Abstract
It has been reported that lipid biosynthesis in plant host root cells plays critical roles in legume‐fungal or ‐rhizobial symbioses, but little is known about its regulatory mechanism in legume–rhizobia interaction. Soybean WRINKLED1 (WRI1) a and b, with their alternative splicing (AS) products a’ and b’, are highly expressed in developing seeds and nodules, but their functions in soybean nodulation are not known. GmWRI1a and b differently promoted triacylglycerol (TAG) accumulation in both Arabidopsis wild‐type and wri1 mutant seeds and when they ectopically expressed in the soybean hairy roots. Transcriptome analysis revealed that 15 genes containing AW boxes in their promoters were targeted by GmWRI1s, including genes involved in glycolysis, fatty acid (FA) and TAG biosynthesis. GmWRI1a, GmWRI1b and b’ differentially transactivated most targeted genes. Overexpression of GmWRI1s affected phospholipid and galactolipid synthesis, soluble sugar and starch contents and led to increased nodule numbers, whereas GmWRI1 knockdown hairy roots interfered root glycolysis and lipid biosynthesis and resulted in fewer nodules. These phenomena in GmWRI1 mutants coincided with the altered expression of nodulation genes. Thus, GmWRI1‐regulated starch degradation, glycolysis and lipid biosynthesis were critical for nodulation. GmWRI1 mutants also altered auxin and other hormone‐related biosynthesis and hormone‐related genes, by which GmWRI1s may affect nodule development. The study expands the views for pleiotropic effects of WRI1s in regulating soybean seed filling and root nodulation.
Keywords: carbon partitioning, lipid biosynthesis, transcriptional regulation, glycolysis, alternative splicing, nodulation
Introduction
Plant seed TAG and oils are synthesized in the endoplasmic reticulum (ER) and stored in the form of lipid bodies assembled from TAGs, polar lipids and lipid droplet‐associated proteins (Bates et al., 2013). In sink tissues, TAG precursors are derived from plastid glycolysis, involving pyruvate kinase (PKP), phosphoenolpyruvate carboxylase (PPC) and pyruvate dehydrogenase complex (PDHC) that convert photosynthesis‐derived carbohydrates into metabolites that serve as substrates for the biosynthesis of FAs in chloroplasts (Ohlrogge and Chapman, 2011). Genes involved in the early steps of fatty acyl biosynthesis include ketoacyl‐ACP synthase (KAS), biotin carboxyl carrier protein (BCCP), malonyltransferase, acyl carrier protein (ACP) and acyl‐CoA binding protein (ACBP) in plastids (Ohlrogge and Chapman, 2011). Fatty acids are exported from plastids to give rise to fatty acyl‐CoAs in the cytoplasm (Manan et al., 2017b). Acyl‐CoAs and glycolysis‐derived glycerol‐3‐phosphate (G3P) serve as precursors for TAG assembly at the ER, which starts with the stepwise acylation by G3P acyltransferase (GPAT) at sn‐1, lysophosphatidic acid acyltransferase (LPAAT) at sn‐2 and diacylglycerol (DAG) acyltransferase (DGAT) or phospholipid:DAG acyltransferase (PDAT) at sn‐3 (Voelker and Kinney, 2001; Zhang et al., 2009). Finally, TAGs are stored in specialized lipid droplets coated with oleosins (Manan et al., 2017b). Phosphatidylcholine:diacylglycerol cholinephosphotransferase (PDCT) catalyses the interconversion of phosphatidylcholine (PC) and DAG, which is involved in soybean TAG biosynthesis (Ohlrogge and Chapman, 2011).
During oilseed development and seed filling, partitioning of photosynthetic carbons into different storage compounds, such as starch, oil and proteins, is highly regulated, and several transcription factors (TFs), including WRI1, LEAFY COTYLEDON 1 (LEC1) and LEC2, have been implicated in the regulation (Manan et al., 2017b). WRI1, encoding an APETALA2 (AP2) TF, has been identified as a key regulator of the carbon partitioning into oils and proteins. Overexpression of WRI1s from Arabidopsis, maize (Zea mays) and rapeseed (Brassica napus) increased seed oil production (Cernac and Benning, 2004; Shen et al., 2010; Wu et al., 2014). WRI1 targeted genes include those encoding glycolytic and FA biosynthetic enzymes (Baud et al., 2007; Ma et al., 2013; Maeo et al., 2009; Pouvreau et al., 2011; Sanjaya et al., 2011; Shen et al., 2010). WRI1 directly binds to AW box or other motifs in the promoters of genes involved in the late steps of glycolysis and early steps of FA biosynthesis (Baud et al., 2009; Maeo et al., 2009). WRI1 orthologues from different species rescued the phenotypes of the Arabidopsis atwri1 mutant (Cernac and Benning, 2004; Ma et al., 2013; Pouvreau et al., 2011), suggesting that the regulatory function of WRI1 in oil biosynthesis is highly conserved in plants. Meanwhile, other AtWRI1 homologs, such as AtWRI3 and AtWRI4 that were expressed primarily in stems and flowers, could also activate FA biosynthesis in seeds and leaves, and genetically complement the atwri1 mutant (Ma et al., 2013; Pouvreau et al., 2011; Shen et al., 2010; Yang et al., 2015).
Despite the conserved function in lipid production, species‐specific functions exist for different WRI1s. For example, ectopic expression of a BdWRI1 ortholog promoted free FA accumulation in both storage and vegetative tissues and triggered leaf cell death in Brachypodium distachyon (Yang et al., 2015), which is not the case for Arabidopsis or maize. AtWRI1 and BnWRI1 promoted plastid glycolysis, but ZmWRI1 did not (Shen et al., 2010). Therefore, how soybean WRI1 regulates the biosynthesis of oils and proteins during soybean seed development and filling cannot simply be predicted. The allotetraploid soybean genome contains more than 15 WRI homologues (Figure S1a). Moreover, some of them are also highly expressed in roots and nodules, raising the question of whether the nodule WRIs may be responsible for carbon partitioning for rhizobial growth and nodule development. Amino acids and dicarboxylic acids from root cells are usually regarded as carbon resources for rhizobia, but other carbon forms likely exist in legume roots to support rhizobial growth (Clarke et al., 2014; Udvardi and Poole, 2013). Recent studies indicated that a soybean GmACP and a stearoyl‐ACP desaturase (GmSACPD‐C) were critically required for nodulation and normal nodule development (Krishnan et al., 2016; Lakhssassi et al., 2017; Wang et al., 2014). Moreover, nodule‐specific lipid transfer proteins, Medicago truncatula MtN5 and Chinese milk vetch (Astragalus sinicus) AsE246, were shown to be required for transport of plant‐synthesized lipids to the symbiosomes for nodule organogenesis (Lei et al., 2014; Pii et al., 2009). Recent discoveries also revealed that monoacylglycerol (MAG) is a major carbon supply provided by host plants for growth and development of arbuscular mycorrhizal fungi (AMF) in exchange for phosphorus (Keymer et al., 2017; Jiang et al., 2017; Luginbuehl et al., 2017). Recent studies further showed that Medicago WRI5a and Lotus WRI1 homolog CTTC Motif‐Binding Transcription Factor1 (CBX1) regulated genes involved in MAG biosynthesis and transport, such as FA thioesterase M (FatM), GPAT, or Stunted Arbuscule 1 (STR1) and were indispensable for successful Legume–AMF symbioses (Jiang et al., 2018; Xue et al., 2018). It is thus of great basic biological and agricultural significance to know whether lipids are carbon sources in rhizobia‐soybean interactions required for nitrogen fixation and how they are regulated.
Here, we report the functional characterization of soybean GmWRI1s in regulating carbohydrate partitioning in soybean seeds and nodules. We characterized pleiotropic effects of GmWRI1s in seed filling and root nodulation and their regulatory mechanisms.
Results
The soybean genome has multiple WRI1‐like and alternative splicing forms
To identify WRI1 orthologs in soybean (Glycine max L), a BLASTP analysis was performed with a strict e‐value threshold limit in Phytozome (http://www.phytozome.net/) using both monocot maize and dicot Arabidopsis and rapeseed WRI1 sequences as queries against the soybean proteome database. Fifteen AP2 domain‐containing WRI1‐like proteins were identified in the soybean genome (Figure S1a). To clarify the relationship between GmWRI1s and their functionally characterized WRI1 orthologs from other plant species, as well as AtWRI2, AtWRI3 and AtWRI4, we constructed a phylogeny tree based on amino acid sequences of these closely related AP2 TFs. Two proteins encoded by Glyma08g227700 (GmWRI1a) and Glyma15g221600 (GmWRI1b) genes, ZmWRI1‐1, ZmWRI1‐2, GhWRI1 and BnWRI1, fell into the same clade as AtWRI1. Other WRIs were present in the other clades defined by AtWRI2 or AtWRI3/AtWRI4 (Figure S1). Thus, GmWRI1a and GmWRI1b are designated as orthologs of AtWRI1. GmWRI1a is annotated to have an alternative splicing (AS) transcript, GmWRI1a'; however, for unknown reason we were not able to clone this transcript. Meanwhile, we cloned an AS form of GmWRI1b, encoding GmWRI1b’, in which three amino acids, VYL, at the N‐terminus have been deleted, similar to the predicted AS form GmWRI1a' encoded by GmWRI1a. These VYL‐encoding 9 nucleotides in this gene form the 3rd exon, which is similar to Arabidopsis WRI1 exon 3 (Figure S1b,c, Ma et al., 2013).
GmWRI1a and b proteins share more than 90% amino acid sequence identity with those of AtWRI1 and BdWRI1. GmWRI1a shares 55% identity with AtWRI1, whereas GmWRI1b shares 56% identity with AtWRI1 (Figure S1). The VYL sequence is present in the first AP2 domain of these full‐length GmWRI1s but is missing in their AS forms GmWRI1a' and b’ (Figure 1a and Figure S2).
Figure 1.
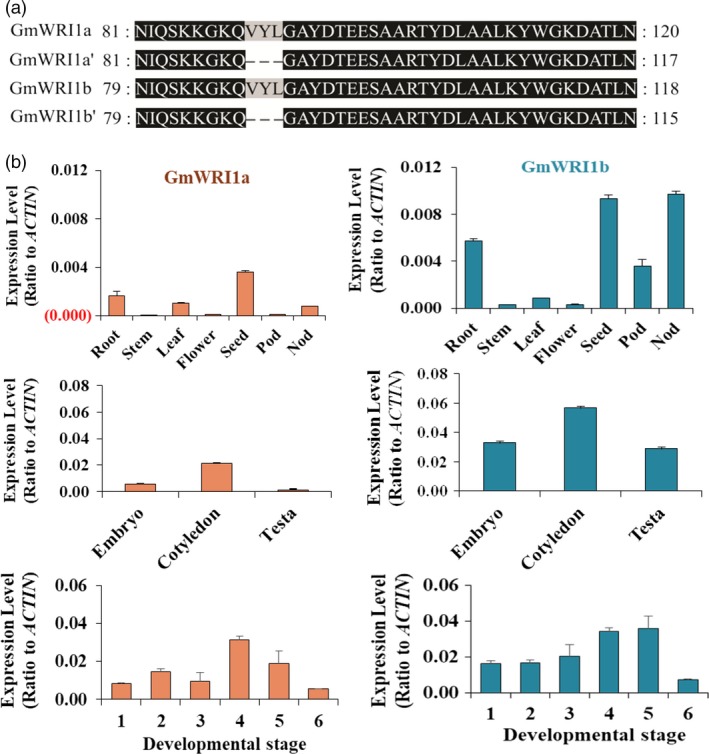
Analysis of GmWRI1 genes from soybean and their expression patterns. (a) Alignment of protein sequences of GmWRI1a, b and their AS forms missing the “VYL” motif. (b) The tissue expression patterns of GmWRI1a and GmWRI1b. qRT‐PCR was performed with GmACTIN as an internal standard. Six seed developmental stages with fresh weights are: stage 1. 40–70 mg; stage 2. 80–100 mg; stage 3.150–200 mg; stage 4. 250–300 mg; stage 5. 320–350 mg; stage 6. 350–430 mg.
