Abstract
The global food chain may significantly promote the dissemination of bacteria resistant to antibiotics around the world. This study was aimed at determining the prevalence and genetic characteristics of Enterobacteriaceae with mcr-mediated colistin (CT) resistance in retail meat of different origins. Bacteria of the Enterobacteriaceae family carrying the mcr-1 gene were detected in 21% (18/86) of the examined samples, especially in turkey meat and liver originating from EU and non-EU countries (19%) and in rabbit meat imported from China (2%). The examined samples of the meat and liver of chicken and other poultry and of pork and beef were negative for the presence of bacteria carrying the mcr-1 to mcr-5 genes. A huge number of isolates belonging to Escherchia coli (n = 54), Klebsiella pneumoniae (n = 6), and Citrobacter braakii (n = 1) carrying the mcr-1 gene were obtained. Despite the high heterogeneity of the tested isolates, the mcr-1 gene was localized on only three types of plasmids (IncX4, IncHI2, and IncI2). The most frequent type of plasmid was IncX4, which carried the mcr-1 gene in 77% of E. coli and K. pneumoniae isolates from turkey meat and liver samples from the Czechia, Germany, Poland, and Brazil. Our findings indicate highly probable interspecies transfer of IncX4 and IncI2 plasmids within one meat sample. The co-resistance of plasmid-mediated CT resistance encoded by the mcr-1 and ESBL genes was detected in 18% of the isolates. Another noteworthy finding was the fosA3 gene coding for fosfomycin resistance in a multidrug-resistant isolate of E. coli from rabbit meat imported from China. The observed high level of Enterobacteriaceae with plasmids carrying the mcr-1 gene in retail meat reflects the need for Europe-wide monitoring of mcr-mediated CT resistance throughout the whole food chain.
Keywords: colistin, mcr gene, resistance, Enterobacteriaceae, meat, retail
Introduction
The dissemination of plasmid-mediated colistin (CT) resistance poses a substantial health concern to both human and veterinary medicine. Plasmid-mediated CT resistance encoded by the mcr-1 gene was first reported in China in Enterobacteriaceae from various sources (Liu et al., 2016). Since then, there has been a worldwide spread of gram-negative bacteria with the mcr-1 gene, especially Escherichia coli, in animals including food animals, food, the environment, and humans (Skov and Monnet, 2016; Al-Tawfiq et al., 2017). Another mobile CT resistance gene, mcr-2, has been described in E. coli isolated from pigs and cattle in Belgium (Xavier et al., 2016). However, its occurrence in other countries has been reported only rarely (Zhang et al., 2018). In 2017, the following four genes were described: (i) mcr-3 in E. coli from pigs in China (Yin et al., 2017); (ii) mcr-4 in Salmonella and E. coli isolated from pigs in Italy, Spain, and Belgium (Carattoli et al., 2017); (iii) mcr-5 in Salmonella Paratyphi B from poultry in Germany (Borowiak et al., 2017); and (iv) mcr-6 (originally termed mcr-2.2 variant) in Moraxella pluranimalium from pigs at slaughter in the United Kingdom (AbuOun et al., 2018). The next year, mcr-7 was detected in Klebsiella pneumoniae from chickens and mcr-8 in K. pneumoniae from pigs, chickens, and humans in China (Wang X. et al., 2018; Yang et al., 2018). In 2019, the mcr-9 gene was described in CT-susceptible Salmonella Typhimurium isolated from a patient in the United States in 2010. However, expression of mcr-9 in E. coli resulted in phenotypic resistance to CT (Carroll et al., 2019). The most recently found mcr gene is mcr-10 in Enterobacter roggenkampii (GenBank, accession number MN179494.1).
Extensive use of CT in veterinary medicine for infection control or prophylaxis contributed to a significant increase in the prevalence of CT-resistant bacteria (Kempf et al., 2016). In human medicine, CT is used as an antibiotic of last resort in patients infected with multidrug-resistant gram-negative bacteria (Al-Tawfiq et al., 2017). E. coli isolates with both chromosomal and plasmid-mediated CT resistance in patients without a history of CT treatment have been detected in China. The fact that more than half of them were farmers suggested the possibility of resistant bacteria transmission to humans in association with agricultural use of antimicrobials (Luo et al., 2017). The level of total sales of antimicrobials to treat farm animals in the Czechia is low compared to in Spain, Italy, Germany, and France (ESVAC, 2018). However, the sales and consumption of antibiotics by farm animals may not correspond to the occurrence of resistant bacteria in foods of animal origin in a given country. The global animal and food market can also contribute to the spread of bacteria resistant to the main classes of antibiotics. The spread of mcr-1 is associated with various types of plasmids, including IncI2, IncX4, IncF, IncHI1, IncHI2, IncP, and IncY. The most prevalent plasmid types are IncI2 and IncX4 (Wang R. et al., 2018). The high similarity of plasmids carrying mcr-1 in E. coli isolated from patients and from poultry meat in Switzerland indicates a role of certain types of “epidemic” plasmids (IncI2 and IncX4) in mcr-1 gene dissemination along the food chain and in humans (Zurfluh et al., 2017).
In a number of European countries, the prevalence of gram-negative bacteria with mcr genes was found to be higher in poultry, especially turkeys, compared to pigs and cattle (Irrgang et al., 2016; Perrin-Guyomard et al., 2016; Alba et al., 2018). In 11 European countries, the prevalence of the mcr-1 gene between 2002 and 2014 was shown to be 1.2% in CT-resistant E. coli from chickens and 0.7% in isolates from pigs. By contrast, mcr-1 was not detected in any isolates from cattle. These E. coli isolates with the mcr-1 gene were detected mainly in Spain and Germany and also in Netherlands and France. In Germany, all E. coli isolates with the mcr-1 gene were detected in poultry, whereas in Spain, they originated from pigs (El Garch et al., 2018). On the other hand, in Germany, Roschanski et al. (2017) described the occurrence of E. coli with mcr-1 in 9.9% of pig samples from 25.9% of pig farms examined. However, direct PCR testing of genes without sample enrichment showed a high prevalence of mcr-1 in China and the presence of mcr-2, mcr-3, mcr-4, and mcr-5 in rectal/cloacal and nasal/oropharyngeal swabs from pigs and poultry (Chen et al., 2018; Zhang et al., 2018). The mcr-1 gene was identified in up to 79.2% samples from pigs and in 31.8% of poultry samples (Zhang et al., 2018). Overall though, it is difficult to compare data on the prevalence of plasmid-mediated CT resistance in individual countries due to differences in the methodological approaches used in various studies.
