Abstract
In recent years, systematic analyses of the subcellular distribution of microRNAs (miRNAs) suggest that the majority of miRNAs are present in both nuclear and cytoplasmic compartments. However, the full extent of nuclear miRNA function in cardiomyocytes is currently unknown. Here, subcellular fractionation, followed by the miRNA microarray, revealed that most miRNAs were detectable in both nuclear and cytoplasmic fractions of cardiomyocytes. We employed miR-320 as an example to explore the function of nucleus-localized miRNAs, finding that CRISPR-Cas9-mediated Ago2 knockdown abolished miR-320-induced transcriptional remodeling. Furthermore, nuclear Ago2 re-expression restored the effects of miR-320 in the nucleus. Moreover, liquid chromatography-mass spectrometry (LC-MS) analysis revealed the association of nuclear Ago2 with transcription factors YLP motif-containing protein 1 (Ylpm1) and single-stranded DNA binding protein 1 (Ssbp1). Intersection of the data of transcriptome-sequencing (seq) with Ago2-chromatin immunoprecipitation (ChIP)-seq revealed that the binding of Ago2 with the target promoter DNA may require promoter RNAs. Specifically, Cep57 was upregulated, whereas Fscn2 was downregulated by miR-320, and a similar effect was also observed by knockdown of their promoter RNA, respectively. Chromatin isolation by RNA purification (ChIRP) analysis showed decreased binding of the Cep57 and Fscn2 promoter RNA on their promoter DNA by miR-320 overexpression.Our work provided a preliminary idea that promoter RNA transcripts act as “pioneers” to disrupt chromatin that permits Ago2/miR-320 complexes to target Cep57 or Fscn2 promoter DNA for transcriptional regulation. miRNAs are naturally located in both cytoplasm and nucleus; however, their pathophysiological functions are largely unknown. Our work provided a theoretical basis for developing nuclear miRNA-based therapeutics against various diseases in the future.
Keywords: nuclear microRNA, cardiomyocytes, promoter RNA
Introduction
MicroRNAs (miRNAs) are a class of endogenous small noncoding RNAs (ncRNAs) that maintain gene-network balance mainly by inhibiting target mRNAs expression through binding to their 3′ UTR region.1,2 miRNAs play important roles in many essential biological and pathological processes, such as cardiovascular diseases, metabolism diseases, and cancers.3, 4, 5, 6 The prevailing view is that miRNAs function to regulate mRNA stability and translation in the cytoplasm; however, multiple studies have detected miRNAs in the nuclear compartment.7 For example, as early as 2004, the Tuschl lab8 showed that up to 20% of mature miR-21 was localized to the nucleus. Subsequently, Hwang et al.9 showed that miR-29b was predominantly nuclear localized, whereas miR-29a was mainly located in the cytoplasm, and the key difference between these two miRNAs was the presence of a hexanucleotide sequence (5′-AGUGUU-3′) at the 3′ terminus of miR-29b. In fact, systematic analyses of the subcellular distribution of miRNAs even suggested that the majority of miRNAs were present in both nuclear and cytoplasmic compartments.10,11 These studies brought out the notion that miRNAs might have general functional roles in the nucleus.7
Functionally, nucleus-localized miRNAs were suggested to participate in epigenetic regulation. For example, miR-205 directed transcriptional activation of the interleukin (IL) genes IL-24 and IL-32 though interactions with their promoter region.12 Paradoxically, small RNAs could also induce transcriptional gene silencing (TGS) by recruiting RNA-induced transcriptional silencing (RITS) complex to promoter-associated transcripts, which triggers alterations in chromatin structure and promoter CpG methylation.13 These studies have revealed the activation or repression effects of nuclear miRNAs on gene transcription, but the mechanistic details remain unclear, and the full extent of nuclear miRNA function is currently unknown.
We have previously investigated the role of miRNAs in various cardiovascular diseases.1,4,14, 15, 16, 17 For example, miR-320a mediated doxorubicin-induced cardiotoxicity by targeting a vascular endothelial growth factor (VEGF) signal pathway.1 miR-217 promoted cardiac hypertrophy and dysfunction by targeting the phosphatase and tensin homolog (PTEN).15 miR-21 protected against diabetic cardiomyopathy-induced diastolic dysfunction by targeting gelsolin.16 miR-30c overexpression repressed BECN1 expression by direct binding to the BECN1 3′ UTR, leading to improved cardiac function in diabetic mice.17 However, these studies only focused on the canonical role of miRNAs in cytoplasm; whether these miRNAs had regulatory function in nucleus was unclear. Because we recently detected abundant expression of miRNAs (including miR-320, miR-30, and miR-21) in the nucleus, as well as in cytoplasm, we here utilized miR-320 as an example to explore its potential targets in nucleus and answer the following questions: (1) Whether the regulation of miR-320 on transcription was mainly in nucleus? (2) Whether miR-320 directly binds to DNA or promoter-associated transcripts? (3) Whether the strand (sense or antisense) targeted by miR-320 determined its activation or repression effect?
Here, by using Ago2-chromatin immunoprecipitation-sequencing (ChIP-seq), RNA immunoprecipitation-seq (RIP-seq), and whole-transcriptome-seq, we tried to get a glimpse of the universal working model for nucleus-localized miRNAs and bring out a potential working model for the function of nuclear miRNAs.
Results
Subcellular Localization of miRNAs in Cardiomyocytes
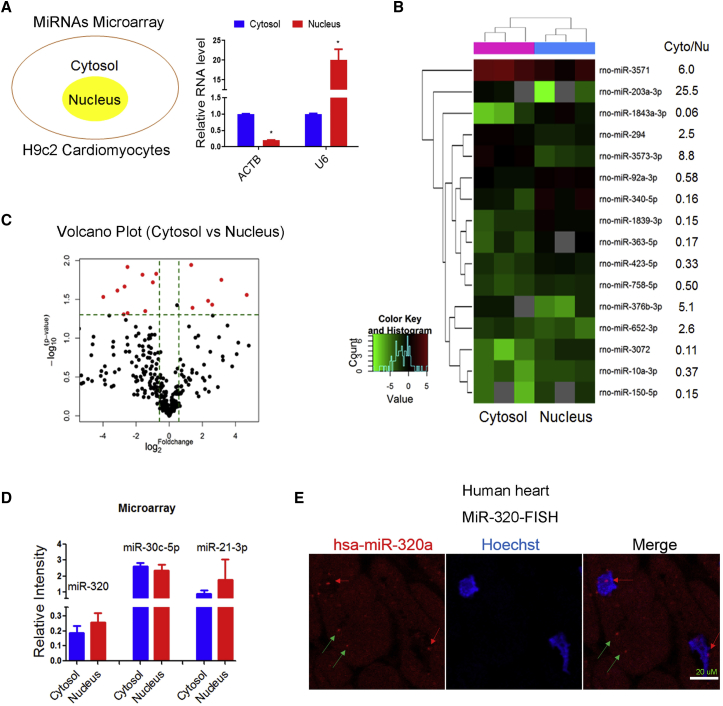
First, cell fractionation, followed by miRNA microarrays, was performed to identify nuclear and cytoplasmic-enriched miRNAs in H9c2 cardiomyocytes. The nuclear fraction was highly purified, as evidenced by the lack of cytoplasmic marker ACTB mRNA and the presence of nuclear marker U6 RNA (Figure 1A). With the use of the cutoff value of 2-fold change, only a limited number of miRNAs were relatively abundant in cytosol or nucleus (Figure 1B). The majority of miRNAs were both expressed in nucleus and cytosol (Figure 1C), which was in line with a previous finding that ∼75% of cellular miRNAs were present in both nucleus and cytoplasm.11 These data suggested that generally, miRNAs were widely distributed in both nucleus and cytoplasm; however, the nuclear functions of these miRNAs were largely unknown.
