Summary
Clustered regularly interspaced short palindromic repeat (CRISPR) and Cas9‐associated protein systems provide a powerful genetic manipulation tool that can drive plant research forward. Nuclease‐dead Cas9 (dCas9) is an enzymatically inactive mutant of Cas9 in which its endonuclease activity is non‐functional. The applications of CRISPR/dCas9 have expanded and diversified in recent years. Originally, dCas9 was used as a CRISPR/Cas9 re‐engineering tool that enables targeted expression of any gene or multiple genes through recruitment of transcriptional effector domains without introducing irreversible DNA‐damaging mutations. Subsequent applications have made use of its ability to recruit modifying enzymes and reporter proteins to DNA target sites. In this paper, the most recent progress in the applications of CRISPR/dCas9 in plants, which include gene activation and repression, epigenome editing, modulation of chromatin topology, live‐cell chromatin imaging and DNA‐free genetic modification, will be reviewed. The associated strategies for exploiting the CRISPR/dCas9 system for crop improvement with a dimer of the future of the CRISPR/dCas9 system in the functional genomics of crops and the development of traits will be briefly discussed.
Keywords: CRISPR/dCas9, sgRNAs, transcriptional regulation, transcriptional activation, epigenome editing, chromatin topology, chromatin imaging, CRISPR/dCas9 ribonucleoproteins
Introduction
It is well known that adverse environmental conditions caused by biotic (e.g. microbial pathogens and insect pests) and abiotic (e.g. extreme temperature, water limitation, excessive salt, high radiation and chemical pollution) factors alter the expression of genes in eukaryotic organisms in a well‐coordinated manner at both spatial and temporal levels. The outcome of transcriptional regulation under stress conditions influences the subsequent steps (translational and post‐translational) before the manifestation of the final phenotype (Zhang, 2015) and may consequently lead to a decrease in plant quality, yield and biomass production. Living organisms have clearly defined strategies for responding to stress that are dependent on a complex regulatory network of molecular interactions. Although, in most cases, the plant response to stress based on the mechanisms of tolerance, resistance and avoidance involves well‐described metabolic pathways, the ability to acclimatize/adapt after single‐generation exposure previously observed in several studies (Boyko and Kovalchuk, 2008; Mittler et al., 2012; Ohama et al., 2017; Zhao et al., 2014) represents an interesting phenomenon that cannot be explained by Mendelian genetics.
Gene expression is a multistep process that is dependent on an accurate and responsive mechanism that regulates gene transcription, transmutation into messenger RNAs (mRNAs) and subsequent translation into proteins (Crick, 1970; Sarkar and Daniels‐Race, 2013). The conversion of DNA to RNA through transcriptional processes with or without epigenetic influence is entirely dependent on accurate gene regulation (Lo and Qi, 2017). Precision in gene regulation is equally critical for achieving objectives in genetic engineering and various applications in synthetic biology (Lowder et al., 2017a,b). The spectrum of external and internal influences experienced during the life span of an organism may lead to the generation of specific changes in gene expression that could be epigenetically (without changing DNA sequence) fixed and passed on to progeny, forming epigenetic memories (Boyko and Kovalchuk, 2008). This can be achieved on several interdependent levels, including reversible methylation of DNA sequences, numerous histone modifications and chromatin remodelling (Adli, 2018; Movahedi et al., 2018).
The emergence of clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 protein and a catalytically inactive or ‘dead’ Cas9 systems provided a powerful genetic manipulation tool that can steer biological studies based on the ability to achieve targeted modifications, which is critical for phenotypic alteration. Levels of gene expression can be changed through the fusion of dCas9 to transcriptional regulators. Recruitment of these transcriptional modulators to the promoter region close to the transcriptional start site alters the expression of the desired downstream genes. In a similar manner, fusing dCas9 to epigenetic modulators such as methylation and deacetylation enzymes will establish dCas9‐based DNA‐chromatin‐modifying enzymes for precise epigenome editing directed by sgRNA. Soon after the emergence of CRISPR/dCas9‐based technology, researchers began attaching regulatory and reporter proteins to harness the targeting abilities of dCas9 for reversible gene activation or repression (Li et al., 2017; Lowder et al., 2017a,b), epigenome editing (Hilton et al., 2015; Zezulin and Musunuru, 2018), modulation of chromatin topology (Guo et al., 2015), live‐cell chromatin imaging (Dreissig et al., 2017; Xue and Acar, 2018) and DNA‐free genetic modification (Liang et al., 2017; Veillet et al., 2019; Woo et al., 2015; Zhang et al., 2016) in plants (Figure 1). However, the success of these applications is highly dependent on choosing the appropriate transcriptional regulators, target genes and specific target sites and the delivery of the CRISPR/dCas9 and sgRNA constructs.
Figure 1.
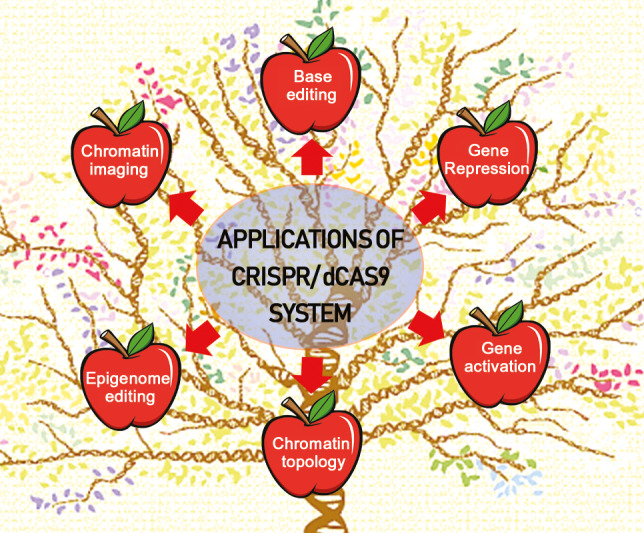
Major applications of the CRISPR/dCas9 system in plant genomics research.
In this paper, the most recent progress in CRISPR/dCas9‐mediated gene regulation in plants will be reviewed. The associated strategies for exploiting the CRISPR/dCas9 system for crop improvement with a dimer of the future of the CRISPR/dCas9 system in the functional genomics of crops and the development of traits will be discussed. We will conclude with future directions and the broader impact of CRISPR technologies that are extending beyond genome editing, as mentioned above. We anticipate that this review will provide a good start for those keen on finding out more about CRISPR/dCas9‐mediated applications. For additional insights into aspects of CRISPR/Cas9 technology not covered here, the reader is referred to a number of earlier reviews (Adli, 2018; Anton et al., 2018; Bortesi et al., 2016; Doudna and Charpentier, 2014; Khatodia et al., 2016; Limera et al., 2017; Ma et al., 2016; Zhang Wen and Guo, 2014).
dCas9 as a re‐engineering CRISPR/Cas9 platform for gene expression regulation in plants
The main advantages of CRISPR/dCas9 over previous genome regulation techniques are its straightforwardness, target specificity, adaptability and reversibility (Gearing, 2016). In the majority of recent reports, CRISPR/dCas9 has been applied through constitutive (non‐conditional) expression systems that do not allow temporal control of induced transcriptional effects. Alternatively, doxycycline‐inducible expression of sgRNA or dCas9 effectors has been applied to provide a certain degree of temporal control over the activity of dCas9 effectors, but the response time might be too slow to dissect fast transcriptional or epigenetic processes. Several recently developed switch systems that act at the protein level instead offer more rapid temporal regulation of the activation or inactivation of dCas9 effectors, as reviewed by Gjaltema and Schulz, 2018.