GmWRI1a and GmWRI1b are expressed highly in the developing seeds and root nodules
To assess functions of GmWRI1genes, we examined expression patterns of GmWRI1a and GmWRI1b in different soybean tissues including seeds at different developmental stages by qRT‐PCR using gene‐specific primers. Both genes showed relatively higher levels of expression in seeds, roots or nodules than in other tissues, with GmWRI1a transcript also high in leaves but GmWRI1b transcript high in pods. The GmWRI1b transcript was more abundant than GmWRI1a in most tissues, particularly in nodules (Figure 1b and Figure S3). The high level of GmWRI1a and GmWRI1b expression in seeds was similar to that of Arabidopsis AtWRI1 in siliques and embryos (Cernac and Benning, 2004). We further separated seed tissues into embryo, cotyledon and testa from developing seeds and found both GmWRI1s were most highly expressed in cotyledons (Figure 1b).
To gain more insights into the spatial expression patterns of GmWRI1 genes at the seed filling stage, qRT‐PCR was performed on RNAs isolated from seeds at different developmental stages. Both genes displayed the highest expression at stages 4–5, about 25–35 days after flowering under the growth condition (Figure 1b and Figure S3). GmWRI1a and GmWRI1b were differentially expressed during the development of soybean seeds and nodules: GmWRI1a transcript levels were higher at early stages, whereas GmWRI1b transcript abundance increased steadily during seed filling and then decreased upon maturation (Figure 1b).
GmWRI1s restore the Arabidopsis wri1 mutant phenotype and promote TAG biosynthesis
We expressed GmWRI1s in Arabidopsis (GmWRI1OE) under the control of the CaMV35S promoter in both wild type (Col‐0) and in wri1 mutant backgrounds, and more than ten independent transgenic lines harbouring the GmWRI1 constructs were generated for each of the individual GmWRI1 construct (Figure S4). Total TAG contents in GmWRI1a‐ and GmWRI1b‐overexpressing Arabidopsis wild‐type seeds were enhanced compared with wild‐type control seeds (Figure S5a). The content of 18:3 (carbons: double bonds) in GmWRI1a‐ and GmWRI1b‐OE lines increased on average by 16% and 19%, respectively, while that of 18:2 was reduced by 16% and 21%, respectively (Figure S5b). GmWRI1a and b also restored the wri1 phenotypes in TAG and protein contents, whereas GmWRI1b' only partly restored the proteins in wri1 mutant seeds (Figure S5c,d and Figure S6). However, GmWRI1b'‐OE in wild type did not result in an increase in seed TAG production or the changes in 18:3 and 18:2 contents in seeds.
Ectopic expression of GmWRI1s promotes plastid glycolysis and FA biosynthesis
To further understand the function of GmWRI1s, we expressed the respective GmWRI1s cDNAs in hairy roots driven by the CaMV 35S promoter. As previously described, hairy root transformation derived from soybean cotyledons is an efficient approach to verify the functions of soybean genes since the generation of soybean transgenic plants is still challenging (Chen et al., 2016). Ectopic expression of three GmWRI1s in hairy roots, as confirmed by RT‐PCR and qRT‐PCR (Figure 2a and Figure 3b), significantly promoted TAG content compared to the control (P < 0.05) (Figure 2b, c). More than 5 independent transgenic hairy root lines for each GmWRI1 cDNA were analysed for TAG accumulation. The ectopic expression of GmWRI1s increased TAG contents by about twofold on average compared to the control, indicating that GmWRI1s promotes TAG production (Figure 2). The increased expression of GmWRI1s also led to an increase in TAGs with linolenic acid (18:3) acyl chains, but a slight decrease in oleic acid (18:1) and linoleic acid (18:2) acyl chains. We also analysed the contents of soluble sugars in these GmWRI1 transgenic hairy root lines (Figure 2e) and observed significantly decreased contents of total soluble sugars, mainly rhamnose, arabinose, xylose, mannose, glucose and galactose as compared to the control (P < 0.01) (Figure 2f). These results suggest that GmWRI1s promote carbohydrate conversion to lipid production in GmWRI1OE hairy roots.
Figure 2.
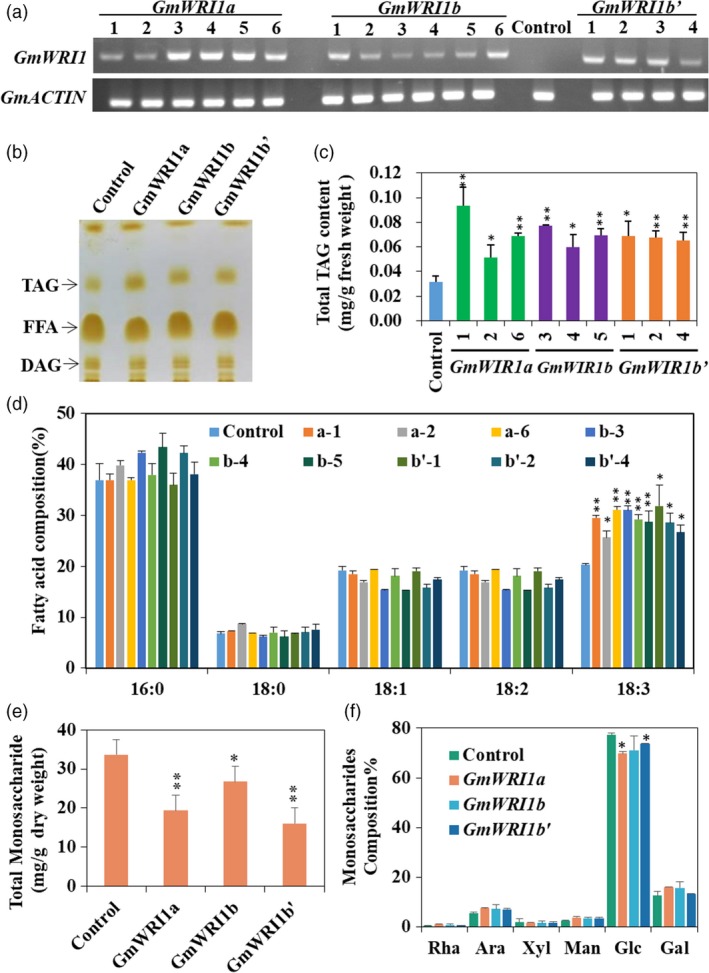
Ectopic expression of GmWRI1s in soybean hairy roots. Soybean hairy roots overexpressing GmWRI1a, GmWRI1b or GmWRI1b’ were analysed for TAG production, with GFP‐expressing hairy roots as a control. Total neutral lipids were extracted and separated by thin‐layer chromatography (TLC) for GC analysis of FA contents and compositions. (a) qRT‐PCR verification of GmWRI1s expression in soybean transgenic hairy root lines. (b) TLC analysis of neutral lipids extracted from transgenic hairy root lines. (c) Total TAG contents in hairy root lines overexpressing GmWRI1a, GmWRI1b, GmWRI1b’ or GFP. (d) TAG FA compositions in hairy root lines. (e) Total soluble sugars in transgenic hairy root lines. (f) Sugar compositions from hairy roots expressing GmWRI1s and GFP (control). Three representative lines from more than 10 transgenic hairy root lines were analysed. All data are expressed as means ± SD (n > 4). *P < 0.05 and **P < 0.01 by Student’s t‐test for significant difference.
Figure 3.
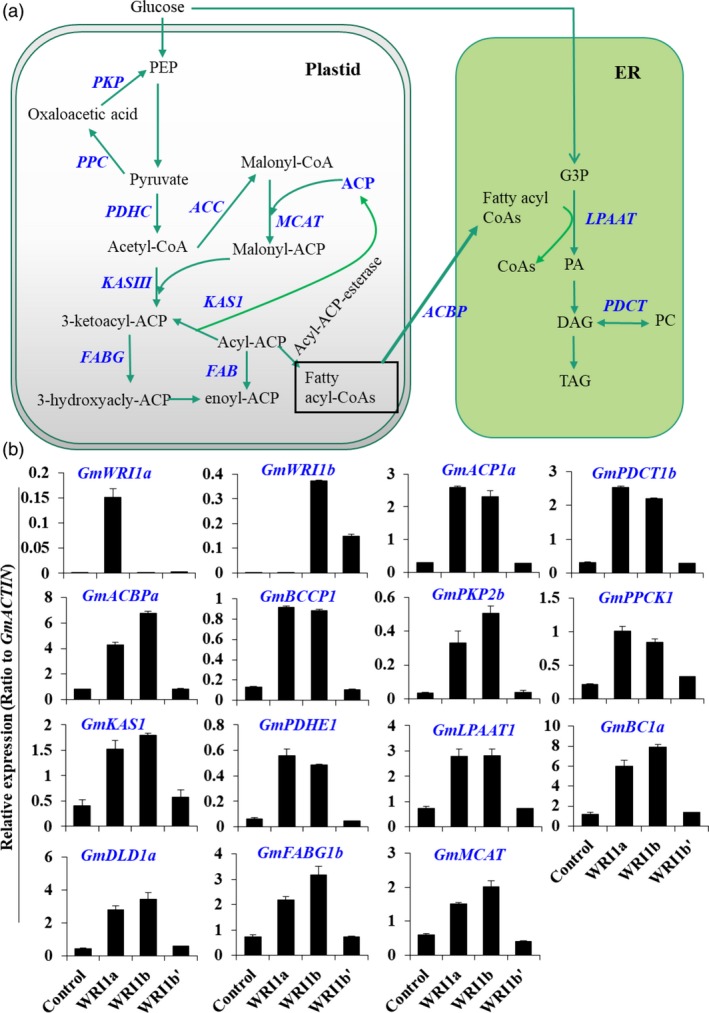
Up‐regulation of metabolic genes by GmWRI1a overexpression in hairy roots as revealed by RNA‐Seq analysis. (a) Schematic display the GmWRI1‐up‐regulated pathways and metabolic genes involved in the plastidic glycolysis, FA biosynthesis and ER TAG biosynthesis according to transcriptome data. GmWRI1a transgenic hairy roots were used for RNA‐Seq analyses, with GFP‐expressing hairy roots as controls. All up‐regulated genes compared with GFP control were marked in blue. (b) qRT‐PCR verification of transcript levels of GmWRI1‐targeted genes involved in glycolysis, FA synthesis and TAG production were analysed on transgenic hairy root lines overexpressing GmWRI1a, b or b’. Data are from three biological replicates and expressed as means ± SD. Two‐tailed Student's t‐test, *P < 0.05 and **P < 0.01 by two‐tailed Student's t‐test for significant differences.
GmWRI1s regulate target genes in plastid glycolysis, FA and TAG biosynthesis in hairy roots
We then conducted RNA‐Seq analysis on these transgenic hairy roots to detect potential changes in global gene expression as affected by ectopic expression of GmWRI1s. As compared with control hairy roots, overexpression of GmWRI1a up‐regulated more than 169 genes, but down‐regulated 93 genes at significant levels (Log 2 > or < 1 as cutoff, P < 0.001). Except for some genes related to cell wall expansion, cell wall polysaccharide hydrolysis, dehydration and water stress, several genes involved in the late steps of plastid glycolysis and early steps of FA and oil biosynthesis were up‐regulated (Table S2, S3). This up‐regulation of genes was further confirmed by qRT‐PCR (Figure 3b and Figure S7). Three genes encoding PKP, PPC and PDHC (composed of PDHE, BC1 and DLD) involved in late steps of glycolysis in soybean plastids were up‐regulated in GmWRI1 transgenic hairy roots (Figure 3). The genes typically involved in early steps of FA biosynthesis in soybean seeds, such as those encoding KAS, BCCP, ACP, ACBP, LPAAT and PDCT, were also up‐regulated by ectopic expression of GmWRI1a. Compared with GmWRI1a, the transgenic hairy roots expressing GmWRI1b displayed significant up‐regulation of similar sets of genes in plastid glycolysis and FA biosynthesis, and TAG biosynthesis in the ER (Figure 3b).