Figure 1.
Nuclear and Cytoplasmic miRNAs Microarrays
(A) Subcellular fractionation followed by cytoplasmic marker ACTB mRNA and nuclear marker U6 RNA detection by qRT-PCR; n = 3. ACTB was enriched in cytosol, whereas U6 was highly expressed in the nucleus. (B) Heatmap of miRNA microarray performed in cytoplasmic and nuclear fractions from H9c2 cardiomyocytes. (C) Volcano plot of miRNA profiles in cytoplasmic and nuclear fractions from H9c2 cardiomyocytes. (D) Expression of miR-320, miR-30c-5p, and miR-21-3p in the nucleus relative to cytoplasm by microarray; n = 3, t test was used to compare miRNA levels in the cytosol group and nucleus group. These miRNAs were highly expressed both in the cytosol and nucleus. (E) Representative image of miR-320 in human heart section by in situ hybridization. Scale bar: 20 μm.
Specifically, we have previously characterized the cytoplasmic/canonical roles of miR-320, miR-30c, and miR-21-3p in cardiovascular diseases.1,17,18 Interestingly, these miRNAs were also detected in the nucleus (Figure 1D). Furthermore, the cytoplasmic and nuclear miR-320 expression was confirmed by fluorescence in situ hybridization (FISH) in normal human heart samples (Figure 1E). In this specific study, by using miR-320 as an example, we tried to investigate the potential mechanistic insights into the nuclear roles of miRNAs.
Identification of Nuclear miR-320 Targets
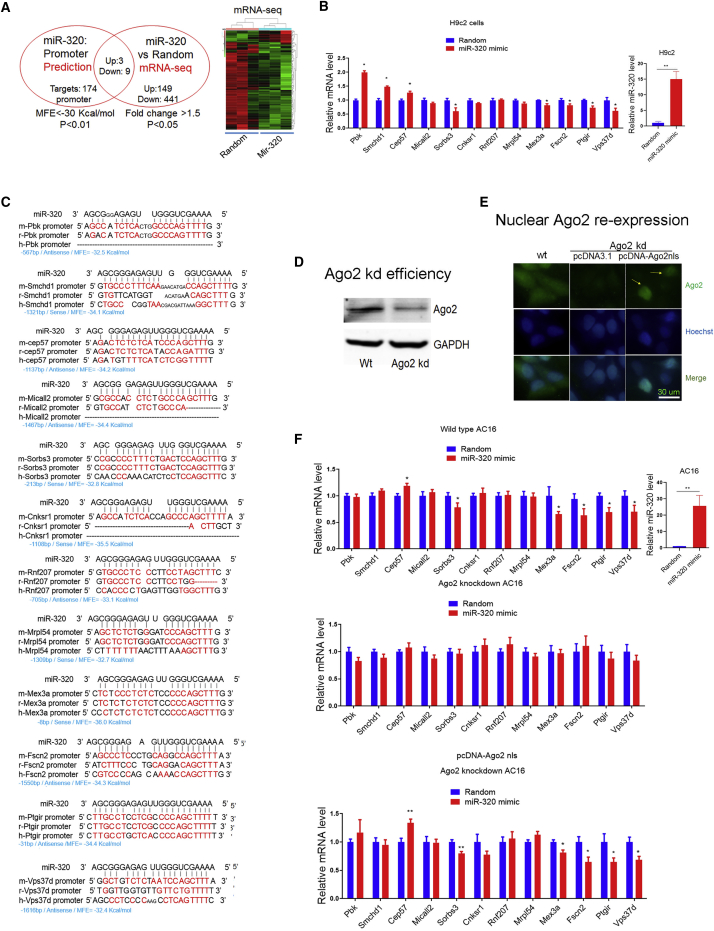
As nuclear small RNAs were suggested to regulate gene transcription by binding to gene promoters,19,20 first, the microRNA-Promoter Interaction Resource (microPIR) database was utilized to identify potential targets of miR-320 in the nucleus. With the use of bioinformatic strategies, following a set of strict filtering criteria (minimum free energy [MFE] < −30 Kcal/mol and p < 0.01), 174 potential miR-320 binding sites were identified on the mouse genome, 12 of which were functional, as evidenced by mRNA-seq analyses in miR-320-treated H9c2 cardiomyocytes (Figure 2A; Supplemental Excel). Among the 12 genes, 3 genes (Pbk, Smchd1, and Cep57) were upregulated, whereas 9 genes (Micall2, Sorbs3, Cnksr1, Rnf207, Mrpl54, Mex3a, Fscn2, Ptgir, and Vps37d) were downregulated by miR-320 (Figure 2A). Most of these genes were successfully validated by qRT-PCR in H9c2 cells (Figure 2B). In terms of conservation, most of these promoter binding sites were conserved among mouse, rat, and human species, except for a few genes, such as Pbk, Micall2, and Cnksr1 (Figure 2C). With the use of rat H9c2, human AC16, and mouse HL-1 cell lines, we tried to generate Ago2 knockout cells by using the CRISPR/Cas9 methods, because Ago2 was crucial for all miRNA functioning. However, probably due to some unknown defects in Ago2−/− cardiomyocytes, only Ago2+/− AC16 cells were obtained so far. Although a half knockout (80 bp deletion and 3 bp deletion in two chromosomes, respectively), Ago2 was significantly decreased compared to wild-type AC16 cells (Figure 2D). Then in Ago2 knockdown (kd) cells, we re-expressed Ago2 that carried a nuclear targeting signal peptide (Ago2nls), which translocated into the nucleus (Figure 2E) and restored miR-320-mediated nuclear target gene regulation (Cep57, Sorbs3, Mex3a, Fscn2, Ptgir, and Vps37d) (Figure 2F). These data, derived from Ago2+/− cardiomyocytes, strongly supported the direct action of nuclear Ago2/miR-320 in transcription control.
Figure 2.
Identification of miR-320 Targets in the Nucleus
(A) Strategy to identify miR-320 targets in the nucleus by using bioinformatics and mRNA-seq. Twelve targeting genes (up: Pbk, Smchd1, and Cep57; down: Micall2, Sorbs3, Cnksr1, Rnf207, Mrpl54, Mex3a, Fscn2, Ptgir, and Vps37d) were regulated by miR-320 in H9c2 cardiomyocytes. (B) qRT-PCR validation of genes regulated by miR-320; n = 3, *p < 0.05 versus random by t test. (C) Sequence alignment of miR-320 on gene promoter regions among species. (D) CRISPR-Cas9-mediated Ago2 kd in human AC16 cardiomyocytes. (E) Nuclear Ago2 re-expression in Ago2 kd AC16 cardiomyocytes. Scale bar: 30 μm. (F) Regulation of miR-320 on targeting genes in wild-type (WT), Ago2 kd, and nuclear Ago2 re-expression AC16 cells. qRT-PCR revealed that re-expression restored miR-320-mediated regulation on Cep57, Mex3a, Fscn2, Ptgir, and Vps37d; n = 8, *p < 0.05 versus random by t test.
