Abstract
Introduction
The study objective was to build a machine learning model to predict incident mild cognitive impairment, Alzheimer's Disease, and related dementias from structured data using administrative and electronic health record sources.
Methods
A cohort of patients (n = 121,907) and controls (n = 5,307,045) was created for modeling using data within 2 years of patient's incident diagnosis date. Additional cohorts 3–8 years removed from index data are used for prediction. Training cohorts were matched on age, gender, index year, and utilization, and fit with a gradient boosting machine, lightGBM.
Results
Incident 2-year model quality on a held-out test set had a sensitivity of 47% and area-under-the-curve of 87%. In the 3-year model, the learned labels achieved 24% (71%), which dropped to 15% (72%) in year 8.
Discussion
The ability of the model to discriminate incident cases of dementia implies that it can be a worthwhile tool to screen patients for trial recruitment and patient management.
Keywords: Prediction, Machine learning, Onset of dementia, Gradient boosting machine, Alzheimer's disease
1. Introduction
Accurate prediction of future onset of Alzheimer's disease or related dementias (ADRD) has several important practical applications. In particular, it facilitates the identification of individuals who are at high risk of developing ADRD to support the clinical development of novel treatments. Commonly, patients are identified after they have already experienced significant neurodegeneration. Predictive models that can identify patients who will subsequently develop dementia might create the opportunity for such persons to be evaluated for potential contributors to cognitive impairment (e.g., high-risk medications, depression, medical illnesses), participate in prevention-based interventions, as well as identify candidates for clinical trials. Persons predicted to be at risk can also be offered the opportunity for more thoughtful planning for the future. Finally, predicting the onset of ADRD can be very helpful for identifying individuals who are at risk of institutionalization. Community support services may help to delay institutionalization, as well as help families to cope with the host of planning issues raised by the loss of independence resulting from dementia [1].
A number of prior dementia risk models have been published in the peer-reviewed literature [[2], [3], [4], [5], [6], [7], [8], [9], [10]]. These have been shown to have moderate discriminatory capabilities (area under the curve ranging from 0.60 to 0.78). Most of these studies used clinical data for model estimation that limits their generalizability to other settings. Additionally, these prior studies do not directly address a key assumption of supervised machine learning methods—are the labels on the data correct? For ADRD, we know that the rules used to label disease onset are error prone due to undercoding, provider variation, and other factors. This raises numerous questions for the application of machine learning. Would a model cement in place current non-optimal diagnostic practices if it uses the raw labels? To what extent is moderate model quality due to erroneous labels versus other limitations of the modeling process? What can be done to enhance the quality of the labels before applying the machine learning models?
This paper extends previous research by basing model estimation on a very large integrated dataset of medical claims and electronic health record data, as well as the use of more sophisticated machine learning estimation methods. In particular, we utilize label learning methods designed to address the inaccuracy of diagnostic coding typically found in administrative claims and electronic health record (EHR) datasets. We hypothesized that the use of larger, more complex data, label learning, and sophisticated machine learning methods lead to substantial improvements in predictive algorithms for dementia, that is, raising sensitivity to 25% or more over the 16% result of Nori [10].
2. Methods
This study used de-identified administrative claims and EHR data between 2007 and 2017 from the OptumLabs® Data Warehouse (OLDW) [11]. The database contains longitudinal health information on enrollees and patients, representing a diverse mixture of ages, ethnicities, and geographical regions across the United States. The claims data in OLDW includes medical and pharmacy claims, laboratory results, and enrollment records for commercial and Medicare Advantage enrollees. Since this study involved the analysis of preexisting, de-identified data, it was exempt from Institutional Review Board approval.
The study dates from 1/1/2007 to 12/31/2017 coincide with the earliest date that EHR data is available, and the end of the latest year after the project was initiated. Patients are 45 years old or more on their confirmation date.
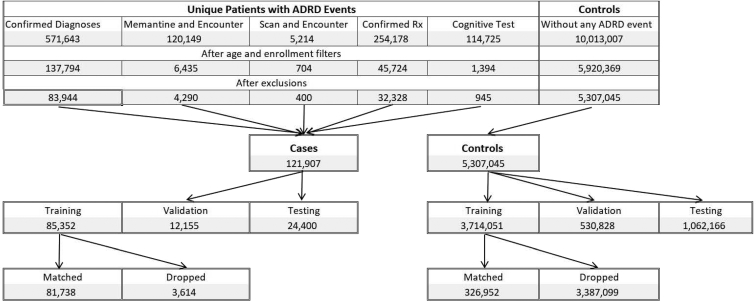
The outcome variable of all the cohorts is an incident diagnosis of ADRD, including mild cognitive impairment (MCI). The study chose this mixed set of diagnoses because of the confusion in patient presentation among these conditions that leads to inaccurate or incomplete coding of incident disease [12]. In this paper, we adopt a label learning methodology designed to address this issue [13]. Full details on the medical coding and rules used to establish the outcome variable and the cohorts are in the Supplementary Material A. Fig. 1 provides the attrition table for the structural EHR (SEHR) cohort in the label learning phase. Table 1 provides sample sizes and summary statistics for each of the data sets. The SEHR, EHR-only subset is less than 0.2% of the data (N = 8644) and omitted in Table 1 as a non-representative subset but included in the analysis. The remainder of this section is an overview of the data sets.
