Abstract
Long non-coding RNAs (lncRNAs) play critical roles in tumorigenesis and tumor progression. However, the biological function of most lncRNAs remains unknown in human gastric cancer. This study here aims to explore the unknown function of lncRNA MAGI2-AS3 in gastric cancer. First, bioinformatics analysis showed that lncRNA MAGI2-AS3 was overexpressed in gastric cancer tissues, and the overexpression of MAGI2-AS3 has been shown to be associated with poor prognosis in all three independent gastric cancer cohorts (The Cancer Genome Atlas stomach cancer [TCGA_STAD], GEO: GSE62254 and GSE15459). The multivariate analysis indicated that lncRNA MAGI2-AS3 was an independent prognostic factor for both overall survival and disease-free survival of gastric cancer patients. Moreover, MAGI2-AS3 was identified to be an epithelial-mesenchymal transition (EMT)-related lncRNA and was highly co-expressed with ZEB1/2 in both gastric cancer tissues and normal stomach tissues. Loss-of-function and gain-of-function studies showed that lncRNA MAGI2-AS3 could positively regulate ZEB1 expression and the process of cell migration and invasion in gastric cancer. Subcellular location assay showed that lncRNA MAGI2-AS3 was mainly located in the cytoplasm of gastric cancer cells. Bioinformatics analysis and functional experiments revealed that lncRNA MAGI2-AS3 was negatively correlated with miR-141/200a expression and negatively regulated miR-141/200a-3p expression in gastric cancer. Therefore, we speculate that lncRNA MAGI2-AS3 promotes tumor progression through sponging miR-141/200a and maintaining overexpression of ZEB1 in gastric cancer. Nevertheless, we identified that BRD4 is a transcriptional regulator of lncRNA MAGI2-AS3 in gastric cancer. Additionally, our findings highlight that lncRNA MAGI2-AS3 is an ideal biomarker and could be a potential therapeutic target for gastric cancer.
Keywords: lncRNA MAGI2-AS3, ZEB1, miR-141, miR-200a, BRD4, gastric cancer
Introduction
Gastric cancer is the fourth most diagnosed type of cancer and the third most common cause of cancer-related death worldwide.1,2 Approximately 989,600 cases are diagnosed with gastric cancer worldwide annually. More than half of those cases are diagnosed in East Asia.3 However, many gastric cancer patients are diagnosed at advanced stages, presenting with malignant proliferation, extensive invasion, and distant metastasis. So, the 5-year survival rate of gastric cancer patients is still unsatisfactory. Characteristic progressive tumorigenesis and distant metastasis may contribute a lot to the overall poor prognosis for gastric cancer.4 Therefore, it is essential to develop novel biomarkers that could accurately predict cancer stage or reflect an individual’s cancer risk.
LncRNAs are more than 200 nt in length with no or limited protein-coding capacity.5 At first, lncRNAs are considered to be useless sequences generated during transcription and have no function. However, as more and more studies have recently discovered that lncRNA plays a role in the regulation of a variety of cellular processes, including stem cell pluripotency, apoptosis, cell differentiation, and cell invasion, the past understanding of lncRNA has been proved to be absolutely wrong.6 Nowadays, more and more lncRNAs are found to play a very important role in tumorigenesis via affecting multiple cellular processes, such as epithelial-mesenchymal transition (EMT), cell invasion, cell growth, and so on.7 For example, lncRNA PTAR (pro-transition associated RNA) promotes EMT and invasion-metastasis in ovarian cancer via sponging miR-101 to maintain ZEB1 overexpression.8 In another study, lncRNA TRERNA1 functions as an enhancer of SNAI1 to promote gastric cancer progression via regulating the EMT process.2 Therefore, identifying key lncRNAs involved in gastric cancer progression is important for understanding the mechanism of this disease.
LncRNA MAGI2-AS3 is rarely studied in cancers. Recently, only a few studies have reported its anti-cancer effects in breast cancer, bladder cancer, and liver cancer. In breast cancer, lncRNA MAGI2-AS3 inhibited breast cancer progression via positively regulating the Fas/FasL signal pathway.9 In bladder cancer, Wang et al.10 reported that lncRNA MAGI2-AS3 functioned as a competing endogenous RNA (ceRNA) to suppress bladder cancer progression through negatively regulating the miR-15b-5p/CCDC19 signal axis. In hepatocellular carcinoma, lncRNA MAGI2-AS3 prevents the development of cancer via recruiting KDM1A and promoting H3K4me2 demethylation of the RACGAP1 promoter.11 However, the function of lncRNA MAGI2-AS3 in gastric cancer remains unknown to date.
Here, according to the analysis of lncRNA MAGI2-AS3 expression in The Cancer Genome Atlas (TCGA) stomach cancer (TCGA-STAD) cohort and Asian Cancer Research Group (ACRG) cohort, we identified that lncRNA MAGI2-AS3 was overexpressed in gastric cancer tissues and predicted a poor prognosis for gastric cancer. Moreover, lncRNA MAGI2-AS3 was an EMT-related lncRNA in gastric cancer due to it being highly co-expressed with ZEB1, and we found that lncRNA MAGI2-AS3 promoted gastric cancer progression by sponging miR-141/200a and maintaining ZEB1 overexpression. Additionally, BRD4 might be a transcriptional regulator of lncRNA MAGI2-AS3. Knockdown of BRD4 expression or treatment with JQ1 to prevent BRD4 from recognizing H3K27AC both remarkably decreased MAGI2-AS3 expression in gastric cancer.
Results
LncRNA MAGI2-AS3 Is Overexpressed in Gastric Cancer Tissues
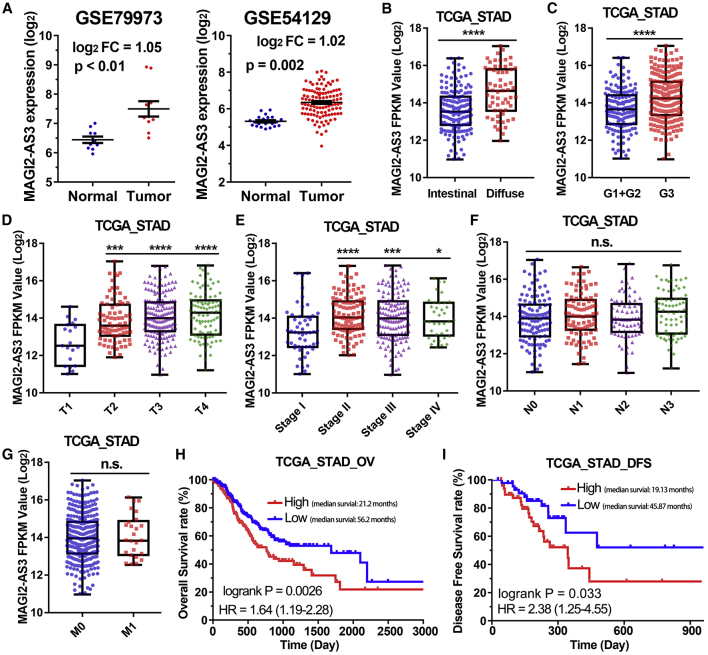
To explore the lncRNA MAGI2-AS3 expression profile in human gastric cancer tissues, we downloaded two microarray gene profiling datasets (GEO: GSE54129 and GSE79973) from public GEO datasets of NCBI. The GSE54129 dataset consists of 21 normal and 111 gastric cancer samples; GSE79973 dataset consists of 10 paired gastric cancer samples. After analysis of lncRNA MAGI2-AS3 expression data of gastric cancer patients from GSE54129 and GSE79973 cohorts, we found that the lncRNA MAGI2-AS3 expression level in tumor tissues was higher than that of the corresponding normal stomach tissues (Figure 1A).
Figure 1.
The Clinical Significance of lncRNA MAGI2-AS3 Expression Was Analyzed in TCGA-STAD Cohort
(A) Difference in expression levels of MAGI2-AS3 between GSE79973 and GSE54129 stomach cancer cohorts. (B) Difference in expression levels of MAGI2-AS3 between intestinal gastric cancer tissues and diffuse gastric cancer tissues. (C) Difference in expression levels of MAGI2-AS3 in gastric cancer tissues with different degrees of differentiation. (D–G) MAGI2-AS3 expression level in different T stages (D), pathologic stages (E), N stages (F), and M stages (G) of gastric cancer. (H and I) Kaplan-Meier analysis of overall survival time (H) and disease-free time (I) of patients with gastric cancer according to the expression of lncRNA MAGI2-AS3. ****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05.
