contrasting mitochondrial and nuclear population structures inferred here and warrant further life‐history studies of other Nycteris species. However, analyses using microsatellites or SNPs to exclude other possible explanations for this mitonuclear discordance would be necessary to establish this.
4.2. Phylogenetic relationships
Our analyses conflict with earlier efforts to resolve the phylogenetic relationships of Nycteris. The tree of Shi and Rabosky (2015) recovered the pair N. hispida and N. thebaica as sister to all Nycteris species; the remainder were arranged as N. javanica + N. tragata as sister to N. grandis + N. arge, with N. macrotis subtending this group. In contrast, Amador et al. (2018) recovered N. macrotis as the earliest diverging lineage of Nycteris, which was sister to a pair of clades, one containing the Asian species N. tragata and N. javanica and the other containing the African species N. grandis and N. arge as sisters, joined successively by N. hispida and N. thebaica. The two studies used the same 7 Nycteris species (arge, grandis, hispida, javanica, macrotis, thebaica, and tragata), but Amador et al. (2018) partitioned cytb and the two nuclear genes included in their analysis (vWF and BRCA) by codon position, whereas Shi and Rabosky partitioned their dataset by gene. All 7 Nycteris species in the concatenated ML analysis of Shi and Rabosky had BS support ≥70%, whereas the concatenated ML tree of Amador et al. (2018) more weakly supported N. macrotis as sister to the remaining Nycteris clades at 60%.
In contrast to both studies, we found strong support (PP 1.0) for two major subclades within the genus (Figures 4 and 5), each comprised of two groups of species. In the first subclade, N. thebaica and the three N. cf. thebaica clades form one group (Clade 1A), while N. arge clade 2 and the two N. nana clades comprise their sister (Clade 1B). In the second subclade, three N. macrotis clades comprise one group (Clade 2B) and N. tragata, N. grandis, N. hispida/aurita, and N. cf. hispida/aurita comprise the other (Clade 2A). Less securely placed in the latter group is N. arge 1 (PP = 0.84). Additional highly informative nuclear markers for bats (e.g., Dool et al., 2016; Demos et al., 2018) are likely responsible for improved resolution although better taxonomic and geographic sampling in this study may also contribute. To some extent, comparisons with these earlier investigations are limited by our conservative approach in withholding species assignment for specimens deemed cryptic and/or subtly differentiated from named taxa. That said, expanded taxonomic coverage alone, regardless of names assigned to terminals in the study, could be expected to result in conflicting topologies, as would possible incorrect species identifications from previous studies that relied on GenBank data.
Comparing the mitochondrial (Figure 2a), concatenated nuclear (Figure 4), and species trees (Figure 5) in our analyses, the only major inconsistency concerns the position of N. arge 2 + N. nana 1 + N. nana 2. The cytb gene tree analyses strongly support this clade as sister to N. macrotis, but the high genetic distances in this dataset raise the specter of substitutional saturation. In turn, the concatenated gene tree analyses infer poor support for the clade as sister to N. thebaica, whereas the species tree analyses strongly support the clade as sister to the N. thebaica group (PP = 1.0). Examination of relationships in both the concatenated nuclear and species trees, along with their substantial branch lengths, provide strong support for two major and four subordinate clades of species within Nycteris. The subordinate groupings represent species groups, as discussed below. The major clades have not previously been recognized, and the use of subgenera for these clades may be appropriate. As discussed by Teta (2019), there are several advantages of applying the category of subgenus to well‐supported clades. The category is recognized in zoological nomenclature at a rank intermediate between genus and species and regulated by the zoological code. Its use preserves binomial usage, and thus nomenclatural stability, and by joining closely related species it can be used to generate phylogenetic predictions (e.g., Teta, Cañón, Patterson, & Pardiñas, 2017; Voss, Gutiérrez, Solari, Rossi, & Jansa, 2014). Proposals to formally name these groups of Nycteris species should include the compilation of comprehensive morphological diagnoses, which is outside the purview of this study.
4.3. Species groups of Nycteris
The four subordinate clusters in the two subclades have been recognized since Andersen's (1912) first generic synopsis. Except for the position of the Asian taxa, they roughly correspond to his four species groups as they are currently defined (e.g., Happold, 2013b). All are separated by cytb distances of at least 16%, and their clade membership is strongly supported in the species tree. First, the cluster comprising Nycteris thebaica + N. cf. thebaica 1–3 (Clade 1A) is strongly supported as monophyletic in the species tree and is >17% cytb diverged from its sister. This group is distributed in northeastern, eastern, and southern Africa and, by definition, corresponds to the N. thebaica species group, although other assigned group members N. gambiensis and N. vinsoni were not explicitly included in our analyses. Second, and sister to the N. thebaica species group, is a cluster comprising N. arge 2 + N. nana 1 and 2 (Clade 1B), which is strongly supported as monophyletic and genetically distant (>17% cytb) from all other Nycteris. Distributed across western, Central, and eastern Africa, this grouping corresponds to the arge species group, although our analyses failed to include other group members N. intermedia and N. major (unless the former is in fact represented but mislabeled as N. nana 1 or N. nana 2). Third, the cluster comprising N. hispida/aurita, N. cf. hispida/aurita, N. grandis, and N. tragata (Clade 2A) is strongly supported as monophyletic and is >16% cytb diverged from the N. macrotis lineages that comprise its sister. This group is widely distributed; its African members correspond to the N. hispida species group but there is strong support for the additional membership of N. tragata from SE Asia. Although we did not sequence N. javanica for nuclear loci, the close relationship of N. javanica to N. tragata is well established (Amador et al., 2018; Shi & Rabosky, 2015; Figure 2a). Previous morphological indications that N. javanica and N. tragata were sister to the N. thebaica, N. hispida, and N. macrotis species groups (Griffiths, 1997) were clearly homoplasious. The relationship of N. arge 1 is uncertain, although it is weakly supported as sister to clade 2A in the species tree. Fourth, a final cluster comprises N. macrotis clades 1–3 (Clade 2B) and is strongly supported as monophyletic. It is >16% cytb diverged from its sister clade and includes members from South Sudan to Malawi and Mozambique east of the Albertine Rift and Congo Basin. It corresponds to the macrotis group, although our samples did not include identified representatives of N. madagascariensis, N. parisii, and N. woodi.