Fig. 1.
Attrition of the two-year cohort into the training, validation and test data.
Table 1.
Data source sample sizes and summary statistics
| Cohort | Subset | N | Age mean (SD) | Encounters mean (SD) | Case prevalence, % | Female, % | Cardiovascular disease prevalence, % | Mood disorder prevalence, % |
|---|---|---|---|---|---|---|---|---|
| Claims | ClaimsOnly | 5,640,637 | 60.0 (10.7) | 10.7 (21.6) | 2.1 | 52.8 | 46.2 | 14.6 |
| SEHR | ClaimsOnly | 4,810,730 | 59.8 (10.6) | 10.6 (21.0) | 2.1 | 52.3 | 45.1 | 13.9 |
| SEHR | Mixed | 609,578 | 61.7 (11.3) | 11.3 (29.4) | 3.5 | 56.2 | 55.1 | 19.3 |
| Open-World | ClaimsOnly | 8,348,496 | 60.4 (10.7) | 10.7 (24.1) | 2.7 | 54.7 | 43.9 | 14.4 |
| Open-World | EHROnIy | 7,276,426 | 62.6 (11.4) | 11.4 (17.4) | 3.7 | 59.0 | 34.2 | 14.1 |
| Open-World | Mixed | 1,602,898 | 60.6 (10.7) | 10.7 (27.4) | 4.1 | 57.6 | 47.7 | 19.1 |
This study compared models estimated using three test data sets to understand better how clinical data adds information to the diagnosis. The simplest data set is claims-only. This data set uses 2 years of continuous enrollment in medical and pharmacy benefits. It uses only administrative data, not EHR data. The SEHR data set also uses 2 years of continuous enrollment exactly as the claims-only data, but it adds the cognitive testing inclusion rule and adds the structured EMR diagnoses and prescriptions written as data for analysis. The last cohort is an Open-World dataset that uses administrative data and EMR data without continuous enrollment. In place of continuous enrollment from a benefits file, this cohort uses a health care encounter in a calendar year as an indication of a patient's ability to access health care services. Consecutive years of encounters were used in place of 2 years of continuous enrollment. Everyone in the Open-World cohort must have two encounters (one in each year), but the other two cohorts only have one or more. Within this Open-World cohort are three distinct sets of patients; those with only claims data, those with only EHR data, and those with mixed data sources. Complete descriptive tables on each of the prediction cohorts are provided in the Supplementary Material B.
In all cohorts, the data included claims' diagnoses (ICD-10 and ICD-9 codes), NDC codes for pharmacy claims, procedure codes, and demographics (age, gender, and utilization measured as unique service days). The study needed to use ICD-10 and ICD-9 codes because the time frame included the transition from ICD-9 to ICD-10. All ICD-10 codes were mapped back to ICD-9 codes before creating the variables using the CMS mappings [14]. The CPT codes were limited to radiology (7*), psychiatric interviews (908*), or neurological testing (961*).
In the SEHR and Open-World cohorts, the EHR data provided diagnosis codes, procedure codes, and prescriptions written in a structured format; no clinical notes were used. These EHR data are merged with the claims data without differentiation.
In the SEHR 2-year incident cohort, there are 13,586 unique medical codes available for the analytic models. Table 1 shows that the Open-World data skew older, as expected, due to more Medicare-eligible patients. The Open-World data subsets show diversity in the prevalence of cardiovascular disease and mood disorders due to the combination of 2 required visits (more than Claims or SEHR) and missingness of some visits in the OW-EHR and OW-Mixed sets.
2.1. Analytic Methods
The study uses two stages of prediction to predict ADRD over a range of 3 to 8 years prior to incident diagnosis, which was determined by having a 2-year clean period. This process is inspired by semi-supervised learning, where a small set of labeled data is used to create a model to predict data labels [13]. This model is applied to a much larger set of unlabeled data to create labels, and then the larger data set with predicted labels are used to create more accurate predictive models. The additional size of the second model allows more accuracy on the same task.
We adopt this two-stage method not to use more unlabeled data, but to improve the incomplete and error-prone case and control labels created from claims and EHR data [12]. These data sources underreport patients with the disease, implying there are controls that should be cases; in other instances, cases may be mislabeled. By modeling the cases and controls at the time of diagnosis, we create a consistent probability label for all patients—consistent, meaning that two patients with the same data profile have the same fitted value from the model. This overcomes a key issue of provider variation in recording diagnoses for the patients. This provider variation can undermine the quality of the machine learning by embedding too much noise in the outcome.