Plasmid-mediated CT resistance in Enterobacteriaceae with a predominance of mcr-1 has also been detected in retailed meat, particularly in chicken and turkey meat (Hasman et al., 2015; Irrgang et al., 2016; Kluytmans-van den Bergh et al., 2016; Monte et al., 2017). Furthermore, the occurrence of Enterobacteriaceae with mcr-1 has been reported in pork (Liu et al., 2016) and beef (Mulvey et al., 2016). However, there is still a lack of comprehensive information concerning the occurrence of plasmid-mediated CT resistance in foodborne bacteria. Therefore, this study aimed at determining the prevalence and characteristics of bacteria in the Enterobacteriaceae family carrying mcr genes that had been isolated from retail meat from different animal species (poultry, pork, beef, and rabbit).
Materials and Methods
Examined Samples
Randomly selected meat samples were collected from those currently on offer at retail markets in the Czechia. A total of 86 meat and liver samples from various animal species were examined that originated from the Czechia (n = 29), Poland (n = 19), Hungary (n = 8), Germany (n = 6), Slovakia (n = 4), France (n = 4), Austria (n = 2), Spain (n = 1), Netherlands (n = 1), Belgium (n = 1), Great Britain (n = 1), Brazil (n = 8), and China (n = 2). These samples included pork (n = 10), beef (n = 19), mixed ground pork and beef (n = 5), turkey meat and liver (n = 24), the meat and liver of chicken (n = 15) and other poultry [goose (n = 2), duck (n = 4), quail (n = 1)], and rabbit meat (n = 6). Five sub-units of 25 g were collected from each sample to increase the detection rate. The samples were cultivated overnight under aerobic conditions in buffered peptone water at 37°C. After enrichment, 1 ml was collected from each sample, and DNA was isolated using a Blood and Tissue Kit according to the manufacturer’s instructions (Qiagen, Germany). The presence of the mcr-1 to mcr-5 genes was verified by PCR (Liu et al., 2016; Xavier et al., 2016; Borowiak et al., 2017; Carattoli et al., 2017; Yin et al., 2017). The artificially synthesized positive controls for mcr-1 to mcr-5 (GeneArt Strings synthesis, Thermo Fisher Scientific, United States) were prepared according to the original sequences (Liu et al., 2016; Xavier et al., 2016; Borowiak et al., 2017; Carattoli et al., 2017; Yin et al., 2017) and were used together with a negative control (sterilized ultrapure water instead of DNA) in PCR reactions. PCR-positive samples were subsequently inoculated on agar medium and cultured, thus allowing selective detection of CT-resistant bacteria and subsequent identification of mcr genes by PCR as described below.
Detection of Colistin-Resistant Bacterial Isolates Carrying mcr Genes
In order to detect CT-resistant Enterobacteriaceae, Brilliance UTI Clarity agar (Oxoid, United Kingdom) with the addition of CT sulfate (Discovery Fine Chemicals, United Kingdom) was used at a final concentration of 3.5 μg/mL and incubated overnight at 37°C. In morphologically different colonies (1–8 per plate), the respective mcr gene was detected with the PCR method according to the results of PCR detection in meat samples after enrichment, as mentioned above.
Species Identification of Bacterial Isolates
The mcr-positive isolates obtained from the UTI Clarity agar supplemented with CT were identified by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) with the use of Biotyper software (version 3.1, Bruker Daltonics GmbH, Germany).
Colistin Susceptibility Testing
The minimum inhibitory concentration (MIC) of CT was determined by the microdilution method recommended by EUCAST (2018). Microtitration plates (LabMediaServis, CZ) were designed to test 15 different concentrations of CT: 0.25, 0.5, 1, 2, 4, 8, 16, 32, 64, 128, 256, 512, 1024, 2048, and 4096 mg/L.
Pulsed-Field Gel Electrophoresis
The Enterobacteriaceae isolates were subjected to DNA macrorestriction analysis followed by pulsed-field gel electrophoresis (PFGE) using restriction endonuclease XbaI according to the PulseNet Europe (2013) protocol for E. coli (2013). BioNumerics v5.1 (Applied Maths), with the settings Dice (Opt: 1.10%) (Tol 1.0–1.0%) (H > 0.0% S > 0.0%) (0.0–100.0%), was used to construct an unweighted pair group method with arithmetic mean (UPGMA) dendrogram.
Whole-Genome Sequencing
For whole-genome sequencing (WGS), strains of different pulsotypes (one band difference) were selected from each positive meat sample. Genomic DNA was isolated using the Blood and Tissue Kit according to the manufacturer’s instructions (Qiagen, Germany). The preparation of DNA libraries and sequencing on the Illumina platform were carried out by Eurofins Genomics (Miseq 2 × 300 bp, n = 9), Macrogen Korea (Hiseq X Ten 2 × 300 bp, n = 20), and CEITEC VFU (Miseq 2 × 250 bp, n = 32). Assembly was performed with SPAdes 3.9 software1. The E. coli sequence type (ST) was determined by the Achtman MLST scheme2, whereas the Pasteur MLST scheme was used for K. pneumoniae strains3. Plasmid types and resistance genes were evaluated by PlasmidFinder (Carattoli et al., 2014) and ResFinder (Zankari et al., 2012)4. Screening for the presence of mcr-1 genes on a particular plasmid type was performed by in silico analysis of the generated contigs.
The sequence data obtained were assembled using Velvet version 1.1.04 on Ridom SeqSphere+ (version 3.5.0; Ridom GmbH, Münster, Germany). The isolates were compared using MLST core genome (cgMLST) analyses comprising 2513 loci5 (for E. coli) and 2568 loci6 (for K. pneumoniae). Subsequently, phylogenetic trees for the given bacterial strain were constructed by UPGMA analysis. The threshold cluster identification was ≤10 alleles for E. coli and ≤15 alleles for K. pneumoniae according to the Ridom SeqSphere+ software.
Conjugation Assay of Plasmids Carrying the mcr-1 Gene
Conjugation assays were performed to determine the transferability of mcr genes into plasmid-free sodium azide-resistant E. coli J53 K12 recipient cells using the filter-mating method (Borowiak et al., 2019). The transconjugants (TCs) were selected on LB agar plates (LB) with sodium azide (100 mg/L) and CT (1 mg/L). In selected strains, the TCs were obtained on LB with sodium azide (100 mg/L) and one of the following antibiotics: sulfonamide (512 mg/L), tetracycline (16 mg/L), or ampicillin (16 mg/L). The antibiotics used for the selection were chosen according to the resistance profiles of the donor strains and WGS-based in silico analysis of mcr-carrying plasmids. In particular, the selection of TCs only on medium with CT was performed for isolates for which the confirmed mcr location was on either IncX4 or IncI2. If no TCs were obtained, the selection was repeated using LB with the above-mentioned antibiotics according to the donor’s resistance profile. For isolates carrying the gene on IncHI2, selections were performed using all four main antibiotics (Supplementary Table 1). Successful transfer of the resistance plasmid was confirmed by PCR targeting the mcr-1 gene (Lescat et al., 2018) and by specific PCR for identification of the E. coli J53 K12 (Bauer et al., 2007). The size of plasmids transferred to recipient cells was estimated by S1 nuclease PFGE analysis (Jamborova et al., 2015), and plasmids were classified into incompatibility (Inc) groups by PCR-based replicon typing (PBRT; Carattoli, 2013).
