Summary
Development of marker‐free and transgene insertion site‐defined (MFTID) transgenic plants is essential for safe application of transgenic crops. However, MFTID plants have not been reported for wheat (Triticum aestivum). Here, we prepared a RNAi cassette for suppressing lipoxygenase (LOX) gene expression in wheat grains using a double right border T‐DNA vector. The resultant construct was introduced into wheat genome via Agrobacterium‐mediated transformation, with four homozygous marker‐free transgenic lines (namely GLRW‐1, ‐3, ‐5 and ‐8) developed. Aided by the newly published wheat genome sequence, the T‐DNA insertion sites in GLRW‐3 and GLRW‐8 were elucidated at base‐pair resolution. While the T‐DNA in GLRW‐3 inserted in an intergenic region, that of GLRW‐8 inactivated an endogenous gene, which was thus excluded from further analysis. Compared to wild ‐type (WT) control, GLRW‐1, ‐3 and ‐5 showed decreased LOX gene expression, lower LOX activity and less lipid peroxidation in the grains; they also exhibited significantly higher germination rates and better seedling growth after artificial ageing treatment. Interestingly, the three GLRW lines also had substantially increased contents of several fatty acids (e.g., linoleic acid and linolenic acid) in their grain and flour samples than WT control. Collectively, our data suggest that suppression of grain LOX activity can be employed to improve the storability and fatty acid content of wheat seeds and that the MFTID line GLRW‐3 is likely of commercial value. Our approach may also be useful for developing the MFTID transgenic lines of other crops with enhanced grain storability and fatty acid content.
Keywords: common wheat, fatty acid content, lipoxygenase, marker‐free transgenic plants, RNA interference, storability
Introduction
Transgenic breeding offers a powerful compliment to plant genetic improvement technologies (Christou, 2013; Parisi et al., 2016). Transgenic crop varieties have played important roles in reducing the damages caused by biotic and abiotic stresses and are anticipated to provide more and more high‐quality agricultural products to global consumers (Chen and Lin, 2013; ISAAA, 2017; Parisi et al., 2016). However, the biosafety of transgenic cultivars has attracted increasingly more public concerns (Ahmad and Mukhtar, 2017; Chen and Lin, 2013; Gilbert, 2013). It is now generally accepted that for developing a transgenic variety the marker gene used to aid the preparation of transgenic plants must be eliminated (Ahmad and Mukhtar, 2017; Privalle et al., 2012). Furthermore, the insertion site of transgene, which is T‐DNA integration site in the plant derived from Agrobacterium‐mediated transformation, and its potential influence on the expression of neighbouring genes should be defined (Harwood, 2012; Ladics et al., 2015; Privalle et al., 2012).
A number of methods have been reported for producing marker‐free transgenic plants, for example, co‐transformation of two T‐DNAs, use of a double right border (DRB) T‐DNA vector, Cre/lox site‐specific recombination, FLP/FRT site‐specific recombination or multi‐auto‐transformation (Ling et al., 2016; Wang et al., 2016a,2016b). A DRB binary vector, with the T‐DNA region organized as left border (LB)‐gene of interest (GOI)‐right border 2 (RB2)‐selectable marker gene (SMG)‐right border 1 (RB1), is relatively easy to construct (Lu et al., 2001; Matheka et al., 2013). When transformed with a DRB construct, some of the positive T0 transformants can carry two independent T‐DNA insertions, that is, LB‐GOI‐RB2‐SMG‐RB1 and LB‐GOI‐RB2. In the selfed T1 population, the progenies carrying only LB‐GOI‐RB2, which are free of marker gene cassette, can be identified. At present, DRB constructs have been successfully used for producing marker‐free transgenic rice and tobacco plants (Lu et al., 2001; Matheka et al., 2013; Xu et al., 2008). For detecting T‐DNA insertion sites in the transgenic lines, thermal asymmetric interlaced PCR (TAIL‐PCR) and adapter ligation‐mediated PCR are two convenient methods and have been successfully used to identify the chromosomal sequences flanking T‐DNA integration sites in Arabidopsis, rice and several other plant species (Liu and Chen, 2007; Liu et al., 1995; O'Malley et al., 2007; Tsuchiya et al., 2009).
Wheat (Triticum aestivum L.) is the most widely cultivated staple food crop in the world and provides approximately 20% of the proteins and calories consumed by humans (International Wheat Genome Sequencing Consortium et al., 2018). Although conventional breeding has contributed greatly to the enhancement of wheat agronomic traits, further and more efficient improvement of wheat performance requires the combination of multiple breeding technologies including transgenic breeding (Bhalla, 2006; Harwood, 2012). Many studies have demonstrated that transgenic overexpression or suppression of endogenous genes are effective for improving wheat agronomic traits (Altenbach et al., 2014; Altpeter et al.,1996; Barro et al., 1997; Blechl and Anderson, 1996; Chen et al., 2018; Fu et al., 2007; Gao et al., 2018; Gil‐Humanes et al., 2010; Mega et al., 2019). In addition, plant‐mediated RNA interference of insect genes has been successfully used to engineer the resistance to crop pests, including wheat defence to the aphid Sitobion avenae (Abdellatef et al., 2015; Mao et al., 2007; Sun et al., 2019). However, there are still no transgenic wheat varieties that have been cultivated in the field in a large scale. This may be caused by a number of factors, one of which has been the difficulty to develop environmentally safe and commercially valuable transgenic wheat lines. Wheat is considerably more recalcitrant to Agrobacterium‐mediated transformation (Harwood, 2012; Jones, 2005); only recently have some wheat genotypes been found transformable by Agrobacterium with relatively high efficiencies (Ishida et al., 2014; Richardson et al., 2014; Wang et al., 2016a, 2016b). Consequently, the majority of past transgenic wheat studies have used the lines transformed through particle gun bombardment, which usually yields complex integration sites containing multiple copies of transgenes (Altpeter et al., 2005; Han et al.,2015). This has hindered the development of marker‐free and transgene insertion site‐defined (MFTID) wheat plants. Nevertheless, several recent studies have succeeded in developing marker‐free transgenic wheat (Richardson et al., 2014; Wang et al., 2016a, 2016b). The wheat transformation efficiencies achieved by Richardson et al. (2014) and Wang et al. (2016b) were much higher because of using the PureWheat Agrobacterium transformation technique (Ishida et al., 2014). However, in neither study, the T‐DNA insertion sites were characterized at base‐pair resolution.
Lipoxygenases (EC1.13.11.12, LOXs) are conserved fatty acid dioxygenases in eukaryotes, which catalyse the oxidative breakdown of polyunsaturated fatty acids (PUFAs) such as linoleic acid (LA) and linolenic acid (LNA) (Hayward et al., 2017; Liavonchanka and Feussner, 2006). Higher plants usually contain a complex family of LOX genes, which play important roles in their growth, development and responses to environmental stimuli. In general, multiple LOX isozymes are expressed in the grains of dicot and monocot plants (Feng et al., 2010; Liavonchanka and Feussner, 2006). Their peroxidation of PUFAs can lead to decomposition of cell structures and formation of cytotoxic products, such as the volatile carbonyl compounds associated with the stale flavour of stored grains (Hayward et al., 2017; Robinson et al., 1995; Shirasawa et al., 2008; Suzuki et al., 1999). Consequently, a number of conventional and transgenic studies have shown that down‐regulation of LOX gene expression is beneficial for improving the storability of grains (Dong et al., 2015; Gayen et al., 2014; Huang et al., 2014; Ma et al., 2015; Shen et al., 1996; Xu et al., 2015). In these studies, controlled reduction of LOX gene expression in the grain tissues is considered to be vital because constitutive silencing of LOX genes may impair their function in other organs (e.g., leaves) and thus obstruct normal plant growth. In our previous study, we demonstrated that grain‐specific reduction of LOX accumulation improves flour colour quality and storability of wheat (Dong et al., 2015). However, the transgenic lines analysed were prepared using particle gun bombardment, which had complex transgene integration sites and not marker‐free.
The main objective of this study was to develop and analyse MFTID transgenic wheat with reduced LOX activity in the grains. Towards this end, we conducted Agrobacterium‐mediated wheat transformation using a DRB binary vector carrying a RNAi cassette designed to silence LOX gene expression in the endospermic tissues (i.e., with the cassette driven by an endosperm specific gene promoter). The latest genome sequence information, published for the hexaploid wheat landrace Chinese Spring (CS) (International Wheat Genome Sequencing Consortium et al., 2018), facilitated the determination of transgene insertion sites in specific chromosomal locations. Our wheat transformation experiment was accomplished before the publication of PureWheat technique (Ishida et al., 2014), but it produced a couple of positive T0 transformants that enabled the development of four independent Grain LOX‐Reduced Wheat (GLRW) lines. The four lines (namely GLRW‐1, ‐3, ‐5 and ‐8) were all marker‐free, and the T‐DNA insertion sites in two of them (GLRW‐3 and ‐8) were elucidated at base‐pair resolution. In addition to showing improved storability, the GLRW lines also accumulated higher contents of fatty acids (FAs) than wild‐type (WT) control. These findings, together with the field trial data, suggest that the MFTID line GLRW‐3 can potentially be further developed into a commercial line with enhanced storability and increased grain FA content.