The CRISPR/dCas9 system consists of three major components: a nuclease‐dead Cas9, a single‐stranded guide RNA (sgRNA) and transcriptional activators. This section will briefly explain these three components.
dCas9
In 2013, Qi et al. mutated the nuclease domains of Cas9 from S. pyogenes. He introduced an H840A mutation in the HNH domain and a D10A mutation in the RuvC domain to generate a nuclease‐deficient dCas9 (Qi et al., 2013), also referred to as a dCas9 null mutant. Although this dead version of Cas9 is no longer able to cleave DNA, it can still target and bind DNA with the same precision when guided by sgRNA. However, instead of irreversibly altering the genome, binding of dCas9 interferes with transcription at the target site, resulting in reversible silencing of the gene. In contrast, the standard CRISPR/Cas9 system depends on the introduction of double‐stranded breaks (DSBs) in DNA through the activity of the Cas9 endonuclease, followed by manipulation of the DNA repair mechanisms for gene editing. dCas9 activation systems deploy transcriptional activators at the protein N and C termini (Li et al., 2017; Piatek et al., 2015; Polstein and Gersbach, 2015) to regulate the expression of gene of interest. Hence, dCas9 is able to regulate the expression of an endogenous gene(s) without permanently modifying the genome. The CRISPR/dCas9 system can be used for genetic screening, overexpression of proteins of interest and exploitation of a varied array of transcriptional activators and repressors in regulating the expression of target genes (Gearing, 2016).
sgRNA
A guide RNA (gRNA) is a single chimeric RNA formed by the fusion of tracrRNA and crRNA (Jinek et al., 2012). gRNAs containing two major regions of importance for CRISPR systems, the scaffold and spacer regions, are used to direct dCas9 to their targets. The spacer region contains nucleotides that are complimentary to those found in the target genes, often in the promoter region. The scaffold region is responsible for the formation of a complex with dCas9. Together, they bind dCas9 and direct it to the gene(s) of interest. Since the spacer region of a gRNA can be modified for any potential sequence, CRISPR systems exhibit great flexibility, as any genes or nucleotides with a sequence complimentary to the spacer region can become possible targets. The sequence within the sgRNA that is responsible for Cas9 binding is similarly responsible for dCas9 complex formation. The CRISPR/Cas9 systems possess high flexibility compared to other genome engineering tools because any genes or nucleotides with a sequence complimentary to the spacer region can become possible targets for editing through modification of the sgRNA spacer region for any potential sequence (Gearing, 2016).
Multiple sgRNAs
CRISPR/dCas9 platforms potentially offer unparalleled simplicity and multiplexability because synthetic guide RNAs can be easily modified to achieve new targeting specificities. dCas9 guided by multiple sgRNAs can simultaneously bind to several different target loci (Didovyk et al., 2016). Li et al. (2017) evaluated the multiplexability of dCas9 transcriptional activation system in cell‐based assays by co‐expressing it with three sgRNAs targeting WRKY30, RLP23 and CDG1. As quantified by RT‐qPCR, the endogenous gene expression was dramatically induced for all three genes. Lowder et al. (2017a,b) tested multiple strategies for dCas9‐based transcriptional activation and found that simultaneous recruitment of VP64 by dCas9 and a modified guide RNA scaffold gRNA 2.0 led to stronger activation of the transcriptional activity than the dCas9‐VP64 system (Figure 3).
Figure 3.
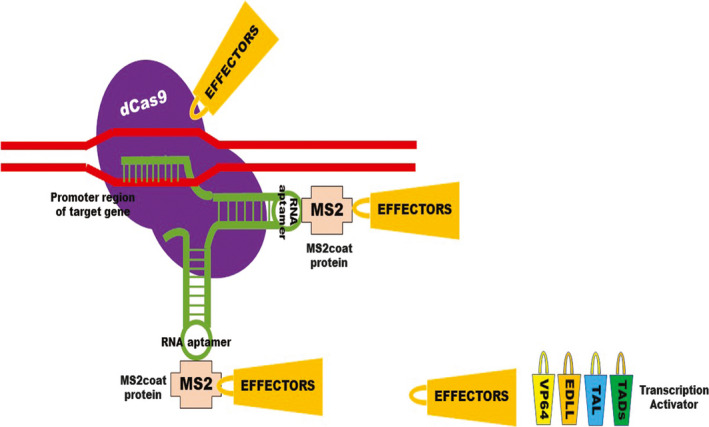
Modified strategy for CRISPR/dCas9‐mediated gene activation in plants. The modified dCas9 system consists of a dCas9 that can be easily fused to transcriptional activators such as VP64, EDLL, TAL and other TADs. The accompanying sgRNA can also be modified to include two RNA aptamers for binding with MS2 coat proteins that are also fused to transcriptional activators.
Recruitment of multiple gRNAs that bind to the sense strand increased the overall transcriptional activation when compared to the levels shown in single gRNA experiments. While, using multiple gRNAs that bind to sense and antisense strands resulted in a lower level of transcriptional activation when compared with the levels observed using gRNAs targeting the sense strand (Baazim, 2014). Although multiple sgRNAs tiling the proximal promoter of the target gene can synergistically boost the dCas9–VP64‐mediated gene activation (Li et al., 2017; Lowder et al., 2017a,b), this strategy reduces the scalability of the system (Konermann et al., 2015) and may increase the risk of dCas9‐mediated transcriptional perturbation at off‐target non‐promoter loci (Cheng et al., 2013).
Transcriptional effectors
Recent advancements in molecular tools for regulating gene functions are moving towards the utilization of novel bioengineering strategies to improve yield and other traits of economic importance in crops (Piatek et al., 2015). Among the emerging molecular techniques for enhancing the expression of desired genes is the use of artificial transcriptional regulators. These transcriptional regulators are chimeric proteins with a DNA‐binding domain attached to a functional domain that controls the transcriptional machinery (Piatek et al., 2015). Transcriptional effectors or synthetic transcriptional regulators, including transcriptional activators or repressors, are proteins or domains of proteins that are fused to dCas9 or sgRNAs that facilitate the recruitment of key cofactors as well as RNA polymerase for the transcription of the gene(s) whose expression is to be manipulated. Transcriptional effectors have a dual function. First, they specify the gene to be regulated through its DNA‐binding domains. Second, they possess the ability to stimulate transcription. RNA polymerases or general transcription factors can be recruited to gene sequences with activation or repression domains to facilitate and tweak transcription. The activation domain in eukaryotes may loosen DNA nucleosome interactions or modify histones in the nucleosome to facilitate gene transcription (Ma, 2011).
Moreover, sgRNAs play central roles in CRISPR activation (CRISPRa) and CRISPR interference (CRISPRi) because dCas9 activators or repressors directed via gRNA(s) can increase or repress the transcription of a target gene. To activate or repress a target gene with dCas9 activators or repressors, sgRNAs should be targeted to the promoter region of the GOI (Baazim, 2014). CRISPR‐dCas9‐based transcriptional regulation experiments in plants also typically require individual gRNAs and dCas9‐effector fusion proteins to be expressed from a single T‐DNA (Lowder et al., 2017a,b).
Synthetic transcriptional regulators may include the transactivation domain of a zinc‐finger protein, transcription activator‐like effector (TALE), other sequence‐unrelated portable TADs, the Herpes simplex viral protein 16 (VP16), multiple tandem copies of VP16, such as VP64 or VP160 (Cheng et al., 2013; Gao et al., 2014), or strong repressor such as Kruppel‐associated Box (KRAB) (Adli, 2018). A number of recent studies have reported the activation or repression of reporters or endogenous genes through these simple fusion proteins, although the effectiveness of these assemblies in transcriptional activation is limited to an average of 2‐ to 5‐fold by using a single gRNA. In the CRISPR/dCas9 system, transcriptional effectors can be introduced into the system by fusion to the sgRNA or dCas9. It has also been reported that the capacity for transcriptional up‐regulation can be modulated using multiple sites for activator fusion in one experiment (Li et al., 2017) or using different combinations of activators at once in a given experiment or sample (Lowder et al., 2016, 2017a,b).
Artificial transcriptional activators offer an advantageous alternative gene activation approach by targeting an autonomous transcriptional activation domain and endogenous gene promoter at a specific genomic locus by using a programmable DNA‐binding module (Li et al., 2017). Among the available synthetic gene activators, dCas9‐transcription activation domains potentially offer unparalleled simplicity and multiplexability compared with zinc‐finger protein – transcription activation domains and transcription activator‐like effector (TALE)‐transcription activation domains because synthetic guide RNAs can be easily modified to achieve new targeting specificities, and dCas9 guided by multiple sgRNAs can simultaneously bind to several different target loci (Didovyk et al., 2016). Thus, dCas9 activators or repressors should be recruited to promoter regions for maximal transcriptional activation.
