Abstract
Epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs), such as gefitinib, have been established as first-line treatments for non-small cell lung cancer (NSCLC) patients and have exhibited notable clinical efficacy. However, resistance to TKIs has become one of the major obstacles in improving the therapeutic efficacy of patients with NSCLC. This study aims to investigate the role of the long non-coding RNA (lncRNA) LINC01116 in gefitinib resistance of NSCLC and explore its underlying mechanism. In this study, we found that LINC01116 is upregulated in the gefitinib-resistant NSCLC cells and tissues. Loss- and gain-of-function assays uncovered that LINC01116 downregulation sensitized gefitinib resistance, whereas the overexpression of LINC01116 conferred PC9/R cells to gefitinib treatment. Moreover, LINC01116 silencing increased IFI44 expression. Overexpression of IFI44 reversed the resistance to gefitinib in PC9/R cells, and rescue experiments confirmed that LINC01116 affects the gefitinib resistance of PC9/R cells partly dependent on regulating IFI44 expression. Moreover, downregulation of LINC01116 increased the sensitivity of PC9/R cells to gefitinib in vivo. Our study demonstrates that LINC01116 plays a critical role in gefitinib resistance of NSCLC cells by affecting IFI44 expression, providing a novel therapeutic target to overcome TKI resistance in NSCLC.
Keywords: NSCLC, TKI resistance, gefitinib, LINC01116, IFI44
Introduction
Lung cancer is one of the most malignant cancer types, with high incidence and mortality rates worldwide. NSCLC is the predominant subtype of lung cancer, which accounts for ∼85% of all lung cancer cases.1,2 The main treatment options for NSCLC patients include surgery, chemotherapy, and radiotherapy. Although conventional chemotherapy following surgery showed limited effects for patients with advanced NSCLC, molecular targeted therapy bright great promise for these patients’ treatment.3 NSCLC patients with sensitive epidermal growth factor receptor (EGFR) mutations are responsive to EGFR-tyrosine kinase inhibitors (TKIs) such as gefitinib, resulting in longer median survival time than in patients with wild-type EGFR.4 Although the EGFR TKIs have been first‐line drugs and have shown measurable efficacy at early stages of treatment, patients eventually develop resistance to those drugs after several months, leading to limited clinical application and treatment failure. Therefore, a better understanding of the mechanism of EGFR TKI resistance is of significant importance.
Long non-coding RNAs (lncRNAs) are a recently discovered major class of non-coding RNAs with a length of more than 200 nt.5,6 A growing number of studies have demonstrated that lncRNAs involve in a variety of cellular biological functions, including chromatin imprinting, cell differentiation, tumorigenesis, and cancer cell drug resistance.7,8 For example, Sun et al.9 reported that overexpression of lncRNA HOXA11-AS promotes tumorigenesis and progression of gastric cancer through scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. In addition, SP1-induced lncRNA AGAP2-AS1 promotes chemoresistance of breast cancer cells by interacting with CBP and increasing the enrichment of H3K27ac at the promoter region of MyD88, thus leading to the upregulation of MyD88.10 Moreover, YiRen and colleagues11 found that downregulation of MALAT1 could sensitize gastric cancer cells to chemotherapy by blocking chemotherapy-induced autophagy via competitively sequestering miR-23b-3p and relieving its inhibitory effect on ATG12.
In recent years, emerging evidence has uncovered the critical roles and molecular mechanism of lncRNAs in NSCLC development and evaluating drug resistance.12,13 For instance, Shi et al.14 reported that upregulated lncRNA PCAT6 promotes the NSCLC tumorigenesis through interacting with EZH2 and repressing tumor suppressor LATS2 expression. Our previous studies revealed that lncRNA SNHG20 promotes cell proliferation and migration by epigenetically silencing P21 expression in NSCLC.15 Additionally, a great number of researches are focusing on identifying the gefitinib-resistance-associated lncRNAs in NSCLC. For example, Cheng et al.16 identified 1,731 upregulated and 2,936 downregulated lncRNAs in EGFR TKI-resistant human lung cancer cells by performing lncRNA microarray profiling analysis. In addition, LINC00460 facilitates the gefitinib resistance in NSCLC through sponging miR-769-5p and antagonizing its repression of EGFR.17 In the present study, we identified that LINC01116 is significantly upregulated in gefitinib-resistant NSCLC cells. Moreover, LINC01116 facilitates the gefitinib resistance of NSCLC cells through regulating IFI44 expression. Taken together, our findings indicate that LINC01116 plays a critical role in gefitinib resistance of NSCLC and might be a potential therapeutic target for NSCLC patients.
Results
LINC01116 Is Upregulated in Gefitinib-Resistant PC9/R Cells
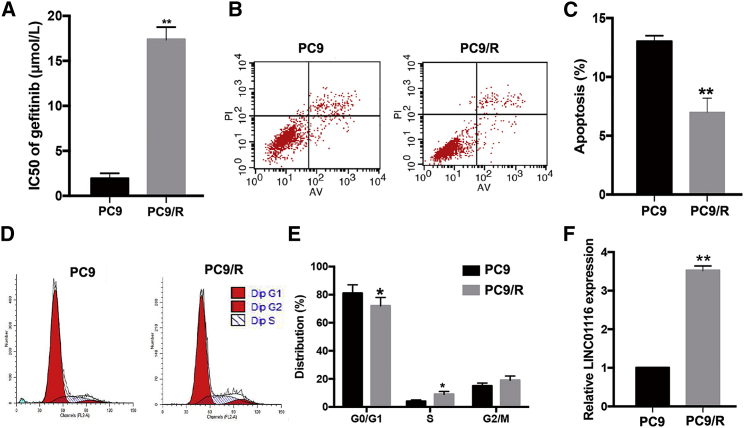
An MTT (3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide) assay was used to determine the IC50 of the gefitinib-resistant PC9/R and its parental PC9 cell line when treated with different concentrations of gefitinib for 24 h. As shown in Figure 1A, the Inhibitory concentration (IC50) value of gefitinib in PC9/R cells (17.35 ± 1.40 μmol/L) was significantly higher than that in parental PC9 cells (1.91 ± 0.62 μmol/L; p < 0.01). Moreover, 24 h after initiation of exposure to gefitinib (1 μmol/L), the apoptosis rate in PC9 cells was significantly higher than that in PC9/R cells (p < 0.01; Figures 1B and 1C). The percentage of PC9/R cells in G0/G1 phase was significantly lower than that of PC9 cells (p < 0.05), while the percentage of PC9/R cells in S phase was significantly higher than that of parental cells (p < 0.05; Figures 1D and 1E). Additionally, using genetic sequencing, we identified the mechanism of acquired resistance to gefitinib of PC9/R cells was a secondary T790M mutation (Figure S1). The results of quantitative real-time PCR revealed that LINC01116 was upregulated ∼3.52-fold in gefitinib-resistant PC9/R cells compared with in parental PC9 cells under gefitinib treatment (Figure 1F; p < 0.01). These data suggest that LINC01116 upregulation might play an important role in the development of gefitinib resistance in lung adenocarcinoma (LA) cells.
