Abstract
Macrophages are an important component of the innate immune response. Priming and activation of macrophages is stimulated by cytokines (i.e IFNγ). However, growth hormone (GH) can also stimulate macrophage activation. Based on these observations, the goal of this work was to 1) to compare the transcriptome profile of macrophages activated in vitro with GH and IFNγ, and 2) to assess the impact of GH on key macrophage functional properties like reactive oxygen species (ROS) production and phagosomal proteolysis. To assess the global transcriptional and functional impact of GH on macrophage programming, bone marrow derived macrophages were treated with GH or IFNγ. Our data strongly support a potential link between GH, which wanes with age, and impaired macrophage function. The notable overlap of GH with IFNγ-induced pathways involved in innate immune sensing of pathogens and antimicrobial responses argue for an important role for GH in macrophage priming and maturation. By using functional assays that report on biochemical activities within the lumen of phagosomes, we have also shown that GH alters physiologically relevant processes such as ROS production and proteolysis. These changes could have far reaching impacts on antimicrobial capacity, signaling, and antigen presentation.
Subject terms: Gene regulation in immune cells, Monocytes and macrophages, Ageing
Introduction
Growth hormone (GH) is well recognized for promoting somatic body growth1, and its main actions are mediated by liver produced insulin-like growth factor (IGF-I), through binding to the GH receptor (GHR)2. GHR is a transmembrane protein member of the cytokine receptor family3, which includes receptors for interleukin 2, 3, 4, 5, 6 and 73,4. Besides the liver, several cell types in the body including macrophages express GHR on their surface and are directly responsive to GH3. Pituitary GH secretion declines with age during adult life5, and has been implicated as a key factor in the development of age related diseases. This decline in GH secretion with age is paralleled by a generalized decline of immune function, a process known as immunosenescence6.
Growth hormone deficient Ames dwarf mice (df/df) mice have a genetic mutation that leads to impaired anterior pituitary gland development7. As a consequence, these mice have very low circulating GH and IGF-I levels, reduced adult body size8, and can live up to 65% longer than wild-type (WT) littermates9. The df/df mice have been extensively characterized as having a reduced pro-inflammatory profile as they age, which may represent the major mechanism promoting extended lifespan10. On the contrary, this phenotype might reflect impairment of the innate immune system, which could increase susceptibility to infectious disease. Mice with disruption of the GH releasing hormone gene have lower proportion of lung macrophages than WT mice, however in the spleen more macrophages were observed11. These mice also exhibited increased susceptibility to S. pneumoniae infection with a time-dependent increase in lung bacterial load11. GH-deficient Snell dwarf mice, despite having absent GH pituitary secretion, do have expression of GH in bone marrow cells12. Therefore, it is of interest to define the pathways and genes regulated by GH in key cells of the innate immune system such as macrophages. If a strong link is established between GH and optimal function of innate immune cells, these mice would serve as a valuable model to explore the role of GH in resistance to infectious diseases.
Macrophages are an important component of the innate immune response, and are central to pathogen recognition and response13. Priming and activation of macrophages by cytokines (i.e IFNγ) and encounters with pathogens exert profound effects on their antimicrobial and immune modulatory capacity. This can reprogram gene expression and cytokine production, trigger oxidative and nitrosative bursts, change the makeup of the phagosome lumen (pH, ions, hydrolytic enzymes), and alter trafficking of pathogens and antigen processing14–21. Interestingly, it was demonstrated that macrophages can be primed by GH similarly to IFNγ, a well-characterized macrophage activator22–24. Treatment of monocytes in vitro with GH increased secretion of pro-inflammatory cytokines, interleukin (IL)-1α, IL-6 and TNFα25 and enhanced macrophage function in vivo by increasing superoxide and TNFα release23,24. This suggests that GH has functions that at least partially overlap with IFNγ and may impact the ability of the host to respond to an infection. Although it is known that GH can prime macrophages similarly to IFNγ22 the specific pathways activated by GH and the extent of this response are not well understood.
Based on the presented evidence, the goal of this work was to 1) to compare the transcriptome profile of macrophages derived from WT and GH-deficient df/df mice, 2) to compare the transcriptome profile of macrophages activated in vitro with GH or IFNγ, and 3) to assess the impact of GH on biochemical processes within macrophage phagosomes such as reactive oxygen species production and phagosomal proteolysis that are critical for their role in innate immunity. Given that earlier studies only determined the GH-dependent regulation of a few select genes25, we chose to apply next generation sequencing to generate global RNASeq transcriptome profiles of GH versus IFNγ activated macrophages from WT and df/df mice. Although there were minimal differences in the transcriptome of untreated macrophages derived from WT versus df/df mice, GH treatment induced a significant transcriptional reprogramming that included pathways that overlapped with IFNγ as well as unique GH specific pathways. Our data showing the ability of GH to impact phagosomal processes involved in antimicrobial defense and antigen presentation corroborate the RNAseq results and suggest an important role for GH in macrophage priming and activation.
Results
Real time RT-PCR for Tnf mRNA and sample selection
We assessed the levels of Tnf mRNA by real-time RT-PCR to determine the optimal GH dose and treatment duration before processing samples for a more in depth RNASeq analysis. GH treatment at 500 ng/mL and IFNγ at 10 ng/mL at the 6 h time point produced significant changes in Tnf mRNA expression in comparison to Control samples independent of genotype (P < 0.05; Suppl. Fig. 1). Therefore, WT and df/df samples from Control, GH at 500 ng/mL and IFN at 10 ng/mL, 6 h after treatment, were selected for further analysis by RNASeq.
RNA sequencing results
As a result of the RNASeq technique on average 20,850,687 reads per library were obtained. Two samples were removed due to very low transcript numbers after sequencing and pre-processing (<3,000,000). Using PCA (Fig. 1A) and unsupervised hierarchical clustering for all genes detected (Fig. 1B), we observed that samples grouped in similar pattern by treatment and showed a very low level of sample variability between genotypes. Only one sample was removed from the study due to an anomalous profile of gene expression (from GH treated cells). The grouping pattern indicate that samples are more affected by the in vitro treatment than the genotype of the mice.
Figure 1.
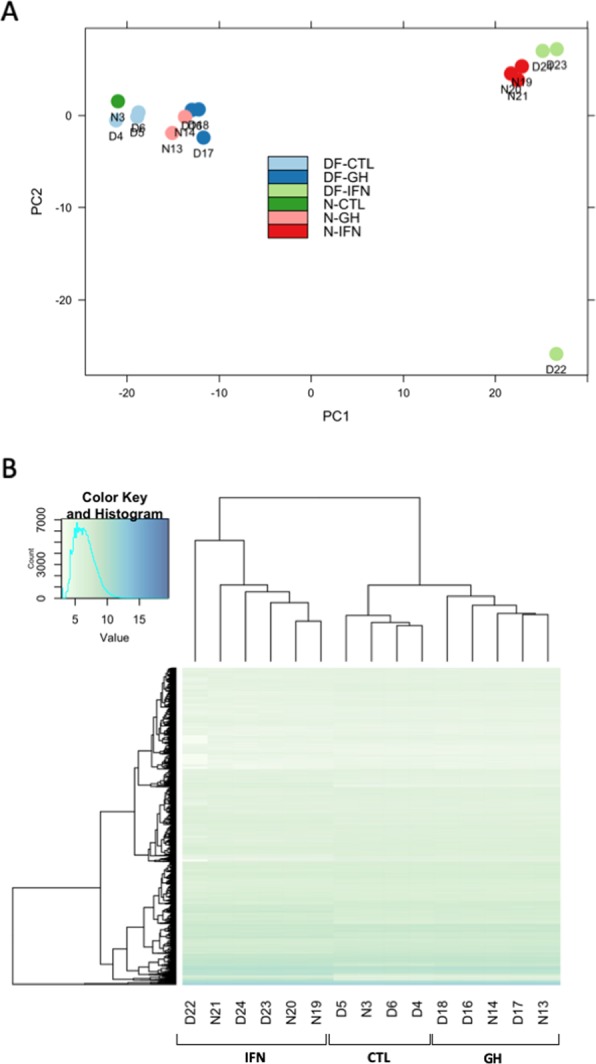
Principal components analysis of the 500 most variable expressed mRNAs in macrophages of Ames dwarf (D) and wild-type mice (WT) in the control, GH and IFN treatments (Panel A). Unsupervised hierarchical clustering of expression levels for the top 200 most expressed genes in macrophages of Ames dwarf (D) and Wild-type mice (WT) in the control, GH and IFN treatments (Panel B).