Results
Preparation of transformation construct and production of T0 transformants
To facilitate the development of MFTID transgenic lines, we prepared a DRB binary construct (pDRB‐LOXRNAi) for Agrobacterium‐mediated wheat transformation. As shown in Figure 1a, the LOXRNAi cassette was located between LB and RB2; it was driven by the promoter (pGlu) of a high‐molecular‐weight glutenin gene, which had been successfully used for silencing wheat gene expression in the endospermic tissues by past studies (Dong et al., 2015; Yue et al., 2008). The sequence used to construct the LOXRNAi hairpin was 365 bp long; it was highly conserved in the LOX genes from wheat, barley, rice and Brachypodium distachyon, but exhibited no significant identities to the six functional human LOX genes (Dong et al., 2015). Additional bioinformatic searching of NCBI database (www.ncbi.nlm.nih.gov/blast) with the 365 bp RNAi‐inducing fragment as query did not find contiguous 21‐nucleotide matches with the genes of a wide range of species, including livestock animals, insects, fungi and bacteria, indicating that the LOXRNAi hairpin is likely safe for nontarget organisms (Sun et al., 2019). The marker gene cassette, expressing Bar gene under the promoter of rice Actin 1 (McElroy et al., 1995), was located between RB2 and RB1 (Figure 1a).
Figure 1.
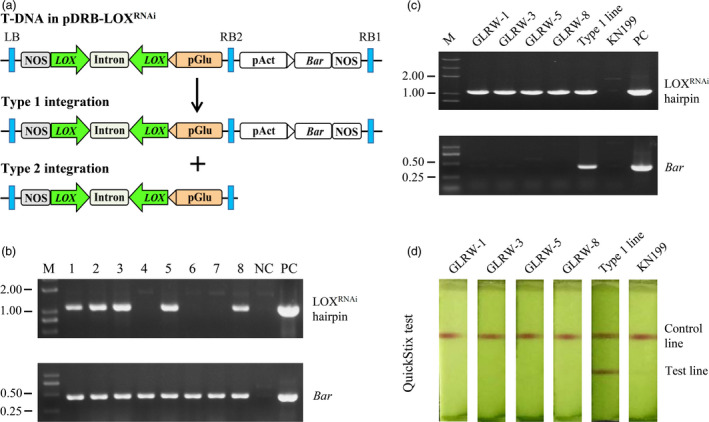
Development of transgenic GLRW lines. (a) Illustration of pDRB‐LOXRNAi T‐DNA region and its two types of integration events. (b) Detection of LOXRNAi hairpin and Bar gene in eight T0 transformants. (c) Amplification of LOXRNAi hairpin and Bar gene by PCR in the four homozygous marker‐free transgenic GLRW lines. (d) Detection of Bar protein using QuickStix strip test by checking band presence or absence in the test line. GLRW‐1, ‐3, ‐5 and ‐8: four homozygous grain LOX‐reduced wheat lines; KN199: Kenong 199; LB: left border; M: DNA size marker (kb); NC: negative control (without template); PC: positive control (with pDRB‐LOXRNAi as template); RB1: right border 1; RB2: right border 2; Type 1 line: a transgenic wheat line carrying both LOXRNAi and Bar gene expression cassettes.
Kenong 199 (KN199), a nationally certified elite winter wheat cultivar and widely cultivated in the northern plain of China (Xu et al., 2017), was used as the recipient for Agrobacterium‐mediated transformation. A total of 300 immature embryos were inoculated with the Agrobacterium cells carrying pDRB‐LOXRNAi, with eight herbicide resistant T0 seedlings obtained from different calli. By PCR screening, Bar gene was detected in all eight seedlings, but the LOXRNAi hairpin was found in only five of them (T0‐1, ‐2, ‐3, ‐5 and ‐8) (Figure 1b). The transformation frequency calculated based on the detection of Bar gene (2.7%) was comparable to that reported by Wang et al. (2016a). The co‐transformation frequency based on the finding of both Bar gene and LOXRNAi hairpin (1.7%) was much lower. The failure of detecting of RNAi hairpin in some of the T0 transformants might be caused by aberrant T‐DNA integration, which led to the insertion of marker gene cassette but not that of LOXRNAi hairpin. This phenomenon has also been observed in a previous plant transformation study involving the T‐DNA construct carrying RNAi cassette (Sunitha et al., 2012).
Identification of homozygous marker‐free lines
The T1 progenies derived from T0‐1, ‐2, ‐3, ‐5 and ‐8 were examined for marker‐free individuals by identifying those positive for LOXRNAi hairpin but negative for Bar gene through PCR. Marker‐free plants were obtained from the progenies of T0‐1, ‐3, ‐5 and ‐8, with the frequencies ranging from 3.2 to 9.7%. However, no such plants were found among the progenies of T0‐2. From the selfed progenies of the marker‐free plants, four independent T4 homozygous transgenic lines containing LOXRNAi hairpin but not Bar gene, that is, GLRW‐1, ‐3, ‐5 and ‐8, were developed (Figure 1c). The lack of Bar protein in GLRW‐1, ‐3, ‐5 and ‐8 was validated using the QuickStix strip test (Figure 1d).
Determination of T‐DNA insertion sites
Thermal asymmetric interlaced PCR (TAIL‐PCR) was employed to determine T‐DNA integration sites in the four GLRW lines. Using an optimized TAIL‐PCR protocol (Liu and Chen, 2007), T‐DNA integration sites in GLRW‐3 and ‐8 were determined, but not in the other two lines despite repeated efforts. The T‐DNA in GLRW‐3 was located on chromosome 1D; the integration caused several indels, including a 14 bp deletion in LB region, a 21 bp deletion in RB2 region, a 19 bp deletion of chromosomal DNA after RB2, followed by a 12 bp nucleotide insertion (Figure 2). The T‐DNA sequences retained in LB and RB2 regions were 16 and 9 bp, respectively, which flanked the 2289 bp LOXRNAi cassette (Figure 2). To assess potential influence of this T‐DNA insertion on the expression of flanking wheat genes, we examined the transcript levels of the two genes (TraesCS1D02G296900.1 and TraesCS1D02G297000.1) upstream of the insertion and the two genes (TraesCS1D02G297100.1 and TraesCS1D02G297200.1) downstream of the insertion (Figure 2). The four genes had been annotated as uncharacterized genes (International Wheat Genome Sequencing Consortium et al., 2018) and occupied a genomic region of more than 610 kb (Figure 2). In repeated qRT‐PCR assays, no significant differences were detected between KN199 (WT control) and GLRW‐3 in the expression of the four genes in the root, stem, leaf and grain tissues (Figure S1).
Figure 2.
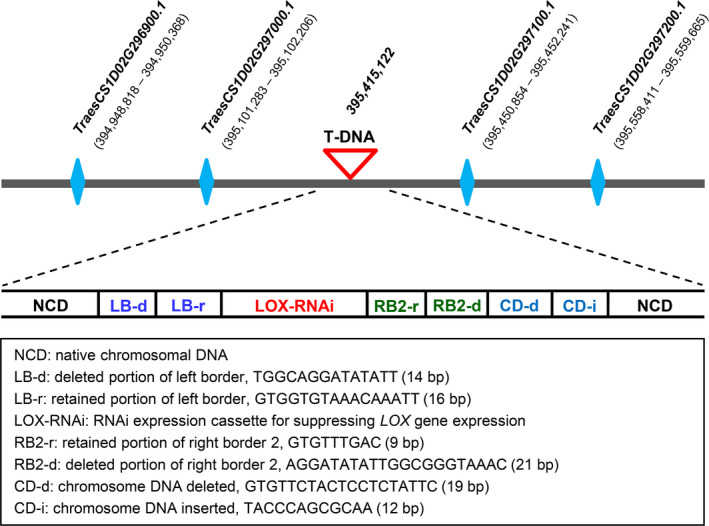
Analysis of T‐DNA integration site in GLRW‐3. The T‐DNA was inserted in an intergenic region on chromosome 1D. The genes upstream (TraesCS1D02G296900.1 and TraesCS1D02G297000.1) and downstream (TraesCS1D02G297100.1 and TraesCS1D02G297200.1) of the T‐DNA insertion site are shown with coordinates based on the annotation deposited in Ensembl Plants (http://plants.ensembl.org/Triticum_aestivum/Info/Index). Various changes occurred to the T‐DNA or wheat chromosomal DNA are summarized in the box at the bottom.
In GLRW‐8, the T‐DNA insertion site was found on chromosome 2B; the integration also caused several indels, including a 14 bp deletion in LB, a 21 bp deletion in RB2, a 40 bp deletion of chromosomal DNA after RB2, followed by an A to G nucleotide replacement (Figure S2a). Remarkably, the LB and RB2 sequences deleted in this site were identical to those happened to the T‐DNA site in GLRW‐3 (Figure 2). But the changes of chromosomal DNA occurred to this site (40 bp deletion and an A/G nucleotide replacement, Figure S2a) differed from those detected in the T‐DNA insertion site in GLRW‐3 (19 bp deletion and 12 bp insertion, Figure 2).
