Abstract
Live-cell imaging of RNA has remained a challenge because of the lack of naturally fluorescent RNAs. Recently developed RNA aptamers that can light-up small fluorogenic dyes could overcome this limitation, but they still suffer from poor brightness and photostability. Here, we propose a concept of cell-permeable fluorogenic dimer of sulforhodamine B dyes (Gemini-561) and corresponding dimerized aptamer (o-Coral) that can drastically enhance performance of the current RNA imaging method. The unprecedented brightness and photostability together with high affinity of this complex allowed, for the first time, direct fluorescence imaging in live mammalian cells of RNA polymerase-III transcription products as well as messenger RNAs labelled with a single copy of the aptamer, i.e. without tag multimerization. The developed fluorogenic module enables fast and sensitive detection of RNA inside live cells, while the proposed design concept opens the route to new generation of ultrabright RNA probes.
Introduction
Cells constantly adapt their content to their needs, to changing environment or to pre-determined cell-cycles and differentiation programs by tuning their gene expression landscape. Moreover, live-cell imaging experiments demonstrated that significant cell-to-cell variation in gene expression occurs even within population of isogenic cells within the same environment1. Currently, imaging of gene expression in live cells relies mainly on proteins genetically modified with either fluorescent proteins2 or tags for specific chemical labeling3. Since no naturally fluorescent RNA has been discovered yet, extensive efforts were devoted to the development of RNA imaging technologies4.
A first breakthrough came with the use of RNA-binding proteins (RBP) fused to fluorescence proteins (FP)5,6. In these completely genetically encoded systems, an array (tens of repeats) of the RNA RBP-binding motif is incorporated into the 3’ untranslated region of the target messenger RNA (mRNA). Co-expressing the RBP-FP-coding gene in the same cell allowed for tracking mRNA (upon decoration by FP), which has become a reference method for collecting data on gene expression and RNA trafficking7. Substantial simplification is possible by using RNA-based fluorogenic modules8 in which bulky FPs are substituted by small fluorogens,9 i.e. dyes lighting up their fluorescence upon interaction with a target (bio)molecule10. In this scheme, the fluorogen is activated upon specific binding to a light-up RNA aptamer11.
The capacity of RNA to light-up fluorogenic dyes was first established with an aptamer activating Malachite Green,12 a dye known for its phototoxicity. Yet, the real breakthrough8,13 came with the introduction of the cell-permeable and non-toxic GFP-mimicking fluorogen, 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI), together with its activating aptamer Spinach14, later followed by improved aptamers (Spinach215, Broccoli16) and fluorogen derivatives17. Unfortunately, these systems suffered from limited brightness and photostability compromising the detection of low-abundant RNAs and extended imaging time.18 Substantial gain in photostability and brightness was achieved using fluorogens based on organic dyes (cyanines19–22 and rhodamines23–25), including those operating by Photoinduced Electron Transfer (PET)26 or Forster Resonance Energy Transfer (FRET)20,24,27 mechanisms. For instance, conjugates of sulforhodamine B (SRB) dye with dinitroaniline (DN) PET quencher recover their fluorescence upon association with an aptamer binding the SRB24,25,28 (e.g. SRB-2 aptamer) or the DN23 moiety. An alternative strategy, which could significantly improve fluorogen brightness, is to use dye homo-dimers that self-quench in aqueous solution but become fluorescent upon dimer opening after binding to the target biomolecule, a concept that yielded probes for detecting ligand-receptor interaction29 or DNA hybridization30.
Brightness and photostability also rely on the aptamer itself as illustrated by Corn31,32 pointing the key role of the selection strategy used. Light-up RNA aptamers are usually isolated by Systematic Evolution of Ligand by EXponential enrichment (SELEX)33,34, which allows selecting aptamers with high affinity and selectivity for their target, as exemplified by Mango RNA, a light-up aptamer binding its fluorogen (TO1-biotin) with nM affinity21. Yet, since no selection pressure is applied for the fluorogenic capacity, these aptamers often suboptimally activate their fluorogen. This limitation can be overcome by functional screening35–37 such as the microfluidic-assisted in vitro compartmentalization (μIVC)38 we recently used to identify brighter mutants of Spinach36 and Mango37.
Here, we propose a new concept for preparation of bright and photostable fluorogen for RNA imaging in cells by exploiting dimerization-induced self-quenching of SRB dyes, which yielded the fluorogen Gemini-561. Following a new selection scheme combining SELEX and μIVC we developed o-Coral, an aptamer of unprecedented compact dimeric structure, able to form a high affinity, bright and photostable complex with Gemini-561, enabling live-cell imaging of mRNAs labelled with a single copy of the aptamer.
Results
Design and synthesis of Gemini-561 fluorogen
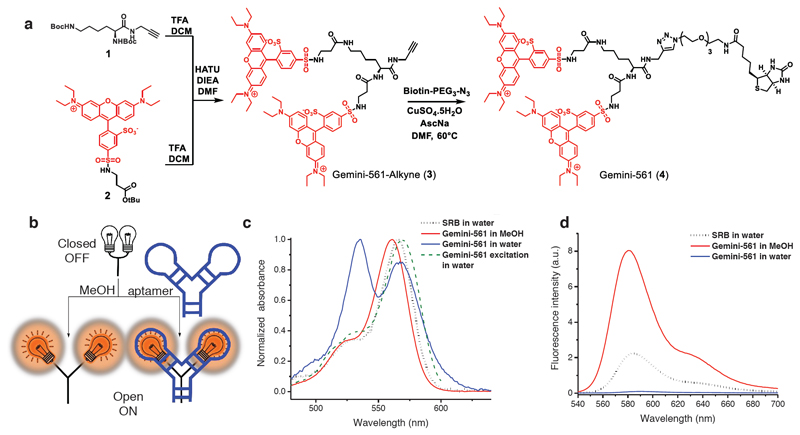
We intended to develop a red emitting fluorogen efficiently excited with a common laser (532-561 nm), allowing multicolor imaging in combination with eGFP-tagged proteins. Rhodamine fluorophores like SRB fulfill this requirement and possess numerous advantages. First, due to their tendency to form H-aggregates39 and their ability to be quenched by different systems (e.g. spirolactamization, PET), rhodamines constitute efficient platforms to develop reliable fluorogenic sensors40,41. SRB bears two sulfonate groups and upon functionalization becomes zwitterionic. Compared to cationic rhodamines or non-charged fluorophores, this feature should increase the polarity of the molecule, thus enhancing its water solubility and preventing non-specific interactions in biological media. Finally, SRB features optimal photophysical properties including elevated quantum yield, good photostability, sharp emission peak and high molar absorption coefficient (~100,000 M-1.cm-1). Gemini-561 was designed to promote the dimerization-induced self-quenching of two SRBs. For this purpose, lysine was chosen as a connector to provide a small distance between the dyes and thus ensure efficient π-stacking upon dimerization. Lysine (1) and SRB (2) derivatives were deprotected and coupled to lead to Gemini-561-alkyne (3) (Fig. 1a). The latter was clicked to biotin-PEG-N3 (the biotin moiety was later used to immobilize the dye during selection experiments) to yield Gemini-561 (4).
Figure 1. Design, synthesis and fluorogenicity of Gemini-561.
(a) Synthesis of Gemini-561. (b) Concept of fluorogenic response of Gemini-561 to environment (organic solvent) and aptamer. (c) Absorption and excitation spectra of Gemini-561 (200 nM) in water and methanol and SRB (200 nM) in water. Results were found identical in n = 3 independent experiments. (d) Fluorescence spectra of Gemini-561(200 nM) in water and methanol and SRB (200 nM) in water. Results were found similar in n = 3 independent experiments.
Spectroscopic properties of Gemini-561
Gemini-561 fluorogenicity was first assessed by spectroscopy. In water, Gemini-561 displayed weak fluorescence intensity with a quantum yield value of 0.01. Moreover, a blue shifted band (530 nm) appeared in the absorption spectrum indicating the formation of dimeric H-aggregate (Fig. 1c), in line with earlier report on the squaraine dimer29. Additionally, excitation spectrum showed that this band did not correspond to emissive specie (Fig. 1d), thus confirming a dimerization-induced quenching phenomenon. However, upon solubilization in methanol, the dimer opened and Gemini-561 displayed absorption and emission spectra similar to free SRB (Fig. 1b to d) along with an impressive increase in the quantum yield value (0.31, Table 1). In a second step, the non-specific opening of the dimer was evaluated, revealing that Gemini-561 conserves its quenched form in various physiological media and is not involved in non-specific interactions with proteins and lipoproteins (Supplementary Fig. 1). Altogether, these experiments demonstrate that Gemini-561 is an effective fluorogenic molecule compatible with biological media, making it a promising candidate for selection of the RNA aptamer.