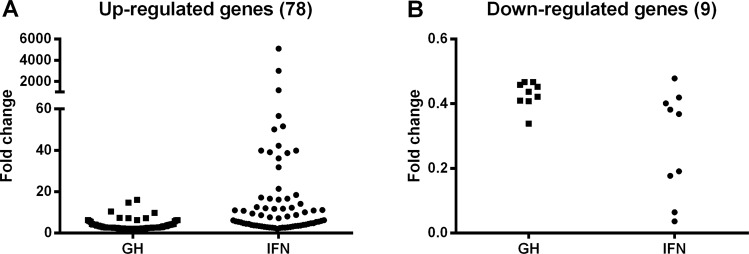
For analysis of differential gene expression (DEG) we considered initially the genes that were regulated with genotype (WT vs df/df) for control, GH or IFNγ treated cells. Since there was no gene regulation between genotypes at FDR < 0.02 and FC > 2.0 or FC < 0.5 for each treatment, we only considered genes that changed between treatments (Control vs. GH and Control vs. IFN) independently of genotype. We observed nine genes commonly down-regulated between GH and IFNγ treated cells, 458 genes exclusively down-regulated by IFNγ and 7 genes exclusively down-regulated in GH treated cells (Fig. 2A). Additionally, we observed 79 genes up-regulated by both treatments, 634 exclusively by IFNγ and 35 exclusively by GH (Fig. 2B). The FC distribution of genes commonly regulated in GH and IFNγ treated cells indicate stronger regulation (P < 0.05) for IFNγ (median of 8.7 and 0.44 for up and down-regulated genes, respectively) than GH (median of 2.7 and 0.36 for up and down-regulated genes, respectively) treated cells (Fig. 3). A detailed list with all the DEGs is provided in Suppl. Table 1. It is important to mention that there were no detectable levels of expression for the Ifn gene in any of the treatments. The top ten common up-regulated genes were Nos2, Iigp1, Gbp5, Gbp2, Cxcl10, Gbp6, Gbp3, Gbp7, Cd40 and Gm12250. The enrichment for genes linked to macrophage activation induced by both IFNγ and GH, including inducible nitric oxide synthase and multiple guanylate-binding proteins, is notable. The top ten common down-regulated genes were Eif4a2, Dab2, Rasgrp3, 1810011H11Rik, Dhrs3, Rnf150, St6gal1, Snhg5, Slco4a1 and Cxcr4.
Figure 2.
Number of differentially expressed genes (DEGs) down-regulated (A) or up-regulated (B) between GH and IFN treatments. Number of regulated Kegg pathways down-regulated (A) or up-regulated (B) between GH and IFN treatments.
Figure 3.
Fold change of expression for common down-regulated (A) or up-regulated (B) genes between GH and IFN treatments.
Considering the genes disturbed by the treatment using GAGE pathway analysis we identified nine commonly down-regulated Kegg pathways, six exclusively in IFNγ and five exclusively in GH treated cells (Fig. 2C). There were 10 Kegg pathways commonly up-regulated between GH and IFNγ treated cells, seven exclusively in IFNγ and eight exclusively in GH treated cells (Fig. 2D). The regulated Kegg pathways for each treatment are presented with details in Table 1. Additionally, we provided detailed figures of regulated genes in each selected pathway in Suppl. Figs. 2–4 for the Jak-Stat signaling, TLR signaling and cell cycle pathways for GH and IFNγ treated cells.
Table 1.
Regulated pathways between GH and IFN treated macrophages.
| Kegg Pathways | GH | IFN | Set size | ||
|---|---|---|---|---|---|
| Stat Mean | P-Value | Stat Mean | P-Value | ||
| Common up-regulated pathways | |||||
| mmu04621 NOD-like receptor signaling pathway | 3.54 | 0.0003 | 1.92 | 0.0294 | 47 |
| mmu04622 RIG-I-like receptor signaling pathway | 3.23 | 0.0009 | 2.91 | 0.0024 | 45 |
| mmu04620 Toll-like receptor signaling pathway | 3.17 | 0.0010 | 2.40 | 0.0091 | 73 |
| mmu04630 Jak-STAT signaling pathway | 2.69 | 0.0040 | 4.03 | 0.0000 | 75 |
| mmu04380 Osteoclast differentiation | 2.23 | 0.0136 | 2.64 | 0.0045 | 92 |
| mmu04623 Cytosolic DNA-sensing pathway | 2.22 | 0.0153 | 2.59 | 0.0062 | 36 |
| mmu04210 Apoptosis | 2.13 | 0.0176 | 2.48 | 0.0071 | 71 |
| mmu04510 Focal adhesion | 2.03 | 0.0220 | 2.43 | 0.0079 | 113 |
| mmu04666 Fc gamma R-mediated phagocytosis | 1.86 | 0.0327 | 2.19 | 0.0151 | 72 |
| mmu04514 Cell adhesion molecules (CAMs) | 1.77 | 0.0404 | 2.64 | 0.0051 | 52 |
| IFN up-regulated pathways | |||||
| mmu04145 Phagosome | — | — | 2.89 | 0.0021 | 104 |
| mmu04612 Antigen processing and presentation | — | — | 2.93 | 0.0022 | 42 |
| mmu04650 Natural killer cell mediated cytotoxicity | — | — | 1.98 | 0.0249 | 71 |
| mmu00760 Nicotinate and nicotinamide metabolism | — | — | 2.01 | 0.0273 | 17 |
| mmu04512 ECM-receptor interaction | — | — | 1.82 | 0.0373 | 28 |
| mmu04912 GnRH signaling pathway | — | — | 1.70 | 0.0457 | 58 |
| mmu04973 Carbohydrate digestion and absorption | — | — | 1.70 | 0.0485 | 21 |
| GH up-regulated pathways | |||||
| mmu04670 Leukocyte transendothelial migration | 2.65 | 0.0045 | — | — | 62 |
| mmu04920 Adipocytokine signaling pathway | 2.21 | 0.0152 | — | — | 46 |
| mmu04722 Neurotrophin signaling pathway | 2.12 | 0.0177 | — | — | 104 |
| mmu04660 T cell receptor signaling pathway | 2.04 | 0.0217 | — | — | 73 |
| mmu04062 Chemokine signaling pathway | 1.88 | 0.0310 | — | — | 114 |
| mmu04662 B cell receptor signaling pathway | 1.76 | 0.0405 | — | — | 64 |
| mmu04664 Fc epsilon RI signaling pathway | 1.72 | 0.0440 | — | — | 52 |
| mmu04520 Adherens junction | 1.68 | 0.0486 | — | — | 49 |
| Common down-regulated pathways | |||||
| mmu00190 Oxidative phosphorylation | −5.80 | 0.0000 | −2.32 | 0.0108 | 98 |
| mmu03030 DNA replication | −4.64 | 0.0000 | −2.07 | 0.0219 | 35 |
| mmu03010 Ribosome | −5.89 | 0.0000 | −1.68 | 0.0483 | 80 |
| mmu03430 Mismatch repair | −4.06 | 0.0001 | −2.14 | 0.0200 | 22 |
| mmu04110 Cell cycle | −3.18 | 0.0008 | −3.31 | 0.0005 | 113 |