Results
Prevalence of Enterobacteriaceae With mcr Genes in Raw Meat and Liver
Bacteria of the Enterobacteriaceae family carrying the mcr-1 gene were detected in 21% (18/86) of the examined samples of originally packaged raw meat and liver of various animal species retailed in the Czechia. The mcr-2, mcr-3, mcr-4, and mcr-5 genes were not detected in Enterobacteriaceae in the tested samples. The occurrence of Enterobacteriaceae with the mcr-1 gene was found to be largely predominant in turkey meat and liver samples from the EU and non-EU countries. The presence of bacteria with the mcr-1 gene has also been sporadically reported in rabbit meat, but only in meat imported from China. Samples of pork and beef, plus samples of the meat and liver of chicken and other poultry, were all found to be negative for the presence of bacteria carrying mcr-1 to mcr-5 genes (Table 1).
TABLE 1.
Number of examined samples and samples positive for the presence of Enterobacteriaceae with mcr-1 to mcr-5 genes according to the type of meat/liver.
| Detection of mcr genes in BPW | |||||||
| after enrichment | |||||||
| Type of meat/liver | Examined samples (n) | mcr-1 (n) | mcr-2 (n) | mcr-3 (n) | mcr-4 (n) | mcr-5 (n) | Bacterial species carrying mcr-1 (number of isolates) |
| Pork | 10 | 0 | 0 | 0 | 0 | 0 | ND |
| Beef | 19 | 0 | 0 | 0 | 0 | 0 | ND |
| Mixed pork/beef | 5 | 0 | 0 | 0 | 0 | 0 | ND |
| Turkey meat and liver | 24 | 16 | 0 | 0 | 0 | 0 | E. coli (51) |
| K. pneumoniae (6) | |||||||
| Chicken meat and liver | 15 | 0 | 0 | 0 | 0 | 0 | ND |
| Other poultry meat and liver (goose, duck, and quail) | 7 | 0 | 0 | 0 | 0 | 0 | ND |
| Rabbit | 6 | 2 | 0 | 0 | 0 | 0 | E. coli (3) |
| C. braakii (1) | |||||||
BPW, buffered peptone water; ND, not detected.
Distribution of Bacterial Species With the mcr-1 Gene and Their Origin
Three bacterial species, E. coli (n = 54), K. pneumoniae (n = 6), and Citrobacter braakii (n = 1) carrying the mcr-1 gene were identified in the present study. E. coli isolates with the mcr-1 gene were mainly detected in samples of turkey meat and liver from animals kept and slaughtered in Poland (23 isolates/6 samples positive for the mcr-1 gene), Germany (9/3), Brazil (12/4) and also in the Czechia (7/3). K. pneumoniae with the mcr-1 gene was isolated from samples of frozen turkey liver imported from Brazil (3/1) and turkey meat from the Czechia (3/1) in which co-occurrence of mcr-1-positive E. coli was also found. In addition to turkey meat, the mcr-1 gene was found in E. coli (3/2) and C. braakii (1/1) isolated from frozen rabbit meat imported from China.
MIC of Colistin
The phenotypic CT resistance was confirmed by the microdilution method. MICs for CT were found to be between 4 and 8 mg/L in E. coli isolates carrying the mcr-1 gene. Most of the E. coli isolates (40/54) exhibited a CT MIC value of 4 mg/L, which was similar to C. braakii, whereas K. pneumoniae isolates usually showed higher MIC values for CT (8–64 mg/L).
PFGE in E. coli and K. pneumoniae Isolates Carrying the mcr-1 Gene
Pulsed-field gel electrophoresis was used to distinguish between strains of one species obtained from one sample. In the same sample of meat and liver, E. coli was classified in one to eight different pulsotypes, and K. pneumoniae belonging to two or three different pulsotypes was isolated. High diversity was demonstrated not only between isolates from the same sample but also between E. coli and K. pneumoniae isolates from meat and liver samples of different origins.
Assignment of E. coli and K. pneumoniae Isolates With the mcr-1 Gene to Sequence Types
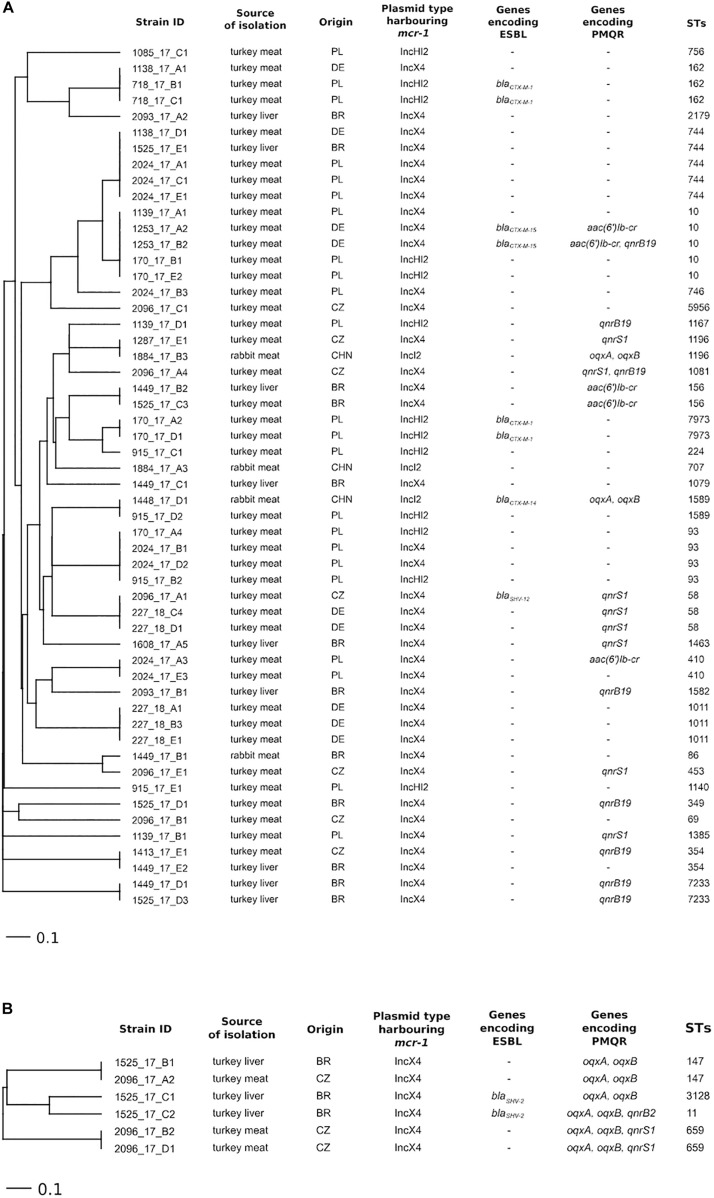
Escherichia coli isolates (n = 54) were assigned to 30 different STs and K. pneumoniae (n = 6) to 4 (Figures 1A,B and Supplementary Table 1). The results did not demonstrate a correlation between the occurrence of a specific ST and the country of origin of the tested samples or the presence of a particular type of plasmid with the mcr-1 gene. Seven STs (ST10, ST58, ST162, ST354, ST744, ST1196, and ST1589) were detected in E. coli from samples of raw meat and liver originating from two or more countries (Figure 1A).