A close inspection revealed that, in GLRW‐8, the T‐DNA inserted into the single exon of TraesCS2B02G181300.1 (Figure S2b), which has been annotated as a high confidence gene (International Wheat Genome Sequencing Consortium et al., 2018). Searching wheat transcriptome databases expVIP and WheatExp reveals that TraesCS2B02G181300.1 is expressed at multiple developmental stages and in both vegetative and reproductive organs. Furthermore, it has a highly conserved ortholog (TRIDC2BG023520.1) in wild emmer wheat (http://plants.ensembl.org/Triticum_dicoccoides/). Thus, TraesCS2B02G181300.1 is likely a functionally important gene. Inactivation of TraesCS2B02G181300.1 by T‐DNA insertion in GLRW‐8 may have negative consequences on wheat growth and development. Therefore, GLRW‐8 was excluded from further experiments.
Comparison of LOX gene expression among WT control and transgenic lines
To investigate if the LOXRNAi hairpin may suppress the expression of endogenous LOX genes, it was necessary to identify the LOX member that is highly expressed in the developing grains of KN199, the WT progenitor of the GLRW lines. Previously, we isolated TaLOX1 (GenBank accession GQ166692), which is located on chromosome 4DS and highly expressed in the developing grains of the winter wheat cultivar Xiaoyan 54 (Feng et al., 2010). Here, we cloned the TaLOX1 allele (TaLOX1‐KN199) of KN199. Through searching the draft genome sequence of common wheat, the counterpart of TaLOX1 was also identified in the landrace variety CS, which was named as Traes_4DS_7868A8C2E.1 (IWGSC, 2014) or TraesCS4D02G035200.1 (International Wheat Genome Sequencing Consortium et al., 2018). To facilitate the analysis in this study, it was designated as TaLOX1‐CS. The three LOX alleles exhibited 95.4–100% identities in coding sequence, and their deduced proteins were 96.1–100% identical (Figure S3a). Based on the expression data deposited at WheatExp and expVIP databases, TaLOX1‐CS was highly expressed in the developing grains, especially at 30 days after anthesis (DAA) (Figure S3b). The 365 bp fragment used to construct LOXRNAi hairpin was 98.9% identical to its counterpart in TaLOX1‐KN199 (Figure S3c). Together, these data indicated that TaLOX1‐KN199 was a highly expressed LOX gene in KN199 grains and could be silenced by the LOXRNAi hairpin transcribed from the integrated transgene in the GLRW lines.
Therefore, a pair of gene‐specific primers was designed for TaLOX1‐KN199 (Figure 3a) and used to compare LOX gene expression in the developing grains among KN199 and the GLRW lines. In KN199, the expression level of TaLOX1‐KN199 was gradually up‐regulated as grain development progressed, with the highest expression level detected at 30 DAA (Figure 3b), which was consistent with the expression feature of TaLOX1‐CS in the developing grains of CS (Figure S3b). The expression levels of TaLOX1‐KN199 in the three transgenic lines were generally similar to that of KN199 at 10 DAA, but became lower than that of KN199 afterwards, especially at 30 DAA (Figure 3b).
Figure 3.
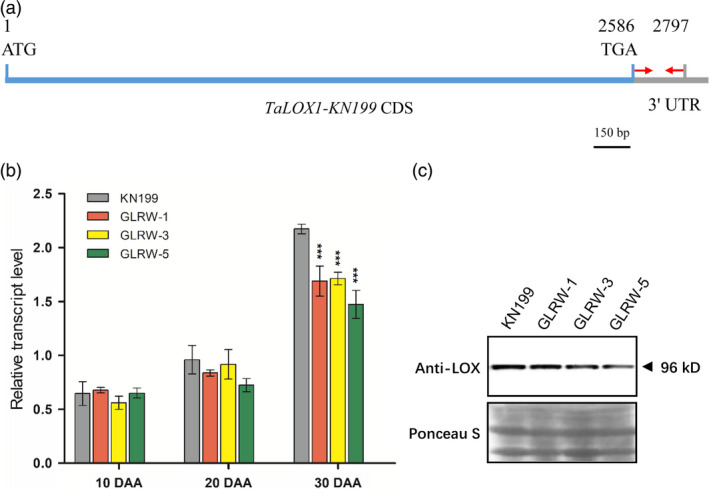
Suppression of LOX transcript and protein accumulation in transgenic GLRW lines. (a) A diagram illustrating the coding sequence (CDS) of TaLOX1‐KN199 and the primer pair, indicated by two red arrows in 3′ UTR, used in the qRT‐PCR assay. (b) Comparison of TaLOX1‐KN199 transcript level between KN199 (WT control) and three transgenic lines (GLRW‐1, ‐3 and ‐5) in the developing grains collected at 10, 20 or 30 days after anthesis (DAA) by qRT‐PCR assay. Each data point was the mean (±SD) of three biological replicates. ***P < 0.001 (Student's t‐test). (c) Immunoblot analysis of TaLOX1‐KN199 protein accumulation in the fresh mature grains (30 DAA) of KN199 and the transgenic lines GLRW‐1, ‐3 and ‐5. The predicted molecular mass of TaLOX1‐KN199 is approximately 96 kilodalton (kD). The result shown was representative of three independent experiments.
Further to the qRT‐PCR experiment, the accumulation of LOX protein in KN199 and the GLRW lines were examined using immunoblot analysis. A polyclonal antibody, raised against a bacterially expressed polypeptide of TaLOX1, was employed. The molecular mass predicted for the protein product of TaLOX1‐KN199 was 96 kDa. This protein was readily detected in the fresh mature grains of WT control; it was also found in the grain tissues of the three GLRW lines but with reduced amounts (Figure 3c). Together, the above results indicated that the LOXRNAi hairpin was effective in suppressing TaLOX1‐KN199 expression in the grains of the three GLRW lines.
Comparison of LOX enzyme activity and malondialdehyde content among WT and transgenic lines
Total LOX enzyme activity was assayed for the flour samples of KN199 and three GLRW lines. In the flour samples derived from the grains harvested from the greenhouse in 2016, the levels of total LOX enzyme activity measured for the three transgenic lines were all significantly lower than that recorded for KN199, with the percentage of reduction ranging from 26.80% (GLRW‐5) to 33.35% (GLRW‐1) (Figure 4a). In the flour samples milled from the grains collected from the field in 2017 (i.e., the first field trial, see below), the values of total LOX enzyme activity obtained for the transgenic lines were again significantly lower than that of KN199, with the percentage of decrease ranging from 26.33% (GLRW‐3) to 31.68% (GLRW‐1) (Figure 4b). Malondialdehyde (MDA), the peroxidation product of PUFAs (Draper and Hadley, 1990), was determined for the flour samples of WT and GLRW lines. For either 2016 or 2017 samples, the MDA contents of the three GLRW lines were all substantially decreased relative to that of KN199 (Figure 4c,d).
Figure 4.
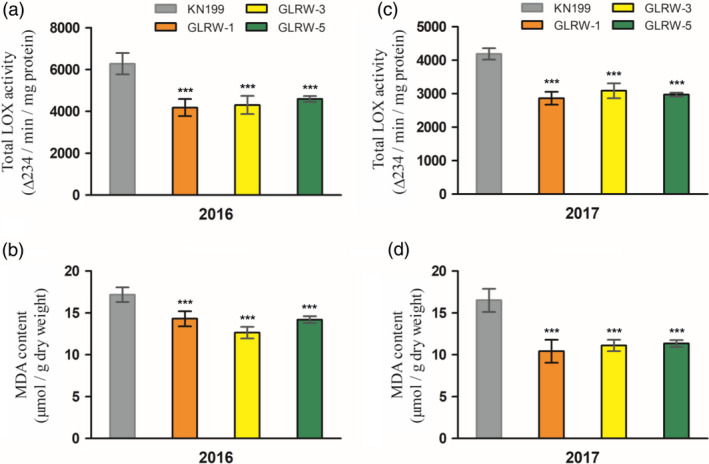
Investigation of total LOX activity and MDA content in the dehydrated mature grains of KN199 (WT control) and three transgenic lines (GLRW‐1, ‐3 and ‐5). (a,c) Total LOX activity levels determined using the grain samples harvested in 2016 and 2017, respectively. (b,d) MDA contents obtained with the grain samples harvested in 2016 and 2017, respectively. Each data point (mean ± SD) was calculated from three measurements conducted with separate grain samples. **P < 0.01; ***P < 0.001 (Student's t‐test).
Effects of artificial ageing on the germination and seedling growth of KN199 and transgenic lines
Artificial ageing (AA) treatment was conducted to examine if the three GLRW lines might exhibit improved storability relative to WT control. AA substantially decreased LOX activity in both KN199 and GLRW lines, but either before or during AA, the levels of LOX activity in the three GLRW lines were generally and significantly lower than that of KN199 (Figure 5a). MDA was gradually elevated by AA in both KN199 and GLRW lines, but for all three GLRW lines their MDA contents were consistently lower than that of KN199 (Figure 5b). Before AA, no significant difference was observed in seed germination between KN199 and GLRW lines, but at either 5 or 10 days post‐AA (DPAA), the germination rates of the three GLRW lines were all clearly higher than that of KN199 (Figure 5c). Neither KN199 nor GLRW seeds germinated after 15 DPAA (Figure 5c). When the seedlings developed from AA‐treated grains were compared, a much better growth performance was found for all three GLRW lines relative to that of KN199 (Figure 5d).
Figure 5.