Plant‐specific transcriptional effectors
In plants, the ethylene response factor from the ERF/EREBP family plays a leading role in boosting the response to different abiotic and biotic stresses (Tiwari et al., 2012). These transcriptional regulators exhibit the characteristic APETALA2 (AP2) DNA‐binding domain and various unspecified motifs (Abdullah et al., 2017; Azzeme et al., 2017; Ebrahimi et al., 2016). Among these unspecified motifs, Tiwari et al., 2012, characterized a short motif consisting of 24 amino acids called EDLL based on the conserved glutamic acid (E), aspartic acid (D) and leucine (L) residues for the first time. It was reported that EDLL could be found in AtERF98/TDR1 and other clade members of the AP2 subfamily. Moreover, the EDLL motif was revealed to be a functionally strong activation domain because of its unique arrangement of hydrophobic leucines and acidic amino acids, and it is also transferable to other TADs and active at both proximal and distal positions from the TATA‐box or GCC‐box of the target gene, with an activation strength that varies with the position of binding to the promoter region (Baazim, 2014; Fukao and Bailey‐Serres, 2008). Several studies demonstrated that the EDLL motif may be employed as a powerful new tool to confer transcriptional activation ability on heterologous DNA‐binding proteins (Lowder et al., 2017a,b; Piatek et al., 2015; Tiwari et al., 2012). The SRDX domain is also found in plant transcription factors associated with the ERF/EREBP family of transcriptional regulators. SRDX is derived from the transcriptional repressor domain referred to as the ERF‐associated amphiphilic repression domain (EAR) (Baazim, 2014; Hiratsu et al., 2003). The SRDX motif is a potent plant repression domain capable of maintaining its activity even in the presence of strong activators such as VP16 (Baazim, 2014). This domain has been used in several studies as a powerful tool for achieving transcriptional repression (Baazim, 2014; Heyl et al., 2008; Mahfouz et al., 2012; Piatek et al., 2015; Tiwari et al., 2012).
Strategies for boosting CRISPR transcriptional activation in plants
Three main strategies are employed to boost transcriptional activation by CRISPR in plants. The first approach involves the fusion of various activators in tandem with dCas9 (Figure 2). The second strategy recruits transcriptional activators by using modified gRNA scaffolds (Figure 3). The first two major strategies have been employed in mammalian systems to boost transcriptional activation through CRISPR/dCas9 systems. The third strategy is multiplexing, which uses a multiplex transcription activator system for synchronized activation of multiple genes in plants (Lowder et al., 2017a,b).
Figure 2.
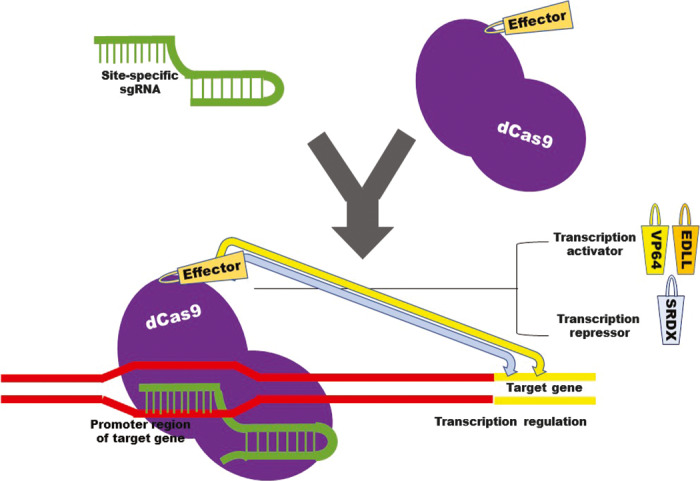
Schematic illustration of the use of dCas9 as a modular system for transcriptional regulator attachment. dCas9 is fused to effectors, transcription activators or repressors, for targeted gene regulation.
First strategy: fusion of various activators in tandem with dCas9
The first strategy, which involves fusing different activators in tandem with dCas9, has been reported in numerous works (Li et al., 2017; Lowder et al., 2017a,b; Piatek et al., 2015) on plants. In this section, we briefly review the gene regulation efficiency of different dCas9 systems using the first strategy (Figure 2).
dCas9‐VP64
Several studies have shown the possibility of transcriptional activation by fusing dCas9 proteins to a tetramer of VP16, termed dCas9‐VP64 (Beerli et al., 1998; Li et al., 2017; Lowder et al., 2017a,b; Piatek et al., 2015). When dCas9‐VP64 is directed to the promoter sequence of a target gene through a gRNA, this complex is generally able to recruit transcription factors that regulate transcription in mammalian cells (Paul and Qi, 2016). Furthermore, a dCas9‐VP64 system may be used in plants for transcriptional activation of endogenous genes. Transcriptional silencing of the FIS2 gene caused by CpG methylation in its promoter region in Arabidopsis was overcome using dCas9‐VP64. dCas9‐VP64 binding to methylated C activated the transcription of the AtFIS2 gene (Lowder et al., 2015).
However, low transcriptional activation activity of dCas9‐VP64 in plant cells has been observed in several recent reports (Lowder et al., 2017a,b). Li et al., 2017 evaluated the transcriptional activation activity of dCas9‐VP64 in activating a luciferase (LUC) reporter gene, and moderate transcriptional up‐regulation (2.4‐fold) of LUC was detected. Several other reports have also confirmed that dCas9‐VP64, as a frequently used TAD (Beerli et al., 1998), inadequately activates target genes using only one sgRNA in plant and mammalian cells (Cheng et al., 2013; Li et al., 2017; Mali et al., 2013; Piatek et al., 2015; Vazquez‐Vilar et al., 2016).
dCas9‐EDLL
Several recent studies have discussed the fusion of different EDLL activators in tandem with dCas9 in plants and evaluated the efficiency of gene regulation by dCas9‐EDLL (Li et al., 2017; Lowder et al., 2017a,b; Piatek et al., 2015; Tiwari et al., 2012). Baazim, 2014, previously reported that the dCas9‐EDLL activator guided by gRNAs complementary to promoter elements of target gene(s) was able to strongly induce the transcriptional activation of an endogenous genomic target and episomal targets in plant cells. Piatek et al., 2015, also effectively controlled the expression of different genes in Nicotiana benthamiana leaves using dCas9 fused to the activation domains of EDLL at its C‐terminus. Li et al., 2017, efficiently up‐regulated the expression of the LUC gene by 12.6‐fold in Arabidopsis protoplasts by enhancing its transcriptional activity by fusing dCas9‐VP128 to four copies of a series of ERF2m–EDLL motifs that form sequence‐unrelated portable transcription activation domains. Based on the above examples, dCas9 fused to an EDLL domain (Tiwari et al., 2012) appears to be a strong inducer for the activation of artificial reporters and endogenous genomic loci.
dCas9‐VP64‐EDLL
The application of two different TAD domains, EDLL and VP64, fused in tandem with dCas9 has also been reported in plants. The transcriptional activation efficiency of different genes has been evaluated and discussed. Lowder et al. (2017a,b) evaluated the activation ability of dCas9‐VP64‐EDLL guided by three identical gRNAs targeting Fertilization‐Independent Seed2 (FIS2). A second gene, Production of Anthocyanin Pigment1 (PAP1), was also targeted in the same Arabidopsis plant. The authors observed that dCas9‐VP64‐EDLL poorly activated the PAP1 and FIS2 genes and that the activation fold was notably less than the dCas9‐VP64 activation system (Lowder et al., 2017a,b). The reports using the first strategy, fusion of various activators in tandem with dCas9 for regulating gene expression in plants, are summarized in Table 1.
Table 1.
Summary of reports using the first strategy: fusion of various activators in tandem with dCas9 for regulating gene expression in plants
| Plant species | Design | Delivery | Detection | References | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | dCas9 expression cassette | sgRNA expression cassette | Method | Type of regulation | Relative gene expression (fold) | Assay | ||||
| Plasmid | Promoter of dcas9 | Plasmid | Promoter of sgRNA | |||||||
| Arabidopsis | LexA | dCas9‐VP64 | 35S | sgRNA‐LexA‐5′G | AtU6‐1 | PEG‐mediated protoplast transfection | Activation | 2.4 | RT‐qPCR | Li et al. (2017) |
| AtWRKY30 | dCas9‐TV | sgRNA‐WRKY30 | 138.8 | |||||||
| AtRLP23 | sgRNA‐ RLP23 | AtU6‐26 | 44.2 | |||||||
| pFGC‐dCas9‐VP64 | sgRNA‐RLP23‐4 | AtU6‐26 | Flora dip Agrobacterium‐mediated | 0.5 | ||||||
| pFGC‐dCas9‐TV | sgRNA‐RLP23‐4 | 32.3 | ||||||||
| Oryza sativa | OsGW7 | LUC/dCas9‐TV | sgRNA‐OsGW7‐2 | OsU6a | PEG‐mediated protoplast transfection | 79 | ||||
| OsER1 | sgRNA‐OsER1‐2 | 62 | ||||||||
| dCas9‐TV‐6 × His | T7 | sgRNA‐OsER1‐2 | ‐ | RNP‐mediated | 13 | |||||
| Arabidopsis | AtCDG1 | dCas9‐TV‐6 × His | sgRNA‐CDG1 | 191.7 | ||||||
| AtWRKY30 | sgRNA‐WRKY30 | 80.3 | ||||||||
| AtRLP23 | sgRNA‐RLP23‐4 | 37.3 | ||||||||
| HEK 293T human cells | HsASCL1 | dCas9‐TV | 35S | sgRNA‐HsASCL1 | human U6 | Co‐transfection with Lipofectamine LTX with PLUS | 46 | |||
| HsOCT4 | sgRNA‐HsOCT4 | 14.6 | ||||||||
Second strategy: recruitment of transcriptional activators by modified gRNA scaffolds
dCas9‐VP64‐MS2‐VP64 (CRISPR‐ Act2.0)
dCas9‐VP64‐MS2‐VP64 was constructed based on the second strategy of improving CRISPR transcriptional activation in plant systems by recruiting an additional MS2‐VP64 fused to dCas9‐VP64, with a dimer of the expression of gRNAs in the gRNA2.0 scaffold with MS2‐binding aptamers. A gRNA2.0 is generated by the insertion of MS2‐binding aptamers (MS2 coat protein of bacteriophage and its scaffold containing the cognate RNA stem‐loop aptamer) into stem‐loop 2 positions of a sgRNA tetraloop. Lowder et al., 2017b, robustly activated the expression of the PAP1 gene by 30‐ to 45‐fold, that of the FIS2 gene by up to 1500‐fold and that of the ULC1 gene by up to 40‐fold in Arabidopsis through dCas9‐VP64‐MS2‐VP64 guided by the gRNA2.0 scaffold. dCas9‐VP64‐MS2‐VP64 and its gRNA2.0 scaffold with MS2‐binding aptamers (termed CRISPR‐ Act2.0) proved to be a more powerful activation system compared to the first type of dCas9‐VP64 system.