As shown in Figure 4a–i, all forms of GmWRI1a, b and b' are nucleus‐located. To identify genes targeted directly by GmWRI1s, we identified one or more AW box DNA sequences at the promoter regions of glycolysis and FA biosynthesis genes, such as PKP, PPC, PDHC,KAS, BCCP, ACP, ACBP, LPAAT and PDCT (Figure 4j,Table S4). We used the yeast one‐hybrid assay to examine the binding activities of the WRI1 transcription factors to these target genes. We observed that GmWRI1a, b and b' bound to the promoter DNA sequence of all the above genes (Figure 4k). However, our in planta transactivation assays using a dual‐luciferase activation system in Arabidopsis protoplasts indicated that GmWRI1a, but not b and b', could efficiently activate these promoters with the exception of PKP (Figure 4l,m). These results suggest that they have different target specificities in planta.
Figure 4.
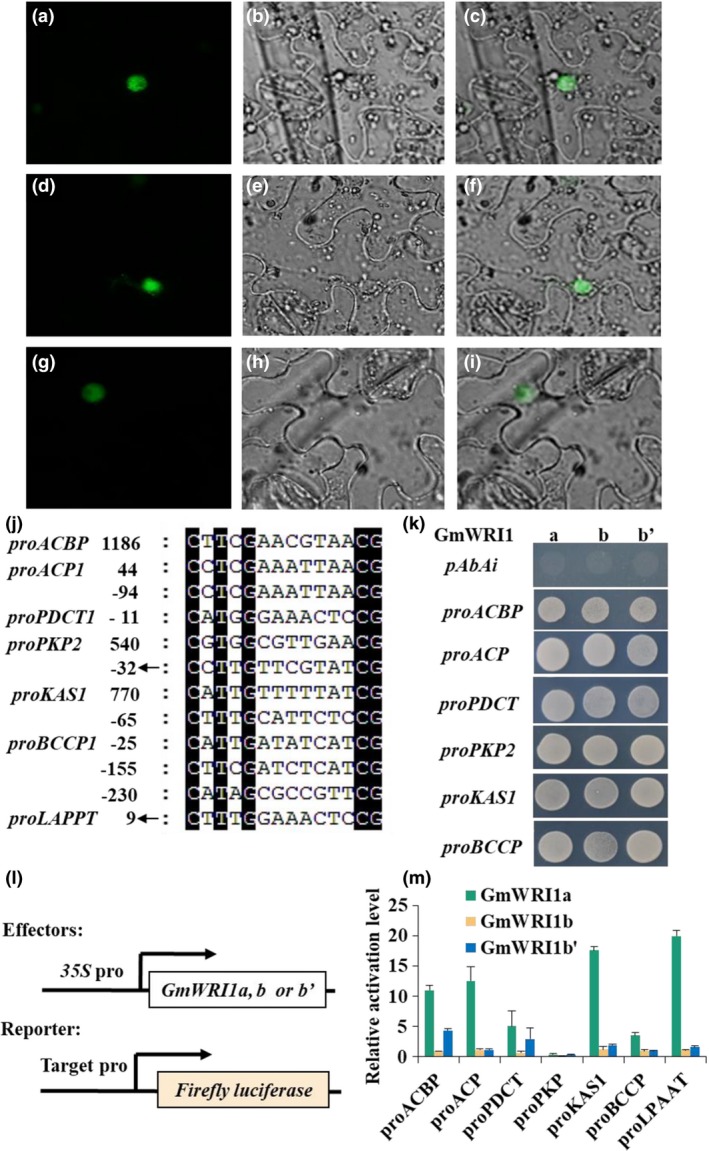
Binding and transactivation of the promoters of target genes by GmWRI1s. (a–i) Subcellular localization of GmWRI1s. The fluorescence (a), bright field (b) and merge (c) images of GmWRI1a‐GFP; the fluorescence (d), bright field (e) and merge (f) images of GmWRI1b‐GFP; the fluorescence (g), bright field (h) and merge (i) images of GmWRI1b’‐GFP. Bars = 30 μm. (j) AW boxes present in genes involved in glycolysis and FA synthesis in plastids. The numbers indicate the positions of the first nucleotide C in the AW box, taking the first base of the most abundant transcription start site (TSS) as +1. Arrows indicate the complementary sequences. (k) Binding of GmWRI1s to the target gene promoters in yeast one‐hybrid assays. The transformants harbouring the empty pAbAi and pGADT7‐GmWRI1 were used as a negative control. (l) Constructs for effecter expression and promoter‐driven reporter gene system for transactivation assays. (m) Transactivation of target genes by GmWRI1s in Arabidopsis leaf protoplasts with renilla luciferase activity as reference. All data are from three biological replicates and expressed as means ± SD.
Overexpression of GmWRI1s results in increased nodule numbers
Both GmWRI1a and GmWRI1b were strongly expressed in soybean nodules as indicated by GUS activity in proGmWRI1a::GUS and proGmWRI1b::GUS transgenic hairy roots (Figure 5a). Transcripts of GmWRI1a and b were up‐regulated upon rhizobia infection after 24 h (Figure 5b). To examine the functions of GmWRI1s in symbiosis, we generated chimeric soybean plants with hairy root lines overexpressing GmWRI1a and GmWRI1b driven by the CaMV35S promoter (GmWRI1‐OE), as verified by using qRT‐PCR (Figure 5c). The GmWRI1a‐ and GmWRI1b‐OE hairy roots produced more nodules (Figure 5d). Hair roots of the GUS vector control produced 15.3 nodules, but GmWRI1a‐OE and GmWRI1b‐OE hairy roots produced on average 24.5 and 26.4, respectively, nodules per gram fresh weight of roots (Figure 5e). GmWRI1‐OE soybean nodules were further examined for the expression of nodulation‐related genes. Transcript levels of a subset of early nodulation genes, including these encoding Nod Factor Receptor 1 (NFR1) kinase, Symbiosis Receptor‐like Kinase (SYMRK), Calcium‐/Calmodulin‐dependent protein Kinase (CCaMK), Nodulation Signalling Pathway1/2 (NSP1/2) and Nodule Inception (NIN), were elevated in both GmWRI1a‐ and b‐OE lines (Figure 5f and Figure S8). GmWRI1a‐ and GmWRI1b transcripts increased markedly over the nodule development stages (Figure S9). Their expression levels peaked at mature nodules with the highest nitrogen fixation activity and then quickly decreased to low levels.
Figure 5.
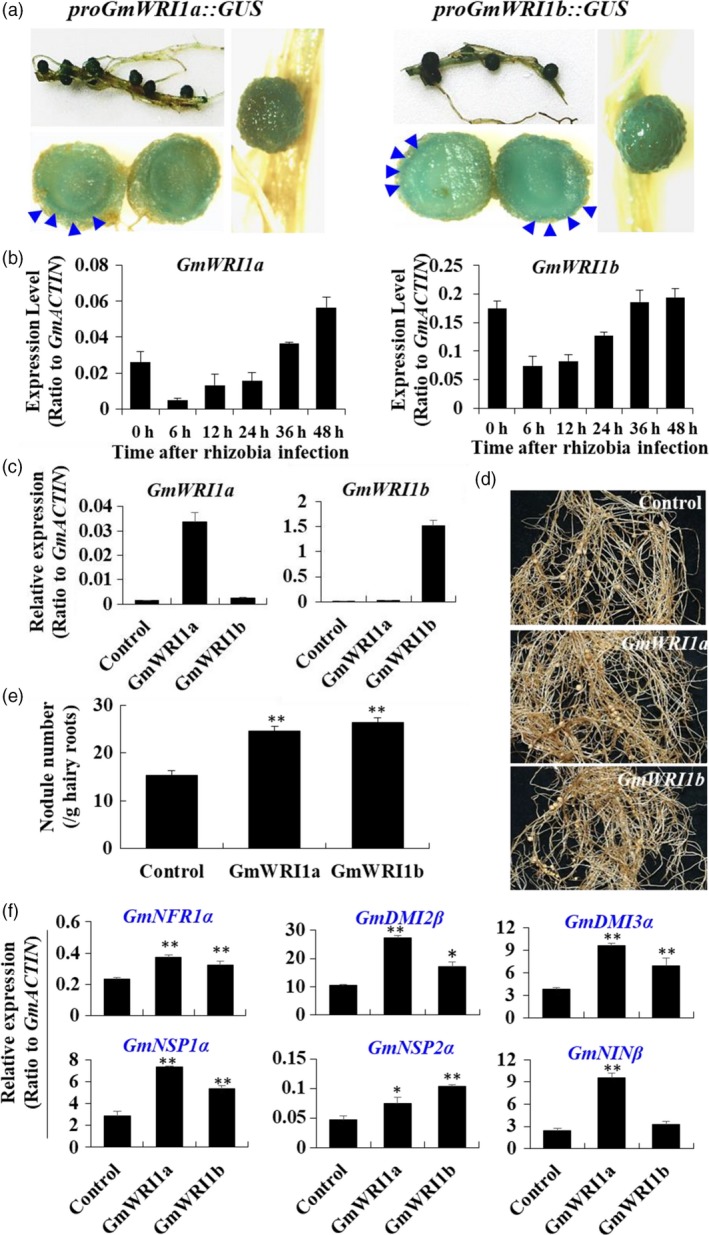
GmWRI1 modulates soybean nodulation. (a) GUS activity assay in root nodules of proGmWRI1a::GUS and proGmWRI1b::GUS transgenic hairy roots after inoculated with rhizobia for 28 days. (b) Expression of GmWRI1a and b in roots upon the infection with rhizobia. (c) qRT‐PCR verification of GmWRI1 transcript levels in soybean nodules. (d) Nodulation photos of 35Spro:GmWRI1a, 35Spro:GmWRI1b and GFP (as a control) transgenic hairy roots after inoculated with rhizobia for 28 days. (e) Quantification of the nodule numbers in these transgenic lines. (f) Examination of early nodulation genes in these transgenic lines. Data were expressed as means ± SD from at least three biological duplicates. *P < 0.05 and **P < 0.01 by Student’s t‐test for significant difference.
The role of GmWRI1s in nodulation was further tested by analysis of carbohydrates and lipids from both hairy roots and nodules of GmWRI1‐OE hairy roots. Similar with above results in hairy roots, the contents of TAG and MAG in GmWRI1‐overexpressing nodules tended to increase, more significantly in GmWRI1b‐OE lines (Figure 6a–d and Figure S9). Total PC and monogalactosyldiacylglycerol (MGDG) contents in the nodule of GmWRI1 a and b‐OE lines were significantly higher than those in the control nodules (P < 0.05 and 0.01, respectively), consistent with the up‐regulation of their biosynthetic genes GmLPCATs and GmMGD2s by nearly 2‐folds (Figure S9). Consistently, the content of free FAs was increased (Figure S9). GmWRI1a and b‐OE lines were lower in soluble sugar and starch contents in nodules than the GUS control (Figure 6e,f).
Figure 6.
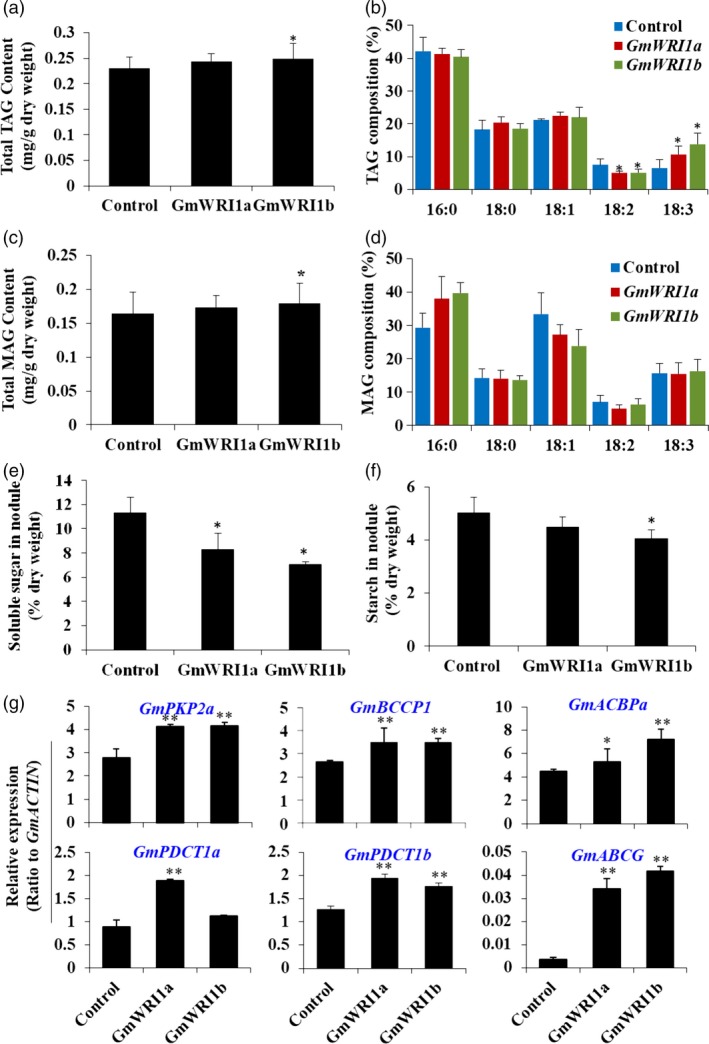
Effects of GmWRI1s on carbon partitioning and lipid biosynthesis in soybean nodules. (a–d) TAG and MAG contents and their FA compositions in nodules from transgenic hairy roots overexpressing GmWRI1a and b. (e–f) The contents of soluble sugars and starches in transgenic nodules overexpressing GmWRI1a or GmWRI1b, as compared with GFP (as a control). (g) Expression of glycolysis and lipid biosynthesis genes expression in GmWRI1s nodules expressing GmWRI1a, GmWRI1b, or GFP (as a control). Expression levels were expressed as means ± SD from three biological replicates. *P < 0.05 and **P < 0.01 by Student’s t‐test labelled for significant difference.