FIGURE 1.
Ridom SeqSphere+ UPGMA tree based on MLST (no missing values) analyses in (A) E. coli (Achtman MLST scheme) and (B) K. pneumoniae (scheme from the Pasteur Institute) isolated from meat and liver of different origin indicating genes encoding ESBL and PMQR and plasmid types harboring the mcr-1 gene.
cgMLST in E. coli and K. pneumoniae Strains Carrying the mcr-1 Gene
Based on cgMLST, high heterogeneity was confirmed among strains of E. coli and K. pneumoniae belonging to the same STs. There was no link between E. coli (ST10, ST58, ST162, ST354, ST744, ST1196, and ST1589) and K. pneumoniae (ST147) isolated from samples of meat and liver from different countries (Figures 2A,B). Only two strains of E. coli ST58 from one turkey meat sample from Poland were assigned to one cluster (seven allelic differences). According to PFGE analysis, these strains differed in only one band and showed a similarity of >90%. The comparison of the detected plasmids showed a difference in the presence of one additional plasmid between the strains (Supplementary Table 1). Similarly, strains of K. pneumoniae ST659 isolated from one turkey meat sample from the Czechia contained 16 allelic differences (close to the cluster threshold of ≤15 different alleles) and otherwise demonstrated a similarity of >90% according to PFGE analysis and differences in blaSHV genes (Supplementary Table 1).
FIGURE 2.
Ridom SeqSphere+ UPGMA tree based on cgMLST (missing values were considered as an own category) analyses in (A) E. coli (scheme based on 2513 loci) and (B) K. pneumoniae (scheme based on 2358 loci) strains of repeatedly detected STs isolated from meat and liver of different origin.
Plasmids Harboring the mcr-1 Gene in Enterobacteriaceae Isolated From Raw Meat and Liver
Based on the analysis of whole-genome sequencing data, three types of plasmids (IncX4, IncHI2, and IncI2) carrying the mcr-1 gene were detected in the tested isolates of E. coli, K. pneumoniae, and C. braakii. In all isolates of K. pneumoniae (6/6) and the majority of E. coli isolates (38/51) from turkey meat and liver, the occurrence of mcr-1 on the IncX4 plasmid type prevailed (Figures 1A,B). Samples with isolates carrying mcr-1 on the IncX4 plasmid originated from the Czechia (seven isolates of E. coli and three isolates of K. pneumoniae from three samples), Poland (10 E. coli isolates from two samples), Germany (nine E. coli isolates from three samples), and Brazil (12 E. coli isolates and 3 K. pneumoniae isolates from four samples). E. coli isolates (n = 13) carrying the mcr-1 gene on an IncHI2-type plasmid were detected in five out of six positive samples of turkey meat from Poland. One sample of turkey meat imported from Poland contained E. coli isolates (n = 2) with the mcr-1 gene located on an IncHI2 plasmid and also an E. coli isolate with mcr-1 on an IncX4 plasmid. The IncI2-type plasmid carrying the mcr-1 gene was detected in only three E. coli isolates and one C. braakii isolate from two rabbit meat samples imported from China.
Conjugative Transfer of Plasmids Carrying the mcr-1 Gene
Conjugative transfer was revealed in all three plasmid types (IncX4, IncHI2, and IncI2) in this study. However, conjugative transfer was not confirmed in all tested isolates under the set experimental conditions. Conjugative transfer of plasmids carrying the mcr-1 gene into recipient cells of E. coli was successful in 73% (27/37) of selected isolates. TCs carrying IncX4 plasmids were obtained on LB with CT (12/17). All TCs with IncI2 plasmids were selected on medium with CT (4/4). On the other hand, most TCs with IncHI2 plasmids were selected on medium with sulfonamides (7/13), ampicillin (6/17), or tetracycline (5/17); selection on LB with CT was successful only in case of one TC. S1 PFGE analysis showed that the mcr-1 gene was carried by IncX4 of size 35–45 kb (17/37), IncHI2 of size 250–260 kb (13/37), or IncI2 of size 55–60 kb (4/37) conjugative plasmids (Supplementary Table 1). The results showed a highly probable interspecies transfer of IncX4 plasmid between E. coli and K. pneumoniae within one sample and of IncI2 between E. coli and C. braakii in another meat sample (Supplementary Table 1).
Occurrence of Other Resistance Genes in Enterobacteriaceae Isolates Carrying the mcr-1 Gene
Regarding the distribution of antimicrobial resistance genes, most of the E. coli, K. pneumoniae, and C. braakii isolates with mcr-1 carried additional genes encoding resistance to macrolides (92%), beta-lactams (88%), tetracycline (85%), and aminoglycosides (79%) (Supplementary Table 1). Importantly, some bacterial isolates with plasmid-mediated CT resistance also carried genes coding for ESBL and PMQR. In four E. coli isolates and two K. pneumoniae isolates, the simultaneous presence of ESBL- and PMQR-encoding genes was detected (Figures 1A,B). None of the isolates carried genes encoding carbapenem resistance. Genes for ESBL production were detected in 15% (8/54) of E. coli isolates and in two of six K. pneumoniae isolates with the mcr-1 gene. CTX-M-type ESBL was detected in E. coli isolates with mcr-1 from turkey meat from Poland and Germany and rabbit meat from China. On the other hand, E. coli isolates from turkey meat from the Czechia carried the blaSHV–12 gene. K. pneumoniae (n = 2) isolated from a sample of turkey liver from Brazil carried the blaSHV–2 gene. All six obtained K. pneumoniae isolates with mcr-1 and 39% (21/54) of E. coli isolates with mcr-1 carried the PMQR-encoding genes. The qnrS and qnrB genes predominated in E. coli isolates from turkey meat and liver originating from different countries, whereas all K. pneumoniae isolates carried the oqxA and oqxB genes. Moreover, the qnrS gene was detected in two K. pneumoniae isolates from the Czechia and the qnrB gene in one isolate from Brazil. Only one C. braakii isolate obtained from rabbit meat from China carried both the ESBL and PMQR genes besides mcr-1.