The fact that every newly sequenced Nycteris is associated with an identifiable museum voucher specimen means that forging linkages between genetic and morphological patterns is possible and because Nycteris taxa were all proposed on morphological grounds, this linkage enables sound nomenclature. Had the same genetic work been accomplished with biopsies from bats that were subsequently released, which is now technically possible, it would be impossible to confirm the identities and characterize the distinctive features of these lineages. As a case in point, lineages designated N. arge clades 1 and 2 (Figures 4 and 5) were each identified as N. arge in the field but clearly represent distinct lineages that likely belong to different species groups. Resolving the relationships of cryptic lineages is greatly expedited by comprehensive voucher material that preserves a broad array of biological characters, in the case of bats including skeletal and soft‐part anatomy, genitalia, vocalizations, and parasites, in addition to their genetic attributes (Gippoliti, 2018). Currently, 16 species of Nycteris are accepted as valid species, but several of these lack tissue samples in repositories or GenBank accessions and many lack vouchers with genetic material from near their type localities, hindering efforts to specify names (see Figure S1 in Supporting Information). Based on the number of well‐supported and deeply diverged lineages inferred here using multiple datasets and phylogenetic inference methods, it is likely that our analyses have uncovered several undescribed taxa.
The next steps in elucidating Nycteridae relationships will be in reconciling the phylogenetic patterns described in this paper with the extensive morphological analyses developed around Nycteris types and throughout their geographic distributions by Van Cakenberghe and de Vree (1985, 1993a, 1993b, 1998). Only then will it be possible to replace the various annotations on our figures with a robust binomial nomenclature.
Supporting information
Table S1. List of locality data for specimens used in genetic analyses of Nycteris.
Figure S1. Geographic sampling of genetic data used in this study. Plotting symbols denote the locations of one or more individuals represented by mitochondrial sequence (cytb) downloaded from GenBank (+), those represented only by cytb data newly generated for this study (open circles), and those where both mitochondrial and nuclear sequences were newly generated (filled circles). Taxon, localities, and coordinates for these points are included in Supporting Information Table S1.
Figure S2. Species tree inferred in StarBEAST for Nycteris for 21 clades, including Nycteris thebaica clades 1 to 6. Nodes are labeled with posterior probabilities.
ACKNOWLEDGEMENTS
Our analysis was strengthened with samples collected by the late W. T. Stanley, Carl Dick, Ruth Makena, David Wechuli, Richard Yego, and Aziza Zuhura. We acknowledge with special thanks the assistance of Jake Esselstyn and Donna Dittmann (LSUMNS), Caleb Phillips and Heath Garner (Museum of Texas Tech University), Mark Omura and Hopi Hoekstra (Harvard Museum of Comparative Zoology), and Jacqui Miller and Burton Lim (ROM) for loans of material. We thank Erwin Lagadec, Gildas Le Minter, Ara Monadjem, and Corrie Schoeman for collaboration in collecting specimens from Mozambique. We also salute the efforts of curators and collection managers in all the institutions cited in the Appendix 1 for maintaining the museum voucher specimens that enable subsequent integrative taxonomic studies needed to assign names confidently. We thank the Grainger Bioinformatics Center for partial funding of this study. Field collections in eastern and southern Africa were funded by a variety of agencies in cooperation with the Field Museum of Natural History, especially the JRS Biodiversity Foundation. Field Museum's Council on Africa, Marshall Field III Fund, and Barbara E. Brown Fund for Mammal Research were critical to fieldwork and analyses, as was the support of Bud and Onnolee Trapp and Walt and Ellen Newsom. Thanks to the John D. and Catherine T. MacArthur Foundation, Fulbright Program of US Department of State, Wildlife Conservation Society, and the Centers for Disease Control and Prevention sponsored and assisted in providing samples from DRC, Malawi, Mozambique, and Uganda. WWF Gabon supported fieldwork in Gabon, as did the Partenariat Mozambique‐Réunion dans la recherche en santé: pour une approche intégrée d'étude des maladies infectieuses à risque épidémique (MoZaR; Fond Européen de Développement Régional, Programme Opérationnel de Coopération Territoriale) in Mozambique. Comments from the reviewers, one of them exceptionally helpful on details, helped us improve the final draft of the manuscript.
APPENDIX 1.
List of specimens used in genetic analyses of Nycteris. Taxon names, voucher numbers, and GenBank accession numbers of sampled individuals of Nycteris: FMNH — Field Museum of Natural History, Chicago; LSUMZ — Louisiana State University, Museum of Natural Science; MHNG — Muséum d'Histoire Naturelle, Genève; NMK — National Museums of Kenya, Nairobi; ROM — Royal Ontario Museum, Toronto; TTU — Museum of Texas Tech University, Lubbock.