| mmu03420 Nucleotide excision repair | −2.61 | 0.0054 | −1.78 | 0.0398 | 42 |
| mmu03040 Spliceosome | −2.53 | 0.0061 | −1.72 | 0.0439 | 115 |
| mmu03440 Homologous recombination | −2.42 | 0.0097 | −1.82 | 0.0384 | 27 |
| mmu00982 Drug metabolism - cytochrome P450 | −1.90 | 0.0338 | −1.85 | 0.0391 | 16 |
| IFN down-regulated pathways | |||||
| mmu00280 Valine, leucine and isoleucine degradation | — | — | −1.98 | 0.0261 | 38 |
| mmu00100 Steroid biosynthesis | — | — | −2.00 | 0.0281 | 14 |
| mmu04710 Circadian rhythm - mammal | — | — | −1.93 | 0.0313 | 19 |
| mmu03008 Ribosome biogenesis in eukaryotes | — | — | −1.87 | 0.0318 | 68 |
| mmu04114 Oocyte meiosis | — | — | −1.82 | 0.0353 | 84 |
| mmu00650 Butanoate metabolism | — | — | −1.80 | 0.0431 | 14 |
| GH down-regulated pathways | |||||
| mmu03410 Base excision repair | −3.49 | 0.0005 | — | — | 30 |
| mmu04260 Cardiac muscle contraction | −2.69 | 0.0046 | — | — | 32 |
| mmu00240 Pyrimidine metabolism | −2.30 | 0.0114 | — | — | 81 |
| mmu00230 Purine metabolism | −1.90 | 0.0291 | — | — | 116 |
| mmu00980 Metabolism of xenobiotics by cytochrome P450 | −1.95 | 0.0308 | — | — | 16 |
The number of enriched GO Terms for biological processes commonly up-regulated between GH and IFNγ treated cell was 261 (top ten: defense response, response to other organism, response to biotic stimulus, innate immune response, immune response, multi-organism process, immune effector process, response to bacterium, cellular response to interferon-beta, defense response to virus), with 277 exclusively up-regulated in IFNγ and 96 exclusively up-regulated in GH treated cells (top ten: macromolecule catabolic process, actin filament-based process, negative regulation of protein transport, modification-dependent macromolecule catabolic process, myeloid cell activation involved in immune response, negative regulation of cysteine-type endopeptidase activity, response to unfolded protein, membrane protein ectodomain proteolysis, response to hormone stimulus, vasoconstriction). There were 50 GO Terms for biological processes commonly down-regulated in IFNγ and GH treated cells (top 10: M phase, cell cycle phase, DNA repair, nuclear division, mitosis, M phase of mitotic cell cycle, DNA replication, organelle fission, cell division, mitotic cell cycle), 54 exclusively in IFNγ and 32 exclusively in GH treated cells (top 10: base-excision repair, proton transport, DNA integrity checkpoint, hydrogen transport, purine ribonucleoside biosynthetic process, purine nucleoside biosynthetic process, DNA damage checkpoint, ribosomal small subunit biogenesis, electron transport chain, G2/M transition of mitotic cell cycle). The enriched GO Terms for biological processes are presented with details in Suppl. Tables 3 and 4 for GH and IFNγ treated cells, respectively.
Confirmation of sequencing results by real time PCR
The validation of RNASeq of ten of the top up-regulated genes (Nos2, Gbp2, Gbp3, Gbp5, Gbp7, Lipg1, Igr1, Cxcl10, CD40, and Tnf) by real time qRT-PCR confirmed our RNA-Seq findings and is presented in Suppl. Fig. 5.
Reactive oxygen species production
Next, we set out to examine whether GH induced transcriptional changes measurably impacted macrophage functions such as antimicrobial effector mechanisms. The GH-mediated induction of multiple guanylate-binding proteins (Gbp), in particular Gpb7 which is known to enhance the oxidative burst through recruitment of NADPH oxidase to the phagosome26 prompted us to assess whether GH triggered production of reactive oxygen species. Consistent with this hypothesis, after exposure of macrophages to GH for 18 hours, we noted significantly increased H2O2 production to levels comparable with IFNγ treatment (P < 0.05) (Fig. 4A).
Figure 4.
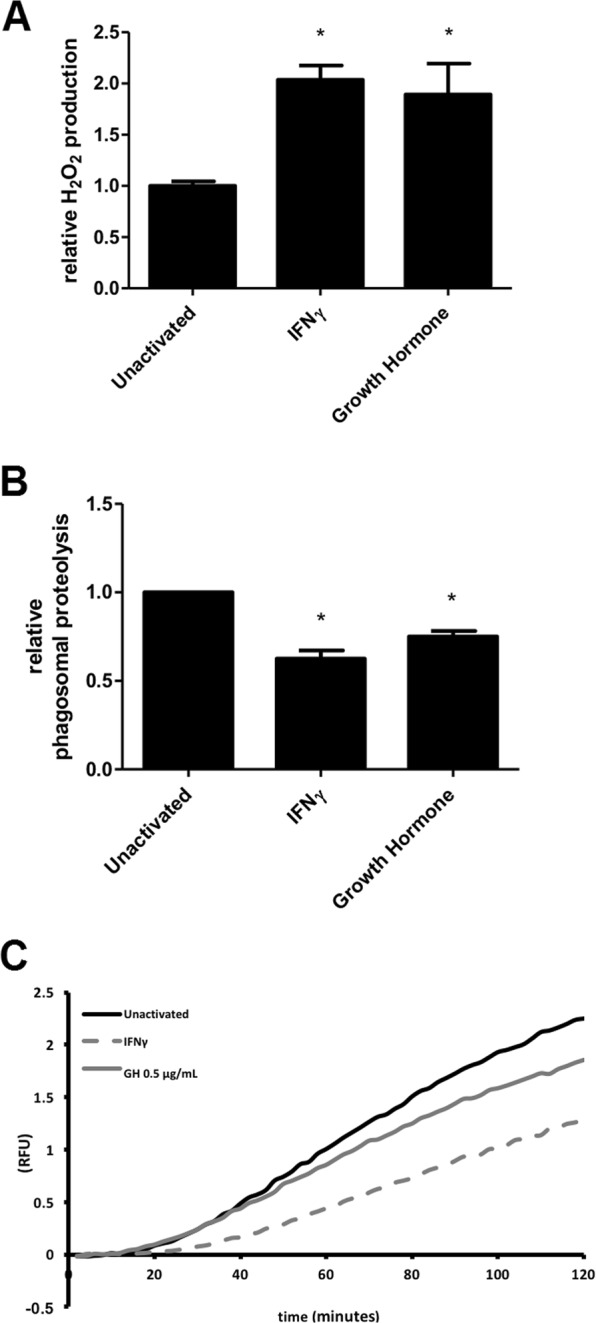
Effect of GH on functional properties of macrophages. For data shown in all panels, BMMØs from C57BL/6 mice were incubated for 18 hours in the presence of either 100 U/mL of IFNγ or 500 ng/mL porcine growth hormone. (A) Cells were then stimulated for 1.5 hours with serum-opsonized zymosan particles. Relative amounts of H202 within supernatants were determined by measuring the reaction of Amplex Red in the presence of horseradish peroxidase. Hydrogen peroxide production is presented as relative to unactivated BMMØs from 3 independent experiments. Error bars denote SEM. *p < 0.05. (B) Phagosomal proteolysis was measured as fluorescence liberation of DQ-Green BSA from IgG opsonized experimental particles that have been phagocytosed by unactivated/IFNγ/growth hormone activated BMMØs. Averaged rate of phagosomal proteolysis relative to unactivated control from 5 independent experiments. Rate was calculated between 80–100 minutes post phagocytosis. Error bars denote SEM. *p < 0.05. (C) Real time representative traces of phagosomal proteolysis.
Phagosomal proteolysis
Based on our previous report that NADPH oxidase (NOX2) negatively regulates phagosomal proteolysis27 we measured the the proteolysis of a quenched fluorescent protein substrate within phagosomes of unstimulated macrophages or macrophages stimulated with either GH or IFNγ. Similar to macrophages treated with IFNγ, those treated with GH displayed lower levels of phagosomal proteolysis compared to unactivated samples (P < 0.05) (Fig. 4B,C).