Discussion
Studies conducted on different continents report the detection of bacteria with plasmid-mediated CT resistance, but the true prevalence remains unknown. The current prevalence of bacteria with mcr genes in food animals and in food originating from the Czechia is unknown, because systematic and proportional studies of this type have not yet been performed. In the Czechia, the first detection of E. coli harboring the mcr-1 gene was reported in 2017 in turkey meat from Poland (Karpíšková et al., 2017). A follow-up pilot study of turkey meat originating from the EU and purchased in the Czechia’s market network between 2017 and 2018 identified mcr-1-positive Enterobacteriaceae in 70.6% of samples (Gelbíčová et al., 2019). In the present study, 67% of turkey meat and liver samples contained mcr-1-positive bacteria. These samples originated from Poland, Germany, and Brazil, and also from the Czechia. Of the total mcr-1-positive isolates obtained from turkey meat and liver, 51 were of E. coli and 6 of K. pneumoniae. Hence, our data showed overall that turkey meat and liver produced in Poland is a major source of bacteria with plasmid-mediated CT resistance, with the highest prevalence given to E. coli with mcr-1.
These findings are consistent with a German study where the prevalence of E. coli with the mcr-1 gene was higher in turkey (8.4%) than in chicken meat (4.3%) (Irrgang et al., 2016). Furthermore, this is in accordance with the higher observed prevalence of mcr-1-positive bacteria in turkeys from Poland, Germany, France, and Italy in comparison with broilers (Irrgang et al., 2016; Perrin-Guyomard et al., 2016; Alba et al., 2018; Zaja̧c et al., 2019).
Even though no Enterobacteriaceae species carrying mcr genes were found in any sample of chicken meat originating either from the EU countries (Czechia, Slovakia, Poland, Great Britain, and France) or non-EU countries (Brazil), other studies have described the occurrence of E. coli with mcr-1 in retailed chicken meat. In a Swiss study, the mcr-1 gene was detected in 25.8% of retail poultry meat (chicken and turkey) from Germany (28 samples) and Italy (5 samples). The mcr-1 gene has not been found in any of a set of chicken and turkey meat samples from Switzerland, Denmark, Austria, and Hungary (Zurfluh et al., 2016). In Brazil, the presence of E. coli with mcr-1 was detected in 19.5% of the tested retailed chicken meat and liver samples (Monte et al., 2017).
The investigated samples of retailed pork and beef from the Czechia and from other European countries have been shown to be safe regarding plasmid-mediated CT resistance. This is in spite of the fact that the presence of bacteria with plasmid-mediated CT resistance has been reported in pigs from European (El Garch et al., 2018) and from Asian (Chen et al., 2018; Zhang et al., 2018) countries, though their occurrence in pork has been described only rarely, e.g., in China (Liu et al., 2016). Similarly, the occurrence of mcr-positive bacteria in beef has been described only occasionally, e.g., in ground beef in Canada (Mulvey et al., 2016). The prevalence of plasmid-mediated CT resistance in bacteria isolated from cattle in Europe (Xavier et al., 2016; Hernández et al., 2017; Haenni et al., 2018) and in Asia (Kawanishi et al., 2016) is lower in comparison to poultry and pigs. CT is recommended for the treatment of gastrointestinal infections caused by non-invasive E. coli, mainly in pigs, chickens, turkey, calves, and sheep. However, extensive studies combining antibiotic consumption and resistance are limited because either CT resistance is not tested at all or the tests are not fully reliable (EMA/AMEG, 2016).
The dissemination of mcr genes in gram-negative bacteria isolated from food-producing animals and from food in Asian countries (Kawanishi et al., 2016; Liu et al., 2017; Wu et al., 2018) was confirmed by the detection of E. coli and C. braakii with the mcr-1 gene in two samples of rabbit meat imported from China. The examined rabbit meat samples from Hungary and the Czechia did not contain bacteria with mcr genes. Currently, there is a paucity of studies screening for the presence of bacteria with plasmid-mediated CT resistance in rabbits and rabbit meat. In Italy, mcr-1 was detected in 15.6% (50/320) of E. coli isolated from rabbits originating from 25 out of 32 investigated farms (Agnoletti et al., 2018). In Portugal, the mcr-1 gene was detected in three E. coli isolates from rabbit meat (Freitas-Silva et al., 2018).
The high diversity of the mcr-1-positive E. coli and K. pneumoniae isolates detected in the present study corresponds to the results for mcr-positive Enterobacteriaceae isolated from different sources (food, animals, and humans) in other studies (Zurfluh et al., 2016; Luo et al., 2017; Roschanski et al., 2017; Zurfluh et al., 2017; El Garch et al., 2018). However, some studies have described an increased prevalence of E. coli with mcr-1 belonging to ST10 obtained from food animals, food, and humans (Zurfluh et al., 2016; El Garch et al., 2018; Shen et al., 2018; Wu et al., 2018). Similarly, E. coli isolates found here belonging to ST10 (n = 5) and ST744 (n = 5) were most often detected. The results of cgMLST analysis provided for a more detailed differentiation compared to MLST, revealing significant differences between isolates belonging to an identical ST-type originating from one sample/country (Figures 2A,B).
Regardless of the high heterogeneity of the tested isolates, the mcr-1 gene was localized on only three types of plasmids (IncX4, IncHI2, and IncI2), which have previously been described in association with the occurrence of mcr-1 (Wang R. et al., 2018). The most common type of plasmid was IncX4, which carried the mcr-1 gene in 77% (44/57) of E. coli and K. pneumoniae isolates from turkey meat and liver samples (n = 16) from the Czechia, Germany, Poland, and Brazil. IncX4 plasmids carrying mcr-1 have also been described in E. coli isolated from meat and humans in China (Liu et al., 2017; Luo et al., 2017; Shen et al., 2018), chicken meat from Brazil (Monte et al., 2017), chicken and turkey meat from the EU (Hasman et al., 2015; Alba et al., 2018), and pigs in Germany (Roschanski et al., 2017). The conjugation assay showed that the majority of IncX4 plasmids carrying mcr-1 were transferable to recipient E. coli cells (14/17). The different origin of these isolates suggesting that these plasmids have circulated worldwide. Sun et al. (2017) also reported successful transport of the mcr-1 gene by IncX4 plasmids (9/11) in Enterobacteriaceae. The second most frequently identified plasmid was IncHI2 carrying the mcr-1 gene in 21% (13/61) of E. coli isolated from turkey meat samples from Poland (n = 5). These plasmids were transferable in 8 out of 13 isolates tested. The IncI2-type plasmid carrying mcr-1 was detected occasionally, and only in E. coli (3) and C. braakii (1) isolates from rabbit meat samples from China (n = 2). The conjugation with IncI2 plasmid was successful in all these isolates. The mcr-1 gene in E. coli isolated from rabbit meat in Portugal was also localized on an IncI2-type plasmid (Freitas-Silva et al., 2018).
