Summary
Heading date 1 (Hd1) is an important gene for the regulation of flowering in rice, but its variation in major cultivated rice varieties, and the effect of this variation on yield and quality, remains unknown. In this study, we selected 123 major rice varieties cultivated in China from 1936 to 2009 to analyse the relationship between the Hd1 alleles and yield‐related traits. Among these varieties, 19 haplotypes were detected in Hd1, including two major haplotypes (H8 and H13) in the japonica group and three major haplotypes (H14, H15 and H16) in the indica group. Analysis of allele frequencies showed that the secondary branch number was the major aimed for Chinese indica breeding. In the five major haplotypes, SNP316(C‐T) was the only difference between the two major japonica haplotypes, and SNP495(C‐G) and SNP614(G‐A) are the major SNPs in the three indica haplotypes. Association analysis showed that H16 is the most preponderant allele in modern cultivated Chinese indica varieties. Backcrossing this allele into the japonica variety Chunjiang06 improved yield without decreasing grain quality. Therefore, our analysis offers a new strategy for utilizing these preponderant alleles to improve yield and quality of japonica varieties for cultivation in the southern areas of China.
Keywords: major Chinese cultivated rice varieties, Hd1 preponderant allele, association analysis, heading date, yield‐ and quality‐related traits improvement
Introduction
Rice (Oryza sativa L.) is an important cereal crop and provides the staple food for more than half of the world's population (Tian et al., 2009). Achieving increases in grain yield in rice is a major focus in agriculture (Ren et al., 2018; Yuan, 2014). Heading date (flowering time) is one of the most important agronomic traits for regional adaptation and grain yield, and is affected by genetic and environmental factors (Li et al., 2015). Selection for the optimal flowering time for a particular region will make maximum use of temperature and sunlight conditions to improve yield potential (Izawa, 2007; Zheng et al., 2016). Therefore, detailed knowledge of the genetic factors for heading date will increase our understanding of the adaptive mechanisms of rice varieties and enable breeders to design appropriate genotypes for specific environments (Putterill et al., 2004; Zheng et al., 2016).
Rice is a short‐day (SD) flowering plant; its heading is promoted under short photoperiod conditions. In recent years, the molecular genetic pathway for SD photoperiodic regulation in cultivated rice has been well characterized. OsGI (a homolog of Arabidopsis thaliana GIGANTEA) is responsible for perceiving light signals and circadian clocks, and regulates the expression of Heading date 1 (Hd1, an ortholog of CONSTANS in Arabidopsis) and OsMADS51 (Hayama et al., 2002, 2003; Kim et al., 2007; Takahashi et al., 2009). Hd1, which encodes zinc‐finger‐type transcriptional activators with CCT domains, promotes flowering under SD conditions and represses flowering under long‐day (LD) conditions by regulating the expression of Heading date 3a (Hd3a, an ortholog of FLOWERING LOCUS T in Arabidopsis; Kojima et al., 2002; Yano et al., 2000). OsMADS51, which encodes a type I MADS‐box protein and functions upstream of Early heading date 1 (Ehd1), promotes flowering under SD conditions (Kim et al., 2007). Ehd1 is a B‐type response regulator, which promotes flowering by regulating the expression of Hd3a and independently from Hd1 pathway (Doi et al., 2004). RICE FLOWERING LOCUS T1 (RFT1) functions as a floral activator, belongs to the rice FT‐like gene family and is the closest homolog of Hd3a (Komiya et al., 2008). RFT1 and Hd3a are both mobile flowering signals but RFT1 functions under LD conditions, whereas Hd3a functions under SD conditions (Komiya et al., 2008). Recent research demonstrated that RFT1 and Hd3a functionally diverged to control flowering time under LD and SD conditions, partly via a fine‐tuned epigenetic mechanism (Li et al., 2015; Sun et al., 2012).
Natural genetic variation provides an opportunity to identify key alleles associated with traits that can be selected to improve agronomic characteristics of crops (Lu et al., 2013; Zhu et al., 2008). For example, the pleiotropic gene Ghd7, which affects flowering time, plant height and spikelet number per panicle, was shown to contain an important single nucleotide polymorphism (SNP) that affects these three related traits in rice (Lu et al., 2012). Two SNPs in maize (Zea mays) Dwarf8 were shown to be independently associated with flowering time and plant height (Thornsberry et al., 2001). As an important heading date gene, Hd1 alleles have been widely used in traditional rice breeding and are also good targets for molecular marker‐assisted selection (MAS) breeding.
China has a long history of rice cultivation and a broad range of rice cultivation regions (from 18°N to 53°N), which contribute to a diversity of flowering time variants. However, the utilization of preponderant alleles of Hd1 in modern cultivated Chinese varieties is still unclear. In this study, we selected 123 major rice varieties cultivated in China from 1936 to 2009 in order to: (i) evaluate their genetic diversity and population structure; (ii) identify the key Hd1 alleles affecting yield‐related traits; and (iii) utilize the preponderant alleles of Hd1 in rice breeding to improve yield and quality. The results of this study will provide valuable data and a new strategy for generating varieties more suited to specific regions, potentially improving yield.
Results
Wide variation of yield‐related traits was noted in the 123 major rice varieties cultivated in China
We selected 123 major rice varieties cultivated in China from 1936 to 2009, based on these varieties were the main cultivars at that time. These 123 varieties, comprising 54 japonica and 69 indica varieties, were selected to analyse the relationship between the Hd1 alleles and eight yield‐related traits (Figure 1a, Table S1). A wide variation in heading date was observed among the 123 varieties, ranging from 54 to 117 days in Hangzhou (a LD site) and 66 to 100 days in Hainan (a SD site; Figure 1b).
Figure 1.
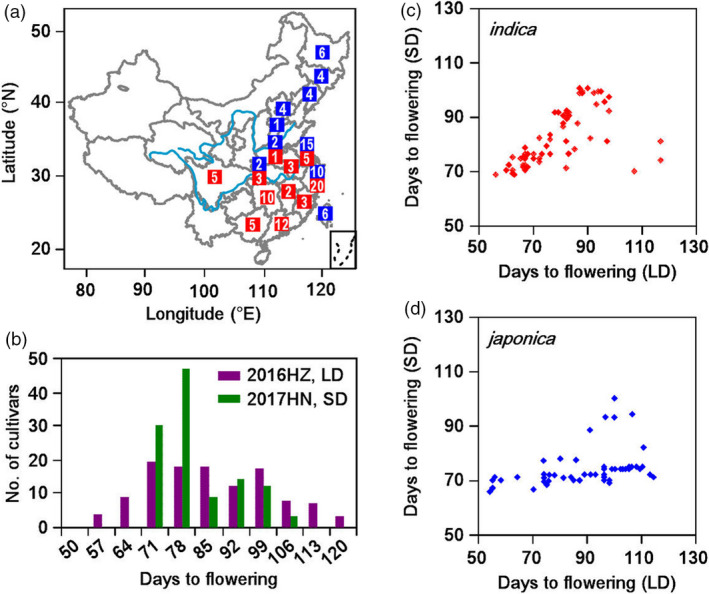
Variations in heading date and seven yield‐related traits among 123 major rice varieties cultivated in China in two conditions. (a) Geographical distribution of 123 major rice varieties cultivated in China. Blue indicates japonica, and red indicates indica. The number within each bar indicates the number of cultivars at this region. (b) Phenotypic variation of heading date in 2016 Hangzhou (long day, violet) and 2017 Hainan (short day, green). (c) Correlation analysis of heading date in indica varieties. (d) Correlation analysis of heading date in japonica varieties.