MAGI2-AS3 Overexpression Predicts Poor Prognosis of Patients from TCGA Gastric Cancer Cohort
To understand the correlation between MAGI2-AS3 overexpression and clinicopathological characteristics of patients with gastric cancer, we performed a comprehensive transcriptome analysis of RNA-seq data from 373 gastric cancer tissues with complete clinical information. The results showed that the expression level of lncRNA MAGI2-AS3 was significantly increased in diffuse gastric cancer tissues compared with intestinal gastric cancer tissues (p < 0.0001; Figure 1B). Moreover, lncRNA MAGI2-AS3 was overexpressed in poorly differentiated gastric cancer tissues compared with moderately or highly differentiated gastric cancer tissues (p < 0.0001; Figure 1C). Furthermore, lncRNA MAGI2-AS3 was more highly expressed in tumors extending beyond the gastric mucosa layer (T2+T3+T4) compared with tumors detected only in the gastric mucosa (T1), and more highly expressed in tumors with a high degree of malignancy (stage II+III+IV) compared with tumors with a low degree of malignancy (stage I; Figures 1D and 1E). However, there was no significant difference in the expression of MAGI2-AS3 in gastric cancer tissues with or without lymph node metastasis or distant metastasis (Figures 1F and 1G).
To evaluate the association between lncRNA MAGI2-AS3 expression and the prognosis of patients with gastric cancer, we assigned 373 patients to the high MAGI2-AS3 expression groups (n = 192) and the low MAGI2-AS3 expression group (n = 181) according to their different expression levels of lncRNA MAGI2-AS3 (Table S1). The cutoff value was determined from the median for the analysis of the 373 patients described below (Figure S1A; Table S1). Then we found that patients who possessed higher expression level of MAGI2-AS3 had both a shorter overall survival (OV) time and a shorter disease-free survival (DFS) time than those with lower MAGI2-AS3 expression level (Figures 1H and 1I). Additionally, the results of multivariate analysis for OV and DFS suggested that lncRNA MAGI2-AS3 expression level was an independent prognostic factor for OV and DFS of gastric cancer patients in TCGA-STAD cohort (hazard ratio, 1.24 and 2.51, respectively; 95% confidence interval [CI]: 1.07–1.42 and 1.17–5.38, respectively; p = 0.0031 and p = 0.0181, respectively; Table 1). Collectively, these results suggested that higher expression of lncRNA MAGI2-AS3 predicted a poorer prognosis in gastric cancer.
Table 1.
The Cox Regression Analysis for Overall Survival (OVS) and Disease-free Survival (DFS) in TCGA Gastric Cancer Cohort
| Factors | OVS |
DFSa |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate Cox |
Multivariate Cox |
Univariate Cox |
Multivariate Cox |
|||||||||
| HR | 95% CI | p Value | HR | 95% CI | p Value | HR | 95% CI | p Value | HR | 95% CI | p Value | |
| MAGI2-AS3 | 1.18 | 1.04–1.34 | 0.0102b | 1.24 | 1.07–1.42 | 0.0031b | 2.54 | 1.21–5.29 | 0.0133b | 2.51 | 1.17–5.38 | 0.0181b |
| Sex | 0.81 | 0.57–1.15 | 0.2442 | N/A | N/A | N/A | 1.22 | 0.56–2.66 | 0.6096 | N/A | N/A | N/A |
| Age | 1.64 | 1.16–2.32 | 0.0051b | 1.98 | 1.38–2.83 | 0.0002b | 1.01 | 0.97–1.05 | 0.6087 | N/A | N/A | N/A |
| T stage | 1.39 | 1.14–1.71 | 0.0014b | 1.28 | 1.02–1.59 | 0.0297b | 0.9 | 0.55–1.49 | 0.6907 | N/A | N/A | N/A |
| N stage | 1.3 | 1.13–1.49 | 0.0002b | 1.26 | 1.09–1.45 | 0.0018b | 0.9 | 0.67–1.23 | 0.5159 | N/A | N/A | N/A |
| M stage | 1.44 | 1.05–1.99 | 0.0237b | 1.36 | 0.99–1.86 | 0.0555b | 0.56 | 0.08–4.15 | 0.5731 | N/A | N/A | N/A |
| Histological type | 0.95 | 0.64–1.41 | 0.7936 | N/A | N/A | N/A | 0.93 | 0.44–1.98 | 0.8502 | N/A | N/A | N/A |
CI, confidence interval; HR, hazard ratio, N/A, not available.
Only 97 gastric cancer patients in TCGA cohort possessed detailed DFS events and time information.
P < 0.05.
MAGI2-AS3 Overexpression Predicts a Poor Prognosis of Gastric Cancer Patients in the ACRG Cohort
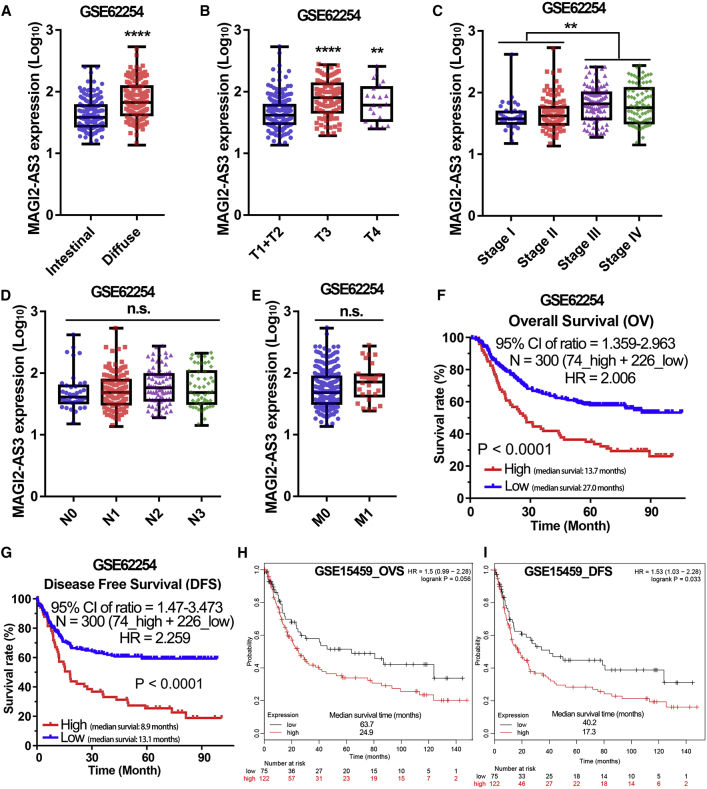
To further verify the oncogenic role of lncRNA MAGI2-AS3 in gastric cancer, we introduced another gastric cancer cohort (GEO: GSE62254) into our study. The ACRG (GEO: GSE62254) cohort contains 300 gastric cancer tissues with very detailed clinical information,12 and the GSE62254 microarray gene profiling dataset based on GPL570 platform contained lncRNA MAGI2-AS3 expression data. After re-annotation and analysis of the gene expression profile data in GSE62254 cohort, we found that MAGI2-AS3 expression level was higher in the diffuse gastric cancer tissues than in intestinal gastric cancer tissues (Figure 2A). Furthermore, MAGI2-AS3 was found to be overexpressed in tumors with a high degree of malignancy compared with the tumors with a low degree of malignancy (Figures 2B and 2C). Additionally, we also noted that lncRNA MAGI2-AS3 had no significant correlation with lymph node metastasis or distant metastasis in the GSE62254 cohort (Figures 2D and 2E).
Figure 2.
The Clinical Significance of lncRNA MAGI2-AS3 Expression Was Analyzed in the GSE62254 and GSE15459 Cohorts
(A) Difference in expression levels of MAGI2-AS3 between intestinal gastric cancer tissues and diffuse gastric cancer tissues. (B–E) MAGI2-AS3 expression level in different T stages (B), pathologic stages (C), N stages (D), and M stages (E) of gastric cancer tissues. (F and G) Kaplan-Meier analysis of overall survival time (F) and disease-free time (G) of gastric cancer patients in the GES62254 cohort according to the expression of lncRNA MAGI2-AS3. (H and I) Kaplan-Meier analysis of overall survival time (H) and disease-free survival time (I) in the gastric cancer cohort of GSE15459 according to the expression of lncRNA MAGI2-AS3. ****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05.
Similarly, we further evaluated the correlation between lncRNA MAGI2-AS3 expression and the prognosis of gastric cancer patients in the GSE62254 cohort. According to the MAGI2-AS3 expression data, the 300 patients in the ACRG cohort were divided into the high MAGI2-AS3 expression groups (n = 74) and the low MAGI2-AS3 expression group (n = 226; Table S1). The cutoff value was determined from the median for the analysis of the 300 patients described below (Figure S1B). After analysis of the survival curve, we found that high expression of lncRNA MAGI2-AS3 was also significantly associated with both poor overall survival rate and disease-free survival rate in the GSE62254 cohort (p < 0.0001; Figures 2F and 2G). Moreover, the results of multivariate analysis for OV and DFS also suggested that lncRNA MAGI2-AS3 was an independent prognostic factor for the OV and DFS of gastric cancer patients in the GSE62254 cohort (hazard ratio, 3.67 and 4.00, respectively; 95% CI: 1.19–11.33 and 1.15–13.92, respectively; p = 0.0235 and 0.0293, respectively; Table 2). In addition, in the third independent gastric cancer cohort (GEO: GSE15459), patients with higher expression of MAGI2-AS3 also had a shorter OV time and DFS time (Figures 2H and 2I). These results strongly suggested that lncRNA MAGI2-AS3 is a very reliable biomarker that can accurately predict the prognosis of gastric cancer patients.