The majority of E. coli as well as K. pneumoniae and C. braakii isolates harboring the mcr-1 gene carried genes encoding co-resistance to several classes of antimicrobial agents (Supplementary Table 1). The high level of resistance to beta-lactams (88%) and tetracycline (85%) in Enterobacteriaceae with mcr-1 from meat that was found in this study is consistent with the results of antibiotic resistance in food-producing animals in other European countries (El Garch et al., 2018). In particular, the presence of plasmid-mediated CT resistance in multidrug-resistant bacteria, such as ESBL producers, poses a significant threat to public health. Indeed, in our study, ESBL-encoding genes were detected in 18% (11/61) of isolates carrying the mcr-1 gene and belonging to E. coli, K. pneumoniae, and C. braakii isolated from meat from Poland, Germany, Czechia, Brazil, and China. The occurrence of plasmid-mediated CT resistance in ESBL producers isolated from meat is also described in other studies. In China, the mcr-1 gene was found in 27% of ESBL-producing E. coli isolated from retailed meat and shrimps (Liu et al., 2017). In Denmark, the mcr-1 gene was detected in 1.5% of ESBL-producing E. coli isolated from imported retailed chicken meat without declaration of origin (Kluytmans-van den Bergh et al., 2016). In a Brazilian study, most E. coli isolates (7/8) from chicken meat and liver were shown to carry some of the ESBL-encoding genes (Monte et al., 2017).
In our study, the fosfomycin resistance gene fosA3 and genes for the production of ESBL (blaCTX–M–14) and PMQR (oqxA, oqxB) were also detected in one E. coli isolate carrying the mcr-1 gene, which was obtained from rabbit meat from China. The use of fosfomycin is advised for the treatment of both urinary and systemic infections caused by multidrug-resistant E. coli. The chromosomal fosA gene is often detected in K. pneumoniae (99.7%) but only occasionally in E. coli (4.6%). The plasmid-mediated fosA3 gene variant is the most commonly reported in E. coli and other Enterobacteriaceae in Asia (Ito et al., 2017). Liu et al. (2017) detected the fosA3 gene in seven ESBL-producing E. coli strain carrying the mcr-1 gene that were isolated from food of animal origin. The occurrence of fosfomycin resistance in Asia, but also its sporadic detection in strain of animal origin in Europe (Lupo et al., 2018), represents another public health concern.
Conclusion
The results of this study confirm the impact of global trade in raw meat on the spread of plasmid-mediated CT resistance. Turkey meat and liver were the most common sources of Enterobacteriaceae carrying the mcr-1 gene when compared to other meats (other poultry, pork, beef, and rabbit). Despite the high heterogeneity of the isolates tested, the mcr-1 gene was primarily localized on an IncX4-type plasmid, regardless of ST or the country of meat origin. The co-resistance of plasmid-mediated CT resistance encoded by the mcr-1 and ESBL genes was 18%. Another noteworthy finding was the fosA3 gene coding for fosfomycin resistance in a multidrug-resistant isolate of E. coli from rabbit meat imported from China. These results point to the threat posed by the dissemination of multidrug-resistant bacteria through the food chain. The increased prevalence of E. coli and K. pneumoniae isolates with the mcr-1 gene in turkey meat support the need for Europe-wide monitoring of plasmid-mediated CT resistance in farm animals, especially in poultry, and in slaughterhouses.
Data Availability Statement
The datasets generated for this study can be found in the European Nucleotide Archive (ENA) at 585 EMBL-EBI under accession number PRJEB34874 586 (https://www.ebi.ac.uk/ena/data/view/PRJB34874).
Author Contributions
TG and RK performed the experimental planning and prepared the manuscript. LP and IK collected the isolates and carried out the experiments. AB and MF analyzed the whole-genome sequencing data. IJ performed the whole-genome sequencing, and MZ performed the conjugation assays in selected isolates. All authors discussed the results.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Adam Valcek and Monika Dolejska from the University of Veterinary and Pharmaceutical Sciences Brno for their help with data analysis. We also thank Tomas Nohejl for his assistance in the laboratory and Prof. Andrew D. Miller for language proofreading.
Funding. This study was supported by the Ministry of Health of the Czech Republic grant no. NV 18-09-00254, the Ministry for Regional Development grant no. CZ.1.05./2.1.00/19.0385 (Prague, Czechia), and CEITEC 2020 – Central European Institute of Technology from the European Regional Development Fund (LQ1601) from the Czech Ministry of Education, Youth, and Sports within the National Programme for Sustainability II, and Ministry of Agriculture grant no. RO0518.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02824/full#supplementary-material
References
- AbuOun M., Stubberfield E. J., Duggett N. A., Kirchner M., Dormer L., Nunez-Garcia J., et al. (2018). mcr-1 and mcr-2 (mcr-6.1) variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J. Antimicrob. Chemother. 73:2904. 10.1093/jac/dky272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agnoletti F., Brunetta R., Bano L., Drigo I., Mazzolini E. (2018). Longitudinal study on antimicrobial consumption and resistance in rabbit farming. Int. J. Antimicrob. Agents 51 197–205. 10.1016/j.ijantimicag.2017.10.007 [DOI] [PubMed] [Google Scholar]
- Alba P., Leekitcharoenphon P., Franco A., Feltrin F., Ianzano A., Caprioli A., et al. (2018). Molecular epidemiology of mcr-encoded colistin resistance in Enterobacteriaceae from food-producing animals in Italy revealed through the EU harmonized antimicrobial resistance monitoring. Front. Microbiol. 9:1217. 10.3389/fmicb.2018.01217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Tawfiq J., Laxminarayan R., Mendelson M. (2017). How should we respond to the emergence of plasmid-mediated colistin resistance in humans and animals? Int. J. Infect. Dis. 54 77–84. 10.1016/j.ijid.2016.11.415 [DOI] [PubMed] [Google Scholar]
- Bauer A. P., Dieckmann S. M., Ludwig W., Schleifer K. H. (2007). Rapid identification of Escherichia coli safety and laboratory strain lineages based on Multiplex-PCR. FEMS Microbiol. Lett. 269 36–40. 10.1111/j.1574-6968.2006.00594.x [DOI] [PubMed] [Google Scholar]