Figure 1.
Characterization of Gefitinib-Resistant Cell Line PC9/R and Parental Cell Line PC9
(A) MTT analysis of the IC50 values of gefitinib in PC9/R and parental PC9 cells. (B) Flow cytometric analysis of apoptosis in PC9/R and parental PC9 cells combined with gefitinib (1 μmol/L). (C) Flow cytometric analysis of cell cycle in PC9/R and parental PC9 cells combined with gefitinib (1 μmol/L). (D) quantitative real-time PCR detection of LINC01116 expression in PC9 and PC9/R cells under gefitinib treatment. Data are expressed as the mean ± SD of three individual experiments. *p < 0.05; **p < 0.01.
Effects of LINC01116 Expression on the In Vitro Sensitivity of LA Cells to Gefitinib
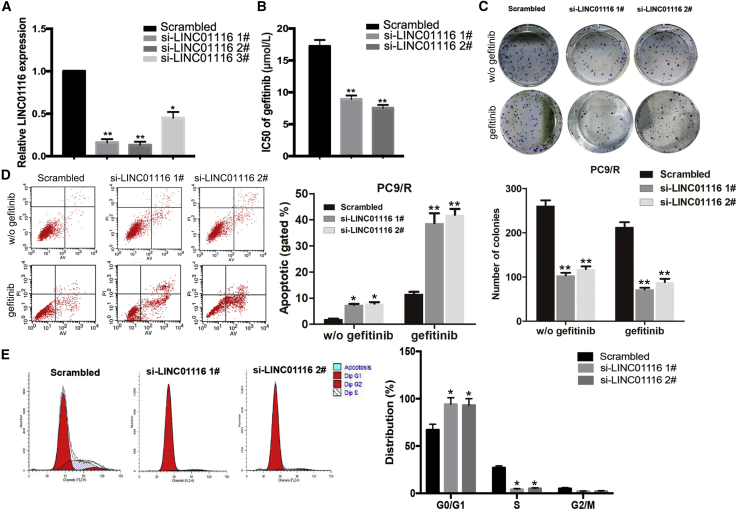
To investigate the effects of LINC01116 on the sensitivity of LA cells to gefitinib, LINC01116-specific small interfering RNA (siRNAs) were transfected into PC9/R cells to downregulate its expression. quantitative real-time PCR results indicated that expression levels of LINC01116 in si-LINC01116-transfected PC9/R cells were significantly inhibited (Figure 2A). As shown in Figure 2B, the IC50 value of gefitinib in si-LINC01116-transfected PC9/R cells was significantly decreased by 48.26% and 56.40% compared with control cells. Next, we determined the effect of LINC01116 on the colony formation ability of PC9/R cells with or without gefitinib treatment. The results showed that colony formation capacity of si-LINC01116-transfected PC9/R cells was significantly reduced compared with that of negative control siRNA (si-NC)-transfected cells, and the effect was much stronger under gefitinib treatment (p < 0.01; Figure 2C). Additionally, si-LINC01116 significantly increased the gefitinib-induced apoptosis rate of PC9/R cells compared with that without gefitinib treatment (Figure 2D). Furthermore, flow cytometry was used to analyze the effects of LINC01116 on the cell cycle progression in PC9/R cells exposed to gefitinib treatment. Compared with control cells, the percentage of si-LINC01116-transfected PC9/R cells in G0/G1 phase of the cell cycle increased, and the percentage in S phase decreased (Figure 2E).
Figure 2.
Downregulation of LINC01116 Significantly Increases the Sensitivity of PC9/R Cells to Gefitinib
(A) quantitative real-time PCR detection of LINC01116 expression in PC9/R cells transfected with si-LINC01116 (1#, 2#, 3#) or siRNA-NC; GAPDH was used as an internal control. (B) MTT analysis of the IC50 values of gefitinib in si-LINC01116- or siRNA-NC-transfected PC9/R cells. (C) Colony-formation assays of the proliferation in PC9/R cells transfected with si-LINC01116 or siRNA-NC combined with gefitinib (5 μmol/L). (D) Flow cytometric analysis of cell apoptosis in PC9/R transfected with si-LINC01116 or siRNA-NC combined with gefitinib (5 μmol/L) or not. (E) Flow cytometric analysis of cell cycle in PC9/R transfected with si-LINC01116 or siRNA-NC combined with gefitinib (5 μmol/L). Data are expressed as the mean ± SD of three individual experiments. *p < 0.05; ** p < 0.01.
