Abstract
The application of nanoparticles rose steeply in the last decade, where they have become a common ingredient used in processed human food, improving food properties such as shelf life and appearance. Nanoparticles have also attracted considerable interest to the livestock industry, due to their efficacy in intestinal pathogen control, with the regulatory and consumer driven push for the removal of antibiotic growth promoters. The influence of selenium (Se) nanoparticles was investigated on a diverse and mature broiler caecal microbiota using in vitro culturing and 16S rRNA gene sequencing methods for microbiota characterisation. Caecal microbiota was collected from 4 traditionally grown heritage roosters and grown for 48 h, in the presence and absence of Se nanoparticles, with 2 technical replicates each. The effect of rooster as a biological variable strongly overpowered the effects of nano-Se in the media, resulting in moderate effects on the structure and diversity of the caecal microbial community. However the nanoparticles showed a significant reduction (P < 0.05) in the abundance of an emerging poultry pathogen, Enterococcus cecorum identical operational taxonomic units (OTU), which could be of notable interest in poultry production for targeted E. cecorum control without significant disturbance to the total microbial community.
Keywords: Nanoparticle, Microbiota, Enterococcus cecorum, Short-chain fatty acids, Livestock
1. Introduction
The application of nano-scaled materials, 1 to 100 nm, has rapidly expanded across various disciplines, including but not limited to, electronics, technology, consumer goods, biomedical science, agriculture and microbiology. They display unique physicochemical properties due to high surface energy and increased surface area to volume ratio (Regan et al., 2012). Nanoparticles are used in everyday applications such as self-cleaning surfaces (Mueller and Nowack, 2008), topical products (Goyal et al., 2016) and food preservation (Espitia et al., 2012). They have excellent antimicrobial properties through the disruption of microbial cell membranes (Hajipour et al., 2012, Thill et al., 2006) and oxidation (Le Ouay and Stellacci, 2015). Culture studies have been used to examine their biocidal interaction (Jia et al., 2017, Teodoro et al., 2011) towards both prokaryotic, bacterial pathogens and eukaryotic cells such as tumour and stem cells (Arora et al., 2008, Greulich et al., 2009, Kaul and Amiji, 2005). They have also been intensively investigated for their use in joint and bone reconstruction therapies (Gangadoo et al., 2015), agricultural products (Gangadoo et al., 2016) and delivery of drugs and other substances to the body (Gupta and Curtis, 2004), as they exhibit high biocompatibility (Lu et al., 2010, Naahidi et al., 2013) and biodegradability (Mahapatro and Singh, 2011, Panyam and Labhasetwar, 2003).
Nanoparticles can be used as vehicles to transport substances to the body effectively and fast, by avoiding complex pathways and defence mechanisms as compared to their bulk counterparts (Desai et al., 1996, Mohanraj and Chen, 2006). Many studies have shown the positive effect of NP formulations delivered to the gut microbiota, focussing primarily on reducing the pathogenic load with an antibiotic based approach, by inhibiting the growth of harmful microbes (Karavolos and Holban, 2016). Since various materials have successfully targeted detrimental microbes through NP delivery, it was proposed that a NP-based system, using metal salts and complex, could also be used to enhance beneficial bacteria by delivering essential nutrients to the gut microbiota. Nanoparticles have been used to increase the abundance of beneficial species such as Lactobacillus and Bifidobacteria and reduce pathogenic bacteria and coliform counts (Gangadoo et al., 2018, Han et al., 2010, Yausheva et al., 2018). The gut ecology is a complex community and it is necessary to consider the complex interactions of multiple bacterial species, the chemistry of their growth environment and the metabolites produced.
Selenium (Se) is an important trace element required by the body for the proper functioning and development of the immune system, and it is routinely supplemented to poultry rations to prevent detrimental effects of Se deficiency in birds (Gangadoo et al., 2016). About one quarter of the gut microbiome has the ability to express selenoproteins and Se availability in microbiological media affects their expression (Kasaikina et al., 2011). These proteins play an important role in both bacteria and mammalian host where they are essential in numerous bodily functions (Labunskyy et al., 2014). We have previously investigated the ability of selenium nanoparticles (nanoSe) to improve the delivery of Se to birds and have characterised the resulting modifications of the intestinal microbiota (Gangadoo et al., 2018). We found increased abundance of some beneficial bacteria, for example Lactobacillus sp. and Faecalibacterium prausnitzii, however, without significant pathogen reduction. The quantity of butyric acid in different gut sections was increased. Butyric acid is a primary energy source for intestinal colonocytes and can promote good gut health (Van Immerseel et al., 2017).
Traditionally raised free range chickens generally show higher diversity of intestinal microbiota compared to intensively reared birds (Chen et al., 2018, Cui et al., 2017). Their microbiota may contain bacterial species that are not commonly encountered in the microbiota of birds raised in modern high-density production systems. We are interested to study such novel microbiotas and determine the effects of feed additives; with the expectation that some novel members of the microbiota may have application in modern chicken production (e.g. novel probiotics). However, it is difficult to obtain sufficient numbers of such traditionally raised birds to carry out statistically powerful in vivo studies. Here we present an in vitro study that has investigated the effect of nanoSe on in vitro cultured chicken, caecal microbial communities. We used a growth medium specifically developed to support a range of unknown and uncultured species as well as more routinely cultured bacteria, to culture caecal material from traditionally raised free range chickens.
2. Materials and methods
2.1. Animal ethics
Collection of chicken caecal and intestinal material from backyard growers was approved by the Animal Ethics Committee at Central Queensland University with the approval number A1409-318. All animal ethics procedures were in agreement with the Australian Animal Welfare Standards and Guidelines.