Discussion
In this study, global transcriptomic analyses corroborated by macrophage functional assays provided detailed insight into GH mediated priming of macrophages. We noted a significant overlap between pathways activated by GH and IFNγ, although the total number of genes activated by IFNγ treatment was much higher. If we consider the 50 highest induced genes in each treatment, we can observe 21 common genes among them. The higher fold-change induction of these shared genes in IFNγ versus GH treated cells suggests a stronger effect of IFNγ compared to GH. This is in agreement with previous suggestions that the dose of GH required to prime macrophages is higher than that of IFNγ22. This does not necessarily minimize the physiological relevance of GH as IFNγ levels in healthy mice are very low (0 to 15 pg/mL)28 whereas GH is present at higher levels (4 to 25 ng/mL)29. It is important to mention that previous studies showed that blocking of the IFNγ receptor did not block the effects of GH22, suggesting that GH effects are independent of IFNγ produced by possible bystander lymphoid cells. We did not detect expression of the Ifn gene in macrophages from any treatment, in agreement with previously observations at the protein level for unstimulated macrophages30. Therefore, we can exclude that the observations could be due to GH-induced IFNγ production. On the other hand, it should be considered that cells were cultured in the presence of 10% fetal calf serum, which may contain both GH and cytokines. However, as cells from both groups were cultivated in the presence of serum, we understand it has no impact on the identification of differentially regulated genes among groups.
The top regulated biological terms for both GH and IFNγ treated cells include response to bacterium, response to virus, response to other organism, defense response and phagocytosis, consistent with a role for GH in immune responses to infections. Additionally, terms like biological adhesion and cell adhesion were also up-regulated in GH and IFNγ treated cells, supporting a role for GH in regulating the ability of monocytes to adhere to cells in the presence of IFNγ31. Consistent with our observation that cell migration was also one of the up-regulated GO Terms, severe reduction of macrophage infiltration into specific tissues, in df/df mice has been reported32. Furthermore, a defective expression of genes in the myeloid cell differentiation pathway was demonstrated in peripheral leukocytes of df/df mice compared to N mice33, further suggesting this inability of monocytes to migrate to tissues in the absence of GH. Conversely, Kumar and Menon34 reported that in vivo GH treatment also enhances monocyte migration to tissue, which can improve the ability to fight infection in local tissues. This evidence, combined with our current findings, indicated that GH may exert a significant impact on the maturation, activation, and functionality of effector macrophages.
Global RNAseq profiling of GH- and IFNγ- induced genes also highlighted several biological terms for pattern recognition and activation of the immune response among the most enriched terms in both treatments. Toll-like receptors (TLRs) receive signals from the extracellular space, while members of the RIG-I-like receptors (RLRs) and NOD-like receptor (NLR) families are sensors of infection and stress in the intracellular space35. These pathways are involved in the response to endogenous molecules released by host cells resulting from necrosis or damage36. These three pathways were highly up-regulated in both GH and IFNγ treated cells. The activation of signaling pathways via TLRs, RLRs and NLRs culminate in the activation of nuclear factor κB (NF-κB)36, which is essential for propagation of the inflammatory response, release of pro-inflammatory cytokines and expression of adhesion molecules37. We detected several biological terms referring to increase release of inflammatory cytokines up-regulated for both GH and IFNγ treated cells, including cytokine production, regulation of cytokine production, positive regulation of cytokine production, cytokine secretion, positive regulation of cytokine secretion, regulation of cytokine production involved in immune response and cytokine biosynthetic process. This suggests that GH, like IFNγ, enhances pattern recognition and pro-inflammatory processes that are critical to the innate immunity role of macrophages. More studies assessing the effect of GH on the response of cells to LPS or stimuli are necessary to further elucidate this response.
No differences in the gene expression profile were observed between macrophages derived from bone marrows of WT or GH-deficient df/df mice. This finding is very interesting and suggests that, after extraction of bone marrow precursors and differentiation into macrophages in vitro, the GH-deficient bone marrow environment in which they developed does not affect their profile at all. As mentioned before, GH-deficient Snell dwarf mice, despite having absent GH pituitary secretion, do have expression of GH in bone marrow cells12. Although there is still controversy about the functionality of the immune system of df/df mice, it is well established that df/df mice have an anti-inflammatory profile when compared to WT mice, with reduced secretion of pro-inflammatory cytokines33 and reduced macrophage infiltration in tissues10. Nevertheless, it has been shown that df/df mice have increased myelopoeisis in the bone marrow38. Additionally, some reports indicate that df/df mice have a deficient immune response, which can be restored by GH treatment, but not by treatment with bone marrow cells from a normal mice39. This suggests that the presence of GH is essential for the maturation of these cells and bone marrow cells from normal mice do not retain the ability to mature in the absence of the pituitary hormones. Future studies to assess the functionality of tissue macrophages from df/df mice will be important to fully elucidate the impact of GH on these key cells of the innate immune system.
The overlap between GH- and IFNγ-dependent genes can be attributed, at least in part, to intersecting downstream signaling pathways. GH binds to the growth hormone receptor (GHR) in the cell surface, which is a transmembrane protein of the cytokine receptor family, the same receptor family as many interleukin receptors3,4. Activation of GHR leads to intracellular activation of the Jak-STAT and transduction of GH signals40. IFNγ also binds to its receptor in the cell surface activating mainly the Jak-STAT signaling pathway41. The Jak-STAT pathway in the macrophage is critical to allow its activation and adequate immune response42. In our study, the Jak-STAT pathway was among the highest up-regulated pathway by both GH and IFNγ. One important target of STATs is the Nos2 gene43, which in our study was the top-regulated gene for both treatments (64 and 467-fold up-regulated by GH and IFNγ, respectively). Nos2 encodes the inducible nitric oxide synthase (iNOS) which is directly responsible for production of reactive nitrogen species (RNS) that exert potent antimicrobial effects on many pathogens, including Mycobacterium tuberculosis44–47. Corroborating with this, among the common up-regulated GO terms between IFNγ and GH were regulation of nitric oxide biosynthetic pathways, nitric oxide metabolic process and positive regulation of nitric oxide biosynthetic pathways.
In addition to polarizing macrophages toward a pro-inflammatory state, IFNγ also alters the lumenal contents and functional capacity of their phagosomes which serves to optimize their response to pathogens27,48–51. One of the major changes to the phagosomal environment following IFNγ stimulation is the increase in ROS through increased expression and activation of the phagocyte NADPH oxidase 252. Increased ROS production by the macrophage constitutes an important effector mechanism for bacterial killing53,54 and is an important marker of macrophage activation55. To complement our global transcriptomics view of macrophage priming by GH, we also demonstrated that GH triggered increased ROS production comparable to IFNγ treatment, despite the subtler GH-induced changes in gene expression. This corroborates earlier observations in a rat model showing enhanced macrophage ROS production and antimicrobial activity against Pasteurella and Salmonella both in vitro and in vivo22,23. Additionally, among the GO Terms up-regulated for both GH and IFNγ treated macrophages was regulation of oxidoreductase activity, which is involved in the release of ROS and RNS. In addition to directly killing phagocytosed pathogens, the long half-life and membrane permeability of H2O2 enables it to also act as a secondary messenger. Thus, the induction of ROS production by GH may impact numerous other processes including phagocytosis, cell cycle and cell death, monocyte recruitment, and monocyte-to-macrophage differentiation56.
In addition to the production of ROS, activation of macrophages with IFNγ has also been reported to modify the degradative capacity of the phagosome, particularly the degradation of proteins51. Our findings revealed that GH stimulation of macrophages reduced phagosomal proteolysis to a similar extent as IFNγ, highlighting a novel facet of GH-mediated macrophage priming and activation. It is known that IFNγ limits the efficiency of phagosomal proteolysis in part through oxidative inhibition of cysteine proteases, and can result in modified processing of MHC-II-restricted antigens27,57. The mechanism by which GH dampens the proteolytic capacity of macrophage phagosomes and the effect on antigen presentation remain to be determined.