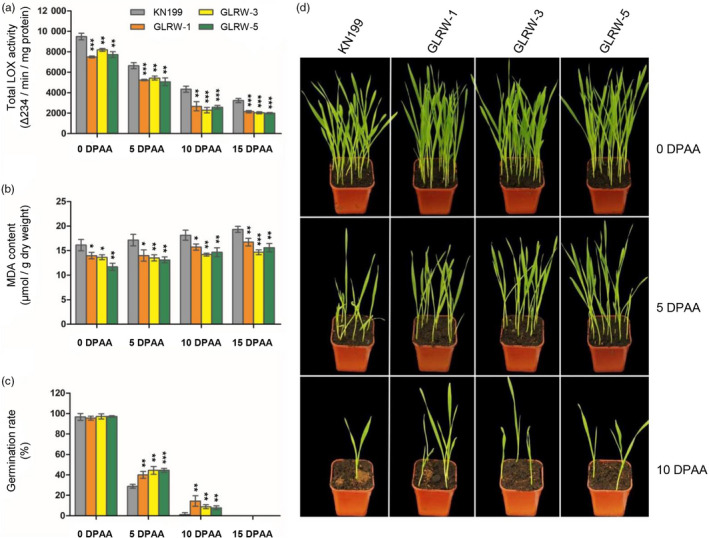
Analysis of the effects of artificial ageing on the grains of KN199 and three transgenic GLRW lines. The grain samples of the four lines were subjected to various assays at 5, 10 or 15 days post artificial ageing treatment (DPAA), with their untreated grains (0 DPAA) used as controls. (a–c) Total LOX activity levels (a), MDA contents (b), and germination rates (c) determined for the grain samples of the four lines at different DPAA points. (d) Growth performance of the seedlings that were germinated from the grains of KN199 and the three transgenic lines at 5 or 10 DPAA or from the untreated controls (0 DPAA). The numerical data points (means ± SD) were each calculated from three measurements conducted with separate grain samples. *P < 0.05; **P < 0.01; ***P < 0.001 (Student's t‐test). The result shown in (d) was representative three separate germination trials.
Investigation of fatty acid contents in KN199 and GLRW lines
The data presented above prompted us to investigate if the accumulation of FAs might be altered in the GLRW lines relative to that in KN199. Two sets of grain samples were analysed using gas chromatography–mass spectrometry (GC‐MS). In addition to LA (18:2) and LNA (18:3), palmitic acid (C16:0, PA), stearic acid (18:0, SA) and oleic acid (18:1, OA) were also included in the analysis. In the 2016 greenhouse samples, the contents of five FAs and their combined amounts were generally higher in the grain samples of the three GLRW lines than in those of KN199 (WT control) (Table 1). Relative to KN199, the contents of PA, SA, OA, LA and LNA in the three GLRW lines were increased by 0.40–7.48%, 2.72–27.24%, 21.11–32.22%, 3.04–7.53% and 11.39–18.99%, respectively; the combined values of the five FAs in the three transgenic lines were 3.70%, 10.31% and 3.86% higher than that of KN199 (Table 1).
Table 1.
Analysis of grain FA contents of KN199 and three transgenic lines cultivated in the greenhouse in 2016
| Palmitic acid (C16:0, PA) | Stearic acid (C18:0, SA) | Oleic acid (C18:1, OA) | Linoleic acid (C18:2, LA) | Linolenic acid (C18:3, LNA) | Total | |
|---|---|---|---|---|---|---|
| KN199 | 12.56 ± 0.54 | 3.67 ± 0.36 | 0.90 ± 0.03 | 18.05 ± 0.19 | 0.79 ± 0.06 | 35.97 ± 0.10 |
| GLRW‐1 | 12.61 ± 0.49 [0.40%] † | 3.77 ± 0.50 [2.72%] | 1.19 ± 0.05*** [32.22%] | 18.85 ± 0.33* [4.43%] | 0.88 ± 0.03* [11.39%] | 37.30 ± 0.39** [3.70%] |
| GLRW‐3 | 13.50 ± 0.34* [7.48%] | 4.67 ± 0.61* [27.24%] | 1.15 ± 0.06** [27.78%] | 19.41 ± 0.40** [7.53%] | 0.94 ± 0.04* [18.99%] | 39.68 ± 0.98** [10.31%] |
| GLRW‐5 | 12.61 ± 0.59 [0.40%] | 4.13 ± 0.21 [12.53%] | 1.09 ± 0.13* [21.11%] | 18.60 ± 0.30* [3.04%] | 0.92 ± 0.03* [16.46%] | 37.36 ± 0.89* [3.86%] |
Each value (μg/mg grain) was the mean ± SD of three different determinations with separate grain samples; *, ** and *** indicate statistical difference from KN199 (WT control) at P < 0.05, 0.01 and 0.001, respectively.
The value in the square brackets indicates the percentage of increase over KN199.
In the 2017 field samples, the contents of grain PA, SA, OA, LA and LNA in the GLRW lines were up‐regulated by 3.20–7.51%, 4.93–11.33%, 55.71–58.57%, 17.86–21.64% and 50.91–65.45%, respectively, relative to those of KN199; the combined amounts of the five FAs were also substantially higher in the three transgenic lines (Table 2). The fine flour samples prepared from the second set of grain materials were also assayed for FA contents because wheat grains are generally milled into flour for human consumption. Relative to WT control, the contents of flour PA, SA, OA, LA and LNA in the three GLRW lines were lifted by 17.30–35.34%, 1.57–15.97%, 66.67–73.33%, 15.32–28.45% and 19.23–53.85%, respectively, and the combined amounts of the five FAs in the three transgenic lines were 25.74% (GLRW‐1), 27.56% (GLRW‐3) and 14.13% (GLRW‐5) higher than that of KN199 (Table S1).
Table 2.
Evaluation of grain FA contents of KN199 and three transgenic lines cultivated in the field in 2017
| Palmitic acid (C16:0) | Stearic acid (C18:0) | Oleic acid (C18:1) | Linoleic acid (C18:2) | Linolenic acid (C18:3) | Total | |
|---|---|---|---|---|---|---|
| KN199 | 11.58 ± 0.45 | 4.06 ± 0.25 | 0.70 ± 0.05 | 13.77 ± 0.80 | 0.55 ± 0.05 | 30.66 ± 1.60 |
| GLRW‐1 | 11.95 ± 0.41 [3.20%] † | 4.52 ± 0.30 [11.33%] | 1.09 ± 0.11** [55.71%] | 16.23 ± 0.25** [17.86%] | 0.87 ± 0.06*** [58.18%] | 34.66 ± 0.33** [13.05%] |
| GLRW‐3 | 12.45 ± 0.45* [7.51%] | 4.26 ± 0.35 [4.93%] | 1.09 ± 0.12** [55.71%] | 16.41 ± 0.60** [19.17%] | 0.83 ± 0.05*** [50.91%] | 35.04 ± 1.56* [14.29%] |
| GLRW‐5 | 12.39 ± 0.36* [6.99%] | 4.45 ± 0.45 [9.61%] | 1.11 ± 0.12** [58.57%] | 16.75 ± 0.69** [21.64%] | 0.91 ± 0.04*** [65.45%] | 35.61 ± 0.22** [16.14%] |
Each value (μg/mg grain) was the mean ± SD of three different determinations with separate grain samples; *, ** and *** indicate statistical difference from KN199 (WT control) at P < 0.05, 0.01 and 0.001, respectively.
The value in the square brackets indicates the percentage of increase over KN199.
Field assessment of GLRW lines
Two field trials were carried out to compare the main agronomic traits between the GLRW lines and KN199. In the first trial, artificially vernalized seedlings were transplanted in the field in March 2017, with the grains harvested in June 2017. This assessment showed that the GLRW lines did not differ significantly from KN199 in plant height (PH), tiller number per plant (TNPP), grain number per spike (GNPS), grain length (GL), grain width (GWH) and thousand grain weight (TGW), although their spike length (SL) appeared to be higher (Table S2). The second field trial was conducted between October 2017 and June 2018, with each line sown in three replicated plots. The GLRW lines were found to behave similarly as KN199 in PH, TNPP, SL, GL, GWH, TGW and plot yield (PY), though their GNPS tended to be higher (Table S3).
Discussion
Improving seed storability through decreasing LOX activities has been achieved in soybean, rice, wheat and maize (Dong et al., 2015; Gayen et al., 2014; Huang et al., 2014; King et al., 1998; Ma et al., 2015; Shen et al., 1996; Xu et al., 2015), indicating the wide applicability of this approach in grain crops. However, prior to this work, no MFTID transgenic lines with reduced LOX activities in the grains have been reported, which hinders the enhancement of seed storability through transgenic breeding. Here, we demonstrated for the first time that MFTID transgenic lines with reduced grain LOX activity can be developed from the progenies descended from Agrobacterium‐mediated genetic transformation. Importantly, seed storability of the three GLRW lines is significantly enhanced as evidenced by better germination rate and more vigorous seedling growth after AA treatment. Furthermore, the three transgenic lines accumulate substantially more FAs in both grain and flour samples. Because FAs are important nutrients for human health, and beneficial for breadmaking quality (see below), we suggest that development of transgenic GLRW lines brings multiple benefits to common wheat grains.