Promoter RNAs Were Potentially Involved in Ago2-DNA Binding
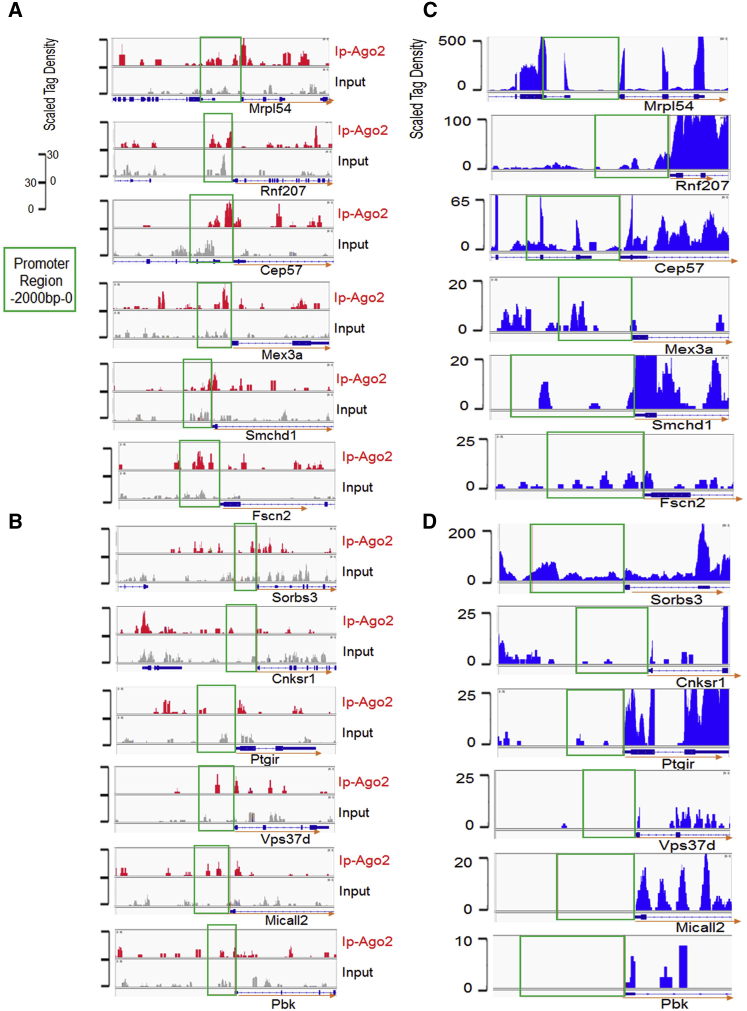
To characterize further the DNA-Ago2/miRNA binding events, Ago2-ChIP-seq was performed on normal mice heart samples in vivo. Clear Ago2 binding peaks on the promoter regions of Mrpl54, Rnf207, Cep57, Mex3a, Smchd1, and Fscn2 genes were observed, respectively (Figure 3A). However, for other genes, such as Sorbs3, Cnksr1, Ptgir, Vps37d, Micall2, and Pbk, the Ago2 binding peak was not significant (Figure 3B). We then asked what might account for the differences in Ago2 binding events on these predicted targeting gene promoters. Interestingly, abundant promoter RNA expressions were observed for Ago2-targeting promoters by whole-transcriptome sequencing on a normal mice heart (Figure 3C). In contrast, less abundant or even undetectable promoter RNAs were observed for poor Ago2 binding genes (Figure 3D), except for Sorbs3, a poor Ago2 binding gene but still detectable with abundant promoter RNA, suggesting that promoter RNA might be necessary but insufficient for Ago2 binding on the Sorbs3 promoter (Figure 3D). These preliminary data suggested the potential involvement of promoter RNAs in Ago2-promoter binding events.
Figure 3.
Ago2-ChIP-seq and Transcriptome-seq
(A and B) Ago2-ChIP-seq profiles of input DNA and immunoprecipitation in mouse heart. ChIP-seq scales were in reads per million unique mapped reads. Input samples were shown on the same scale relative to respective immunoprecipitation. (A) Gene promoter with clear Ago2 binding peaks. (B) Gene promoter with poor Ago2 binding peaks. (C and D) Whole-transcriptome-seq of mRNAs and related more (C) or less (D) abundant promoter lncRNAs in mouse heart.
Ago2/miR-320 Working Model
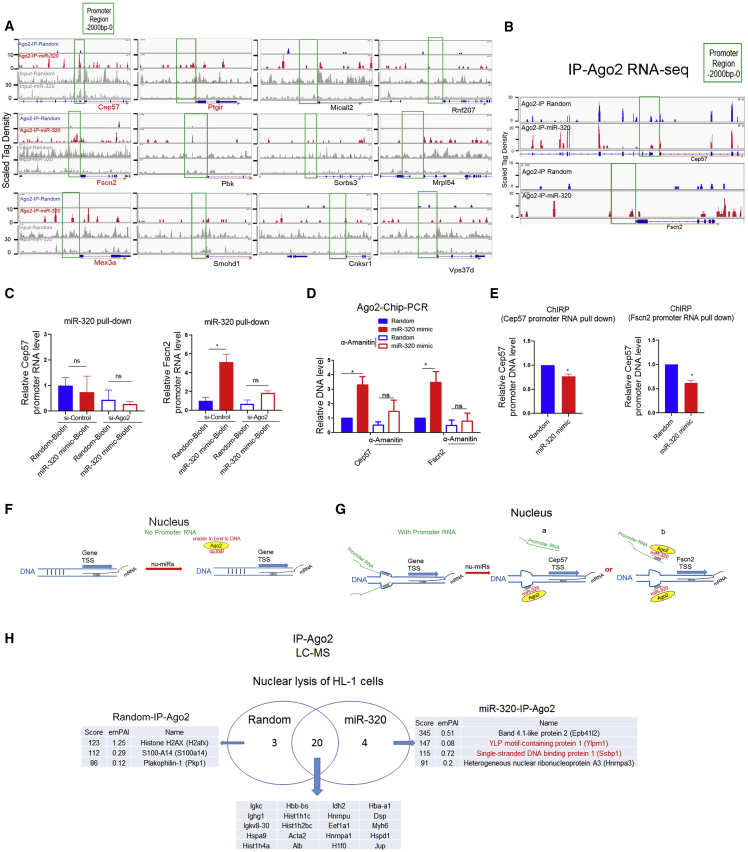
Furthermore, Ago2-ChIP-seq was performed on cardiomyocytes to explore the Ago2/miR-320-promoter binding mechanism in vitro, and increased Ago2 binding on Cep57, Fscn2, Mex3a, and Ptgir promoters by miR-320 mimic treatment in HL-1 cells was detected (Figure 4A). To determine whether Ago2/miR-320 targeted to DNA or promoter RNA, RIP-seq was performed to determine the association between Ago2 and gene promoter RNAs. Interestingly, miR-320 increased an Ago2 binding event on Fscn2 promoter RNA but not on Cep57 promoter RNA (Figure 4B). Correspondingly, an miR-320 pulldown assay revealed direct binding of miR-320 on Fscn2 promoter RNA, which was largely abolished in Ago2 kd cells. Meanwhile, Cep57 promoter RNA was not directly targeted by miR-320 (Figure 4C). These data suggested that for a specific gene, such as Cep57, Ago2/miR-320 might not necessarily/directly bind to promoter RNAs. However, we utilized α-amanitin, an inhibitor of RNA polymerase II and III, to block promoter RNA synthesis, finding that miR-320-mediated Ago2 binding on Cep57 and Fscn2 promoter DNA was significantly decreased (Figure 4D). These data suggested that the binding of Ago2/miR-320 on the gene promoter was promoter RNA dependent, even though promoter RNA itself might not be directly targeted by Ago2/miR-320. Moreover, chromatin isolation by RNA purification (ChIRP) analysis revealed decreased binding events of Cep57 and Fscn2 promoter RNA on their promoter DNA in miR-320-treated cardiomyocytes (Figure 4E). According to these data, we proposed that when there is no promoter RNA, miRNAs might not be able to bind to DNA (Figure 4F). If the promoter RNA existed, then the DNA promoter might be unstable and accessible for miRNA binding (Figure 4G). In this case, if miRNAs have no binding site on promoter RNA, then Ago2/miRNA might directly bind to DNA by competing shared binding sites with promoter RNA (Figure 4Ga); if miRNAs have a binding site on promoter RNA, then Ago2/miRNA might bind to both DNA and promoter RNA (Figure 4Gb). In either case, Ago2/miRNA binding would lead to promoter RNA dissociation from the DNA promoter (Figures 4E and 4G).