In addition, we suppressed GmWRI1 expression by generating GmWRI1‐RNAi soybean hairy roots for nodulation assay (Figure 7). GmWRI1RNAi knockdown lines produced only about two‐third nodules of those of the GUS control (Figure 7a,b). Metabolite analyses showed that while MAG contents were affected significantly (Figure 7c), TAG contents were reduced in GmWRI1RNAi nodules compared with the GUS control (Figure 7d). Both starch and soluble sugar contents in nodules of GmWRI1RNAi lines were significantly higher than the GUS control, suggesting a reduced starch breakdown and glycolysis (Figure 7e,f). The early nodulation genes were also mostly down‐regulated in GmWRI1RNAi nodules compared with the GUS control (Figure 7g).
Figure 7.
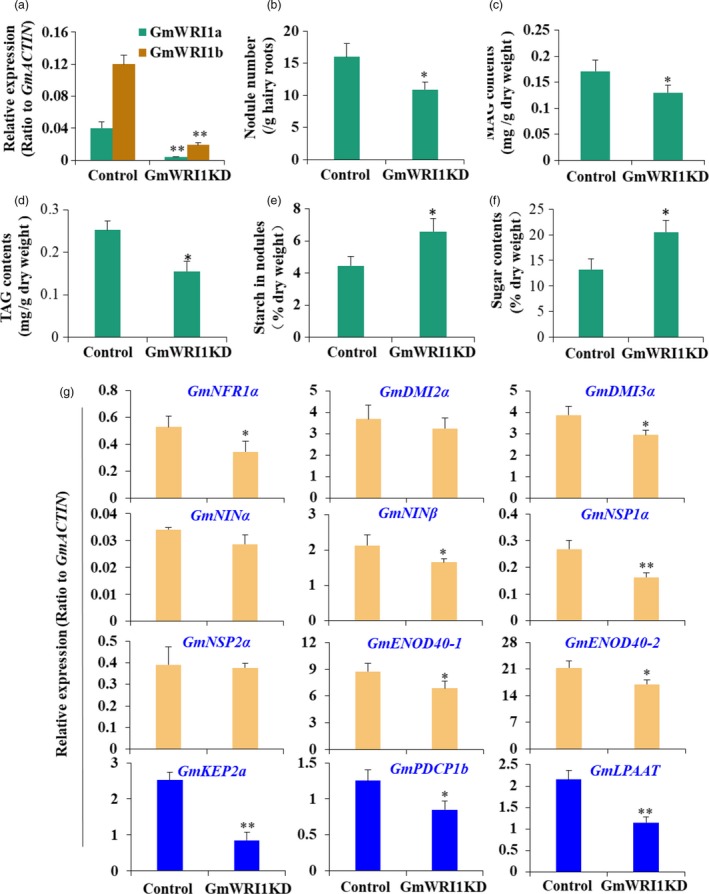
Suppression of GmWRI1 expression affected nodulation and carbon partitioning in soybean nodules. The GmWRI1 knockdown hairy roots were generated by GmWRI1RNAi repression, and used for nodulation assay, metabolite and gene expression analysis. (a‐f) Characterization and metabolite analyses of GmWRI1RNAi knockdown hairy roots. qRT‐PCR confirmation of GmWRI1a and b expression (a), nodule numbers (b), the contents of TAG (c), DAG (d), soluble sugar (e) and starch (f) of GmWRI1RNAi hairy root lines compared with GFP line (as a control). (g) Expression of nodulation genes, glycolysis and lipid biosynthesis genes in GmWRI1RNAi hairy root lines compared with GFP line (as a control). Expression levels were expressed as means ± SD from three biological replicates. *P < 0.05 and **P < 0.01 by Student’s t‐test labelled for significant difference.
Histochemical study on carbohydrate and lipid distributions in nodules showed that Nile red‐stained lipids and I‐KI‐stained starch were mostly in the vascular tissues around out layer of nodules, coinciding with tissue regions where GmWRI1a and b were expressed (Figure 8a, 5a). Sugars detected by ruthenium red‐staining resided mostly in the central nodules where bacteroides stayed (Figure 8a). Quantification of soluble sugars and starch indicated that nodules had significantly lower starch than did roots, indicating that starch was hydrolysed into sugars triggered by GmWRI1 (Figure 8b). By contrast, soluble sugars in nodules were about 2.5‐fold higher than in roots, indicating that nodule development required more sugars, which could be generated from starch degradation (Figure 8b). Together, these data from altered GmWRI1 expression indicated that WRI1 plays a role in promoting glycolysis and glycerolipid production in nodules.
Figure 8.
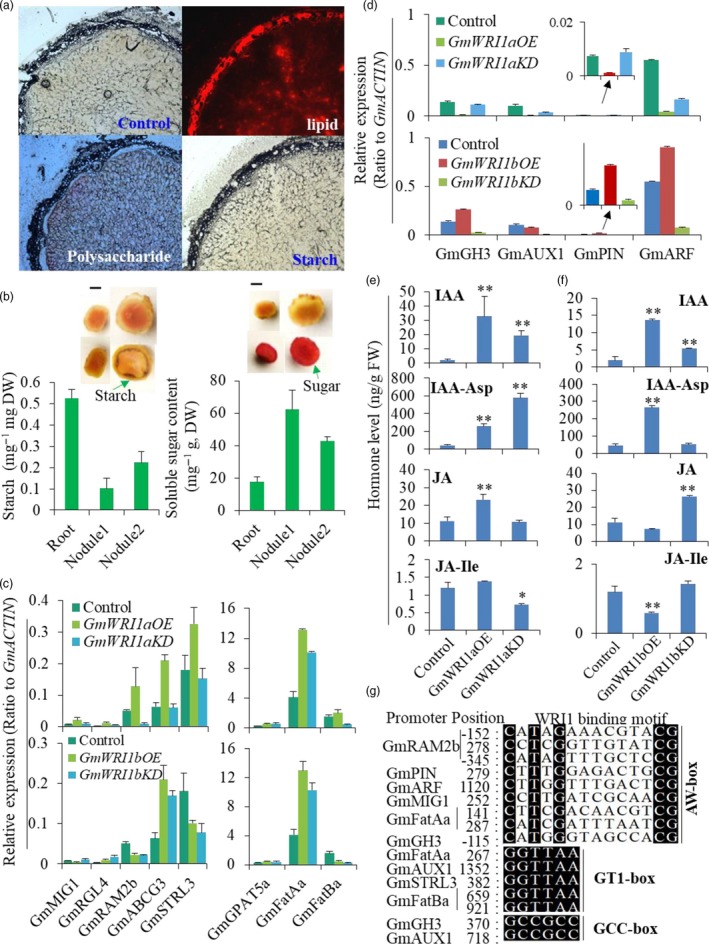
Effects of GmWRI1s on auxin and lipid biosynthesis in root nodules. (a) Distribution of lipids, polysaccharides and starch in nodules. Cross‐section of stage‐5 nodules was stained with Nile Red, ruthenium red and I2‐KI solutions, respectively. (b) Quantification of starches and total soluble sugars in roots and nodules at stages 1 and 2. (c, d) qRT‐PCR analysis of auxin transport and signalling genes in GmWRI1a‐ (c) or GmWRI1b (d)‐overexpression (OE) and ‐GmWRI1RNAi knockdown (KD) hairy root lines. (e, f) The IAA, JA and their conjugation level in GmWRI1a‐ (e) or GmWRI1b (f)‐ overexpression (OE) and ‐GmWRI1RNAi knockdown (KD) hairy root lines. (g) GmWRI1‐targeted genes containing WRI‐binding cis‐elements involved in auxin signalling or 2‐MAG synthesis and transport and alignment of their WRI‐binding motifs. Expression levels were expressed as means ± SD from three biological replicates. *P < 0.05 and **P < 0.01 by Student’s t‐test labelled for significant difference.
We further analysed the expression of soybean genes homologous to Medicago‐AMF symbiosis‐employed genes, such as FatB gene FatM, Gibberellic Acid Insensitive, Repressor of GAI, and Scarecrow (GRAS) TF Required for Arbuscular Mycorrhization 1 (RAM1), GPAT gene RAM2 and ATP binding cassette G‐type (ABCG) transporter STR1/2, in nodules at early‐stage nodules and roots. We observed that their soybean homolog genes RAM1a, RAM2b, FatMb and STR1a were up‐regulated in nodules compared with in roots, even at low expression levels (Figures S10–S12). The genes were expressed higher in GmWRI1a‐ or b‐OE hairy roots than in control included GmRAM2b, GmFatBa, GmFatBa, GmFatAa, GmSTR2b and GmSTR‐like 3 (GmSTRL3) (Figure 8c and Figure S10–S12). Other homolog genes, such as FatBs, Mycorrhiza‐Induced GRAS 1 (MIG1), and RGA‐LIKE1 (RGL1), GmGPAT4,6s, as well as GmABCG transporter homolog genes, were up‐regulated in nodules by more than fivefold to 10‐fold higher than in roots (Figure S10–S12). We further examined the auxin‐related genes in GmWRI1‐KD and ‐OE hairy roots. GmWRI1a‐ or GmWRI1b ‐OE and GmWRI1RNAi had opposite effects on the expression of GmGH3, GmPIN, GmARF and GmAUX1 genes (Figure 8d). In addition, overexpression of GmWRI1b activated whereas that of GmWRI1a repressed the expression of these genes (Figure 8d). Hormone profiling on these transgenic hairy roots showed that GmWRI1b‐OE lines had increased, whereas GmWRI1b‐KD lines had decreased levels of IAA and IAA‐Asp conjugates, which could be the results of GH3.3 enzymes (Figure 8f). GmWRI1a‐OE lines had increased IAA and decreased IAA‐Asp conjugates level (Figure 8e). Interestingly, the effects of GmWRI1b‐OE or GmWRI1RNAi on JA and JA‐Ile conjugate contents were opposite to their effects on IAA and its Asp conjugate contents (Figure 8f), indicating that GmWRI1a and b can modulate GH3 family gene expression (Figure S13, S14). Indeed, genes that were also up‐regulated in GmWRI1bOE lines own one or more AW boxes or other WRI1‐binding cis‐elements, such as AW, GT1 and GCC boxes in their promoter regions (Figure 8g and Figure S10a,b).