| Taxon | Voucher No. | cytb | ACOX2 | COPS7A | ROGDI | STAT5A |
|---|---|---|---|---|---|---|
| Coleura afra | FMNH 220403 | MK837103 | MK837325 | MK837394 | MK837464 | MK837534 |
| Nycteris arge 1 | FMNH 167763 | MK837079 | MK837329 | MK837398 | MK837468 | MK837538 |
| Nycteris arge 1 | FMNH 222429 | MK837077 | MK837327 | MK837396 | MK837466 | MK837536 |
| Nycteris arge 1 | FMNH 226934 | MK837078 | MK837328 | MK837397 | MK837467 | MK837537 |
| Nycteris arge 1 | FMNH 227433 | MK837076 | MK837326 | MK837395 | MK837465 | MK837535 |
| Nycteris arge 1 | FMNH 232918 | MK837080 | ||||
| Nycteris arge 2 | FMNH 149405 | MK837081 | MK837330 | MK837399 | MK837469 | MK837539 |
| Nycteris arge 2 | FMNH 215539 | MK837083 | ||||
| Nycteris arge 2 | FMNH 215540 | MK837084 | ||||
| Nycteris arge 2 | FMNH 222430 | MK837082 | ||||
| Nycteris arge 2 | FMNH 224102 | MK837088 | MK837332 | MK837401 | MK837471 | MK837541 |
| Nycteris arge 2 | FMNH 224103 | MK837089 | ||||
| Nycteris arge 2 | FMNH 224104 | MK837090 | ||||
| Nycteris arge 2 | NMK 184961 | MK837085 | MK837331 | MK837400 | MK837470 | MK837540 |
| Nycteris arge 2 | NMK 184967 | MK837086 | ||||
| Nycteris arge 2 | NMK 187405 | MK837087 | ||||
| Nycteris cf. hispida/aurita | FMNH 187139 | MK837094 | MK837335 | MK837404 | MK837474 | MK837544 |
| Nycteris cf. hispida/aurita | FMNH 220978 | MK837091 | MK837333 | MK837402 | MK837472 | MK837542 |
| Nycteris cf. hispida/aurita | FMNH 220979 | MK837092 | MK837334 | MK837403 | MK837473 | MK837543 |
| Nycteris cf. hispida/aurita | FMNH 220982 | MK837093 | ||||
| Nycteris cf. thebaica 1 | FMNH 195603 | MK837096 | MK837337 | MK837406 | MK837476 | MK837546 |
| Nycteris cf. thebaica 1 | FMNH 195604 | MK837097 | ||||
| Nycteris cf. thebaica 1 | FMNH 195605 | MK837098 | MK837338 | MK837407 | MK837477 | MK837547 |
| Nycteris cf. thebaica 1 | FMNH 195606 | MK837099 | ||||
| Nycteris cf. thebaica 1 | FMNH 226239 | MK837095 | MK837336 | MK837405 | MK837475 | MK837545 |
| Nycteris cf. thebaica 2 | NMK 184231 | MK837100 | MK837339 | MK837408 | MK837478 | MK837548 |
| Nycteris cf. thebaica 2 | NMK 184384 | MK837101 | MK837340 | MK837409 | MK837479 | MK837549 |
| Nycteris cf. thebaica 3 | FMNH 158336 | MK837102 | MK837341 | MK837410 | MK837480 | MK837550 |
| Nycteris grandis | FMNH 150065 | MK837111 | ||||
| Nycteris grandis | FMNH 151187 | MK837112 | MK837346 | MK837415 | MK837485 | MK837555 |
| Nycteris grandis | FMNH 151188 | MK837113 | ||||
| Nycteris grandis | FMNH 151189 | MK837114 | ||||
| Nycteris grandis | FMNH 151190 | MK837115 | ||||
| Nycteris grandis | FMNH 151416 | MK837116 | ||||
| Nycteris grandis | FMNH 168092 | MK837117 | ||||
| Nycteris grandis | FMNH 192814 | MK837118 | ||||
| Nycteris grandis | FMNH 192815 | MK837119 | ||||
| Nycteris grandis | FMNH 192816 | MK837120 | ||||
| Nycteris grandis | FMNH 192882 | MK837121 | ||||
| Nycteris grandis | FMNH 192883 | MK837122 | MK837347 | MK837416 | MK837486 | MK837556 |
| Nycteris grandis | FMNH 192884 | MK837123 | ||||
| Nycteris grandis | FMNH 192885 | MK837124 | ||||
| Nycteris grandis | FMNH 192936 | MK837125 | ||||
| Nycteris grandis | FMNH 213625 | MK837110 | MK837345 | MK837414 | MK837484 | MK837554 |
| Nycteris grandis | FMNH 219603 | MK837107 | MK837343 | MK837412 | MK837482 | MK837552 |
| Nycteris grandis | FMNH 222427 | MK837108 | MK837344 | MK837413 | MK837483 | MK837553 |
| Nycteris grandis | FMNH 222428 | MK837109 | ||||
| Nycteris grandis | FMNH 227439 | MK837104 | ||||
| Nycteris grandis | FMNH 227441 | MK837105 | MK837342 | MK837411 | MK837481 | MK837551 |
| Nycteris grandis | FMNH 227442 | MK837106 | ||||
| Nycteris hispida/aurita | FMNH 137625 | MK837140 | ||||
| Nycteris hispida/aurita | FMNH 137626 | MK837141 | ||||
| Nycteris hispida/aurita | FMNH 151191 | MK837139 | MK837352 | MK837421 | MK837491 | MK837561 |
| Nycteris hispida/aurita | FMNH 165131 | MK837142 | ||||
| Nycteris hispida/aurita | FMNH 195607 | MK837138 | MK837351 | MK837420 | MK837490 | MK837560 |
| Nycteris hispida/aurita | FMNH 215546 | MK837130 | ||||
| Nycteris hispida/aurita | FMNH 215547 | MK837131 | ||||
| Nycteris hispida/aurita | FMNH 215548 | MK837132 | ||||
| Nycteris hispida/aurita | FMNH 220746 | MK837133 | ||||
| Nycteris hispida/aurita | FMNH 220980 | MK837127 | ||||
| Nycteris hispida/aurita | FMNH 225217 | MK837135 | ||||
| Nycteris hispida/aurita | FMNH 225218 | MK837136 | ||||
| Nycteris hispida/aurita | FMNH 225240 | MK837137 | MK837350 | MK837419 | MK837489 | MK837559 |
| Nycteris hispida/aurita | FMNH 225445 | MK837134 | ||||
| Nycteris hispida/aurita | FMNH 227445 | MK837126 | MK837348 | MK837417 | MK837487 | MK837557 |
| Nycteris hispida/aurita | FMNH 232892 | MK837143 | ||||
| Nycteris hispida/aurita | FMNH 232893 | MK837144 | ||||