Upregulation of LINC01116 Facilitates the Gefitinib Resistance of PC9 Cells In Vitro
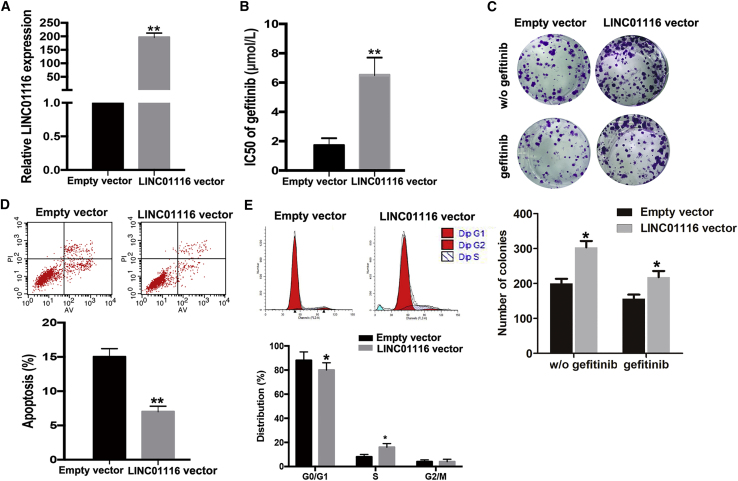
To determine whether LINC01116 could induce acquired resistance to gefitinib in parental PC9 cells, pcDNA3.1-LINC01116 was transfected into PC9 cells to upregulate LINC01116 expression. The results of quantitative real-time PCR indicated that LINC01116 levels were increased in PC9 cells transfected with the pcDNA3.1-LINC01116 vector compared with control cells (p < 0.01; Figure 3A). As shown in Figure 3B, the IC50 value of gefitinib in PC9 cells ectopically expressing LINC01116 was increased by 70.59% (p < 0.01). Moreover, the colony-formation capacity of PC9 cells ectopically expressing LINC01116 was enhanced compared with that of empty-vector-transfected PC9 cells with or without gefitinib treatment (Figure 3C). Next, the results of flow cytometry revealed that the percentage of cells in G0/G1 phase of the cell cycle decreased and the percentage in S phase increased among gefitinib-treated PC9 cells ectopically expressing LINC01116 compared with control cells under gefitinib treatment (Figure 3D). Additionally, the apoptosis rate of gefitinib-treated PC9 cells ectopically expressing LINC01116 was significantly decreased compared with that of control cells exposed to gefitinib (Figure 3E).
Figure 3.
Upregulation of LINC01116 Significantly Reduces the Sensitivity of PC9 Cells to Gefitinib
(A) quantitative real-time PCR detection of LINC01116 expression in PC9 cells transfected with pcDNA-LINC01116 or empty vector; GAPDH was used as an internal control. (B) MTT analysis of the IC50 values of gefitinib in pcDNA-LINC01116- or empty vector-transfected PC9 cells. (C) Colony-formation assays of the proliferation in PC9 transfected with pcDNA-LINC01116 or empty vector combined with gefitinib (0.5 μmol/L) or not. (D) Flow cytometric analysis of apoptosis in PC9 transfected with pcDNA-LINC01116 or empty vector combined with gefitinib (0.5 μmol/L). (E) Flow cytometric analysis of cell cycle in PC9 transfected with pcDNA-LINC01116 or empty vector combined with gefitinib (0.5 μmol/L). Data are expressed as the mean ± SD of three individual experiments. *p < 0.05; **p < 0.01.
IFI44 Is an Underlying Target of LINC01116
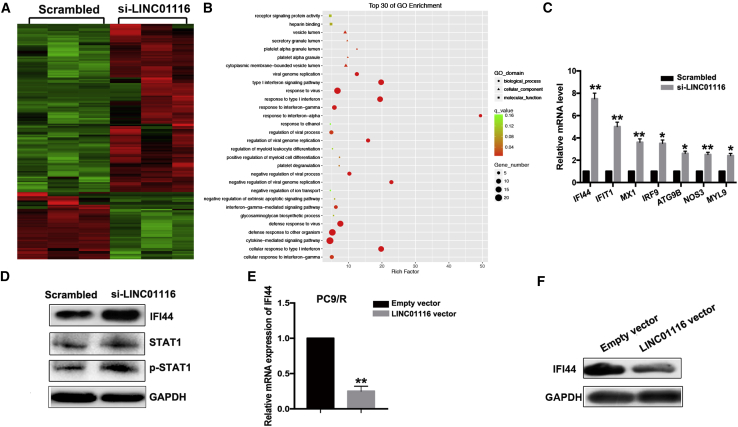
To explore the underlying targets of LINC01116, we used RNA sequencing to identify genes that were differentially expressed between LINC01116-depleted PC9/R cells and control cells. Of 169 differentially expressed transcripts, 121 were upregulated and 48 were downregulated (Figure 4A). Gene Ontology (GO) and pathway analysis showed that these altered genes are enriched in the cytokine-mediated signaling pathway, interferon signaling pathway, etc. (Figure 4B). Next, we selected seven representative genes verified as tumor suppressors in PC9/R cells for further verification. Among these, IFI44 showed the highest fold-upregulation in LINC01116-depleted PC9/R cells (p < 0.01; Figure 4C). Furthermore, quantitative real-time PCR analysis showed that the expression of IFI44 mRNA was significantly downregulated in LINC01116-overexpressing PC9 cells and significantly upregulated in si-LINC01116-transfected PC9/R cells compared with control cells (p < 0.01; Figure 4E). The results of western blot data confirmed the results of quantitative real-time PCR analysis (Figures 4D and 4F). Meanwhile, we found that downregulation of LINC01116 also increased the p-STAT1 protein levels, not the total STAT1 (Figure 4D). These results indicate that IFI44, which is involved in the interferon (IFN)/STAT1 pathway, might be a target of LINC0116.
Figure 4.
IFI44 Was a Functional Target of LINC01116
(A) RNA transcriptome sequencing identified differentially expressed genes between PC9/R/si-LINC01116 and PC9/R/siRNA-NC cells. (B) Top 30 of GO enrichment of differentially expressed genes between PC9/R/si-LINC01116 and PC9/R/siRNA-NC cells. (C) quantitative real-time PCR detection of seven representative mRNAs’ expression in PC9/R cells transfected with si-LINC01116 or siRNA-NC. (D) Western blotting detection of IFI44, STAT1, and p-STAT1 protein expression in PC9/R cells transfected with si-LINC01116 or siRNA-NC. (E ) quantitative real-time PCR detection of IFI44 mRNA expression in PC9 cells transfected with pcDNA-LINC01116 or empty vector, respectively. Data are expressed as the mean ± SD of three individual experiments. (F) Western blotting detection of IFI44 protein expression in PC9 cells transfected with pcDNA-LINC01116 or empty vector, respectively. *p < 0.05; **p < 0.01.