Figure 4.
Promoter ncRNAs Were Involved in miR-320-DNA Binding
(A) Ago2-ChIP-seq profiles of input DNA and immunoprecipitations in murine HL-1 cardiomyocytes transfected with miR-320 mimic or random. ChIP-seq scales were in reads per million unique mapped reads. Ago2 binding events were increased in promoter regions of Cep57, Fscn2, Mex3a, and Ptgir by miR-320. (B) Ago2-immunoprecipitation (IP) RNA-seq analysis of immunoprecipitations in H9c2 cardiomyocytes transfected with miR-320 mimic or random. RIP-seq scales were in reads per million unique mapped reads. miR-320 increased Ago2 binding on Fscn2 promoter RNA but not Cep57 promoter RNA. (C) RNA pulldown assays using streptavidin-labeled magnetic beads in HL-1 cardiomyocytes treated with the biotinylated miR-320 mimic. miR-320 pulldown showed direct targeting of miR-320 on Fscn2 promoter RNA; n = 3, *p < 0.05 by t test. (D) α-Amanitin treatment decreased miR-320-mediated Ago2 binding on gene promoters detected by Ago2-ChIP-PCR. (E) ChIRP analysis using biotinylated antisense probes to capture promoter RNAs and associated chromatin in HL-1 cells. miR-320 treatment led to detachment of Cep57 and Fscn2 promoter RNAs from their promoter DNA; n = 3, *p < 0.05 versus random by t test. (F and G) Promoter RNAs might be required for miR-320 binding on DNA. Schematic diagrams of Ago2/miRNA complexes binding to DNA without (F) or with (G) promoter RNA. Promoter RNA transcripts act as “pioneers” to disrupt chromatin that permits Ago2/miRNA complexes to target specific single-stranded DNA. (H) LC-MS analysis of Ago2-associated proteins in the nucleus.
Moreover, liquid chromatography-mass spectrometry (LC-MS) was performed to determine Ago2/miR-320 cofactors in the nucleus. Interestingly, four proteins were potentially associated with nuclear Ago2 by miR-320 overexpression (Figure 4H). Among these proteins, YLP motif-containing protein 1 (Ylpm1) and single-stranded DNA (ssDNA) binding protein 1 (Ssbp1) were transcription factors, which might collectively participate in Ago2/miR-320-mediated transcription control.
The Activation/Inhibition Effects of miR-320 Were Likely Promoter RNA Dependent
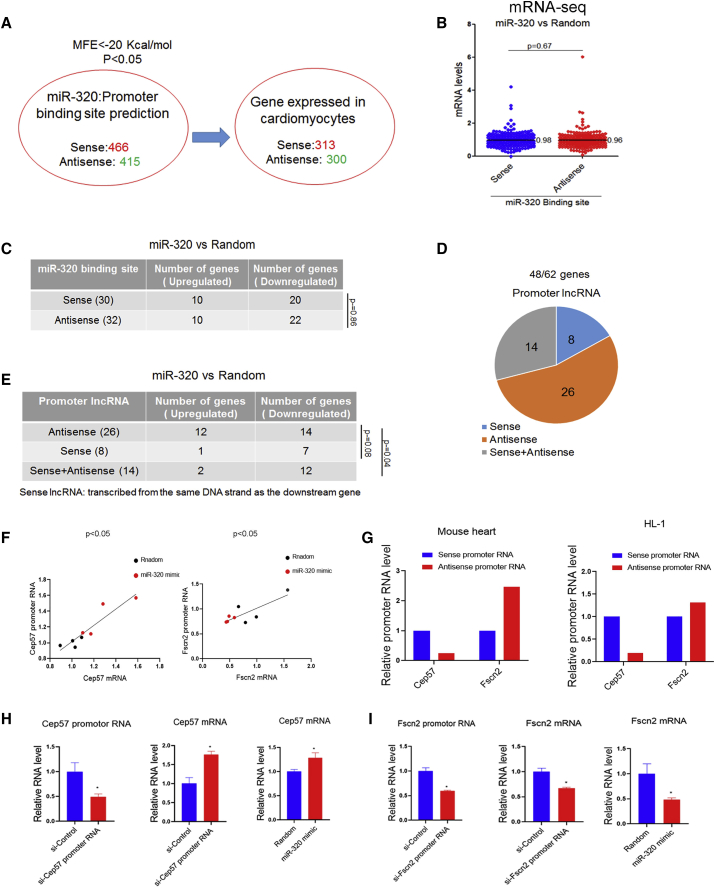
We then tried to explore whether the DNA strand selected by miR-320 would determine the activation or repression of its target gene. The sense strand was termed as the strand of DNA that has the same sequence as the mRNA. To gain a more global binding pattern, a less strict filtering criterion (MFE < −20 Kcal/mol and p < 0.05) was used for miR-320-target prediction on the mouse genome. As a result, 466 sense and 415 antisense miR-320 targeting promoter regions were identified, among which 613 promoter region-located genes were detectable in cardiomyocytes in comparison to ∼200 undetectable genes (Figure 5A). Specifically, miR-320 was predicted to target 313 sense promoters and 300 antisense promoters. However, no differences in activation or repression tendency were observed between sense and antisense binding by miR-320 mimic treatment (Figure 5B). Moreover, among 613 cardiomyocyte-expressed miR-320 targets, 62 genes were significantly regulated by miR-320 with a p value <0.05 (Figure 5C). Very interestingly, sense or antisense binding, miR-320 could lead to activation of some genes although repression of some other genes (Figure 5C). These data suggested that the selection of the miR-320 binding strand (sense or antisense) might not be able to determine the activation or repression effects of miR-320.
Figure 5.