2.2. Media preparation
LYHBHI medium (Brain-heart infusion medium supplemented with yeast extract [5 g/L, Alfa Aesar], cellobiose [1 g/L, BD], hemin [5 mg/L, BD], cysteine [0.5 g/L, Alfa Aesar]) and resazurin (0.5 mg/L, Alfa Aesar)] (Fenn et al., 2017, Zhang et al., 2014) was enriched with a multivitamin mix (1 mL), trace element mix (1 mL), feed extract (100 g/L) and bacterial ferment (100 mL). The multivitamin mix was prepared with 5 capsules of 50 + MULTI Vitamins & Minerals (CENOVIS) and 5 capsules of vitamin K (Caruso's Natural Health, Queensland, Australia), dissolved in 50 mL of Milli-Q water, and the resulting mixture was filtered twice through a 0.45-μm and a 0.2-μm syringe filter. Appendix Table 1 shows the resulting vitamin concentrations in the medium. The trace element mix was obtained from Youngevity (California, USA) and contains plant-derived minerals. The feed extract was prepared by mixing 100 g of poultry feed (Red Hen Chick premium micro starter, Lauke Mills, Daveystone SA, Australia) to 1 L of Milli-Q water using a 1,500 W blender (Nutri Ninja Duo Auto-iQ) and left to soak overnight. The mixture was then autoclaved, centrifuged and 100 mL of the supernatant extract was added to the medium. The bacterial ferment was prepared by aerobically growing cultures of Lactobacillus plantarum (ATCC BAA-793) and Lactobacillus rhamnosus (ATCC 53103) to mid-stationary phase in LYHBHI media. The supernatant of the bacterial ferments were then mixed at a 1:1 ratio with a final volume of 50 mL, filter sterilised and added to the media. The volume of water was adjusted to ensure that the original LYHBHI medium was not diluted. The enriched LYHBHI was purged for 30 min prior to inoculation of caecal content with anaerobic gas mix (80%N2/10%CO2/10%H2, BOC, Queensland, Australia).
NanoSe was prepared as previously described (Gangadoo et al., 2017). Briefly, selenium tetrachloride was reduced with ascorbic acid to Se atoms, to which a protecting agent, polystyrene-4-sulfonate, was added to allow the formation of nanoparticle clusters. The synthesised, dark red solution was washed by multiple centrifugation with Milli-Q water and a full characterisation, including size, shape, morphology and crystallinity, was conducted. The nanoSe was then diluted with Milli-Q water to 0.9 mg/kg.
2.3. Cecum starter cultures
Caeca, from 4 roosters, were donated by a local heritage breeder. The roosters were raised with organic feed without antibiotics and had exclusive outdoor access, including overnight outdoor roosting, which provided intensive contact with wild flora and fauna. The whole intestine of each rooster was removed and placed immediately into an anaerobic gas pack (Cat. #260683, BD GasPak EZ Pouch Systems) and stored at −20 °C. The caecal samples were slowly allowed to defrost at 4 °C for 30 min. The contents of the whole ceca, for each rooster separately, was squeezed out and diluted in 50 mL of enriched LYHBHI media with 15% glycerol in an anaerobic work station (A35, Whitley, Shipley, UL). The caecal starter cultures for each rooster's caecal content were then aliquoted as 50 × 1 mL stock and stored at −80 °C until the start of the experiment. This would eliminate cold sensitive species and allow the reproducible use of each 1 mL stock for the future in vitro experiments.
2.4. In vitro growth cultures
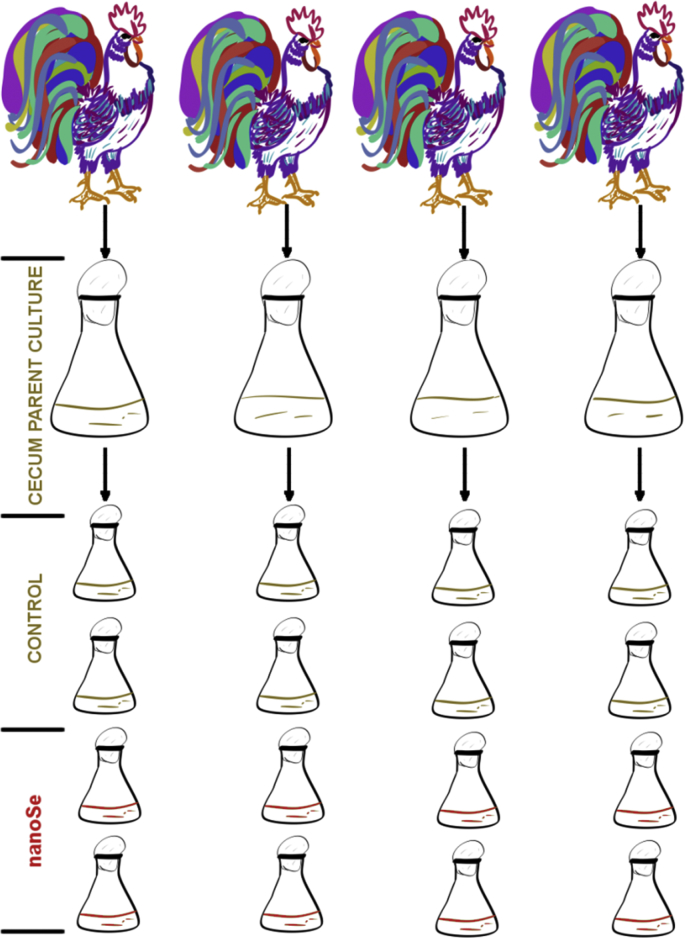
On the day of the experiment, a single glycerol stock for each one of the 4 roosters was thawed and inoculated into 50 mL of enriched LYHBHI media to grow parent cultures for the experimental inoculation. The experimental cultures were prepared in 20 mL of media in 50 mL Erlenmeyer flask with a cotton stopper, allowing for gas exchange, and incubated at 37 °C on a digital orbital shaker (Heathrow Scientific), shaking at a speed of 0.21 × g in an anaerobic hood (Whitley A35 Anaerobic Workstation, UK) running on a nitrogen rich gas mix (80% N2/10% CO2/10% H2). Four cultures were prepared from each rooster's caecal content, 2 as control and 2 with 0.9 mg/kg of nanoSe, by inoculating late exponential growing parental culture to achieve a starting culture OD620 of 0.1. Thus, the final experiment was performed on 16 cultures; n = 8 for control and the nanoSe treatment each, on 4 biological replicates (rooster's caecal content) and 2 technical replicates each, as shown in Fig. 1. Sampling of the cultures was done at 24 and 48 h and the samples were centrifuged at 18,500 × g at 4 °C for 10 min. The pellets and the supernatants were used for microbial and metabolite analysis, respectively.