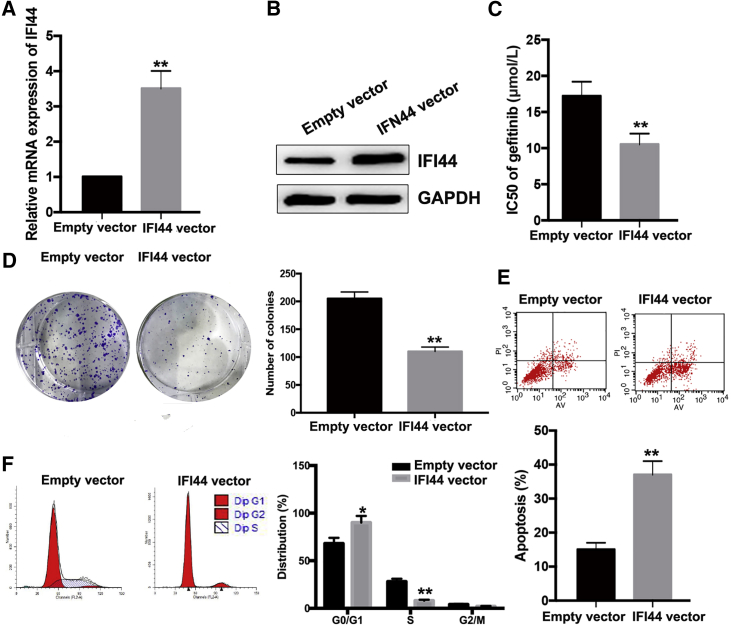
Upregulation of IFI44 Increases PC9/R Cell Sensitivity to Gefitinib
To further investigate the roles of IFI44 in PC9/R cells, pcDNA-IFI44 or pcDNA-control was transfected into PC9/R cells. quantitative real-time PCR and western blot assays revealed that the expression of IFI44 mRNA and protein were significantly upregulated in PC9/R cells ectopically expressing IFI44 in comparison with control cells (Figures 5A and 5B). Compared with that in control PC9/R cells (17.25 ± 1.92 μmol/L), the IC50 value of gefitinib in PC9/R cells ectopically expressing IFI44 (10.47 ± 1.15 μmol/L) was decreased by approximately 39.5% (p < 0.01; Figure 5C). Moreover, colony-formation assay showed that proliferation in the presence of gefitinib of PC9/R cells ectopically expressing IFI44 was significantly reduced compared with that of control cells (p < 0.01; Figure 5D). Flow cytometric analysis of the cell cycle indicated that the percentage of PC9/R cells ectopically expressing IFI44 in G0/G1 phase increased and the percentage of cells in S phase decreased upon treatment with gefitinib (Figure 5E). An increased apoptosis rate was observed in PC9/R cells ectopically expressing IFI44 that were exposed to gefitinib treatment (p < 0.01; Figure 5F). These data indicate that ectopic IFI44 expression increased the sensitivity of PC9/R cells to gefitinib.
Figure 5.
Overexpression of IFI44 Could Mimic the Effect of si-LINC01116 on the Sensitivity of PC9/R Cells to Gefitinib
(A) Quantitative real-time PCR detection of IFI44 mRNA in stably transfected empty vector or pcDNA-IFI44 PC9/R cells. (B) Western blotting detection of IFI44 protein expression in stably transfected empty vector or pcDNA-IFI44 PC9/R cells. (C) MTT analysis of the IC50 values of gefitinib in PC9/R/NC or PC9/R/IFI44 cells. (D) Colony-formation assays of proliferation in PC9/R/NC or PC9/R/IFI44 cells combined with gefitinib (5 μmol/L). (E) Flow cytometric analysis of apoptosis in PC9/R/NC or PC9/R/IFI44 cells combined with gefitinib (5 μmol/L). (F) Flow cytometric analysis of cell cycle in PC9/R/NC or PC9/R/IFI44 cells combined with gefitinib (5 μmol/L). Data are expressed as the mean ± SD of three individual experiments. *p < 0.05; **p < 0.01.
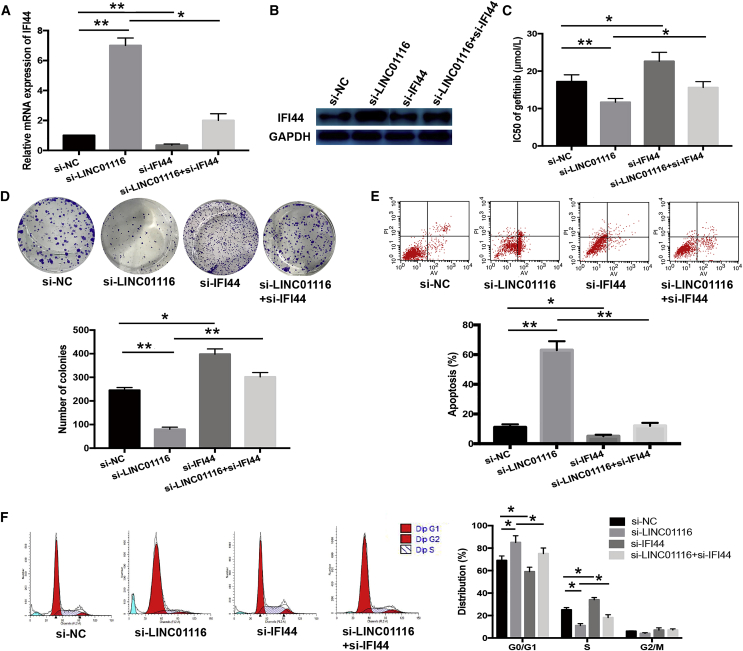
LINC01116 Contributes to PC9/R Cell Gefitinib Resistance Partly Dependent on Regulating IFI44
We next investigated whether suppressing IFI44 expression would reverse the effect of LINC01116 downregulation on the sensitivity of PC9/R cells to gefitinib. siRNAs targeting IFI44 and LINC01116 were co-transfected into PC9/R cells; quantitative real-time PCR and western blot assays showed that siRNA targeting IFI44 reversed the si-LINC01116-induced increases in IFI44 mRNA and protein expression (p < 0.05; Figures 6A and 6B). Moreover, siRNA targeting IFI44 reversed the si-LINC01116-induced decrease in the IC50 value of gefitinib in PC9/R cells (p < 0.05; Figure 6C) and abrogated the effects of si-LINC01116 on colony-formation capacity, G1/S transition, and gefitinib-induced apoptosis of PC9/R cells (Figures 6D–6F). Thus, IFI44 suppression mimicked the effect of LINC01116 overexpression on the sensitivity of PC9 cells to gefitinib and reversed the effect of si-LINC01116 on the gefitinib sensitivity of PC9/R cells.
Figure 6.