The Activation/Inhibition Effects of miR-320 Were Likely ncRNA Dependent
(A) miR-320 binding strand analysis. With the use of the microPIR database, miR-320 was predicted to target 466 sense and 415 antisense promoters. (B) Regulation of miR-320 on sense-strand binding genes and antisense binding genes (all targeting genes). miR-320 binding on gene sense or antisense promoters had no differences in affecting gene expression. (C) Regulation of miR-320 on sense-strand binding genes and antisense binding genes (significantly regulated genes, with p < 0.05). (D) Promoter RNA classification for significantly regulated genes by miR-320. (E) Regulation of miR-320 on genes with sense or antisense promoter RNAs. (F) Coordinated expression of Cep57 and Fscn2 mRNA and their promoter RNAs was observed in HL-1 cells treated with miR-320. (G) Strand-specific qRT-PCR revealed a higher abundance of sense promoter RNA for Cep57 and antisense promoter RNA for Fscn2 in mice heart and HL-1 cell. (H) Cep57 promoter RNA-specific siRNA and miR-320 both increased Cep57 mRNA levels, as determined by qRT-PCR. (I) Fscn2 promoter RNA-specific siRNA and miR-320 both decreased Fscn2 mRNA levels, as determined by qRT-PCR; n = 3, *p < 0.05 versus random by t test.
We then tried to determine whether the strand of promoter RNA would determine the activation or repression effects of miRNAs. Among the 62 genes significantly regulated by miR-320 (Figure 5C), 48 genes were detectable with promoter RNAs by whole-transcriptome-seq on mice hearts. Promoter RNAs are classified as “sense” if they are transcribed from the same DNA strand as the downstream gene. By strand-specific RNA-seq, these 48 promoter RNAs were divided into three classes that were sense (8), antisense (26), and sense and antisense (14), due to their downstream genes (Figure 5D). Notably, a previous study demonstrated that intragenic long non-coding RNAs (lncRNAs) constituted 64% of all lncRNAs, with the majority (40%) classified as antisense lncRNAs. Meanwhile, sense lncRNAs comprised a fraction of 13%, and 11% fell in both sense and antisense categories.21 This study was in consistent with our study that the majority of promoter RNAs were antisense RNAs.
Interestingly, when antisense promoter RNAs were detectable, miR-320 treatment led to the activation of 12 genes and inhibition of 14 genes. However, when sense promoter RNAs were presented, 7/8 and 12/14 genes were downregulated by miR-320 (Figure 5E). These data suggested a possibility that the selectivity of the promoter RNA strand might determine the activation or repression effects mediated by nuclear miR-320. With Cep57 and Fscn2, for example, coordinated expression of mRNAs and promoter RNAs under miR-320 mimic treatment in cardiomyocytes was observed. Interestingly, Cep57 sense promoter RNA was more abundant than its antisense promoter RNA, whereas Fscn2 promoter RNA was more antisense abundant (Figure 5G). Functionally, Cep57 mRNA was increased by Cep57 promoter RNA kd, whereas Fscn2 mRNA was decreased by Fscn2 promoter RNA kd (Figures 5H and 5I). Importantly, miR-320 showed similar effects on Cep57 and Fscn2 mRNA as their promoter RNAs specific small interfering RNAs (siRNAs) (Figures 5H and 5I). These data indicated that promoter RNAs were indeed functional, and different promoter RNAs might have very distinct regulation patterns. miR-320-mediated gene activation (Cep57) or repression (Fscn2) seems to be dependent on the functional property (activation or repression) of the promoter RNA itself (Figure 6).
Figure 6.
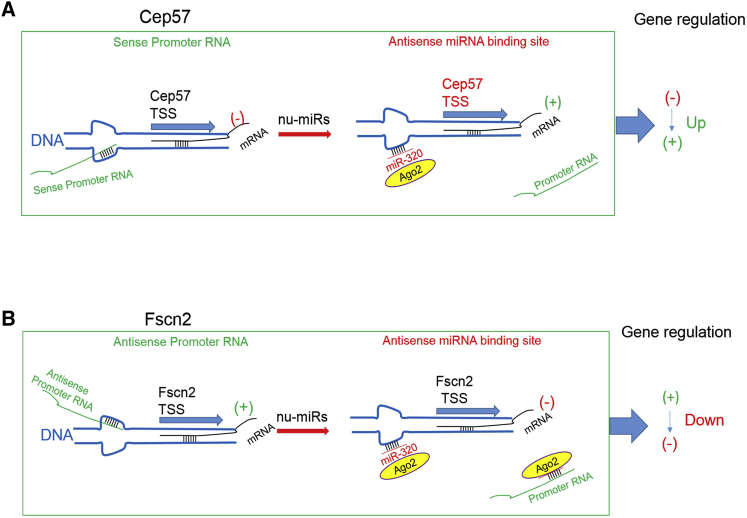
Nuclear miR-320 Working Model
(A) For Cep57, this sense strand-derived promoter RNA did not have a miR-320 binding site. Instead, Cep57 promoter RNA competed for the shared binding site on Cep57 promoter DNA. Therefore, miR-320 led to increased binding of Ago2 on Cep57 promoter DNA but not promoter RNA. (B) Fscn2 promoter RNA (antisense transcript) harbored a miR-320 binding site; in this case, miR-320 seems to bind directly on its promoter RNA, as well as promoter DNA. In either case, miR-320 overexpression led to Cep57 and Fscn2 promoter RNA detachment from the ssDNA promoter; as such, the original effects mediated by promoter RNA were compromised. The activation or repression effect mediated by miRNAs was dependent on the functional properties of promoter RNAs themselves.
Discussion
Previous systematic analyses of the subcellular distribution of miRNAs suggested that the majority of miRNAs were present in both nuclear and cytoplasmic compartments.8 The canonical action of miRNAs in cytosol via targeting 3′ UTR of mRNA have been well studied for decades. In recent years, many studies have revealed the functions of nuclear miRNAs in regulating gene transcription, for example, miR-320 mimics targeted to the POLR3D locus, leading to heterochromatinization and TGS.22 Meanwhile, miRNAs can also induce transcriptional gene activation (TGA).19,23 The biggest challenge for this field was that previous studies usually focused on an isolated miRNA; the common or universal mechanisms underlying nuclear miRNAs were largely unknown.
Our study, based on Ago2-ChIP-seq, RIP-seq, and whole-transcriptome-seq data, has brought some of the following insights, although still preliminary but quite new to this field: (1) miR-320-mediated transcription control was nuclear Ago2/miR-320 dependent; (2) Ylpm1 and Ssbp1 were closely associated with nuclear Ago2, which might collectively contribute to transcriptional remodeling mediated by nuclear miRNAs; (3) the selection of sense or antisense promoter targeted by miR-320 might not determine the activation or repression effects; (4) Ago2 binding on promoter DNA seemed to require the presence of promoter RNAs but not necessarily/directly bind to promoter RNAs; (5) for genes with sense promoter RNAs, most of them were downregulated by miR-320. With antisense promoter RNAs, one-half of the genes was upregulated and one-half was downregulated by miR-320; and (6) specifically, miR-320-mediated gene activation (Cep57) or repression (Fscn2) seems to be dependent on the functional property (activation or repression) of the promoter RNA itself.