dCas9‐VP64‐MS2‐EDLL
dCas9‐VP64‐MS2‐EDLL was constructed to enhance CRISPR transcriptional activation systems in plants with the aid of MS2‐EDLL by using gRNA2.0 linked to dCas9‐VP64. Lowder et al., 2017a,b, successfully evaluated the activation of the PAP1 and FIS2 genes in Arabidopsis using dCas9‐VP64‐MS2‐EDLL. dCas9‐VP64‐MS2‐EDLL notably activated the PAP1 gene by 30‐fold (higher than dCas9‐VP64) and weakly activated the FIS2 gene by 30‐fold (lower than dCas9‐VP64).
SAM‐modified sgRNA
SAM‐modified sgRNA was also generated based on the second strategy for improving CRISPR transcriptional activation in plants. The SAM sgRNA enables the recruitment of a chimeric TAD consisting of the MS2 phage coat protein (MCP) (Konermann et al., 2015). Li et al., 2017, reported targeting of the WRKY30 promoter with the SAM sgRNA‐WRKY30‐2, pairing dCas9–VP64 with MCP–4EE–VP128 or MCP–TV induced WRKY30–LUC more vigorously than the pairing of dCas9–VP64 and a normal sgRNA. MCP–4EE–VP128 and MCP–TV were shown to be active transcription activators that cause addition or synergistic activation of the target gene linked with the target promoter. However, the three‐component activation system consisting of dCas9‐TV with SAM sgRNA and MCP–4EE–VP128 or MCP‐TV did not enhance the induction of WRKY30–LUC and even reduced the induction of RLP23–LUC in contrast to the two‐component system of dCas9–TV and normal sgRNAs. The reports using the second strategy, recruitment of transcriptional activators by modified gRNA scaffolds for the regulation of gene expression in plants, are summarized in Table 2 and illustrated in Figure 2.
Table 2.
Summary of reports using the second strategy: recruitment of transcriptional activators via modified gRNA scaffolds for regulating gene expression in plants
| Plant species | Design | Delivery | Detection | References | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | dCas9 expression cassette | sgRNA expression cassette | Multiplex strategy | Method | Type of regulation | Relative gene expression (fold) | Assay | ||||
| Plasmid | Promoter of dcas9 | Plasmid | Promoter of sgRNA | ||||||||
| Arabidopsis | AtWRKY30‐LUC | dCas9‐TV | 35S | Normal sgRNA‐ | AtU6‐1 | Yes, co‐expressing dCas9–TV with three sgRNAs | PEG‐mediated transfection | Activation | 369.3 | RT‐qPCR | Li et al. (2017) |
| dCas9‐TV MCP‐4EE‐VP128 | SAM sgRNA‐ | 359.1 | |||||||||
| dCas9‐MCP‐TV | SAM sgRNA‐ | 359.1 | |||||||||
| AtPAP1 | pco‐dCas9‐VP64 | AtUBQ10 | AtPAP1‐gR1‐gR2‐gR3 | AtU6 | Multiplex gRNAs by Golden Gate Assembly & MultiSite Gateway | Flora dip Agrobacterium‐mediated | 2–7 | RT‐qPCR | Lowder et al. (2015) | ||
| AtmiR319 | pco‐dCas9‐VP64 | miR319‐gR1‐gR2‐gR3 | 3–7.5 | ||||||||
| AtFIS2 | pco‐dCas9‐VP64 | AtFIS2‐gR1‐gR2‐gR3 | Activation by RNA‐guided targeting of promoter methylation sites. | 400 | |||||||
| AtCSTF64 | pco‐dCas9‐3X(SRDX) | AtCSTF64‐gR1‐gR2‐gR3 | Repression | −0.6 | |||||||
| AtPAP1 | dCas9‐VP64‐ EDLL | AtPAP1‐gR1‐gR2‐gR3 | Activation | 200 | Kaufmann and Walker (2017) | ||||||
| AtFIS2 | dCas9‐VP64‐EDLL | AtFIS2‐gR1‐gR2‐gR3 | 2–3 | ||||||||
| AtPAP1 | dCas9‐VP64‐T2A‐MS2‐EDLL | AtPAP1‐gR1‐gR2‐gR3 | 30 | ||||||||
| AtFIS2 | dCas9‐VP64‐T2A‐MS2‐EDLL | AtFIS2‐gR1‐gR2‐gR3 | 35 | ||||||||
| AtPAP1 | dCas9‐VP64‐T2A‐MS2‐VP64 | AtPAP1‐gR1‐gR2‐gR3 | 50 | ||||||||
| AtFIS2 | dCas9‐VP64‐T2A‐MS2‐VP64 | AtFIS2‐gR1‐gR2‐gR3 | 1500 | ||||||||
| Oryza sativa | Os03g01240, Os04g39780 & Os11g35410 | dCas9‐VP64 | Maize Ubi1 | Os03g01240‐gR1 (N21), Os04g39780‐gR1 (N23) & Os11g35410‐gR1 (N25) | AtU6 | Golden Gate Assembly & MultiSite Gateway | PEG‐mediated transfection | Activation. | 1–2.5 | RT‐qPCR | Lowder et al. (2017a,b) |
| Os03g01240, Os04g39780 and Os11g3541 | dCas9‐VP64‐T2A‐MS2‐VP64‐N21‐23‐25 | Os03g01240‐gR1 (N21), Os04g39780‐gR1 (N23) & Os11g35410‐gR1 (N25) | 2.5–7 | ||||||||
| Nicotiana benthamiana | Target telomere DNA sequence | sp/sa‐dCas9 | Ubi4 | sgRNA‐telomere | AtU6‐26 | Gibson Assembly cloning | Agroinfiltration | Visualization of tandem repeats in live plant cells | 21–42 | Immunofluorescence analysis and fluorescence in situ hybridization and Microscopic analyses | Dreissig et al. (2017) |
Strategy three: multiplex transcription activator system for synchronized activation of multiple genes for regulating gene expression in plants
CRISPR offers great convenience in multiplexing that simultaneously targets multiple loci. Because CRISPR is such a potent system, the efficiency of editing or labelling does not change when multiple gRNAs are added. The CRISPR/dCas9 system possesses advantages over other existing platforms due to its amenability to multiplexing and the possibility of using viruses to systemically deliver gRNAs (Piatek et al., 2015). The CRISPR system has the ability to utilize multiple special sgRNAs encoded in the same vector to target different sites within a genome, providing an alternative to the tedious task of performing multiple crosses of plants with single mutations (Schaeffer and Nakata, 2015). Robust compounded transcriptional activation in plants through CRISPR/dCas9 and full TALE‐based transcriptional activation systems was recently reported (Lowder et al., 2017b).
Multiplexable CRISPR/dCas9 expression systems
The assembly of several gRNAs is made possible by Golden Gate Assembly methods for multiplexing the CRISPR/dCas9 expression system in plants. Golden Gate Assembly applies type II restriction enzymes, which cleave outside of their recognition sequence, to generate flanking overhangs (Moradpour and Abdulah, 2017). Furthermore, these overhangs will be modified to link together multiple fragments and enable ordered assembly of multiple components into a destination vector.