Downregulation of IFI44 Could Rescue the Effect of si-LINC01116 on the Sensitivity of PC9/R Cells to Gefitinib
(A) Quantitative real-time PCR detection of IFI44 mRNA expression in transfected PC9/R/control, PC9/R/si-LINC01116, PC9/R/si-IFI44, and PC9/R/si-LINC01116+si-IFI44 cells. (B) Western blotting detection of IFI44 protein expression in transfected PC9/R/control, PC9/R/si-LINC01116, PC9/R/si-IFI44, and PC9/R/si-LINC01116+si-IFI44 cells. (C) MTT analysis of the IC50 values of gefitinib in PC9/R/control, PC9/R/si-LINC01116, PC9/R/si-IFI44, and PC9/R/si-LINC01116+si-IFI44 cells. (D) Colony-formation assays of proliferation in PC9/R/control, PC9/R/si-LINC01116, PC9/R/si-IFI44, and PC9/R/si-LINC01116+si-IFI44 cells combined with gefitinib (5 μmol/L). (E) Flow cytometric analysis of apoptosis in PC9/R/control, PC9/R/si-LINC01116, PC9/R/si-IFI44, and PC9/R/si-LINC01116+si-IFI44 cells combined with gefitinib (5 μmol/L). (F) Flow cytometric analysis of cell cycle in PC9/R/control, PC9/R/si-LINC01116, PC9/R/si-IFI44, and PC9/R/si-LINC01116+si-IFI44 cells combined with gefitinib (5 μmol/L). Data are expressed as the mean ± SD of three individual experiments. *p < 0.05; **p < 0.01.
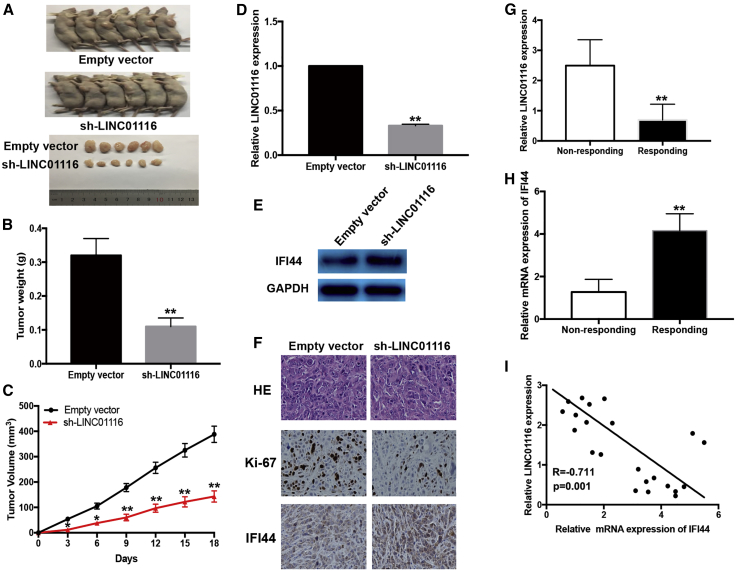
Effects of LINC01116 Expression on the Sensitivity of PC9/R Cells to Gefitinib In Vivo
We further investigated the effect of LINC01116 on the in vivo sensitivity of LA cells to gefitinib. PC9/R cells transfected with sh-LINC01116 or empty vector were injected into nude mice, which were then treated with gefitinib. The tumors that developed from the sh-LINC01116-transfected PC9/R cells appeared to be smaller than those formed from the empty vector-transfected PC9/R cells (Figure 7A). After gefitinib treatment, the average volume (and weight) of tumors formed from empty-vector-transfected and sh-LINC01116-transfected PC9/R cells was 388.5 mm3 (0.32 g) and 143.7 mm3 (0.11 g) (Figures 7B and 7C). Next, tumor homogenates were subjected to quantitative real-time PCR to detect LINC01116 and western blotting to detect IFI44. These assays revealed that the expression of LINC01116 was significantly downregulated and the expression of IFI44 protein was significantly increased in tumor tissues formed from sh-LINC01116-transfected PC9/R cells (Figures 7D and 7E). Immunostaining revealed significantly enhanced positive staining for IFI44 protein in tumors from sh-LINC01116-transfected PC9/R cells compared with tumors from empty vector-transfected PC9/R cells (Figure 7F). Taken together, these findings suggest that LINC01116 downregulation enhanced the in vivo sensitivity of PC9/R cells to gefitinib.
Figure 7.
Downregulation of LINC01116 Reduces the In Vivo Sensitivity of PC9/R Cells to Gefitinib and the Expression of LINC01116 in LA Tissues Was Negatively Correlated with IFI44
Mice were treated with gefitinib (10.0 mg/kg) or with 1% Tween 80. (A) Representative features of tumors 18 days after inoculation using PC9/R/sh-LINC01116 or PC9/R/Empty vector cells treated with 1% Tween 80 or gefitinib. (B and C) Tumor volume and weight at day 18 after the inoculation. (D) Quantitative real-time PCR detection of relative LINC01116 expression in tumors developed from PC9/R/shRNA-LINC01116 or PC9/R/Empty vector cells treated with 1% Tween 80 or gefitinib. (E) Western blotting detection of IFI44 protein expression in tumors developed from PC9/R/shRNA-LINC01116 or PC9/R/Empty vector cells treated with 1% Tween 80 or gefitinib. (F) Immunostaining of IFI44 and ki-67 protein expression in tumors developed from PC9/R/shRNA-LINC01116 or PC9/R/Empty vector cells treated with 1% Tween 80 or gefitinib. Upper, H&E staining. Intermediate and lower, immunostaining. Bars, 100 μm. (G) quantitative real-time PCR detection of relative LINC01116 expression in responding (n = 11) and non-responding (n = 14) LA tissues (p < 0.0001). Abundance of LINC01116 was normalized to U6 RNA. (H) quantitative real-time PCR detection of relative IFI44 mRNA expression in responding (n = 11) and non-responding (n = 14) LA tissues (p < 0.0001). Abundance of IFI44 was normalized to GAPDH. (I) A statistically significant inverse correlation between LINC01116 and IFI44 mRNA levels in 25 cases of LA tissues (Pearson’s correlation analysis, R = −07112; p < 0.0001). Data are expressed as the mean ± SD of three individual experiments. *p < 0.05; **p < 0.01.