Table 1. Spectral and biochemical properties of Gemini-561 alone or in complex with SRB-2 or o-Coral aptamers.
| λAbs max (nm) |
ε (M-1.cm-1) |
λEm max (nm) |
Q.Y. | Fluorescence Enhancement |
KD (nM) |
Relative Brightness | |
|---|---|---|---|---|---|---|---|
| eGFP | 490 | 39,200 | 508 | 0.68 | / | / | 1.00 |
| Buffer | 535 | 100,000 | 590 | 0.02 | 1.0 | / | 0.08 |
| MeOH | 561 | 162,000 | 580 | 0.31 | 10.3 | / | 1.88 |
| SRB-2 | 567 | 98,000 | 597 | 0.04 | 1.2 | 441 ± 167.5 | 0.15 |
| o-Coral | 580 | 141,000 | 596 | 0.58 | 12.8 | 73 ± 1.5 | 3.07 |
Measures were performed in selection buffer (40 mM phosphate buffer pH7.5, 100 mM KCl, 1mM MgCl2 and 0.05% Tween-20). Fluorescence quantum yield (QY) and enhancement were obtained at an excitation wavelength of 530 nm. KD values are the mean of n = 3 independent experiments ± 1 SD.
Isolation of Gemini-561 lighting-up aptamers
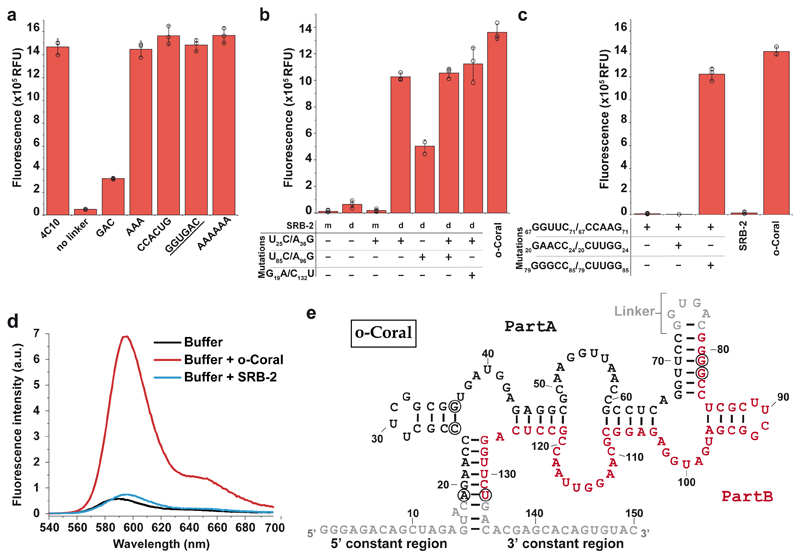
We first studied SRB-2 aptamer, reported to specifically interact with sulforhodamine B28 and its derivatives24 However, it was a poor activator of Gemini-561 fluorescence (Fig. 2a, Supplementary Fig. 2). We assumed that this weak effect might be attributed to inhibition of dye-aptamer interaction by the dye-dye dimerization.
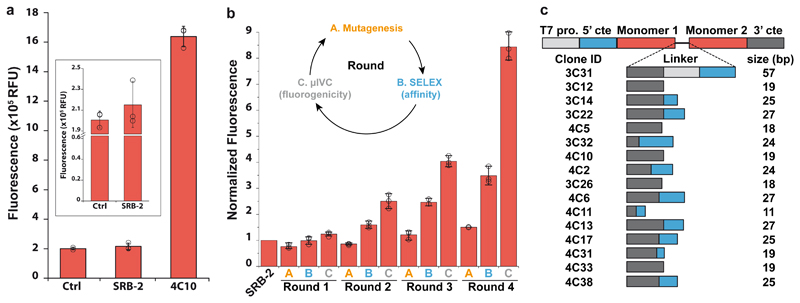
Figure 2. Isolation of Gemini-561 lighting-up aptamers by in vitro evolution.
(a) Gemini-561 activation capacity of the parental SRB-2 and the evolved 4C10 variant. 500 nM of RNAs were incubated with 50 nM of Gemini-561 and the fluorescence was measured at λ ex/em = 560/600 nm. The values are the mean of n = 3 independent experiments and the error bars correspond to ± 1 standard deviation. (b) Monitoring of the evolution process. For each round, the enriched library was transcribed in vitro in the presence of 100 nM of Gemini-561 and the fluorescence monitored. The fluorescence apparition rate was computed for each library and normalized to that of the parental SRB-2 aptamer. The inset schematizes the different steps (A, B and C) of an evolution round. The values are the mean of n = 3 independent experiments, each measurement being shown as an open circle. The error bars correspond to ± 1 standard deviation. (c) Schematic representation of genes coding for the 16 dimerized variants found among the 19 best aptamers at the end of the evolution process. For each variant, the width and the color of the box respectively inform on linker length (numerical value given on the right) and the nature of the sequence (light gray: T7 promoter, blue: 5'constant, dark grey: 3' constant). Red boxes correspond to SRB-2-derived core. The clone ID refers to the round of selection from which the clone was extracted (first number) and the clone number assigned during the final screening.
We therefore started an in vitro evolution of SRB-2 using a strategy combining SELEX and μIVC (Fig. 2b, Supplementary Fig. 3) to isolate RNAs endowed with both high affinity and high fluorogenic capacity. SRB-2 mutant library (~ 3.4 mutations per gene) was first generated by error-prone PCR and subjected to a first round of SELEX during which RNAs were challenged to bind bead-immobilized Gemini-561. Upon stringent wash, bound aptamers were recovered and reverse transcribed into cDNAs carrying T7 RNA polymerase promoter. Resulting genes were then subjected to a round of μIVC screening37. Genes encoding fluorogenic RNAs were finally recovered and used to prime a new round of error-prone PCR. A total of four such evolution cycles (mutagenesis, SELEX, μIVC) were performed and allowed to gradually improve the average fluorogenic capacity of the population (Fig. 2b), where ~40 % of the obtained sequences were significantly more fluorogenic than the parental SRB-2 aptamer. Surprisingly 16 out of the 19 best mutants also displayed a doubled size resulting from complete duplication (dimerization) of SRB-2 sequence upon recombination at a variable position between the 5’ and 3’ constant regions of two aptamers (Fig. 2c and Supplementary Fig. 4). The exact mechanism of this spontaneous recombination will require a dedicated study. In addition to this duplication, each optimized variant displayed 1 to 6 point mutations concentrated on P1 and P2 regions of the SRB-2 (Supplementary Fig. 4), while leaving region J2/3, P3 and L3 largely intact, in agreement with their proposed involvement in SRB recognition28.
Among the different clones, 4C10 had a remarkably high activation capacity by forming with Gemini-561 a complex an order of magnitude more fluorescent than with SRB-2 (Fig. 2a). 4C10 improvement correlates with a significant increase of affinity (~ 73±1.5 nM and 441±167 nM for 4C10 and SRB-2 respectively, Table 1 and Supplementary Table 1). By further engineering 4C10, we successfully reduced the 19-nucleotide long linker spacing the repeats down to 6 nucleotides while preserving intact fluorogenicity (Fig. 3a). Further reducing this length down to 3 nucleotides made aptamer sensitive to the linker sequence. Therefore, we pursued the study of a 6-nucleotide long linker 4C10 derivative, further named “o-Coral” (Supplementary Table 2). The duplication event by itself accounts only partly for the high performances of o-Coral since a molecule made of two wild-type SRB-2 modules displays only 6% of o-Coral fluorescence (Fig. 3b). Progressive reimplantation of o-Coral mutations showed that all the mutations contributed to o-Coral function, with the double mutant U25C/A36G (a mutation found in a third of the 19 best mutants, Supplementary Fig. 4) having the predominant effect. Remarkably, simple introduction of U25C/A36G mutation into SRB-2 did not yield any improvement of the monomer, indicating a synergic effect of the mutation in the dimer (Fig. 3b). Furthermore, U25C/A36G had a higher effect when present in the 5’ monomer, whereas introducing it in both monomers did not further improve the aptamer.
Figure 3. Characterization and engineering of the evolved molecule.