Fig. 1.
A schematic of the in vitro experiment performed to examine the effect of nanoSe on growth cultures of rooster caeca samples.
2.5. DNA extraction
DNA was extracted from the centrifuged pellets of the microbial cultures. The lysis step was based on the method suggested by Yu and Morrison (2004), followed by a DNA purification step. The lysis buffer (0.5 mL) and 0.1 g of sterile zirconia beads (Cat. #11079101, BioSpec Products) were added prior to bead-beating (Mini-beadbeater, BioSpec Products) for 5 min. Following a 15-min incubation at 85 °C, the samples were centrifuged for 5 min, and binding buffer (0.8 mL) was added to the supernatant and placed through DNA Silica Membrane Mini Spin Column (Cat. #1920-250, Epoch Life Science, Inc.), followed by a two-step washing with wash buffer (0.7 mL). The washed and dried column was then eluted with 50 μL of elution buffer. The composition of the buffers is included in Appendix Table 2.
2.6. DNA amplification and sequencing
Sequencing of 16S rRNA gene DNA amplicons was performed on the Illumina MiSeq platform using 2 × 300 bp paired-end sequencing. Primers were selected to amplify the V3 – V4 region of 16S rRNA genes: forward 5′-ACTCCTACGGGAGGCAGCAG-3′ and reverse 5′-GGACTACHVGGGTWTCTAAT-3′. The primers contained barcodes, spaces and Illumina sequencing linkers as previously described (Fadrosh et al., 2014). Two samples, one from the rooster 2 and one from the rooster 4, failed the sequencing process, and were thus excluded from the analysis.
2.7. Statistical analysis
The analysis of microbial communities was performed in QIIME v.1.9.1 (Caporaso et al., 2010). Paired end sequences were joined using the Fastq-Join algorithm and no allowed mismatches using only sequences with Phred quality threshold higher than 20. Operational taxonomic units were picked at 97% similarity using Uclust (Edgar, 2010) and inspected for chimeric sequences using Pintail (Ashelford et al., 2005). Taxonomic assignments were performed against the GreenGenes (DeSantis et al., 2006) database and QIIME default arguments. Further data exploration was done using Calypso (Zakrzewski et al., 2016). Total sum normalisation and a square root transformation was performed prior to statistical analysis. Student's t-test was used to detect the significance of the differences between the groups. Analysis of similarities (ANOSIM) was performed using Calypso on weighed and unweighed UniFrac distance matrices calculated in QIIME, each with 99,999 permutations. Calypso was also used to implement the supervised multivariate Redundancy Analysis (RDA) using 999 permutations and linear regression analysis using Pearson correlation.
The complete annotated sequence dataset is publicly available on the MG-RAST database under library ID (Id pending).
2.8. Short-chain fatty acid analysis
The supernatants from the caecal cultures were diluted with 70% ethanol, filtered through a 0.45 μm syringe filter (Cat. #54504-RC, ThermoFisher) and analysed on the Gas Chromatography-Mass Spectrometry (GCMS) system. A standard stock solution (100 mg/kg) was used to construct calibration curves and stored as a method processing parameter in scan mode for the following short-chain fatty acids (SCFA), acetic, n-butyric, isobutyric, propionic and n-valeric acid.
The GCMS system used for metabolite analysis was a Shimadzu QP2010-Plus, fitted with a high-polarity column SH-Rxi-5Sil-MS (30 m × 0.25 mm × 0.25 μm, Restek) and equipped with an AOC-20i + s autosampler. The GC temperature programme started at 100 °C and was held for 1 min, increased to 12 °C per min to a temperature of 170 °C, and ramped at 100 °C per min, until a final temperature of 260 °C was reached and held for 1 min (a total of an 8.73-min programme). The GC oven temperature was set as presented in Appendix Table 3. A sample of 1 μL was injected at 250 °C using helium (5.0, Coregas, Australia) as a carrier gas at 1.97 mL/min in a split injection mode. The pressure was held at 143.3 kPa, with a total He flow of 103.4 mL/min and using a split ratio of 5. The mass spectrometer was operated in the electron ionisation mode at 0.2 kV with a source temperature of 220 °C where scan mode was used from 33 to 150 m/z. The peaks were identified by matching the mass spectra with the National Institute of Standards and Technology (NIST) library, http://chemdata.nist.gov/.