The photoperiod analysis showed that most of the indica varieties were photoperiod insensitive, while most japonica varieties were not (Figure 1c, d). The other seven yield‐related traits were highly diverse among the collection. For example, tiller number (TN) ranged from 5.6 to 14.2 in Hangzhou and 5.5 to 12.0 in Hainan. Grain number per plant (GNPP) ranged from 59.5 to 247.3 in Hangzhou and 71.4 to 207.2 in Hainan. Thousand grain weight (TGW) ranged from 17.6 to 28.8 in Hangzhou and 19.6 to 33.4 in Hainan (Table 1).
Table 1.
Descriptive statistics of eight yield‐related traits of 123 major rice varieties cultivated in China in 2016 (Hangzhou, LD) and 2017 (Hainan, SD)
| Trait | Year | Japonica (n = 54) | Indica (n = 69) | ||
|---|---|---|---|---|---|
| Range | Mean ± SD | Range | Mean ± SD | ||
| HD | 2016 LD | 54.0–114.0 | 87.8 ± 17.51 | 56.0–117.0 | 78.1 ± 13.34 |
| 2017 SD | 66.0–100.0 | 73.9 ± 7.12 | 68.0–100.0 | 81.3 ± 9.88 | |
| TN | 2016 LD | 5.6–14.2 | 9.2 ± 1.76 | 5.4–16.3 | 9.7 ± 2.11 |
| 2017 SD | 5.5–12.0 | 8.2 ± 1.51 | 4.5–10.8 | 6.7 ± 1.55 | |
| PBN | 2016 LD | 6.3–16.3 | 11.4 ± 2.22 | 6.0–16.5 | 11.3 ± 2.18 |
| 2017 SD | 6.8–12.0 | 8.8 ± 1.21 | 5.2–15.8 | 10.3 ± 1.92 | |
| SBN | 2016 LD | 9.0–51.0 | 24.8 ± 8.58 | 14.3–72.5 | 35.8 ± 12.10 |
| 2017 SD | 10.4–41.2 | 21.4 ± 6.00 | 13.6–62.0 | 32.6 ± 11.00 | |
| GNPP | 2016 LD | 59.5–247.3 | 138.9 ± 37.60 | 84.0–370.8 | 186.7 ± 54.57 |
| 2017 SD | 71.4–207.2 | 116.9 ± 27.18 | 80.0–312.6 | 176.4 ± 53.85 | |
| TGW | 2016 LD | 17.6–28.8 | 23.9 ± 2.76 | 13.5–31.4 | 23.8 ± 3.36 |
| 2017 SD | 19.6–33.4 | 27.0 ± 2.37 | 15.0–34.9 | 27.1 ± 3.74 | |
| GWPP | 2016 LD | 15.0–46.8 | 29.5 ± 6.50 | 22.0–68.5 | 40.6 ± 9.28 |
| 2017 SD | 16.5–30.9 | 25.0 ± 3.14 | 15.0–43.1 | 30.2 ± 5.73 | |
| GWSP | 2016 LD | 1.5–5.4 | 3.3 ± 0.87 | 2.1–7.1 | 4.4 ± 1.24 |
| 2017 SD | 1.9–5.2 | 3.1 ± 0.59 | 2.3–7.8 | 4.7 ± 1.42 | |
GNPP, grain number per plant; GWPP, grain weight per plant; GWSP, grain weight per single panicle; HD, heading date; LD, long day; n, number of cultivars tested; PBN, primary branch number; SBN, secondary branch number; SD, short day; SD, standard deviation; TGW, 1000‐grain weight; TN, tiller number.
SSR diversity and population structure
Twenty‐eight polymorphic SSR markers, randomly distributed on the 12 rice chromosomes, were selected to evaluate the genetic diversity of the 123 cultivated varieties. A total of 128 alleles were amplified and individual SSR marker contained between 2 and 9 alleles with an average of 4.5714 alleles for each marker. The average gene diversity was 0.5835, ranging from 0.4489 to 0.7980. The average polymorphism information content (PIC) value was 0.5027, ranging from 0.3586 to 0.7735 (Table S2).
The population structure of the 123 cultivated varieties based on the SSR genotype showed that the highest log‐likelihood scores of the population structure were observed when the number of populations was set at two (K = 2), suggesting that these varieties can be classified into two subpopulations (Figure S1a). A neighbour‐joining tree of 123 cultivated varieties was constructed based on Nei's genetic distance, which indicated that the rice accessions could be well differentiated into japonica and indica groups (Figure S1a).
Hd1 exhibited a high degree of nucleotide polymorphism and protein diversity
We analysed the Hd1 coding region (1825 bp, which includes one intron and two exons) in the 123 cultivated varieties. The Hd1 sequence exhibited a high degree of nucleotide polymorphism; Thirteen SNPs and 10 insertions/deletions (InDels) were detected in the coding region of Hd1, of which 3 SNPs were found in the zinc finger domain and 1 InDel was found in the CCT domains (Figure 2). The Hd1 sequence of Nipponbare (NPB, haplotype 1, H1) was defined as the reference sequence. A total of 19 haplotypes, named H2 to H20, were identified in the 123 cultivated varieties (Figure 2). Haplotypes H4, H5, H8‐13, H17 and H20 belong to the japonica subpopulation, while haplotypes H2, H3, H6, H7, H14‐16, H18 and H19 belong to the indica subpopulation. The most prevalent haplotypes were H8, H13, H14, H15 and H16, which included 30, 9, 12, 26 and 24 varieties, respectively. Of the remaining haplotypes, each was found in four or fewer varieties (Figure 2, Table S1). When comparing the mean values of the eight target traits in plants with the five major Hd1 haplotypes, significant differences were detected among the five haplotypes except for TN in Hangzhou and TGW in both locations (Table 2).
Figure 2.