Although many lines of evidence point to an important role for GH in macrophage priming and innate immunity, the relevance of GH and changes in GH levels (i.e. during aging) to immune senescence and specific susceptibilities remains unclear. On the one hand, studies in humans with congenital GH deficiency indicate no difference in the incidence of several viral and bacterial infections as well as response to vaccination58. On the other hand, reduced macrophage function is evident in older individuals and is associated with increased susceptibility to infections59. Absence of GHR signaling can prevent activation of the NLPR3 inflammasome in macrophages, reducing the inflammatory response to endogenous ligands from cellular necrosis60. Some reports also suggest that GH drives macrophages through a M2 anti-inflammatory phenotype characterized by low Nos2 and high arginase-1 expression61. However, our data indicate high levels of Nos2 and no changes in arginase-1 expression for both GH and IFN treatments, suggesting an M1 pro-inflammatory phenotype. Our data strongly support a potential link between GH, which wanes with age, and impaired macrophage function. This study has provided the first global view of the scope of GH-mediated changes in macrophage gene expression. The notable overlap with IFNγ-induced pathways involved in innate immune sensing of pathogens and antimicrobial responses argue for an important role for GH in macrophage priming and maturation. Additional studies will be required to confirm that these effects observed with bone marrow-derived macrophages are generalizable to specific differentiated macrophage subtypes. By using functional assays that report on biochemical activities within the lumen of phagosomes, we have shown that GH alters physiologically relevant processes such as ROS production and proteolysis. These changes could have far reaching impacts on antimicrobial capacity, signaling, and antigen presentation. Additional work beyond the scope of this study is required to fully characterize the role of GH on innate immunity and host reponses to infection.
Methods
Tissue collection and macrophage culture
All animal procedures employed in our present work were approved by and performed in accordance with the guidelines from the Laboratory Animal Care and Use Committee at the University of Central Florida or Animal Care and Use Committee at the University of Calgary. A total of six mice were used for this study. Wild-type (WT; n = 3) and Ames dwarf mice (df/df; n = 3) were bred and maintained under temperature- and light-controlled conditions (22 ± 2 °C, 12 hour light/12 hour dark cycle)62. The animals were anesthetized and euthanized for collection of femur samples for bone marrow extraction.
Bone-marrow derived macrophages (BMMØs), were derived from bone marrow isolated from the femurs, tibias and ilia of mice and cultured in DMEM supplemented with 10% fetal calf serum, 1% L-glutamine, 1% sodium pyruvate, 1% penicillin/streptomycin and 20% L-cell conditioned media from M-CSF producing L929 cells. BMMØs were maintained at 37 °C and 7% CO2, and were considered fully differentiated ten days after bone marrow isolation. This protocol yields >95% CD11b + F4/80+ macrophages63,64.
Fully-differentiated macrophages (from 3 WT and 3 df/df mice) were cultured in macrophage media as above, incubated at 37 °C and 5% CO2 divided into four treatments (samples of each mice were represented in each treatment): 1) Control (no GH and no IFN), 2) 50 ng/mL of recombinant porcine GH (Reporcin, Alpharma, Inc., Victoria, Australia), 3) 500 ng/mL of GH (Reporcin), and 4) 50 U/mL of IFNγ (Peprotech, Canada). Samples from each mouse/treatment (n = 48 samples) were collected for RNA extraction at 6 h and 24 h after the beginning of treatments. The doses of GH tested were based on previous studies25,65,66 and pilot experiments.
RNA extraction and real time PCR
The macrophages were removed from culture and homogenized with 3 mL of Qiazol (Qiagen, Valencia, CA, USA) and gDNA Eliminator (RNeasy Mini Kit, Qiagen). Next, chloroform was added, and the samples were homogenized and incubated for 3 min at room temperature. Samples were then centrifuged at 4 °C at 12,000 xg for 15 min and the clear upper phase transferred to a new tube with cold ethanol. The solution was transferred to columns (RNeasy Mini Kit, Qiagen) following manufacturer’s instructions. The quantity of RNA was determined using a spectrophotometer (Epoch Microplate Spectrophotometer, Biotek, Winooski, VT, USA). Reverse transcription reactions were performed with 1 μg of RNA (5 μL) using iScript Synthesis Kit (Biorad, Hercules, CA, USA) in a 20 μL volume incubated for 5 min at 25 °C, 20 min at 46 °C and 1 min at 95 °C (MJ Mini Personal Thermal Cycler, Biorad). The final cDNA solution was diluted to 10 ng/μL before use.
Real-time PCR using SYBR Green dye was used to evaluate gene expression. β2 microglobulin (B2m) expression was used as an internal control and Tnf as an initial target gene for assay validation and sample selection, based on previous reports of GH-mediated Tnf induction24. The primer sequences are listed in Table 2 and included genes differentially regulated and used to validate the results of the RNASeq analysis. Validation of RNASeq analysis was performed with a group samples from nine N mice processed as previously described. The PCR reactions were performed in duplicate in a 20 μL volume using 5 μL of Fast SYBR Green Mastermix (Applied Biosystems, Foster City, CA, USA), 0.4 μL of each primer (10 μM stock) and 20 ng of cDNA. Fluorescence was quantified with the ABI Prism 7500 Fast Real Time PCR System (Applied Biosystems). For each assay, 45 PCR cycles were run (95 °C for 3 sec and 60 °C for 30 sec) and a dissociation curve was included at the end of the reaction to verify the amplification of a single PCR product. Analyses of amplification plots were performed with the 7500 Software (Applied Biosystems). Each assay plate included a negative control. Relative expression was calculated from the 2−ΔΔCt equation67.
Table 2.
Primer sequences used in the study.
| Gene | Accession | Sequence 5′-3′ | Product size (bp) | |
|---|---|---|---|---|
| Nos2 | NM_010927.4 | Forward | GTGAAAAGTCCAGCCGCACC | 206 |
| Reverse | CCAGTAGCTGCCGCTCTCAT | |||
| Tnf | NM_013693.3 | Forward | CCACGCTCTTCTGTCTACTG | 145 |
| Reverse | GCTACAGGCTTGTCACTCG | |||
| Iipg1 | NM_001146275.1 | Forward | GACACAGGAGTTTCTGTGCCTTT | 102 |
| Reverse | ACCAGTAAAGCTGGAGGGCA | |||
| Cxcl10 | NM_021274.2 | Forward | CCACGTGTTGAGATCATTGCCA | 144 |
| Reverse | TGCGTGGCTTCACTCCAGTT | |||
| Igr1 | NM_008392.1 | Forward | ACCAAAGAGATTCCACCCTCCC | 153 |
| Reverse | TGAGTGGCAGCGTTCGCTAT | |||
| Cd40 | NM_011611.2 | Forward | CCCTGGACAAGCTGTGAGGA | 147 |
| Reverse | CACCCCGAAAATGGTGATGAGG | |||
| Gbp2 | NM_010260.1 | Forward | CCAGCTGCACTATGTGACG | 160 |
| Reverse | GGGTTTTCCGTTAACCTCCAG | |||
| Gbp3 | NM_001289492.1 | Forward | TAGTGTTCCCTGACGCTGCC | 113 |
| Reverse | GCCACAAGACCCTGTGAGGT | |||
| Gbp5 | NM_153564.2 | Forward | TTTTGACGCTCCTGCGCTTG | 177 |
| Reverse | AGGCTTTCTAGACGAGGTCCG | |||
| Gbp7 | NM_013506.3 | Forward | TTGTGGCTTCCGAAAGGGGA | 169 |
| Reverse | GCCGCCATGTTCTCTGTTGTAA | |||
| B2m | NM_009735.3 | Forward | AAGTATACTCACGCCACCCA | 217 |
| Reverse | CAGCGCTATGTATCAGTCTC |
RNA sequencing analyses
Transcriptomic profile of individual samples was performed using commercial RNA-sequencing kits (NEBNext mRNA Library Prep Master Mix and NEBNext Multiplex Oligos for Illumina, New England Biolabs, Ipswich, MA, USA) and adapted according to previous descriptions68,69 using 5 µg of total RNA per sample as initial input. Only samples from the Control (n = 3 WT and n = 3 df/df), 500 ng/mL of GH (n = 3 WT and n = 3 df/df) and 10 ng/mL of IFN (n = 3 WT and n = 3 df/df) at 6 h time point were used for RNA-Seq. Briefly, Poly A RNA enrichment was performed using the Magnetic mRNA Isolation kit (New England Biolabs) according to manufacturer instructions. Subsequently, samples were processed using the NEBNext mRNA Library Prep Master Mix (New England Biolabs) with slight modifications from the manufacturer’s instructions. After the RNA fragmentation step, RNA samples were cleaned and concentrated using RNA Clean & Concentrator™-5 (Zymo Research, Irvine, CA, USA). After this initial step, first and second strand cDNA were generated, samples were subjected to end-repair, dA tailing and finally adaptor ligation. Between each step samples were cleaned up using the Qiagen MinElute kit (Qiagen). Fragments between 150–300 bp were recovered after 2% agarose gel electrophoresis (QIAquick Gel Extraction Kit, Qiagen) in order to remove adapter dimers by size selection. Nine different indexes (NEBNext Multiplex Oligos for Illumina, New England Biolabs) were added to individual libraries during 12 cycles of PCR amplification. After that, samples were combined in two mixtures (nine in each mix), and submitted to sequencing on two flowcell lanes on a HiSeq. 2500 instrument (Illumina Inc., San Diego, CA, USA). All RNA-Seq data are available at the Sequence Read Archive (SRA) at NCBI under accession number SRP142633.