3. Results
3.1. Sample origin influences overall microbiota composition and abundance
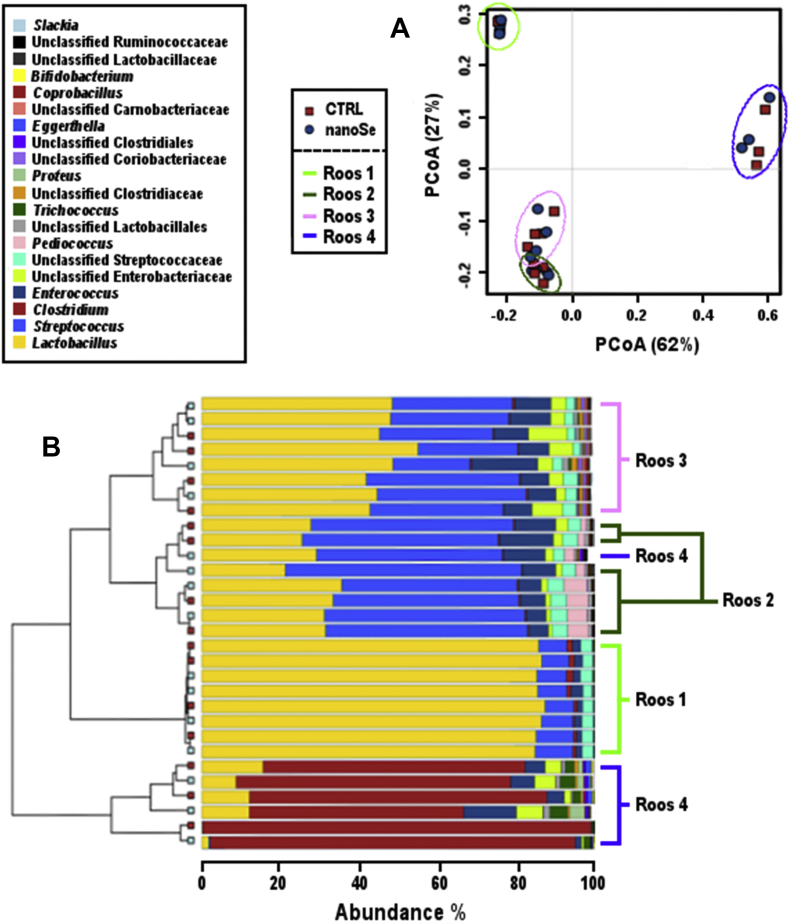
The origin of caeca greatly influenced the microbiota community of the samples, showing great biological variation. The abundance of phyla Firmicutes, Bacteroidetes, Proteobacteria and Actinobacteria differed significantly between the 4 roosters (ANOVA, P < 0.001). Lactobacillus (>20%) was the dominant genus in all roosters combined, followed by Streptococcus (>15%), Enterococcus and Clostridium (>5%) (Appendix Table 4). Principal coordinates analysis (PCoA), as shown in Fig. 2A, was performed on both unweighed and weighed UniFrac matrices and shows similarities between roosters 2 and 3, while roosters one and 4 were very distinctive (Fig. 2B). The microbiota of roosters 2 and 3 included multiple genera not found in roosters one or 4, such as Collinsella, Coprobacillus, Slackia, and unclassified families of Burkholderiales and Ruminococcaceae. Rooster 4 had the most distinctive microbiota; dominated by Clostridium, with lower abundance of Lactobacillus compared to the other roosters, and higher amounts of Trichococcus, Proteus and unclassified families comprising of Clostridiales and Burkholderiales.
Fig. 2.
Gut microbiota profile was clustered based on the bird sample. (A) PCoA analysis performed on weighed UniFrac matrices shows gut community profiles clustered by bird origin. (B) The multiple genera present in the samples confirm similarities and differences between rooster's gut communities. PCoA = Principal coordinates analysis.
3.2. NanoSe influence on microbial composition and metabolite production
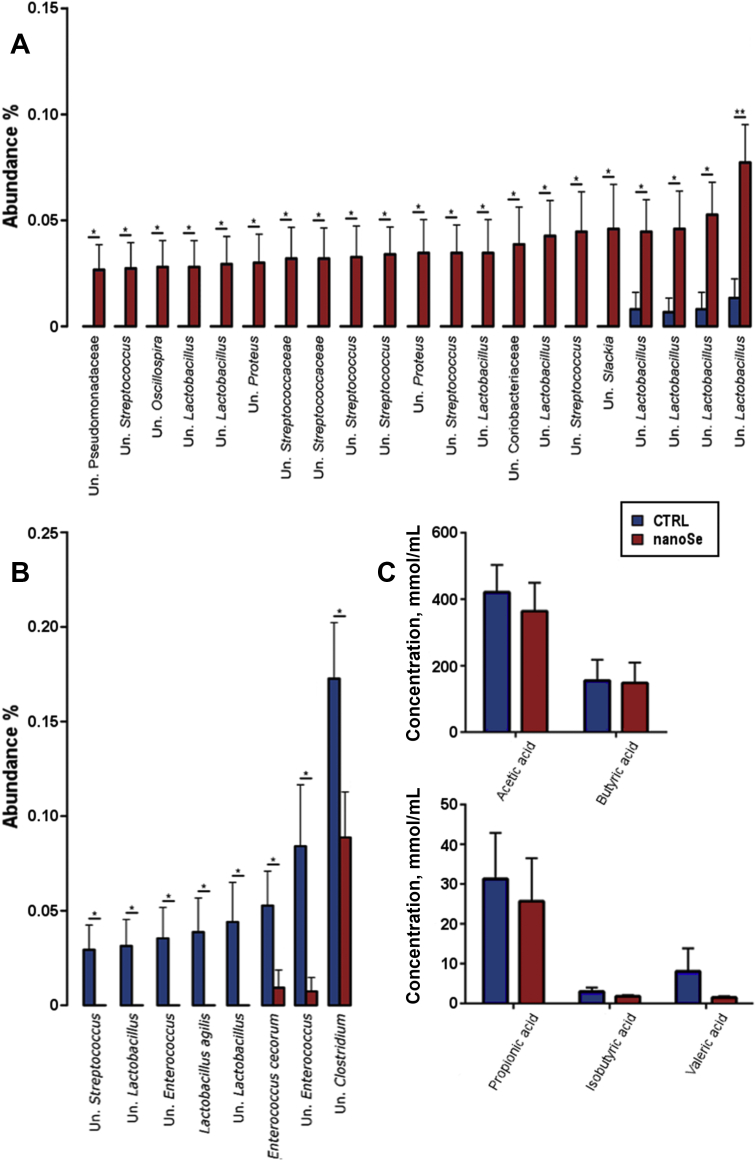
The enriched LYHBHI supported the growth of a diverse range of genera comprising of multiple previously uncultured species as shown in Appendix Table 3. NanoSe supplementation significantly (P < 0.05) increased 20 operational taxonomic units (OTU), as shown in Fig. 3A and reduced 8 OTU (P < 0.05), one of which was identified as 100% identical across the amplified region to Enterococcus cecorum, followed by 2 other Enterococcus OTU significantly reduced by nanoSe (Fig. 3B). Enterococcus OTU, including pathogenic E. cecorum, were exclusively reduced by nanoSe while genera Lactobacillus and Streptococcus were rearranged with some OTU significantly reduced and other significantly increased. Although nanoSe treatment was correlated with changes in abundance of some specific OTU an ANOSIM multivariate analysis of group similarities showed that the overall gut microbial composition was not affected by nanoSe (P = 0.991) or an additional 24 h of incubation (P = 0.55). Furthermore, alpha diversity indices (Shannon's index, richness and evenness) were also not affected by nanoSe or time of incubation (P > 0.05). The supplementation of nanoSe had no effect (using t-test) on SCFA production, as shown in Fig. 3C.