A recent report on the application of CRISPR/dCas9 in plants by Lowder et al., 2017b, described the establishment of a multiplex system of transcription activator‐like effector activation (mTALE‐Act) to simultaneously activate up to four genes in plants based on simplified, PCR‐independent and efficient Gateway and Golden Gate cloning strategies. This approach enables the application of transcriptional activation in both dicots and monocots.
CRISPR‐Act2.0 system and multiplexed activation of multiple genes in plants
A powerful method for investigating biological and biotechnological systems is the customizable and synchronized activation of multiple genes in vivo. Lowder et al., 2017a,b, effectively evaluated the efficiency of the CRISPR‐Act2.0 system in simultaneously activating three independent endogenous genes, Os11g35410, Os03g01240 and Os04g39780, in rice protoplasts. The analysis of transformants derived from rice protoplasts demonstrated that CRISPR‐Act2.0 is an effective system for gene activation compared to the dCas9‐VP64 system. The effective demonstration of the real‐time activation of multiple genes with CRISPR‐Act2.0 suggests promising applications for future plant research.
mTALE‐Act System and multiplexed activation of multiple genes in plants
In a recent report by Lowder et al. (2017a,b), the multiplexed TALE‐activation (mTALE‐Act) system was introduced as a strong transcriptional activation tool for the up‐regulation of multiple genes in monocots and dicots. mTALE‐Act with a dimer of the gRNA2.0 system efficiently regulated the expression of the PAP1 promoter by 40‐ to 150‐fold with a dimer of TALE‐VP64 in Arabidopsis. The development of the mTALE‐Act system for multiplexed transcriptional activation in plants enables instant assembly of up to four TALE‐VP64 genes into a single T‐DNA vector for synchronized activation of this series of genes in plants. Practically, mTALE‐Act shows stronger efficiency than CRISPR‐Act2.0 as a transcriptional activator. The reports using the third strategy, employing a multiplex transcription activator system for synchronized activation of multiple genes for regulating gene expression in plants, are summarized in Table 3.
Table 3.
Summary of reports using the third strategy: multiplex transcription activator system for synchronized activation of multiple genes for regulating gene expression in plants
| Plant species | Design | Delivery | Detection | References | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | dCas9 expression cassette | sgRNA expression cassette | Multiplex strategy | Method | Type of regulation | Relative gene expression (fold) | Assay | ||||
| Plasmid | Promoter of dcas9 | Plasmid | Promoter of sgRNA | ||||||||
| Arabidopsis | AtWRKY30 AtRLP23 AtCDG1 | dCas9‐TV | 35S | sgRNA‐WRKY30sgRNA‐RLP23 sgRNA‐CDG1‐4 | AtU6‐1 AtU6‐26 AtU6‐26 | Yes, co‐expressing dCas9–TV with three sgRNAs | PEG‐mediated transfection |
Simultaneous Activation |
9–13 | RT‐qPCR | Li et al. (2017) |
| CSTF64 | mTALE‐ | 2 × 35S | CSTF64 | AtU6 | Flora dip Agrobacterium‐mediated |
CSTF 15‐20‐fold GL1 500–700‐fold |
Lowder et al. (2017a,b) | ||||
| GL1 | Act | GL1 | |||||||||
| RBP‐DR1 | RBP‐DR1 | ||||||||||
| Oryza sativa | Os03g01240, Os04g39780 & Os11g35410 | dCas9‐VP64 | Maize Ubi1 | Os03g01240‐gR1 (N21), Os04g39780‐gR1 (N23) & Os11g35410‐gR1 (N25) | AtU6 | Multiplex gRNAs by Golden Gate Assembly & MultiSite Gateway | PEG‐mediated transfection | Activation | 2–2.5 | ||
| Os03g01240, Os04g39780 and Os11g3541 | dCas9‐VP64‐T2A‐MS2‐VP64‐N21‐23‐25 | Os03g01240‐gR1 (N21), Os04g39780‐gR1 (N23) & Os11g35410‐gR1 (N25) | 3–7 | ||||||||
CRISPR–dCas9 for transcriptional repression
dCas9 is a powerful tool not only for transcriptional activation but also for targeted inhibition of gene transcription. This inhibition occurs when binding of dCas9 to the target site sterically interferes with the binding and function of the transcriptional machinery; dCas9 alone functions as a repressor and blocks transcription, possibly by stalling transcriptional elongation, a phenomenon called CRISPR interference (CRISPRi) (Khatodia et al., 2016; Lagana’ et al., 2014; Larson et al., 2013; Qi et al., 2013). CRISPRi has been reported to be used for effective, stable RNA‐guided transcriptional suppression of a target gene (Larson et al., 2013). The suppression of transcription is inducible and reversible, and the recognition of targets only depends on the sgRNA sequence. Recruitment of dCas9 to the recognition complex containing sgRNA inhibits gene expression by interfering with transcriptional elongation, RNA polymerase binding or transcription factor binding (Khatodia et al., 2016; Lagana’ et al., 2014; Qi et al., 2013). The interference mechanism functions through the transcription repression domain of the transcription effector proteins. In plants, modulation of the transcription of a reporter construct and the endogenous PDS gene in Nicotiana benthamiana by fusing the dCas9 C‐terminus to the SRDX domain of the transcription effector to act as a repressor has been reported (Bortesi and Fischer, 2015; Piatek et al., 2015). Fusion of repressor domains such as SRDX to TAL effectors was shown to repress the expression of the RD29‐LUC transgene and the endogenous RD29A gene in Arabidopsis (Mahfouz et al., 2012).
dCas9 as a re‐engineering platform for base editing
Base editing (BE) represents a new dimension of CRISPR/Cas‐mediated precise editing to generate single‐nucleotide changes in DNA or RNA independently of DSBs and homology‐directed repair. It is a new genome‐editing technique that generates mutations at single‐base resolution. All four transition mutations, C to T, G to A, A to G and T to C, can be produced in the genome with the available CRISPR/Cas base editors. The cytosine and adenine base editors (CBE and ABE) are catalytically impaired deaminases that replace a C–G to T–A and A‐T to G‐C mutation, respectively. In RNA, conversion of adenine to inosine is also possible with the RNA base editor (Molla and Yang, 2019). Unlike regular CRISPR/Cas‐mediated genome‐editing techniques, fusion of BEs to either dCas9 or nCas9 (D10A nickase) does not create a DSB, and therefore, indel generation is limited. As a result, BEs offer precise genome editing with much cleaner product output, reducing on‐ and off‐target indels (Komor et al., 2017). The dCas9/nCas9‐based CBEs and ABEs have been successfully employed to create targeted base alterations in several plant species including rice, maize, wheat, tomato, Arabidopsis and Brassica napus (Kang et al., 2018; Lu and Zhu, 2017; Shimatani et al., 2017; Zong et al., 2017).
dCas9 as a re‐engineering platform for epigenome editing
Epigenetic control of the plant response to stress is a complex phenomenon (Boyko and Kovalchuk, 2008). Epigenetic modifications are not only stress inducible but can also cause changes in gene expression, which may remain over many generations. The formation of paramutations and epialleles is a consequence of several epigenetically mediated changes that are generated in response to stress. Such changes that are heritable cause divergence in plant ecotypes (Liu and Moschou, 2018; Liu et al., 2015). Thus, epigenetic marks may regulate gene evolution in a stress‐directed manner, allowing the rapid generation of new adaptive alleles at genetic and epigenetic levels (Boyko and Kovalchuk, 2008).