Table 2.
The Cox Regression Analysis for Overall Survival (OVS) and Disease-free Survival (DFS) in the GSE62254 Gastric Cancer Cohort
| Factors | OVS |
DFS |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate Cox |
Multivariate Cox |
Univariate Cox |
Multivariate Cox |
|||||||||
| HR | 95% CI | p Value | HR | 95% CI | p Value | HR | 95% CI | p Value | HR | 95% CI | p Value | |
| MAGI2-AS3 | 6.73 | 2.43–18.66 | 0.0002a | 3.67 | 1.19–11.33 | 0.0235a | 8.33 | 2.71–25.55 | 0.0002a | 4 | 1.15–13.92 | 0.0293a |
| Sex | 1.11 | 0.79–1.55 | 0.5587 | N/A | N/A | N/A | 1.04 | 0.72–1.51 | 0.8303 | N/A | N/A | N/A |
| Age | 1.01 | 1–1.03 | 0.1812 | N/A | N/A | N/A | 1.47 | 1.03–2.09 | 0.0331a | N/A | N/A | N/A |
| T stage | 1.78 | 1.43–2.22 | 0.0000a | 1.29 | 1.01–1.65 | 0.0438a | 1.75 | 1.37–2.22 | 0.0000a | 1.24 | 0.95–1.62 | 0.1094 |
| N stage | 1.96 | 1.63–2.34 | 0.0000a | 1.72 | 1.42–2.09 | 0.0000a | 2.08 | 1.71–2.53 | 0.0000a | 1.81 | 1.46–2.23 | 0.0000a |
| M stage | 3.84 | 2.48–5.94 | 0.0000a | 2.29 | 1.45–3.6 | 0.0003a | 4.04 | 2.51–6.51 | 0.0000a | 2.11 | 1.28–3.47 | 0.0034a |
| Lauren | 0.58 | 0.42–0.8 | 0.0010a | 0.87 | 0.61–1.24 | 0.4354 | 0.59 | 0.41–0.85 | 0.0041a | 0.9 | 0.61–1.34 | 0.6090 |
CI, confidence interval; HR, hazard ratio; N/A, not available.
P < 0.05.
MAGI2-AS3 Is an EMT-Related lncRNA and Is Highly Co-expressed with ZEB1
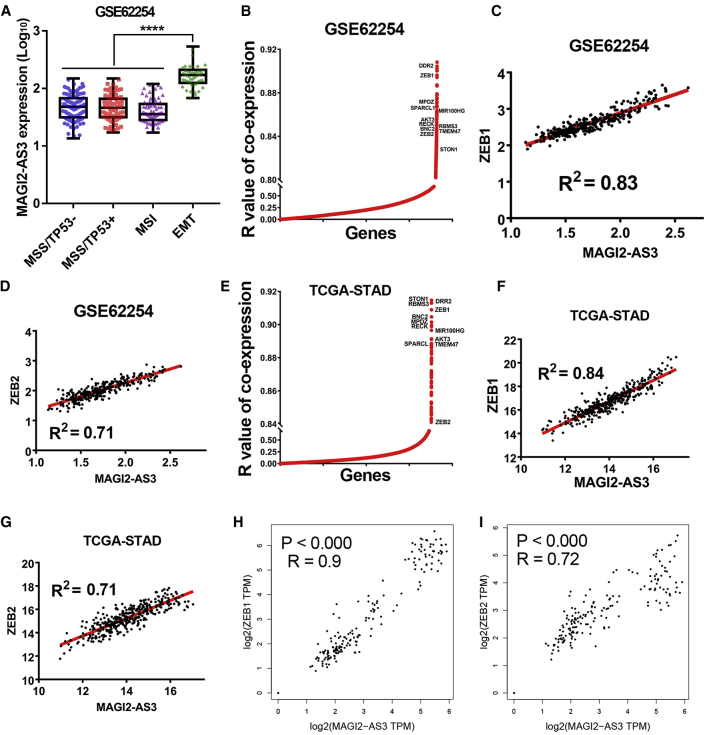
Previous studies had reported that gastric cancer could be further divided into four subtypes, including microsatellite stablity (MSS)/TP53−, MSS/TP53+, MSI, and MSS/EMT subtypes, according to their molecular subtypes.12 Therefore, lncRNA MAGI2-AS3 expression level in the four subtypes of gastric cancer was analyzed. Interestingly, lncRNA MAGI2-AS3 showed an extremely high expression level in the MSS/EMT subtype of gastric cancer tissues, which indicated that MAGI2-AS3 is an EMT-related lncRNA (p < 0.0001; Figure 3A).
Figure 3.
MAGI2-AS3 Was an EMT-Related lncRNA and Highly Co-expressed with ZEB1/2 in Normal Gastric Tissues and Gastric Cancer Tissues
(A) Expression level of lncRNA MAGI2-AS3 in the four subtypes (MSS/TP53−, MSS/TP53+, MSI, and EMT) of gastric cancer in the GSE62254 cohort. (B) R values of co-expression between different genes and lncRNA MAGI2-AS3 were analyzed in the GSE62254 cohort. (C and D) lncRNA MAGI2-AS3 was highly co-expressed with ZEB1 (C) and ZEB2 (D) in the GSE62254 cohort. (E) R values of co-expression between different genes and lncRNA MAGI2-AS3 were analyzed in the TCGA_STAD cohort. (F and G) lncRNA MAGI2-AS3 was highly co-expressed with ZEB1 (F) and ZEB2 (G) in TCGA-STAD cohort. (H and I) lncRNA MAGI2-AS3 was highly co-expressed with ZEB1 (H) and ZEB2 (I) in normal stomach tissues from the GTEx dataset. ****p < 0.0001.
To figure out why lncRNA MAGI2-AS3 was so highly expressed in the MSS/EMT subtype of gastric cancer, we first analyzed the expression relationship between all the different mRNAs and MAGI2-AS3 in the ACRG gastric cancer cohort. The results revealed that ZEB1 and ZEB2, the two widely accepted core regulators of EMT process,13 were both highly co-expressed with MAGI2-AS3 in the GSE62254 gastric cancer cohort (Figures 3B–3D); ZEB1 was one of the top three genes that showed the highest co-expression with MAGI2-AS3 (R > 0.92); and the high co-expression relationship between ZEB1/ZEB2 and MAGI2-AS3 was further confirmed in TCGA-STAD cohort (Figures 3E–3G). In addition, ZEB1/ZEB2 and MAGI2-AS3 were also highly co-expressed in the normal gastric tissues by analyzing their expression level in the genotype-tissue expression (GTEx) datasets through using the GEPIA online web tool (Figures 3H and 3I). Those results together indicated that lncRNA MAGI2-AS3 was highly co-expressed with ZEB1/2 transcripts in both normal stomach tissues and gastric cancer tissues.
LncRNA MAGI2-AS3 Mainly Locates in Cytoplasm and Promotes Cell Migration and Invasion in gastric cancer
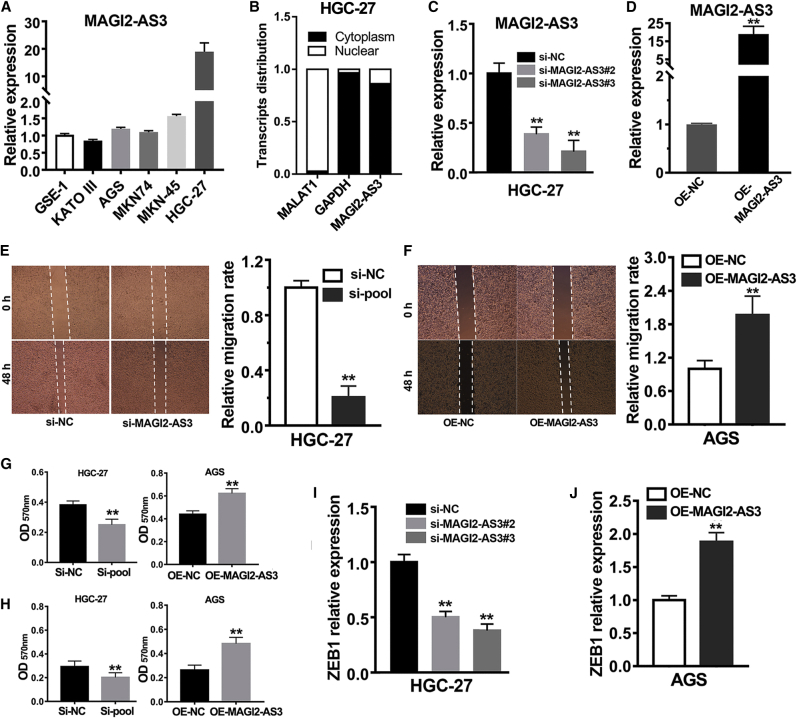
Due to our above results strongly suggesting that lncRNA MAGI2-AS3 might be an oncogene in gastric cancer, we then considered verifying the oncogenic role of lncRNA MAGI2-AS3 in GC cell lines. First, we examined the expression of lncRNA MAGI2-AS3 in different gastric cancer cell lines by performing qRT-PCR assays. The result showed that MAGI2-AS3 was highly expressed in the HCG-27 cell line but hardly expressed in other studied cell lines (Figure 4A). The subcellular location of lncRNA plays an important role in helping us interpret and understand the mechanisms of lncRNA function.14,15 Therefore, we used a one-step method to simultaneously separate cytoplasm RNA and nucleus RNA. After reverse transcription and determination of transcripts level of lncRNA MAGI2-AS3 by qPCR, we noted that lncRNA MAGI2-AS3 transcripts were mainly located in cytoplasm of HGC-27 cells (Figure 4B).