In this study, we alter the standard semi-supervised machine learning method by adding a time component. In this work, the first stage of label learning creates a propensity model of a patient being like the incident diagnosed patients using data in a two year window ending at the index date; the second stage of prediction then uses these propensity scores to update the outcome for a model using data in windows for years 3 and 4, 4 and 5, up to 8 and 9 years removed from the patient's index date (i.e., 6 different new models).
We set the new outcome labels based on the details of the inter-quartile range of the case's calibrated scores in the label learning model. Patients scoring below the 25th percentile of the case's scores are set to controls, and those above the 75th percentile are set to cases. More simply, we reset the labels of the least likely cases to be controls, and most likely controls to be cases. Because this label change is performed based on the percentiles of the cases, the thresholds wind up changing approximately 1% of all scores (50% in 2% prevalent cases). In a clinical setting, one could choose the lower and upper threshold of scores to reflect the economics of these choices using the profile of the learned label distribution.
All model fitting is performed with LightGBM [15]. LightGBM is an open-source implementation of a gradient boosting framework that uses a sequence of trees to solve classification or regression models. Details of the variable selection and model fitting and post-fitting calibration processes are in the Supplementary Material C. Special care was taken, so the process is automated and simple. The same fitting steps are applied to the label learning and prediction stages.
The label learning model is trained only on the SEHR data. This data set was chosen because it is nearly the same as the claims-only data (so it is simple to collect), and its use of cognitive tests may increase the accuracy of the index date versus the claims only data. This trained model is used to score the Claims-only and Open-World data sets to create their learned labels.
Predictive models are fit to the SEHR, Claims-Only, and Open-World cohorts separately. For the Open-World cohort model, quality assessments are computed on the subsets of patients who have claims-only data, EHR-only data, and mixed data. These subsets provide additional data to understand how the model may generalize to various data inputs.
To evaluate the results, we used an array of comparisons. First, we used a baseline of predicting the original label from each data set, that is, each cohort, and each time period, 18 models in all. Second, we fitted each model to the learned labels using the same process.
A direct comparison of the results is not obvious. The two models for a single data set have changed their outcomes and the prevalence of their outcomes. The degree of change depends on the survivorship of patients from the label learning period into the time period of the prediction. While many comparisons could be computed, in the end, changing the labels in this data-driven way would be similar to changing the way the inclusion rules are computed, then comparing model fits.
3. Results
In the label learning model, test data held out from the same data source shows an excellent fit of 87% area-under-the-curve (AUC) and a sensitivity of 0.47 (Table 2). Since AUC can be a misleading indicator of fit as it tends to perform very well in highly unbalanced samples, we also computed the sensitivity of the models. The study uses multiple thresholds to compute the sensitivity within the model because of the wide range in prevalence in the outcome across the age groups. In each age group, the model chooses the thresholds to be the age-based case prevalences. Choosing the threshold by case prevalence forces false positives and false negatives to be equal, which in turn forces sensitivity to equal the positive predictive value (Supplementary Material C). From these age group assignments, the global model quality measures can be computed. Table 2 shows the label learning model quality per age-group as fit to the original labels. The model performs best in the elderly (0.53 sensitivity, 81% AUC), where the prevalence and health care utilization is highest and has the least sensitivity in the youngest. However, model discrimination, as measured by AUC, is highest in the populations less than 70 years old, and least in those over 70.
Table 2.
Label Learning Model results by age group
| Age group | Sensitivity | AUC | Lift | True positives | False positives | True negatives | False negatives | Case, % | Case count | Total count |
|---|---|---|---|---|---|---|---|---|---|---|
| 45,55 | 0.29 | 0.89 | 94.0 | 403 | 977 | 444,939 | 977 | 0.31 | 1380 | 447,296 |
| 55,60 | 0.34 | 0.90 | 63.8 | 325 | 622 | 175,999 | 622 | 0.53 | 947 | 177,568 |
| 60,64 | 0.39 | 0.90 | 48.8 | 374 | 583 | 117,945 | 583 | 0.80 | 957 | 119,485 |
| 64,70 | 0.38 | 0.88 | 24.0 | 844 | 1396 | 139,148 | 1396 | 1.57 | 2240 | 142,784 |
| 70,75 | 0.43 | 0.85 | 10.7 | 1499 | 1998 | 81,507 | 1998 | 4.02 | 3497 | 87,002 |
| 75,80 | 0.49 | 0.83 | 5.2 | 2722 | 2818 | 49,961 | 2818 | 9.50 | 5540 | 58,319 |
| 80,99 | 0.53 | 0.81 | 2.9 | 5205 | 4634 | 39,639 | 4634 | 18.18 | 9839 | 54,112 |
| Summary | 0.47 | 0.87 | 20.9 | 11,372 | 13,028 | 1,049,138 | 13,028 | 2.25 | 24,400 | 1,086,566 |
Abbreviation: AUC, area-under-the-curve.