The epigenome is a second level of genomic regulation that involves DNA methylation and covalent post‐translational modifications of histone proteins in the nucleosome. Histone modifications alter the net charge of nucleosomes, which influences the interactions between DNA and histones and between histones themselves at inter‐ and intranucleosomal levels. Epigenetic regulation functions by influencing the structure of a stretch of chromatin, either via compression into a compact transcriptionally inactive state (heterochromatin) or by opening to allow access of the transcriptional machinery. Although endeavours in functional genomics have enabled the characterization and mapping of numerous genome modifications for epigenetic regulation in different tissues and cell types, existing approaches for single‐locus probing are tedious, costly and toxic to living cells. CRISPR/dCas9 technology now makes epigenome engineering possible, which enables scientists to better understand how specific phenotypes are influenced by epigenetic regulation. For example, the acetylation of histone H3 at lysine 27 was targeted within a gene of interest at the enhancer and promoter to increase gene expression by using dCas9 fused to a mammalian acetyltransferase (p300) (Hilton et al., 2015). The establishment of dCas9‐based DNA‐/chromatin‐modifying enzymes will enable precise epigenome editing to control genome‐wide gene regulation and chromatin status. In addition to histone acetyltransferases, other enzymes, such as DNA methyltransferases, methylcytosine deoxygenases, ubiquitin ligases and poly‐ADP ribosyltransferases, can be employed (Liu and Moschou, 2018; Osakabe et al., 2016). The role of different chromatin states induced through DNA methylation in abiotic stress responses and acclimation in plants could be determined by fusing dCas9 to epigenetic modulators. Various histone post‐translational effectors and domains that can be introduced via dCas9 could control methylation, phosphorylation or deacetylation of histones to regulate gene expression in eukaryotic cells (Jain, 2015). Moreover, the growth of new regulatory modules (s) from naturally existing components for epigenetic modification enables the engineering of signalling, regulatory and metabolic processes to modify plant stress tolerance(Jain, 2015). In the future, it is conceivable that such strategies may be employed to achieve greater control of plant epigenomes.
dCas9 as a re‐engineering platform for chromatin topology
Genome organization and the folding of chromatin within the nucleus are believed to be the key determinants of gene expression programs (Morgan et al., 2017). The existence of artificially engineered chromatin loops in the centre of genomic regulatory regions facilitates the manipulation of endogenous chromatin structures to enhance their functions in contributing to gene expression (Morgan et al., 2017). Such efforts may form new enhancer–promoter connections to offset‐specific genetic deficiencies (Adli, 2018).
A new technology has been developed to employ dCas9‐based platforms to target and manipulate chromatin structure and DNA loop formation. An innovative study by Morgan et al., 2017, used two dimerizable protein domains, PYL1 and ABI1, from the plant abscisic acid signalling pathway (Liang et al., 2011) to enhance gene expression in selected human cell lines harbouring the β‐globin locus. An artificial chromatin loop was formed between the distal enhancer and promoter regions by tethering these protein dimerization systems to two separate orthologues of dCas9, which resulted in an increase in gene expression. DNA‐looping mechanisms are a component of networks that regulate all aspects of DNA activities, including transcription, replication and recombination (Matthews, 1992). Such studies indicate the potency of CRISPR as a targeted chromatin structure‐rewiring tool.
dCas9 as a re‐engineering platform for live‐cell chromatin imaging in plants
Light intensity, temperature, microbial infection and hormonal signals for cell differentiation are some examples of the many environmental and developmental factors that are able to trigger global rearrangement of chromatin in plants (Liu and Weigel, 2015). The spatial and temporal organization of genomes is crucial for preserving and regulating cell functions such as DNA replication and repair, gene expression and proper chromosomal separation during cell division (Dreissig et al., 2017). Traditionally, cytological tools have been used to complement sequencing‐based methods, through which the behaviour of chromatin loci in the nucleus can be visualized and monitored. For example, small chromatin loops could be detected by increasing the resolution of photoactivated localization microscopy (PALM) (Schubert and Weisshart, 2015) using traditional padlock fluorescent in situ hybridization (FISH) (Feng et al., 2014). In plants, using a live imaging system, effects on the mobility and subnuclear localization of local gene expression were revealed using visually trackable T‐DNA insertions. The application of LacO arrays to enable specific recognition by the bacterial LacI protein labelled with fluorescent proteins for visualization of plant chromatin was also described (Liu and Weigel, 2015).
Today, more sophisticated genome‐editing techniques such as CRISPR/dCas9 would allow the non‐random insertion of LacO arrays into the genome. A CRISPR/dCas‐based chromatin imaging method has already been used in plant cell lines for the visualization of non‐repetitive genomic loci (Liu and Weigel, 2015). In a recent study, robust visualization of telomere repeats in live leaf cells of Nicotiana benthamiana was carried out by fusing eGFP/mRuby2 to dCas9 (Dreissig et al., 2017). Understanding chromosome dynamics in live plant cells or the regulation of genes or non‐coding sequences during development and environmental changes and approaches such as chromatin imaging through CRISPR imaging present the potential to bridge the long‐standing gap between sequencing studies, which reveal genomic information, and imaging studies, which provide spatial and temporal information on defined genomic regions (Dreissig et al., 2017).
dCas9‐FokI as a re‐engineering platform for reducing the off‐target effects
Unintended editing (off‐target) happens due to the nonspecific interaction of nucleases at sites other than the targeted site in DNA or RNA. Partial homology of the guide RNA sequence with nontarget sequence and recognition of unrelated PAM may result in off‐target editing (Molla and Yang, 2019). Dimeric instead of monomeric nucleases are employed to overcome the high levels of off‐target effects. Both nucleases in the dimer system bind to their specific targets or half‐sites and subsequently interact and dimerize for cleavage initiation with significant reduction in off‐target effects. The reliability of dimerization‐dependent FokI nuclease domains, ZFNs and TALENs, and the simplicity of CRISPR‐cas9 have been combined to develop an effective system. The FokI nuclease from Flavobacterium okeanokoites will only cleave DNA following dimerization activation. This nuclease when fused to a CRISPR complex with an inactivated Cas9 nuclease (dCas9‐FokI) (Tsai et al., 2014) enables the gRNA to direct the CRISPR complex to the target site for the dimerized FokI to carry out DNA excision. The Fok1‐dCas9 strategy reduces detectable off‐target effects by 10,000‐fold, excellent for highly precise and specific genome editing (Guilinger et al., 2014; Tsai et al., 2014). Application in Arabidopsis significantly reduced the off‐target cleavage in comparison with Cas9, however with a large trade‐off in efficiency (Paul and Qi, 2016).
Conclusion and future direction
This paper reviews the different strategies developed in the last a few years using the CRISPR/dCas9 activation system for various applications in both fundamental and applied plant research. The dCas9 activation system enables the enhancement of the expression of any gene or multiple genes together in the same cell. It allows the control of any gene without introducing any mutations in the host genome. Thus, it does not face problems associated with creating mutations, which may include irreversible DNA damage or undesirable effects due to off‐target mutations (Olsson, 2013). dCas9 activation systems have proved to be an efficient and robust transcriptional activation tool in plants involving single or several genes that already exhibit modest levels of expression. Fusing modified dCas9 to different TADs can be a useful strategy for genetic screening of desired traits in plants, which can be facilitated by protoplast‐based gain‐of‐function screening for gene regulation studies in a targeted signalling pathway employing common reporter genes such as green fluorescent protein (GFP) and β‐glucuronidase (GUS). The CRISPR/dCas9 activation system was demonstrated to be valuable for studying the functions of transcriptional regulators based on their effects on the transcriptome. It can also be employed in metabolic engineering through manifold activation of enzymes that can provide important insights about metabolic pathways and how to increase the production of valuable metabolites. This system may also be applied for the up‐regulation of key crop genes that control important traits such as resistance to abiotic and biotic stresses.
Moreover, dCas9‐based DNA‐/chromatin‐modifying enzymes can be employed to achieve greater control of plant epigenomes. The application of dCas9‐based platforms to modify chromatin structure and DNA loop formation for targeting and robust manipulation showed the potential of CRISPR as a targeted chromatin structure‐rewiring tool for future plant research. Understanding chromosome dynamics in living plant cells and chromatin imaging may facilitate CRISPR‐based imaging for deciphering genomic activities. CRISPR/dCas9 RNPs, referred to as the next‐generation DNA‐free CRISPR/Cas9, which may overcome GMO‐related regulations, would be valuable when introduced with a dimer of a protein‐stabilizing polypeptides for the engineering of genome modifications.
Conflict of interest
The authors declare that they have no conflict of interest.
Author Contributions
M. M. and S. N. A. jointly developed the conceptual structure of manuscript. M. M was involved in the compilation of relevant literature and drafting the manuscript. S. N. A. provided a critical feedback and edited the final manuscript.
Acknowledgements
The authors would like to thank Universiti Putra Malaysia and Ministry of Education, Malaysia, for providing Trans Disciplinary Research Grant Scheme (TRGS) (TRGS/1/2016/UPM/01/6/1) and Putra Grant (UPM/700‐2/1/GP/2017/9551500) for the study.