It is still unclear whether nuclear miRNAs directly target promoter RNAs or DNA. Because α-amanitin decreased Ago2 binding on gene promoters, the Ago2 binding event on DNA seemed to require the presence of promoter RNAs. However, according to our Ago2-ChIP-seq and RIP-seq data, miR-320 increased Ago2 targeting to Fscn2 and Cep57 promoter DNA (Figure 4A) but only increased Ago2 targeting to Fscn2 promoter RNA (Figure 4D), suggesting the Ago2/miR-320 complex might not necessarily/directly bind to promoter RNAs. Therefore, the idea might be that promoter proximal lncRNA transcripts act as “pioneers” to disrupt chromatin, which permits small RNA-guided ribonucleoprotein complexes to target specific DNA (Figures 4F and 4G). Interestingly, previous studies have observed the widespread formation of RNA–DNA ⋅ DNA triples in situ by polypurine/polypyrimidine DNA probes and anti-triplex antibodies.24 An RNA third strand within the range of 12–16 bases in length could bind to a target DNA duplex.25 Whether miRNAs bound to ssDNA or double-strand DNA remained to be determined; however, our LC-MS data did suggest the association of Ago2 with Ssbp1, which was an intriguing subject for further studies.
Ago2 was generally regarded as a post-transcriptional regulator. However, our data showed that Ago2 kd prevented miR-320-mediated gene regulation at transcriptional levels. It is intriguing to ask how Ago2, an RNA-binding protein (RBP), directly acts on chromatin. Interestingly, a recent study revealed widespread RBP (including Ago2) presence in the active chromatin regions in the human genome. This study proposed that various RBPs, such as Ago2, may enhance network interaction through harnessing regulatory RNAs to control transcription.26 Moreover, Ago2 was suggested to have physical interactions with RBPs and transcriptional factors (TFs), such as Ago1, HNRNPL, RBM22, POLR2G, and POL2S2, at protein levels.26 It is possible that Ago2 and colocalized RBPs/TFs, for example. Ago1, have concerted functions at the chromatin levels. Interestingly, Ago1 had also been shown to target promoter regions by ChIP-seq.27 In the future, the detailed functions of Ago2 and its interactions with other Agos in transcriptional control need further investigation.
Since the binding of Ago2/miR-320 on gene promoters were promoter RNA dependent, it is intriguing to ask how promoter RNAs regulate the transcription of downstream genes. Enormous studies have revealed the function of antisense promoter RNAs in activating or repressing gene transcription.28 Mechanically, for gene repression, ncRNAs may (1) regulate transcription by virtue of RNA–DNA triplex formation, preventing the formation of the transcription-initiation complex at promoters and (2) act as decoys by titrating transcription factors away from their cognate promoters. For gene activation, ncRNAs may regulate transcription through the targeting of TFs to promoters or acting as cofactors involved in TF activity.29 Compared to antisense promoter RNAs, sense promoter RNAs studies were very limited, probably due to their limited number. In these studies, promoter RNAs usually (although not always) activated gene transcription (Table S2). For example, kd of COX-2 and doublesex1 (dsx1) sense promoter RNAs reduced their downstream mRNA levels.19,30 dsx1 promoter RNA overlapped dsx1 5′ UTR, which was suggested to be required for dsx1 activation.30 As such, nuclear miRNAs might compete for a binding site on DNA (Figure 6A) or directly target promoter RNA (Figure 6B). In either case, nuclear Ago2/miRNA functioning would lead to detachment of promoter RNA from DNA. This might explain why the selection of binding strand (sense or antisense) alone was not able to determine the activation or repression effects of miR-320 (Figure 5C).
Therefore, we proposed a working model for miR-320 functioning in the nucleus (Figure 6) in which nuclear miR-320 targeting to the sense or antisense promoter both leads to promoter RNA detachment from the ssDNA promoter; as such, the original effects mediated by promoter RNA were compromised. The activation or repression effect mediated by miR-320 was dependent on the functional properties of promoter RNAs themselves.
Still, there are some limitations in this study. (1) The observations and hypothesis are based on data from a limited number of miRNAs, genes, and promoter RNAs, which might not be universal in all cases. (2) Apart from nested promoter RNAs, trans-acting ncRNAs might also be involved in miRNA-mediated transcription control, which is not discussed here. (3) Whether these promoter RNAs have the ability to produce peptide or protein are still known. (4) In vitro and in vivo experiments are needed to validate further the working model of nuclear miRNAs. (5) The detail mechanism underlying sense or antisense promoter RNA was still unknown. In the future, instead of the examination of one promoter RNA at one time, large-scale bioinformatics, followed by systematic biological experiments, are needed to reveal the universal mechanism for sense and antisense promoter RNA functioning.
In summary, our work provided a possible model, although preliminary but quite new, that promoter RNA transcripts act as pioneers to disrupt chromatin, which permits Ago2/miR-320 complexes to target specific ssDNA. Ago2/miR-320 targeting (sense or antisense both) resulted in the detachment of Cep57 and Fscn2 promoter RNAs from their promoter DNA, which further leads to compromised promoter RNA functioning and transcription remodeling. This model could be used to manipulate the expression of disease-related genes and present exciting possibilities for novel therapeutic interventions in the future.
Materials and Methods
Ethics statement Human heart samples were collected at Tongji Hospital (Wuhan, China) between January 2012 and October 2014. This study was approved by the Ethics Review Board of Tongji Hospital and Tongji Medical College, and conformed to the principles outlined in the Declaration of Helsinki. Written, informed consent was obtained from individual subjects or their immediate family members in cases of incapacitation.
Cell Culture, Transfection, and Reagents
Rat H9c2 cells and human A16 cardiomyocytes from American Type Culture Collection (ATCC) were maintained in DMEM with 10% fetal bovine serum (FBS; Life Technologies, Carlsbad, CA, USA). Murine HL-1 cardiomyocytes were cultured in Claycomb medium, supplemented with 100 μM norepinephrine stock (consisting of 10 mM norephinephrine [Sigma, Shanghai, China]), and dissolved in 0.3 mM L-ascorbic acid (Sigma, Shanghai, China), 4 mM L-glutamine (Gibco, the Netherlands), and 10% FBS (Life Technologies, Carlsbad, CA), as described.31 miR-320 mimic (5′-AAAAGCUGGGUUGAGAGGGCGA-3′, 50 nM) and random small RNA control (5′-UUUGUACUACACAAAAGUACUG-3′, 50 nM) were designed by Riobio (Guangzhou, China) and transfected by Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA). α-Amanitin (Cayman Chemical; 10 μg/mL, 6 h) was used to slow the rate of transcription and transiently block RNA production.32 Plasmid (pcDNA-Ago2nls) was constructed by Tianyi Huiyuan (Wuhan, China).
Subcellular Fractionation
Cytoplasmic and nuclear fractions were prepared using the Cell Fractionation kit (Cell Signaling Technology, Danvers, MA, USA; Cat. #9038), according to the manufacturer’s protocol. Real-time PCR analysis was performed to determine cytoplasmic marker ACTB and nuclear markers U6.
miRNA Microarray
RNA samples from cytoplasmic and nuclear fractions were labeled with the Exiqon miRCURY Hy3/Hy5 power labeling kit (Exiqon, Vedbaek, Denmark) and hybridized on the miRCURY LNA microRNA array (7th generation, which contains 3,100 capture probes, covering all human, mouse, and rat miRNAs annotated in miRBase 19.0). Scanning was performed with the Axon GenePix 4000B microarray scanner (Axon Instruments, Foster City, CA, USA). Scanned images were then imported into GenePix Pro 6.0 software (Axon Instruments) for grid alignment and data extraction.