| Nycteris hispida/aurita | FMNH 232902 | MK837145 | ||||
| Nycteris hispida/aurita | FMNH 232903 | MK837146 | MK837353 | MK837422 | MK837492 | MK837562 |
| Nycteris hispida/aurita | FMNH 232904 | MK837317 | ||||
| Nycteris hispida/aurita | FMNH 232905 | MK837147 | ||||
| Nycteris hispida/aurita | FMNH 232906 | MK837148 | ||||
| Nycteris hispida/aurita | FMNH 232908 | MK837149 | ||||
| Nycteris hispida/aurita | MHNG 1971.039 | HQ693722 | ||||
| Nycteris hispida/aurita | MHNG 1971.04 | HQ693723 | ||||
| Nycteris hispida/aurita | NMK 184937 | MK837128 | ||||
| Nycteris hispida/aurita | NMK 184976 | MK837129 | MK837349 | MK837418 | MK837488 | MK837558 |
| Nycteris javanica | ROM 101970 | EF584225 | ||||
| Nycteris macrotis 1 | FMNH 192937 | MK837170 | ||||
| Nycteris macrotis 1 | FMNH 215541 | MK837150 | MK837354 | MK837423 | MK837493 | MK837563 |
| Nycteris macrotis 1 | FMNH 216029 | MK837151 | ||||
| Nycteris macrotis 1 | FMNH 216030 | MK837152 | MK837355 | MK837424 | MK837494 | MK837564 |
| Nycteris macrotis 1 | FMNH 216031 | MK837153 | ||||
| Nycteris macrotis 1 | FMNH 216032 | MK837154 | ||||
| Nycteris macrotis 1 | FMNH 216033 | MK837155 | ||||
| Nycteris macrotis 1 | FMNH 216034 | MK837160 | ||||
| Nycteris macrotis 1 | FMNH 216035 | MK837161 | MK837356 | MK837425 | MK837495 | MK837565 |
| Nycteris macrotis 1 | FMNH 216036 | MK837158 | ||||
| Nycteris macrotis 1 | FMNH 219068 | MK837171 | ||||
| Nycteris macrotis 1 | FMNH 219069 | MK837172 | MK837359 | MK837428 | MK837498 | MK837568 |
| Nycteris macrotis 1 | FMNH 219239 | MK837173 | ||||
| Nycteris macrotis 1 | FMNH 220492 | MK837156 | ||||
| Nycteris macrotis 1 | FMNH 220494 | MK837157 | ||||
| Nycteris macrotis 1 | FMNH 220742 | MK837318 | ||||
| Nycteris macrotis 1 | FMNH 220744 | MK837162 | ||||
| Nycteris macrotis 1 | FMNH 220745 | MK837319 | ||||
| Nycteris macrotis 1 | FMNH 220977 | MK837163 | MK837357 | MK837426 | MK837496 | MK837566 |
| Nycteris macrotis 1 | FMNH 220981 | MK837159 | ||||
| Nycteris macrotis 1 | FMNH 223200 | MK837174 | ||||
| Nycteris macrotis 1 | FMNH 223660 | MK837175 | ||||
| Nycteris macrotis 1 | FMNH 232911 | MK837176 | ||||
| Nycteris macrotis 1 | FMNH 232912 | MK837177 | MK837360 | MK837429 | MK837499 | MK837569 |
| Nycteris macrotis 1 | FMNH 232913 | MK837178 | ||||
| Nycteris macrotis 1 | FMNH 232914 | MK837179 | ||||
| Nycteris macrotis 1 | FMNH 232915 | MK837180 | ||||
| Nycteris macrotis 1 | FMNH 232916 | MK837320 | ||||
| Nycteris macrotis 1 | FMNH 232917 | MK837181 | MK837361 | MK837430 | MK837500 | MK837570 |
| Nycteris macrotis 1 | FMNH HB115 | MK837166 | MK837358 | MK837427 | MK837497 | MK837567 |
| Nycteris macrotis 1 | FMNH HB121 | MK837167 | ||||
| Nycteris macrotis 1 | FMNH HB122 | MK837168 | ||||
| Nycteris macrotis 1 | FMNH HB124 | MK837169 | ||||
| Nycteris macrotis 1 | FMNH HB61 | MK837164 | ||||
| Nycteris macrotis 1 | FMNH HB62 | MK837165 | ||||
| Nycteris macrotis 2 | FMNH 220739 | MK837182 | ||||
| Nycteris macrotis 2 | FMNH 220740 | MK837321 | MK837362 | MK837431 | MK837501 | MK837571 |
| Nycteris macrotis 3 | FMNH 226237 | MK837183 | MK837363 | MK837432 | MK837502 | MK837572 |
| Nycteris macrotis 3 | FMNH 228897 | MK837184 | MK837364 | MK837433 | MK837503 | MK837573 |
| Nycteris macrotis 3 | FMNH 228898 | MK837185 | MK837365 | MK837434 | MK837504 | MK837574 |
| Nycteris macrotis 3 | FMNH 228899 | MK837186 | ||||
| Nycteris macrotis 3 | FMNH 228900 | MK837187 | ||||
| Nycteris macrotis 3 | FMNH 228901 | MK837188 | ||||
| Nycteris nana 1 | FMNH 227448 | MK837189 | MK837366 | MK837435 | MK837505 | MK837575 |
| Nycteris nana 2 | FMNH 167764 | MK837191 | MK837368 | MK837437 | MK837507 | MK837577 |
| Nycteris nana 2 | FMNH 227446 | MK837190 | MK837367 | MK837436 | MK837506 | MK837576 |
| Nycteris thebaica 1 | FMNH 225210 | MK837236 | ||||
| Nycteris thebaica 1 | FMNH 225241 | MK837237 | MK837372 | MK837441 | MK837511 | MK837581 |
| Nycteris thebaica 1 | FMNH 225242 | MK837238 | MK837373 | MK837442 | MK837512 | MK837582 |
| Nycteris thebaica 1 | FMNH 225243 | MK837239 | ||||
| Nycteris thebaica 1 | FMNH 225244 | MK837240 | ||||
| Nycteris thebaica 1 | FMNH 225245 | MK837241 | MK837374 | MK837443 | MK837513 | MK837583 |
| Nycteris thebaica 1 | FMNH 225406 | MK837192 | ||||
| Nycteris thebaica 1 | FMNH 225407 | MK837193 | ||||
| Nycteris thebaica 1 | FMNH 225408 | MK837194 | ||||
| Nycteris thebaica 1 | FMNH 225409 | MK837195 | ||||
| Nycteris thebaica 1 | FMNH 225410 | MK837196 | ||||
| Nycteris thebaica 1 | FMNH 225411 | MK837197 | ||||
| Nycteris thebaica 1 | FMNH 225412 | MK837198 | ||||
| Nycteris thebaica 1 | FMNH 225413 | MK837199 | ||||
| Nycteris thebaica 1 | FMNH 225420 | MK837200 | ||||
| Nycteris thebaica 1 | FMNH 225421 | MK837201 | ||||