Important to this work is the utilization of the reference genome sequence of common wheat (International Wheat Genome Sequencing Consortium et al., 2018). It enabled us to precisely elucidate the T‐DNA insertion sites in two MFTID lines. To our knowledge, no previous studies have succeeded in elucidating T‐DNA insertion sites at single base‐pair resolution in transgenic wheat, although this has been achieved in the transgenic plants with relatively small genome size, such as Agrobacterium‐transformed Arabidopsis, Brachypodium distachyon and rice (Chen et al., 2003; Kim et al., 2007; Kleinboelting et al., 2015; Thole et al., 2010). Notably, complex DNA sequence changes were observed in the two T‐DNA insertion sites characterized in this work. Some of the alterations, that is, deletions in the LB and RB2 regions, were conserved, whereas others (changes to chromosomal DNA) differed completely between the two sites, indicating similarities and differences among different T‐DNA insertion sites. Nevertheless, these changes did not affect the intactness and function of the LOXRNAi cassette. The occurrence of indels in the LB and RB regions of T‐DNA and plant chromosomal DNA, as found in the T‐DNA integration sites in GLRW‐3 and GLRW‐8, has also been observed in previous studies (Gambino et al., 2009; Kleinboelting et al., 2015; Ling et al., 2016). For example, the T‐DNA insertion site characterized in an insect resistant‐transgenic rice line displayed a 15 bp deletion in the LB region, a complete missing of the RB, and a 7 bp deletion of rice chromosomal DNA downstream of RB, but the cry2A* cassette expressing the insecticidal Bt protein remains intact (Ling et al., 2016). Lastly, Wu et al. (2006) discovered incomplete T‐DNA integration and vector backbone integration in their study of Agrobacterium‐transformed wheat lines using Southern hybridization and PCR methods. We did not find evidence for vector backbone integration in GLRW‐3 and GLRW‐8 using a similar PCR amplification approach.
Wheat grains accumulate a large variety of lipid compounds, many of which have been found to affect the nutritional and end‐use properties of flour (González‐Thuillier et al., 2015; Pareyt et al., 2011; Tosi et al., 2018). FAs constitute a major component of the total lipids in wheat flour and may contribute significantly to breadmaking quality (Gerits et al., 2015; González‐Thuillier et al., 2015). There are now increasing interests in understanding and manipulating the accumulation of FAs in cereal grains by genetic means (Kraic et al., 2018; Mihálik et al., 2014, 2015; Wang et al., 2011). Transgenic expression of a fungal Δ6 desaturase gene in barley and wheat leads to the production of γ‐linolenic acid and stearidonic acid in the grains, both of which are beneficial for human health but not naturally present in cereal plants (Mihálik et al., 2014, 2015). Here, we demonstrate for the first time that grain‐specific reduction of LOX gene expression leads to substantial increases of PA, SA, OA, LA and LNA in both the grain and flour samples (Tables 1 and 2; Table S1), thus providing a new option for manipulating FA accumulation in cereal grains. Based on this work, it becomes necessary and important to examine if other lipid components may also be altered in the three GLRW lines, and the potential consequence of altered lipid accumulation on wheat end‐use properties in further research.
Concomitant to the improvement in grain storability and FA contents, the three GLRW lines maintained their agronomic performance as the elite cultivar KN199 under field conditions (Tables S2 and S3), indicating that suppression of LOX gene expression has not brought apparent adverse effects to the three transgenic lines. Considering the important function of LOX genes in plant growth and development and responses to environmental changes, we deliberately limited the silencing effect of LOXRNAi to the endospermic tissues by using the promoter pGlu (Dong et al., 2015; Yue et al., 2008). As anticipated, the reduction of LOX enzyme activity in the grains of the three GLRW lines was not excessive (33.35% at most, Figure 4), which may have assisted the three GLRW lines to maintain their normal agronomic performance in the field. Nevertheless, more vigorous tests, including multi‐year and multilocation trials, are still necessary to fully evaluate the agronomic traits and economic benefits of the GLRW lines in the future.
The CRISPR/Cas‐mediated genome editing, which has been found to be highly efficient and versatile (Chen et al., 2019; Li et al., 2019), may also be potentially useful for decreasing LOX gene expression in the grains. However, complete knockout of important LOX genes by simple genome editing may be deleterious to plant growth and survival. In this regard, base editors, which are novel developments of CRISPR/Cas9 system (Chen et al., 2019; Komor et al., 2016), may be employed to create expression and/or catalytic function reduced LOX alleles. These alleles are probably more suitable for the down‐regulation of grain LOX activity and the improvement of seed storability and fatty acid contents via genome editing.
In summary, this work has demonstrated that transgenic suppression of grain LOX gene expression simultaneously improves wheat seed storability and FA content, and that MFTID transgenic plants can be developed with the aid of wheat genome sequence information. Our approach is also likely applicable for improving the grain storability and FA content of other food crops, for example, rice, maize and barley. Considering that the annual stocks of major cereal grains in the world generally exceed 100 million tons in recent years (https://apps.fas.usda.gov/psdonline/app/index.html#/app/home; Table S4), development and application of wheat, rice and maize cultivars with enhanced storability and nutrient quality are highly desirable. In this respect, the MFTID wheat line GLRW‐3 characterized in this work may have considerable economic value and a good prospect of gaining commercial application. If approved, GLRW3 may be cultivated directly because its agronomic performance resembles highly the nationally certified elite variety KN199 or used as a germplasm to introduce the improved traits (i.e., enhanced grain storability and FA content) to more diverse wheat backgrounds.
Experimental procedures
Plant materials and growth conditions
KN199 and derivative transgenic GLRW lines were used in this study. For growth in the greenhouse, seeds were germinated at room temperature for 7 days, and subsequently maintained at 4 °C for 30 days for vernalization. The vernalized plants were grown to maturity at a temperature and photoperiod regime of 25 °C, 16 h light/18 °C, 8 h dark. Two field trials were carried out in Zhaoxian experimental farm in Hebei province, China, to compare the growth between KN199 and three GLRW lines. The first trial, accomplished from March 2016 to June 2016, used artificially vernalized seedlings. For each line, approximately 1000 seedlings were transplanted to the soil, which were arranged in 10 rows with 10 cm between neighbouring seedlings and 15 cm between adjacent rows. In the second trial conducted from October 2017 to June 2018, KN199 and the three GLRW lines were each sown in three replicated plots. Each plot (10 m × 1.2 m) had four rows of plants with adjacent rows separated by 15 cm. Standard agronomic measures were applied to promote plant growth and seed setting, with several agronomic traits, including PH, TNPP, TGW, GL, GWH and PY, assessed as described previously (Zhang et al., 2013).
Construction of pDRB‐LOXRNAi and wheat transformation
The Bar gene expression cassette was amplified from the plasmid pAct1‐bar (Dong et al., 2015) using the forward and reverse primers carrying BamHI and XbaI sites (Table S5), respectively. The obtained PCR product was digested with BamHI and XbaI, followed by cloning into the pMNDRBBin6 vector (Lu et al., 2001) cut with the same two enzymes. This resulted in the plasmid pDRB‐bar. The DNA fragment carrying the LOXRNAi cassette, prepared by cutting the plasmid pGlu‐LOXRNAi (Dong et al., 2015) with XhoI and HpaI, was cloned into pDRB‐bar digested with the same two enzymes, thus giving rise to the wheat transformation construct pDRB‐LOXRNAi. The Agrobacterium tumefaciens cells (strain AGL1) carrying pDRB‐LOXRNAi were used to transform 300 immature embryos of KN199 following the protocol detailed previously (He et al., 2010; Wang et al., 2016a). The resultant T0 transformants and their selfed progenies were employed to develop marker‐free GLRW lines.
Identification of transgenic plants by PCR and QuickStix test
Genomic DNA was extracted from KN199, T0 transformants and their progenies at two‐leaf stage using the CTAB method (Saghai‐Maroof et al., 1984). The resultant DNA samples were used for PCR to amplify the LOXRNAi hairpin and the Bar gene, respectively, with gene‐specific primers (Table S5). The QuickStix test for detecting Bar protein was carried out using LibertyLink® strip (QuickStix; Envirologix, Portland, ME) according to the manufacturer's instructions.
Determination of T‐DNA integration sites
TAIL‐PCR was adopted for isolating genomic sequences flanking T‐DNA integration sites in the GLRW lines. Three nested specific primers (RB550, RB402 and RB322) near T‐DNA RB and 17 degenerate primers (AD1‐17) were designed (Table S5). The PCR conditions were based on those described by Liu and Chen (2007). The tertiary PCR amplification products were sequenced, with the sequence reads mapped to the reference genome of CS (International Wheat Genome Sequencing Consortium et al., 2018). Based on the results obtained, additional specific primers (Table S5) were designed and used to confirm the T‐DNA integration site in GLRW‐3 (primer pair GT38F and GT3R) or GLRW‐8 (primer pair GT38F and GT8R).
RNA isolation and qRT‐PCR analysis
Total RNA was extracted from the developing grains of KN199 and GLRW‐1, ‐3 and ‐5 at 10, 20, 30 DAA, respectively, using the RNAiso Plus Kit (TaKaRa, Otsu, Japan). After DNaseI treatment, the RNA was reverse‐transcribed into cDNA using the GoScript™ Reverse Transcription System (Promega, Madison, WI). The resulting cDNA was diluted 100 times and used in the qRT‐PCR assay of TaLOX1‐KN199 expression with the Roche LightCycler 480 system (Roche Applied Science, Indianapolis, IN). The assay used SYBR Green qPCR Master Mix (Fermentas Inc., MD) and the primer pair specific for TaLOX1‐KN199 (Figure 3a; Table S5). Amplification of a wheat Actin gene (Table S5) served as the internal control. Three biological repeats, each involving the sampling of 3 to 5 developing grains, were assayed for each line at each stage.