Discussion
Results of this study indicate that soybean GmWRI1s are TFs governing oil biosynthesis by transactivation of several key genes involved in late glycolysis and early FA biosynthesis likely by binding to an AW box‐cis‐element in their promoter regions. GmWRI1s also activate genes that are required for ER‐based TAG assembly, and all these targets have AW cis‐elements in their promoters. While the role of WRI1s in storage lipid production has been extensively studied, our results show that GmWRI1s have a legume‐specific feature in nodules, where GmWRI1 is involved in carbon partitioning by which host cells provide carbon and other sources to rhizobia bacteria. GmWRI1‐OE promoted nodulation with enhanced nodule numbers, likely through promoting starch breakdown and glycolysis, and glycerolipid biosynthesis. The requirement of GmWRI1 for nodule growth and development was also reflected by that, GmWRI1a‐ and GmWRI1b transcripts continued increasing over nodule development and decreased till nodule starting senescing. Our data support that GmWRI1‐regulated MAG and other lipid production are required for soybean nodulation, similar to MAG in legume–AMF symbiosis, which also involves a Lotus WRI1‐ homolog CBX1 or Medicago WRI5a‐ (Jiang et al., 2018; Luginbuehl et al., 2017; Xue et al., 2018). In addition, we observed different functions of GmWRI1a and b in modulating auxin‐related genes, and the AS form of GmWRI1b did not completely lose its function.
Soybean GmWRI1s have a conserved function in glycolysis, FA biosynthesis and TAG accumulation
GmWRI1a, b and b' restored to a different degree the defects in FA synthesis and development in the Arabidopsis wri1‐1 mutant, and all showed TF activity. Different from rapeseed BnWRI1 and ZmWRI1s, overexpression of GmWRI1s re‐programmed carbon fluxes from storage carbohydrates to TAG and protein in both soybean hairy roots and Arabidopsis (Shen et al., 2010; Yang et al., 2015). Gene expression profiling and quantitative reverse transcription‐PCR data allowed us to identify a set of putative targets of GmWRI1s, including genes in late glycolysis and early FA biosynthetic pathways, as was shown for other WRI1s (Baud et al., 2007; Maeo et al., 2009; Shen et al., 2010). Unlike ZmWRI1 and WRI1s from other plants (Shen et al., 2010), overexpression of GmWRI1a and b also up‐regulated genes involved in the late steps of TAG assembly in the ER, such as PDCT and LPAAT. Furthermore, our study provides evidence that TAG synthesis in GmWRI1 transgenic tissues likely activated acyl editing and the production of more highly unsaturated FA‐containing TAGs because significant up‐regulation of LPAAT and PDCT was observed. So far, it has not been reported for any other plant that WRI1 directly targets genes involved in TAG assembly in the ER (Manan et al., 2017b). Transgenic expression of GmWRI1 in Arabidopsis increased seed oil content with changes in the FA composition of TAG, suggesting a potential use for oil engineering.
GmWRI1a, b and b' differently regulate target metabolic genes
Unlike GmWRI1a, which fully activates target genes in Arabidopsis protoplasts, GmWRI1b and b' may require legume‐specific partners in forming a transcriptional complex to activate target genes. Thus, GmWRI1b and b' could not activate promoters of genes that were targeted and activated by GmWRI1a in promoter activation assay. The few amino acids in GmWRI1b' different from GmWRI1b may be required for activity, since GmWRI1b′ is less active as compared to GmWRI1a and b in soybean hairy roots. The less active AS forms may attenuate the activation activity of full‐length GmWRI1s. Overexpression of GmWRI1a, b and b' had activated late glycolysis and early FA biosynthesis and TAG biosynthesis in both Arabidopsis seeds and soybean hairy roots to different extents. GmWRI1a, b and b' also displayed different transactivation activities on target genes in vitro and in vivo, suggesting that differences may be due to the formation of dimers of GmWRI1b and b' with other partners. Functional redundancy of two GmWRI1 homologs and the diversified properties of the AS forms may reflect the regulatory flexibility in soybean. GmWRI1s are highly expressed in the nodule, indicating roles of GmWRI1s in regulating soybean–rhizobia symbiosis. GmWRI1a has an overall lower expression level and different tissue expression patterns compared to GmWRI1b, together with different transactivation activities, indicating that they may differently function in seed and nodule FA biosynthesis. Both GmWRI1a and GmWRI1b are located in the nucleus and directly bind to the AW box at proximal upstream regions of genes involved in FA biosynthesis and TAG assembly, and we observed opposite regulatory roles of GmWRI1a and b in modulating auxin‐related genes and root hormone levels, perhaps indicating their roles in balancing these hormones in roots, which are finely tuned for nodule initiation and formation (Oldroyd and Downie, 2008). Molecular and genetic analyses identified WRI1s as targets of LEC2 and ABI3b in Arabidopsis and soybean (Baud et al., 2007; Manan et al., 2017a; Zhang et al., 2017a). Although rhizobial infection up‐regulated both GmWRI1a and b, it is still not clear how this happened. We found many GRAS TF‐binding motifs in GmWRI1a and b promoters, and thus, we proposed that NSP1 or NSP2 might play a role in Rhizobia infection‐mediated up‐regulation of GmWRI1s, perhaps similar to AMF‐activated RAM1 up‐regulation of WRI5a during Medicago–AMF interaction (Jiang et al., 2018).
Distinguishable functions of the AS forms of GmWRI1 in oil production
AS usually occurs at genes under highly active transcription in certain tissues. It is regarded as an important form of regulation for many functionally important genes since it increases the functional diversity of transcripts in eukaryotic cells (Reddy et al., 2013; Staiger and Brown, 2013). TFs,such as CIRCADIAN CLOCK‐ASSOCIATED1 (CCA1) (Seo et al., 2012) and Jasmonate ZIM‐domain (JAZ) (Zhang et al., 2017b),can generate inhibitory AS forms to regulate the full‐length transcript’s function or stability. An Arabidopsis alternative splice transcript of AtWRI1 was characterized as a less functional WRI1 form (Ma et al., 2016). We determined that the AS forms lack a short 9‐bp exon encoding amino acids ‘VYL,’ as compared to the full‐length WRI1s. However, different from the less active AtWRI1 regarding activation of oil accumulation, GmWRI1b' was less active in stimulating oil biosynthesis in both transgenic Arabidopsis and soybean hairy roots, and it exhibited decreased activity in transactivation assay in Arabidopsis protoplasts. GmWRI1a and b also displayed different roles in transactivation of target genes in the Arabidopsis protoplasts, and regulation of auxin‐related genes in their OE or GmWRI1RNAi knockdown (KD) hairy roots, which are coinciding with their different tissue expression patterns. Although GmWRI1b’, which misses the conserved VYL, still partially restored the oil content in Arabidopsis wri1 mutant seed, the GmWRI1b'‐complemented seedlings did not resemble those for the GmWRI1a‐ and GmWRI1b‐complemented lines under certain conditions, suggesting a compromised function of GmWRI1b'. Transgenic soybean hairy roots overexpressing GmWRI1s all showed enhanced glycolysis and FA biosynthesis and TAG production. However, GmWRI1b' displayed a reduced ability to activate glycolysis and FA biosynthesis genes, both in hairy roots and in a transactivation assay in Arabidopsis protoplasts. This is in part consistent with a previous report that Arabidopsis WRI1‐exon 3 with the same deletion of VYL completely lost activity in transactivation of target genes and complementation of the Arabidopsis wri1 mutant (Ma et al., 2015). Since WRI1s usually function as a dimer, GmWRI1b' may attenuate GmWRIa and b activity by forming heterodimers with them. Nevertheless, the redundant GmWRI1s in the soybean genome may enhance their regulatory flexibility.
Legume‐specific functions of GmWRI1s in carbon partitioning in nodules
Legume–rhizobia interaction and symbiotic nodule growth are an energy‐demanding process, and the supply of carbons and other nutrients to the rhizobia in nodules critically affects nodule development and nitrogen fixation (Clarke et al., 2014; Udvardi and Poole, 2013). Source‐derived sugars are not the direct carbon sources utilized by rhizobia but have to be converted into dicarboxylic acids through glycolysis and the tricarboxylic acid cycle to serve as direct carbon sources (Clarke et al., 2014). It has been recently realized that multiple forms of carbons are likely used by rhizobia during legume–rhizobia symbiosis (Clarke et al., 2014; Udvardi and Poole, 2013). Emerging evidence shows that root biosynthesis and transport of FAs and lipids are essentially required for nodulation (Krishnan et al., 2016; Lakhssassi et al., 2017; Lei et al., 2014; Pii et al., 2009; Wang et al., 2014). However, the underlying mechanisms of these nutrient‐exchanging processes in host cells and rhizobia are not understood. Since GmWRI1 directly regulated late glycolysis and early FA biosynthetic genes in roots, these genes are critically activated to meet the metabolic and energy needs for early nodulation, the rhizobia infection‐induced up‐regulation of GmWRI1a and b genes may thus be required for regulating the process. GmWRI1RNAi hairy roots had reduced FA and MAG biosynthesis required for early rhizobia development in roots and resulted in reduced nodulation. On the contrary, GmWRI1a‐ and b‐OE lines had increased levels of FA and MAG, as well as other membrane lipids, and thus also gave more nodules. Recent studies revealed that the major carbon form legumes provide to AMF is 2‐MAG. A set of legume genes, including RMA1, which regulates WRI5a, GPAT, RAM2 and STR1/2, are required for successful legume–AMF symbioses (Keymer et al., 2017; Jiang et al., 2017, 2018; Luginbuehl et al., 2017). Recently, Lotus WRI1 homolog CBX1‐ and Medicago WRI5a‐regulated FA biosynthesis genes such as FatM, GPAT or STRs appeared to be required for successful legume–AMF symbioses (Jiang et al., 2018; Xue et al., 2018). Here, we show that transcript levels of soybean homologs, FatBa, FatBb, FatAa, RAM2b and STR2b, were markedly higher in nodules than in roots, and they were up‐regulated by GmWR1s (Fig S10‐S12).
Rhizobial lipochitooligosaccharides (Nod‐LCO)‐activated signalling pathways leading to nodulation shared several common components with mycorrhizal lipochitooligosaccharides (Myc‐LCO)‐activated signalling leading to AMF development, such as NSP1/2 (Heckmann et al., 2006). Myc‐LCO activation of gene expression required both RAM1 and NSP1 in presymbiotic transcriptional reprogramming (Hohnjec et al., 2015). As a central and specific regulator of root colonization by AMF, RMA1 and WRIs together regulate 2‐MAG biosynthesis and transport (Heck et al., 2016; Luginbuehl et al., 2017). Here, we show for the first time, to the best of our knowledge, that genes in soybean roots homologous to those required for Medicago‐AMF, including WRIs, FatBs, GPAT4s and GPAT6s, STR1/2, and three STR1‐like (STRL) ABCG transporter genes, were simultaneously up‐regulated in nodules and in GmWRI1‐OE hairy roots compared with control (Fig S10), suggesting that 2‐MAG synthesis is activated and MAG exported. Similar to Myc factors, we observed that Nod factor up‐regulated the GRAS TFs GmNSP1, MtMIG1 and MtRGL homolog genes (Fig S10; Gobbato et al., 2012; Heck et al., 2016). In roots, cutin monomers produced in Arabidopsis by GPAT4 and 6 (Yang et al., 2010) or in Medicago by RAM2 promoted hyphopodia formation in AMF (Luginbuehl et al., 2017). We posited that up‐regulation of these GPAT, STR and STR‐like ABCG genes could be responsible for synthesis and export of 2‐MAGs to the nodule surfaces for cutin synthesis. Alternatively, it could serve rhizobia in the bacteroides as precursor for glycerolipids, similar to those in Medicago‐AMFs (Luginbuehl et al., 2017; Wang et al., 2012). Consistently, we observed higher MGDG and PC contents and 16:0 FA‐acyl chains in an increasing content of MAG in GmWRI1‐OE lines (Fig S9).