References
- Abdullah, S.N.A. , Azzeme, A.M. , Ebrahimi, M. , Ariff, E.A.K.E. and Hanifiah, F.H.A. (2017) Transcription factors associated with abiotic stress and fruit development in oil palm. In: Crop Improvement ( Abdullah, S.N.A. , Chai-Ling, H. and Wagstaff, C. , eds), pp. 71–99. Cham: Springer International Publishing. [Google Scholar]
- Adli, M. (2018) The CRISPR tool kit for genome editing and beyond. Nat. Commun. 9, 1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton, T. , Karg, E. and Bultmann, S. (2018) Applications of the CRISPR/Cas system beyond gene editing. Biol. Methods Protoc. 3, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzeme, A.M. , Abdullah, S.N.A. , Aziz, M.A. and Wahab, P.E.M. (2017) Oil palm drought inducible DREB1 induced expression of DRE/CRT‐ and non‐DRE/CRT‐containing genes in lowland transgenic tomato under cold and PEG treatments. Plant Physiol. Biochem. 112, 129–151. [DOI] [PubMed] [Google Scholar]
- Baazim, H . (2014) RNA‐guided Transcriptional Regulation in Plants via dCas9 Chimeric Proteins. Thesis.
- Beerli, R.R. , Segal, D.J. , Dreier, B. and Barbas, C.F. (1998) Toward controlling gene expression at will: Specific regulation of the erbB‐2/HER‐2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl. Acad. Sci. 95, 14628–14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortesi, L. and Fischer, R. (2015) The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33, 41–52. [DOI] [PubMed] [Google Scholar]
- Bortesi, L. , Zhu, C. , Zischewski, J. , Perez, L. , Bassié, L. , Nadi, R. , Forni, G. et al. (2016) Patterns of CRISPR/Cas9 activity in plants, animals and microbes. Plant Biotechnol. J. 14(12), 2203–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko, A. and Kovalchuk, I. (2008) Epigenetic control of plant stress response. Environ. Mol. Mutagen. 49, 61–72. [DOI] [PubMed] [Google Scholar]
- Cheng, A.W. , Wang, H. , Yang, H. , Shi, L. , Katz, Y. , Theunissen, T.W. , Rangarajan, S. et al. (2013) Multiplexed activation of endogenous genes by CRISPR‐on, an RNA‐guided transcriptional activator system. Cell Res. 23, 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick, F. (1970) Central dogma of molecular biology. Nature, 227, 561–563. [DOI] [PubMed] [Google Scholar]
- Didovyk, A. , Borek, B. , Tsimring, L. and Hasty, J. (2016) Transcriptional regulation with CRISPR‐Cas9: principles, advances, and applications. Curr. Opin. Biotechnol. 40, 177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna, J.A. and Charpentier, E. (2014) The new frontier of genome engineering with CRISPR‐Cas9. Science (80‐.), 346, 1258096. [DOI] [PubMed] [Google Scholar]
- Dreissig, S. , Schiml, S. , Schindele, P. , Weiss, O. , Rutten, T. , Schubert, V. , Gladilin, E. et al. (2017) Live‐cell CRISPR imaging in plants reveals dynamic telomere movements. Plant J. 91, 565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi, M. , Abdullah, S.N.A. , Abdul Aziz, M. and Namasivayam, P. (2016) Oil palm EgCBF3 conferred stress tolerance in transgenic tomato plants through modulation of the ethylene signaling pathway. J. Plant Physiol. 202, 107–120. [DOI] [PubMed] [Google Scholar]
- Feng, C.M. , Qiu, Y. , Van Buskirk, E.K. , Yang, E.J. and Chen, M. (2014) Light‐regulated gene repositioning in Arabidopsis. Nat. Commun. 5, 3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao, T. and Bailey‐Serres, J. (2008) Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc. Natl. Acad. Sci. U. S. A. 105, 16814–16819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, X. , Tsang, J.C.H. , Gaba, F. , Wu, D. , Lu, L. and Liu, P. (2014) Comparison of TALE designer transcription factors and the CRISPR/dCas9 in regulation of gene expression by targeting enhancers. Nucleic Acids Res. 42, e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearing, M. (2016) CRISPR 101: a desktop resource. Addgene's eBook collection. 2016, 125. [Google Scholar]
- Gjaltema, R.A.F. and Schulz, E.G. (2018) CRISPR/dCas9 switch systems for temporal transcriptional control. In: Epigenome Editing. pp. 167–185. New York, NY: Humana Press. [DOI] [PubMed] [Google Scholar]
- Guilinger, J.P. , Thompson, D.B. and Liu, D.R. (2014) Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 32, 577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y. , Xu, Q. , Canzio, D. , Shou, J. , Li, J. , Gorkin, D.U. Jung, I. et al. (2015) CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell, 162, 900–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyl, A. , Ramireddy, E. , Brenner, W.G. , Riefler, M. , Allemeersch, J. and Schmulling, T. (2008) The transcriptional repressor ARR1‐SRDX suppresses pleiotropic cytokinin activities in arabidopsis. Plant Physiol. 147, 1380–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton, I.B. , D'Ippolito, A.M. , Vockley, C.M. , Thakore, P.I. , Crawford, G.E. , Reddy, T.E. and Gersbach, C.A. (2015) Epigenome editing by a CRISPR‐Cas9‐based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 33, 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu, K. , Matsui, K. , Koyama, T. and Ohme‐Takagi, M. (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34, 733–739. [DOI] [PubMed] [Google Scholar]
- Jain, M. (2015) Function genomics of abiotic stress tolerance in plants: a CRISPR approach. Front. Plant Sci. 6, 2011–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek, M. , Chylinski, K. , Fonfara, I. , Hauer, M. , Doudna, J.A. and Charpentier, E. (2012) A programmable dual‐RNA–guided DNA endonuclease in adaptive bacterial immunity. Science (80‐.), 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, B.‐C. , Yun, J.‐Y. , Kim, S.‐T. , Shin, Y. , Ryu, J. , Choi, M. , Woo, J.W. et al. (2018) Precision genome engineering through adenine base editing in plants. Nat. Plants, 4, 427–431. [DOI] [PubMed] [Google Scholar]
- Kaufmann, K. and Walker, J.M. , eds. (2017) Plant Gene Regulatory Networks: Methods and Protocols. New York, NY: Springer. [Google Scholar]
- Khatodia, S. , Bhatotia, K. , Passricha, N. , Khurana, S.M.P. and Tuteja, N. (2016) The CRISPR/Cas genome‐editing tool: application in improvement of crops. Front. Plant Sci. 7, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor, A.C. , Kim, Y.B. , Gaudelli, N.M. , Waterbury, A.L. , Zhao, K.T. , Packer, M.S. , Kim, Y.B. et al. (2017) Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G‐to‐T: A base editors with higher efficiency and product purity. Sci. Adv. 3, eaao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann, S. , Brigham, M.D. , Trevino, A.E. , Joung, J. , Abudayyeh, O. , Barcena, C. , Hsc, P.D. et al. (2015) Genome‐scale transcriptional activation by an engineered CRISPR‐Cas9 complex. Nature, 517, 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagana’, A. , Shasha, D. and Croce, C.M. (2014) Synthetic RNAs for gene regulation: design principles and computational tools. Front. Bioeng. Biotechnol. 2, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson, M.H. , Gilbert, L.A. , Wang, X. , Lim, W.A. , Weissman, J.S. and Qi, L.S. (2013) CRISPR interference (CRISPRi) for sequence‐specific control of gene expression. Nat. Protoc. 8, 2180–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. , Zhang, D. , Xiong, X. , Yan, B. , Xie, W. , Sheen, J. and Li, J.F. (2017) A potent Cas9‐derived gene activator for plant and mammalian cells. Nat. Plants, 3, 930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, F. , Ho, W. and Crabtree, G.R. (2011) Engineering the ABA plant stress pathway for regulation of induced proximity. Sci. Signal. 4, rs2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Z. , Chen, K. , Li, T. , Zhang, Y. , Wang, Y. , Zhao, Q. , Liu, J. et al. (2017) ARTICLE Efficient DNA‐free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Publ. Gr. 8, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limera, C. , Sabbadini, S. , Sweet, J.B. and Mezzetti, B. (2017) New biotechnological tools for the genetic improvement of major woody fruit species. Front. Plant Sci. 8, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C. and Moschou, P.N. (2018) Phenotypic novelty by CRISPR in plants. Dev. Biol. 435, 170–175. [DOI] [PubMed] [Google Scholar]