| Nycteris thebaica 1 | FMNH 225423 | MK837202 | ||||
| Nycteris thebaica 1 | FMNH 225424 | MK837203 | ||||
| Nycteris thebaica 1 | FMNH 225425 | MK837204 | ||||
| Nycteris thebaica 1 | FMNH 225426 | MK837205 | ||||
| Nycteris thebaica 1 | FMNH 225442 | MK837206 | MK837369 | MK837438 | MK837508 | MK837578 |
| Nycteris thebaica 1 | FMNH 225443 | MK837207 | ||||
| Nycteris thebaica 1 | FMNH 225444 | MK837208 | ||||
| Nycteris thebaica 1 | FMNH 225446 | MK837209 | ||||
| Nycteris thebaica 1 | FMNH 225447 | MK837210 | ||||
| Nycteris thebaica 1 | FMNH 225448 | MK837211 | ||||
| Nycteris thebaica 1 | FMNH 225449 | MK837212 | ||||
| Nycteris thebaica 1 | FMNH 225450 | MK837213 | ||||
| Nycteris thebaica 1 | FMNH 225451 | MK837214 | ||||
| Nycteris thebaica 1 | FMNH 225452 | MK837215 | ||||
| Nycteris thebaica 1 | FMNH 225453 | MK837216 | ||||
| Nycteris thebaica 1 | FMNH 225454 | MK837217 | ||||
| Nycteris thebaica 1 | FMNH 225455 | MK837218 | ||||
| Nycteris thebaica 1 | FMNH 225456 | MK837219 | ||||
| Nycteris thebaica 1 | FMNH 225457 | MK837220 | ||||
| Nycteris thebaica 1 | FMNH 225458 | MK837221 | MK837370 | MK837439 | MK837509 | MK837579 |
| Nycteris thebaica 1 | FMNH 225459 | MK837222 | ||||
| Nycteris thebaica 1 | FMNH 225460 | MK837223 | ||||
| Nycteris thebaica 1 | FMNH 225461 | MK837224 | ||||
| Nycteris thebaica 1 | FMNH 225462 | MK837225 | ||||
| Nycteris thebaica 1 | FMNH 225463 | MK837226 | ||||
| Nycteris thebaica 1 | FMNH 225464 | MK837227 | ||||
| Nycteris thebaica 1 | FMNH 225465 | MK837322 | ||||
| Nycteris thebaica 1 | FMNH 225466 | MK837228 | ||||
| Nycteris thebaica 1 | FMNH 225467 | MK837229 | ||||
| Nycteris thebaica 1 | FMNH 225468 | MK837230 | ||||
| Nycteris thebaica 1 | FMNH 225469 | MK837323 | ||||
| Nycteris thebaica 1 | FMNH 225470 | MK837231 | ||||
| Nycteris thebaica 1 | FMNH 225471 | MK837232 | ||||
| Nycteris thebaica 1 | FMNH 225472 | MK837233 | MK837371 | MK837440 | MK837510 | MK837580 |
| Nycteris thebaica 1 | FMNH 225473 | MK837234 | ||||
| Nycteris thebaica 1 | FMNH 225474 | MK837235 | ||||
| Nycteris thebaica 2 | FMNH 147220 | MK837242 | MK837375 | MK837444 | MK837514 | MK837584 |
| Nycteris thebaica 2 | FMNH 198085 | MK837243 | MK837376 | MK837445 | MK837515 | MK837585 |
| Nycteris thebaica 2 | FMNH 198086 | MK837244 | ||||
| Nycteris thebaica 2 | FMNH 198087 | MK837245 | ||||
| Nycteris thebaica 2 | FMNH 198088 | MK837246 | MK837377 | MK837446 | MK837516 | MK837586 |
| Nycteris thebaica 2 | FMNH 198089 | MK837247 | ||||
| Nycteris thebaica 3 | FMNH 215536 | MK837324 | ||||
| Nycteris thebaica 3 | FMNH 215537 | MK837249 | ||||
| Nycteris thebaica 3 | FMNH 215538 | MK837250 | MK837378 | MK837447 | MK837517 | MK837587 |
| Nycteris thebaica 3 | FMNH 215542 | MK837259 | ||||
| Nycteris thebaica 3 | FMNH 215543 | MK837260 | ||||
| Nycteris thebaica 3 | FMNH 215544 | MK837261 | ||||
| Nycteris thebaica 3 | FMNH 215545 | MK837262 | ||||
| Nycteris thebaica 3 | FMNH 220741 | MK837258 | ||||
| Nycteris thebaica 3 | FMNH 225400 | MK837263 | ||||
| Nycteris thebaica 3 | FMNH 225401 | MK837264 | ||||
| Nycteris thebaica 3 | FMNH 225402 | MK837265 | ||||
| Nycteris thebaica 3 | FMNH 225403 | MK837266 | MK837381 | MK837450 | MK837520 | MK837590 |
| Nycteris thebaica 3 | FMNH 225404 | MK837267 | ||||
| Nycteris thebaica 3 | FMNH 225405 | MK837268 | ||||
| Nycteris thebaica 3 | FMNH 232109 | MK837277 | ||||
| Nycteris thebaica 3 | FMNH 232429 | MK837278 | MK837383 | MK837452 | MK837522 | MK837592 |
| Nycteris thebaica 3 | FMNH 232430 | MK837279 | ||||
| Nycteris thebaica 3 | FMNH 232431 | MK837280 | ||||
| Nycteris thebaica 3 | FMNH 232919 | MK837281 | ||||
| Nycteris thebaica 3 | NMK 184407 | MK837253 | MK837379 | MK837448 | MK837518 | MK837588 |
| Nycteris thebaica 3 | NMK 184520 | MK837254 | MK837380 | MK837449 | MK837519 | MK837589 |
| Nycteris thebaica 3 | NMK 184521 | MK837255 | ||||
| Nycteris thebaica 3 | NMK 184522 | MK837256 | ||||
| Nycteris thebaica 3 | NMK 184636 | MK837257 | ||||
| Nycteris thebaica 3 | NMK 184658 | MK837248 | ||||
| Nycteris thebaica 3 | NMK 184759 | MK837269 | ||||
| Nycteris thebaica 3 | NMK 184854 | MK837270 | ||||
| Nycteris thebaica 3 | NMK 184855 | MK837271 | ||||
| Nycteris thebaica 3 | NMK 185133 | MK837252 | ||||
| Nycteris thebaica 3 | NMK 187337 | MK837272 | MK837382 | MK837451 | MK837521 | MK837591 |
| Nycteris thebaica 3 | NMK 187338 | MK837273 | ||||
| Nycteris thebaica 3 | NMK 187339 | MK837274 | ||||
| Nycteris thebaica 3 | NMK 187340 | MK837275 | ||||
| Nycteris thebaica 3 | NMK 187341 | MK837276 | ||||
| Nycteris thebaica 3 | NMK 187450 | MK837251 | ||||
| Nycteris thebaica 4 | FMNH 216037 | MK837282 | MK837386 | MK837455 | MK837525 | MK837595 |
| Nycteris thebaica 4 | FMNH 216039 | MK837283 | MK837456 | MK837526 | MK837596 | |