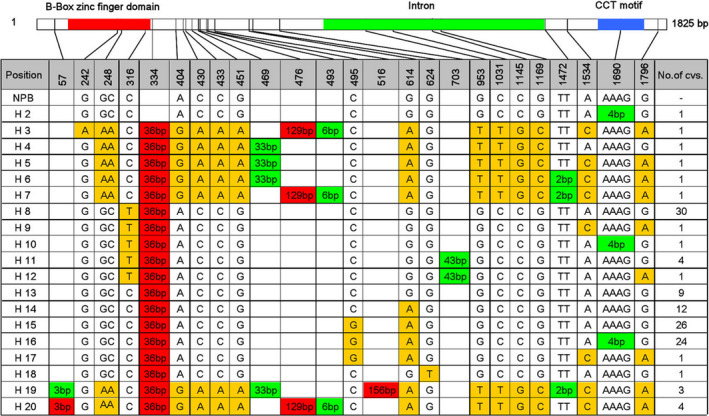
A high degree of polymorphism in the Hd1 coding sequence in 123 major rice varieties cultivated in China. Hd1 contains two exons (indicated in white rectangles) and two domains (red rectangle indicates the B‐Box zinc finger domain, and blue rectangle indicates the CCT motif). The Hd1 nucleotide sequences of the 123 varieties were compared with that of cv. Nipponbare. Polymorphic nucleotides are indicated by different colours. Base substitution, deletion and insertion sites are indicated by yellow, green and red background, respectively. The number of cultivars with each type of sequence (haplotypes 2–20) is shown in the column at the right.
Table 2.
Comparison of means of eight traits among the five major haplotypes
| Trait | Year | H 8 (n = 30) | H 13 (n = 9) | H 14 (n = 12) | H 15 (n = 26) | H 16 (n = 24) | F | P |
|---|---|---|---|---|---|---|---|---|
| Means ± SD | Means ± SD | Means ± SD | Means ± SD | Means ± SD | ||||
| HD | 2016 LD | 91.8 ± 13.34b | 102.3 ± 9.08a | 70.0 ± 6.38c | 74.5 ± 16.93c | 85.5 ± 8.41b | 3.53 | <0.0001 |
| 2017 SD | 75.3 ± 8.09b | 72.3 ± 1.66b | 74.6 ± 1.68b | 75.0 ± 5.89b | 91.3 ± 7.22a | 5.71 | <0.0001 | |
| TN | 2016 LD | 8.9 ± 1.24b | 10.11 ± 1.71ab | 9.0 ± 1.18b | 10.5 ± 2.26a | 9.0 ± 1.83b | 1.48 | 0.0891 |
| 2017 SD | 8.0 ± 1.39b | 9.5 ± 1.24a | 6.5 ± 1.09 cd | 7.3 ± 1.63bc | 6.1 ± 1.45d | 2.47 | 0.0009 | |
| PBN | 2016 LD | 12.1 ± 1.60a | 11.2 ± 1.29ab | 10.1 ± 1.50b | 10.7 ± 2.36b | 12.4 ± 1.61a | 1.71 | 0.0323 |
| 2017 SD | 8.8 ± 1.21 cd | 8.0 ± 0.41d | 9.2 ± 1.03bc | 9.9 ± 2.04b | 11.5 ± 1.14a | 2.68 | 0.0003 | |
| SBN | 2016 LD | 26.9 ± 7.67 cd | 21.6 ± 4.42d | 36.3 ± 9.32ab | 30.2 ± 11.44bc | 42.7 ± 11.66a | 2.07 | 0.0059 |
| 2017 SD | 22.0 ± 5.96c | 17.0 ± 3.98c | 32.5 ± 6.62ab | 29.5 ± 11.67b | 38.3 ± 10.54a | 2.95 | <0.0001 | |
| GNPP | 2016 LD | 149.2 ± 31.57bc | 128.5 ± 17.20c | 178.2 ± 39.31b | 161.2 ± 50.87b | 220.5 ± 52.43a | 2.11 | 0.0048 |
| 2017 SD | 119.1 ± 27.29c | 96.6 ± 16.36c | 164.5 ± 32.03b | 163.5 ± 60.68b | 206.4 ± 46.19a | 3.33 | <0.0001 | |
| TGW | 2016 LD | 23.4 ± 3.00ab | 25.1 ± 2.63a | 25.6 ± 2.64a | 24.5 ± 1.51ab | 22.6 ± 4.64b | 0.92 | 0.5939 |
| 2017 SD | 26.7 ± 2.61bc | 29.2 ± 1.75a | 29.0 ± 2.45ab | 27.5 ± 2.31ab | 26.2 ± 5.21c | 1.37 | 0.1393 | |
| GWPP | 2016 LD | 30.5 ± 5.32b | 32.1 ± 4.67b | 40.0 ± 6.90a | 39.6 ± 9.53a | 43.1 ± 9.79a | 1.90 | 0.0135 |
| 2017 SD | 24.5 ± 2.75b | 26.4 ± 2.75b | 30.6 ± 5.21a | 30.5 ± 5.84a | 31.3 ± 5.12a | 1.98 | 0.0089 | |
| GWSP | 2016 LD | 3.5 ± 0.75c | 3.2 ± 0.55c | 4.6 ± 1.06ab | 3.9 ± 1.21bc | 4.9 ± 1.19a | 1.86 | 0.0160 |
| 2017 SD | 3.1 ± 0.55c | 2.8 ± 0.40c | 4.8 ± 1.02ab | 4.4 ± 1.46b | 5.3 ± 1.41a | 3.54 | <0.0001 |
H, haplotype; LD, long day; N, number of cultivars tested; SD, short day; SD, standard deviation.
Letters are ranked by Duncan test at P < 0.05. The same letter within the same column represents no significant difference. F ratio and probability based on one‐way analysis of variance.
Hd1 proteins also displayed high diversity in the present study (Figure 3). A total of 17 distinct proteins were identified, which include 10 functional proteins (types 3–5, 11–16 and 18) and 7 nonfunctional proteins (types 2, 6, 7, 8–10 and 17). Haplotypes H11 and H12, and H13 and H18 shared the same protein type. Among the 17 distinct proteins, five major protein types (types 9, 11, 12, 14 and 15) were found, types 11 and 12 belong to the japonica group and types 9, 14 and 15 belong to the indica group (Figure 3).
Figure 3.
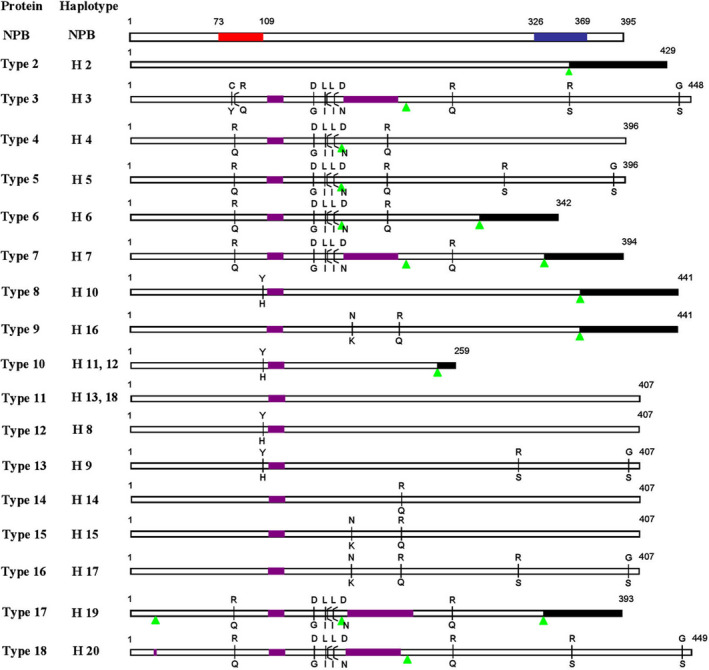
Hd1 protein diversity in 123 major rice varieties cultivated in China. The Hd1 protein sequences of the 123 major rice varieties cultivated in China were compared with that of NPB. The B‐Box zinc finger domain and CCT motif are indicated by red and blue rectangles, respectively. Purple rectangles and green triangles indicate amino acid insertion and deletion sites, respectively. Black rectangles indicate altered amino acid sequences caused by frame shifts. The letter above the vertical lines indicates NPB amino acids, and the letter below indicates the amino acid substitution.