Table 3 shows how the model fit to the original labels can be used with different thresholds and measures in the test data. The thresholds are for calibrated scores; they reflect the fraction of cases in the raw score neighborhoods. Adjacent to the original label results is the result for changing only the outcomes to the learned labels while keeping the fitted values the same. This demonstrates the large improvement in the quality measures when changing the labels. This is not to say a model fit to the learned labels would have this performance; the data for the prediction models is in Table 4.
Table 3.
Comparison of onset model quality for original versus learned labels
| Original labels |
Learned labels |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Prediction threshold | Sensitivity of ADRD | Specificity of ADRD | Positive predictive value of ADRD | Proportion of cohort over threshold | Prediction threshold | Sensitivity of ADRD | Specificity of ADRD | Positive predictive value of ADRD | Proportion of cohort over threshold |
| Choosing by threshold greater than | |||||||||
| 0.75 | 0.060 | 1.000 | 0.857 | 0.002 | 0.75 | 0.075 | 1.000 | 1.000 | 0.002 |
| 0.50 | 0.180 | 0.998 | 0.681 | 0.006 | 0.50 | 0.283 | 1.000 | 1.000 | 0.006 |
| 0.20 | 0.388 | 0.987 | 0.405 | 0.021 | 0.20 | 0.619 | 0.991 | 0.604 | 0.021 |
| Choosing by sensitivity | |||||||||
| 0.102 | 0.50 | 0.971 | 0.282 | 0.040 | 0.328 | 0.50 | 0.999 | 0.926 | 0.011 |
| 0.007 | 0.90 | 0.545 | 0.043 | 0.465 | 0.040 | 0.90 | 0.921 | 0.196 | 0.096 |
| 0.004 | 0.95 | 0.325 | 0.031 | 0.681 | 0.031 | 0.95 | 0.893 | 0.160 | 0.124 |
| Choosing by specificity | |||||||||
| 0.064 | 0.572 | 0.95 | 0.209 | 0.061 | 0.061 | 0.830 | 0.95 | 0.262 | 0.066 |
| 0.223 | 0.353 | 0.99 | 0.448 | 0.018 | 0.189 | 0.634 | 0.99 | 0.576 | 0.023 |
| 0.610 | 0.128 | 0.999 | 0.747 | 0.004 | 0.326 | 0.503 | 0.999 | 0.916 | 0.012 |
Abbreviation: ADRD, Alzheimer's disease or related dementias.
Table 4.
Sensitivity (area-under-the-curve) scores over different time windows
| Time window | Outcome label | SEHR | OW-C | OW-E | OW-M | Claims |
|---|---|---|---|---|---|---|
| Label Learning | Original | 0.47 (0.87) | 0.49 (0.87) | 0.41 (0.83) | 0.50 (0.86) | 0.46 (0.87) |
| 3 year | Original | 0.26 (0.70) | 0.29 (0.70) | 0.26 (0.67) | 0.29 (0.68) | 0.23 (0.69) |
| 3 year | Learned | 0.24 (0.71) | 0.28 (0.73) | 0.27 (0.72) | 0.30 (0.72) | 0.24 (0.71) |
| 4 year | Original | 0.27 (0.67) | 0.29 (0.68) | 0.26 (0.66) | 0.29 (0.66) | 0.25 (0.69) |
| 4 year | Learned | 0.21 (0.68) | 0.27 (0.72) | 0.26 (0.72) | 0.29 (0.71) | 0.20 (0.71) |
| 5 year | Original | 0.25 (0.64) | 0.27 (0.63) | 0.24 (0.61) | 0.26 (0.62) | 0.25 (0.67) |
| 5 year | Learned | 0.22 (0.66) | 0.24 (0.71) | 0.21 (0.71) | 0.25 (0.70) | 0.23 (0.68) |
| 6 year | Original | 0.26 (0.68) | 0.27 (0.65) | 0.23 (0.64) | 0.27 (0.64) | 0.25 (0.69) |
| 6 year | Learned | 0.22 (0.67) | 0.24 (0.69) | 0.23 (0.69) | 0.25 (0.69) | 0.23 (0.69) |
| 7 year | Original | 0.25 (0.65) | 0.25 (0.67) | 0.21 (0.64) | 0.26 (0.66) | 0.26 (0.68) |
| 7 year | Learned | 0.22 (0.67) | 0.21 (0.69) | 0.20 (0.67) | 0.22 (0.68) | 0.18 (0.68) |
| 8 year | Original | 0.25 (0.63) | 0.25 (0.63) | 0.22 (0.60) | 0.28 (0.62) | 0.24 (0.59) |
| 8 year | Learned | 0.15 (0.72) | 0.21 (0.65) | 0.21 (0.63) | 0.25 (0.66) | 0.18 (0.70) |
Abbreviations: SEHR, structured electronic health record data; OW-C, Open World claims only data; OW-E, Open World EHR data; OW-M Open World mixed data.