Fig. 3.
NanoSe influence on the gut microbiota of roosters at OTU level and the subsequent effect on SCFA production. (A) Increased and (B) decreased OTU from several unclassified genera and families. (C) SCFA production was not significantly affected by the addition of nanoSe in in vitro cultures. OUT = operational taxonomic units; SCFA = short-chain fatty acids; Un. = unclassified.
3.3. Interaction between gut community and short-chain fatty acid
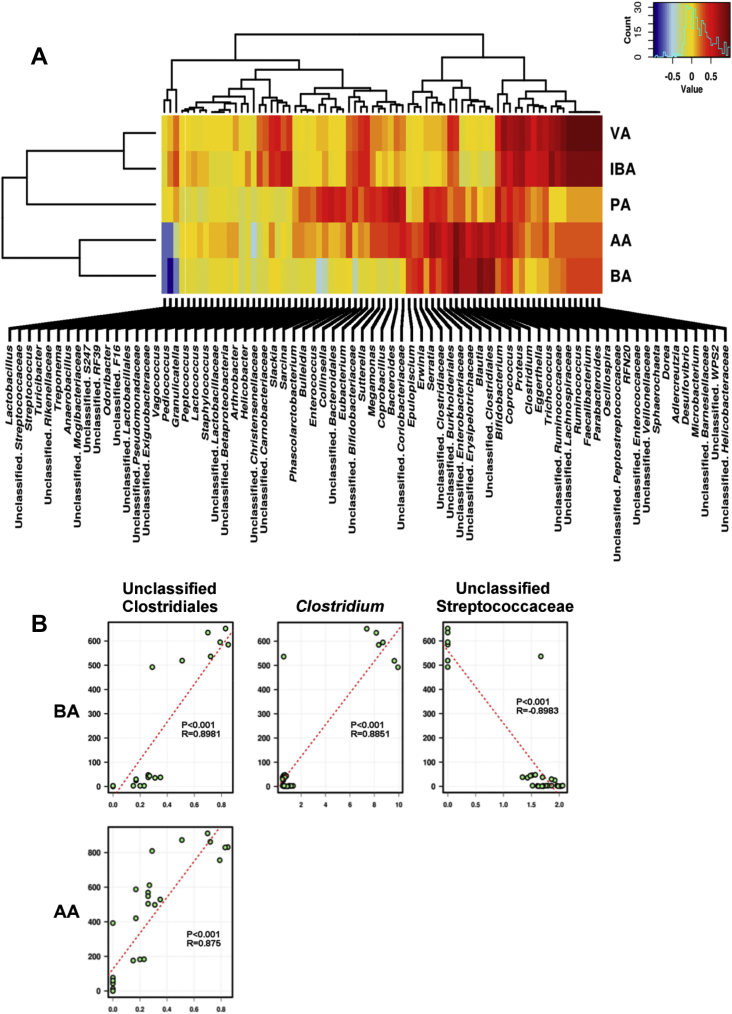
PERMANOVA showed that SCFA production was significantly related to the microbiota composition (P = 0.00067) and RDA demonstrated that the microbial composition was significantly related to the SCFA (P < 0.01). The 5 SCFA, acetic acid, butyric acid, isobutyric acid (IBA), propanoic acid and valeric acid correlated with the abundance of a number of taxa (Fig. 4A). Valeric acid and IBA strongly correlated (P < 0.001; R > 0.85) with the same genera, including Adlercreutzia, Desulfovibrio, Microbacterium, unclassified Barnesiellaceae, unclassified Helicobacteraceae and unclassified WPS2, (Fig. 4B). Butyric acid and acetic acid shared one genus, an unclassified Clostridiales, exhibiting a strong correlation (P < 0.001; R > 0.85). Butyric acid additionally had a positive correlation (P < 0.001; R = 0.86) with Clostridium and an inverse correlation (P < 0.001; R = −0.90) with an unclassified Streptococcaceae.
Fig. 4.
Short-chain fatty acids (SCFA) interaction effect with the microbial community. (A) overall interaction effect of SCFA with genera yielded by enriched LYHBHI media (Brain-heart infusion medium supplemented with yeast extract [5 g/L, Alfa Aesar], cellobiose [1 g/L, BD], hemin [5 mg/L, BD], cysteine [0.5 g/L, Alfa Aesar]) and resazurin (0.5 mg/L, Alfa Aesar)]. (B) Highest correlation (R > 0.85) of SCFA with generated bacteria. AA = acetic acid; BA = butyric acid; PA = propanoic acid; IBA = isobutyric acid; VA = valeric acid.