The Hd1 ind haplotypes displayed signatures of artificial selection
In order to identify which Hd1 haplotypes were the major types used in different years, we divided the 123 varieties into three groups according to the period of time of their release: years 1936–1969, 1970–1993 and 1993–2009 (Figure 4). Among the five major haplotypes, the Hd1 jap haplotype H8 maintained a high selection frequency in all 3 groups, with allele frequencies of 50%, 61% and 60%, respectively (Figure 4a, b). Moreover, the expression level of H8 changed very little among three groups (Figure 4c). For Hd1 ind haplotypes, distinct differences were noted among the three groups. For example, H15 was the major haplotype from 1936 to 1969, but H14 and H16 were the major haplotypes from 1993 to 2009 (Figure 4a). The allele frequencies of H14, H15 and H16 were 0%, 67% and 10% in the 1936–1969 group, and 31%, 15% and 50% in the 1993–2009 group, respectively (Figure 4b). Interestingly, the expression of Hd1 ind alleles decreased in H15 and H16 (Figure 4c). These results indicated that the preponderant Hd1 allele has changed in recent decades in Chinese indica varieties.
Figure 4.
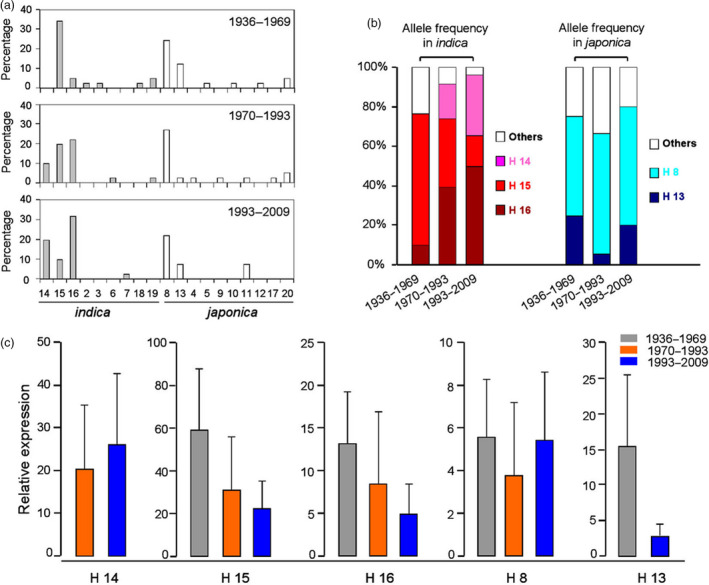
Frequency and expression of Hd1 haplotypes in 123 major rice varieties cultivated in China. (a) Allele frequencies of 19 Hd1 haplotypes among the three time period groups. Darker grey rectangles indicate indica haplotypes, and white rectangles indicate japonica haplotypes. (b) Allele frequencies of five major haplotypes among the three stages. Pink, red, brown, light blue, deep blue and white indicate haplotypes 14, 15, 16, 8, 13, and all other haplotypes, respectively. (c) The expression of five major haplotypes among the three stages.
We further compared these 3 indica haplotypes to the mean values of the eight target traits; significant differences were detected in secondary branch number (SBN), indicating that the secondary branch number was the major aim for breeding of Chinese indica cultivars (Table 3).
Table 3.
Comparison of eight yield‐related traits among three major Hd1 ind haplotypes in varieties cultivated in 1936–1969 and 1993–2009
| Trait | Year | 1936–1969 | 1993–2009 | F | P | |
|---|---|---|---|---|---|---|
| H 15 (n = 14) | H 14 (n = 8) | H 16 (n = 13) | ||||
| HD | 2016 LD | 78.5 ± 20.84ab | 69.4 ± 6.00b | 86.9 ± 9.01a | 1.35 | 0.2660 |
| 2017 SD | 74.3 ± 5.21b | 74.6 ± 1.41b | 90.9 ± 6.69a | 7.63 | <0.0001 | |
| TN | 2016 LD | 11.3 ± 2.13a | 8.7 ± 1.00b | 8.8 ± 1.96b | 1.37 | 0.2548 |
| 2017 SD | 7.9 ± 1.32a | 6.7 ± 1.30ab | 5.8 ± 1.58b | 1.81 | 0.1104 | |
| PBN | 2016 LD | 10.6 ± 2.a | 10.4 ± 1.47a | 12.1 ± 1.30a | 1.02 | 0.4781 |
| 2017 SD | 9.4 ± 2.03b | 9.2 ± 1.22b | 11.3 ± 0.93a | 1.23 | 0.3301 | |
| SBN | 2016 LD | 23.7 ± 5.93b | 37.9 ± 9.84a | 45.1 ± 13.38a | 2.62 | 0.0248 |
| 2017 SD | 24.3 ± 7.35c | 33.1 ± 7.67b | 42.0 ± 11.68a | 2.46 | 0.0332 | |
| GNPP | 2016 LD | 134.9 ± 28.59b | 185.4 ± 40.99a | 223.9 ± 60.18a | 1.95 | 0.0857 |
| 2017 SD | 137.2 ± 41.11b | 164.1 ± 38.19b | 217.78 ± 52.65a | 1.90 | 0.0932 | |
| TGW | 2016 LD | 24.7 ± 1.50a | 24.7 ± 1.74a | 23.2 ± 5.38a | 1.00 | 0.4902 |
| 2017 SD | 28.0 ± 1.31a | 27.9 ± 1.61a | 26.5 ± 6.11a | 1.07 | 0.4353 | |
| GWPP | 2016 LD | 36.7 ± 6.28a | 39.2 ± 7.41a | 43.2 ± 9.28a | 1.00 | 0.4934 |
| 2017 SD | 29.3 ± 6.44a | 29.8 ± 6.18a | 31.5 ± 5.62a | 1.46 | 0.2177 | |
| GWSP | 2016 LD | 3.3 ± 0.68b | 4.6 ± 1.08a | 5.0 ± 1.12a | 1.58 | 0.1717 |
| 2017 SD | 3.8 ± 1.09b | 4.6 ± 1.16b | 5.7 ± 1.53a | 2.38 | 0.0384 | |
H, haplotype; LD, long day; N, number of cultivars tested; SD, short day; SD, standard deviation. Letters are ranked by Duncan test at P < 0.05. The same letter within the same column represents no significant difference. F ratio and probability based on one‐way analysis of variance.