The mapping of sequencing reads to the mouse transcriptome (Illumina iGenomes annotation for UCSC mm10, http://support.illumina.com/sequencing/sequencing_software/ igenome.html) was performed using Tophat 2.1.0.0 and Bowtie 2.2.670. mRNA abundance was calculated using Cufflinks 2.1.171 and is presented as Fragments Per Kb of exon per Million reads mapped to mRNAs (FPKM). The number of reads aligned to its corresponding gene was calculated by HTSeq. 0.6.172. Genes with an average FPKM lower than 1/100,000th of the total aligned reads in more than 50% of the samples were eliminated from further analyses (10,117 transcripts selected with these criteria from 23,457 transcripts available in the mm10 database). mRNAs were further processed for pathway analysis using the Generally Applicable Gene-set Enrichment (GAGE), which uses log- based fold changes as per gene statistics, and Pathview packages in R73,74, using KEGG molecular pathways database75,76. Enrichment of gene ontology (GO) terms (biological processes, molecular function and cellular component) was also performed using the GAGE package using same criteria described for pathways analysis. P values lower than 0.05 were considered as significant for pathways and GO Terms analysis.
Reactive oxygen species production
Macrophages derived from C57BL/6 mice as described above, were incubated for 18 hours in the presence of either 10 ng/mL of IFNγ or 500 ng/mL of porcine GH. Immediately prior to the addition of experimental particles, growth medium was removed from cells and replaced with assay medium with low autofluorescence (DPBS containing 1 mM CaCl2, 2.7 mM KCl, 0.5 mM MgCl2, 5 mM glucose, 0.1% calf skin gelatin, and 10 mM HEPES). Cells were stimulated for 1.5 hours with 50 μg/mL of serum-opsonized zymosan particles (Sigma Aldrich). Relative amounts of H2O2 within supernatants were determined by measuring the reaction of 20 μM Amplex Red (Thermo Fisher Scientific) in the presence of horse radish peroxidase using a FLUOstar Optima fluorescence plate reader (BMG Labtech) as previously described48,77. Hydrogen peroxide production is presented as relative to unactivated macrophages from 3 independent experiments.
Phagosomal proteolysis
For in vitro phagosomal assessment, BMMØs from C57BL/6 mice were plated in 96 µClear® black microplates (Greiner Bio-One) 16 h prior to fluorometric analyses. The hydrolytic capacity of the phagosome was assessed using 3.0 µM silica experimental particles bearing the proteolytic substrate, DQ-Green BSA (Thermo Fisher) and a calibration fluor (Alexa Fluor 594 SE (succinimidyl ester)) in live macrophages as previously described14,50,78. Briefly, phagosomal proteolysis was determined by measuring the fluorescence liberated by the fluorogenic substrate DQ-Green BSA from IgG-opsonized experimental particles that had been phagocytosed by macrophages cultured in the presence of either 10 ng/mL of IFNγ or 500 ng/mL porcine GH for 18 hours prior to assessment. The average rates of phagosomal proteolysis were calculated as the average rate of increase in substrate fluorescence relative to the calibration fluorescence between 80–100 minutes post phagocytosis. Data is presented as relative to unactivated macrophage samples from 5 independent experiments.
Statistical analyses
The results are presented as mean ± standard error of the mean (SEM) for real time PCR results and in vitro assessment of phagosomal characteristic. All statistical analyses for these parameters were performed using GraphPad Prism 5.0 (GraphPad Software Inc., La Jolla, CA, USA) using a one-way ANOVA with a Tukey post-test. P values lower than 0.05 were considered as significant.
Statistical analyses for differentially expressed mRNAs was performed using the software R (3.2.2) and the Bioconductor package DESeq (1.2.0)79 using the HTSeq output count. Read counts were normalized for library depth, and pairwise comparisons between genotypes and treatment, measuring fold change, uncorrected P-values from the negative binomial distribution, and adjusted P values (false discovery rate; FDR) were obtained. Principal components analysis (PCA) including all genes was also performed using R to observe sample distribution in a two-dimensional plot and eliminate outliers. Unsupervised hierarchical clustering, included all genes, was performed also using the DESeq package to observe sample clustering. Genes with a FDR <0.02 and fold change (FC) >2.0 were considered as up-regulated; and with FDR <0.02 and FC <0.5 were considered as down-regulated.
Supplementary information
Acknowledgements
This work was supported by Universidade Federal de Pelotas internal funds to AS; National Institute on Aging of the National Institutes of Health to MM (grant numbers R03AG059846, R15AG059190), and Competitive Research Grant from University of Central Florida College of Medicine to KR and MM. The authors are thankful to the Kegg Database Project team from Kanehisa Laboratories for providing permission to use the pathway images.
Author contributions
A.S., H.N.W., R.M.Y., S.J.G., M.M.M. and K.H.R. designed experiments. A.S., H.N.W., S.G., S.J.G., R.M.Y., M.M.M. and K.H.R. performed experiments and data analysis. A.S. and K.H.R. wrote the manuscript. H.N.W., S.G., C.J.G., R.M.Y. and M.M.M. revised the manuscript.
Data availability
All RNA-Seq data are available at the Sequence Read Archive (SRA) at NCBI under accession number SRP142633. Other datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Michal M. Masternak, Email: michal.masternak@ucf.edu
Kyle H. Rohde, Email: kyle.rohde@ucf.edu
Supplementary information
is available for this paper at 10.1038/s41598-019-56017-6.