4. Discussion
The human and animal microbiota is continuously altered with different lifestyles and environmental changes, and has undergone major rearrangements since the introduction of industrialised, large-scale food production in the last few centuries (Flandroy et al., 2018). This change in eating habits and the subsequent changes in gut microbiota has led to the modern age being described as an age of “microbiota genocide” (Sonnenburg et al., 2016). The lifestyle and eating habits of hunter-gatherer societies are very different to that of modern western societies and the difference drive profound changes when comparing ancient and modern human microbiotas (Davenport et al., 2017, Kumar and Forster, 2017, Warinner et al., 2015). This effect spills over to livestock and birds with characteristics microbiota changes occurring because of altered husbandry and feeding practices. Industrial scale grown birds experience very different growth conditions compared to their ancestors; the eggs are hatched under highly clean conditions, removing the influence of parental microbiota passage to the next generation (Donaldson et al., 2017). This results in aberrant microbiotas and high microbiota variation from one batch of hatchlings to another (Stanley et al., 2013). Microbiota analyses of chicken caeca across various projects have displayed an enormous discrepancy between bacterial species present in industrial birds and those present in birds grown in traditional low density open housing ways such as that found with “village chickens” or “backyard chickens” as called in Australia. Here we used the caeca of backyard chickens to investigate the effects of Se nanoparticles (nanoSe) on gut microbiota. The gut microbiota of an industrially grown domestic chicken, Gallus gallus domesticus, is typically comprised of 4 main phyla; Firmicutes, Bacteroidetes, Proteobacteria and a low amount of Actinobacteria (Oakley et al., 2014, Waite and Taylor, 2014, Wei et al., 2013). The high number of unclassified genera, presented in this study, is possibly indicative of the influence of non-industrialised housing and other environmental conditions (Kers et al., 2018), such as access to pasture, live plant and insect food content, full free range, and exposure to wild birds and animals.
Culturable genera, Lactobacillus, Streptococcus, Clostridium and Enterococcus strongly dominated (>60%) the rooster's caecal community, while numerous uncultured genera remained in low abundance. The 80% to 90% sequence similarities render it impossible to infer function, pathogenicity or probiotic potential of these unidentified species to known cultured bacteria. The unknown and uncultured species often require metabolic feedback from other bacteria and can be cultured only in a complex community rather than as a single culture. The caecal microbiota communities were more diverse and different to the ones previously investigated with live birds treated with different concentrations of nanoSe (Gangadoo et al., 2018), and consequently the in vitro response of cultured caecal microbiota to nanoSe proved dissimilar to that seen in the microbiota of treated birds, including the lack of SCFA and Lactobacillus genus stimulation. It is not clear whether the different test systems or the different starting microbiotas have more pronounced influence in producing the different outcomes.
In contrast, the reduction of an emerging avian pathogen, E. cecorum, and 2 unknown enterococcus species, was observed with nanoSe at a concentration as low as 1 mg/kg. E. cecorum has been linked to enterococcal spondylitis and femoral head necrosis, resulting in symptoms such as hind limb weakness (Borst et al., 2017, Dolka et al., 2016) and lameness in poultry (McNamee and Smyth, 2000). Other symptoms observed include arthritis and spinal lesions (Dolka et al., 2017), with E. cecorum infection leading to a marked increase in flock mortality among all poultry types. Additionally, the ability to carry and spread antimicrobial resistance among other Enterococcus spp. has been observed from an analysis of retail meat samples (Jung et al., 2018). It can be deduced from this current study that nanoSe may exert targeted antimicrobial activity against pathogenic bacteria such as E. cecorum within the complex environment of caecal microbiota without causing significant alteration to the rest of the community. Further investigations should focus on the mechanisms by which the nanoparticles may inhibit the growth of various pathogens.
5. Conclusion
The data presented in this study suggests an immense untapped potential for microbiota manipulation in unconventionally grown birds and could reveal useful information for future attempts in standardising the microbiome of industrial poultry. The application of nanoparticles, with careful optimisation, could help uncover a range of unknown bacterial species and their role in the expression of beneficial microbial products. Nanoparticles have rapidly emerged in the food and agricultural industry, and it is of vital importance to understand their gut microbiome interaction, while modifying their properties to our best advantage.
Conflict of interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Acknowledgements
We wish to acknowledge and appreciate help from Jason Bell provided in all aspects of high performance computing. We also thank Corine Ting and Charmaine Elder for their assistance in molecular methods. This research was funded internally by Central Queensland University. Sheeana Gangadoo received the scholarship from the Poultry CRC established by the Australian Government.