Haplotype 16 was the preponderant allele in Chinese indica breeding
An association analysis between Hd1 haplotypes and eight yield‐related traits was conducted to identify SNP‐trait associations separately using a general linear model (GLM), which accounted for population structure data (Table S1). Five major haplotypes, H8, H13, H14, H15 and H16, were analysed. Other haplotypes were excluded because of a limited number of varieties (four or fewer). Three SNPs (SNP316(C‐T), SNP495(C‐G) and SNP614(G‐A)) were found in the five types (Figure 2). SNP316(C‐T) is a C/T mutation located at 316 bp downstream of the ATG initiation site, which was found in the japonica group (Figure 2). SNP316(C‐T) showed a significant association with HD, SBN, GNPP, TGW and GWSP in the 2016 LD experiment (Table 4). SNP495(C‐G) and SNP614(G‐A) are major SNPs in the indica group. SNP495(C‐G) only showed a significant association with HD in the 2016 LD experiment, while SNP614(G‐A) exhibited a significant association with SBN, GNPP and GWSP in the 2016 LD experiment and with TGW in the LD and SD conditions (Table 4). SNP495(C‐G) is the only difference between haplotypes H14 and H15, but there was no significant difference among the eight yield‐related traits between these haplotypes. However, a significant difference was found between haplotype H16 (one base substituted and four base (AAAG) deficiency with H14, four base (AAAG) deficiency with H15) and H14 or H15 among the eight yield‐related traits (Table S3).
Table 4.
Hd1 haplotype associations with eight agronomic traits
| Site | Year | C316T | C495G | G614A | |||
|---|---|---|---|---|---|---|---|
| Traits | P | R 2 | P | R 2 | P | R 2 | |
| HD | 2016 LD | 0.0495 | 0.2149 | 0.0154 | 0.3543 | n.s. | – |
| 2017 SD | n.s. | – | n.s. | – | n.s. | – | |
| TN | 2016 LD | n.s. | – | n.s. | – | n.s. | – |
| 2017 SD | n.s. | – | n.s. | – | n.s. | – | |
| PBN | 2016 LD | n.s. | – | n.s. | – | n.s. | – |
| 2017 SD | n.s. | – | n.s. | – | n.s. | – | |
| SBN | 2016 LD | 0.0275 | 0.2115 | n.s. | – | 0.0403 | 0.1868 |
| 2017 SD | n.s. | – | n.s. | – | n.s. | – | |
| GNPP | 2016 LD | 0.0146 | 0.2653 | n.s. | – | 0.0132 | 0.2717 |
| 2017 SD | n.s. | – | n.s. | – | n.s. | – | |
| TGW | 2016 LD | 0.0354 | 0.2509 | n.s. | – | 0.0025 | 0.4458 |
| 2017 SD | n.s. | – | n.s. | – | 0.0387 | 0.2187 | |
| GWPP | 2016 LD | n.s. | – | n.s. | – | n.s. | – |
| 2017 SD | n.s. | – | n.s. | – | n.s. | – | |
| GWSP | 2016 LD | 0.0263 | 0.1733 | n.s. | – | 0.0263 | 0.1733 |
| 2017 SD | n.s. | – | n.s. | – | n.s. | – | |
Result of structure‐based association mapping (P < 0.05) of haplotypes 8 and 13–16 by GLM analysis of TASSEL. R 2, the total variation explained by the SNP.
Improvement of Hd1 alleles in indica‐japonica breeding
Due to the influence of photosensitivity, the heading date of japonica cultivars was gradually shortened in the process of extending rice cultivation to the south area of China, which resulted in a decrease in grain yield. Therefore, it is necessary to introduce the heading‐related genes of indica varieties into japonica varieties to prolong the heading date. According to the above results of preponderant Hd1 alleles of indica and japonica varieties, we hybridized and screened the progeny of the japonica variety Chunjiang06 (CJ06, the same Hd1 alleles as haplotype H8) and the indica variety Taichung native 1 (TN1, the same Hd1 alleles as haplotype H16; Figure 5a). A BC4F5 line (Q77) containing the Hd1 TN1 fragment in the CJ06 background was selected (Figure 5b, c). In a comparison of yield‐related traits among CJ06, Q77 and TN1 plants, we found that the HD of Q77 was 7 days longer than that of CJ06 (receptor parent), the number of GNPP, PBN and SBN were significantly increased by 29.50, 2.31 and 3.77, respectively, and the weight of grain weight per plant (GWPP) and GWSP were significantly increased by 18.50 and 0.54, respectively. In contrast, there was no significant difference in these traits between Q77 and TN1 (Figure 5d–i).
Figure 5.
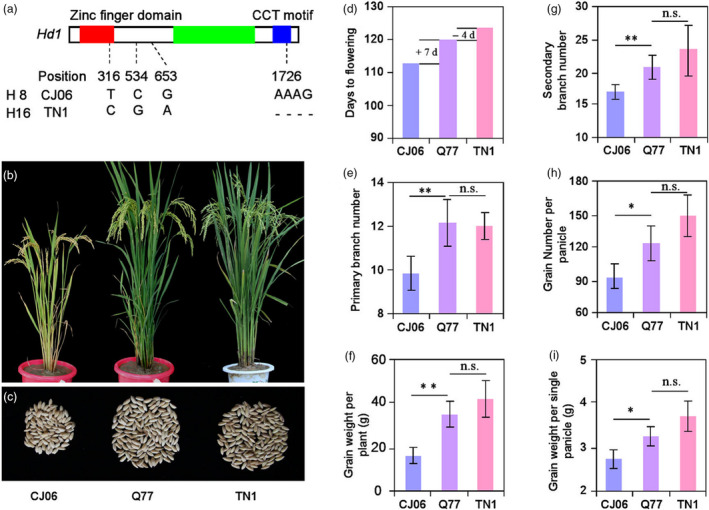
Improvement of Hd1 alleles between japonica and indica. (a) Sequence alignment in Hd1 between CJ06 and TN1. The zinc finger domain, CCT motif and intron are indicated by red, blue and green rectangles, respectively. The number and letter indicate the base position and difference, respectively. (b) Phenotypes of CJ06, Q77 and TN1 plants at the heading stage in the SD condition. (c) Phenotypes of grain weight per plant in CJ06, Q77 and TN1 plants. The comparison of heading date (d), primary branch number (e), grain weight per plant (f), secondary branch number (g), grain number per panicle (h) and grain weight per single panicle (i) in CJ06, Q77 and TN1 plants. Error bars indicate SD; *P < 0.05, **P < 0.01 (Student's t test).