Figure 4.
MAGI2-AS3 Positively Regulated Gastric Cancer Cell Migration and Invasion, and Positively Regulated ZEB1 Expression in gastric cancer
(A) lncRNA MAGI2-AS3 expression level in different gastric cancer cell lines was determined by qRT-PCR assay. (B) Relative lncRNA MAGI2-AS3 expression level in cytoplasm and nuclear of the HGC-27 cell line was determined by qRT-PCR assay. (C and D) The efficiency of knockdown (C) and overexpression (D) of lncRNA MAGI2-AS3 was determined by qRT-PCR assay. (E and F) The wound healing assays were performed in gastric cancer cells after knockdown (E) and overexpression (F) of lncRNA MAGI2-AS3. (G) The Transwell migration assays were performed in gastric cancer cells after knockdown and overexpression of lncRNA MAGI2-AS3. (H) The Transwell invasion assays were performed in gastric cancer cells after knockdown and overexpression of lncRNA MAGI2-AS3. (I and J) Relative ZEB1 expression level in gastric cancer cells after knockdown (I) and overexpression (J) of lncRNA MAGI2-AS3 was determined by qRT-PCR assay. **p < 0.01.
Based on the MAGI2-AS3 expression level in different gastric cancer cell lines, we knocked down the expression of lncRNA MAGI2-AS3 in the HCG-27 cell line and overexpressed lncRNA MAGI2-AS3 in the AGS cell line. After examination of the transcript abundance of lncRNA MAGI2-AS3 by qPCR, we found that the RNAi experiments of MAGI2-AS3 in the HGC-27 cell line and the overexpression experiments of MAGI2-AS3 in the AGS cell line were successful (Figures 4C and 4D). The EMT process is an essential step for tumor cells to acquire metastasis.16,17 Considering that MAGI2-AS3 is an EMT-related lncRNA, we further evaluated the effects of MAGI2-AS3 knockdown and overexpression on gastric cancer cell migration and cell invasion. The wound healing assays, the Transwell migration assays, and the Transwell invasion assays were conducted in different gastric cancer cell lines. The wound healing assays and the Transwell migration assays showed that knockdown expression of MAGI2-AS3 significantly suppressed gastric cancer cell migration, whereas overexpression of MAGI2-AS3 remarkably promoted gastric cancer cell migration (Figures 4E–4G). The Transwell invasion assays showed that knockdown expression of MAGI2-AS3 significantly suppressed gastric cancer cell invasion, whereas overexpression of MAGI2-AS3 remarkably promoted gastric cancer cell invasion (Figure 4H). These results together suggested that lncRNA MAGI2-AS3 positively regulated the process of cell migration and invasion in gastric cancer.
LncRNA MAGI2-AS3 Positively Regulated ZEB1 Expression but Negatively Regulated miR-141 and miR-200a Expression in gastric cancer
Due to MAGI2-AS3 being highly co-expressed with ZEB1, we further investigated ZEB1 transcripts level after knockdown and overexpression of MAGI2-AS3 in gastric cancer cell lines. The results showed that knockdown expression of MAGI2-AS3 remarkably decreased ZEB1 expression (Figure 4I), whereas overexpression of MAGI2-AS3 remarkably increased ZEB1 expression (Figure 4J). These results suggested that lncRNA MAGI2-AS3 could positively regulate ZEB1 expression in gastric cancer.
However, it is unclear how lncRNA MAGI2-AS3 positively regulates ZEB1 expression in gastric cancer. MicroRNA (miRNA)-141 and miR-200a were known to be negative regulators of ZEB1 gene and the EMT pathway.18,19 Also, our results have shown that lncRNA MAGI2-AS3 was mainly located in the cytoplasm of gastric cancer cells. Given these factors, we thought that lncRNA MAGI2-AS3 might regulate ZEB1 expression through two mechanisms: one is the ceRNA mechanism, and the other is that lncRNA MAGI2-AS3 directly binds on ZEB1 mRNA and protects ZEB1 mRNA stability from degradation by miRNAs.
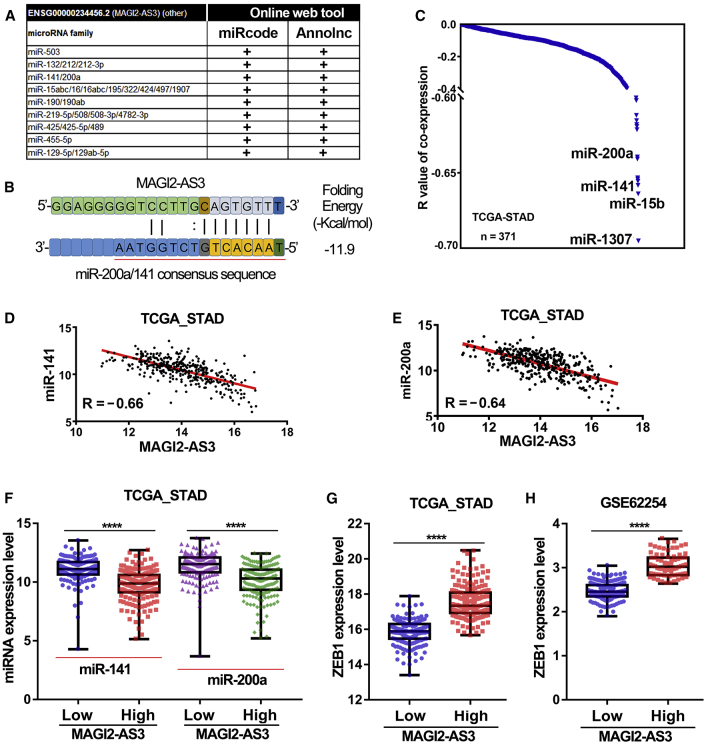
So, we predicted the RNA-RNA interactions between lncRNA MAGI2-AS3 and 3′ UTR of ZEB1 using the online tool IntaRNA.20 According to the prediction results, we found the interaction region could not mask the binding sites of miR-141 and miR-200a (Figure S2). Therefore, it is highly probable that lncRNA MAGI2-AS3 promoted gastric cancer progression through ceRNA mechanism. To further verify this possibility, we explored the possible miRNAs that interacted with lncRNA MAGI2-AS3 by using two different online web tools: miRcode and Annolnc. The results showed that a total of nine miRNA families have potential ability to interact with MAGI2-AS3 (Figure 5A). Among them, one of the predicted miRNA families was the miR-141/200a family. Then, we further verified their binding site on lncRNA MAGI2-AS3 and folding energy by using the RNA22 online web server (Figure 5B). On the other hand, we downloaded miRNA expression data in stomach cancer from TCGA dataset and analyzed the expression correlation of all miRNAs to MAGI2-AS3. The results showed that lncRNA MAGI2-AS3 was highly negatively correlated with miR-141-3p and miR-200a-3p expression (Figures 5C–5E).
Figure 5.
LncRNA MAGI2-AS3 Was Negatively Correlated with miR-141/200a Expression in gastric cancer Tissues
(A) Possible miRNA families that could interact with lncRNA MAGI2-AS3 were predicted by two different online web tools. (B) The binding region and the folding energy of miR-141/200a family and MAGI2-AS3 were predicted by the RNA22 web server. (C) R values of co-expression between different miRNAs and lncRNA MAGI2-AS3 were analyzed in the TCGA_STAD cohort. (D and E) lncRNA MAGI2-AS3 showed a negative correlation with miR-141 (D) and miR-200a (E) expression in the TCGA_STAD cohort. (F) The difference in expression level of miR-141 and miR-200a between the high MAGI2-AS3 expression group and low MAGI2-AS3 expression group in the TCGA_STAD cohort. (G and H) The difference in expression level of ZEB1 between high MAGI2-AS3 expression group and low MAGI2-AS3 expression group in the TCGA_STAD cohort (G) and GSE62254 cohort (H). ****p < 0.0001.