- Liu, C. and Weigel, D. (2015) Chromatin in 3D: Progress and prospects for plants. Genome Biol. 16, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, R. , Chen, L. , Jiang, Y. , Zhou, Z. and Zou, G. (2015) Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov. 1, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, A. and Qi, L. (2017) Genetic and epigenetic control of gene expression by CRISPR–Cas systems. F1000Research, 6, 747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder, L.G. , Zhang, D. , Baltes, N.J. , Paul, J.W. , Tang, X. , Zheng, X. , Voytas, D.F. et al. (2015) A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 169, 971–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder, L. , Malzahn, A. and Qi, Y. (2016) Rapid evolution of manifold CRISPR systems for plant genome editing. Front. Plant Sci. 7, 1–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder, L.G. , Zhou, J. , Zhang, Y. , Malzahn, A. , Zhong, Z. , Hsieh, T.‐F. , Voytas, D.‐F. et al. (2017a) Robust transcriptional activation in plants using multiplexed CRISPR‐Act2.0 and mTALE‐Act systems. Mol. Plant, 11, 245–256. [DOI] [PubMed] [Google Scholar]
- Lowder, L.G. , Paul, J.W. and Qi, Y. (2017b) Multiplexed transcriptional activation or repression in plants using CRISPR‐dCas9‐based systems. Plant Gene Regulatory Networks, pp. 167–184. New York, NY: Humana Press. [DOI] [PubMed] [Google Scholar]
- Lu, Y. and Zhu, J.K. (2017) Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Mol. Plant, 10, 523–525. [DOI] [PubMed] [Google Scholar]
- Ma, J. (2011) Transcriptional activators and activation mechanisms. Protein Cell, 2, 879–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X. , Zhu, Q. , Chen, Y. and Liu, Y. (2016) CRISPR/Cas9 platforms for genome editing in plants: developments and applications. Mol. Plant, 9, 961–974. [DOI] [PubMed] [Google Scholar]
- Mahfouz, M.M. , Li, L. , Piatek, M. , Fang, X. , Mansour, H. , Bangarusamy, D.K. and Zhu, J.K. (2012) Targeted transcriptional repression using a chimeric TALE‐SRDX repressor protein. Plant Mol. Biol. 78, 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali, P. , Aach, J. , Stranges, P.B. , Esvelt, K.M. , Moosburner, M. , Kosuri, S. , Yang, L. et al. (2013) CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31, 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, K.S. (1992) DNA looping. Microbiol. Rev. 56, 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler, R. , Finka, A. and Goloubinoff, P. (2012) How do plants feel the heat? Trends Biochem. Sci. 37, 118–125. [DOI] [PubMed] [Google Scholar]
- Molla, K.A. and Yang, Y. (2019) CRISPR/Cas‐mediated base editing: technical considerations and practical applications. Trends Biotechnol.. 10.1016/j.tibtech.2019.03.008 [DOI] [PubMed] [Google Scholar]
- Moradpour, M. and Abdulah, S.N.A. (2017) Evaluation of pEASY‐uni seamless cloning and assembly kit to clone multiple fragments of Elaeis guineensis DNA. Meta Gene, 14, 134–141. [Google Scholar]
- Morgan, S.L. , Mariano, N.C. , Bermudez, A. , Arruda, N.L. , Wu, F. , Luo, Y. , Shankar, G. et al. (2017) Manipulation of nuclear architecture through CRISPR‐mediated chromosomal looping. Nat. Commun. 8, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movahedi, A. , Sang, M. , Zhang, J. , Mohammadi, K. , Sun, W. , Yaghuti, A.A.Z. , Wei, H. et al. (2018) Functional analyses of PtROS1‐RNAi in poplars and evaluation of its effect on DNA methylation. J. Plant Biol. 61, 227–240. [DOI] [PubMed] [Google Scholar]
- Ohama, N. , Sato, H. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2017) Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 22, 53–65. [DOI] [PubMed] [Google Scholar]
- Olsson, S. (2013) The competitive effects of adopting modern biotechnical methods in plant breeding programs an economic and regulatory evaluation of the agrobiotech industry.
- Osakabe, Y. , Sugano, S.S. and Osakabe, K. (2016) Genome engineering of woody plants: past, present and future. J. Wood Sci. 62, 217–225. [Google Scholar]
- Paul, J.W. and Qi, Y. (2016) CRISPR/Cas9 for plant genome editing: accomplishments, problems and prospects. Plant Cell Rep. 35, 1417–1427. [DOI] [PubMed] [Google Scholar]
- Piatek, A. , Ali, Z. , Baazim, H. , Li, L. , Abulfaraj, A. , Al‐Shareef, S. , Aouida, M. et al. (2015) RNA‐guided transcriptional regulation in planta via synthetic dCas9‐based transcription factors. Plant Biotechnol. J. 13, 578–589. [DOI] [PubMed] [Google Scholar]
- Polstein, L.R. and Gersbach, C.A. (2015) A light‐inducible CRISPR‐Cas9 system for control of endogenous gene activation. Nat. Chem. Biol. 11, 198–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, L.S. , Larson, M.H. , Gilbert, L.A. , Doudna, J.A. , Weissman, J.S. , Arkin, A.P. and Lim, W.A. (2013) Repurposing CRISPR as an RNA‐γuided platform for sequence‐specific control of gene expression. Cell, 152, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar, A. and Daniels‐Race, T. (2013) Electrophoretic deposition of carbon nanotubes on 3‐amino‐propyl‐triethoxysilane (APTES) surface functionalized silicon substrates. Nanomaterials, 3, 272–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer, S.M. and Nakata, P.A. (2015) CRISPR/Cas9‐mediated genome editing and gene replacement in plants: transitioning from lab to field. Plant Sci. 240, 130–142. [DOI] [PubMed] [Google Scholar]
- Schubert, V. and Weisshart, K. (2015) Abundance and distribution of RNA polymerase II in Arabidopsis interphase nuclei. J. Exp. Bot. 66, 1687–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimatani, Z. , Kashojiya, S. , Takayama, M. , Terada, R. , Arazoe, T. , Ishii, H. , Teramura, H. et al. (2017) Targeted base editing in rice and tomato using a CRISPR‐Cas9 cytidine deaminase fusion. Nat. Biotechnol. 35(5), 441–443. [DOI] [PubMed] [Google Scholar]
- Tiwari, S.B. , Belachew, A. , Ma, S.F. , Young, M. , Ade, J. , Shen, Y. , Marion, C.M. et al. (2012) The EDLL motif: a potent plant transcriptional activation domain from AP2/ERF transcription factors. Plant J. 70, 855–865. [DOI] [PubMed] [Google Scholar]
- Tsai, S.Q. , Wyvekens, N. , Khayter, C. , Foden, J.A. , Thapar, V. , Reyon, D. , Goodwin, M.J. et al. (2014) Dimeric CRISPR RNA‐guided FokI nucleases for highly specific genome editing HHS Public Access Author manuscript. Nat. Biotechnol. 32, 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez‐Vilar, M. , Bernabé‐Orts, J.M. , Fernandez‐del‐Carmen, A. , Ziarsolo, P. , Blanca, J. , Granell, A. and Orzaez, D. (2016) A modular toolbox for gRNA‐Cas9 genome engineering in plants based on the GoldenBraid standard. Plant Methods, 12, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillet, F. , Perrot, L. , Chauvin, L. , Kermarrec, M.‐P. , Guyon‐Debast, A. , Chauvin, J.‐E. , Nogué, F. et al. (2019) Transgene‐free genome editing in tomato and potato plants using agrobacterium‐mediated delivery of a CRISPR/Cas9 cytidine base editor. Int. J. Mol. Sci. 20, 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, J.W. , Kim, J. , Kwon, S.Il , Corvalán, C. , Cho, S.W. , Kim, H. , Kim, S.G. et al. (2015) DNA‐free genome editing in plants with preassembled CRISPR‐Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164. [DOI] [PubMed] [Google Scholar]
- Xue, Y. and Acar, M. (2018) Live‐cell imaging of chromatin condensation dynamics by CRISPR. iScience, 4, 216–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zezulin, A. and Musunuru, K. (2018) Turning up the heat with therapeutic epigenome editing. Cell Stem Cell, 22, 10–11. [DOI] [PubMed] [Google Scholar]
- Zhang, B. (2015) MicroRNA: a new target for improving plant tolerance to abiotic stress. J. Exp. Bot. 66, 1749–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Wen, Y. and Guo, X. (2014) CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum. Mol. Genet. 23, 39–46. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Liang, Z. , Zong, Y. , Wang, Y. , Liu, J. , Chen, K. , Qiu, J.L. et al. (2016) Efficient and transgene‐free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7, 12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X.X. , Huang, L.K. , Zhang, X.Q. , Li, Z. and Peng, Y. (2014) Effects of heat acclimation on photosynthesis, antioxidant enzyme activities, and gene expression in orchardgrass under heat stress. Molecules, 19, 13564–13576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong, Y. , Wang, Y. , Li, C. , Zhang, R. , Chen, K. , Ran, Y. , Qiu, J.L. et al. (2017) Precise base editing in rice, wheat and maize with a Cas9‐cytidine deaminase fusion. Nat. Biotechnol. 35, 438–440. [DOI] [PubMed] [Google Scholar]