(a) Impact of linker size and sequence on the capacity of 4C10 aptamer to activate Gemini-561 fluorescence. 500 nM of RNAs were incubated with 50 nM of Gemini-561 and the fluorescence was measured at λ ex/em = 560/600 nm. The underlined sequence corresponds to o-Coral linker. (b) Contribution of the dimerization and the mutations to o-Coral functionality. SRB-2 aptamer was used as scaffold either in its monomeric (m) or dimeric (d) form containing o-Coral linker. Indicated mutations were then implemented and the different constructs tested as above. (c) Identification of interacting regions. A destabilized mutant (67GGUUC71/67CCAAG71) of o-Coral and two potential compensatory mutants (1: 67GGUUC71/67CCAAG71_20GAACC24/20CUUGG24 and 2: 67GGUUC71/67CCAAG71_79GGGCC85/79CUUGG85) were prepared and tested as above. The values (a-c) are the mean of n = 3 independent experiments, each measurement being shown as an open circle. The error bars correspond to ± 1 standard deviation. (d) Fluorescence emission spectra of Gemini-561 (200 nM) in absence and in the presence of RNA aptamers (600 nM). Excitation wavelength was 530 nm. Results were found similar in n = 3 independent experiments. (e) Model of secondary structure for o-Coral aptamer. This model was established based on enzymatic probing experiments (Supplementary Fig. 8) and mutagenesis experiments shown on c. SRB-2 derived sequences (Part A and B) are shown in black or red whereas the constant regions and the linker are shown in grey. Acquired mutations found to contribute to o-Coral function are circled in black.
o-Coral structural characterization
The role of the RNA module duplication was further studied at a structural level. Enzymatic structural probing characterization42 (Supplementary Fig. 5) was in agreement with an overall conservation of the structure initially proposed for the SRB-2 aptamer made of 3 stems and 3 unpaired regions28. Yet, two RNA folding models could account for the probing signal observed: i) a model in which both SRB-2 derived modules fold independently (Supplementary Fig. 6a) and ii) an intertwined folding (Supplementary Fig. 6b). To discriminate both models, we generated a mutant with a destabilized stem (67GGUUC71 changed for 67CCAAG71, Supplementary Fig. 7) leading to the complete loss of fluorogenic capacity (Fig. 3c). We then tested compensatory mutants relevant either to the independent folding model (20GAACC24 changed for 20CUUGG24) or to the intertwined structure (79GGGCC85 changed for 79CUUGG85). Interestingly, only the second compensatory mutant rescued o-Coral function (Fig. 3c), suggesting that o-Coral adopts an intertwined folding (Fig. 3e). This model received further support from another mutant (69UU70 changed for 69CC70) expected to stabilize only the intertwined folding (Supplementary Fig. 8). Finally, it can be noticed that two of the mutations selected during the evolution process (G19A and C132U) compensate each other in the intertwined model (Supplementary Fig. 6).
In our model, unpaired regions corresponding to J2/3 and L3 in the original SRB-2 aptamer, proposed to form the SRB-binding site28, are preserved (nucleotides 37-43, 49-60, 97-103 and 109-120). The three-dimensional structure of SRB-2 aptamer has not been established yet. However, even though pairs of G bases are found in SRB-2 and o-Coral, the existence of a G-quartet structure, common to many other light-up aptamers43, is unlikely since the complex was found to be insensitive to the nature of the monovalent cation added in the medium (Supplementary Fig. 9a). o-Coral also preserved the low magnesium dependency of SRB-2 aptamer25 by reaching its maximum fluorogenicity at 1 mM magnesium (Supplementary Fig. 9b), a value close to that in cell.
Gemini-561/o-Coral characterization in solution
Among studied dye/aptamer pairs, only combination of Gemini-561 and o-Coral resulted in significant fluorescence enhancement accompanied by disappearance of the short-wavelength band of the non-emissive H-aggregate (Supplementary Fig. 10 a-b). Remarkably, a parent analogue of Gemini-561, bearing alkyne group instead of biotin (Gemini-561-alkyne (3), Fig. 1a), showed very similar fluorogenic response to o-Coral (Supplementary Fig. 10 c-d), indicating that biotin is not required for the recognition of the aptamer (Supplementary Table 1). Detailed spectroscopic characterization of Gemini-561/o-Coral complex revealed a significant red-shifted absorption and fluorescence emission (by 19 and 16 nm, respectively) compared to those in an “activating” solvent methanol (Fig. 3d and Supplementary Figs. 10 to 12), presumably because of interaction between the dyes and RNA nucleobases. Additionally, we showed that the interaction of Gemini-561 (100 nM) with o-Coral (300 nM) was fast, reaching plateau after 400 s (Supplementary Fig. 10e). Interestingly, the quantum yield of the complex reached a higher value than Gemini-561 in MeOH (Table 1), showing that the dye is well confined within the aptamer. Moreover, estimating the brightness (i.e. extinction coefficient × quantum yield) of the complex indicated that a single copy of o-Coral associated with Gemini-561 is more than 3-fold brighter than eGFP (Table 1), making it the brightest aptamer-based module described so far8. Moreover, Fluorescence Correlation Spectroscopy (FCS) confirmed high brightness of Gemini-561/o-Coral complex (at least 1.14-fold brighter than single tetramethyl rhodamine, a close analogue of SRB), suggesting a 1:1 complex composed of a single molecule of Gemini-561 bound to a single copy of o-Coral (Supplementary Table 3). We finally investigated the effect of physiological media on the Gemini-561/o-Coral complex and found that, whereas 10% FBS slightly interferes with the aptamer-fluorogen interaction (Supplementary Fig. 13), SRB-2 aptamer, BSA or DNA did not challenge the fluorescence of the complex, highlighting the compatibility of Gemini-561/o-Coral module with complex cellular environment.
Imaging Gemini-561/o-Coral in live cells
After having ruled out potential cytotoxicity of Gemini-561 (Supplementary Fig. 14), we validated the functionality of Gemini-561/o-Coral complex in live HeLa cells by micro-injecting the preformed complex either directly into their nucleus or into their cytoplasm of living HeLa cells (Supplementary Fig. 15a). In both cases, an intense red fluorescence was readily observed in the presence of the module, whereas the injection of Gemini-561 alone or mixed with SRB-2 aptamer did not yield significant fluorescent signal, confirming the specific activation of Gemini-561 by o-Coral and the suitability of the system for live cell applications. Cell permeability of Gemini-561 was also assessed by incubating HeLa cells with 200 nM fluorogen prior to washing them and microinjecting o-Coral along with Dextran-Alexa-647 conjugate. Both o-Coral and SRB-2 were successfully injected in the cells as attested by Alexa-647 fluorescence, but cytosolic fluorescence of Gemini-561 was observed only in the presence of o-Coral (Supplementary Fig. 15b), demonstrating both cell permeability of the fluorogen and its capacity to detect o-Coral inside live cells.
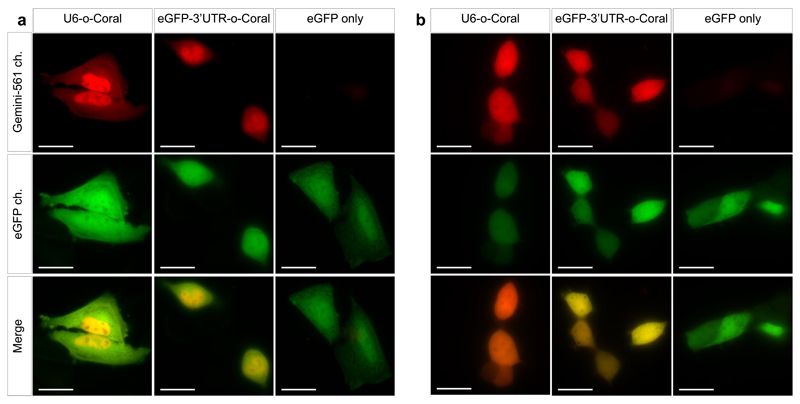
We next evaluated the performances of our system with aptamers synthesized by the cell machinery. o-Coral was inserted into a F30 scaffold44 and expressed from a U6 truncated promoter45 allowing the gene to be transcribed by RNA polymerase III (pol III). The plasmid also contained eGFP-coding region produced from a RNA polymerase II (pol II) promoter and used for identifying transfected green fluorescent cells. Taking advantage of Gemini-561 cell-permeability, we expressed o-Coral gene in live Hela and HEK293T cells and incubated them with 200 nM Gemini-561 for 5 minutes prior to imaging. Remarkably, green fluorescent transfected cells also displayed intense red (Gemini-561) fluorescence, especially in the nucleus (Fig. 4, Supplementary Fig. 16a) as expected for U6-driven transcripts, confirming that o-Coral preserves RNA sub-cellular localization. Moreover, the sensitivity of this red fluorescence to actinomycin D treatment, a known inhibitor of RNA polymerases, confirmed the requirement of the RNA component. Interestingly, during these live-cell imaging experiments, we noticed cell-to-cell variations in the fluorescence intensity of the Gemini-561/o-Coral (Supplementary Fig. 17), suggesting different transcription states as reported recently with a different aptamer9. Taken together, these data show that, owing to elevated brightness and affinity of Gemini-561/o-Coral complex, a single copy of pol III-expressed o-Coral aptamer is sufficient for RNA detection in living mammalian cells.
Figure 4. Live-cell imaging of o-Coral expressed from pol. II and pol. III promoter.
Live cell imaging of HeLa (a) and HEK293T (b) cells expressing U6-o-Coral, the egfp mRNA eGFP-3’UTR-o-Coral, or eGFP only. Cells were incubated with Gemini-561 (200 nM) for 5 min before imaging. Hoechst was used to stain the nucleus (5 μg/mL). The images were acquired using a 500 ms exposure time. Gemini-561 in red (ex: 550 nm, em: 595±40 nm) and eGFP in green (ex: 470 nm, em: 531±40 nm). Results were found similar in n = 3 independent experiments. Scale bar is 30 μm.