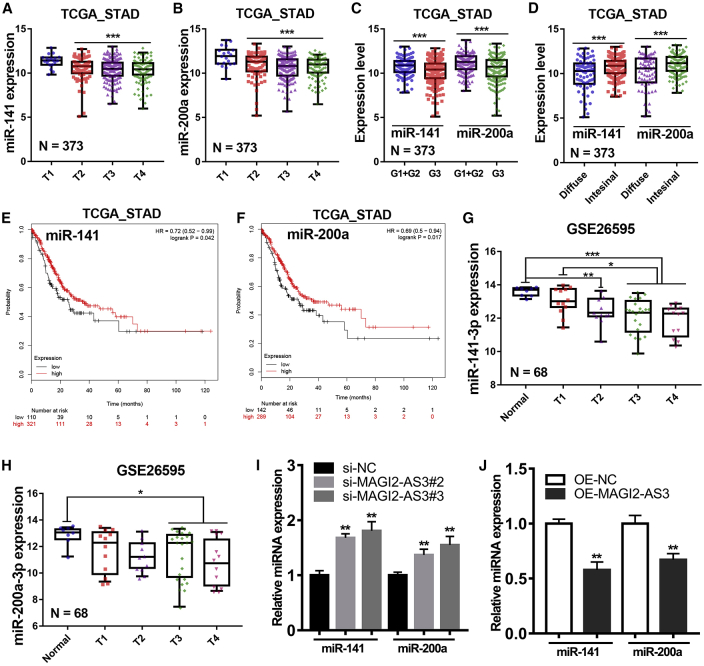
Besides, we further analyzed the expression of miR-141, miR-200a, and ZEB1 in the high MAGI2-AS3 expression groups and the low MAGI2-AS3 expression group in TCGA cohort and GSE62254 cohort. The results showed that the patients in high MAGI2-AS3 expression groups possessed lower expression of miR-141 and miR-200a, but had higher expression of ZEB1 compared with patients in the low MAGI2-AS3 expression group (Figures 5F–5H). Additionally, as mentioned above, MAGI2-AS3 was lowly expressed in gastric mucosa (T1), but highly expressed in poorly differentiated (G3) or diffuse type gastric cancers. After analysis of the relationship between miR-141/200a expression level and clinical pathology of gastric cancer in TCGA, we found that miR-141/200a highly expressed in gastric mucosa (T1), but lowly expressed in poorly differentiated (G3) or diffuse type gastric cancers (Figures 6A–6D). That is, the expression pattern of miR-141 and miR-200a in gastric cancer is completely opposite to that of lncRNA MAGI2-AS3. Besides, gastric cancer patients with lower expression level of miR-141 or miR-200a had a poorer overall survival time compared with those patients with higher expression level of miR-141 or miR-200a, respectively (Figures 6E and 6F). Moreover, the fourth independent gastric cancer cohort (GEO: GSE26596, n = 68) was introduced into our study. After reannotation and analysis of these data, we found that miR-141 and miR-200a were lowly expressed in tumors compared with normal gastric tissues (Figures 6G and 6H). These results together suggested that miR-141 and miR-200a showed negative correlation with lncRNA MAGI2-AS3 in gastric cancer.
Figure 6.
LncRNA MAGI2-AS3 Negatively Regulated miR-141 and miR-200a Expression in gastric cancer
(A and B) Difference in expression levels of miR-141 (A) and miR-200a (B) in different T stages of gastric cancer tissues in TCGA cohort. (C) Difference in expression levels of miR-141/200a in gastric cancer tissues with different degrees of differentiation. (D) Difference in expression levels of miR-141/200a between intestinal gastric cancer tissues and diffuse gastric cancer tissues from TCGA cohort. (E and F) Kaplan-Meier analysis of overall survival time in TCGA cohort according to the expression of miR-141 (E) and miR-200a (F). (G and H) Difference in expression levels of miR-141 (G) and miR-200a (H) in normal stomach tissues and different T stage of gastric cancer tissues from the GSE26595 cohort. (I and J) Relative miR-141 and miR-200a expression levels in gastric cancer cells after knockdown (I) and overexpression (J) of lncRNA MAGI2-AS3 was determined by qRT-PCR assay. ***p < 0.001; **p < 0.01; *p < 0.05.
Transcription factor ZEB1 was known to be a target gene of miR-141/200a in diverse kinds of cancers.21 Therefore, we further examined the expression levels of miR-141-3p and miR-200a in gastric cancer cell lines after knockdown and overexpressed lncRNA MAGI2-AS3 expression. As expected, miR-141-3p and miR-200a expression levels in the HGC-27 cells silencing MAGI2-AS3 expression were significantly higher than those in the negative control HGC-27 cells, whereas overexpression of lncRNA MAGI2-AS3 significantly decreased miR-141 and miR-200a expression in AGS cells (Figures 6I and 6J). These results strongly indicated that lncRNA MAGI2-AS3 could negatively regulate mature miR-141/200a-3p expression in gastric cancer.
MAGI2-AS3 Is Transcriptionally Regulated by BRD4
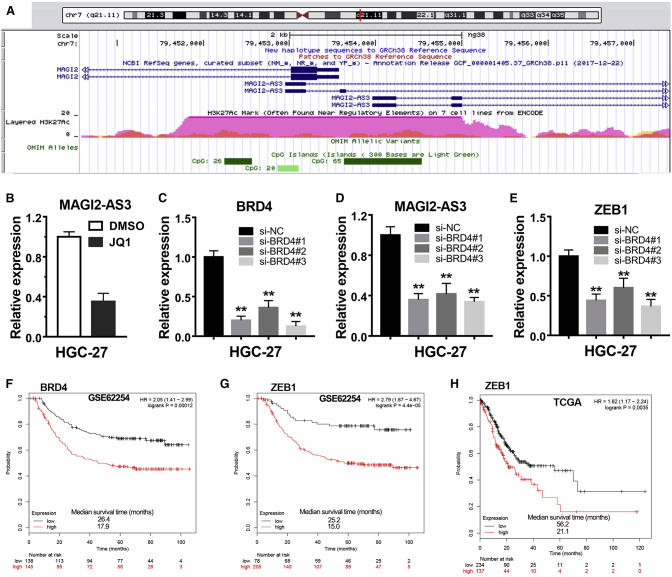
In addition, we noted the presence of abundant histone H3K27ac modifications in the promoter region of lncRNA MAGI2-AS3 using a University of California Santa Cruz (UCSC) online web tool (Figure 7A). Mountains of evidence showed that histone modifications were one of the manners to regulate gene expression.22 Among diverse kinds of the histone modifications, H3K27ac mark was especially recognized by the BET family of bromodomain proteins, such as BRD4.23,24 Therefore, we first considered verifying whether lncRNA MAGI2-AS3 expression level was changed or not after treatment of HGC-27 cells with JQ1, a potent inhibitor of the BET family of bromodomain proteins.25 A significant decrease of MAGI2-AS3 expression was observed in the HGC-27 cells treated with JQ1 compared with the negative control team (Figure 7B).
Figure 7.
BRD4 Transcriptionally Regulated lncRNA MAGI2-AS3 Expression in gastric cancer
(A) Abundant H3K27Ac marks were observed in the promoter region of MAGI2-AS3 according to the UCSC web server. (B) lncRNA MAGI2-AS3 expression remarkably decreased in the HGC-27 cells treated with 2.5 μM JQ1 compared with negative control. (C) Knockdown efficiency of BRD4 by RNA interference in HGC-27 cell line. (D and E) Knockdown of BRD4 expression reduced lncRNA MAGI2-AS3 expression (D) and ZEB1 expression (E) in gastric cancer. (F and G) Kaplan-Meier analysis of overall survival time in the gastric cancer cohort of GSE62254 according to BRD4 (F) and ZEB1 (G) expression levels. (H) Kaplan-Meier analysis of overall survival time in the gastric cancer cohort of TCGA according to ZEB1 expression. **p < 0.01.
Then, we wanted to know which bromodomain protein of the BET family had played a role in regulating MAGI2-AS3 expression. The ToolKit application in the Cistrome web server helped us a lot. By using this application to survey potential transcriptional regulators, we found that BRD4 was the most probable bromodomain protein that could regulate MAGI2-AS3 expression because BRD4 binding peaks were observed in five different samples (Table S2). To verify this possibility, we knocked down the expression level of BRD4 in the HGC-27 cells by transfection with small interfering RNAs (siRNAs) that targeted to BRD4 (Figure 7C). The results showed that knockdown of BRD4 significantly downregulated MAGI2-AS3 expression and suggested that BRD4 was a transcriptional regulator of MAGI2-AS3 in gastric cancer (Figure 7D). Besides, we further investigated ZEB1 expression level after knockdown of BRD4 in HGC-27. As expected, knockdown of BRD4 significantly decreased ZEB1 expression level (Figure 7E). In addition, we further analyzed the association between survival rate of gastric cancer patients and the expression level of BRD4 and ZEB1 in gastric cancer tissues by using Kaplan-Meier Plotter tools. The results showed that gastric cancer patients with high expression level of BRD4 or ZEB1 both had a shorter survival time than gastric cancer patients with low expression of BRD4 and ZEB1 (Figures 7F–7H).
Discussion
Gastric cancer, a leading cause of cancer-related deaths, is a heterogeneous disease with many kinds of subtypes. Mountains of evidence have shown that different subtypes of gastric cancer usually possess distinct clinical outcomes.26,27 Hence diverse kinds of biomarkers are used to distinguish subtypes of gastric cancer. Nowadays, long non-coding RNAs are found to be functional and could act as novel biomarkers to predict clinical outcomes in all kinds of cancers.28 Therefore, in order to provide a rational treatment option and acquire a better outcome for stomach cancer patients, more and more lncRNAs needed to be developed and identified as diagnostic and prognostic biomarkers in gastric cancer.