Tables 2 and 4 also show the model lift statistic. Lift is the ratio of sensitivity to the prevalence and better highlights improvements in sensitivity for low prevalence models.
The label learning model is also used to score data from claims-only and three subsets of the Open-World data, claims-only, EHR-only and mixed (OW-C, OW-E, and OW-M) (Table 4). These 4 tests show the sensitivity degrades only slightly for OW-E (0.41, 83%) and claims-only (0.46, 87%), and improves in OW-C (0.49, 87%) and OW-M (0.50, 86%). The OW-E cohort has the fewest encounters per member due to missingness, so the lower quality of that model is understandable.
As the time horizon grows from 3 to 8 years, the model accuracy falls as the patient population size declines as fewer incident patients are available, and the number of medical encounters falls.
Fitting to the learned labels generally results in slightly lower sensitivity but better AUC values (Table 4). This holds true in each data set and across the years.
Table 5 reports the top ten features in the label learning model. These include several codes indicating neurological testing, presence of neurological diseases, and diagnoses of altered mental status and presence of psychotic and schizophrenic disorders. Details on other features in the model are provided in the Supplementary Materials A–D.
Table 5.
Top 10 Features that explain the model prediction in Label Learning∗
| Type of variable | Code | Time window (days) | Code description | Percent gain | Cumulative gain |
|---|---|---|---|---|---|
| icd9 | 78097 | 730 | Altered Mental Status | 7.2 | 7.2 |
| cpt4 | 70551 | 730 | Magnetic Resonance (e.g., Proton) Imaging, Brain (including Brain Stem); Without Contrast Material | 6.4 | 13.6 |
| etg | 319900 | 60 | Neurological Diseases Signs & Symptoms | 4.4 | 18.0 |
| cpt4 | 70450 | 730 | Computed Tomography, Head Or Brain; Without Contrast Material | 4.2 | 22.3 |
| cpt4 | 70551 | 60 | Magnetic Resonance (e.g., Proton) Imaging, Brain (including Brain Stem); Without Contrast Material | 4.0 | 26.3 |
| cpt4 | 96118 | 730 | Neuropsychological Testing (e.g., Halstead-reitan Neuropsychological Battery, Wechsler Memory Scales And Wisconsin Card Sorting Test), Per Hour Of The Psychologist's Or Physician's Time, Both Face-to-face Time Administering Tests To The Patient And Time Interpreting These Test Results And Preparing The Report | 3.4 | 29.7 |
| etg | 319900 | 730 | Neurological Diseases Signs & Symptoms | 2.4 | 32.1 |
| cpt4 | 96116 | 730 | Neurobehavioral Status Exam (clinical Assessment Of Thinking, Reasoning And Judgment, e.g., Acquired Knowledge, Attention, Language, Memory, Planning And Problem Solving, And Visual Spatial Abilities), Per Hour Of The Psychologist's Or Physician's Time, Both Face-to-face Time With The Patient And Time Interpreting Test Results And Preparing The Report | 2.4 | 34.5 |
| etg | 239300 | 730 | Psychotic & Schizophrenic Disorders | 2.3 | 36.8 |
| cpt4 | 96118 | 60 | Neuropsychological Testing (e.g., Halstead-Reitan Neuropsychological Battery, Wechsler Memory Scales And Wisconsin Card Sorting Test), Per Hour Of The Psychologist's Or Physician's Time, Both Face-to-face Time Administering Tests To The Patient And Time Interpreting These Test Results And Preparing The Report | 2.3 | 39.1 |
Additional features reported in Supplementary Material D.
4. Discussion
The results presented in this paper contribute to a growing literature on dementia risk models, although relatively few have used claims or EHR data. A recent systematic review of dementia risk prediction models [16] found models that could be grouped into five categories: (1) demographic factors only; (2) cognitive-based (cognitive test scores); (3) health variables and risk factors; (4) genetic risk scores; and (5) multivariable models that combined demographic with health and lifestyle factors. However, of the 21 models reviewed, only four used model validation techniques by testing the models on different data than was used for estimation. Consequently, it is difficult to know how the models would perform in other settings.
Previously, traditional regression modeling techniques have been applied to clinical data to successfully identify early cases of ADRD [17], to cluster patients into fast versus slow progression sub-types [18], to distinguish mild cognitive impairment or normal aging from early dementia [19], and to assist in the interpretation and clinical significance of findings from neuroimaging studies [[20], [21], [22], [23], [24]].