To better understand Gemini-561 cell permeability, we monitored Gemini-561/o-Coral fluorescence in cells and found that internalization kinetics accelerated while increasing Gemini-561 concentration (200 vs 50 nM), reaching a plateau in <10 min with both HeLa and HEK293T cells (Supplementary Fig. 18). Yet, further increasing Gemini-561 concentration to 800 nM led to significant background and dye aggregation (Supplementary Fig. 19). Moreover, since Gemini-561 internalization was still observed at 4 °C in various cell types (Supplementary Fig. 20), the fluorogen likely reaches the cytosol by direct diffusion through the cell plasma membrane rather than by endocytosis.
To extend our technology to Pol II transcripts, we inserted o-Coral/F30 directly into the 3’ untranslated region of eGFP gene carried on the plasmid. Excitingly, we found that labelling eGFP mRNA with a single copy of o-Coral was sufficient to detect an intense and homogeneous red fluorescence in eGFP transfected cells (Fig. 4, Supplementary Fig. 16b), while not significantly altering eGFP mRNA level (Supplementary Fig. 21). The mRNA detection with Gemini-561/o-Coral module showed its efficiency in HeLa, HEK293T and U87 cells (Supplementary Fig. 20 and 22), revealing fine difference in mRNA distribution. Indeed, in HEK cells the signal of mRNA was evenly distributed between nucleus and cytosol, similarly to eGFP, whereas in HeLa cells the mRNA signal was slightly stronger in the nucleus (Fig. 4). Additionally, 5S RNA was also successfully imaged in all three studied cell lines (Supplementary Fig. 23), showing universality of our RNA detection approach. It is noteworthy to highlight that, to the best of our knowledge, it is the first time that a mRNA labelled with a single copy of a light-up aptamer can be visualized without the need for extensive image processing46.
Importantly, the eGFP fluorescence signal correlated well with that of Gemini-561/o-Coral in egfp-3’UTR-o-Coral expressing HeLa cells (Supplementary Fig. 24). As the protein expression correlates to some extent with corresponding mRNA expression47, we concluded that Gemini-561 signal reflects semi-quantitatively the abundance of corresponding mRNA. Interestingly, the same correlation was also observed with cells in which o-Coral and eGFP were produced from genes colocalized on the same plasmid and controlled by constitutive promoters (U6 and CMV, respectively).
Comparison of Gemini-561/o-Coral to other systems
To further demonstrate the advantage of our new fluorogenic system, we systematically compared its performances with other systems using commercially available fluorogens: Broccoli16, Mango III37 and Corn31. To properly compare our data with those reported in the literature, aptamers were embedded into the RNA scaffold as they were previously characterized in (i.e. tRNA scaffold for Corn31 and F30 scaffold for Broccoli37, Mango III37 and o-Coral). Photostability was first assessed in cuvettes where the entire solution of a fluorogenic dye (0.2 μM) and aptamer (1 μM) was irradiated with a laser light (Fig. 5a). Moreover, to ensure that each system absorbed similar amount of photons, the applied irradiation power density (irradiance) was inversely proportional to the molar absorption coefficient of the corresponding dye at the excitation wavelength used. DFHBI-1T/Broccoli complex showed very poor photostability as its intensity vanished within <1 s, as reported before.31,37 Significantly higher photostability was observed for DFHO/Corn and TO1-biotin/Mango III, as their emission changed to much lower extent within 200 s of irradiation, in line with earlier studies.31,37 Excitingly, emission intensity of Gemini-561/o-Coral did not show any change in the fluorescence intensity in these conditions, indicating that it is significantly more photostable than the three other systems. Similar experiments performed with RFP and mCherry, also revealed a much higher photostability of Gemini-561/o-Coral (Supplementary Fig. 25), highlighting its advantage over approaches employing FPs (e.g. MS2 system).
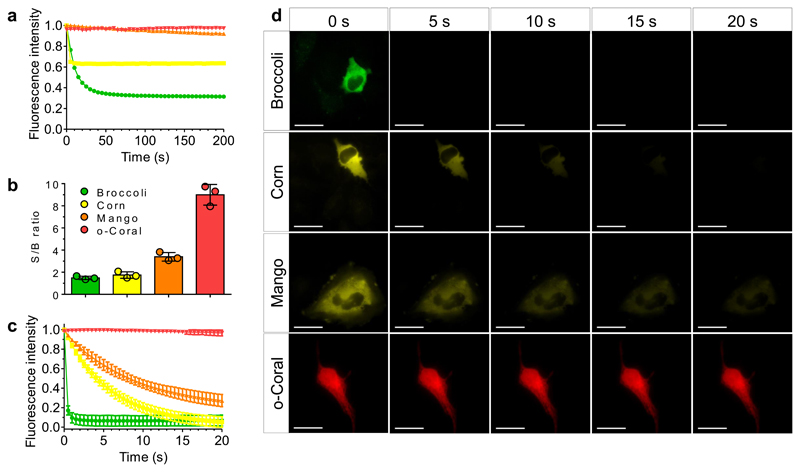
Figure 5. Comparative analysis of photostability by fluorescence microscopy and spectroscopy.
(a) Photostability of G561/o-Coral (0.2 μM/1 μM) compared to Broccoli+DFHBI-1T (0.2 μM/1 μM), Corn+DFHO (0.2 μM/1 μM), Mango + TO1-Biotin (0.2 μM/1 μM). Each complex was excited at the same molar extinction coefficient value: 30,000 M-1 cm-1. Broccoli, Corn and Mango were excited using 488 nm laser (7.75 mW cm-2, 11 mW cm-2, 10 mW cm-2 respectively) and o-Coral was excited using 532 nm laser (7 mW cm-2). Fluorescence intensity was monitored at 507 nm for Broccoli, 545 nm for Corn, 535 nm for Mango and 596 nm for o-Coral. (b-d) Photostability and signal to background noise ratio measurement in live Hela cells. In vitro transcribed and purified aptamers were preincubated with respective fluorogenic dyes for 10 min in selection buffer to form complex. Complexes were microinjected in live HeLa cells using 5 μM dye and 20 μM aptamer concentration. Microinjection parameters: Pi=90 [hPa]; Ti=0.3 [s]; Pc=10 [hPa]. Consecutive images were acquired, each using a 500 ms exposure time. The excitation power was adjusted for the fluoromodules to absorb similar amount of photons. Broccoli (ex: 470 nm, em: 475±50 nm); Corn (ex: 470 nm, em: 531±40 nm); Mango (ex: 470 nm, em: 531±40 nm); Coral (ex: 550 nm, em: 595±40 nm). (b). Signal to background noise ratio of the first acquired image depicting the brightness of the system and the quality of obtain images. Signal to background noise ratios were calculated from fluorescence intensity values extracted from images using same region of interest from n = 3 independent injections. The value of each measurement is shown as a colored dot. The error bars correspond to ± 1 standard deviation. (c) Fluorescence intensity decay curves over the time. Data represent average values ± 1 standard deviation extracted from images from n = 3 independent experiments. (d) Representative micrographs taken during the experiment. Results were found similar in n = 3 independent experiments. Scale bar is 30μm.
Second, we compared brightness of the complexes at the single-molecule level using FCS (Mango system was not characterized because of incompatibility with our setup). Single-molecule brightness of DFHO/Corn and DFHBI-1T/Broccoli was correspondingly 0.62 and 0.18 (Supplementary Table 3) with respect to that of reference dye fluorescein (at pH 9), confirming the higher brightness of DFHO/Corn31. On the other hand, the single molecule brightness of Gemini-561/o-Coral was 1.14 that of reference dye tetramethyl rhodamine in water. Taking into account that fluorescein and TMR exhibit similar brightness in our FCS setup, the new fluorogenic module, Gemini-561/o-Coral, is ~2-fold brighter than DFHO/Corn in the single molecule experiments. In order to compare brightness of our module with MS2-based technology5,6, we measured FCS for representative fluorescent proteins used in MS2 labelling. In comparison to RFP, mCherry and eGFP, our module was respectively 19-, 7- and 7-fold brighter, explaining why our technology does not require multimerization and allows mRNA detection using a single copy of the aptamer tag. Nevertheless, we have to admit that multimerized tandem of 30-copies of these proteins, used in MS2 method, is expected to be brighter than a single copy of our module.
Finally, we microinjected the different fluorogen/aptamer complexes into live cells right before taking images (Fig. 5b to d) and measured the signal/background ratio (S/B). Gemini-561/o-Coral system provided images with a S/B value significantly higher than those obtained with other studied systems (Fig. 5b). Image analysis also showed that fluorescence intensity within the cells decayed much slower for Gemini-561/o-Coral (Fig. 5c and d), confirming superior photostability of our system. In an alternative experiment, cells were incubated in the presence of the fluorogen followed by microinjection of the corresponding aptamer. This experiment further confirmed that Gemini-561/o-Coral was significantly more photostable than reference aptamer-based systems (Supplementary Fig. 26), allowing >20 s continuous imaging with only a minor loss of fluorescence intensity. Remarkably, Gemini-561/o-Coral fluorescence can still be detected upon several hours of constant illumination (Supplementary Fig. 27).