Expression of LINC01116 in LA Tissues Is Negatively Associated with IFI44 Expression and LA Patient Response to Gefitinib
To investigate the correlation between LINC01116/IFI44 dysregulation and response to gefitinib in LA patients, the expression levels of LINC01116 and IFI44 mRNA were examined in the tumors from 25 patients with LA (stage IIIB or IV) who received gefitinib. Patients were divided into two groups: responding (complete response + partial response) and non-responding (stable disease + progressive disease). quantitative real-time PCR revealed that the relative level of LINC01116 expression in the responding group (n = 11) was significantly lower than that in the non-responding group (n = 14) (p < 0.0001; Figure 7G). However, the relative level of IFI44 mRNA expression in the responding group was significantly higher than that in the non-responding group (p < 0.0001; Figure 7H). Moreover, IFI44 expression was significantly negatively correlated with LINC01116 expression (Figure 7I).
Discussion
Gefitinib is an EGFR inhibitor that interrupts EGFR-mediated signaling in target cells. Gefitinib has been the standard first-line treatment for patients with advanced NSCLC harboring EGFR-activating mutations.18,19 In more than 50% of these cases in which gefitinib treatment fails, it is due to T790M mutation in EGFR. Emerging evidence has shown that lncRNAs play important roles in the pathogenesis of malignant tumors by regulating genes involved in cell growth, apoptosis, metastasis, and chemo- or radioresistance.20,21 However, the mechanisms underlying lncRNA-induced gefitinib therapy resistance have not been clearly demonstrated.
In the current study, we established a gefitinib-resistant cell line termed PC9/R, which has been identified with a T790M mutation in EGFR, to examine lncRNA-mediated molecular mechanisms of gefitinib resistance. Recent studies showed that LINC01116 functions as an oncogene in tumors by inducing cell growth and inhibiting apoptosis and that its overexpression is associated with poor prognosis.22, 23, 24 In the present study, we found that gefitinib treatment upregulated LINC01116 expression in the gefitinib-resistant PC9/R cells but had no effects on its expression in the parental PC9 cell line. These findings led us to explore in-depth the roles of LINC01116 upregulation in the formation of a gefitinib-resistant phenotype in LA cells.
To date, the biological function and underlying mechanism of LINC01116 in cancer cell drug resistance has not been reported. Here, we investigated the emerging roles of LINC01116 in gefitinib resistance in human LA cells. Loss- and gain-of-function studies revealed that suppressing LINC01116 expression reversed the gefitinib resistance of gefitinib-resistant PC9/R cells, while ectopically expressing LINC01116 reduced the sensitivity of parental PC9 cells to gefitinib by regulating cell proliferation, G1/S cell cycle transition, and apoptosis. Furthermore, analysis of LINC01116 expression in LA tissues collected from patients who received gefitinib treatment revealed that the relative level of LINC01116 expression in responding patients was significantly lower than that in non-responding patients, suggesting that LINC01116 expression was negatively correlated with LA patient response to gefitinib. To further explore the molecular mechanisms by which LINC01116 exerts its effect, we performed RNA transcriptome sequencing. This revealed that more than 100 genes were predicted to be potential targets of LINC01116. Of these differentially expressed genes, IFI44 showed the most meaningful fold upregulation in LINC01116-depleted PC9/R cells, which was further confirmed by quantitative real-time PCR and western blot. Our data suggest that LINC01116 might promote gefitinib resistance in LA cells by regulating IFI44, which is involved in the IFN/STAT1 pathway.
IFN/STAT1 signaling is traditionally associated with antiviral response and pro-apoptotic tumor-suppressor functions. Emerging evidence has suggested that evaluating the induction of genes associated with IFN/STAT1 signaling could represent a novel means of predicting chemotherapy response. In KRAS wild-type squamous carcinomas, cetuximab-induced activation of the IFN/STAT1 appeared to switch from inhibiting growth in acutely treated cells to promoting survival in chronically adapted cells.25,26 The IFI44 gene is induced by alpha/beta-IFN and participates in many biological processes through IFN signaling pathways, such as antiviral response, pathogenesis of systemic lupus erythematosus, radiation resistance, and anti-proliferative activity in tumors.27 IFI44 has been shown to abolish extracellular signal-regulated kinase signaling by binding intracellular GTP, thus ultimately resulting in cell-cycle arrest.28 Importantly, some studies have suggested an association between constitutive IFN/STAT1 pathway activation and aggressive tumor phenotype, manifested through characteristics such as ionizing radiation resistance, drug resistance, and poor patient survival.29,30 Here, we also found that overexpression of IFI44 mimicked the effect of LINC01116 suppression in PC9/R cells, whereas silencing IFI44 partially reversed the effect of LINC01116 suppression in PC9/R cells.
In summary, our data reveals that LINC01116 promoted gefitinib resistance in LA cells by regulating cell proliferation, G1/S transition, and apoptosis via effecting IFI44 expression, which is involved in the IFN/STAT1 pathway. LINC01116/IFI44 signature may predict the responses of LA patients to gefitinib and represent potential targets for therapeutic intervention. This study has several limitations; for example, further studies are needed to identify the precise mechanism by which LINC01116 regulates IFI44.
Materials and Methods
Cell Culture
The human non-small cell lung cancer cell line PC9 was purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China). PC9 cells were cultured with DMEM supplied with 10% fetal bovine serum and 100 IU/mL penicillin and streptomycin (Gibco-BRL) and maintained in a humidified atmosphere with 5% CO2 at 37°C. The gefitinib-resistant PC9 cell line (PC9/R) was established and kept in our lab.
Quantitative Real-Time PCR
The total RNA of PC9 and PC9/R cell lines were extracted by using TRIzol reagent (Invitrogen) according to the manufacturer’s manual. Then, 1 μg total RNA was reverse transcribed into cDNA with the PrimeScript RT reagent kit according to the manufacturer’s manual (Takara, Dalian, China). SYBR pre-mix Ex Taq (Takara) was used to conduct the real-time PCR assay based on the manufacturer’s protocol. The data of quantitative real-time PCR were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and then calculated according to the comparative cycle threshold (CT) (2−ΔΔCT) method. The primer sequences of LINC01116 and its targets are listed in Table S1.
Cell Transfection
Three LINC01116 siRNAs (si-LINC01116 1#, 2#, and 3#) and one scrambled control siRNA were purchased from Invitrogen. The sequence of LINC01116 was amplified and cloned into pcDNA3.1 vector. Then, PC9 and PC9/R cells were seeded into 6-well plates and transfected with LINC01116, scrambled siRNAs, LINC01116 overexpression vector, or empty vector using Lipofectamine 2000. The siRNA sequences of LINC01116 are listed in Table S1. 48 h post-transfection, the cells were collected for western blot or qRT-PCR analyses.