In this study, we identified the lncRNA MAGI2-AS3 as a novel and reliable biomarker that could accurately predict the prognosis of patients with gastric cancer. A variety of different independent gastric cancer cohorts were used to verify accuracy and reliability of the prediction by MAGI2-AS3, including TCGA stomach cancer (STAD) cohort, gastric cancer cohort of the ACRG (GEO: GSE62254, Korea), and the Singapore gastric cancer cohort (GEO: GSE15459). Also, the phenomenon that higher expression of MAGI2-AS3 predicts poorer prognosis of gastric cancer patients existed in all of the studied gastric cancer cohorts. Besides, our multivariate analysis for OV and DFS in two independent gastric cancer cohorts (TCGA_STAD and GEO: GSE62254) both suggested that lncRNA MAGI2-AS3 was an independent prognostic marker of gastric cancer (Tables 1 and 2).
As described above, lncRNA MAGI2-AS3 is an EMT-related lncRNA and is highly co-expressed with the core EMT regulators ZEB1 and ZEB2 (Figure 3). Given that EMT is an essential process for tumor cells to acquire invasion,29 we thought lncRNA MAGI2-AS3 may play a role in gastric cancer cell migration and invasion. Loss-of-function and gain-of-function studies in different gastric cancer cell lines showed that lncRNA MAGI2-AS3 positively regulated the process of cell migration and invasion in gastric cancer (Figures 4E–4H). These results strongly indicated that MAGI2-AS3 indeed functioned as an oncogene in gastric cancer progression.
However, the reason why MAGI2-AS3 are highly co-expressed with ZEB1 is still unknown. Considering that ZEB1 was a transcription factor, the first possible reason for explaining the high co-expression of ZEB1 and MAGI2-AS3 is that ZEB1 can directly regulate the expression of lncRNA MAGI2-AS3. However, after analysis of the genetic alterations of ZEB1 gene in TCGA-STAD samples, we noted that lncRNA MAGI2-AS3 was still highly expressed in some ZEB1 mutant samples (Figure S3). It suggested that the high co-expression between MAGI2-AS3 and ZEB1 may not be due to ZEB1-regulated MAGI2-AS3, but MAGI2-AS3-regulated ZEB1, and the subsequent studies showed that MAGI2-AS3 could positively regulate the expression of ZEB1 in gastric cancer (Figures 4I and 4H), which further confirms our hypothesis.
Subcellular location assay showed that lncRNA MAGI2-AS3 was mainly located in cytoplasm of gastric cancer cells (Figure 4B). As a cytoplasm lncRNA, there are only two reasons (the ceRNA mechanism or lncRNA-mRNA interactions) that can be used to explain how MAGI2-AS3 regulates ZEB1 expression.20,30 Because the interaction region between lncRNA MAGI2-AS3 and ZEB1 mRNA could not mask the binding sites of miR-141 and miR-200a (Figure S2), it is highly probable that lncRNA MAGI2-AS3 may regulate ZEB1 gene expression through ceRNA mechanism. Consistent with this inference, it has been found that the roles of lncRNA MAGI2-AS3 in bladder cancer,10 breast cancer,31 lung cancer,32 and liver cancer33 were achieved through the ceRNA mechanism.
Hence we further predicted probable miRNAs that may interact with lncRNA MAGI2-AS3 by bioinformatics analysis (Figures 5A and 5B), given that lncRNA MAGI2-AS3 was negatively correlated with miR-141 and miR-200a expression in gastric cancer tissues (Figures 5C–5E). So, we further examined ZEB1 transcripts level and miR-141/200a-3p expression level in the HGC-27 cells after knockdown of MAGI2-AS3. The result shows that MAGI2-AS3 negatively regulates miR-141/200a-3p expression. Besides, the clinical outcome and the expression patterns of MAGI2-AS3 and miR-141/200a are almost exactly the opposite (Figures 6A–6H). That is, lncRNA MAGI2-AS3 positively controls ZEB1 overexpression in gastric cancer through sponging miR-141/200a-3p. Consistent with this speculation, in the ZEB1 upregulated samples of TCGA, MAGI2-AS3 usually possessed the highest expression level, accompanying very low expression levels of miR-141 and miR-200a (Figure S3).
However, the reason why MAGI2-AS3 was overexpressed in gastric cancer remains unclear. During investigating the function of MAGI2-AS3 in gastric cancer, we note that there are abundant H3K27Ac marks existing in the promoter region of MAGI2-AS3 by using the UCSC web server. As we all know, histone modifications are an important manner to regulate gene expression. So, we wonder whether the overexpression of MAGI2-AS3 is related to this modification. To verify this possibility, we conducted a series of experiments in gastric cancer. Both the JQ1 treatment and the BRD4 knockdown significantly decrease MAGI2-AS3 expression in HGC-27. Besides, the ZEB1 transcripts in the BRD4 knockdown cells are also reduced. Additionally, previous studies have proved that BRD4 and ZEB1 function as oncogenic roles in gastric cancer,34,35 whereas miR-141-3p functions as a tumor suppressor role in gastric cancer.36 Therefore, we speculate that the overexpression of MAGI2-AS3 in gastric cancer is caused by overexpression of BRD4. Collectively, our results prove that BRD4 is a transcriptional regulator of MAGI2-AS3 in gastric cancer.
Conclusions
Our findings reveal that MAGI2-AS3 is an EMT-related lncRNA, and overexpression of MAGI2-AS3 predicts a poor prognosis in gastric cancer. lncRNA MAGI2-AS3 was mainly located in cytoplasm and promotes tumor progression through sponging miR-141/200a to maintain overexpression of ZEB1 in gastric cancer. Additionally, we prove that BRD4 is a transcriptional regulator of MAGI2-AS3 in gastric cancer. Besides, our results highlight that lncRNA MAGI2-AS3 might be an ideal therapeutic target in gastric cancer.
Materials and Methods
RNA-Seq Data Analysis of Stomach Cancer in TCGA
RNA-seq data of 407 gastric cancer samples, miRNA-seq data of 491 gastric cancer samples, and the correlated clinical information of 443 gastric cancer samples were downloaded from The Cancer Genome Atlas (TCGA) by using SangerBox software (http://sangerbox.com/) developed by ShengXinRen (https://shengxin.ren/). Expression level per gene was calculated from log2 of its upper quartile Fragments Per Kilobase of transcript per Million mapped reads (FPKM-UQ) value. Expression level per miRNA was calculated from log2 of its TPM (transcripts per million) value.. The genetic alterations and the mutations of ZEB1 gene in TCGA-STAD samples were analyzed by using a web-based tool: cBioPortal web server (http://www.cbioportal.org/).37
Microarray Data Analysis
Public gastric cancer microarray gene profiling datasets used in this study (GEO: GSE79973, GSE54129, GSE62254, and GSE26595) were downloaded from the GEO in the NCBI web server. After passing the quality-control check, the expression values of each GEO dataset were normalized using the robust multi-array analysis (RMA). The RMA normalized expression data were further annotated using the R package in SangerBox software. The GPL 570 platform file was used to replace the probe name with corresponding gene name, which can be downloaded from the GEO dataset. The clinical information of GSE62254 gastric cancer cohort was obtained as we described previously.38 The expression data of normal stomach tissue was obtained from GTEx by using GEPIA web server (http://gepia.cancer-pku.cn/index.html). Kaplan-Meier survival plot of overall survival time of gastric cancer in the datasets of GSE22377 and GSE51105 was analyzed by the web tool Kaplan-Meier Plotter (http://kmplot.com/analysis/index.php?p=service&cancer=gastric). The possible miRNA families that interacted with lncRNA MAGI2-AS3 were predicted by miRcode (http://www.mircode.org) and Annolnc server (http://annolnc.cbi.pku.edu.cn/index.jsp). The folding energy of the interaction between MAGI2-AS3 and miR-141/200a was predicted by the RNA22 web server (https://cm.jefferson.edu/rna22/Interactive/).
Cell Culture, Transfection, and Treatment
The human gastric cancer cell lines (MKN74, MKN45, KATO III, AGS, and HGC27) and the normal gastric cell line GES-1 were purchased from the Shanghai Cell Bank of Chinese Academy of Sciences (Shanghai, China). All of the gastric cancer cell lines were cultured in DMEM containing 10% fetal bovine serum (FBS), 100 U/mL penicillin, 100 U/mL streptomycin, and 0.03% glutamine at 37°C in 5% CO2. For siRNA transfection: the siRNAs were designed and synthesized by Genepharma (Shanghai, China) and then used to knock down MAGI2-AS3 and BRD4 expression in HGC-27 (si-MAGI2-AS3#2: 5′-CCTTCACACTTCCTGCTAT-3′, si-MAGI2-AS3#3: 5′-CCACAGACACUUAACACAA-3′, si-BRD4#1: 5′-AGCUGAACCUCCCUGAUUA-3′; si-BRD4#2: 5′-UAAAUGAGCUACCCACAGA-3′, si-BRD4#3: 5′-CGGAGCCCAAGACCACCAA-3′; si-NC: 5′-UUCU-CCGAACGUGUCACGU-3′). gastric cancer cells were grown in six-well plates and transfected by Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. At 48 h post-transfection, cells were harvested for qPCR analysis. For lncRNA MAGI2-AS3 overexpression, full-length cDNA of MAGI2-AS3 (NR_038346) was linkage into pcDNA3.1 vector. The recombinant vector and empty vector were transfected into gastric cancer cells using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. For JQ1 treatment, HGC-27 cells were seeded overnight at 1 × 105 cells/well of a six-well plate. The next day, medium was changed and HGC-27 growing cells were treated with 2.5 μM JQ1 (Selleck Chemicals) and DMSO. After 24 h, cells were harvested for qPCR analysis.