Discussion
Light-up aptamers gained their niche as powerful genetically encoded RNA imaging tools. Due to high modularity of nucleic acids and available SELEX methodologies, a palette of fluorogen-aptamer systems was discovered to shine light on complex cell machinery. Unfortunately, limited brightness and photostability of aptamer-based fluorogenic modules developed to date narrow their broad application.
In this work we developed and characterized Gemini-561/o-Coral, a new RNA-based fluorogenic module displaying high brightness, affinity and photostability making it, to the best of our knowledge, the brightest module described so far in the literature8. We reached this goal by combining two key innovations. First, the fluorogen Gemini-561 was designed based on two copies of the bright and photostable sulforhodamine B dye that self-quenches in water and is able to enter the live cells within a few minutes. This quenching mode is important since, upon activation, both fluorophores become strongly fluorescent making such a probe brighter than any monomeric probe described to date. Second, we developed the light-up RNA aptamer o-Coral using an integrated in vitro evolution strategy combining rounds of mutagenesis and SELEX in tandem with μIVC screening. In this scheme, the SELEX step allows isolating RNAs with high affinity for Gemini-561, whereas the μIVC isolates the most fluorogenic sequences. The obtained o-Coral efficiently opens the self-quenched dimer Gemini-651, thus activating its fluorescence. Using the same overall strategy, it should be possible to further expand RNA imaging toolbox by developing new orthogonal modules made of dimeric (Gemini) fluorophores of any desired color while selecting new specific Coral aptamers.
The intertwined dimerized structure of o-Coral may also be advantageous for the future engineering of aptamers. Indeed, both L2 and L2’ loops together with the linker region are highly amenable to sequence modification and are attractive sites for inserting other sequences (e.g. sensing aptamers). By doing so, o-Coral could be converted into complex multi-inputs logic gates or biosensors.
o-Coral is readily expressed in mammalian cells where it forms a bright complex with Gemini-561 that is otherwise not activated by cell components. Similarly to Malachite Green48 and DIR2s aptamers49, o-Coral does not seem to possess the G-quadruplex organization shared by most of the other structurally characterized light-up aptamers43. This is of particular interest when considering a recent report indicating that most of the RNA G-quadruplex domains would be kept globally unfolded in mammalian cells50, suggesting that G-quadruplex-based RNA may not be optimal for being used in living cells.
Direct comparison of Gemini-561/o-Coral with the most representative aptamer-based fluorogenic modules in live cells showed clear advantages of the new module in terms of brightness and photostability. Moreover, its significantly higher brightness (7-20-fold according to FCS) and photostability compared to fluorescent proteins explain the capacity to detect RNA using a single copy of the aptamer tag, whereas in case of MS2-eGFP module5,6 a tandem of ~30 copies should be used. Imaging of RNA by integrating just a single copy of the aptamer, achieved with Gemini-561/o-Coral, has remained a challenge so far in this technology. However, since dedicated experiments are yet to be performed, it is too early to conclude if the single molecule resolution can be reached without tag multimerization. Overall, Gemini-561/o-Coral system significantly strengthens the toolbox for RNA imaging and shows a new direction in the development of ultrabright fluorogenic aptamer-based modules.
Online Methods
Synthetic procedures
Synthetic procedures of Gemini-561-alkyne (3) and Gemini-561 (4) are given as a Supplementary Note available online.
Optical Spectroscopy
Optical spectroscopy was performed with Milli-Q water (Millipore®). All the solvents were spectroscopic grade. Absorption and emission spectra were recorded on a Cary 4000 Scan ultraviolet–visible spectrophotometer (Varian) and a FluoroMax-4 spectrofluorometer (Horiba Jobin Yvon) equipped with a thermostated cell compartment, respectively. For standard recording of fluorescence spectra, the emission was collected 10 nm after the excitation wavelength. All the spectra were corrected from wavelength-dependent response of the detector and measured at room temperature (25°C). Absorbance values of all solutions were systematically below 0.1 at the maximum. Quantum yields were determined using a reference dye (Rhodamine B in water).
Gene library generation
The sequence coding for the SRB aptamer28 was flanked with constant regions at 5' (GGGAGACAGCTAGAGTAC) and 3' (GACACGAGCACAGTGTAC) ends to allow DNA amplification and RNA reverse transcription. Mutant libraries were generated by error prone polymerase chain reaction (PCR) by subjecting 10 fmoles of DNA (diluted in 200 μg/ml of yeast total RNA solution (Ambion)) to 4 amplification cycles in the presence of Fwd (CTTTAATACGACTCACTATAGGGAGACAGCTAGAGTAC) and Rev (GACACGAGCACAGTGTAC) primers as well as nucleotide analogues (JBS dNTPMutagenesis Kit, Jena Bioscience) as described before38. 1 ng of PCR products was amplified in a second PCR mixture containing 10 pmoles of each primer (Fwd and Rev), 0.2 mM of each dNTPs, 5 U of PhireII® (Fermentas) and the corresponding buffer (Fermentas). The mixture was thermocycled starting with an initial step of denaturation of 30 sec at 95°C followed by 25 cycles of: 5 sec at 95°C and 30 sec at 60°C. The PCR products were purified following the "Wizard® SV Gel and PCR Clean-up System" (Promega) kit instructions and the quantity of DNA was determined by NanoDrop™ measurement.
In vitro transcription and RNA purification
Genes coding for aptamers were PCR amplified with the same procedure used before (25 cycles of PCR using PhireII enzyme). 1 μg of PCR products was then in vitro transcribed in 500 μl of mixture containing 2 mM of each NTP (Larova), 25 mM MgCl2, 44 mM Tris-HCl pH 8.0 (at 25°C), 5 mM DTT, 1 mM Spermidine and 17.5 μg/ml T7 RNA polymerase (prepared in the laboratory). After 3h of incubation at 37°C, 1000 units of Baseline-Zero™DNase (Epicentre) and the corresponding buffer were added to the mixture and a second incubation of 1 h at 37°C was performed. RNA was then recovered by phenol extraction. In vitro transcribed RNA was purified using ion exchange chromatography (FastFlow DEAE sepharose, GE Healthcare) by loading the RNA in and washing the resin with bind/wash buffer (50 mM NaCl, 50 mM Tris-HCl pH 7.5 and 10% Glycerol) and eluting it with elution buffer (600 mM NaCl and 50 mM Tris-HCl pH 7.5). Alternatively, RNA was gel purified by ethanol precipitating transcription mixture and dissolving the pellet into loading buffer (0.05% bromophenol blue, 20% glycerol, TBE 1x, 8M urea). The solution was then loaded on a 12% denaturing 8 M urea acrylamide/bisacrylamide gel. The piece of gel containing RNA was identified by UV shadowing, and the RNA electroeluted as described before38. Eluted RNA was then ethanol precipitated, the washed pellets were dissolved in DEPC treated water and quantified with Nanodrop (Thermo Scientific).
SELEX
100 μL of streptavidin-agarose beads (Sigma-Aldrich) were washed with 200 μL of activation buffer (100 mM NaOH, 50 mM NaCl). The beads were then centrifuged 5 minutes at 5000 g and room temperature and the supernatant was removed by pipetting. This procedure was repeated with 200 μL of pre-wash buffer (40 mM potassium phosphate buffer pH 7.5, 100 mM KCl, 1 mM MgCl2 and 0.05% Tween 20) and finally 200 μL of wash buffer (pre-wash buffer supplemented with 1 mg/mL BSA (New England Biolabs), 0.1 mg/mL sodium heparin (Sigma-Aldrich) and 200 μg/ml yeast total RNA (Ambion). The resin was loaded into a cartridge (Plastic small column CS-20 ABT) previously equilibrated by an overnight incubation with wash buffer at 4°C. Then, 500 μL of wash buffer supplemented with 10 nmoles of biotinylated Gemini-561 dye were added on the beads at a controlled flow-rate of 10 mL/h using a syringe pump (PhD 2000, Harvard Apparatus). Afterward, the unbound fluorophore was washed away by 20 mL of wash buffer (20 mL / h). ~ 50 μg of purified RNA were introduced in 250 μL of ultra-pure DEPC-treated water and renatured by 2 min at 85°C followed by 5 min at 25°C. Then, 250 μL of twice concentrated washing buffer were added and the mixture was infused through the Gemini-561 substituted resin at a flow-rate of 1.5 mL/h. Unbound RNAs were eliminated per 20 mL of wash buffer infused at 20 mL/h. This initial wash was followed by three additional washes of 15 mL of buffer while reducing the ionic strength (respectively 100 mM, 10 mM and 1 mM KCl). The selection pressure was further increased during the last round by introducing 5 μM of free Gemini-561 dye during the last wash of the column. The beads were then collected using a Pasteur pipette, centrifuged and placed in 100 μL of elution buffer (95% formamide and 25 mM EDTA). After 2 minutes of heating at 90°C, the beads were centrifuged, the supernatant was recovered and the RNA precipitated as above. RNA was pelleted, washed and resuspended in 50 μL of 2 μM Rev primer solution. The mixture was heated for 2 min at 85°C and cooled at 25°C for 5 min. 50 μL of reaction mixture containing 0.5 mM of each dNTP, 400 U of reverse transcriptase (Maxima H Minus, ThermoFisher) and the corresponding 2x concentrated buffer were added and the mixture and incubated 1h00 at 55°C. The resulting cDNA was then extracted with a mixture Phenol / Chloroform / Isoamyl alcohol 25/24/1 (Roth) and precipitated. cDNA was recovered by 30 minutes of centrifugation at 21000 g and 4°C, washed, dried, resuspended in 250 μL of PhireII PCR mixture and amplified by PCR as described above.