Although studies had shown that the rhizobia can synthesize their membrane lipids, and the mutants defective in synthesis of PC or PE partly impaired their symbiotic nodulation with legume hosts, the membrane lipids in rhizobia bacteria, such as PC and galactolipids, are partly depending on host membrane lipid turnovers (Gaude et al., 2004; López‐Lara et al., 2003). The metabolically active symbiotic space of soybean nodules, periplasm, was found to contain almost equal amounts of proteins from B. diazoefficiens and host cells, both contributing to the biosynthesis of fatty acids for bacteroides (Strodtman et al., 2017). Previous study has shown that the FA, phospholipid, DGDG and MGDG biosynthesis in host root cells and their transport were required for nodulation (Gaude et al., 2004; Krishnan et al., 2016; Lakhssassi et al., 2017; Lei et al., 2014; Pii et al., 2009; Wang et al., 2014). Our study here showed that GmWRI1s was involved in the regulating the biosynthesis of FA, and the membrane and neutral lipids for nodulation. Although it is still not clear exactly how these lipids were used by rhizobia for nodulation, the available pieces of evidence point to carbon supplying for symbiosome development, rhizobia growth and nutrient exchanges in bacteroids for nitrogen fixation. Similar to RAM1 regulation of WRI5a, the promoters of GmWRI1a and b also contain many GRAS‐binding motifs. Soybean nodules did not express wax synthesis genes, but ABCG11‐like wax transporter‐encoding genes were up‐regulated in nodules compared to those in roots (Figure S10, S11, S15).
Upon rhizobia infection, sucrose synthase and transporter genes were markedly induced, indicating that nodulation requires photosynthesis‐derived carbohydrates (Baier et al., 2007; Breakspear et al., 2014). As a key regulator of glycolysis and FA biosynthesis, GmWRI1s were also induced by rhizobia infections and played a key role in regulating carbohydrate partitioning or modifying auxin signalling during nodule development. GmWRI1‐promoted starch breakdown and further glycolysis, as well as FA biosynthesis, provided carbon sources for rhizobia proliferation and growth and nodule development.
GmWRI1 modulate soybean nodulation
GmSACPD‐C mutant soybean plants developed abnormal nodules with defective nitrogen fixation (Krishnan et al., 2016; Lakhssassi et al., 2017). Transgenic soybean roots with a silenced plastid GmACP showed reduction in 16:0 and 18:0 contents, accompanied by a reduced nodule number compared to the control (Wang et al., 2014). Furthermore, nodule‐specific lipid transfer proteins participated in transport of plant‐synthesized lipids to the symbiosomal membrane and were essential for nodule organogenesis (Lei et al., 2014; Pii et al., 2009). These studies support that legume root cells indeed synthesize and provide lipids to rhizobia inside symbiosomes of nodules, and these are essential for nodulation. Nodules, particularly the inner parts of the nodules where rhizobia are present, require carbon sources for bacterial growth, including the biosynthesis of symbiosomal membrane and endomembrane expansion, as proposed in Figure S15. We showed that overexpression of GmRWI1 promoted soybean nodulation by increasing the nodule number, whereas suppression of GmWRI1s reduced the nodule numbers. The data suggest that GmWRI1 plays an important role in nodulation, possibly through affecting two aspects of nodulation as described below.
First, GmWRI1s directly regulate late glycolysis and early FA biosyntheses for metabolic and energy needs in nodulation, the rhizobia infection up‐regulates GmWRI1a and b genes may thus be required for this purpose. GmWRI1RNAi hairy roots had reduced FA and MAG biosynthesis, followed by a reduced nodulation, whereas GmWRI1a‐ and b‐OE lines had enhanced FA and lipid biosynthesis capability, and thus also had increased nodulation numbers. Inoculation with B. japonicum altered the fatty acid biosynthesis in soybean root hairs (Brechenmacher et al., 2010), and de novo phospholipid biosynthesis was essential for the establishment of infection thread and symbiosomal membranes, which originate from the host infection‐thread membrane, ER and de novo synthesis of membranes (Krishnan et al., 2016). Studies on M. truncatula nodulation indicated that the infection thread formation and endocytosis of rhizobium in ZII requires efficient starch breakdown and glycolysis for production of carboxylic acids, FAs and membrane lipids such as MGDGs and PCs to support rhizobia growth and symbiosome or endomembrane expansion (Gaude et al., 2004; Larrainzar et al., 2014; Ogden et al., 2017). Likely, starch breakdown, glycolysis, FA, MAG, MGDG and phospholipid biosynthesis are essential for rhizobia development and soybean nodulation, and GmWRI1 is involved in the regulation of these processes (Figure S15). We observed that the outer parts of nodules contain more starch than roots and require WRI1‐activated glycolysis and lipid biosynthesis to provide carbons for the synthesis of MGDGs and PCs, which are the primary components of symbiosomal membranes in the inner nodules, where bacteroid differentiation and nitrogen fixation take place (Gaude et al., 2004).
Second, WRI1 was reported to affect primary root growth and auxin homeostasis in Arabidopsis roots, by specifically modifying GH3.3 and auxin carrier‐protein PIN genes (Kong et al., 2017). Auxin biosynthesis and gradient distribution in roots and nodules, and signalling are critically required for nodulation development (Roy et al., 2017). GmWRI1a and b mutants also displayed different expression patterns of auxin‐related genes, particularly GH3.3, which were consistent with IAA, IAA‐Asp, JA and JA‐Ile contents in their ‐OE or ‐ KD hairy roots. These data strongly support that GmWRI1a and b indeed modulated auxin and JAs and thereby affected nodulation in complicated ways (Figs 8e,f, S13‐14). The opposite regulatory effects of GmWRI1a and b on these genes and hormones may indicate their roles in balancing these hormones, which are finely tuned for nodule initiation and formation (Oldroyd and Downie, 2008). Thus, GmWRI1s may corporately modulate soybean nodule organogenesis through affecting auxin and other hormone signalling and carbohydrate and lipid metabolic pathways, which adds another layer to WRI1 regulatory mechanisms (Roy et al., 2017).
In conclusion, our study indicates important roles of GmWRI1s in regulating carbohydrate partitioning and lipid biosynthesis, as well as auxin signalling, in soybean developing seeds and in root–rhizobia interaction for nodulation. This provides new insights into the not only complex roles of various GmWRI1 isoforms and alternative splicing forms, but also the pleiotropic effects of GmWRI1s on soybean seed oil and protein biosyntheses and root nodulation. The study, thus, expands our understanding of the diverse functions of WRI1s in the regulation of carbon partitioning and modulating of auxin and other hormone signalling, which could guide future investigation into the other roles these TFs play in plants.
Materials and methods
Plant materials
Soybean (Glycine max L.) seeds (“Tianlong No. 1” a soybean cultivar) were germinated in three‐gallon pots containing soils. Seedlings were grown in a growth chamber (26/20°C day/night temperature, photoperiod of 14/10 h, 800 μmol m‐2 s−1 light intensity and 60% humidity). Roots, leaves, stems, flowers, seeds and drupes at different developmental stages were harvested. Arabidopsis thaliana wild‐type and the wri1 mutant were used in this study. Arabidopsis and Nicotiana benthamiana plants were grown in a growth chamber at 22°C with a 16‐h light/8‐h dark photoperiod (Li et al., 2016). Arabidopsis transformation, plant growth on plates, seed surface sterilization and transgenic seedling selection were performed as described previously (Haq et al., 2017).
Plasmid construction
The open reading frames (ORFs) of GmWRI1 (including AS GmWRI1 variants) were amplified by PCR using primer pairs as listed in Table. S1 and cloned into a T‐easy vector and sequenced (Promega). The genes were then subcloned into destination vectors, such as overexpressing vector pB2GW7 or promoter::luciferase reporter vector p2GW7, or RNA interference (RNAi) repression vector pB7GWIWGII for transgenic studies by using BP and LR recombinases (Life Technologies, Carlsbad, CA, USA). The promoters (about 1.5–2k bp) of putative target genes, including proACBP, proACP, proPDCT, proPKP2, proKAS1, proBCCP and proLPAAT containing at least one AW box, were amplified from soybean genomic DNA using their corresponding primers (Table S1) and subcloned into p2GW7 and to produce promoter::luciferase reporter constructs. The overexpression constructs were transformed into Agrobacterium rhizogenes strains K599 and GV3101. Resulting constructs were used for transformation of soybean hairy roots and Arabidopsis wri1 mutant and wild type. For GmWRI1a and GmWRI1b promoter analysis, a 1.5‐kb promoter region was amplified and subcloned into pDONR221 and subsequently pHGWFS7 with a β‐glucuronidase (GUS) reporter by using BP and LR recombinases (Life Technologies).
Yeast one‐hybrid and transactivation assay
The full‐length cDNA of GmWRI1s was cloned by using a pair of cloning primers (Table S1), and then, DNA fragments were ligated into pGADT7 vectors that were digested with EcoRI and BamHI. The promoters of target genes, including proACBP, proACP, proPDCT, proPKP2, proKAS1 and proBCCP, were cloned with primers listed in Table S1, and these promoters were ligated to the yeast vector pAbAi to generate MatchmakerTM Gold Yeast One‐Hybrid Library screening system. Y1H Gold strain was used for transformation of above constructs for one‐hybrid assays with URA3 as a selectable marker. The positive transformants were further texted on YNB medium lacking uracil and leucine and containing 600 ng/mL Aureobasidin A (AbA). Empty vector was used as a control for excluding the self‐activation and false‐positive results. Promoter transactivation was assayed as described previously (Li et al., 2016). The Dual‐Luciferase Reporter Assay Kit (Promega, Madison, WI, USA) was used to quantify the luciferase activities using a microplate luminometer according to the manufacturer’s instructions.
Subcellular localization
The ORFs of GmWRI1s were cloned into pK7WGF2 in frame fusion at the N‐terminus of GFP by using the Gateway recombination cloning. The construct containing GmWRI1s‐GFP was transformed into A. tumefaciens stain GV3010. The agrobacteria transformants were infiltrated into N. benthamiana leaves and incubated for 4 days before the leaves being used for observation of the green fluorescence protein signals, as described previously (Li et al., 2016).
Soybean hairy root transformation and nodulation assay
The transgenic chimeric soybean plants that are composites of wild‐type shoots and transgenic hairy roots were generated as described previously (Haq et al., 2017). In detail, A. rhizogenes strains K599 harbouring pB2GW7‐GmWRI1a or b for overexpression, or pB7GWIWGII‐GmWRI1a and b cDNA fragments for RNAi knockdown, or GUS were grown on LB‐agar medium at 28°C with spectinomycin and streptomycin as selection markers. The overnight agrobacteria cultures were used for transformation of soybean cultivar “Tianlong No. 1” grown in pots (10*3*10 cm) containing a 3:1 mixture of vermiculite and perlite and grown for 1 week (16 h of light, 25°C, and 50% RH). The one‐week‐old soybean seedlings were pounced and wounded at hypocotyls for incubating with A. rhizogenes harbouring target constructs for 24 h in high humidity. The infected seedlings were grown in autoclaved soils at 25°C for 1 week till hairy roots appeared on the wounding sites. About 1 week after hairy root emergence when the transgenic hairy roots were about to support the plants, the chimeric soybean plants were examined for transgene in hairy roots, and the main nontransgenic roots are removed, before being inoculated with rhizobia strain Bradyrhizobium japonicum strain USDA110 grown in the YMA on 28°C. The transformation frequency is usually around 85‐95% with a transgene positive frequency of ~ 70% within a short transformation period (Kereszt et al., 2007; Chen et al., 2016). The prepared rhizobia bacteria solution (OD600nm was adjusted at 0.8–1.0) was applied about 25 ml to each plant in soil. After 4 weeks of rhizobium application, the hairy roots and nodules were examined and collected for RNA and hormone analyses. For each binary vector including GUS control, at least three independent in vitro transformation experiments with at least 10 individual transgenic lines under the identical treatments and growth conditions were examined. We counted and calculated the nodulation rate data as nodule number per g roots for normalization purposes.
Quantitative RT‐PCR (qRT‐PCR) analysis of gene expression
Total RNA from tissues of soybean plants was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA) or RNA isolation kit (Biotech, Beijing, China) according to the manufacturer's instructions. For each sample, 10 μg of total RNA was digested with RNase‐free DNase I (Promega, Madison, WI) to remove genomic DNA contamination. After DNase I treatment, RNA concentration was determined again using a NanoDrop ND‐2000 UV spectrophotometer (Thermo Scientific, Wilmington, DE, USA). First‐strand cDNA was synthesized from 2 μg total RNA using the Superscript III first‐strand synthesis system (Life Technologies, Carlsbad, CA, USA). All cDNA samples were diluted 50‐fold in sterile water for qRT‐PCR. Gene‐specific primers are listed in Table S1. Soybean GmACTIN (Glyma.15g034000) was used as internal control. qRT‐PCR data were generated using an iQ5 Real Time PCR machine (Bio‐Rad) in 40 cycles (94°C for 30 s, 58°C for 30s and 72°C for 30 s).