Changing the heading date in rice may affect grain qualities (Cho et al., 2013). Therefore, we detected the quality‐related traits (amylose content; AC, gel consistency; GC, gelatinization temperature; GT and eating and cooking qualities; ECQ index) of CJ06 (high quality), Q77 and TN1 (low quality). There were no significant differences between Q77 and CJ06 for AC, GC and the ECQ index, but there was a significant difference between Q77 and TN1 for these traits (Figure 6a, c, d). There were no significant differences among CJO6, Q77 and TN1 for GT (Figure 6b). The results indicated that the introduction of the Hd1 ind alleles of haplotype H16 into the CJ06 did not change the quality of the grain. Therefore, it is feasible to use the Hd1 ind alleles of haplotype H16 to prolong the heading date of a japonica variety, thereby increasing grain yield without changing quality.
Figure 6.
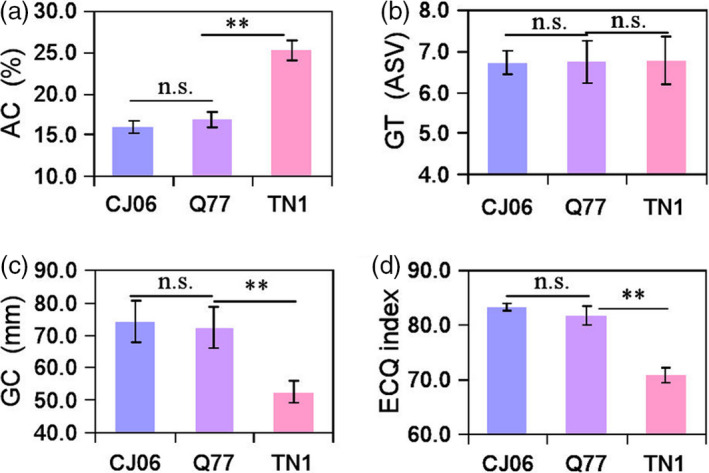
Comparison of eating and cooking quality‐related traits among CJ06, Q77 and TN1. Comparison of amylose content (a), gelatinization temperature (b), gel consistency (c) and ECQs index (d) among CJ06, Q77 and TN1 plants. Error bars indicate SD; **P < 0.01 (Student's t test).
Discussion
Rice has an estimated 8000‐ to 10 000‐year history of domestication and breeding (Doebley et al., 2006; Takahashi et al., 2009). Human selection and adaptation to diverse environments have resulted in numerous cultivars (Khush, 1997). Natural genetic variation for heading date has been largely documented, which provides a valuable material for improving adaptation to local environments (Goretti et al., 2017). Despite the different alleles of heading date was application in rice traditional breeding programmes, the best gene pyramiding at the molecular level is still unclear. In this study, we selected and sequenced Hd1 from 123 major rice varieties cultivated from 1936 to 2009 in China to identify the key SNPs in Hd1 that affect yield‐related traits. Then, we utilized the preponderant alleles of Hd1 for heading date‐, yield‐ and quality‐related trait improvement.
The diverse Hd1 alleles display significant indica‐japonica differentiation
Hd1 is an important gene for the control of flowering time, and its sequence has evolved a high degree of polymorphism during rice domestication (Takahashi et al., 2009; Wei et al., 2014; Zheng et al., 2016). Takahashi et al. (2009) identified 17 Hd1 allele types using 64 core rice cultivars worldwide. Zheng et al. (2016) reported 39 allele types from 154 rice germplasms. However, these reports did not point out whether Hd1 has preponderant alleles in indica‐japonica rice breeding. Our results show that there are obvious differences in the preponderant Hd1 alleles between indica and japonica varieties (Figure 4a). Furthermore, our results clearly demonstrated that the preponderant alleles of Hd1 jap did not change much in the breeding of modern Chinese varieties, but did change notably in Hd1 ind (Figure 4a, b). In addition, we found that the increase in grain yield in modern indica varieties is mostly due to the selection for SBN (Table 3). This result indicates that the rice breeding model in China has focused on the high‐yield model with large panicles, and increasing the number of secondary branches is the key way to generate rice with large panicles.
Association analysis and utilization of preponderant alleles provide a critical strategy for rice improvement
Candidate gene‐based association mapping takes advantage of recombination events in a natural population to resolve complex trait variation to individual nucleotides (Zhu et al., 2008). The selection and utilization of preponderant alleles have become an important way to cultivate high‐yielding varieties. For example, Lu et al. (2012) identified a preponderant allele (S_555) of Ghd7 using 104 rice accessions. Utilization of the alleles (S_555) can decrease plant height and allow the plant to be more resistant to lodging, while not influencing heading date and yield traits in indica subspecies. In our study, we successfully used the preponderant alleles of Hd1 ind haplotype H16 to prolong the heading date of a japonica variety CJ06 (Figure 5), indicating that this is a feasible approach to improving the heading date in japonica varieties.
Japonica varieties are mainly cultivated in the middle and low Yangtze region and northeast China, but this area is only one‐fourth of the total rice planting area of China. The biggest problem with growing japonica varieties in lower latitudes is that in lower latitudes they will flower too early and not reach their full potential yield and quality. Therefore, it is an important aim for rice breeding to prolong the heading date of japonica varieties appropriately without affecting grain yield and quality. Our results demonstrated that the introduction of the preponderant Hd1 ind alleles of haplotype H16 into a japonica cultivar significantly increased the yield per plant but did not change the quality‐related traits (Figures 5 and 6). This result implied that cultivating japonica varieties in the south of China can be achieved by introducing the Hd1 ind allele of haplotype H16 to improve heading date, yield and quality traits.
Experimental procedures
Plant materials and plant growth condition
A total of 123 major rice (O. sativa L.) varieties comprising 69 indica and 54 japonica varieties were selected from the germplasm centres of the China National Rice Research Institute. Most accessions are leading varieties cultivated in China from 1936 to 2009. The basic information for each germplasm appears in Table S1. The experiments were performed at the experimental farm of the China National Rice Research Institute in Hangzhou (HZ, 36°30′N) and Hainan (HN, 18°48′N) during the 2016 and 2017 rice‐growing seasons, respectively. The plants were grown in ten rows with six plants in each row at a planting density of 20 × 20 cm2. Field management, including irrigation, fertility and pest control, followed normal agricultural practices.
Evaluation of traits
Heading date (HD) was defined as the days from sowing to the appearance of the first panicle. Tiller number (TN), primary branch number (PBN), secondary branch number (SBN) and grain number per plant (GNPP) were measured 25 days after heading. GNPP was selected the highest panicle as the grain number. Except for two marginal plants in each side, ten independent plants were used to score the phenotypic data sets. Thirty‐five days after flowering, seeds from each plant were harvested and weighed after physicochemical properties had stabilized. Grain weight was calculated on the basis of 200 grains and converted to TGW. Grain weight per plant (GWPP) was the weight of all seeds harvested from a single plant. Grain weight per single panicle (GWSP) was the weight of the seeds harvested from a single tiller. For quality‐related traits, 15 grains of milled rice were selected for measuring gelatinization temperature (GT), and 15 g of grain was ground to flour to measure amylose content (AC) and gel consistency (GC). AC, GC and GT were measured according to the procedures in Leng et al. (2013). The eating and cooking qualities (ECQs) index was measured according to the procedure in Zeng et al. (2017).