Droplet-based microfluidics
Microfluidic chips were made of polydimethylsiloxane (PDMS) as described before38.
i. Droplet digital PCR
DNA mutant libraries were diluted in 200 μg/mL yeast total RNA solution (Ambion) down to ≈ 8 template DNA molecules per picoliter. 1 μL of this dilution was then introduced in 100 μL of PCR mixture containing 0.2 μM of each primer (Fwd and Rev), 0.2 mM of each dNTPs, 20 μM of coumarin, 0.1% Pluronic F68 (Sigma), 5 U of PhireII enzyme (Fermentas) and the corresponding buffer (Fermentas). The mixture was loaded in a length of PTFE tubing and infused into droplet generator microfluidic chip where it was dispersed in 2.5 pL droplets (production rate of ≈12 000 droplets/s) carried by Novec 7500 fluorinated oil (3M) supplemented with 3% of a fluorosurfactant (proprietary synthesis). Droplet production frequency was monitored in real time using an optical device and software developed by the team38 and used to determined droplet volume. 2.5 pL droplets were generated by adjusting pumps flowrates (MFCS, Fluigent). The emulsion was collected in 0.2 mL tubes and subjected to an initial denaturation step of 2 min at 98°C followed by 30 cycles of: 10 sec at 98°C, 30 sec at 55°C, 30 sec at 72°C. Droplets were then reinjected into a droplet fusion microfluidic device.
ii. Droplet fusion
PCR droplets were reinjected and spaced into a fusion device at a rate of ~1500 droplets/s. Each PCR droplet was then synchronized with a 16 pL in vitro transcription (IVT) droplet containing 2 mM each NTP (Larova), 25 mM MgCl2, 44 mM Tris-HCl pH 8.0 (at 25°C), 5 mM DTT, 1 mM Spermidine, 0.1% of Pluronic F68 (Sigma), 1 μg of pyrophosphatase (Roche), 500 nM Gemini-561, 1 μM coumarin acetate (Sigma) and 17.5 μg/mL T7 RNA polymerase (prepared in the laboratory). IVT mixture was loaded in a length of PTFE tubing and kept on ice during all experiment. PCR droplets were spaced and IVT droplets produced using a single stream of Novec 7500 fluorinated oil (3M) supplemented with 2% (w/w) of fluorinated. Flow-rates (MFCS, Fluigent) were adjusted to generate 16 pL IVT droplets and maximize synchronization of 1 PCR droplet with 1 IVT droplet. Pairs of droplets were then fused with an AC field (400 V at 30 kHz) and the resulting emulsion was collected off-chip and incubated for 2 h at 37°C.
iii. Droplet analysis and sort
The emulsion was finally reinjected into an analysis and sorting microfluidic device at a frequency of ~ 150 droplets/s and spaced with a stream of surfactant-free Novec 7500 fluorinated oil (3M). The orange fluorescence (Gemini-561 in complex with the aptamer) of each droplet was analyzed and the 1–2% most orange fluorescence droplets were sorted. The gated droplets were deflected into collecting channel by applying an AC fields (1000 V 30 kHz) and collected into a 1.5 mL tube. Sorted droplets were recovered from the collection tubing by flushing 200 μL of Novec fluorinated oil (3M). 100 μL of 1H, 1H, 2H, 2H-perfluoro-1-octanol (Sigma-Aldrich) and 200 μL of 200 μg/mL yeast total RNA solution (Ambion) were then added and the droplets broken by vortexing the mixture. DNA-containing aqueous phase was recovered, and the DNA recovered by PCR.
TA cloning
The DNA contained in the libraries obtained after the last two rounds of μIVC screening were PCR-amplified as described above but using the DreamTaq® enzyme and buffer (Fermentas) instead of PhireII (Fermentas). PCR products were purified ("Wizard® SV Gel and PCR Clean-Up System" kit (Promega)) and inserted into the cloning vector of the "insTAclone PCR Cloning" kit (Thermo-Scientific) following the manufacturer's recommendations by overnight ligation at 4°C. ElectroTEN Blue bacteria (Agilent) were transformed by electroporation and plated onto a 2YT / agar / Ampicillin (100 μg / mL) plate.
Individual colonies were used to seed 20 μL of PhireII PCR mixture (see above) while the rest of the colony was introduced in 3 mL of liquid medium 2YT / Ampicillin (100 μg / ml) overnight at 37°C under agitation. Upon thermocycling, 2 μL of PCR product were added to 18 μL of in vitro transcription mixture (see above) supplemented with 100 nM of Gemini-561. The mixture was then incubated at 37 °C in a thermocycler (Stratagene Mx3005P, Agilent Technologies) and the fluorescence (ex/em 575/602 nm) was monitored for 2 hours. Finally, plasmid DNA was extracted from bacteria of interest using "GenElute Plasmid Miniprep" kit (ThermoFisher) and sequenced by the Sanger method (GATC Biotech).
Real-time IVT measurements
PCR products of each selection cycle were purified ("Wizard® SV Gel and PCR Clean-up System" (Promega)) kit and quantified (NanoDrop™). 50 ng of pure DNA was introduced into 38 μL of in vitro transcription mixture (see above) supplemented with 100 nM of Gemini-561. This mixture was then incubated at 37 °C in a real-time thermocycler (Stratagene Mx3005P, Agilent Technologies) and the fluorescence was monitored as above.
Fluorescence measurement on purified RNA
1 μM purified RNA was heated for 1 min at 90 °C and cooled at 4°C for 1 min. The solution was then supplemented with 1 volume of a twice concentrated mixture containing 80 mM potassium phosphate buffer pH 7.5, 2 mM MgCl2, 0.1% Tween 20, 200 mM of salt (KCl, NaCl, LiCl or CsCl) and 100 nM Gemini-561. The mixture was then incubated for 10 min at 25°C prior to measuring the fluorescence at 25°C on a real-time thermocycler (ex/em 575/602 nm, Mx 3005P, Agilent) or on microplate reader (ex/em 560/600 nm, SpectraMax iD3, Molecular Devices).
RNA probing
20 μg of RNA were first dephosphorylated for 20 min at 37 °C using 1 U FastAP (Fermentas) per μg of RNA. Upon phenol/chlorophorm extraction and RNA precipitation, dephosphorylated RNA was 5’ labelled by incubating 5 μg of dephosphorylated RNA with 50 μCi of [P32]γATP and 10 U of T4 polynucleotide kinase, with T4 PNK 1× buffer in final volume of 15 μL during 1h00 at 37 °C prior to be phenol/chloroform extracted, precipitated, pelleted. Labeled RNA was then gel-purified and eluted from the gel by an overnight incubation at 4 °C and gently mixing in RNA Elution Buffer (500 mM of ammonium acetate and 1 mM of EDTA). Eluted radiolabeled RNA was extracted by phenol/chloroform treatment, precipitated in ethanol and pelleted as described above. The RNA is resuspended in nuclease-free water. The specific activity (cpm/μL) was calculated by measuring 1 μL in a radioactivity counter “Multi-Purpose Scintilliator Counter” (Beckman) by Cerenkov counting. Labeled RNA (200,000 cpm) was renatured by heating it for 1 min at 90°C then 1 min on ice and then pre-incubated at 20 °C for 15 min in a buffer containing 20 mM of this-HCl pH7.5, 1mM of MgCl2 and 150 mM of KCl. 1 μg of total RNA was then added to the preparation and RNAs were incubated with T1 enzyme (0.25 U, 0.5 U, 1 U), T2 enzyme (0.0125 U, 0.025 U, 0.05 U) and V1 enzyme (0.001 U, 0.002 U, 0.004 U) for 5 min at 20°C (T1 and V1) or 10 min at 20°C (T2) or water (Ctrl lane). The same amount of digested product was loaded on a 10% denaturing 8 M urea acrylamide/bisacrylamide gel in parallel to an alkaline hydrolysis ladder and a denaturing T1 as described previously42. The radiolabeled RNAs were then visualized on autoradiographic film.