| Nycteris thebaica 4 | FMNH 216040 | MK837284 | ||||
| Nycteris thebaica 4 | FMNH 216042 | MK837285 | ||||
| Nycteris thebaica 4 | FMNH 216043 | MK837286 | ||||
| Nycteris thebaica 4 | FMNH 220446 | MK837288 | ||||
| Nycteris thebaica 4 | FMNH 220447 | MK837287 | ||||
| Nycteris thebaica 4 | FMNH 220448 | MK837289 | ||||
| Nycteris thebaica 4 | FMNH 220449 | MK837290 | ||||
| Nycteris thebaica 4 | FMNH 220450 | MK837291 | ||||
| Nycteris thebaica 4 | FMNH 220451 | MK837292 | ||||
| Nycteris thebaica 4 | FMNH 220452 | MK837293 | ||||
| Nycteris thebaica 4 | FMNH 220453 | MK837294 | MK837384 | MK837453 | MK837523 | MK837593 |
| Nycteris thebaica 4 | FMNH 220455 | MK837295 | ||||
| Nycteris thebaica 4 | FMNH 220484 | MK837296 | MK837385 | MK837454 | MK837524 | MK837594 |
| Nycteris thebaica 4 | FMNH 220485 | MK837297 | ||||
| Nycteris thebaica 4 | FMNH 220486 | MK837298 | ||||
| Nycteris thebaica 4 | FMNH 220487 | MK837299 | ||||
| Nycteris thebaica 4 | FMNH 220488 | MK837300 | ||||
| Nycteris thebaica 5 | FMNH 213626 | MK837301 | MK837387 | MK837457 | MK837527 | MK837597 |
| Nycteris thebaica 5 | FMNH 213627 | MK837302 | MK837388 | MK837458 | MK837528 | MK837598 |
| Nycteris thebaica 5 | FMNH 213628 | MK837303 | MK837389 | MK837459 | MK837529 | MK837599 |
| Nycteris thebaica 5 | FMNH 213629 | MK837304 | ||||
| Nycteris thebaica 5 | FMNH 213630 | MK837305 | ||||
| Nycteris thebaica 5 | FMNH 213631 | MK837306 | ||||
| Nycteris thebaica 5 | FMNH 213632 | MK837307 | ||||
| Nycteris thebaica 6 | FMNH 187360 | MK837310 | ||||
| Nycteris thebaica 6 | FMNH 187361 | MK837311 | MK837391 | MK837461 | MK837531 | MK837601 |
| Nycteris thebaica 6 | FMNH 187412 | MK837312 | ||||
| Nycteris thebaica 6 | FMNH 193210 | MK837313 | ||||
| Nycteris thebaica 6 | FMNH 219066 | MK837314 | ||||
| Nycteris thebaica 6 | FMNH 219067 | MK837315 | MK837392 | MK837462 | MK837532 | MK837602 |
| Nycteris thebaica 6 | FMNH 226238 | MK837308 | MK837390 | MK837460 | MK837530 | MK837600 |
| Nycteris thebaica 6 | FMNH 226240 | MK837309 | ||||
| Nycteris tragata | LSUMZ 4413 | MK837316 | MK837393 | MK837463 | MK837533 | MK837603 |
| Nycteris tragata | TTU 108180 | EU21624 |
Demos TC, Webala PW, Kerbis Peterhans JC, Goodman SM, Bartonjo M, Patterson BD. Molecular phylogenetics of slit‐faced bats (Chiroptera: Nycteridae) reveal deeply divergent African lineages. J Zool Syst Evol Res. 2019;57:1019–1038. 10.1111/jzs.12313
Contributing authors: Paul W. Webala (paul.webala@gmail.com), Julian C. Kerbis Peterhans (jkerbis@fieldmuseum.org), Steven M. Goodman (sgoodman@fieldmuseum.org), Michael Bartonjo (abartonjo@yahoo.com), Bruce D. Patterson (bpatterson@fieldmuseum.org)
REFERENCES
- Aellen, V. (1959). Chiroptères nouveaux d'Afrique. Archives Des Sciences, Genève, 12(2), 217–235. [Google Scholar]
- Amador, L. I. , Moyers Arévalo, R. L. , Almeida, F. C. , Catalano, S. A. , & Giannini, N. P. (2018). Bat systematics in the light of unconstrained analyses of a comprehensive molecular supermatrix. Journal of Mammalian Evolution, 25(1), 37–70. 10.1007/s10914-016-9363-8 [DOI] [Google Scholar]
- Andersen, K. (1912). Brief diagnoses of eight new Petalia, with a list of the known forms of the genus. Annals & Magazine of Natural History, Series, 8(10), 546–550. [Google Scholar]
- Bouckaert, R. , Heled, J. , Kühnert, D. , Vaughan, T. , Wu, C.‐H. , Xie, D. , … Drummond, A. J. (2014). BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Computational Biology, 10(4), e1003537 10.1371/journal.pcbi.1003537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalquest, W. W. (1965). Mammals from the Save River, Mozambique, with descriptions of two new bats. Journal of Mammalogy, 46(2), 254–264. 10.2307/1377845 [DOI] [Google Scholar]
- Demos, T. C. , Webala, P. W. , Bartonjo, M. , & Patterson, B. D. (2018). Hidden diversity of African Yellow house bats (Vespertilionidae, Scotophilus): Insights from multilocus phylogenetics and lineage delimitation. Frontiers in Ecology and Evolution, 6, 86 10.3389/fevo.2018.00086 [DOI] [Google Scholar]
- Demos, T. C. , Webala, P. W. , Goodman, S. M. , Kerbis Peterhans, J. C. , Bartonjo, M. , & Patterson, B. D. (2019). Molecular phylogenetics of the African horseshoe bats (Chiroptera: Rhinolophidae): expanded geographic and taxonomic sampling of the Afrotropics. BMC Evolutionary Biology. 10.1186/s12862-019-1485-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dool, S. E. , Puechmaille, S. J. , Foley, N. M. , Allegrini, B. , Bastian, A. , Mutumi, G. L. , … Jacobs, D. S. (2016). Nuclear introns outperform mitochondrial DNA in inter‐specific phylogenetic reconstruction: Lessons from horseshoe bats (Rhinolophidae: Chiroptera). Molecular Phylogenetics and Evolution, 97, 196–212. 10.1016/j.ympev.2016.01.003 [DOI] [PubMed] [Google Scholar]