Recently, administrative claims data have been used to develop dementia risk models with performance similar to other models in the published literature [9,10]. Models using claims data are more widely available for large populations, offering the potential for their practical use in screening and identifying patients. These studies also included validation components. Our study used label learning methods that address dementia undercoding and miscoding. In one study, Alzheimer's disease and related dementias were recorded as a diagnosis for less than 25% of patients with moderate to severe cognitive impairment [25]; and in another, physicians were unaware of cognitive impairment in more than 40% of their cognitively impaired patients [26]. Among participants in a Medicare Alzheimer's Disease Demonstration, less than 20% of participants were classified with dementia of the Alzheimer type based on a year's worth of claims data, although 68% carried that diagnosis upon referral [27]. A review of seven studies examining the extent to which dementia is omitted as a cause of death, found that the reporting on death certificates ranged from 7.2% to 41.8% [28].
This prior work [9,10,[17], [18], [19], [20], [21], [22], [23], [24]] does not address the issue of errors in labeling the patients as cases and controls. When a patient's diagnoses are incorrectly assigned, the machine learning algorithm will learn the wrong patterns. As this study shows, assigning a learned label can improve some measures of model quality, but there remains an issue of ground-truth about the assignment of the new labels. Improving methods to assess diagnostic accuracy, as well as promoting cognitive data collection during clinical encounters, should be an area of further active research.
Table 3 shows that different thresholds can serve different purposes. A wide-ranging public service announcement can target 90% of at-risk people by contacting less than half the population (46.5%), or if one wishes to use the learned labels, the same 90% outreach can be done by reaching out to 9.6% of the population. A much more targeted outreach with 99.9% specificity would have 74.7% positive predictive value, or 91.6% PPV in the learned labels.
As with recent studies using claims data, the current study includes a validation component. The use of the two-step label learning technique, combined with more sophisticated machine learning estimation methods [13,15], results in a model with a sensitivity of 47% and AUC of 87% (Table 2). Model performance is strong across all age groups, but the lift is greatest in the youngest age groups (e.g., improvement of 94.7%). This is particularly important in using the models to identify potential patients for recruitment into clinical studies as this results in very substantial improvements in the efficiency with which preclinical-early stage disease potential study subjects can be identified, as well as potentially detecting acute progression. Model performance is consistent across datasets and time horizons (Table 4), with the exception of years 7 and 8, where the reduction in data volume results in deterioration in performance (Supplementary Material D).
Table 5 and Supplementary Material D identify the top predictive features. The top ten predictors were dominated by several codes indicating neurological testing, presence of neurological diseases, signs and symptoms, and diagnoses of altered mental status, and the presence of psychosis. Details on other features in the model are provided in the Supplementary Material D.
Although not appearing in the top ten features reported in Table 5, evidence of vascular disease was represented among the features explaining the top 80% of model prediction (Supplementary Material D), which supports clinical data suggesting an overlap of risk factors for cardiovascular disease with vascular dementia [29,30]. Mental health diagnoses and treatments were also prominent in both the top 10 features and the extended feature set (Supplementary Table D.1). However, the association of dementia risk with diabetes mellitus found in previous studies was not evident among the top predictive features [30]. Anticholinergic drug exposure has also been identified as a risk factor for dementia [31]. Anticholinergic drugs figured prominently in the list of top predictive features in our models as well (Supplementary Material D).
Fitting the model to different outcome variables was performed to address variation in diagnostic quality. The fits show that the process can generate similar results at the population level, but that the learned labels are not a silver bullet to creating excellent models. Changing the outcome variable cannot address the issues of lack of precision in data collection that limit model feature quality, and therefore, model performance. We call upon the clinical profession to place greater emphasis on the accuracy of diagnostic information that they enter into EHR systems and submit to payers for reimbursement. Accurate diagnostic data is fundamental for identifying patients for early clinical intervention, community support, financial planning, recruitment in clinical trials, and other purposes.
Model performance can be improved in the following areas. More precise rules for an incident disease that use provider specialty or more comprehensive confirmation rules could increase label accuracy. Calibration of the fitted scores can be enhanced using a smoothing model. Grid search for optimal fits is inefficient. This grid search chose to optimize to the sensitivity, but other objectives could give different results. Deep learning models can be used to fit the data, enhancing the fit by exploiting the nonlinear nature of the deep learning networks. Adding clinical data, like notes and cognitive testing results, from the EHR system, can add nuance that providers do not enter into structured codes.
Research in context.
-
1.
Systematic review: A recent survey by Tang 2015 found that there are no dementia risk models that rely solely on structured claims and EHR data, nor do any use advanced machine learning methods. Since that report, Albrecht 2018 and Nori 2018 have published regression models using claims data, but not advanced machine learning models.
-
2.