DNA extraction, PCR and sequence analysis
Genomic DNA was extracted from fresh leaves of each plant using the cetyltrimethylammonium bromide (CTAB) method (Murray and Thompson, 1980). The polymorphic simple sequence repeat (SSR) markers, randomly distributed across the 12 rice chromosomes, are listed in Table S2. PCRs for amplification of the SSRs were conducted according to the methods in Jin et al. (2010) and the products were run on an 8% denaturing polyacrylamide gel at 120 V for 100 min and the gel was visualized using silver staining. Hd1, including the 1825‐bp coding region, was amplified from genomic DNA using KOD plus (TOYOBO, Tokyo, Japan). PCRs were conducted using standard PCR protocols. The primers used for PCR and sequencing are listed in Table S4. The initial genomic sequence of Hd1 was assembled using DNA star software (DNAStar Inc., Madison, WI).
Genetic diversity and population structure analysis
PowerMarker V3.25 was used to analyse the genetic diversity including the number of alleles per locus, major allele frequency, gene diversity and polymorphism information content (PIC) values (Liu and Muse, 2005). Nei's distance (Nei et al., 1983) was calculated and used for the unrooted phylogeny reconstruction using the neighbour‐joining method as implemented in PowerMarker with the tree viewed using MEGA 4.0 (Tamura et al., 2007). The population structure among the 123 rice varieties, based on the genotype data, was performed using STRUCTURE V 2.3.4 (Pritchard and Wen, 2004). The number of populations (K) was selected from 2 to 10 and five independent runs of a burn‐in of 10 000 iterations followed by 100 000 iterations for each value of K. The optimum structure number of K was selected based on the report of Evanno et al., 2005.
RNA preparation and qRT‐PCR analysis
Total RNA was extracted using the RNeasy plant mini kit (Qiagen, Valencia, CA) following the manufacturer's instructions. RNA preparations were treated with DNase I (Takara, Tokyo, Japan) to remove traces of DNA contamination. Reverse transcription was conducted with the ReverTra Ace qPCR‐RT Kit (TOYOBO). After synthesis, the cDNA was diluted fivefold in TE buffer, and 1 μL was used for quantitative PCR using the Fast SYBR Green Master Mix (Applied Biosystems, Carlsbad, CA) and gene‐specific primers (Table S4) in an ABI7900 analyser (Applied Biosystems, Foster City, CA).
The development of chromosome segment substitution lines
The TN1/CJ06 chromosome segment substitution lines (CSSLs) were obtained according to Su et al. (2011). The plants carrying the TN1 genotype at the flanking region of Hd1 were selected to cross with the CJ06 variety for five rounds of backcrossing. A total of 71 SSR markers were used to screen for the genetic background (Ren et al., 2016). The SSR markers RM539 and RM454 were used to identify the plants containing the TN1 genotype in backcross progeny.
Statistical analysis
Analysis of variance was performed using Microsoft Excel 2003. Duncan's multiple comparison was performed by SAS 8.0 software. Hd1 sequences were aligned by Clustalx 2.1, and the alignment results were input into TASSEL. A general linear model (GLM) was performed in TASSEL V 3.0 for association analysis, which accounted for population structure (Q).
Author contributions
YJL and YHG performed most of the research. LC, LCH, LPD, DYR, QKX, YZ, KP, LS and GJD performed the trait investigation. YLY, JH, GHZ, GC, ZYG, LBG and GYY analysed the data. YJL wrote the article. YJL, LZ, QQ and DLZ designed the research. DLZ revised the article.
Competing financial interests
The authors declare that they have no competing financial interests to disclose.
Supporting information
Table S1 Basic information for 123 major rice varieties cultivated in China.
Figure S1 Population structure and unrooted neighbor‐joining trees of 123 major rice varieties cultivated in China.
Table S2 Summary statistics for the 28 SSR markers used in this study.
Table S3 Comparison of means of eight traits among the three major indica haplotypes.
Table S4 The primer sequences used in this study.
Acknowledgements
We thank Dr. J.Z. Wu (Institute of Crop Science, National Agriculture and Food Research Organization, Japan) for his kindly suggestion for improving the manuscript. This work was supported by the National Natural Science Foundation of China (Grant Nos. 91735303, 31661143006, 31801337), the Ministry of Agriculture of China for transgenic research (Grant No. 2016ZX08009003‐001) and the ‘Science and Technology Innovation Project’ of the Chinese Academy of Agriculture Sciences.
Contributor Information
Qian Qian, Email: dalizeng@126.com.
Li Zhu, Email: zhuli05@caas.cn.
Dali Zeng, Email: qianqian188@hotmail.com.
References
- Cho, Y.C. , Suh, J.P. , Yoon, M.R. , Baek, M.K. , Won, Y.J. , Lee, J.H. , Park, H.S. et al. (2013) QTL mapping for paste viscosity characteristics related to eating quality and QTL‐NIL development in Japonica rice (Oryza sativa L.). Plant Breed. Biotechnol. 1, 333–346. [Google Scholar]
- Doebley, J.F. , Gaut, B.S. and Smith, B.D. (2006) The molecular genetics of crop domestication. Cell, 127, 1309–1321. [DOI] [PubMed] [Google Scholar]
- Doi, K. , Izawa, T. , Fuse, T. , Yamanouchi, U. , Kubo, T. , Shimatani, Z. , Yano, M. et al. (2004) Ehd1, a B‐type response regulator in rice, confers short‐day promotion of flowering and controls FT‐like gene expression independently of Hd1 . Genes Dev. 18, 926–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno, G. , Regnaut, S. and Goudet, J. (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. [DOI] [PubMed] [Google Scholar]
- Goretti, D. , Martignago, D. , Landini, M. , Brambilla, V. , GoAmez‐Ariza, J. , Gnesutta, N. , Galbiati, F. et al. (2017) Transcriptional and post‐transcriptional mechanisms limit Heading Date1 (Hd1) function to adapt rice to high latitudes. PLoS Genet. 13, e1006530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama, R. , Izawa, T. and Shimamoto, K. (2002) Isolation of rice genes possibly involved in the photoperiodic control of flowering by a fluorescent differential display method. Plant Cell Physiol. 43, 494–504. [DOI] [PubMed] [Google Scholar]
- Hayama, R. , Yokoi, S. , Tamaki, S. , Yano, M. and Shimamoto, K. (2003) Adaptation of photoperiodic control pathways produces short‐day flowering in rice. Nature, 422, 719–722. [DOI] [PubMed] [Google Scholar]