In Situ Hybridization
miRNA in situ hybridization was performed using formalin-fixed and paraffin-embedded tissue specimens, as previously described.33
miRNA-Promoter Prediction
The microPIR (http://www4a.biotec.or.th/micropir2/) and miRBase (http://www.mirbase.org/) websites were used for miRNA-promoter prediction. Base pairing to the seed region between miRNA positions 2 and 8 (allowing guanine:uracil [G:U] wobbles) is required for target recognition. Moreover, a MFE of hybridization lower than −20 Kcal/mol is sufficient for formation of a miRNA/mRNA complex,3 and MFE <−30 Kcal/mol was used as a strict cutoff setting.
Transcriptome Sequencing
Strand-specific mRNA and ncRNA sequencing and data analysis were performed by Personal Biotechnology (Shanghai, China).
ChIP-seq
ChIP assay was performed to evaluate Ago2 and targeting at specific promoters. Briefly, cardiomyocytes were fixed with 1% formaldehyde for 10 min at room temperature and then quenched by addition of 125 mM glycine. The samples were homogenized in lysis buffer and sonicated to generate chromatin samples with average fragment sizes of 200–1,000 bp. The samples were incubated with anti-Ago2 (Abnova; Cat. #H00027161-M01) at 4°C overnight on an inverse rotator. Then, Pierce protein A/G Magnetic Beads (Thermo Fisher Scientific; Cat. #88802) were added into the reactions and gently vortexed to mix. After standard washes, the immunoprecipitated DNA was eluted and purified with a PCR purification kit (QIAGEN). DNA sequencing and data analysis were performed by Personal Biotechnology. Coverage maps normalized to the total library size were used to evaluate relative Ago2 enrichment on promoter regions (−2,000 to 0 bp upstream of the transcription start site [TSS]), as described.34
CRISPR-Cas9 kd of Ago2
The PX459 (pSpCas9(BB)-2A-Puro) plasmid (Addgene; #48139) encoding Ago2 single guide RNA (sgRNA; F: 5′-CCTCCACCTAGACCCGACTT-3′) was transfected into AC16 cells by Lipofectamine 2000. At 48 h post-transfection, puromycin selection was applied at a concentration of 1 μg/mL for 72 h. Then, the remaining cells were sorted as a single cell into a 96-well plate and cultured in regular medium for an additional 3 weeks. Cells were genotyped by the PCR assay using primers (F1: 5′-GCGCTTCCTGAGTAAACGAG-3′, R1: 5′-AACTCTCCTCGGGCACTTCT-3′). Ago2+/− AC16 cells were obtained with an 80-bp deletion and a 3-bp deletion in two chromosomes, respectively.
RNA Extraction and qRT-PCR
Total RNA was isolated using TRIzol and reverse transcribed with SuperScript III First Strand Synthesis Kit (Life Technologies, Carlsbad, CA, USA). Real-time PCR assays were performed with the SYBR Select Master Mix (Life Technologies, Carlsbad, CA, USA) on a 7900HT FAST Real-Time PCR System (Life Technologies, Carlsbad, CA, USA).35 Primers used in the present study were listed in Table S1.
RIP-seq
Lysed cell extracts were immunoprecipitated with anti-Ago2 antibody (Abnova, Taiwan, China) or immunoglobulin G (IgG; Santa Cruz Biotechnology, Santa Cruz, CA, USA) using protein A/G Magnetic Beads (Thermo Fisher Scientific; Cat. #88802), as described previously.36 After elution from the beads, bound RNA was extracted with TRIzol. Subsequent RNA sequencing was performed by Personal Biotechnology (Shanghai, China).
miR-320 Pulldown
To identify the direct binding partners of miR-320, we used a tagged miRNA pulldown assay, as described.37 Biotinylated miR-320 mimic or control miRNA (designed and synthesized by Riobio, Guangzhou, China) was transfected into HL-1 cells for 48 h. Cells were then harvested, and biotinylated miR-320 was captured using streptavidin-labeled magnetic beads. After elution from the beads, bound RNA was extracted with TRIzol and quantified by real-time RT-PCR.
ChIRP
Promoter RNA probes were designed using the online probe designer at https://www.biosearchtech.com. Oligonucleotides were biotinylated at the 3′ end. HL-1 cells were transfected with miR-320 or control mimic for 48 h. Cells were collected and subjected to ChIRP using the method described.38
LC-MS Following Ago2-IP
Lysed nuclear extracts were immunoprecipitated with anti-Ago2 antibody using Pierce protein A/G Magnetic Beads. After elution from the beads, bound proteins were extracted for MS by Sangon Biotech (Shanghai).
Statistical Analysis
Data are presented as mean ±SEM. The Student’s t test was used to evaluate statistical significance among treatment groups at p <0.05.
Author Contributions
HL designed the study, analyzed, interpreted the data and drafted the paper; JZ, and YZ performed the experiments, interpreted the data and drafted the paper; JF, SY, ZY, and BD participated in acquiring the data; CC and DWW designed the work and drafted the paper.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank our colleagues in Dr. Wang’s group for technical assistance and stimulating discussions during the course of this investigation. This work was supported by the National Natural Science Foundation of China (grant numbers 81822002, 91439203, 81630010, 81790624, 31771264, and 31800973). The funders had no role in study design, data collection, and analysis, manuscript preparation, or decision to publish.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.11.006.
Contributor Information
Chen Chen, Email: chenchen@tjh.tjmu.edu.cn.
Dao Wen Wang, Email: dwwang@tjh.tjmu.edu.cn.