Interpretation: Model accuracy over 80% area-under-the-curve and sensitivity over 40% is an indication that the model developed has utility to prescreen patients for follow-up diagnosis, or evaluation for clinical trials. Evaluation of the model across data sets with millions of patients and differing enrollment criteria strengthens its generalization to data gathered from other settings. The model quality is a substantial improvement over the prior work using claims data alone.
-
3.
Future directions: Additional progress will require better methods, more patient and family data and better clinical documentation to determine which patients are miss-identified in the source data.
Acknowledgments
The authors thank Pamela Hansen of OptumLabs for her diligent support of the project. This project was supported in part by funding from Optum, Global CEOi Initiative, Biogen, Janssen Pharmaceutical, Merck and computing hardware donated from Dell.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.trci.2019.10.006.
Supplementary data
References
- 1.Prince M.J. World Alzheimer Report 2011: the benefits of early diagnosis and intervention 2011. https://www.alz.co.uk/research/world-report-2011
- 2.Barnes D.E., Beiser A.S., Lee A., Langa K.M., Koyama A., Preis S.R. Development and validation of a brief dementia screening indicator for primary care. Alzheimer's Dement. 2014;10:656–665.e1. doi: 10.1016/j.jalz.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byeon H. A prediction model for mild cognitive impairment using random forests. Int J Adv Comput Sci App. 2015;6:8–12. [Google Scholar]
- 4.Barnes D.E., Covinsky K.E., Whitmer R.A., Kuller L.H., Lopez O.L., Yaffe K. Predicting risk of dementia in older adults: the late-life dementia risk index. Neurology. 2009;73:173–179. doi: 10.1212/WNL.0b013e3181a81636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Exalto L.G., Biessels G.J., Karter A.J., Huang E.S., Katon W.J., Minkoff J.R. Risk score for prediction of 10 year dementia risk in individuals with type 2 diabetes: a cohort study. Lancet Diabetes Endocrinol. 2013;1:183–190. doi: 10.1016/S2213-8587(13)70048-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Exalto L.G., Quesenberry C.P., Barnes D., Kivipelto M., Biessels G.J., Whitmer R.A. Midlife risk score for the prediction of dementia four decades later. Alzheimer's Dement. 2014;10:562–570. doi: 10.1016/j.jalz.2013.05.1772. [DOI] [PubMed] [Google Scholar]
- 7.Kivipelto M., Ngandu T., Laatikainen T., Winblad B., Soininen H., Tuomilehto J. Risk score for the prediction of dementia risk in 20 years among middle aged people: a longitudinal, population-based study. Lancet Neurol. 2006;5:735–741. doi: 10.1016/S1474-4422(06)70537-3. [DOI] [PubMed] [Google Scholar]
- 8.Reitz C., Tang M.-X., Schupf N., Manly J.J., Mayeux R., Luchsinger J.A. A summary risk score for the prediction of Alzheimer disease in elderly persons. Arch Neurol. 2010;67:835–841. doi: 10.1001/archneurol.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albrecht J.S., Hanna M., Kim D., Perfetto E.M. Predicting diagnosis of Alzheimer's disease and related dementias using administrative claims. JMCP. 2018;24:1138–1145. doi: 10.18553/jmcp.2018.24.11.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nori V.S., Hane C.A., Martin D.C., Kravetz A.D., Sanghavi D.M. Identifying incident dementia by applying machine learning to a very large administrative claims dataset. PLoS One. 2019;14:e0203246. doi: 10.1371/journal.pone.0203246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wallace P, Shah N, Dennen T, Bleicher P, Crown W. Optum Labs: building a novel node in the learning health care system. Health Aff (Millwood) 2014;33:1187–1194. doi: 10.1377/hlthaff.2014.0038. [DOI] [PubMed] [Google Scholar]
- 12.Murphy K., O'Connor D.A., Browning C.J., French S.D., Michie S., Francis J.J. Understanding diagnosis and management of dementia and guideline implementation in general practice: a qualitative study using the theoretical domains framework. Implementation Sci. 2014;9:31. doi: 10.1186/1748-5908-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu X., Goldberg A.B. Introduction to semi-supervised learning. Synth Lectures Artif Intelligence Machine Learn. 2009;3:1–130. [Google Scholar]
- 14.CMS ICD10 GEMS. https://www.cms.gov/Medicare/Coding/ICD10/2018-ICD-10-CM-and-GEMs.html
- 15.Ke G., Meng Q., Finley T., Wang T., Chen W., Ma W. LightGBM: a highly efficient gradient boosting decision tree. In: Guyon I., Luxburg U.V., Bengio S., Wallach H., Fergus R., Vishwanathan S., editors. Advances in Neural Information Processing Systems 30. Curran Associates, Inc.; 2017. pp. 3146–3154. [Google Scholar]