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aninu.2019.06.004.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Arora S., Jain J., Rajwade J., Paknikar K. Cellular responses induced by silver nanoparticles: in vitro studies. Toxicol Lett. 2008;179:93–100. doi: 10.1016/j.toxlet.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Ashelford K.E., Chuzhanova N.A., Fry J.C., Jones A.J., Weightman A.J. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol. 2005;71(12):7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst L., Suyemoto M., Sarsour A., Harris M., Martin M., Strickland J., Oviedo E., Barnes H. Pathogenesis of enterococcal spondylitis caused by Enterococcus cecorum in broiler chickens. Vet Pathol. 2017;54:61–73. doi: 10.1177/0300985816658098. [DOI] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Xiang H., Zhu X., Zhang H., Wang D., Liu H., Wang J., Yin T., Liu L., Kong M. Free dietary choice and free-range rearing improve the product quality, gait score, and microbial richness of chickens. Animal. 2018;8:84. doi: 10.3390/ani8060084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Wang Q., Liu S., Sun R., Zhou Y., Li Y. Age-related variations in intestinal microflora of free-range and caged hens. Front Microbiol. 2017;8:1310. doi: 10.3389/fmicb.2017.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport E.R., Sanders J.G., Song S.J., Amato K.R., Clark A.G., Knight R. The human microbiome in evolution. BMC Biol. 2017;15:127. doi: 10.1186/s12915-017-0454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M.P., Labhasetwar V., Amidon G.L., Levy R.J. Gastrointestinal uptake of biodegradable microparticles: effect of particle size. Pharm Res. 1996;13:1838–1845. doi: 10.1023/a:1016085108889. [DOI] [PubMed] [Google Scholar]
- Dolka B., Chrobak-Chmiel D., Czopowicz M., Szeleszczuk P. Characterization of pathogenic Enterococcus cecorum from different poultry groups: broiler chickens, layers, turkeys, and waterfowl. PLoS One. 2017;12 doi: 10.1371/journal.pone.0185199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolka B., Chrobak-Chmiel D., Makrai L., Szeleszczuk P. Phenotypic and genotypic characterization of Enterococcus cecorum strains associated with infections in poultry. BMC Vet Res. 2016;12:129. doi: 10.1186/s12917-016-0761-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson E.E., Stanley D., Hughes R.J., Moore R.J. The time-course of broiler intestinal microbiota development after administration of cecal contents to incubating eggs. PeerJ. 2017;5:e3587. doi: 10.7717/peerj.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Espitia P.J.P., Soares NdFF., dos Reis Coimbra J.S., de Andrade N.J., Cruz R.S., Medeiros E.A.A. Zinc oxide nanoparticles: synthesis, antimicrobial activity and food packaging applications. Food Bioprocess Technol. 2012;5:1447–1464. [Google Scholar]
- Fadrosh D.W., Ma B., Gajer P., Sengamalay N., Ott S., Brotman R.M., Ravel J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2:6. doi: 10.1186/2049-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenn K., Strandwitz P., Stewart E.J., Dimise E., Rubin S., Gurubacharya S., Clardy J., Lewis K.J.M. Quinones are growth factors for the human gut microbiota. 2017;5:161. doi: 10.1186/s40168-017-0380-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flandroy L., Poutahidis T., Berg G., Clarke G., Dao M.-C., Decaestecker E., Furman E., Haahtela T., Massart S., Plovier H. The impact of human activities and lifestyles on the interlinked microbiota and health of humans and of ecosystems. Sci Total Environ. 2018;627:1018–1038. doi: 10.1016/j.scitotenv.2018.01.288. [DOI] [PubMed] [Google Scholar]
- Gangadoo S., Dinev I., Chapman J., Hughes R.J., Van T.T.H., Moore R.J., Stanley D. Selenium nanoparticles in poultry feed modify gut microbiota and increase abundance of Faecalibacterium prausnitzii. Appl Microbiol Biotechnol. 2018;102:1455–1466. doi: 10.1007/s00253-017-8688-4. [DOI] [PubMed] [Google Scholar]
- Gangadoo S., Stanley D., Hughes R.J., Moore R.J., Chapman J. Nanoparticles in feed: progress and prospects in poultry research. Trends Food Sci Technol. 2016;58:115–126. [Google Scholar]
- Gangadoo S., Stanley D., Hughes R.J., Moore R.J., Chapman J. The synthesis and characterisation of highly stable and reproducible selenium nanoparticles. Inorg Nano Metal Chem. 2017;47:1568–1576. [Google Scholar]
- Gangadoo S., Taylor-Robinson A.W., Chapman J. From replacement to regeneration: are bio-nanomaterials the emerging prospect for therapy of defective joints and bones. J Biotechnol Biomater. 2015;5:2. [Google Scholar]
- Goyal R., Macri L.K., Kaplan H.M., Kohn J. Nanoparticles and nanofibers for topical drug delivery. J Control Release. 2016;240:77–92. doi: 10.1016/j.jconrel.2015.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greulich C., Kittler S., Epple M., Muhr G., Köller M. Studies on the biocompatibility and the interaction of silver nanoparticles with human mesenchymal stem cells (hMSCs) Langenbeck's Arch Surg. 2009;394:495–502. doi: 10.1007/s00423-009-0472-1. [DOI] [PubMed] [Google Scholar]
- Gupta A.K., Curtis A.S. Surface modified superparamagnetic nanoparticles for drug delivery: interaction studies with human fibroblasts in culture. J Mater Sci Mater Med. 2004;15:493–496. doi: 10.1023/b:jmsm.0000021126.32934.20. [DOI] [PubMed] [Google Scholar]
- Hajipour M.J., Fromm K.M., Ashkarran A.A., de Aberasturi D.J., de Larramendi I.R., Rojo T., Serpooshan V., Parak W.J., Mahmoudi M. Antibacterial properties of nanoparticles. Trends Biotechnol. 2012;30:499–511. doi: 10.1016/j.tibtech.2012.06.004. [DOI] [PubMed] [Google Scholar]
- Han X.Y., Du W.L., Fan C.L., Xu Z.R. Changes in composition a metabolism of caecal microbiota in rats fed diets supplemented with copper-loaded chitosan nanoparticles. J Anim Physiol Anim Nutri (Berl) 2010;94:e138–e144. doi: 10.1111/j.1439-0396.2010.00995.x. [DOI] [PubMed] [Google Scholar]
- Jia Y.-P., Ma B.-Y., Wei X.-W., Qian Z.-Y. The in vitro and in vivo toxicity of gold nanoparticles. Chin Chem Lett. 2017;28:691–702. [Google Scholar]
- Jung A., Chen L.R., Suyemoto M.M., Barnes H.J., Borst L.B. A review of Enterococcus cecorum infection in poultry. Avian Dis. 2018;62:261–271. doi: 10.1637/11825-030618-Review.1. [DOI] [PubMed] [Google Scholar]