Wound Healing Assay, Cell Migration, and Invasion Assay
For wound healing assay, cells were seeded in individual wells of a six-well culture plate. Transfection was performed as above. Before transfection, a sterile 100-μL pipette tip was used to longitudinally scratch a constant-diameter stripe in the confluent monolayer. The medium and cell debris were aspirated away and replaced with 2 mL of fresh medium. Photographs were taken at 0 and 48 h after wounding. For statistical analysis, 10 randomly selected fields along each wound were marked, the area of the wound was measured, and the average was calculated as the wound area of this wound. The cell suspension was prepared by digesting the cultured cells with trypsin, washing twice with serum-free medium, grinding the cells into a single-cell suspension, and adjusting the cell concentration to 1 × 105 cells/mL.
For Transwell assay, the upper chamber was placed into a 24-well plate containing 500 μL of medium containing 20% FBS. A cell suspension (200 μL) was added to the upper chamber of the Transwell module and incubated for 16 h at 37°C in 5% CO2. The Transwell module was then washed twice with PBS and fixed with pre-chilled methanol for 10 min at −20°C. The upper chamber of the Transwell module was washed twice with PBS. The cells remaining on the top surface of the upper chamber were removed with a wet cotton swab. The upper chamber was then washed three times with PBS and air-dried at the inverted position. The chamber membrane was stained with a 0.1% crystal violet staining solution at 500 μL per well for 30 min at 37°C, washed three times, and air-dried. The cell migration rate through the Transwell membrane was measured by eluting the crystal violet stained on the Transwell with 33% acetic acid and measuring the optical density (OD) 570 nm value of the eluate by a microplate reader. The cell invasion rate through the Transwell membrane, which has been incubated with Matrigel in the upper chamber side of the membrane, was measured by eluting the crystal violet stained on the Transwell with 33% acetic acid and measuring the OD 570 nm value of the eluate by a microplate reader.
Subcellular Location of lncRNA MAGI2-AS3
HGC-27 cells were seeded in individual wells of a six-well culture plate and cultured in DMEM containing 10% fetal bovine serum (FBS), 100 U/mL penicillin, 100 U/mL streptomycin, and 0.03% glutamine at 37°C in 5% CO2. The next day, when the cell plating density reached 80%–90%, cells were harvested for isolation of cytoplasm and nuclear using the Nuclear/cytoplasmic separation kit (BB-36021-2; BestBio, China) according to the manufacturer’s instructions. The obtained cytoplasmic components and cytoplasmic components were added to 500 μL of TRIzol and then extracted RNA using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions. For qRT-PCR, 1 μg of cytoplasm RNA and 1 μg of nuclear RNA were reversed to obtain cDNA using the PrimeScript RT reagent kit (Perfect Real Time, Takara). Relative fold changes of gene expression were calculated using the comparative ΔCt method. The primers for quantitation of GAPDH and lncRNA MALAT-1 transcripts level were listed below: GAPDH qF, 5′-TCACCAGGGCTGCTTTTA-3′, qR, 5′-AAGGTCATCCCTGAGCTGAA-3′; MALAT1 qF, 5′-AAAGCAAGGTCTCCCCACAAG-3′, qR, 5′-GGTCTGTGCTAGATCAAAAGGCA-3′. Each gene was run in triplicate with different cDNAs synthesized from three biological replicates.
qRT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen, USA). For detection of the mRNA level by real-time qPCR, reverse transcription was performed to obtain cDNA using the PrimeScript RT Reagent Kit (Perfect Real Time, Takara). The mRNA expressions of MAGI2-AS3, BRD4, and ZEB1 were determined by using the specific primers synthesized by Wcgene Biotech (Shanghai, China): MAGI2-AS3 qF, 5′-tgggtctgtgcagagttgag-3′, qR, 5′-agggagtctaggccccttct-3′; BRD4 qF, 5′-gtggtgcacatcatccagtc-3′, qR, ttgacttcggagccatctct-3′; ZEB1 qF, 5′-ACCTCTTCACAGGTTGCTCCT-3′, qR, 5′-AGTGCAGGAGCTGAGAGTCA-3′; ACTIN qF, 5′-ATCGTCCACCGCAAATGCTTCTA-3′, qR, 5′-AGCCATGCCAATCTCATCTTGTT-3′. For detection of the mature miRNAs level by qRT-PCR, we used miRNA-specific stem-loop primers to obtain cDNA using the PrimeScript RT reagent Kit (Perfect Real Time, Takara). For miR-141-3p stem-loop reverse primer: 5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCCATCTT-3′; miR-141-3p qPCR primer qF: 5′-ggggTAACACTGTCTGGTAAAG-3′; for mir-200a-3p stem-loop reverse primer: 5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACAC ATCGT-3′; qPCR primer qF: 5′-gggTAACACTGTCTGGTAACG; the universal primer qR: 5′-CAGTGCGTGT CGTGGAGT-3′; U6 qF: 5′-CTCGCTTCGGCAGCACA-3′; U6 qR: 5′-AACGCTTCACGAATTTGCGT-3′. Each gene was run in triplicate with different cDNAs synthesized from three biological replicates. Relative fold changes of gene expression were calculated using the comparative ΔΔCt method.
Statistical Analysis
For gene expression analysis of different subtypes of gastric cancer, the p values were estimated using Mann-Whitney nonparametric test. R values of co-expression were calculated using Pearson’s product-moment correlation coefficient. Survival curves were calculated using the Kaplan-Meier method, and differences between the curves were analyzed using the log rank test. A Cox proportional hazards model was used for multivariate survival analysis. All of the rest of the experiments used unpaired t test or one-way ANOVA test. For in vitro experiments, a minimum of triplicates per group and repetition of at least three times was applied to achieve reproducibility. All tests with p values less than 0.05 were considered statistically significant.
Author Contributions
S.Q. designed the study. S.Q. wrote the paper. D.L. performed most of the experiments. J.W., M.Z., X.H., and J.S. performed part of the experiments. D.L., J.W., Q.X., X.Z., and L.X. carried out initial data analyses. All authors contributed to drafting the manuscript. All authors have read and approved the final submitted manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (grant no. 81802375); the Natural Science Foundation of Hubei Provincial Department of Education (grant nos. Q20172105 and Q20182103); the Foundation of Health and Family Planning Commission of Hubei Province (grant no. WJ2017F066); and the Faculty Development Grants from Hubei University of Medicine (grant nos. 2016QDJZR07 and 2017QDJZR05). In addition, we are very grateful to Dr. JiWei Li for the help of Cox model analysis.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.11.003.
Contributor Information
Ying Liu, Email: ying_liu1002@163.com.
Shanshan Qin, Email: qinss77@163.com.