- 16.Tang E.Y.H., Harrison S.L., Errington L., Gordon M.F., Visser P.J., Novak G. Current developments in dementia risk prediction modelling: an updated systematic review. PLoS One. 2015;10:e0136181. doi: 10.1371/journal.pone.0136181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.So A., Hooshyar D., Park K., Lim H. Early diagnosis of dementia from clinical data by machine learning techniques. Appl Sci. 2017;7:651. [Google Scholar]
- 18.Gamberger D., Lavrač N., Srivatsa S., Tanzi R.E., Doraiswamy P.M. Identification of clusters of rapid and slow decliners among subjects at risk for Alzheimer's disease. Sci Rep. 2017;7:6763. doi: 10.1038/s41598-017-06624-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shankle W.R., Mani S., Pazzani M.J., Smyth P. Detecting very early stages of dementia from normal aging with machine learning methods. In: Keravnou E., Garbay C., Baud R., Wyatt J., editors. Lecture Notes in Artificial Intelligence: Artificial Intelligence in Medicine. Springer; Berlin, Heidelberg: 1997. pp. 71–85. [Google Scholar]
- 20.Dyrba M., Barkhof F., Fellgiebel A., Filippi M., Hausner L., Hauenstein K. Predicting prodromal Alzheimer's disease in subjects with mild cognitive impairment using machine learning classification of multimodal multicenter diffusion-tensor and magnetic resonance imaging data: predicting prodromal AD in MCI using multicenter imaging. J Neuroimaging. 2015;25:738–747. doi: 10.1111/jon.12214. [DOI] [PubMed] [Google Scholar]
- 21.Kloppel S., Stonnington C.M., Chu C., Draganski B., Scahill R.I., Rohrer J.D. Automatic classification of MR scans in Alzheimer's disease. Brain. 2008;131:681–689. doi: 10.1093/brain/awm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lao Z., Shen D., Xue Z., Karacali B., Resnick S.M., Davatzikos C. Morphological classification of brains via high-dimensional shape transformations and machine learning methods. NeuroImage. 2004;21:46–57. doi: 10.1016/j.neuroimage.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 23.Li S., Shi F., Pu F., Li X., Jiang T., Xie S. Hippocampal shape analysis of Alzheimer disease based on machine learning methods. Am J Neuroradiology. 2007;28:1339–1345. doi: 10.3174/ajnr.A0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Callahan C.M., Hendrie H.C., Tierney W.M. Documentation and evaluation of cognitive impairment in elderly primary care patients. Ann Intern Med. 1995;122:422–429. doi: 10.7326/0003-4819-122-6-199503150-00004. [DOI] [PubMed] [Google Scholar]
- 25.Chodosh J., Petitti D.B., Elliott M., Hays R.D., Crooks V.C., Reuben D.B. Physician recognition of cognitive impairment: evaluating the need for improvement. J Am Geriatr Soc. 2004;52:1051–1059. doi: 10.1111/j.1532-5415.2004.52301.x. [DOI] [PubMed] [Google Scholar]
- 26.Newcomer R.J., Clay T., Luxenberg J.S., Miller R.H. Misclassification and selection bias when identifying Alzheimer's disease solely from medicare claims records. J Am Geriatr Soc. 1999;47:215–219. doi: 10.1111/j.1532-5415.1999.tb04580.x. [DOI] [PubMed] [Google Scholar]
- 27.Romero J.P., Benito-León J., Mitchell A.J., Trincado R., Bermejo-Pareja F. Under reporting of dementia deaths on death certificates: a systematic review of Population-Based Cohort Studies. J Alzheimer's Dis. 2014;39:741–748. doi: 10.3233/JAD-131622. [DOI] [PubMed] [Google Scholar]
- 28.Adelborg K., Horváth-Puhó E., Ording A., Pedersen L., Sørensen H.T., Henderson V.W. Heart failure and risk of dementia: a Danish Nationwide Population-based Cohort Study. Eur J Heart Fail. 2017;19:253–260. doi: 10.1002/ejhf.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rusanen M., Kivipelto M., Levälahti E., Laatikainen T.K.M., Tuomilehto J., Soininen H. Heart diseases and long-term risk of dementia and Alzheimer's disease: a Population based CAIDE Study. J Alzheimer's Dis. 2014;42:183–191. doi: 10.3233/JAD-132363. [DOI] [PubMed] [Google Scholar]
- 30.Haan M.N. Therapy insight: type 2 diabetes mellitus and the risk of late-onset Alzheimer's disease. Nat Clin Pract Neurol. 2006;2:159–166. doi: 10.1038/ncpneuro0124. [DOI] [PubMed] [Google Scholar]
- 31.Coupland C.A.C., Hill T., Dening T., Morriss R., Moore M., Hippisley-Cox J. Anticholinergic drug exposure and the risk of dementia: a Nested Case-Control Study. JAMA Intern Med. 2019;179:1084–1093. doi: 10.1001/jamainternmed.2019.0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.