Lipid analysis
For lipid analysis, soybean hairy roots, developing seeds and leaves were harvested and frozen in liquid nitrogen, and dry mature Arabidopsis seeds were directly used for lipid analysis. Frozen tissues were lyophilized prior to lipid analysis. Lipid analysis was performed as described previously (Chen et al., 2016) with slight modifications. Briefly, total lipids extracted from Arabidopsis rosette fresh tissues and soybean hairy roots were separated on a thin‐layer chromatography (TLC) plate (SIL GF254, 0.25 mm) with a developing solvent consisting of hexane/diethyl ether/acetic acid (80:20:1, v/v/v). TAG bands were scrapped and transmethylated for analysis on GC (Agilent 7890A, Santa Clara, CA).
Analyses of soluble sugars, starches, membrane lipids and hormones in hairy roots or in nodules
Sugar contents and composition analysis on hairy roots were performed with GC‐MS as described previously (Peng et al., 2000). Starch was analysed by the I2‐KI methods. TAGs and phospholipids and galactolipids were analysed by using LC‐MS as described previously (Lu et al., 2018). Hormones, such as auxins—indole acetic acid (IAA) and jasmonate (JA), and their amino acid conjugates IAA‐asparagine (IAA‐Asp) or JA‐isoleucine (JA‐Ile) in hairy roots were analysed as described previously (Wang et al., 2017).
RNA‐Seq analysis of gene expression in transgenic hairy roots and developing nodules
RNA isolation and cDNA library construction for Illumina deep sequencing were conducted as described previously (Haq et al., 2017). Briefly, total RNA was isolated from transgenic hairy roots of at least 3 independent lines and developing nodules by using the TRIzol reagent (Invitrogen) or RNA kit (Biotech) following the manufacturer’s instructions. RNA integrity was confirmed by using the 2100 Bioanalyzer. RNA‐Seq, data processing, gene annotation and analysis were done as described previously (Haq et al., 2017). qRT‐PCR was used for further verification of target genes of GmWRI1 and to determine their expression patterns in developing nodules. Gene lists were constituted of differentially expressed genes presenting a minimum 1‐log2 Fold change.
Bioinformatics analysis
GmWRI1a (KY131950), GmWRI1b (KY131951) and GmWRI1b′ (KY131952) were analysed with other references with ClustalW (http://www.ebi.ac.uk/clustalw/). Phylogenetic trees were constructed using MEGA6 with the neighbour‐joining analysis examined by bootstrap testing with 1000 repeats.
Statistical analysis
All experimental data were obtained from three or more independent experiments with replicates and were analysed using Student's t‐test. The significant differences between two sets of data represent 95% confidence limits. For fluorescence imaging experiments, at least two repeat experiments were done, and representatives of photographs or images are shown.
Conflict of interest
The authors declare no conflicts of interest.
Author contributions
JZ conceived and designed the research. B.C. GZ, PL, JY and LG performed experiments and analysed data. BC and GZ conducted bioinformatics analyses. JZ, BC, CB and XW wrote the manuscript.
Supporting information
Figure S1 Phylogenetic (a) and gene structure analysis (b) and sequence alignment (c) of GmWRI1s.
Figure S2 Schematic diagram of the GmWRI1s and AtWRI1 predicted protein sequence illustrating the locations of known domains.
Figure S3 Public data for expression of soybean GmWRI1s.
Figure S4 Confirmation of transgenic atwri1–1 (a) and wild‐type (b) plants over‐expressing GmWRI1s.
Figure S5 Functional expression of GmWRI1a and b in Arabidopsis thaliana seeds.
Figure S5 Functional expression of GmWRI1a and b in Arabidopsis thaliana seeds.
Figure S6 Proteins content of wild type, atwri1 mutant and transgenic plant in mature seeds.
Figure S7 Quantitative PCR confirmation gene regulation by GmWRI1 in soybean hairy roots.
Figure S8 Effects of GmWRI1a and b overexpression on soybean nodulation and glycolysis and lipid metabolism.
Figure S9 Accumulation of lipids and expression patterns of soybean lipid synthesis‐related genes at soybean nodule developmental stages.
Figure S10 Identification and transcriptional analysis of FatB, GPAT, GRAS transcription factors RAM1 and ABCG transporter STR1/2.
Figure S11 The heat map analysis of tissue expression patterns of ortholog genes (a), other homology genes (b) for M. truncatula RAM1, RAM2, WRI1, STR1/2, and FatM.
Figure S12 The phylogenic tree (a) and heat map analysis (b) in roots and nodules of CUS (cutin synthase).
Figure S13 Expression level of GmGH3, GmPIN, and Auxin‐responsive protein SAUR genes in GmWRI1b overexpression (GmWRI1bOE) hairy roots.
Figure S14 Expression level of jasmonate biosynthesis and abscisic acid biosynthesis and catabolic genes in GmWRI1bOE hairy roots.
Figure S15 Proposed functions for GmWRI1 in soybean nodulation.
Table S1 Primers used in this study.
Table S2 The list of up‐regulated genes in GmWRI1 overexpression soybean hairy roots.
Table S3 The list of down‐regulated genes in GmWRI1 overexpression soybean hairy roots.
Table S4 The list of AW‐box genes regulated by GmWRI1.
Acknowledgments
We thank Mr. Qing Li and Mr. Shuai Fang for assistance on lipid and hormone measurements.
Funding
This work was supported by the National Science Foundation of China (31670294), the National Key Research and Development Program of China (2016YFD0101005), and the Ministry of Science and Technology of China (grant 2016YFD0100504). CB was supported by the Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences of the United States Department of Energy (Grant DE‐FG02‐91ER20021) and MSU AgBioResearch.
References
- Baier, M.C. , Barsch, A. , Küster, H. and Hohnjec, N. (2007) Antisense repression of the Medicago truncatula nodule‐enhanced sucrose synthase leads to a handicapped nitrogen fixation mirrored by specific alterations in the symbiotic transcriptome and metabolome. Plant Physiol. 145, 1600–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates, P.D. , Stymne, S. and Ohlrogge, J. (2013) Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol. 16, 358–364. [DOI] [PubMed] [Google Scholar]
- Baud, S. , Mendoza, M.S. , To, A. , Harscoët, E. , Lepiniec, L. and Dubreucq, B. (2007) WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards FA metabolism during seed maturation in Arabidopsis. Plant J. 50, 825–838. [DOI] [PubMed] [Google Scholar]
- Baud, S. , Wuillème, S. , To, A. , Rochat, C. and Lepiniec, L. (2009) Role of WRINKLED1 in the transcriptional regulation of glycolytic and FA biosynthetic genes in Arabidopsis. Plant J. 60, 933–947. [DOI] [PubMed] [Google Scholar]
- Breakspear, A. , Liu, C. , Roy, S. , Stacey, N. , Rogers, C. , Trick, M. , Morieri, G. , et al. (2014) The root hair "infectome" of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for Auxin signaling in rhizobial infection. Plant Cell, 26, 4680–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brechenmacher, L. , Lei, Z. , Libault, M. , Findley, S. , Sugawara, M. , Sadowsky, M.J. , et al. (2010) Soybean metabolites regulated in root hairs in response to the symbiotic bacterium Bradyrhizobium japonicum . Plant Physiol. 153, 1808–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernac, A. and Benning, C. (2004) WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 40, 575–585. [DOI] [PubMed] [Google Scholar]
- Chen, B. , Wang, J. , Zhang, G. , Liu, J. , Manan, S. , Hu, H. and Zhao, J. (2016) Two types of soybean diacylglycerol acyltransferases are differentially involved in triacylglycerol biosynthesis and response to environmental stresses and hormones. Sci. Rep. 6, 28541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, V.C. , Loughlin, P.C. , Day, D.A. and Smith, P.M. (2014) Transport processes of the legume symbiosome membrane. Front. Plant Sci. 5, 699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaude, N. , Tippmann, H. , Flemetakis, E. , Katinakis, P. , Udvardi, M. and Dörmann, P. (2004) The galactolipid digalactosyldiacylglycerol accumulates in the peribacteroid membrane of nitrogen‐fixing nodules of soybean and Lotus. J. Biol. Chem. 279, 34624–34630. [DOI] [PubMed] [Google Scholar]
- Gobbato, E. , Marsh, J.F. , Vernié, T. , Wang, E. , Maillet, F. , Kim, J. , Miller, J.B. , et al. (2012) A GRAS‐type transcription factor with a specific function in mycorrhizal signaling. Curr. Biol. 22, 2236–2241. [DOI] [PubMed] [Google Scholar]
- Haq, B.U. , Ahmad, M.Z. , Ur Rehman, N. , Wang, J. , Li, P. , Li, D. and Zhao, J. (2017) Functional characterization of soybean strigolactone biosynthesis and signaling genes in Arabidopsis MAX mutants and GmMAX3 in soybean nodulation. BMC Plant Biol. 17, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck, C. , Kuhn, H. , Heidt, S. , Walter, S. , Rieger, N. and Requena, N. (2016) Symbiotic fungi control plant root cortex development through the novel GRAS transcription factor MIG1. Curr. Biol. 26, 2770–2778. [DOI] [PubMed] [Google Scholar]
- Heckmann, A.B. , Lombardo, F. , Miwa, H. , Perry, J.A. , Bunnewell, S. , Parniske, M. , Wang, T.L. , et al. (2006) Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 142, 1739–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohnjec, N. , Czaja‐Hasse, L.F. , Hogekamp, C. and Küster, H. (2015) Pre‐announcement of symbiotic guests: transcriptional reprogramming by mycorrhizal lipochitooligosaccharides shows a strict co‐dependency on the GRAS transcription factors NSP1 and RAM1. BMC Genom. 16, 994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y. , Wang, W. , Xie, Q. , Liu, N. , Liu, L. , Wang, D. , Zhang, X. , et al. (2017) Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science, 356, 1172–1175. [DOI] [PubMed] [Google Scholar]
- Jiang, Y. , Xie, Q. , Wang, W. , Yang, J. , Zhang, X. , Yu, N. , Zhou, Y. , et al.(2018) Medicago AP2‐domain transcription factor WRI5a is a master regulator of lipid biosynthesis and transfer during mycorrhizal symbiosis. Molecular Plant, 11, 1344–1359. [DOI] [PubMed] [Google Scholar]
- Keymer, A. , Pimprikar, P. , Wewer, V. , Huber, C. , Brands, M. , Bucerius, S.L. , Delaux, P.M. , et al. (2017) Lipid transfer from plants to arbuscular mycorrhiza fungi. Elife 6, e29107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, Q. , Ma, W. , Yang, H. , Ma, G. , Mantyla, J.J. and Benning, C. (2017) The Arabidopsis WRINKLED1 transcription factor affects auxin homeostasis in roots. J. Exp. Bot. 68, 4627–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan, H.B. , Alaswad, A.A. , Oehrle, N.W. and Gillman, J.D. (2016) Deletion of the SACPD‐C locus alters the symbiotic relationship between Bradyrhizobium japonicum USDA110 and soybean, resulting in elicitation of plant defense response and nodulation defects. Mol. Plant‐Microbe Interact. 29, 862–877. [DOI] [PubMed] [Google Scholar]
