Abstract
Chromatin remodeling proteins of the chromodomain DNA-binding protein family, CHD7 and CHD8, mediate early neurodevelopmental events including neural migration and differentiation. As such, mutations in either protein can lead to neurodevelopmental disorders. How chromatin remodeling proteins influence the activity of mature synapses, however, is relatively unexplored. A critical feature of mature neurons is well-regulated endocytosis, which is vital for synaptic function to recycle membrane and synaptic proteins enabling the continued release of synaptic vesicles. Here we show that Kismet, the Drosophila homolog of CHD7 and CHD8, regulates endocytosis. Kismet positively influenced transcript levels and bound to dap160 and endophilin B transcription start sites and promoters in whole nervous systems and influenced the synaptic localization of Dynamin/Shibire. In addition, kismet mutants exhibit reduced VGLUT, a synaptic vesicle marker, at stimulated but not resting synapses and reduced levels of synaptic Rab11. Endocytosis is restored at kismet mutant synapses by pharmacologically inhibiting the function of histone deacetyltransferases (HDACs). These data suggest that HDAC activity may oppose Kismet to promote synaptic vesicle endocytosis. A deeper understanding of how CHD proteins regulate the function of mature neurons will help better understand neurodevelopmental disorders.
Subject terms: Genetics of the nervous system, Cellular neuroscience, Development of the nervous system
Introduction
Endocytosis and endosomal trafficking are required for the import of required nutrients and macromolecules as well as for cell growth, survival, and both autocrine and paracrine signaling1. Additionally, the strict balance between endocytosis and its complement, exocytosis, allows for precise control of the cell’s interaction with its environment2. An imbalance between the two can quickly lead to cell death. To this effect, mutations in major components of the endocytic pathway are rarely found in human disease, underscoring their importance3. However, mutations in associated and accessory proteins can cause subtle changes in cellular homeostasis that compound and lead to whole organismal dysfunction.
Endocytosis proceeds via two morphologically distinct pathways characterized by the dependence of the scaffolding protein clathrin2–5. Clathrin-mediated endocytosis is the better characterized endocytic pathway, which begins with the recognition of cargo proteins by specific adaptor protein complexes, which recruit clathrin triskelia6 on the inner leaflet of plasma membrane. The curvature induced by the clathrin coats forces the membrane to bend. Scission proteins, like the GTPase Dynamin, mediate membrane fusion and a vesicle is liberated intracellularly3,4,6. Once internalized, the vesicle sheds its clathrin coat and transitions to an early endosome, which can then be trafficked to distinct cellular locations based on the fate of the cargo. Clathrin-independent endocytosis also produces early endosomes but without reliance on clathrin, and sometimes dynamin, for membrane curvature and scission and occur on different time scales2,3. Early endosomes can rapidly recycle cargo proteins back to the plasma membrane where they were internalized, or be trafficked to become recycling endosomes which are slower to return internalized cargo to multiple locations on the cell surface1.
Efficient endocytosis is of particular importance in neurons due to the high volume of neurotransmitter vesicle turnover necessary to illicit postsynaptic responses5. Therefore, neurons have developed a sophisticated network of recycling endosomes to efficiently recapture and reuse membrane for neurotransmitter packaging. Partitioning of endosomes into the recycling or late endosome maturation pathway requires the functions of distinct Rab-family GTPases at each step, which can also be used to track vesicle progress7. For example, Rab5 predominantly functions at early endocytic vesicles and is required for their fusion with early endosomes. As early endosomes mature to late endosomes, the functions of Rab7 and Rab9 are required for cargo sorting into the correct cellular compartments8. Rab11, on the other hand, is mainly involved in targeting recycling endosomes, often containing receptor cargo, to the periactive zones, or sites surrounding areas of neurotransmitter release at synapses7,9.
Disruptions in any components of the endocytic pathway, such as Rab11, Dynamin, Nervous Wreck, or Synaptojanin, tend to produce similar phenotypes at the Drosophila neuromuscular junction (NMJ)9,10. Overgrowth and an excess of satellite synaptic connections, or boutons, have been reported in mutants of most endocytic genes at the Drosophila NMJ5,9–11. These supernumerary connections are thought to be due partially to the defect in membrane internalization of endocytic mutants and partially to defective signaling at the synapse that drives upstream signals for growth and development9,10. The Drosophila NMJ has been the premier choice of model systems to characterize these defects due to its highly stereotypical development and capacity for synaptic plasticity post-differentiation12,13. Additionally, as a glutamatergic system, the Drosophila NMJ is excellent for elucidating molecular and neurotransmission properties of glutamate signaling in vivo14.