For assessing potential influence of T‐DNA insertion in GLRW‐3 on the expression of flanking genes by qRT‐PCR assay, the root, stem and leaf samples were collected from the plants at heading stage, while the seed sample was prepared at 30 DAA. The assay was executed as above with gene‐specific primers (Table S5).
LOX activity and lipid peroxidation assays
Total LOX activity levels in the seeds of KN199 and GLRW‐1, ‐3 and ‐5 were determined as described by Feng et al. (2010). For each line, the assay was carried out three times using enzyme extracts from three separate batches of seeds. Lipid peroxidation was measured by determining seed MDA contents of KN199 and GLRW‐1, ‐3 and ‐5 using a MDA assay kit (Beyotime, Beijing, China). For each line, three independent determinations were conducted with separate seed samples.
Immunoblot assay
The C‐terminal end (561–861 residues) of TaLOX1 (GenBank accession ACS34909.1, Feng et al., 2010) was expressed as a His‐tag protein in Escherichia coli with the aid of the bacterial expression vector pET28a+ (Novagen, Madison, WI). The recombinant polypeptide was purified using nickel affinity chromatography, which was then employed to raise a mouse polyclonal antibody specific for TaLOX1 as reported previously (Qin et al., 2008). The fresh mature grains of KN199 and GLRW‐1, ‐3 and ‐5 were collected at 30 DAA and used for analysing LOX protein accumulation by immunoblotting with the TaLOX1 antibody following the method outlined in Li et al. (2018).
AA treatment
AA treatment was performed according to Dong et al. (2015). Briefly, the dried mature seeds of KN199 and GLRW‐1, ‐3 and ‐5 were incubated in an electronically controlled environmental cabinet set at 40 °C and 90% relative humidity. The samples were taken out from the cabinet at three predetermined time points, that is, 5, 10 and 15 DPAA, for downstream investigations (assaying LOX activity, MDA content and germination rate). The seeds used for each line at each time point was 300 g. Untreated seeds were used as controls.
Germination assay
Germination assay was conducted for AA‐treated KN199 and GLRW seed samples in triplicates. Each replicate included 60 seeds, which were sown in the pot filled with peat moss, and maintained in the greenhouse as described above. Germination rate was defined as the percentage of germinated seeds at the seventh day.
Fatty acid analysis
FAs were extracted and analysed using GC‐MS as described by Song et al. (2010) and Zhang et al. (2014). For measuring grain FA contents in each line, 5 g of seeds were ground into powder, with three separate samples (40 mg each) taken for FA extraction and analysis. For measuring flour FA contents in each line, 200 g of seeds were milled in the Brabender Quadrumat Junior mill (Brabender, Duisburg, Germany) (70% flour yield), with three different flour samples (40 mg each) used for subsequent FA extraction and determination.
Bioinformatic analysis
The nonredundant nucleotide collection in the NCBI database was used in the BLAST search for the presence of contiguous 21‐nt matches in the nontarget organisms to the LOX‐RNAi‐inducing fragment, with the maximum target sequences adjusted to 10 000. Genes flanking the T‐DNA integration site in GLRW‐3 and GLRW‐8 were retrieved from Ensembl Plants (http://plants.ensembl.org/Triticum_aestivum/Info/Index). Wheat gene expression data were searched at expVIP (http://www.wheat-expression.com/) and WheatExp (https://wheat.pw.usda.gov/WheatExp/). The molecular mass of TaLOX1 was predicted using the ProtParam tool (https://web.expasy.org/protparam/).
Statistical analysis
Statistical tests were performed with the GraphPad Prism 5 software. Each result was presented as mean ± SD based on at least three biological replicates.
Conflict of interest
The authors declare no conflict of interests.
Author contributions
D.W., K.Z. and X.C. designed the study. X.C. and Z.D. identified and analysed the marker‐free transgenic lines; D.T. and L.D. conducted bioinformatic analysis; W.Q., J.L., W.Z. and C.G. contributed to T‐DNA construct preparation and performed wheat transformation; X.C., K.Z., X.L. and H.Q. conducted and analysed field trials; D.W., X.C., Z.D. and K.Z. wrote the manuscript.
Supporting information
Figure S1 Analysis of the expression of the four genes flanking the T‐DNA insertion site in GLRW‐3.
Figure S2 Elucidation of the T‐DNA integration site in GLRW‐8.
Figure S3 Characterization of TaLOX1‐KN199 allele.
Table S1 Comparison of flour FA contents of KN199 and three transgenic lines cultivated in the field in 2017.
Table S2 Field assessment of important agronomic traits of three transgenic GLRW lines in 2017.
Table S3 Comparison of important agronomic traits between Kenong 199 and three transgenic GLRW lines in 2017–2018 growth season.
Table S4 Annual stocks of three major cereal grains in the world in recent years.
Table S5 Oligonucleotide primers used in this study.
Acknowledgements
We thank Dr. Yongqiang Li (Sanyou Biopharmaceuticals (Shanghai) Co., Ltd) for assisting the preparation of TaLOX1 specific antibody. This work was supported by the Ministry of Agriculture of China (2016ZX08002003).
Contributor Information
Kunpu Zhang, Email: kpzhang@heanu.edu.cn.
Daowen Wang, Email: dwwang@genetics.ac.cn.
References
- Abdellatef, E. , Will, T. , Koch, A. , Imani, J. , Vilcinskas, A. and Kogel, K.H. (2015) Silencing the expression of the salivary sheath protein causes transgenerational feeding suppression in the aphid Sitobion avenae . Plant Biotechnol. J. 13, 849–857. [DOI] [PubMed] [Google Scholar]
- Ahmad, N. and Mukhtar, Z. (2017) Genetic manipulations in crops: challenges and opportunities. Genomics, 109, 494–505. [DOI] [PubMed] [Google Scholar]
- Altenbach, S.B. , Tanaka, C.K. and Seabourn, B.W. (2014) Silencing of omega‐5 gliadins in transgenic wheat eliminates a major source of environmental variability and improves dough mixing properties of flour. BMC Plant Biol. 14, 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altpeter, F. , Vasil, V. , Srivastava, V. and Vasil, I.K. (1996) Integration and expression of the high‐molecular‐weight glutenin subunit 1Ax1 gene into wheat. Nat. Biotechnol. 14, 1155–1159. [DOI] [PubMed] [Google Scholar]
- Altpeter, F. , Baisakh, N. , Beachy, R. , Bock, R. , Capell, T. , Christou, P. , Daniell, H. et al. (2005) Particle bombardment and the genetic enhancement of crops: myths and realities. Mol. Breed. 15, 305–327. [Google Scholar]
- Barro, F. , Rooke, L. , Békés, F. , Gras, P. , Tatham, A.S. , Fido, R. , Lazzeri, P.A. et al. (1997) Transformation of wheat with high‐molecular‐weight subunit genes results in improved functional properties. Nat. Biotechnol. 15, 1295–1299. [DOI] [PubMed] [Google Scholar]
- Bhalla, P.L. (2006) Genetic engineering of wheat – current challenges and opportunities. Trends Biotechnol. 24, 305–311. [DOI] [PubMed] [Google Scholar]
- Blechl, A.E. and Anderson, O.D. (1996) Expression of a novel high‐molecular‐weight glutenin subunit gene in transgenic wheat. Nat. Biotechnol. 14, 875–879. [DOI] [PubMed] [Google Scholar]
- Chen, H. and Lin, Y. (2013) Promise and issues of genetically modified crops. Curr. Opin. Plant Biol. 16, 255–260. [DOI] [PubMed] [Google Scholar]
- Chen, S. , Jin, W. , Wang, M. , Zhang, F. , Zhou, J. , Jia, Q. , Wu, Y. et al. (2003) Distribution and characterization of over 1000 T‐DNA tags in the rice genome. Plant J. 36, 105–113. [DOI] [PubMed] [Google Scholar]
- Chen, D. , Chai, S. , McIntyre, C.L. and Xue, G.P. (2018) Overexpression of a predominantly root‐expressed NAC transcription factor in wheat roots enhances root length, biomass and drought tolerance. Plant Cell Rep. 37, 225–237. [DOI] [PubMed] [Google Scholar]
- Chen, K. , Wang, Y. , Zhang, R. , Zhang, H. and Gao, C. (2019) CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 28.1‐28.31. [DOI] [PubMed] [Google Scholar]
- Christou, P. (2013) Plant genetic engineering and agricultural biotechnology 1983–2013. Trends Biotechnol. 31, 125–127. [DOI] [PubMed] [Google Scholar]
- Dong, Z. , Feng, B. , Liang, H. , Rong, C. , Zhang, K. , Cao, X. , Qin, H. et al. (2015) Grain‐specific reduction in lipoxygenase activity improves flour color quality and seed longevity in common wheat. Mol. Breed. 35, 1–18. [Google Scholar]