References
- 1.Davidson MB. Effect of growth hormone on carbohydrate and lipid metabolism. Endocrine reviews. 1987;8:115–131. doi: 10.1210/edrv-8-2-115. [DOI] [PubMed] [Google Scholar]
- 2.Jones JI, Clemmons DR. Insulin-like growth factors and their binding proteins: biological actions. Endocrine reviews. 1995;16:3–34. doi: 10.1210/edrv-16-1-3. [DOI] [PubMed] [Google Scholar]
- 3.Waters MJ. The growth hormone receptor. Growth hormone & IGF research: official journal of the Growth Hormone Research Society and the International IGF Research Society. 2016;28:6–10. doi: 10.1016/j.ghir.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Bazan JF. Structural design and molecular evolution of a cytokine receptor superfamily. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:6934–6938. doi: 10.1073/pnas.87.18.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sonntag WE, Steger RW, Forman LJ, Meites J. Decreased pulsatile release of growth hormone in old male rats. Endocrinology. 1980;107:1875–1879. doi: 10.1210/endo-107-6-1875. [DOI] [PubMed] [Google Scholar]
- 6.Ponnappan S, Ponnappan U. Aging and immune function: molecular mechanisms to interventions. Antioxid Redox Signal. 2011;14:1551–1585. doi: 10.1089/ars.2010.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sornson MW, et al. Pituitary lineage determination by the Prophet of Pit-1 homeodomain factor defective in Ames dwarfism. Nature. 1996;384:327–333. doi: 10.1038/384327a0. [DOI] [PubMed] [Google Scholar]
- 8.Chandrashekar V, Bartke A. Induction of endogenous insulin-like growth factor-I secretion alters the hypothalamic-pituitary-testicular function in growth hormone-deficient adult dwarf mice. Biology of reproduction. 1993;48:544–551. doi: 10.1095/biolreprod48.3.544. [DOI] [PubMed] [Google Scholar]
- 9.Brown-Borg HM, Borg KE, Meliska CJ, Bartke A. Dwarf mice and the ageing process. Nature. 1996;384:33. doi: 10.1038/384033a0. [DOI] [PubMed] [Google Scholar]
- 10.Masternak Michal M., Bartke Andrzej. Growth hormone, inflammation and aging. Pathobiology of Aging & Age-related Diseases. 2012;2(1):17293. doi: 10.3402/pba.v2i0.17293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farhat K, et al. Growth Hormone (GH) Deficient Mice With GHRH Gene Ablation Are Severely Deficient in Vaccine and Immune Responses Against Streptococcus pneumoniae. Front Immunol. 2018;9:2175. doi: 10.3389/fimmu.2018.02175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kooijman R, Malur A, Van Buul-Offers SC, Hooghe-Peters EL. Growth hormone expression in murine bone marrow cells is independent of the pituitary transcription factor Pit-1. Endocrinology. 1997;138:3949–3955. doi: 10.1210/endo.138.9.5414. [DOI] [PubMed] [Google Scholar]
- 13.Mogensen T. H. Pathogen Recognition and Inflammatory Signaling in Innate Immune Defenses. Clinical Microbiology Reviews. 2009;22(2):240–273. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balce DR, Greene CJ, Tailor P, Yates RM. Endogenous and exogenous pathways maintain the reductive capacity of the phagosome. J Leukoc Biol. 2016;100:17–26. doi: 10.1189/jlb.2HI0315-083R. [DOI] [PubMed] [Google Scholar]
- 15.Balce DR, et al. Alternative activation of macrophages by IL-4 enhances the proteolytic capacity of their phagosomes through synergistic mechanisms. Blood. 2011;118:4199–4208. doi: 10.1182/blood-2011-01-328906. [DOI] [PubMed] [Google Scholar]
- 16.Balce DR, Rybicka JM, Greene CJ, Ewanchuk BW, Yates RM. Ligation of FcgammaR Alters Phagosomal Processing of Protein via Augmentation of NADPH Oxidase Activity. Traffic. 2016;17:786–802. doi: 10.1111/tra.12396. [DOI] [PubMed] [Google Scholar]
- 17.Cheung S, Greene C, Yates RM. Simultaneous Analysis of Multiple Lumenal Parameters of Individual Phagosomes Using High-Content Imaging. Methods Mol Biol. 2017;1519:227–239. doi: 10.1007/978-1-4939-6581-6_15. [DOI] [PubMed] [Google Scholar]
- 18.Schaible UE, Sturgill-Koszycki S, Schlesinger PH, Russell DG. Cytokine activation leads to acidification and increases maturation of Mycobacterium avium-containing phagosomes in murine macrophages. J Immunol. 1998;160:1290–1296. [PubMed] [Google Scholar]
- 19.Weiss G, Schaible UE. Macrophage defense mechanisms against intracellular bacteria. Immunol Rev. 2015;264:182–203. doi: 10.1111/imr.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.VanderVen BC, Yates RM, Russell DG. Intraphagosomal measurement of the magnitude and duration of the oxidative burst. Traffic. 2009;10:372–378. doi: 10.1111/j.1600-0854.2009.00877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murray HW. Interferon-gamma, the activated macrophage, and host defense against microbial challenge. Ann Intern Med. 1988;108:595–608. doi: 10.7326/0003-4819-108-4-595. [DOI] [PubMed] [Google Scholar]
- 22.Edwards CK, 3rd, et al. The macrophage-activating properties of growth hormone. Cell Mol Neurobiol. 1992;12:499–510. doi: 10.1007/BF00711550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edwards CK, 3rd, et al. In vivo administration of recombinant growth hormone or gamma interferon activities macrophages: enhanced resistance to experimental Salmonella typhimurium infection is correlated with generation of reactive oxygen intermediates. Infection and immunity. 1992;60:2514–2521. doi: 10.1128/iai.60.6.2514-2521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edwards CK, 3rd, et al. Hypophysectomy inhibits the synthesis of tumor necrosis factor alpha by rat macrophages: partial restoration by exogenous growth hormone or interferon gamma. Endocrinology. 1991;128:989–986. doi: 10.1210/endo-128-2-989. [DOI] [PubMed] [Google Scholar]
- 25.Uronen-Hansson H, et al. Growth hormone enhances proinflammatory cytokine production by monocytes in whole blood. Growth hormone & IGF research: official journal of the Growth Hormone Research Society and the International IGF Research Society. 2003;13:282–286. doi: 10.1016/S1096-6374(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 26.Kim BH, et al. A family of IFN-gamma-inducible 65-kD GTPases protects against bacterial infection. Science. 2011;332:717–721. doi: 10.1126/science.1201711. [DOI] [PubMed] [Google Scholar]
- 27.Rybicka JM, Balce DR, Khan MF, Krohn RM, Yates RM. NADPH oxidase activity controls phagosomal proteolysis in macrophages through modulation of the lumenal redox environment of phagosomes. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10496–10501. doi: 10.1073/pnas.0914867107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hillyer LM, Maliwichi HE, Woodward B. Blood serum interferon-gamma bioactivity is low in weanling mice subjected to acute deficits of energy or both protein and energy. Br J Nutr. 2007;97:528–534. doi: 10.1017/S0007114507352409. [DOI] [PubMed] [Google Scholar]
- 29.Sinha YN, Salocks CB, Vanderlaan WP. Prolactin and growth hormone levels in different inbred strains of mice: patterns in association with estrous cycle, time of day, and perphenazine stimulation. Endocrinology. 1975;97:1112–1122. doi: 10.1210/endo-97-5-1112. [DOI] [PubMed] [Google Scholar]
- 30.Darwich L, et al. Secretion of interferon-gamma by human macrophages demonstrated at the single-cell level after costimulation with interleukin (IL)-12 plus IL-18. Immunology. 2009;126:386–393. doi: 10.1111/j.1365-2567.2008.02905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimball ES, Kovacs E, Clark MC, Schneider CR. Activation of cytokine production and adhesion molecule expression on THP-1 myelomonocytic cells by macrophage colony-stimulating factor in combination with interferon-gamma. J Leukoc Biol. 1995;58:585–594. doi: 10.1002/jlb.58.5.585. [DOI] [PubMed] [Google Scholar]
- 32.Matzkin ME, et al. Alterations in oxidative, inflammatory and apoptotic events in short-lived and long-lived mice testes. Aging. 2016;8:95–110. doi: 10.18632/aging.100875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dhahbi J, Li X, Tran T, Masternak MM, Bartke A. Circulating blood leukocyte gene expression profiles: effects of the Ames dwarf mutation on pathways related to immunity and inflammation. Exp Gerontol. 2007;42:772–788. doi: 10.1016/j.exger.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar P, Menon R. New insights into growth hormone’s actions on the macrophage: implications for non-growth-related actions of growth hormone. OA Biochemistry. 2013;1:15. doi: 10.13172/2052-9651-1-2-1122. [DOI] [Google Scholar]