Cell Proliferation Assays
For the cell viability assay, PC9 and PC9/R cells were seeded into 96-well plates. Then, the cell counting kit-8 (CCK8) regent was added into each well and incubated for 2 h. Next, the optical density 450 (OD450) absorbance value was measured every 24 h following the manufacturer’s protocol. For the colony-formation assay, PC9 and PC9/R cells were seeded into 6-well plates (1,000/well) and maintained for 2 weeks. Then, the colonies were fixed with methanol for 15 min and stained with 0.1% crystal violet (Sigma-Aldrich) for 15 min. Finally, the number of visible colonies was calculated.
Flow Cytometric Analysis
PC9 and PC9/R cells were harvested 48 h after transfection by trypsinization (without EDTA). Then, the collected cells were double stained with fluorescein isothiocyanate (FITC)-Annexin V and propidium iodide (PI) by using the FITC Annexin V apoptosis detection kit (BD Biosciences) based on the manufacturer’s instruction. The stained cells were analyzed by flow cytometry (FACScan; BD Biosciences) equipped with CellQuest software (BD Biosciences). Next, the relative number of Annexin V- or PI-positive cells was calculated and compared. For cell cycle assay, PC9 and PC9/R cells were stained with PI by using the CycleTESTTM plus DNA reagent kit (BD Biosciences) following the manufacturer’s instruction, and the stained cells were analyzed by FACScan. The relative percentage of cells in G0/G1, S, and G2/M phase were calculated and compared.
Western Blot Analyses
The RIPA lysis and extraction buffer (Thermo Fisher) is used to lyse PC9/R cells. 40 μg of the total cell lysis was separated by 10% SDS polyacrylamide gel and transferred to 0.22 μm polyvinylidene fluoride (PVDF) membranes (Millipore). Then, the membranes were inhibited with 5% milk and incubated with antibodies. The GAPDH (1:1,000) antibody was purchased from Cell Signaling Technology, and IFI44 antibody (1:1,000) was obtained from Abcam. The densitometry (Quantity One software; Bio-Rad) was used to quantify the protein fragments’ intensities.
In Vivo Tumor Formation Assay
Male athymic BALB/c mice (5 weeks old) were maintained under specific-pathogen-free conditions and used according to protocols approved by the Shanghai Medical Experimental Animal Care Commission. PC9/R cells were transfected with sh-LINC01116 or empty vector harvested from 6-well cell culture plates, washed with PBS, and re-suspended at a concentration of 1 × 108 cells/mL. A total of 100 μL of suspended cells was subcutaneously injected into a single side of the posterior flank of each mouse, and gefitinib treatment was started 5 days after the tumor cell inoculation. Gefitinib was administered by oral gavage every 3 days at a dose of 10 mg/kg in 1% Tween 80 (Sigma). Tumor growth was examined every 3 days, and tumor volumes were calculated using the equation V = 0.5 × D × d2, where V represents volume, D represents longitudinal diameter, and d represents latitudinal diameter. At 18 days post-injection, mice were euthanized, and the subcutaneous growth of each tumor was examined. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the NIH. The protocol was approved by the Committee on the Ethics of Animal Experiments of Nanjing Medical University.
Statistical Analyses
The Student’s t test (two-tailed) and one-way ANOVA test were used to analyze the comparisons between groups. All the data calculation and analysis were conducted by using SPSS 18.0 software (IBM, SPSS, Chicago, IL, USA) and GraphPad Prism 5 (GraphPad Software, San Diego, CA, USA). The differences were defined statistically significant when p < 0.05.
Author Contributions
H. Wang and Z.X. Wang designed and supervised the study, H. Wang, B.B. Lu and S.N. Ren conducted the experiment. F.B. Wu and X.X. Wang contributed to acquisition of results. C.Y. Yan performed the data analysis. He Wang and Z.X. Wang wrote the manuscript. Z.X.W. provided technical and administrative support. All the authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (nos. 81472198 and 81672307 to Z.W. and 81501980 to H.W.); the Medical Innovation Team Foundation of the Jiangsu Provincial Enhancement Health Project (no. CXTDA2017021 to Z.W.); the Science and Technology Development Foundation of Nanjing Medical University (no. 2017NJMUZD137 to H.W.); the Scientific Research Foundation of Jiangsu Province Health Department (no. H201310 to B.L.); and the Key Young Medical Talents of Jiangsu Province (no. QNRC2016662 to B.L.). The data that support the findings of this study are available from the corresponding author upon reasonable request.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.10.039.
Supplemental Information
References
- 1.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer Statistics, 2017. CA Cancer J. Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Wood S.L., Pernemalm M., Crosbie P.A., Whetton A.D. Molecular histology of lung cancer: from targets to treatments. Cancer Treat. Rev. 2015;41:361–375. doi: 10.1016/j.ctrv.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Recondo G., Facchinetti F., Olaussen K.A., Besse B., Friboulet L. Making the first move in EGFR-driven or ALK-driven NSCLC: first-generation or next-generation TKI? Nat. Rev. Clin. Oncol. 2018;15:694–708. doi: 10.1038/s41571-018-0081-4. [DOI] [PubMed] [Google Scholar]
- 5.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Nagano T., Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 7.Quinodoz S., Guttman M. Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends Cell Biol. 2014;24:651–663. doi: 10.1016/j.tcb.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartonicek N., Maag J.L., Dinger M.E. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol. Cancer. 2016;15:43. doi: 10.1186/s12943-016-0530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun M., Nie F., Wang Y., Zhang Z., Hou J., He D., Xie M., Xu L., De W., Wang Z., Wang J. LncRNA HOXA11-AS Promotes Proliferation and Invasion of Gastric Cancer by Scaffolding the Chromatin Modification Factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299–6310. doi: 10.1158/0008-5472.CAN-16-0356. [DOI] [PubMed] [Google Scholar]