The Drosophila protein Kismet (Kis) is an epigenetic chromatin reader previously shown to affect synaptic morphology and neurotransmission at the NMJ15,16. Kis is homologous to the mammalian chromodomain DNA-binding protein 7 (CHD7), which is implicated in the congenital neurodevelopmental disorder, CHARGE syndrome17. Kis functions in the nucleus where its binding to DNA colocalizes with sites of active transcription and is implicated in transcriptional activation and elongation18–20. Kis is of particular importance in nervous system development and maintenance as its depletion in neurons causes axon pruning and migration defects and behavioral abnormalities such as decreased immediate recall memory21. In the CNS specifically, Kis acts to maintain active histone modifications at the ecdysone receptor (EcR) locus to promote developmental axon pruning during metamorphosis22. At the NMJ, decreased Kis leads to overgrowth of the presynaptic motor neuron, which is commonly observed in endocytic mutants, along with impaired neurotransmission and reduced postsynaptic glutamate receptors15,16. Whether Kis has different functions based on cell-type specificity remains to be answered.
Here we show that Kis positively regulates endocytosis possibly by altering gene expression thereby affecting the recycling of endocytic vesicles. Kis mutants exhibit a significant increase in satellite boutons at the NMJ and defective neurotransmission indicative of an endocytic defect. In particular, Kis preferentially affects the recycling pool of vesicles as VGLUT levels are decreased after stimulation but not at rest. In agreement with this, Rab11, a marker of recycling vesicles, is significantly decreased at kis mutant boutons. Using ChIP-qPCR, we found that Kis binding is enriched at the endocytic genes dap160 and endoB. These two endocytic proteins are significantly decreased at the NMJ of kis mutants. Additionally, while mRNA levels of dynamin were slightly reduced, its protein levels at kis mutant boutons were increased and mislocalized. We therefore propose a novel function for the epigenetic reader Kis in the regulation of endocytosis related genes.
Results
Kismet is important for presynaptic endocytosis
We previously showed that chromatin reader, Kis, promotes neurotransmission and the apposition between presynaptic active zones and postsynaptic glutamate receptors at the Drosophila NMJ. The structural defect of kis mutant synapses was associated with a significant reduction in evoked excitatory junctional currents (eEJCs)15. Collectively, these data suggest that Kis’ transcriptional activity may influence the synaptic vesicle cycle. Therefore, we were interested in examining endocytosis in kis mutants. Homozygous kis null mutants are embryonically lethal so we chose to use the adult viable hypomorph, kisk13416 23, and the heteroallelic kisLM27/kisk13416, constructed with the null allele, kisLM27 24.
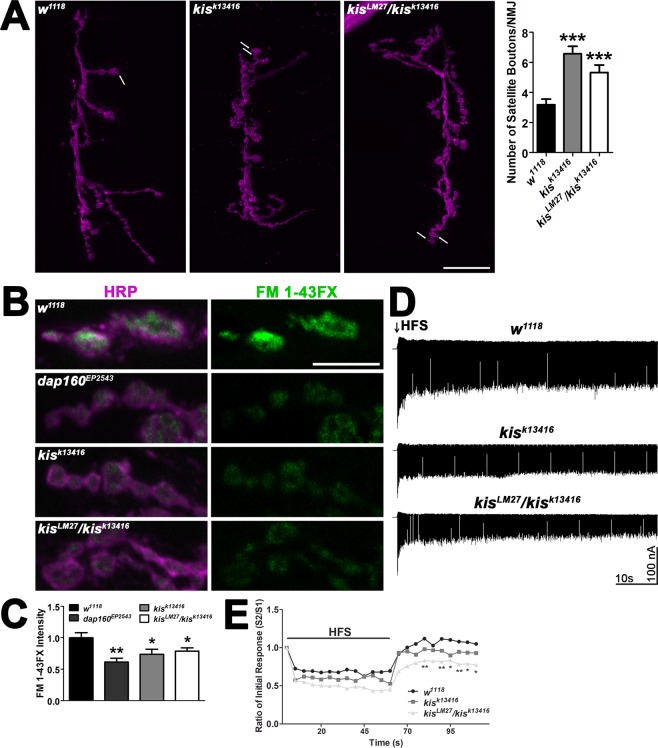
Satellite boutons are small, ectopic functional boutons that are significantly increased in several endocytic mutants including endophilin, dynamin, AP180, and synaptojanin mutants10. Kis mutants possess 2–3x the number of satellite boutons as controls (Fig. 1A). To further explore the synaptic vesicle cycle in kis mutant synapses, we used FM 1–43FX to label newly endocytosed synaptic vesicles after 1 min stimulation with 90 mM KCl25. We compared kis mutants to dap160EP2543, which includes a P-element insertion in the 5′ end of the dap160 gene. Dap160 is the Drosophila ortholog of Intersectin and interacts with synaptic vesicles26, endocytic proteins, and is required for endocytosis27. Both kis and dap160EP2543 mutants showed a significant decrease in FM 1–43FX fluorescence indicative of defective endocytosis compared to w1118 controls (Fig. 1B,C).
Figure 1.