- Draper, H.H. and Hadley, M. (1990) Malondialdehyde determination as index of lipid peroxidation. Methods Enzymol. 186, 421–431. [DOI] [PubMed] [Google Scholar]
- Feng, B. , Dong, Z. , Xu, Z. , An, X. , Qin, H. , Wu, N. , Wang, D. et al. (2010) Molecular analysis of lipoxygenase (LOX) genes in common wheat and phylogenetic investigation of LOX proteins from model and crop plants. J. Cereal Sci. 52, 387–394. [Google Scholar]
- Fu, D. , Uauy, C. , Blechl, A. and Dubcovsky, J. (2007) RNA interference for wheat functional gene analysis. Transgenic Res. 16, 689–701. [DOI] [PubMed] [Google Scholar]
- Gambino, G. , Chitarra, W. , Maghuly, F. , Laimer, M. , Boccacci, P. , Torello Marinoni, D. and Gribaudo, I. (2009) Characterization of T‐DNA insertions in transgenic grapevines obtained by Agrobacterium‐mediated transformation. Mol. Breed. 24, 305–320. [Google Scholar]
- Gao, H. , Wang, Y. , Xu, P. and Zhang, Z. (2018) Overexpression of a WRKY transcription factor TaWRKY2 enhances drought stress tolerance in transgenic wheat. Front. Plant Sci. 9, 997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayen, D. , Ali, N. , Ganguly, M. , Paul, S. , Datta, K. and Datta, S.K. (2014) RNAi mediated silencing of lipoxygenase gene to maintain rice grain quality and viability during storage. Plant Cell Tissue Organ Cult. 118, 229–243. [Google Scholar]
- Gerits, L.R. , Pareyt, B. , Masure, H.G. and Delcour, J.A. (2015) Native and enzymatically modified wheat (Triticum aestivum L.) endogenous lipids in bread making: a focus on gas cell stabilization mechanisms. Food Chem. 172, 613–621. [DOI] [PubMed] [Google Scholar]
- Gilbert, N. (2013) A hard look at GM crops. Nature, 497, 24–26. [DOI] [PubMed] [Google Scholar]
- Gil‐Humanes, J. , Pistón, F. , Tollefsen, S. , Sollid, L.M. and Barro, F. (2010) Effective shutdown in the expression of celiac disease‐related wheat gliadin T‐cell epitopes by RNA interference. Proc. Natl Acad. Sci. USA, 107, 17023–17028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González‐Thuillier, I. , Salt, L. , Chope, G. , Penson, S. , Skeggs, P. , Tosi, P. Power, S.J. et al. (2015) Distribution of lipids in the grain of wheat (cv. Hereward) determined by lipidomic analysis of milling and pearling fractions. J. Agric. Food Chem. 63, 10705–10716. [DOI] [PubMed] [Google Scholar]
- Han, Y. , Blechl, A. and Wang, D. (2015) The distribution of cotransformed transgenes in particle bombardment‐mediated transformed wheat. Transgenic Res. 24, 1055–1063. [DOI] [PubMed] [Google Scholar]
- Harwood, W.A. (2012) Advances and remaining challenges in the transformation of barley and wheat. J. Exp. Bot. 63, 1791–1798. [DOI] [PubMed] [Google Scholar]
- Hayward, S. , Cilliers, T. and Swart, P. (2017) Lipoxygenases: from isolation to application. Compr. Rev. Food Sci. Food Saf. 16, 199–211. [DOI] [PubMed] [Google Scholar]
- He, Y. , Jones, H.D. , Chen, S. , Chen, X.M. , Wang, D. , Li, K.X. , Wang, D.S. et al. (2010) Agrobacterium‐mediated transformation of durum wheat (Triticum turgidum L. var. durum cv Stewart) with improved efficiency. J. Exp. Bot. 61, 1567–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. , Cai, M. , Long, Q. , Liu, L. , Lin, Q. , Jiang, L. , Chen, S. et al. (2014) OsLOX2, a rice type I lipoxygenase, confers opposite effects on seed germination and longevity. Transgenic Res. 23, 643–655. [DOI] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium (2014) A chromosome‐based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science, 345, 1251788. [DOI] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium , Appels, R. , Eversole, K. , Feuillet, C. , Keller, B. , Rogers, J. , Stein, N. et al. (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science, 361, eaar7191. [DOI] [PubMed] [Google Scholar]
- ISAAA (2017) Global Status of Commercialized Biotech/GM Crops in 2017: Biotech Crop Adoption Surges as Economic Benefits Accumulate in 22 Years. ISAAA Brief, No. 53. Ithaca, NY: ISAAA. [Google Scholar]
- Ishida, Y. , Tsunashima, M. , Hiei, Y. and Komari, T. (2014) Wheat (Triticum aestivum L.) transformation using immature embryos. Methods Mol. Biol. 1223, 189–198. [DOI] [PubMed] [Google Scholar]
- Jones, H.D. (2005) Wheat transformation: current technology and applications to grain development and composition. J. Cereal Sci. 41, 137–147. [Google Scholar]
- Kim, S.I. , Veena and Gelvin, S.B. (2007) Genome‐wide analysis of Agrobacterium T‐DNA integration sites in the Arabidopsis genome generated under non‐selective conditions. Plant J. 51, 779–791. [DOI] [PubMed] [Google Scholar]
- King, J.M. , Svendsen, L.K. , Fehr, W.R. , Narvel, J.M. and White, P.J. .(1998) Oxidative and flavor stability of oil from lipoxygenase-free soybeans. J. Am. Oil Chem. Soc. 75, 1121–1126. [Google Scholar]
- Kleinboelting, N. , Huep, G. , Appelhagen, I. , Viehoever, P. , Li, Y. and Weisshaar, B. (2015) The structural features of thousands of T‐DNA insertion sites are consistent with a double‐strand break repair‐based insertion mechanism. Mol. Plant, 8, 1651–1664. [DOI] [PubMed] [Google Scholar]
- Komor, A.C. , Kim, Y.B. , Packer, M.S. , Zuris, J.A. and Liu, D.R. (2016) Programmable editing of a target base in genomic DNA without double‐stranded DNA cleavage. Nature, 533, 420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraic, J. , Mihálik, D. , Klčová, L. , Gubišová, M. , Klempová, T. , Hudcovicová, M. , Ondreičková, K. et al. (2018) Progress in the genetic engineering of cereals to produce essential polyunsaturated fatty acids. J. Biotechnol. 284, 115–122. [DOI] [PubMed] [Google Scholar]
- Ladics, G.S. , Bartholomaeus, A. , Bregitzer, P. , Doerrer, N.G. , Gray, A. , Holzhauser, T. , Jordan, M. et al. (2015) Genetic basis and detection of unintended effects in genetically modified crop plants. Transgenic Res. 24, 587–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, D. , Jin, H. , Zhang, K. , Wang, Z. , Wang, F. , Zhao, Y. , Huo, N. et al. (2018) Analysis of the Gli‐D2 locus identifies a genetic target for simultaneously improving the breadmaking and health‐related traits of common wheat. Plant J. 95, 414–426. [DOI] [PubMed] [Google Scholar]
- Li, J. , Luo, J. , Xu, M. , Li, S. , Zhang, J. , Li, H. , Yan, L. et al. (2019) Plant genome editing using xCas9 with expanded PAM compatibility. J. Genet. Genomics. 10.1016/j.jgg.2019.03.004 [DOI] [PubMed] [Google Scholar]
- Liavonchanka, A. and Feussner, I. (2006) Lipoxygenases: occurrence, functions and catalysis. J. Plant Physiol. 163, 348–357. [DOI] [PubMed] [Google Scholar]
- Ling, F. , Zhou, F. , Chen, H. and Lin, Y. (2016) Development of marker‐free insect‐resistant indica rice by Agrobacterium tumefaciens‐mediated co‐transformation. Front. Plant Sci. 7, 1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.G. and Chen, Y. (2007) High‐efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques, 43, 649–650. [DOI] [PubMed] [Google Scholar]
- Liu, Y.G. , Mitsukawa, N. , Oosumi, T. and Whittier, R.F. (1995) Efficient isolation and mapping of Arabidopsis thaliana T‐DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Lu, H. , Zhou, X. , Gong, Z. and Upadhyaya, N.M. (2001) Generation of selectable marker‐free transgenic rice using double right‐border (DRB) binary vectors. Aust. J. Plant Physiol. 28, 241–248. [Google Scholar]
- Ma, L. , Zhu, F. , Li, Z. , Zhang, J. , Li, X. , Dong, J. and Wang, T. (2015) TALEN‐based mutagenesis of lipoxygenase LOX3 enhances the storage tolerance of rice (Oryza sativa) seeds. PLoS ONE, 10, e0143877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, Y.B. , Cai, W.J. , Wang, J.W. , Hong, G.J. , Tao, X.Y. , Wang, L.J. , Huang, Y.P. et al. (2007) Silencing a cotton bollworm P450 monooxygenase gene by plant‐mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 25, 1307–1313. [DOI] [PubMed] [Google Scholar]
- Matheka, J.M. , Anami, S. , Gethi, J. , Omer, R.A. , Alakonya, A. , Machuka, J. and Runo, S. (2013) A new double right border binary vector for producing marker‐free transgenic plants. BMC Res. Notes, 6, 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy, D. , Chamberlain, D.A. , Moon, E. and Wilson, K.J. (1995) Development of gusA reporter gene constructs for cereal transformation: availability of plant transformation vectors from the CAMBIA molecular genetic resource service. Mol. Breed. 1, 27–37. [Google Scholar]