- 35.Shaw PJ, Lamkanfi M, Kanneganti TD. NOD-like receptor (NLR) signaling beyond the inflammasome. Eur J Immunol. 2010;40:624–627. doi: 10.1002/eji.200940211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int Immunol. 2009;21:317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb Perspect Biol. 2009;1:a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Capitano ML, et al. Ames hypopituitary dwarf mice demonstrate imbalanced myelopoiesis between bone marrow and spleen. Blood Cells Mol Dis. 2015;55:15–20. doi: 10.1016/j.bcmd.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fabris N, Pierpaoli W, Sorkin E. Hormones and the immunological capacity. IV. Restorative effects of developmental hormones or of lymphocytes on the immunodeficiency syndrome of the dwarf mouse. Clin Exp Immunol. 1971;9:227–240. [PMC free article] [PubMed] [Google Scholar]
- 40.Herrington J, Smit LS, Schwartz J, Carter-Su C. The role of STAT proteins in growth hormone signaling. Oncogene. 2000;19:2585–2597. doi: 10.1038/sj.onc.1203526. [DOI] [PubMed] [Google Scholar]
- 41.Bach EA, Aguet M, Schreiber RD. The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu Rev Immunol. 1997;15:563–591. doi: 10.1146/annurev.immunol.15.1.563. [DOI] [PubMed] [Google Scholar]
- 42.Hu X, Chen J, Wang L, Ivashkiv LB. Crosstalk among Jak-STAT, Toll-like receptor, and ITAM-dependent pathways in macrophage activation. J Leukoc Biol. 2007;82:237–243. doi: 10.1189/jlb.1206763. [DOI] [PubMed] [Google Scholar]
- 43.Hagemann T, Biswas SK, Lawrence T, Sica A, Lewis CE. Regulation of macrophage function in tumors: the multifaceted role of NF-kappaB. Blood. 2009;113:3139–3146. doi: 10.1182/blood-2008-12-172825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arias M, et al. Inhibition of virulent Mycobacterium tuberculosis by Bcg(r) and Bcg(s) macrophages correlates with nitric oxide production. J Infect Dis. 1997;176:1552–1558. doi: 10.1086/514154. [DOI] [PubMed] [Google Scholar]
- 45.Garbe TR, Hibler NS, Deretic V. Response of Mycobacterium tuberculosis to reactive oxygen and nitrogen intermediates. Mol Med. 1996;2:134–142. doi: 10.1007/BF03402209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams LB, Dinauer MC, Morgenstern DE, Krahenbuhl JL. Comparison of the roles of reactive oxygen and nitrogen intermediates in the host response to Mycobacterium tuberculosis using transgenic mice. Tuber Lung Dis. 1997;78:237–246. doi: 10.1016/S0962-8479(97)90004-6. [DOI] [PubMed] [Google Scholar]
- 47.MacMicking JD, et al. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc Natl Acad Sci USA. 1997;94:5243–5248. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balce DR, Allan ER, McKenna N, Yates R. M. gamma-Interferon-inducible lysosomal thiol reductase (GILT) maintains phagosomal proteolysis in alternatively activated macrophages. The Journal of biological chemistry. 2014;289:31891–31904. doi: 10.1074/jbc.M114.584391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohde K, Yates RM, Purdy GE, Russell DG. Mycobacterium tuberculosis and the environment within the phagosome. Immunol Rev. 2007;219:37–54. doi: 10.1111/j.1600-065X.2007.00547.x. [DOI] [PubMed] [Google Scholar]
- 50.Yates RM, Hermetter A, Russell DG. The kinetics of phagosome maturation as a function of phagosome/lysosome fusion and acquisition of hydrolytic activity. Traffic. 2005;6:413–420. doi: 10.1111/j.1600-0854.2005.00284.x. [DOI] [PubMed] [Google Scholar]
- 51.Yates RM, Hermetter A, Taylor GA, Russell DG. Macrophage activation downregulates the degradative capacity of the phagosome. Traffic. 2007;8:241–250. doi: 10.1111/j.1600-0854.2006.00528.x. [DOI] [PubMed] [Google Scholar]
- 52.Cassatella MA, et al. Molecular basis of interferon-gamma and lipopolysaccharide enhancement of phagocyte respiratory burst capability. Studies on the gene expression of several NADPH oxidase components. The Journal of biological chemistry. 1990;265:20241–20246. [PubMed] [Google Scholar]
- 53.Zhao X, Drlica K. Reactive oxygen species and the bacterial response to lethal stress. Curr Opin Microbiol. 2014;21:1–6. doi: 10.1016/j.mib.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burton NA, et al. Disparate impact of oxidative host defenses determines the fate of Salmonella during systemic infection in mice. Cell Host Microbe. 2014;15:72–83. doi: 10.1016/j.chom.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 55.Adams DO, Hamilton TA. Molecular transductional mechanisms by which IFN gamma and other signals regulate macrophage development. Immunol Rev. 1987;97:5–27. doi: 10.1111/j.1600-065X.1987.tb00514.x. [DOI] [PubMed] [Google Scholar]
- 56.Tan HY, et al. The Reactive Oxygen Species in Macrophage Polarization: Reflecting Its Dual Role in Progression and Treatment of Human Diseases. Oxid Med Cell Longev. 2016;2016:2795090. doi: 10.1155/2016/2795090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Allan ER, et al. NADPH oxidase modifies patterns of MHC class II-restricted epitopic repertoires through redox control of antigen processing. J Immunol. 2014;192:4989–5001. doi: 10.4049/jimmunol.1302896. [DOI] [PubMed] [Google Scholar]
- 58.Campos VC, et al. Infectious diseases and immunological responses in adult subjects with lifetime untreated, congenital GH deficiency. Endocrine. 2016;54:182–190. doi: 10.1007/s12020-016-1061-z. [DOI] [PubMed] [Google Scholar]
- 59.Linehan E, Fitzgerald D. Ageing and the immune system: focus on macrophages. European Journal of Microbiology and Immunology. 2015;5:14–24. doi: 10.1556/EuJMI-D-14-00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spadaro O, et al. Growth Hormone Receptor Deficiency Protects against Age-Related NLRP3 Inflammasome Activation and Immune Senescence. Cell Reports. 2016;14:1571–1580. doi: 10.1016/j.celrep.2016.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Villares R, et al. Growth hormone prevents the development of autoimmune diabetes. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E4619–4627. doi: 10.1073/pnas.1314985110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Masternak MM, et al. Divergent effects of caloric restriction on gene expression in normal and long-lived mice. The journals of gerontology. Series A, Biological sciences and medical sciences. 2004;59:784–788. doi: 10.1093/gerona/59.8.B784. [DOI] [PubMed] [Google Scholar]
- 63.Lee CM, Hu J. Cell density during differentiation can alter the phenotype of bone marrow-derived macrophages. Cell Biosci. 2013;3:30. doi: 10.1186/2045-3701-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ying, W., Cheruku, P. S., Bazer, F. W., Safe, S. H. & Zhou, B. Investigation of macrophage polarization using bone marrow derived macrophages. J Vis Exp, 10.3791/50323 (2013). [DOI] [PMC free article] [PubMed]
- 65.Warwick-Davies J, Lowrie DB, Cole PJ. Growth hormone activation of human monocytes for superoxide production but not tumor necrosis factor production, cell adherence, or action against Mycobacterium tuberculosis. Infection and immunity. 1995;63:4312–4316. doi: 10.1128/iai.63.11.4312-4316.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Edwards CK, III., Ghiasuddin SM, Schepper JM, Yunger LM, Kelley KW. A newly defined property of somatotropin: priming of macrophages for production of superoxide anion. Science. 1988;239:769–771. doi: 10.1126/science.2829357. [DOI] [PubMed] [Google Scholar]
- 67.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 68.Matkovich SJ, Dorn GW., II Deep sequencing of cardiac microRNA-mRNA interactomes in clinical and experimental cardiomyopathy. Methods in molecular biology. 2015;1299:27–49. doi: 10.1007/978-1-4939-2572-8_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schneider Augusto, Matkovich Scot J., Saccon Tatiana, Victoria Berta, Spinel Lina, Lavasani Mitra, Bartke Andrzej, Golusinski Pawel, Masternak Michal M. Ovarian transcriptome associated with reproductive senescence in the long-living Ames dwarf mice. Molecular and Cellular Endocrinology. 2017;439:328–336. doi: 10.1016/j.mce.2016.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luo W, Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29:1830–1831. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic acids research. 2016;44:D457–462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Balce DR, Yates RM. Redox-sensitive probes for the measurement of redox chemistries within phagosomes of macrophages and dendritic cells. Redox Biol. 2013;1:467–474. doi: 10.1016/j.redox.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yates RM, Russell DG. Real-time spectrofluorometric assays for the lumenal environment of the maturing phagosome. Methods in molecular biology. 2008;445:311–325. doi: 10.1007/978-1-59745-157-4_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-Seq data are available at the Sequence Read Archive (SRA) at NCBI under accession number SRP142633. Other datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.