- Izawa, T. (2007) Adaptation of flowering‐time by natural and artificial selection in Arabidopsis and rice. J. Exp. Bot. 58, 3091–3097. [DOI] [PubMed] [Google Scholar]
- Jin, L. , Lu, Y. , Xiao, P. , Sun, M. , Corke, H. and Bao, J.S. (2010) Genetic diversity and population structure of a diverse set of rice germplasm for association mapping. Theor. Appl. Genet. 121, 475–487. [DOI] [PubMed] [Google Scholar]
- Khush, G.S. (1997) Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 35, 25–34. [PubMed] [Google Scholar]
- Kim, S.L. , Lee, S.Y. , Kim, H.J. , Nam, H.G. and An, G.H. (2007) OsMADS51 is a short‐day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a . Plant Physiol. 145, 1484–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima, S. , Takahashi, Y. , Kobayashi, Y. , Monna, L. , Sasaki, T. , Araki, T. and Yano, M. (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short‐day conditions. Plant Cell Physiol. 43, 1096–1105. [DOI] [PubMed] [Google Scholar]
- Komiya, R. , Ikegami, A. , Tamaki, S. , Yokoi, S. and Shimamoto, K. (2008) Hd3a and RFT1 are essential for flowering in rice. Development, 135, 767–774. [DOI] [PubMed] [Google Scholar]
- Leng, Y.J. , Xue, D.W. , Yang, Y.L. , Hu, S.K. , Su, Y. , Huang, L.C. , Wang, L. et al. (2013) Mapping of QTLs for eating and cooking quality‐related traits in rice (Oryza sativa L.). Euphytica, 197, 99–108. [Google Scholar]
- Li, X.F. , Liu, H.Z. , Wang, M.Q. , Liu, H.L. , Tan, X.J. , Zhou, W.J. , Lü, T.X. et al. (2015) Combinations of Hd2 and Hd4 genes determine rice adaptability to Heilongjiang Province, northern limit of China. J. Integr. Plant Biol. 57, 698–707. [DOI] [PubMed] [Google Scholar]
- Liu, K. and Muse, S.V. (2005) PowerMarker: integrated analysis environment for genetic marker data. Bioinformatics, 21, 2128–2129. [DOI] [PubMed] [Google Scholar]
- Lu, L. , Yan, W.H. , Xue, W.Y. , Shao, D. and Xing, Y.Z. (2012) Evolution and association analysis of Ghd7 in rice. PLoS ONE, 7, e34021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, L. , Shao, D. , Qiu, X.J. , Sun, L. , Yan, W.H. , Zhou, X.C. , Yang, L. et al. (2013) Natural variation and artificial selection in four genes determine grain shape in rice. New Phytol. 200, 1269–1280. [DOI] [PubMed] [Google Scholar]
- Murray, M.G. and Thompson, W.F. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M. , Tajima, F. and Tateno, Y. (1983) Accuracy of estimated phylogenetic trees from molecular data. J. Mol. Evol. 19, 153–170. [DOI] [PubMed] [Google Scholar]
- Pritchard, J.K. and Wen, W. (2004) Documentation for STRUCTURE Software. Chicago, IL: The University of Chicago Press. [Google Scholar]
- Putterill, J. , Laurie, R. and Macknight, R. (2004) It's time to flower: the genetic control of flowering time. BioEssays, 26, 363–373. [DOI] [PubMed] [Google Scholar]
- Ren, D.Y. , Rao, Y.C. , Huang, L.C. , Leng, Y.J. , Hu, J. , Lu, M. , Zhang, G.H. et al. (2016) Fine mapping identifies a new QTL for brown rice rate in rice (Oryza Sativa L.). Rice, 9, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, D.Y. , Yu, H.P. , Rao, Y.C. , Xu, Q.K. , Zhou, T.T. , Hu, J. , Zhang, Y. et al. (2018) ‘Two‐floret spikelet’ as a novel resource has the potential to increase rice yield. Plant Biotechnol. J. 16, 351–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, Y. , Rao, Y.C. , Hu, S.K. , Yang, Y.L. , Gao, Z.Y. , Zhang, G.H. , Liu, J. et al. (2011) Map‐based cloning proves qGC‐6, a major QTL for gel consistency of japonica/indica cross, responds by Waxy in rice (Oryza sativa L.). Theor. Appl. Genet. 123, 859–867. [DOI] [PubMed] [Google Scholar]
- Sun, C. , Fang, J. , Zhao, T.L. , Xu, B. , Zhang, F.T. , Liu, L.C. , Tang, J.Y. et al. (2012) The histone methyltransferase SDG724 mediates H3K36me2/3 deposition at MADS50 and RFT1 and promotes flowering in rice. Plant Cell, 24, 3235–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y. , Teshima, K.M. , Yokoi, S. , Innan, H. and Shimamoto, K. (2009) Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice. Proc. Natl Acad. Sci. USA, 106, 4555–4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Dudley, J. , Nei, M. and Kumar, S. (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Thornsberry, J.M. , Goodman, M.M. , Doebley, J. , Kresovich, S. , Nielsen, D. and Buckler, E.S. IV . (2001) Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 28, 286–289. [DOI] [PubMed] [Google Scholar]
- Tian, Z.X. , Qian, Q. , Liu, Q.Q. , Yan, M.M. , Liu, X.F. , Yan, C.J. , Liu, G.F. et al. (2009) Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc. Natl Acad. Sci. USA, 106, 21760–21765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, X. , Qiao, W.H. , Yuan, N.N. , Chen, Y.T. , Wang, R.S. , Cao, L.R. , Zhang, W.X. et al. (2014) Domestication and association analysis of Hd1 in Chinese mini‐core collections of rice. Genet. Resour. Crop Evol. 61, 121–142. [Google Scholar]
- Yano, M. , Katayose, Y. , Ashikari, M. , Yamanouchi, U. , Monna, L. , Fuse, T. , Baba, T. et al. (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS . Plant Cell, 12, 2473–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, L.P. (2014) Development of hybrid rice to ensure food security. Rice Sci. 21, 1–2. [Google Scholar]
- Zeng, D.L. , Tian, Z.X. , Rao, Y.C. , Dong, G.J. , Yang, Y.L. , Huang, L.C. , Leng, Y.J. et al. (2017) Rational design of high‐yield and superior‐quality rice. Nat. Plants, 3, 17031. [DOI] [PubMed] [Google Scholar]
- Zheng, X.M. , Feng, L. , Wang, J.R. , Qiao, W.H. , Zhang, L.F. , Cheng, Y.L. and Yang, Q.W. (2016) Nonfunctional alleles of long‐day suppressor genes independently regulate flowering time. J. Integr. Plant Biol. 58, 540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, C.S. , Michael, G. , Buckler, E.S. and Yu, J.M. (2008) Status and prospects of association mapping in plants. Plant Genome, 1, 5–20. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Basic information for 123 major rice varieties cultivated in China.
Figure S1 Population structure and unrooted neighbor‐joining trees of 123 major rice varieties cultivated in China.
Table S2 Summary statistics for the 28 SSR markers used in this study.
Table S3 Comparison of means of eight traits among the three major indica haplotypes.
Table S4 The primer sequences used in this study.