Supplemental Information
References
- 1.Tang C.T., Liang Q., Yang L., Lin X.L., Wu S., Chen Y., Zhang X.T., Gao Y.J., Ge Z.Z. RAB31 Targeted by MiR-30c-2-3p Regulates the GLI1 Signaling Pathway, Affecting Gastric Cancer Cell Proliferation and Apoptosis. Front. Oncol. 2018;8:554. doi: 10.3389/fonc.2018.00554. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Wu H., Hu Y., Liu X., Song W., Gong P., Zhang K., Chen Z., Zhou M., Shen X., Qian Y., Fan H. LncRNA TRERNA1 Function as an Enhancer of SNAI1 Promotes Gastric Cancer Metastasis by Regulating Epithelial-Mesenchymal Transition. Mol. Ther. Nucleic Acids. 2017;8:291–299. doi: 10.1016/j.omtn.2017.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russo A.E., Strong V.E. Gastric Cancer Etiology and Management in Asia and the West. Annu. Rev. Med. 2019;70:353–367. doi: 10.1146/annurev-med-081117-043436. [DOI] [PubMed] [Google Scholar]
- 4.Sano T., Coit D.G., Kim H.H., Roviello F., Kassab P., Wittekind C., Yamamoto Y., Ohashi Y. Proposal of a new stage grouping of gastric cancer for TNM classification: International Gastric Cancer Association staging project. Gastric Cancer. 2017;20:217–225. doi: 10.1007/s10120-016-0601-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian T., Wang M., Lin S., Guo Y., Dai Z., Liu K., Yang P., Dai C., Zhu Y., Zheng Y. The Impact of lncRNA Dysregulation on Clinicopathology and Survival of Breast Cancer: A Systematic Review and Meta-analysis. Mol. Ther. Nucleic Acids. 2018;12:359–369. doi: 10.1016/j.omtn.2018.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun M., Nie F.Q., Wang Z.X., De W. Involvement of lncRNA dysregulation in gastric cancer. Histol. Histopathol. 2016;31:33–39. doi: 10.14670/HH-11-655. [DOI] [PubMed] [Google Scholar]
- 7.Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 8.Liang H., Yu T., Han Y., Jiang H., Wang C., You T., Zhao X., Shan H., Yang R., Yang L. LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression. Mol. Cancer. 2018;17:119. doi: 10.1186/s12943-018-0870-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y., Yang H., Xu M., Zhang H., Sun M., Mu P., Dong T., Du S., Liu K. Long non-coding RNA (lncRNA) MAGI2-AS3 inhibits breast cancer cell growth by targeting the Fas/FasL signalling pathway. Hum. Cell. 2018;31:232–241. doi: 10.1007/s13577-018-0206-1. [DOI] [PubMed] [Google Scholar]
- 10.Wang F., Zu Y., Zhu S., Yang Y., Huang W., Xie H., Li G. Long noncoding RNA MAGI2-AS3 regulates CCDC19 expression by sponging miR-15b-5p and suppresses bladder cancer progression. Biochem. Biophys. Res. Commun. 2018;507:231–235. doi: 10.1016/j.bbrc.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 11.Pu J., Wang J., Wei H., Lu T., Wu X., Wu Y., Shao Z., Luo C., Lu Y. Long non-coding RNA MAGI2-AS3 prevents the development of hepatocellular carcinoma via recruiting KDM1A and promoting H3K4me2 demethylation of RACGAP1 promoter. Mol. Ther. Nucleic Acids. 2019;18:351–362. doi: 10.1016/j.omtn.2019.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cristescu R., Lee J., Nebozhyn M., Kim K.M., Ting J.C., Wong S.S., Liu J., Yue Y.G., Wang J., Yu K. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015;21:449–456. doi: 10.1038/nm.3850. [DOI] [PubMed] [Google Scholar]
- 13.Felipe Lima J., Nofech-Mozes S., Bayani J., Bartlett J.M.S. EMT in Breast Carcinoma—A Review. J. Clin. Med. 2016;5:65. doi: 10.3390/jcm5070065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon J.H., Abdelmohsen K., Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J. Mol. Biol. 2013;425:3723–3730. doi: 10.1016/j.jmb.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu K.-P., Ma X.-L., Zhang C.-L. LncRNA ODRUL contributes to osteosarcoma progression through the miR-3182/MMP2 Axis. Mol. Ther. 2017;25:2383–2393. doi: 10.1016/j.ymthe.2017.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Liu B., Wu S., Ma J., Yan S., Xiao Z., Wan L., Zhang F., Shang M., Mao A. lncRNA GAS5 Reverses EMT and Tumor Stem Cell-Mediated Gemcitabine Resistance and Metastasis by Targeting miR-221/SOCS3 in Pancreatic Cancer. Mol. Ther. Nucleic Acids. 2018;13:472–482. doi: 10.1016/j.omtn.2018.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chu J.Y.S., Chau M.K.M., Chan C.C.Y., Tai A.C.P., Cheung K.F., Chan T.M., Yung S. miR-200c Prevents TGF-β1-Induced Epithelial-to-Mesenchymal Transition and Fibrogenesis in Mesothelial Cells by Targeting ZEB2 and Notch1. Mol. Ther. Nucleic Acids. 2019;17:78–91. doi: 10.1016/j.omtn.2019.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Brien S.J., Carter J.V., Burton J.F., Oxford B.G., Schmidt M.N., Hallion J.C., Galandiuk S. The role of the miR-200 family in epithelial-mesenchymal transition in colorectal cancer: a systematic review. Int. J. Cancer. 2018;142:2501–2511. doi: 10.1002/ijc.31282. [DOI] [PubMed] [Google Scholar]
- 19.Korpal M., Lee E.S., Hu G., Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barbagallo C., Brex D., Caponnetto A., Cirnigliaro M., Scalia M., Magnano A., Caltabiano R., Barbagallo D., Biondi A., Cappellani A. LncRNA UCA1, Upregulated in CRC Biopsies and Downregulated in Serum Exosomes, Controls mRNA Expression by RNA-RNA Interactions. Mol. Ther. Nucleic Acids. 2018;12:229–241. doi: 10.1016/j.omtn.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mongroo P.S., Rustgi A.K. The role of the miR-200 family in epithelial-mesenchymal transition. Cancer Biol. Ther. 2010;10:219–222. doi: 10.4161/cbt.10.6312548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berger S.L. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 23.Tasdemir N., Banito A., Roe J.S., Alonso-Curbelo D., Camiolo M., Tschaharganeh D.F., Huang C.H., Aksoy O., Bolden J.E., Chen C.C. BRD4 Connects Enhancer Remodeling to Senescence Immune Surveillance. Cancer Discov. 2016;6:612–629. doi: 10.1158/2159-8290.CD-16-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding N., Hah N., Yu R.T., Sherman M.H., Benner C., Leblanc M., He M., Liddle C., Downes M., Evans R.M. BRD4 is a novel therapeutic target for liver fibrosis. Proc. Natl. Acad. Sci. USA. 2015;112:15713–15718. doi: 10.1073/pnas.1522163112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Filippakopoulos P., Qi J., Picaud S., Shen Y., Smith W.B., Fedorov O., Morse E.M., Keates T., Hickman T.T., Felletar I. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abbas M., Faggian A., Sintali D.N., Khan G.J., Naeem S., Shi M., Dingding C. Current and future biomarkers in gastric cancer. Biomed. Pharmacother. 2018;103:1688–1700. doi: 10.1016/j.biopha.2018.04.178. [DOI] [PubMed] [Google Scholar]
- 27.Yasui W., Oue N., Aung P.P., Matsumura S., Shutoh M., Nakayama H. Molecular-pathological prognostic factors of gastric cancer: a review. Gastric Cancer. 2005;8:86–94. doi: 10.1007/s10120-005-0320-0. [DOI] [PubMed] [Google Scholar]
- 28.Baniak N., Senger J.-L., Ahmed S., Kanthan S.C., Kanthan R. Gastric biomarkers: a global review. World J. Surg. Oncol. 2016;14:212. doi: 10.1186/s12957-016-0969-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pastushenko I., Blanpain C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol. 2019;29:212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Noh J.H., Kim K.M., McClusky W.G., Abdelmohsen K., Gorospe M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip. Rev. RNA. 2018;9:e1471. doi: 10.1002/wrna.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du S., Hu W., Zhao Y., Zhou H., Wen W., Xu M., Zhao P., Liu K. Long non-coding RNA MAGI2-AS3 inhibits breast cancer cell migration and invasion via sponging microRNA-374a. Cancer Biomark. 2019;24:269–277. doi: 10.3233/CBM-182216. [DOI] [PubMed] [Google Scholar]
- 32.Hao X.Z., Yang K. LncRNA MAGI2-AS3 suppresses the proliferation and invasion of non-small cell lung carcinoma through miRNA-23a-3p/PTEN axis. Eur. Rev. Med. Pharmacol. Sci. 2019;23:7399–7407. doi: 10.26355/eurrev_201909_18848. [DOI] [PubMed] [Google Scholar]
- 33.Yin Z., Ma T., Yan J., Shi N., Zhang C., Lu X., Hou B., Jian Z. LncRNA MAGI2-AS3 inhibits hepatocellular carcinoma cell proliferation and migration by targeting the miR-374b-5p/SMG1 signaling pathway. J. Cell. Physiol. 2019;234:18825–18836. doi: 10.1002/jcp.28521. [DOI] [PubMed] [Google Scholar]
- 34.Ba M., Long H., Yan Z., Wang S., Wu Y., Tu Y., Gong Y., Cui S. BRD4 promotes gastric cancer progression through the transcriptional and epigenetic regulation of c-MYC. J. Cell. Biochem. 2018;119:973–982. doi: 10.1002/jcb.26264. [DOI] [PubMed] [Google Scholar]
- 35.Chen D., Chu Y., Zheng Q., Xu Z., Zhou P., Li S. Knock-down of ZEB1 inhibits the proliferation, invasion and migration of gastric cancer cells. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2017;33:1073–1078. [PubMed] [Google Scholar]
- 36.Zhou Y., Zhong J.H., Gong F.S., Xiao J. MiR-141-3p suppresses gastric cancer induced transition of normal fibroblast and BMSC to cancer-associated fibroblasts via targeting STAT4. Exp. Mol. Pathol. 2019;107:85–94. doi: 10.1016/j.yexmp.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 37.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li D., Cheng P., Wang J., Qiu X., Zhang X., Xu L., Liu Y., Qin S. IRF6 Is Directly Regulated by ZEB1 and ELF3, and Predicts a Favorable Prognosis in Gastric Cancer. Front. Oncol. 2019;9:220. doi: 10.3389/fonc.2019.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.