- Borowiak M., Fischer J., Hammerl J. A., Hendriksen R. S., Szabo I., Malorny B. (2017). Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J. Antimicrob. Chemother. 72 3317–3324. 10.1093/jac/dkx327 [DOI] [PubMed] [Google Scholar]
- Borowiak M., Hammerl J. A., Deneke C., Fischer J., Szabo I., Malorny B. (2019). Characterization of mcr-5-harboring Salmonella enterica subsp. enterica serovar Typhimurium isolates from animal and food origin in Germany. Antimicrob. Agents Chemother. 63:e00063-19. 10.1128/AAC.00063-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A. (2013). Plasmids and the spread of resistance. Int. J. Med. Microbiol. 303 298–304. 10.1016/j.ijmm.2013.02.001 [DOI] [PubMed] [Google Scholar]
- Carattoli A., Villa L., Feudi C., Curcio L., Orsini S., Luppi A., et al. (2017). Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill. 22:30589. 10.2807/1560-7917.ES.2017.22.31.30589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Zankari E., García-Fernández A., Voldby Larsen M., Lund O., Villa L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58 3895–3903. 10.1128/AAC.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll L. M., Gaballa A., Guldimann C., Sullivan G., Henderson L. O., Wiedmann M. (2019). Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. mBio 10:e00853-19. 10.1128/mBio.00853-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Zhang J., Wang J., Butaye P., Kelly P., Li M., et al. (2018). Newly identified colistin resistance genes, mcr-4 and mcr-5, from upper and lower alimentary tract of pigs and poultry in China. PLoS One 13:e0193957. 10.1371/journal.pone.0193957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Garch F., de Jong A., Bertrand X., Hocquet D., Sauget M. (2018). mcr-1-like detection in commensal Escherichia coli and Salmonella spp. from food-producing animals at slaughter in Europe. Vet. Microbiol. 213 42–46. 10.1016/j.vetmic.2017.11.014 [DOI] [PubMed] [Google Scholar]
- EMA/AMEG (2016). Updated Advice on the use of Colistin Products in Animals Within the European Union: Development of Resistance and Possible Impact on Human and Animal Health. Available at: http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2016/07/WC500211080.pdf (accessed February 22, 2019). [Google Scholar]
- ESVAC (2018). European Medicines Agency, European Surveillance of Veterinary Antimicrobial Consumption, Sales of veterinary Antimicrobial Agents in 30 European Countries in 2016. Available at: https://www.ema.europa.eu/veterinary-regulatory/overview/antimicrobial-resistance/european-surveillance-veterinary-antimicrobial-consumption-esvac (accessed February 22, 2019). [Google Scholar]
- EUCAST (2018). European Committee on Antimicrobial Susceptibility Testing Clinical Breakpoints – Bacteria (v. 8.0). Available at: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_8.1_Breakpoint_Tables.pdf (accessed January 1, 2018). [Google Scholar]
- Freitas-Silva J., Inácio A. S., Mourao J., Antunes P., Mendes A., de Carvalho A. P., et al. (2018). Occurrence of mcr-1 in Escherichia coli from rabbits of intensive farming. Vet. Microbiol. 227 78–81. 10.1016/j.vetmic.2018.10.020 [DOI] [PubMed] [Google Scholar]
- Gelbíčová T., Koláčková I., Krütová M., Karpíšková R. (2019). The emergence of mcr-1-mediated colistin-resistant Escherichia coli and Klebsiella pneumoniae in domestic and imported turkey meat in the Czech Republic 2017-2018. Folia Microbiol. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Haenni M., Beyrouthy R., Lupo A., Châtre P., Madec J. Y., Bonnet R. (2018). Epidemic spread of Escherichia coli ST744 isolates carrying mcr-3 and blaCTX-M-55 in cattle in France. J. Antimicrob. Chemother. 73 533–536. 10.1093/jac/dkx418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasman H., Hammerum A. M., Hansen F., Hendriksen R. S., Olesen B., Agersø Y., et al. (2015). Detection of mcr-1-encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill. 20:49. 10.2807/1560-7917.ES.2015.20.49.30085 [DOI] [PubMed] [Google Scholar]
- Hernández M., Iglesias M. R., Rodríguez-Lázaro D., Gallardo A., Quijada N., Miguela-Villoldo P., et al. (2017). Co-occurrence of colistin-resistance genes mcr-1 and mcr-3 among multidrug-resistant Escherichia coli isolated from cattle, Spain, September 2015. Euro Surveill. 22:30586. 10.2807/1560-7917.ES.2017.22.31.30586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irrgang A., Roschanski N., Tenhagen B. A., Grobbel M., Skladnikiewicz-Ziemer T., Thomas K., et al. (2016). Prevalence of mcr-1 in E. coli from livestock and food in Germany, 2010-2015. PLoS One 11:e0159863. 10.1371/journal.pone.0159863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito R., Mustapha M. M., Tomich A. D., Callaghan J. D., McElheny C. L., Mettus R. T., et al. (2017). Widespread fosfomycin resistance in gram-negative bacteria attributable to the chromosomal fosA gene. mBio 8:e00749-17. 10.1128/mBio.00749-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamborova I., Dolejska M., Vojtech J., Guenther S., Uricariu R., Drozdowska J., et al. (2015). Plasmid-mediated resistance to cephalosporins and fluoroquinolones in various Escherichia coli sequence types isolated from rooks wintering in Europe. Appl. Environ. Microbiol. 81 648–657. 10.1128/AEM.02459-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpíšková R., Koláčková I., Gelbíčová T., Zobaníková M. (2017). A rare mechanism of resistance to colistin in Escherichia coli isolated from raw poultry meat. Klin. Mikrobiol. Infekc. Lek. 23 58–60. [PubMed] [Google Scholar]
- Kawanishi M., Abo H., Ozawa M., Uchiyama M., Shirakawa T., Suzuki S., et al. (2016). Prevalence of colistin resistance gene mcr-1 and absence of mcr-2 in Escherichia coli isolated from healthy food-producing animals in Japan. Antimicrob. Agents Chemother. 61:e02057-16. 10.1128/AAC.02057-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf I., Jouy E., Chauvin C. (2016). Colistin use and colistin resistance in bacteria from animals. Int. J. Antimicrob. Agents 48 598–606. 10.1016/j.ijantimicag.2016.09.016 [DOI] [PubMed] [Google Scholar]