- Lakhssassi, N. , Colantonio, V. , Flowers, N.D. , Zhou, Z. , Henry, J. , Liu, S. and Meksem, K. (2017) Stearoyl‐Acyl carrier protein desaturase mutations uncover an impact of stearic acid in leaf and nodule structure. Plant Physiol. 174, 1531–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrainzar, E. , Gil‐Quintana, E. , Seminario, A. , Arrese‐Igor, C. and González, E.M. (2014) Nodule carbohydrate catabolism is enhanced in the Medicago truncatula A17‐Sinorhizobium medicae WSM419 symbiosis. Front. Microbiol. 5, 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, L. , Chen, L. , Shi, X. , Li, Y. , Wang, J. , Chen, D. , Xie, F. , et al.(2014) A nodule‐specific lipid transfer protein AsE246 participates in transport of plant‐synthesized lipids to symbiosome membrane and is essential for nodule organogenesis in Chinese milk vetch. Plant Physiol. 164, 1045–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P. , Dong, Q. , Ge, S. , He, X. , Verdier, J. , Li, D. and Zhao, J. (2016) Metabolic engineering of proanthocyanidin production by repressing the isoflavone pathways and redirecting anthocyanidin precursor flux in legume. Plant Biotechnol. J. 14, 1604–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Lara, I.M. , Sohlenkamp, C. and Geiger, O. (2003) Membrane lipids in plant‐associated bacteria: their biosyntheses and possible functions. Molecular Plant‐Microbe Interact 16, 567–579. [DOI] [PubMed] [Google Scholar]
- Lu, S. , Sturtevant, D. , Aziz, M. , Jin, C. , Li, Q. , Chapman, K.D. and Guo, L. (2018) Spatial analysis of lipid metabolites and expressed genes reveals tissue‐specific heterogeneity of lipid metabolism in high‐ and low‐oil Brassica napus L. seeds. Plant J. 94, 915–932. [DOI] [PubMed] [Google Scholar]
- Luginbuehl, L.H. , Menard, G.N. , Kurup, S. , Van Erp, H. , Radhakrishnan, G.V. , Breakspear, A. , Oldroyd, G.E.D. , et al.(2017) FAs in arbuscular mycorrhizal fungi are synthesized by the host plant. Science, 356, 1175–1178. [DOI] [PubMed] [Google Scholar]
- Ma, W. , Kong, Q. , Arondel, V. , Kilaru, A. , Bates, P.D. , Thrower, N.A. , Benning, C. , et al.(2013) Wrinkled1, a ubiquitous regulator in oil accumulating tissues from Arabidopsis embryos to oil palm mesocarp. PLoS ONE, 8, e68887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, W. , Kong, Q. , Grix, M. , Mantyla, J.J. , Yang, Y. , Benning, C. and Ohlrogge, J.B. (2015) Deletion of a C‐terminal intrinsically disordered region of WRINKLED1 affects its stability and enhances oil accumulation in Arabidopsis. Plant J. 83, 864–874. [DOI] [PubMed] [Google Scholar]
- Ma, W. , Kong, Q. , Mantyla, J.J. , Yang, Y. , Ohlrogge, J.B. and Benning, C. (2016) 14‐3‐3 protein mediates plant seed oil biosynthesis through interaction with AtWRI1. Plant J. 88, 228–235. [DOI] [PubMed] [Google Scholar]
- Maeo, K. , Tokuda, T. , Ayame, A. , Mitsui, N. , Kawai, T. , Tsukagoshi, H. , Ishiguro, S. , et al.(2009) An AP2‐type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW‐box sequence conserved among proximal upstream regions of genes involved in FA synthesis. Plant J. 60, 476–487. [DOI] [PubMed] [Google Scholar]
- Manan, S. , Ahmad, M.Z. , Zhang, G. , Chen, B. , Haq, B.U. , Yang, J. and Zhao, J. (2017a) LEAFY COTYLEDON 2 regulates subsets of genes involved in controlling the biosynthesis and catabolism of seed storage substances and seed development. Front. Plant Sci. 8, 1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manan, S. , Chen, B. , She, G. , Wan, X. and Zhao, J. (2017b) Transport and transcriptional regulation of oil production in plants. Crit. Rev. Biotechnol. 37, 641–655. [DOI] [PubMed] [Google Scholar]
- Ogden, A.J. , Gargouri, M. , Park, J. , Gang, D.R. and Kahn, L. (2017) Integrated analysis of zone‐specific protein and metabolite profiles within nitrogen‐fixing Medicago truncatula‐Sinorhizobium medicae nodules. PLoS ONE, 12, e0180894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge, J. and Chapman, K. (2011) The seeds of green energy: expanding the contribution of plant oils as biofuels. Biochemist (Lond), 33, 34–38. [Google Scholar]
- Oldroyd, G.E.D. and Downie, J.A. (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59, 519–546. [DOI] [PubMed] [Google Scholar]
- Peng, L. , Hocart, C.H. , Redmond, J.W. and Williamson, R.E. (2000) Fractionation of carbohydrates in Arabidopsis root cell walls shows that three radial swelling loci are specifically involved in cellulose production. Planta, 211, 406–414. [DOI] [PubMed] [Google Scholar]
- Pii, Y. , Astegno, A. , Peroni, E. , Zaccardelli, M. , Pandolfini, T. and Crimi, M. (2009) The Medicago truncatula N5 gene encoding a root‐specific lipid transfer protein is required for the symbiotic interaction with Sinorhizobium meliloti. Mol. Plant Microbe Interact. 22, 1577–1587. [DOI] [PubMed] [Google Scholar]
- Pouvreau, B. , Baud, S. , Vernoud, V. , Morin, V. , Py, C. , Gendrot, G. , Pichon, J.P. , et al.(2011) Duplicate maize Wrinkled1 transcription factors activate target genes involved in seed oil biosynthesis. Plant Physiol. 156, 674–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, A.S.N. , Marquez, Y. , Kalyna, M. and Barta, A. (2013) Complexity of the alternative splicing landscape in plants. Plant Cell, 25, 3657–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, S. , Robson, F. , Lilley, J. , Liu, C.W. , Cheng, X. , Wen, J. , Walker, S. , et al. (2017) MtLAX2, a functional homologue of the Arabidopsis auxin influx transporter AUX1, is required for nodule organogenesis. Plant Physiol. 174, 326–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjaya, D.T.P. , Weise, S.E. and Benning, C. .(2011) Increasing the energy density of vegetative tissues by diverting carbon from starch to oil biosynthesis in transgenic Arabidopsis. Plant Biotechnol. J. 9, 874–883. [DOI] [PubMed] [Google Scholar]
- Seo, P.J. , Park, M.J. , Lim, M.H. , Kim, S.G. , Lee, M. , Baldwin, I.T. and Park, C.M. (2012) A self‐regulatory circuit of CIRCADIAN CLOCK‐ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell, 24, 2427–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, B. , Allen, W.B. , Zheng, P. , Li, C. , Glassman, K. , Ranch, J. , Nubel, D. , et al.(2010) Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol. 153, 980–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger, D. and Brown, J.W.S. .(2013) Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell, 25, 3640–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strodtman, K.N. , Stevenson, S.E. , Waters, J.K. , Mawhinney, T.P. , Thelen, J.J. , Polacco, J.C. and Emerich, D.W. (2017) The bacteroid periplasm in soybean nodules is an interkingdom symbiotic space. Mol. Plant‐Microbe Interact. 30, 997–1008. [DOI] [PubMed] [Google Scholar]
- Udvardi, M. and Poole, P.S. .(2013) Transport and metabolism in legume‐rhizobia symbioses. Ann. Rev. Plant Biol. 64, 781–805. [DOI] [PubMed] [Google Scholar]
- Voelker, T. and Kinney, A.J. (2001) Variations in the biosynthesis of seed‐storage lipids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 335–361. [DOI] [PubMed] [Google Scholar]
- Wang, E. , Schornack, S. , Marsh, J.F. , Gobbato, E. , Schwessinger, B. , Eastmond, P. , Schultze, M. , et al. (2012) A common signaling process that promotes mycorrhizal and oomycete colonization of plants. Curr. Biol. 22, 2242–2246. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Tóth, K. , Tanaka, K. , Nguyen, C.T. , Yan, Z. , Brechenmacher, L. , Dahmen, J. , et al.(2014) A soybean acyl carrier protein, GmACP, is important for root nodule symbiosis. Mol. Plant Microbe Interact. J. 27, 415–23. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Hou, Q. , Li, P. , Yang, L. , Sun, X. , Benedito, V.A. , Wen, J. , et al.(2017) Diverse functions of multidrug and toxin extrusion (MATE) transporters in citric acid efflux and metal homeostasis in Medicago truncatula . Plant J. 90, 79–95. [DOI] [PubMed] [Google Scholar]
- Wu, X.L. , Liu, Z.H. , Hu, Z.H. and Huang, R.Z. (2014) BnWRI1 coordinates fatty acid biosynthesis and photosynthesis pathways during oil accumulation in rapeseed. J. Integr. Plant Biol. 56, 582–93. [DOI] [PubMed] [Google Scholar]
- Xue, L. , Klinnawee, L. , Zhou, Y. , Saridis, G. , Vijayakumar, V. , Brands, M. , Dörmann, P. , et al.(2018) AP2 transcription factor CBX1 with a specific function in symbiotic exchange of nutrients in mycorrhizal Lotus japonicus. Proc. Natl Acad. Sci. USA 115, E9239–E9246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W. , Pollard, M. , Li‐Beisson, Y. , Beisson, F. , Feig, M. and Ohlrogge, J. (2010) A distinct type of glycerol‐3‐phosphate acyltransferase with sn‐2 preference and phosphatase activity producing 2‐monoacylglycerol. Proc. Natl Acad. Sci. USA, 107, 12040–12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Munz, J. , Cass, C. , Zienkiewicz, A. , Kong, Q. , Ma, W. , Sanjaya, S.J. , et al. (2015) Ectopic expression of WRINKLED1 affects fatty acid homeostasis in Brachypodium distachyon vegetative tissues. Plant Physiol. 169, 1836–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M. , Fan, J. , Taylor, D.C. and Ohlrogge, J.B. (2009) DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell, 21, 3885–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D. , Zhao, M. , Li, S. , Sun, L. , Wang, W. , Cai, C. , Dierking, E.C. , et al. (2017a) Plasticity and innovation of regulatory mechanisms underlying seed oil content mediated by duplicated genes in the palaeopolyploid soybean. Plant J. 90, 1120–1133. [DOI] [PubMed] [Google Scholar]
- Zhang, F. , Ke, J. , Zhang, L. , Chen, R. , Sugimoto, K. , Howe, G.A. , Xu, H.E. , et al. (2017b) Structural insights into alternative splicing‐mediated desensitization of jasmonate signaling. Proc. Natl Acad. Sci. USA, 114, 1720–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Phylogenetic (a) and gene structure analysis (b) and sequence alignment (c) of GmWRI1s.
Figure S2 Schematic diagram of the GmWRI1s and AtWRI1 predicted protein sequence illustrating the locations of known domains.
Figure S3 Public data for expression of soybean GmWRI1s.
Figure S4 Confirmation of transgenic atwri1–1 (a) and wild‐type (b) plants over‐expressing GmWRI1s.
Figure S5 Functional expression of GmWRI1a and b in Arabidopsis thaliana seeds.
Figure S5 Functional expression of GmWRI1a and b in Arabidopsis thaliana seeds.
Figure S6 Proteins content of wild type, atwri1 mutant and transgenic plant in mature seeds.
Figure S7 Quantitative PCR confirmation gene regulation by GmWRI1 in soybean hairy roots.
Figure S8 Effects of GmWRI1a and b overexpression on soybean nodulation and glycolysis and lipid metabolism.
Figure S9 Accumulation of lipids and expression patterns of soybean lipid synthesis‐related genes at soybean nodule developmental stages.
Figure S10 Identification and transcriptional analysis of FatB, GPAT, GRAS transcription factors RAM1 and ABCG transporter STR1/2.
Figure S11 The heat map analysis of tissue expression patterns of ortholog genes (a), other homology genes (b) for M. truncatula RAM1, RAM2, WRI1, STR1/2, and FatM.
Figure S12 The phylogenic tree (a) and heat map analysis (b) in roots and nodules of CUS (cutin synthase).
Figure S13 Expression level of GmGH3, GmPIN, and Auxin‐responsive protein SAUR genes in GmWRI1b overexpression (GmWRI1bOE) hairy roots.
Figure S14 Expression level of jasmonate biosynthesis and abscisic acid biosynthesis and catabolic genes in GmWRI1bOE hairy roots.
Figure S15 Proposed functions for GmWRI1 in soybean nodulation.
Table S1 Primers used in this study.
Table S2 The list of up‐regulated genes in GmWRI1 overexpression soybean hairy roots.
Table S3 The list of down‐regulated genes in GmWRI1 overexpression soybean hairy roots.
Table S4 The list of AW‐box genes regulated by GmWRI1.