- Karavolos M., Holban A. Nanosized drug delivery systems in gastrointestinal targeting: interactions with microbiota. Pharmaceuticals (Basel) 2016;9 doi: 10.3390/ph9040062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasaikina M.V., Kravtsova M.A., Lee B.C., Seravalli J., Peterson D.A., Walter J., Legge R., Benson A.K., Hatfield D.L., Gladyshev V.N. Dietary selenium affects host selenoproteome expression by influencing the gut microbiota. FASEB J. 2011;25:2492–2499. doi: 10.1096/fj.11-181990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul G., Amiji M. Tumor-targeted gene delivery using poly (ethylene glycol)-modified gelatin nanoparticles: in vitro and in vivo studies. Pharm Res. 2005;22:951–961. doi: 10.1007/s11095-005-4590-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kers J.G., Velkers F.C., Fischer E.A., Hermes G.D., Stegeman J.A., Smidt H. Host and environmental factors affecting the intestinal microbiota in chickens. Front Microbiol. 2018;9:235. doi: 10.3389/fmicb.2018.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N., Forster S.C. Genome watch: microbiota shuns the modern world. Nat Rev Microbiol. 2017;15 doi: 10.1038/nrmicro.2017.136. [DOI] [PubMed] [Google Scholar]
- Labunskyy V.M., Hatfield D.L., Gladyshev V.N. Selenoproteins: molecular pathways and physiological roles. Physiol Rev. 2014;94:739–777. doi: 10.1152/physrev.00039.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Ouay B., Stellacci F. Antibacterial activity of silver nanoparticles: a surface science insight. Nano Today. 2015;10:339–354. [Google Scholar]
- Lu J., Liong M., Li Z., Zink J.I., Tamanoi F. Biocompatibility, biodistribution, and drug-delivery efficiency of mesoporous silica nanoparticles for cancer therapy in animals. Small. 2010;6:1794–1805. doi: 10.1002/smll.201000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatro A., Singh D.K. Biodegradable nanoparticles are excellent vehicle for site directed in-vivo delivery of drugs and vaccines. J Nanobiotechnol. 2011;9:55. doi: 10.1186/1477-3155-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamee P.T., Smyth J.A. Bacterial chondronecrosis with osteomyelitis ('femoral head necrosis') of broiler chickens: a review. Avian Pathol. 2000;29:477–495. doi: 10.1080/030794500750047243. [DOI] [PubMed] [Google Scholar]
- Mohanraj V., Chen Y. Nanoparticles – a review. Trop J Pharm Res. 2006;5:561–573. [Google Scholar]
- Mueller N.C., Nowack B. Exposure modeling of engineered nanoparticles in the environment. Environ Sci Technol. 2008;42:4447–4453. doi: 10.1021/es7029637. [DOI] [PubMed] [Google Scholar]
- Naahidi S., Jafari M., Edalat F., Raymond K., Khademhosseini A., Chen P. Biocompatibility of engineered nanoparticles for drug delivery. J Control Release. 2013;166:182–194. doi: 10.1016/j.jconrel.2012.12.013. [DOI] [PubMed] [Google Scholar]
- Oakley B.B., Lillehoj H.S., Kogut M.H., Kim W.K., Maurer J.J., Pedroso A., Lee M.D., Collett S.R., Johnson T.J., Cox N.A. The chicken gastrointestinal microbiome. FEMS Microbiol Lett. 2014;360:100–112. doi: 10.1111/1574-6968.12608. [DOI] [PubMed] [Google Scholar]
- Panyam J., Labhasetwar V. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Adv Drug Deliv Rev. 2003;55:329–347. doi: 10.1016/s0169-409x(02)00228-4. [DOI] [PubMed] [Google Scholar]
- Regan F., Chapman J., Sullivan T. Royal Society of Chemistry; 2012. Nanoparticles in anti-microbial materials: use and characterisation. [Google Scholar]
- Sonnenburg E.D., Smits S.A., Tikhonov M., Higginbottom S.K., Wingreen N.S., Sonnenburg J.L. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529:212. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley D., Geier M.S., Hughes R.J., Denman S.E., Moore R.J. Highly variable microbiota development in the chicken gastrointestinal tract. PLoS One. 2013;8 doi: 10.1371/journal.pone.0084290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teodoro J.S., Simões A.M., Duarte F.V., Rolo A.P., Murdoch R.C., Hussain S.M., Palmeira C.M. Assessment of the toxicity of silver nanoparticles in vitro: a mitochondrial perspective. Toxicol In Vitro. 2011;25:664–670. doi: 10.1016/j.tiv.2011.01.004. [DOI] [PubMed] [Google Scholar]
- Thill A., Zeyons O., Spalla O., Chauvat F., Rose J., Auffan M., Flank A.M. Cytotoxicity of CeO2 nanoparticles for Escherichia coli. Physico-chemical insight of the cytotoxicity mechanism. Environ Sci Technol. 2006;40:6151–6156. doi: 10.1021/es060999b. [DOI] [PubMed] [Google Scholar]
- Van Immerseel F., Eeckhaut V., Moore R.J., Choct M., Ducatelle R. Beneficial microbial signals from alternative feed ingredients: a way to improve sustainability of broiler production? Microb Biotechnol. 2017;10:1008–1011. doi: 10.1111/1751-7915.12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite D.W., Taylor M.W. Characterizing the avian gut microbiota: membership, driving influences, and potential function. Front Microbiol. 2014;5:223. doi: 10.3389/fmicb.2014.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warinner C., Speller C., Collins M.J., Lewis C.M., Jr. Ancient human microbiomes. J Hum Evol. 2015;79:125–136. doi: 10.1016/j.jhevol.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S., Morrison M., Yu Z. Bacterial census of poultry intestinal microbiome. Poult Sci. 2013;92:671–683. doi: 10.3382/ps.2012-02822. [DOI] [PubMed] [Google Scholar]
- Yausheva Е., Miroshnikov S., Sizova Е. Intestinal microbiome of broiler chickens after use of nanoparticles and metal salts. Environ Sci Pollut Res. 2018:1–12. doi: 10.1007/s11356-018-1991-5. [DOI] [PubMed] [Google Scholar]
- Yu Z., Morrison M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques. 2004;36:808–812. doi: 10.2144/04365ST04. [DOI] [PubMed] [Google Scholar]
- Zakrzewski M., Proietti C., Ellis J.J., Hasan S., Brion M.-J., Berger B., Krause L. Calypso: a user-friendly web-server for mining and visualizing microbiome–environment interactions. Bioinformatics. 2016;33:782–783. doi: 10.1093/bioinformatics/btw725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Qiu X., Zhang H., Yang X., Hong N., Yang Y., Chen H., Yu C.J.P.O. Faecalibacterium prausnitzii inhibits interleukin-17 to ameliorate colorectal colitis in rats. 2014;9 doi: 10.1371/journal.pone.0109146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.