Affinity measurements
To measure KD, the concentration of renatured and purified RNAs was progressively increased from 2.45 nM to 40 μM for SRB-2 and from 3.9 nM to 4 μM for 4C10 and o-Coral aptamer with 100 nM (for SRB-2) or 25 nM (for 4C10 and o-Coral) of Gemini-561 in 40 mM potassium phosphate buffer pH 7.5, 100 mM KCl, 1 mM MgCl2 and 0.05% Tween 20. The fluorescence was measured on a microplate reader (ex/em 580/620 nm, SpectraMax iD3, Molecular Devices).
Expression vectors design
The sequences coding for o-Coral or 20 nucleotides from Broccoli aptamer (Ctrl) were introduced downstream a U6 promotor into a F30-scaffold (Supplementary Table 2) contained into a pUC57 vector (Proteogenix) via a restriction (SbfI) / ligation step. The entire sequences (pU6_o-Coral_F30 or pU6_Ctrl_F30) were then introduced into an eGFP-N1 vector (Clontech) using AflII restriction sites. Alternatively, o-Coral-F30 or Ctrl-F30 sequences were introduced directly in the 3’UTR of the eGFP coding sequence under the control of a CMV promotor by a restriction (MfeI) / ligation step.
RNA extraction and RT qPCR
Total RNA was extracted from HeLa cells (transfected with plasmid carrying gfp of gfp-o-Coral genes) using Monarch® Total RNA Miniprep kit (New England Biolabs) according to manufacturer's instructions. 4 μg of total RNA were used for DNAse digestion using dsDNAse enzyme (Thermo Scientific). cDNA was then synthesized using Maxima minus H reverse transcriptase (Thermo Scientific) following manufacturer's instructions and specific cDNAs were then quantified by qPCR: 1 μL of cDNA was introduced into 19 μL of Ssofast evagreen qPCR mixture (Biorad) supplemented in eGFP specific primers or into 19 μL of Ssoadvanced qPCR mixture (Biorad) supplemented with S18 or GAPDH specific primers and Taqman probes (Biorad). Real-time PCR was carried out on an CFX96 Touch™ System (Biorad) starting with an initial denaturation step of 2 min at 95°C followed by 40 cycles of: 5 sec at 95°C and 30 sec at 60°C. Reactions were run in triplicate in three independent experiments. The housekeeping genes GAPDH and S18 were used as internal controls to normalize the variability in expression levels.
Cell culture and transfection
HeLa (ATCC® CCL-2™) and HEK293T (ATCC® CRL-3216™) cells were grown in Dulbecco's Modified Eagle Medium without phenol red (DMEM, Gibco-Invitrogen) supplemented with 10% fetal bovine serum (FBS, Lonza), 1% L-Glutamine (Sigma Aldrich) and 1% antibiotic solution (Penicillin-Streptomycin, Sigma-Aldrich) at 37°C in humidified atmosphere containing 5% CO2. U87MG (ATCC® HTB-14™) were grown in Minimum Essential Medium (MEM, Gibco-Invitrogen) supplemented with 10% FBS, 1% Ultra-Glutamine (Gibco-Invitrogen) and 1% antibiotic solution. RNA-coding constructcs were transfected directly into a 35 mm glass-bottomed imaging dish (IBiDi®) using FuGene HD (Promega) transfecting agent following recommended manufacturer protocol. Imaging experiments were performed 16-48 h post-transfection.
Cellular Imaging
Cells were seeded onto a 35 mm glass-bottomed imaging dish (IBiDi®) at a density of 3-5×104 cells/well 48 h before the microscopy measurement. 16-24 h prior to imaging, cells were transfected with corresponding DNA plasmid. For imaging, the culture medium was removed and the attached cells were washed with Opti-MEM (Gibco–Invitrogen). Next, the cells were incubated in Opti-MEM with Hoechst (5 μg/mL) to stain the nuclei and in the presence of Gemini-561 dye (0.2 μM) for 5 min, the living cells were washed twice with Opti-MEM and visualized in Opti-MEM or were fixed in 4% PFA in PBS for 5 minutes before being washed twice in PBS. The images were acquired in epi-fluorescence mode with a Nikon Ti-E inverted microscope, equipped with CFI Plan Apo × 60 oil (NA = 1.4) objective, and a Hamamatsu Orca Flash 4 sCMOS camera. The acquisition settings were: Hoechst (ex. 395 nm, em. 510±42 nm), eGFP (ex: 470 nm, em: 531±40 nm), G561/o-Coral complex (ex: 550 nm, em: 595±40 nm) and Alexa-647 (ex: 638 nm, em: LP 647 nm). The images were recorded using NIS Elements and then processed with Icy software.
Microinjection experiments
Cells were seeded onto a 35 mm glass-bottomed imaging dish (IBiDi®) at a density of 1×105 cells/well 24 h before the microscopy measurement. For imaging, the culture medium was removed and the attached cells were washed with Opti-MEM (Gibco–Invitrogen). Next, the cells were incubated in Opti-MEM with Hoechst (5 μg/mL) to stain the nuclei. In vitro transcribed and purified aptamers were preincubated with respective fluorogen for 10 min in selection buffer to form complex at corresponding concentrations indicated in figures. Microinjection parameters: Pi=90 [hPa]; Ti=0.3 [s]; Pc=10 [hPa]. The images were acquired in epi-fluorescence mode with a Nikon Ti-E inverted microscope, equipped with CFI Plan Apo × 60 oil (NA = 1.4) objective, and a Hamamatsu Orca Flash 4 sCMOS camera. The acquisition settings were: Hoechst (ex. 395 nm, em. 475±50 nm), Broccoli (ex: 470 nm, em: 531±40 nm); Corn (ex: 470 nm, em: 531±40 nm); Mango (ex: 470 nm, em: 531±40 nm); Coral (ex: 550 nm, em: 595±40 nm). The images were recorded using NIS Elements and then processed with Icy software.
Acknowledgments
We thank Rémi Leblay, Anne-Catherine Helfer, Elsa Gutzwiller and Gautier Lieber for technical assistance as well as Pascale Romby and Isabelle Caldelari for fruitful scientific discussion. We thank Oleksandr Kucherak for synthesis of intermediate 2, Eleonore Real for scientific discussions, Jurga Valanciunaite and Nicolas Humbert for the technical assistance. This work received financial support from Agence Nationale de la Recherche (BrightRiboProbes, ANR-16-CE11-0010-01/02 (M.R. and A.S.K.) and BacNet, ANR-10-BINF_02_02 (M.R.)) and ERC Consolidator grant BrightSens 648528 (A.S.K.). This work has also been published under the framework of the LabEx: ANR-10-LABX-0036_NETRNA (M.R.) and benefits from a funding from the state managed by the French National Research Agency as part of the Investments for the future program. It was also supported by the Université de Strasbourg and the Centre National de la Recherche Scientifique.
Footnotes
Data Availability Statement
The data supporting the findings of this study are available within the paper and its supplementary information file.
Authors Contributions
A.S.K., M.C. and M.R. proposed the concept of this study; M.C. synthesized Gemini-561; F.B. and K.T.F. performed main part of experimental work and data analysis with help and supervision of M.R., M.C., and A.S.K.; F.B. and M.R. developed the aptamer o-Coral, including its analogues, and characterized their structure with the help of S.M; A.A. and F.B. characterized the contribution of the different selected mutations. M.C., K.T.F., F.B. and A.S.K. characterized Gemini-561/o-Coral fluorescence properties in solution; F.B. generated plasmids encoding o-Coral; K.T.F. realized all cellular studies, and, with help of A.S.K. and M.C., he realized all fluorescence imaging of cells.
Competing Interests
F.B., K.T.F., M.C., A.K., M.R., the University of Strasbourg and the CNRS have filed a patent application covering the technology presented in this manuscript.