- Kluytmans-van den Bergh M. F., Huizinga P., Bonten M. J., Bos M., De Bruyne K., Friedrich A. W., et al. (2016). Presence of mcr-1-positive Enterobacteriaceae in retail chicken meat but not in humans in the Netherlands since 2009. Euro Surveill. 21:30149. 10.2807/1560-7917.ES.2016.21.9.30149 [DOI] [PubMed] [Google Scholar]
- Lescat M., Poirel L., Nordmann P. (2018). Rapid multiplex polymerase chain reaction for detection of mcr-1 to mcr-5 genes. Diagn. Microbiol. Infect. Dis. 92 267–269. 10.1016/j.diagmicrobio.2018.04.010 [DOI] [PubMed] [Google Scholar]
- Liu X., Li R., Zheng Z., Chen K., Xie M., Chan E. W.-C., et al. (2017). Molecular characterization of Escherichia coli isolates carrying mcr-1, fosA3, and extended-spectrum-ß-lactamase genes from food samples in China. Antimicrob. Agents Chemother. 61:e00064-17. 10.1128/AAC.00064-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Y., Wang Y., Walsh T. R., Yi L. X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16 161–168. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- Luo Q., Yu W., Zhou K., Guo L., Shen P., Lu H., et al. (2017). Molecular epidemiology and colistin resistance mechanism of mcr-positive and mcr-negative clinical isolated Escherichia coli. Front. Microbiol. 8:2262. 10.3389/fmicb.2017.02262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo A., Saras E., Madec J. Y., Haenni M. (2018). Emergence of blaCTX-M-55 associated with fosA, rmtB and mcr gene variants in Escherichia coli from various animal species in France. J. Antimcrob. Chemother. 73 867–872. 10.1093/jac/dkx489 [DOI] [PubMed] [Google Scholar]
- Monte D. F., Mem A., Fernandez M. R., Cerdeira L., Esposito F., Galvão J. A., et al. (2017). Chicken meat as a reservoir of colistin-resistant Escherichia coli strains carrying mcr-1 genes in South America. Antimicrob. Agents Chemother. 61:e02718-16. 10.1128/AAC.02718-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey M. R., Mataseje L. F., Robertson J., Nash J. H., Boerlin P., Toye B., et al. (2016). Dissemination of the mcr-1 colistin resistance gene. Lancet Infect. Dis. 16 289–290. 10.1016/S1473-3099(16)00067-0 [DOI] [PubMed] [Google Scholar]
- Perrin-Guyomard A., Bruneau M., Houée P., Deleurme K., Legrandois P., Poirier C., et al. (2016). Prevalence of mcr-1 in commensal Escherichia coli from French livestock, 2007 to 2014. Euro Surveill. 21:6. 10.2807/1560-7917.ES.2016.21.6.30135 [DOI] [PubMed] [Google Scholar]
- PulseNet Europe, (2013). Standard Operating Procedure for PulseNet PFGE of Escherichia coli O157:H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri. Stockholm: PulseNet Europe. [Google Scholar]
- Roschanski N., Falgenhauer L., Grobbel M., Guenther S., Kreienbrock L., Imirzalioglu C., et al. (2017). Retrospective survey of mcr-1 and mcr-2 in German pig-fattening farms, 2011-2012. Int. J. Antimicrob. Agents 50 266–271. 10.1016/j.ijantimicag.2017.03.007 [DOI] [PubMed] [Google Scholar]
- Shen Y., Zhou H., Xu J., Wang Y., Zhang Q., Walsh T. R., et al. (2018). Anthropogenic and environmental factors associated with high prevalence of mcr-1 carriage in humans across China. Nat. Microbiol. 3 1054–1062. 10.1038/s41564-018-0205-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skov R. L., Monnet D. L. (2016). Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Euro Surveill. 21:30155. 10.2807/1560-7917.ES.2016.21.9.30155 [DOI] [PubMed] [Google Scholar]
- Sun J., Fang L. X., Wu Z., Deng H., Yang R. S., Li X. P., et al. (2017). Genetic analysis of the IncX4 plasmids: implications for a unique pattern in the mcr-1 acquisition. Sci. Rep. 7:424. 10.1038/s41598-017-00095-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., van Dorp L., Shaw L. P., Bradley P., Wang Q., Wang X., et al. (2018). The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 9:1179. 10.1038/s41467-018-03205-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang Y., Zhou Y., Li J., Yin W., Wang S., et al. (2018). Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microbes. Infect. 7:122. 10.1038/s41426-018-0124-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Wang Y., Shi X., Wang S., Ren H., Shen Z., et al. (2018). Rapid rise of the ESBL and mcr-1 genes in Escherichia coli of chicken origin in China, 2008-2014. Emerg. Microbes Infect. 7:30. 10.1038/s41426-0118-0033-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier B. B., Lammens C., Ruhal R., Kumar-Singh S., Butaye P., Goossens H., et al. (2016). Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June, 2016. Euro Surveill. 21:30280. 10.2807/1560-7917.ES.2016.21.27.30280 [DOI] [PubMed] [Google Scholar]
- Yang Y. Q., Li Y. X., Lei C. W., Zhang A. Y., Wang H. N. (2018). Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 73 1791–1795. 10.1093/jac/dky111 [DOI] [PubMed] [Google Scholar]
- Yin W., Li H., Shen Y., Liu Z., Wang S., Shen Z., et al. (2017). Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio 8:e00543-17. 10.1128/mBio.00543-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaja̧c M. M., Sztromwasser P., Bortolaia V., Leekitcharoenphon P., Cavaco L. M., Ziêtek-Barszcz A., et al. (2019). Occurrence and characterization of mcr-1-positive Escherichia coli isolated from food-producing animals in Poland, 2011-2016. Front. Microbiol. 10:1753. 10.3389/fmicb.2019.01753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67 2640–2644. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Chen L., Wang J., Yassin A. K., Butaye P., Kelly P., et al. (2018). Molecular detection of colistin resistance genes (mcr-1, mcr-2 and mcr-3) in nasal/oropharyngeal and anal/cloacal swabs from pigs and poultry. Sci. Rep. 8:3705. 10.1038/s41598-018-22084-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurfluh K., Buess S., Roger S., Nuesch-Inderbinen M. (2016). Assessment of the occurrence of MCR producing Enterobacteriaceae in Swiss and imported poultry meat. SDRP J. Food Sci. Tech. 1 137–141. 10.15436/JFST.1.4.5 [DOI] [Google Scholar]
- Zurfluh K., Nuesch-Inderbinen M., Klumpp J., Poirel L., Nordmann P., Stephan R. (2017). Key features of mcr-1-bearing plasmids from Escherichia coli isolated from humans and food. Antimicrob. Resist. Incfect. Control 6:91. 10.1186/s13756-017-0250-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in the European Nucleotide Archive (ENA) at 585 EMBL-EBI under accession number PRJEB34874 586 (https://www.ebi.ac.uk/ena/data/view/PRJB34874).