- 10.Dong H., Wang W., Mo S., Chen R., Zou K., Han J., Zhang F., Hu J. SP1-induced lncRNA AGAP2-AS1 expression promotes chemoresistance of breast cancer by epigenetic regulation of MyD88. J. Exp. Clin. Cancer Res. 2018;37:202. doi: 10.1186/s13046-018-0875-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.YiRen H., YingCong Y., Sunwu Y., Keqin L., Xiaochun T., Senrui C., Ende C., XiZhou L., Yanfan C. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol. Cancer. 2017;16:174. doi: 10.1186/s12943-017-0743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L., Ma L., Xu F., Zhai W., Dong S., Yin L., Liu J., Yu Z. Role of long non-coding RNA in drug resistance in non-small cell lung cancer. Thorac. Cancer. 2018;9:761–768. doi: 10.1111/1759-7714.12652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osielska M.A., Jagodziński P.P. Long non-coding RNA as potential biomarkers in non-small-cell lung cancer: What do we know so far? Biomed. Pharmacother. 2018;101:322–333. doi: 10.1016/j.biopha.2018.02.099. [DOI] [PubMed] [Google Scholar]
- 14.Shi X., Liu Z., Liu Z., Feng X., Hua F., Hu X., Wang B., Lu K., Nie F. Long noncoding RNA PCAT6 functions as an oncogene by binding to EZH2 and suppressing LATS2 in non-small-cell lung cancer. EBioMedicine. 2018;37:177–187. doi: 10.1016/j.ebiom.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Z., Chen X., Chen P., Yu S., Nie F., Lu B., Zhang T., Zhou Y., Chen Q., Wei C. Long non-coding RNA SNHG20 promotes non-small cell lung cancer cell proliferation and migration by epigenetically silencing of P21 expression. Cell Death Dis. 2017;8:e3092. doi: 10.1038/cddis.2017.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng N., Li X., Zhao C., Ren S., Chen X., Cai W., Zhao M., Zhang Y., Li J., Wang Q., Zhou C. Microarray expression profile of long non-coding RNAs in EGFR-TKIs resistance of human non-small cell lung cancer. Oncol. Rep. 2015;33:833–839. doi: 10.3892/or.2014.3643. [DOI] [PubMed] [Google Scholar]
- 17.Ma G., Zhu J., Liu F., Yang Y. Long Noncoding RNA LINC00460 Promotes the Gefitinib Resistance of Nonsmall Cell Lung Cancer Through Epidermal Growth Factor Receptor by Sponging miR-769-5p. DNA Cell Biol. 2019;38:176–183. doi: 10.1089/dna.2018.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rawluk J., Waller C.F. Gefitinib. Recent Results Cancer Res. 2018;211:235–246. doi: 10.1007/978-3-319-91442-8_16. [DOI] [PubMed] [Google Scholar]
- 19.Minari R., Bordi P., Tiseo M. Third-generation epidermal growth factor receptor-tyrosine kinase inhibitors in T790M-positive non-small cell lung cancer: review on emerged mechanisms of resistance. Transl. Lung Cancer Res. 2016;5:695–708. doi: 10.21037/tlcr.2016.12.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xue W., Li L., Tian X., Fan Z., Yue Y., Zhang C., Ding X., Song X., Ma B., Zhai Y. Integrated analysis profiles of long non-coding RNAs reveal potential biomarkers of drug resistance in lung cancer. Oncotarget. 2017;8:62868–62879. doi: 10.18632/oncotarget.16444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin C., Yang L. Long Noncoding RNA in Cancer: Wiring Signaling Circuitry. Trends Cell Biol. 2018;28:287–301. doi: 10.1016/j.tcb.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang Y.N., Huang Z.L., Li H., Tan W.B., Zhang Q.G., Wang L., Wu J.L. LINC01116 promotes the progression of epithelial ovarian cancer via regulating cell apoptosis. Eur. Rev. Med. Pharmacol. Sci. 2018;22:5127–5133. doi: 10.26355/eurrev_201808_15707. [DOI] [PubMed] [Google Scholar]
- 23.Zhang B., Yu L., Han N., Hu Z., Wang S., Ding L., Jiang J. LINC01116 targets miR-520a-3p and affects IL6R to promote the proliferation and migration of osteosarcoma cells through the Jak-stat signaling pathway. Biomed. Pharmacother. 2018;107:270–282. doi: 10.1016/j.biopha.2018.07.119. [DOI] [PubMed] [Google Scholar]
- 24.Hu H.B., Chen Q., Ding S.Q. LncRNA LINC01116 competes with miR-145 for the regulation of ESR1 expression in breast cancer. Eur. Rev. Med. Pharmacol. Sci. 2018;22:1987–1993. doi: 10.26355/eurrev_201804_14726. [DOI] [PubMed] [Google Scholar]
- 25.Legrier M.E., Bièche I., Gaston J., Beurdeley A., Yvonnet V., Déas O., Thuleau A., Château-Joubert S., Servely J.L., Vacher S. Activation of IFN/STAT1 signalling predicts response to chemotherapy in oestrogen receptor-negative breast cancer. Br. J. Cancer. 2016;114:177–187. doi: 10.1038/bjc.2015.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khodarev N.N., Beckett M., Labay E., Darga T., Roizman B., Weichselbaum R.R. STAT1 is overexpressed in tumors selected for radioresistance and confers protection from radiation in transduced sensitive cells. Proc. Natl. Acad. Sci. USA. 2004;101:1714–1719. doi: 10.1073/pnas.0308102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitamura A., Takahashi K., Okajima A., Kitamura N. Induction of the human gene for p44, a hepatitis-C-associated microtubular aggregate protein, by interferon-alpha/beta. Eur. J. Biochem. 1994;224:877–883. doi: 10.1111/j.1432-1033.1994.00877.x. [DOI] [PubMed] [Google Scholar]
- 28.Nzeusseu Toukap A., Galant C., Theate I., Maudoux A.L., Lories R.J., Houssiau F.A., Lauwerys B.R. Identification of distinct gene expression profiles in the synovium of patients with systemic lupus erythematosus. Arthritis Rheum. 2007;56:1579–1588. doi: 10.1002/art.22578. [DOI] [PubMed] [Google Scholar]
- 29.Duarte C.W., Willey C.D., Zhi D., Cui X., Harris J.J., Vaughan L.K., Mehta T., McCubrey R.O., Khodarev N.N., Weichselbaum R.R., Gillespie G.Y. Expression signature of IFN/STAT1 signaling genes predicts poor survival outcome in glioblastoma multiforme in a subtype-specific manner. PLoS ONE. 2012;7:e29653. doi: 10.1371/journal.pone.0029653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaston J., Cheradame L., Yvonnet V., Deas O., Poupon M.F., Judde J.G., Cairo S., Goffin V. Intracellular STING inactivation sensitizes breast cancer cells to genotoxic agents. Oncotarget. 2016;7:77205–77224. doi: 10.18632/oncotarget.12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.