- Eick, G. N. , Jacobs, D. S. , & Matthee, C. A. (2005). A nuclear DNA phylogenetic perspective on the evolution of echolocation and historical biogeography of extant bats (Chiroptera). Molecular Biology and Evolution, 22, 1869–1886. 10.1093/molbev/msi180 [DOI] [PubMed] [Google Scholar]
- Flot, J. F. (2010). SEQPHASE: A web tool for interconverting PHASE input/output files and FASTA sequence alignments. Molecular Ecology Resources, 10(1), 162–166. 10.1111/j.1755-0998.2009.02732.x [DOI] [PubMed] [Google Scholar]
- Garrick, R. C. , Sunnucks, P. , & Dyer, R. J. (2010). Nuclear gene phylogeography using PHASE: Dealing with unresolved genotypes, lost alleles, and systematic bias in parameter estimation. BMC Evolutionary Biology, 10(1), 118 10.1186/1471-2148-10-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gippoliti, S. (2018). Natural history collecting and the arrogance of the modern Ark researcher. Bionomina, 13(1), 69–73. 10.11646/bionomina.13.1.6 [DOI] [Google Scholar]
- Griffiths, T. A. (1997). Phylogenetic position of the bat Nycteris javanica (Chiroptera: Nycteridae). Journal of Mammalogy, 78(1), 106–116. 10.2307/1382644 [DOI] [Google Scholar]
- Happold, M. (2013a). Family Nycteridae: Slit‐faced bats In Happold M., & Happold D. C. D. (Eds.), The Mammals of Africa, Vol. IV: Hedgehogs, Shrews and Bats (pp. 438–439). London, UK: Bloomsbury Publishing. [Google Scholar]
- Happold, M. (2013b). Genus Nycteris: Slit‐faced bats In Happold M., & Happold D. C. D. (Eds.), The Mammals of Africa, Vol. IV: Hedgehogs, Shrews and Bats (pp. 440–442). London, UK: Bloomsbury Publishing. [Google Scholar]
- Hassanin, A. , Colombo, R. , Gembu, G. C. , Merle, M. , Tu, V. T. , Görföl, T. , … Ing, R. K. (2018). Multilocus phylogeny and species delimitation within the genus Glauconycteris (Chiroptera, Vespertilionidae), with the description of a new bat species from the Tshopo Province of the Democratic Republic of the Congo. Journal of Zoological Systematics and Evolutionary Research, 56(1), 1–22. 10.1111/jzs.12176 [DOI] [Google Scholar]
- Koopman, K. F. (1993). Order Chiroptera In Wilson D. E., & Reeder D. A. M. (Eds.), Mammal species of the World: A taxonomic and geographic reference, 2nd ed. (pp. 137–242). Washington, DC: Smithsonian Institution Press. [Google Scholar]
- Kumar, S. , Stecher, G. , Li, M. , Knyaz, C. , & Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35(6), 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear, R. , Frandsen, P. B. , Wright, A. M. , Senfeld, T. , & Calcott, B. (2016). PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution, 34(3), 772–773. 10.1093/molbev/msw260 [DOI] [PubMed] [Google Scholar]
- Leigh, J. W. , & Bryant, D. (2015). POPART: Full‐feature software for haplotype network construction. Methods in Ecology and Evolution, 6(9), 1110–1116. 10.1111/2041-210X.12410 [DOI] [Google Scholar]
- Mammal Diversity Database (2019). American Society of Mammalogists. Retrieved from www.mammaldiversity.org. Accessed 2019–01‐10. [Google Scholar]
- Miller, M. A. , Pfeiffer, W. , & Schwartz, T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE). In. New Orleans, LA: IEEE. doi: 10.1109/GCE.2010.5676129 [DOI] [Google Scholar]
- Monadjem, A. (2005). Survival and roost‐site selection in the African bat Nycteris thebaica (Chiroptera : Nycteridae) in Swaziland. Belgian Journal of Zoology, 135(Suppl.), 103–107. [Google Scholar]
- Monadjem, A. , Taylor, P. J. , Cotterill, F. P. D. , & Schoeman, M. C. (2010). Bats of Southern and Central Africa: A biogeographic and taxonomic synthesis. Johannesburg, South Africa: Wits University Press: i‐xii; 1‐596. [Google Scholar]
- Nguyen, L.‐T. , Schmidt, H. A. , von Haeseler, A. , & Minh, B. Q. (2015). IQ‐TREE: A fast and effective stochastic algorithm for estimating maximum‐likelihood phylogenies. Molecular Biology and Evolution, 32(1), 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie, H. A. , Bouckaert, R. R. , & Drummond, A. J. (2017). StarBEAST2 brings faster species tree inference and accurate estimates of substitution rates. Molecular Biology and Evolution, 34(8), 2101–2114. 10.1093/molbev/msx126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, B. D. , & Webala, P. W. (2012). Keys to the bats (Mammalia: Chiroptera) of East Africa. Fieldiana: Life and Earth Sciences, 6, 1–60. 10.3158/2158-5520-12.6.1 [DOI] [Google Scholar]