- Mega, R. , Abe, F. , Kim, J.S. , Tsuboi, Y. , Tanaka, K. , Kobayashi, H. , Sakata, Y. et al. (2019) Tuning water‐use efficiency and drought tolerance in wheat using abscisic acid receptors. Nat. Plants, 5, 153–159. [DOI] [PubMed] [Google Scholar]
- Mihálik, D. , Gubišová, M. , Klempová, T. , Čertík, M. , Ondreičková, K. , Hudcovicová, M. , Klčová, L. et al. (2014) Transgenic barley producing essential polyunsaturated fatty acids. Biol. Plant. 58, 348–354. [Google Scholar]
- Mihálik, D. , Klčová, L. , Ondreičková, K. , Hudcovicová, M. , Gubišová, M. , Klempová, T. , Čertík, M. et al. (2015) Biosynthesis of essential polyunsaturated fatty acids in wheat triggered by expression of artificial gene. Int. J. Mol. Sci. 16, 30046–30060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley, R.C. , Alonso, J.M. , Kim, C.J. , Leisse, T.J. and Ecker, J.R. (2007) An adapter ligation‐mediated PCR method for high‐throughput mapping of T‐DNA inserts in the Arabidopsis genome. Nat. Protoc. 2, 2910–2917. [DOI] [PubMed] [Google Scholar]
- Pareyt, B. , Finnie, S.M. , Putseys, J.A. and Delcour, J.A. (2011) Lipids in bread making: sources, interactions, and impact on bread quality. J. Cereal Sci. 54, 266–279. [Google Scholar]
- Parisi, C. , Tillie, P. and Rodríguez‐Cerezo, E. (2016) The global pipeline of GM crops out to 2020. Nat. Biotechnol. 34, 31–36. [DOI] [PubMed] [Google Scholar]
- Privalle, L.S. , Chen, J. , Clapper, G. , Hunst, P. , Spiegelhalter, F. and Zhong, C.X. (2012) Development of an agricultural biotechnology crop product: testing from discovery to commercialization. J. Agric. Food Chem. 60, 10179–10187. [DOI] [PubMed] [Google Scholar]
- Qin, C. , Qian, W. , Wang, W. , Wu, Y. , Yu, C. , Jiang, X. , Wang, D. et al. (2008) GDP‐mannose pyrophosphorylase is a genetic determinant of ammonium sensitivity in Arabidopsis thaliana . Proc. Natl Acad. Sci. USA, 105, 18308–18313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, T. , Thistleton, J. , Higgins, T.J. , Howitt, C. and Ayliffe, M. (2014) Efficient Agrobacterium transformation of elite wheat germplasm without selection. Plant Cell Tissue Organ Cult. 119, 647–659. [Google Scholar]
- Robinson, D.S. , Wu, Z. , Domoney, C. and Casey, R. (1995) Lipoxygenases and the quality of foods. Food Chem. 54, 33–43. [Google Scholar]
- Saghai‐Maroof, M.A. , Soliman, K.M. , Jorgenson, R.A. and Allard, R.W. (1984) Ribosomal DNA spacer‐length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl Acad. Sci. USA, 81, 8014–8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, N. , Fehr, W. , Johnson, L. and White, P. (1996) Oxidative stabilities of soybean oils that lack lipoxygenases. J. Am. Oil Chem. Soc. 73, 1327–1336. [Google Scholar]
- Shirasawa, K. , Takeuchi, Y. , Ebitani, T. and Suzuki, Y. (2008) Identification of gene for rice (Oryza sativa) seed lipoxygenase‐3 involved in the generation of stale flavor and development of SNP markers for lipoxygenase‐3 deficiency. Breed. Sci. 58, 169–176. [Google Scholar]
- Song, L.Y. , Lu, W.X. , Hu, J. , Zhang, Y. , Yin, W.B. , Chen, Y.H. , Hao, S.T. et al. (2010) Identification and functional analysis of the genes encoding Δ6‐desaturase from Ribes nigrum . J. Exp. Bot. 61, 1827–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y. , Sparks, C. , Jones, H. , Riley, M. , Francis, F. , Du, W. and Xia, L.Q. (2019) Silencing an essential gene involved in infestation and digestion in grain aphid through plant-mediated RNA interference generates aphid-resistant wheat plants. Plant Biotechnol. J. 17, 852–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunitha, S. , Shivaprasad, P.V. , Sujata, K. and Veluthambi, K. (2012) High frequency of T‐DNA deletions in transgenic plants transformed with intron‐containing hairpin RNA genes. Plant Mol. Biol. Rep. 30, 158–167. [Google Scholar]
- Suzuki, Y. , Ise, K. , Li, C.Y. , Honda, I. , Iwai, Y. and Matsukura, U. (1999) Volatile components in stored rice [Oryza sativa (L.)] of varieties with and without lipoxygenase‐3 in seeds. J. Agr. Food Chem. 47, 1119–1124. [DOI] [PubMed] [Google Scholar]
- Thole, V. , Worland, B. , Wright, J. , Bevan, M.W. and Vain, P. (2010) Distribution and characterization of more than 1000 T‐DNA tags in the genome of Brachypodium distachyon community standard line Bd21. Plant Biotechnol. J. 8, 734–747. [DOI] [PubMed] [Google Scholar]
- Tosi, P. , He, J. , Lovegrove, A. , Gonzáles‐Thuillier, I. , Penson, S. and Shewry, P.R. (2018) Gradients in compositions in the starchy endosperm of wheat have implications for milling and processing. Trends Food Sci. Technol. 82, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya, T. , Kameya, N. and Nakamura, I. (2009) Straight walk: a modified method of ligation‐mediated genome walking for plant species with large genomes. Anal. Biochem. 388, 158–160. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Sun, X. , Zhao, Y. , Kong, F. , Guo, Y. , Zhang, G. , Pu, Y.Y. et al. (2011) Enrichment of a common wheat genetic map and QTL mapping for fatty acid content in grain. Plant Sci. 181, 65–75. [DOI] [PubMed] [Google Scholar]
- Wang, G.P. , Yu, X.D. , Sun, Y.W. , Jones, H.D. and Xia, L.Q. (2016a) Generation of marker‐and/or backbone‐free transgenic wheat plants via Agrobacterium‐mediated transformation. Front. Plant Sci. 7, 1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K. , Liu, H. , Du, L. and Ye, X. (2016b) Generation of marker‐free transgenic hexaploid wheat via an Agrobacterium‐mediated co‐transformation strategy in commercial Chinese wheat varieties. Plant Biotechnol. J. 15, 614–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H.X. , Sparks, C.A. and Jones, H.D. (2006) Characterization of T‐DNA loci and vector backbone sequences in transgenic wheat produced by Agrobacterium‐mediated transformation. Mol. Breed. 18, 195–208. [Google Scholar]
- Xu, M.R. , Xia, Z.H. , Zhai, W.X. , Xu, J.L. , Zhou, Y.L. and Li, Z.K. (2008) Construction of double right‐border binary vector carrying non‐host gene Rxo1 resistant to bacterial leaf streak of rice. Rice Sci. 15, 243–246. [Google Scholar]
- Xu, H. , Wei, Y. , Zhu, Y. , Lian, L. , Xie, H. , Cai, Q. , Chen, Q. et al. (2015) Antisense suppression of LOX3 gene expression in rice endosperm enhances seed longevity. Plant Biotechnol. J. 13, 526–539. [DOI] [PubMed] [Google Scholar]
- Xu, H. , Cao, Y. , Xu, Y. , Ma, P. , Ma, F. , Song, L. , Li, L. et al. (2017) Marker‐assisted development and evaluation of near‐isogenic lines for broad‐spectrum powdery mildew resistance gene Pm2b introgressed into different genetic backgrounds of wheat. Front. Plant Sci. 8, 1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, S. , Li, H. , Li, Y. , Zhu, Y. , Guo, J. , Liu, Y. , Chen, Y. et al. (2008) Generation of transgenic wheat lines with altered expression levels of 1Dx5 high‐molecular weight glutenin subunit by RNA interference. J. Cereal Sci. 47, 153–161. [Google Scholar]
- Zhang, K. , Wang, J. , Zhang, L. , Rong, C. , Zhao, F. , Peng, T. , Li, H. et al. (2013) Association analysis of genomic loci important for grain weight control in elite common wheat varieties cultivated with variable water and fertiliser supply. PLoS ONE, 8, e57853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Hao, Q. , Bai, L. , Xu, J. , Yin, W. , Song, L. , Xu, L. et al. (2014) Overexpression of the soybean transcription factor GmDof4 significantly enhances the lipid content of Chlorella ellipsoidea . Biotechnol. Biofuels, 7, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Analysis of the expression of the four genes flanking the T‐DNA insertion site in GLRW‐3.
Figure S2 Elucidation of the T‐DNA integration site in GLRW‐8.
Figure S3 Characterization of TaLOX1‐KN199 allele.
Table S1 Comparison of flour FA contents of KN199 and three transgenic lines cultivated in the field in 2017.
Table S2 Field assessment of important agronomic traits of three transgenic GLRW lines in 2017.
Table S3 Comparison of important agronomic traits between Kenong 199 and three transgenic GLRW lines in 2017–2018 growth season.
Table S4 Annual stocks of three major cereal grains in the world in recent years.
Table S5 Oligonucleotide primers used in this study.