Supplementary Material
References
- 1.Levine JH, Lin Y, Elowitz MB. Functional roles of pulsing in genetic circuits. Science. 2013;342:1193–200. doi: 10.1126/science.1239999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–24. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 3.Xue L, Karpenko IA, Hiblot J, Johnsson K. Imaging and manipulating proteins in live cells through covalent labeling. Nat Chem Biol. 2015;11:917–23. doi: 10.1038/nchembio.1959. [DOI] [PubMed] [Google Scholar]
- 4.Dean KM, Palmer AE. Advances in fluorescence labeling strategies for dynamic cellular imaging. Nat Chem Biol. 2014;10:512–23. doi: 10.1038/nchembio.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertrand E, et al. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–45. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 6.Tutucci E, et al. An improved MS2 system for accurate reporting of the mRNA life cycle. Nat Methods. 2018;15:81–89. doi: 10.1038/nmeth.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buxbaum AR, Haimovich G, Singer RH. In the right place at the right time: visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol. 2015;16:95–109. doi: 10.1038/nrm3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouhedda F, Autour A, Ryckelynck M. Light-Up RNA Aptamers and Their Cognate Fluorogens: From Their Development to Their Applications. Int J Mol Sci. 2018;19:44. doi: 10.3390/ijms19010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li C, Tebo AG, Gautier A. Fluorogenic Labeling Strategies for Biological Imaging. Int J Mol Sci. 2017;18:1473. doi: 10.3390/ijms18071473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klymchenko AS. Solvatochromic and Fluorogenic Dyes as Environment-Sensitive Probes: Design and Biological Applications. Acc Chem Res. 2017;50:366–375. doi: 10.1021/acs.accounts.6b00517. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, et al. Tandem Spinach Array for mRNA Imaging in Living Bacterial Cells. Sci Rep. 2015;5 doi: 10.1038/srep17295. 17295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Babendure JR, Adams SR, Tsien RY. Aptamers switch on fluorescence of triphenylmethane dyes. J Am Chem Soc. 2003;125:14716–7. doi: 10.1021/ja037994o. [DOI] [PubMed] [Google Scholar]
- 13.You M, Jaffrey SR. Structure and Mechanism of RNA Mimics of Green Fluorescent Protein. Annu Rev Biophys. 2015;44:187–206. doi: 10.1146/annurev-biophys-060414-033954. [DOI] [PubMed] [Google Scholar]
- 14.Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science. 2011;333:642–6. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strack RL, Disney MD, Jaffrey SR. A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA. Nat Methods. 2013;10:1219–24. doi: 10.1038/nmeth.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filonov GS, Moon JD, Svensen N, Jaffrey SR. Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J Am Chem Soc. 2014;136:16299–308. doi: 10.1021/ja508478x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song W, Strack RL, Svensen N, Jaffrey SR. Plug-and-Play Fluorophores Extend the Spectral Properties of Spinach. J Am Chem Soc. 2014;136:1198–201. doi: 10.1021/ja410819x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han KY, Leslie BJ, Fei J, Zhang J, Ha T. Understanding the photophysics of the spinach-DFHBI RNA aptamer-fluorogen complex to improve live-cell RNA imaging. J Am Chem Soc. 2013;135:19033–8. doi: 10.1021/ja411060p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Constantin TP, et al. Synthesis of new fluorogenic cyanine dyes and incorporation into RNA fluoromodules. Org Lett. 2008;10:1561–4. doi: 10.1021/ol702920e. [DOI] [PubMed] [Google Scholar]
- 20.Murata A, Sato S, Kawazoe Y, Uesugi M. Small-molecule fluorescent probes for specific RNA targets. Chem Commun (Camb) 2011;47:4712–4. doi: 10.1039/c1cc10393h. [DOI] [PubMed] [Google Scholar]
- 21.Dolgosheina EV, et al. RNA mango aptamer-fluorophore: a bright, high-affinity complex for RNA labeling and tracking. ACS Chem Biol. 2014;9:2412–20. doi: 10.1021/cb500499x. [DOI] [PubMed] [Google Scholar]
- 22.Tan X, et al. Fluoromodules Consisting of a Promiscuous RNA Aptamer and Red or Blue Fluorogenic Cyanine Dyes: Selection, Characterization, and Bioimaging. J Am Chem Soc. 2017;139:9001–9009. doi: 10.1021/jacs.7b04211. [DOI] [PubMed] [Google Scholar]
- 23.Arora A, Sunbul M, Jaschke A. Dual-colour imaging of RNAs using quencher- and fluorophore-binding aptamers. Nucleic Acids Res. 2015;43:e144. doi: 10.1093/nar/gkv718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sunbul M, Jaschke A. Contact-mediated quenching for RNA imaging in bacteria with a fluorophore-binding aptamer. Angew Chem Int Ed Engl. 2013;52:13401–4. doi: 10.1002/anie.201306622. [DOI] [PubMed] [Google Scholar]
- 25.Sunbul M, Jaschke A. SRB-2: a promiscuous rainbow aptamer for live-cell RNA imaging. Nucleic Acids Res. 2018;46:e110. doi: 10.1093/nar/gky543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sparano BA, Koide K. A strategy for the development of small-molecule-based sensors that strongly fluoresce when bound to a specific RNA. J Am Chem Soc. 2005;127:14954–5. doi: 10.1021/ja0530319. [DOI] [PubMed] [Google Scholar]
- 27.Shin I, et al. Live-cell imaging of Pol II promoter activity to monitor gene expression with RNA IMAGEtag reporters. Nucleic Acids Res. 2014;42:e90. doi: 10.1093/nar/gku297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holeman LA, Robinson SL, Szostak JW, Wilson C. Isolation and characterization of fluorophore-binding RNA aptamers. Fold Des. 1998;3:423–31. doi: 10.1016/S1359-0278(98)00059-5. [DOI] [PubMed] [Google Scholar]
- 29.Karpenko IA, et al. Fluorogenic Squaraine Dimers with Polarity-Sensitive Folding As Bright Far-Red Probes for Background-Free Bioimaging. Journal of the American Chemical Society. 2015;137:405–412. doi: 10.1021/ja5111267. [DOI] [PubMed] [Google Scholar]
- 30.Okamoto A. ECHO probes: a concept of fluorescence control for practical nucleic acid sensing. Chem Soc Rev. 2011;40:5815–28. doi: 10.1039/c1cs15025a. [DOI] [PubMed] [Google Scholar]
- 31.Song W, et al. Imaging RNA polymerase III transcription using a photostable RNA-fluorophore complex. Nat Chem Biol. 2017;13:1187–1194. doi: 10.1038/nchembio.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warner KD, et al. A homodimer interface without base pairs in an RNA mimic of red fluorescent protein. Nat Chem Biol. 2017;13:1195–1201. doi: 10.1038/nchembio.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–22. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 34.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–10. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 35.Gotrik M, et al. Direct Selection of Fluorescence-Enhancing RNA Aptamers. J Am Chem Soc. 2018;140:3583–3591. doi: 10.1021/jacs.7b10724. [DOI] [PubMed] [Google Scholar]
- 36.Autour A, Westhof E, Ryckelynck M. iSpinach: a fluorogenic RNA aptamer optimized for in vitro applications. Nucleic Acids Res. 2016;44:2491–500. doi: 10.1093/nar/gkw083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Autour A, et al. Fluorogenic RNA Mango aptamers for imaging small non-coding RNAs in mammalian cells. Nat Commun. 2018;9:656. doi: 10.1038/s41467-018-02993-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryckelynck M, et al. Using droplet-based microfluidics to improve the catalytic properties of RNA under multiple-turnover conditions. RNA. 2015;21:458–69. doi: 10.1261/rna.048033.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shulov I, et al. Fluorinated counterion-enhanced emission of rhodamine aggregates: ultrabright nanoparticles for bioimaging and light-harvesting. Nanoscale. 2015;7:18198–210. doi: 10.1039/c5nr04955e. [DOI] [PubMed] [Google Scholar]
- 40.Collot M, et al. Calcium rubies: a family of red-emitting functionalizable indicators suitable for two-photon Ca2+ imaging. J Am Chem Soc. 2012;134:14923–31. doi: 10.1021/ja304018d. [DOI] [PubMed] [Google Scholar]
- 41.Despras G, et al. H-Rubies, a new family of red emitting fluorescent pH sensors for living cells. Chem Sci. 2015;6:5928–5937. doi: 10.1039/c5sc01113b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duval M, et al. RNA Structure and FoldingBiophysical Techniques and Prediction Methods. De Gruyter; 2013. Footprinting methods for mapping RNA-protein and RNA-RNA interactions; pp. 29–50. [Google Scholar]
- 43.Trachman RJ, 3rd, Truong L, Ferre-D'Amare AR. Structural Principles of Fluorescent RNA Aptamers. Trends Pharmacol Sci. 2017;38:928–939. doi: 10.1016/j.tips.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Filonov GS, Kam CW, Song W, Jaffrey SR. In-gel imaging of RNA processing using broccoli reveals optimal aptamer expression strategies. Chem Biol. 2015;22:649–60. doi: 10.1016/j.chembiol.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Good PD, et al. Expression of small, therapeutic RNAs in human cell nuclei. Gene Ther. 1997;4:45–54. doi: 10.1038/sj.gt.3300354. [DOI] [PubMed] [Google Scholar]
- 46.Guet D, et al. Combining Spinach-tagged RNA and gene localization to image gene expression in live yeast. Nat Commun. 2015;6 doi: 10.1038/ncomms9882. 8882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y, Beyer A, Aebersold R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell. 2016;165:535–50. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 48.Baugh C, Grate D, Wilson C. 2.8 A crystal structure of the malachite green aptamer. J Mol Biol. 2000;301:117–28. doi: 10.1006/jmbi.2000.3951. [DOI] [PubMed] [Google Scholar]
- 49.Shelke SA, et al. Structural basis for activation of fluorogenic dyes by an RNA aptamer lacking a G-quadruplex motif. Nat Commun. 2018;9 doi: 10.1038/s41467-018-06942-3. 4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo JU, Bartel DP. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science. 2016 353 doi: 10.1126/science.aaf5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.