- Patterson, B. D. , Webala, P. W. , Bartonjo, M. , Nziza, J. , Dick, C. W. , & Demos, T. C. (2018). On the taxonomic status and distribution of African species of Otomops (Chiroptera: Molossidae). PeerJ, 6, e4864 10.7717/peerj.4864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut, A. , Drummond, A. J. , Xie, D. , Baele, G. , & Suchard, M. A. (2018). Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Systematic Biology, 67(5), 901–904. 10.1093/sysbio/syy032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist, F. , Teslenko, M. , van der Mark, P. , Ayres, D. L. , Darling, A. , Höhna, S. , … Huelsenbeck, J. P. (2012). MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61(3), 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salicini, I. , Ibáñez, C. , & Juste, J. (2011). Multilocus phylogeny and species delimitation within the Natterer’s bat species complex in the Western Palearctic. Molecular Phylogenetics and Evolution, 61(3), 888–898. 10.1016/j.ympev.2011.08.010 [DOI] [PubMed] [Google Scholar]
- Shi, J. J. , & Rabosky, D. L. (2015). Speciation dynamics during the global radiation of extant bats. Evolution, 69(6), 1528–1545. 10.1111/evo.12681 [DOI] [PubMed] [Google Scholar]
- Sikes, R. S. (2016). 2016 Guidelines of the American Society of Mammalogists for the use of wild mammals in research and education. Journal of Mammalogy, 97(3), 663–688. 10.1093/jmammal/gyw078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons, N. B. (2005). Chiroptera In Wilson D. E., & Reeder D. A. M. (Eds.), Mammal species of the world: A taxonomic and geographic reference, Vol. 1, 3rd ed. (pp. 312–529). Baltimore, MD: Johns Hopkins University Press. [Google Scholar]
- Stephens, M. , Smith, N. J. , & Donnelly, P. (2001). A new statistical method for haplotype reconstruction from population data. The American Journal of Human Genetics, 68(4), 978–989. 10.1086/319501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teta, P. (2019). The usage of subgenera in mammalian taxonomy. Mammalia, 83(3), 209–211. 10.1515/mammalia-2018-0059. [DOI] [Google Scholar]
- Teta, P. , Cañón, C. , Patterson, B. D. , & Pardiñas, U. F. J. (2017). Phylogeny of the tribe Abrotrichini (Cricetidae, Sigmodontinae): Integrating morphological and molecular evidence into a new classification. Cladistics, 33(2), 153–182. 10.1111/cla.12164 [DOI] [PubMed] [Google Scholar]
- Thomas, N. M. , Harrison, D. L. , & Bates, P. J. J. (1994). A study of the baculum in the genus Nycteris (Mammalia, Chiroptera, Nycteridae) with consideration of its taxonomic importance. Bonner Zoologische Beiträge, 45(1), 17–31. [Google Scholar]
- Thorn, E. , Kerbis Peterhans, J. C. , & Baranga, J. (2009). Chiroptera In Thorn E., & Peterhans J. C. K. (Eds.), Small Mammals of Uganda. Bonner Zoologische Monographien 55 (pp. 12–75). Bonn, Germany: Museum Alexander Koenig. [Google Scholar]
- Trujillo, R. G. , Patton, J. C. , Schlitter, D. A. , & Bickham, J. W. (2009). Molecular phylogenetics of the bat genus Scotophilus (Chiroptera: Vespertilionidae): Perspectives from paternally and maternally inherited genomes. Journal of Mammalogy, 90(3), 548–560. 10.1644/08-MAMM-A-239R2.1 [DOI] [Google Scholar]
- Van Cakenberghe, V. , & de Vree, F. (1985). Systematics of African Nycteris (Mammalia: Chiroptera) In Schuchmann K.‐L. (Ed.), Proceedings of the International Symposium on African Vertebrates (pp. 53‐90). Berlin, Germany: Museum Alexander Koenig. [Google Scholar]
- Van Cakenberghe, V. , & De Vree, F. (1993a). The systematic status of Southeast Asian Nycteris (Chiroptera: Nycteridae). Mammalia, 57(2), 227–244. 10.1515/mamm.1993.57.2.227 [DOI] [Google Scholar]
- Van Cakenberghe, V. , & de Vree, F. (1993b). Systematics of African Nycteris (Mammalia: Chiroptera) Part II. The Nycteris hispida group. Bonner Zoologische Beiträge, 44(3–4), 299–332. [Google Scholar]
- Van Cakenberghe, V. , & de Vree, F. (1998). Systematics of African Nycteris (Mammalia: Chiroptera) Part III. The Nyteris thebaica group. Bonner Zoologische Beiträge, 48(2), 123–166. [Google Scholar]
- Voss, R. S. , Gutiérrez, E. E. , Solari, S. , Rossi, R. V. , & Jansa, S. A. (2014). Phylogenetic relationships of mouse opossums (Didelphidae, Marmosa) with a revised subgeneric classification and notes on sympatric diversity. American Museum Novitates, 3817, 1–27. 10.1206/3817.1 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of locality data for specimens used in genetic analyses of Nycteris.
Figure S1. Geographic sampling of genetic data used in this study. Plotting symbols denote the locations of one or more individuals represented by mitochondrial sequence (cytb) downloaded from GenBank (+), those represented only by cytb data newly generated for this study (open circles), and those where both mitochondrial and nuclear sequences were newly generated (filled circles). Taxon, localities, and coordinates for these points are included in Supporting Information Table S1.
Figure S2. Species tree inferred in StarBEAST for Nycteris for 21 clades, including Nycteris thebaica clades 1 to 6. Nodes are labeled with posterior probabilities.