Supplemental Information
References
- 1.Yin Z., Zhao Y., Li H., Yan M., Zhou L., Chen C., Wang D.W. miR-320a mediates doxorubicin-induced cardiotoxicity by targeting VEGF signal pathway. Aging (Albany N.Y.) 2016;8:192–207. doi: 10.18632/aging.100876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li H., Fan J., Yin Z., Wang F., Chen C., Wang D.W. Identification of cardiac-related circulating microRNA profile in human chronic heart failure. Oncotarget. 2016;7:33–45. doi: 10.18632/oncotarget.6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li H., Zhang X., Wang F., Zhou L., Yin Z., Fan J., Nie X., Wang P., Fu X.D., Chen C., Wang D.W. MicroRNA-21 Lowers Blood Pressure in Spontaneous Hypertensive Rats by Upregulating Mitochondrial Translation. Circulation. 2016;134:734–751. doi: 10.1161/CIRCULATIONAHA.116.023926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen C., Wang Y., Yang S., Li H., Zhao G., Wang F., Yang L., Wang D.W. MiR-320a contributes to atherogenesis by augmenting multiple risk factors and down-regulating SRF. J. Cell. Mol. Med. 2015;19:970–985. doi: 10.1111/jcmm.12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Y., Yin Z., Li H., Fan J., Yang S., Chen C., Wang D.W. MiR-30c protects diabetic nephropathy by suppressing epithelial-to-mesenchymal transition in db/db mice. Aging Cell. 2017;16:387–400. doi: 10.1111/acel.12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai M.M., Wang C.S., Tsai C.Y., Huang H.W., Chi H.C., Lin Y.H., Lu P.H., Lin K.H. Potential Diagnostic, Prognostic and Therapeutic Targets of MicroRNAs in Human Gastric Cancer. Int. J. Mol. Sci. 2016;17:945. doi: 10.3390/ijms17060945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts T.C. The MicroRNA Biology of the Mammalian Nucleus. Mol. Ther. Nucleic Acids. 2014;3:e188. doi: 10.1038/mtna.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Hwang H.W., Wentzel E.A., Mendell J.T. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 10.Liao J.Y., Ma L.M., Guo Y.H., Zhang Y.C., Zhou H., Shao P., Chen Y.Q., Qu L.H. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS ONE. 2010;5:e10563. doi: 10.1371/journal.pone.0010563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gagnon K.T., Li L., Chu Y., Janowski B.A., Corey D.R. RNAi factors are present and active in human cell nuclei. Cell Rep. 2014;6:211–221. doi: 10.1016/j.celrep.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majid S., Dar A.A., Saini S., Yamamura S., Hirata H., Tanaka Y., Deng G., Dahiya R. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer. 2010;116:5637–5649. doi: 10.1002/cncr.25488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts T.C., Andaloussi S.E.L., Morris K.V., McClorey G., Wood M.J.A. Small RNA-Mediated Epigenetic Myostatin Silencing. Mol. Ther. Nucleic Acids. 2012;1:e23. doi: 10.1038/mtna.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan J., Li H., Nie X., Yin Z., Zhao Y., Zhang X., Yuan S., Li Y., Chen C., Wang D.W. MiR-665 aggravates heart failure via suppressing CD34-mediated coronary microvessel angiogenesis. Aging (Albany N.Y.) 2018;10:2459–2479. doi: 10.18632/aging.101562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nie X., Fan J., Li H., Yin Z., Zhao Y., Dai B., Dong N., Chen C., Wang D.W. miR-217 Promotes Cardiac Hypertrophy and Dysfunction by Targeting PTEN. Mol. Ther. Nucleic Acids. 2018;12:254–266. doi: 10.1016/j.omtn.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai B., Li H., Fan J., Zhao Y., Yin Z., Nie X., Wang D.W., Chen C. MiR-21 protected against diabetic cardiomyopathy induced diastolic dysfunction by targeting gelsolin. Cardiovasc. Diabetol. 2018;17:123. doi: 10.1186/s12933-018-0767-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C., Yang S., Li H., Yin Z., Fan J., Zhao Y., Gong W., Yan M., Wang D.W. Mir30c Is Involved in Diabetic Cardiomyopathy through Regulation of Cardiac Autophagy via BECN1. Mol. Ther. Nucleic Acids. 2017;7:127–139. doi: 10.1016/j.omtn.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan M., Chen C., Gong W., Yin Z., Zhou L., Chaugai S., Wang D.W. miR-21-3p regulates cardiac hypertrophic response by targeting histone deacetylase-8. Cardiovasc. Res. 2015;105:340–352. doi: 10.1093/cvr/cvu254. [DOI] [PubMed] [Google Scholar]
- 19.Matsui M., Chu Y., Zhang H., Gagnon K.T., Shaikh S., Kuchimanchi S., Manoharan M., Corey D.R., Janowski B.A. Promoter RNA links transcriptional regulation of inflammatory pathway genes. Nucleic Acids Res. 2013;41:10086–10109. doi: 10.1093/nar/gkt777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Portnoy V., Lin S.H.S., Li K.H., Burlingame A., Hu Z.H., Li H., Li L.C. saRNA-guided Ago2 targets the RITA complex to promoters to stimulate transcription. Cell Res. 2016;26:320–335. doi: 10.1038/cr.2016.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goyal A., Myacheva K., Groß M., Klingenberg M., Duran Arqué B., Diederichs S. Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res. 2017;45:e12. doi: 10.1093/nar/gkw883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim D.H., Saetrom P., Snøve O., Jr., Rossi J.J. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc. Natl. Acad. Sci. USA. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Place R.F., Li L.C., Pookot D., Noonan E.J., Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. USA. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohno M., Fukagawa T., Lee J.S., Ikemura T. Triplex-forming DNAs in the human interphase nucleus visualized in situ by polypurine/polypyrimidine DNA probes and antitriplex antibodies. Chromosoma. 2002;111:201–213. doi: 10.1007/s00412-002-0198-0. [DOI] [PubMed] [Google Scholar]
- 25.Roberts R.W., Crothers D.M. Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science. 1992;258:1463–1466. doi: 10.1126/science.1279808. [DOI] [PubMed] [Google Scholar]
- 26.Xiao R., Chen J.Y., Liang Z., Luo D., Chen G., Lu Z.J., Chen Y., Zhou B., Li H., Du X. Pervasive Chromatin-RNA Binding Protein Interactions Enable RNA-Based Regulation of Transcription. Cell. 2019;178:107–121.e18. doi: 10.1016/j.cell.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alló M., Agirre E., Bessonov S., Bertucci P., Gómez Acuña L., Buggiano V., Bellora N., Singh B., Petrillo E., Blaustein M. Argonaute-1 binds transcriptional enhancers and controls constitutive and alternative splicing in human cells. Proc. Natl. Acad. Sci. USA. 2014;111:15622–15629. doi: 10.1073/pnas.1416858111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geisler S., Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Angrand P.O., Vennin C., Le Bourhis X., Adriaenssens E. The role of long non-coding RNAs in genome formatting and expression. Front. Genet. 2015;6:165. doi: 10.3389/fgene.2015.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato Y., Perez C.A.G., Mohamad Ishak N.S., Nong Q.D., Sudo Y., Matsuura T., Wada T., Watanabe H. A 5′ UTR-Overlapping LncRNA Activates the Male-Determining Gene doublesex1 in the Crustacean Daphnia magna. Curr. Biol. 2018;28:1811–1817.e4. doi: 10.1016/j.cub.2018.04.029. [DOI] [PubMed] [Google Scholar]
- 31.Claycomb W.C., Lanson N.A., Jr., Stallworth B.S., Egeland D.B., Delcarpio J.B., Bahinski A., Izzo N.J., Jr. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc. Natl. Acad. Sci. USA. 1998;95:2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X., Farnung L., Wigge C., Cramer P. Cryo-EM structure of a mammalian RNA polymerase II elongation complex inhibited by α-amanitin. J. Biol. Chem. 2018;293:7189–7194. doi: 10.1074/jbc.RA118.002545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaudhuri A.D., Yelamanchili S.V., Fox H.S. Combined fluorescent in situ hybridization for detection of microRNAs and immunofluorescent labeling for cell-type markers. Front. Cell. Neurosci. 2013;7:160. doi: 10.3389/fncel.2013.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park P.J. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H., Chen C., Fan J., Yin Z., Ni L., Cianflone K., Wang Y., Wang D.W. Identification of cardiac long non-coding RNA profile in human dilated cardiomyopathy. Cardiovasc. Res. 2018;114:747–758. doi: 10.1093/cvr/cvy012. [DOI] [PubMed] [Google Scholar]
- 36.Li H., Dai B., Fan J., Chen C., Nie X., Yin Z., Zhao Y., Zhang X., Wang D.W. The Different Roles of miRNA-92a-2-5p and let-7b-5p in Mitochondrial Translation in db/db Mice. Mol. Ther. Nucleic Acids. 2019;17:424–435. doi: 10.1016/j.omtn.2019.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z., Zhang X.J., Ji Y.X., Zhang P., Deng K.Q., Gong J., Ren S., Wang X., Chen I., Wang H. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat. Med. 2016;22:1131–1139. doi: 10.1038/nm.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z., Chao T.C., Chang K.Y., Lin N., Patil V.S., Shimizu C., Head S.R., Burns J.C., Rana T.M. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. USA. 2014;111:1002–1007. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.