Kismet promotes endocytosis of presynaptic vesicles. (A) Confocal micrographs of the 6/7 NMJ as indicated by immunolabeling with HRP (magenta). Arrows denote satellite boutons as quantified in the right histogram. Scale bar = 20 μm. (B) High resolution confocal micrographs showing the presynaptic motor neuron (HRP, magenta) and internalization of the lipophilic dye FM 1–43FX (green) after 90 s stimulation with 90 mM KCl and 2 mM Ca2+ in controls (w1118), the endocytosis mutant dap160EP2543, and kis mutants. Scale bar = 5 µm. (C) Quantification of FM 1–43FX fluorescence intensity relative to controls indicates that kis mutants exhibit a significant reduction in endocytosis as indicated by internalization of FM1–43FX compared with controls. (D) Representative recordings of eEJCs resulting from a 20 Hz, 1 min HFS train in genotypes as indicated. (E) eEJC amplitudes in controls and kis mutants during HFS followed by a recovery period of 0.2 Hz stimulation. kis mutants exhibit a significant reduction in eEJCs evoked after HFS suggesting that vesicle recycling is impaired in kis mutants.
We next recorded eEJCs in 1.0 mM Ca2+ from animals during and after high frequency stimulation (HFS). This stimulation fully expels the readily releasable pool of vesicles to measure the recycling of newly formed synaptic vesicles. eEJC amplitudes were assessed first at 0.2 Hz to establish basal eEJC amplitudes followed by 60 sec of 20 Hz stimulation and 50 sec of 0.2 Hz stimulation28. Using this paradigm, controls show a rapid 27–37% reduction in eEJC amplitudes during HFS compared to pre-stimulation amplitudes (Fig. 1D,E). During the post-stimulation recovery period, when the releasable pool of vesicles is replenished, controls show a 3–29% increase in eEJC amplitudes. Both kis mutants display a reduction in eEJC amplitudes relative to pre-stimulation amplitudes over time during HFS similar to that of controls but the kisLM27/kisk13416 mutant failed to exhibit the potentiating response in eEJC amplitudes during recovery (Fig. 1D,E). Instead, eEJC amplitudes were reduced by 17–31% compared with pre-stimulation amplitudes suggesting synaptic vesicle recycling is deficient in kisLM27/kisk13416 mutants.
Both endo- and exocytosis require local increases in intracellular Ca2+, 29. In order to rule out impaired calcium sensing, which could account for the impaired exocytosis and endocytosis observed in kis mutants, paired pulse recordings were performed (Supplemental Fig. 1). There were no significant differences in paired pulse ratios at any interstimulus interval examined including 100 Hz/10 ms, which is when residual calcium left in the nerve terminal from earlier stimulations provokes a larger response (Supplemental Fig. 1B). This indicates the endocytic phenotype in kis mutants is not likely due to a defect in Ca2+ sensing. Collectively, these data suggest kis mutants exhibit impaired endocytosis at the NMJ and this may be related to deficient recycling of synaptic vesicles.
Kismet differentially affects the recycling pool of synaptic vesicles
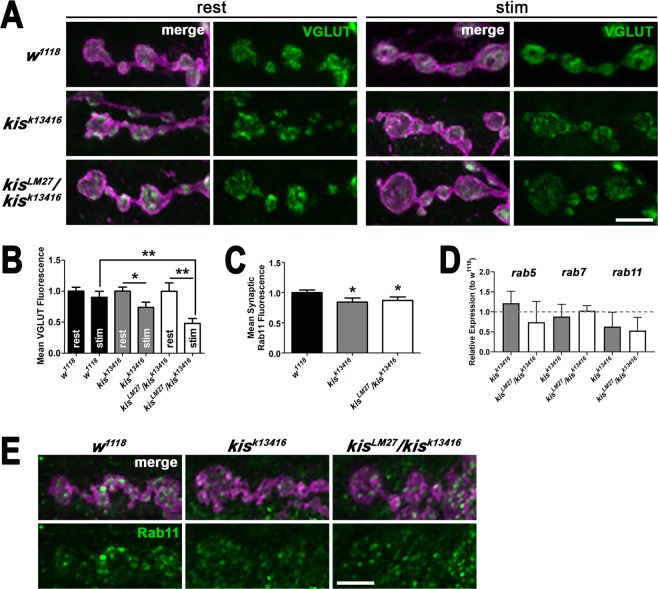
Next, we sought to further examine synaptic vesicle pool dynamics in kis mutants. The Drosophila NMJ synapse contains at least two vesicle pools including the readily releasable pool, which is part of the recycling pool, and a reserve pool30,31. Vesicular glutamate transporters (VGLUTs) are localized to vesicular membranes and responsible for transporting glutamate into vesicles for release32,33. To estimate the number of vesicles at the synapse, we quantified the levels of VGLUT. There were no significant differences in synaptic VGLUT immunofluorescence at rest in kis mutants compared with controls (Fig. 2A,B) suggesting the total number of synaptic vesicles was not affected by mutations in kis. The distribution of VGLUT in kis mutants, however, appeared punctate indicating that Kis may influence processes important for VLUT localization. Next, we examined VGLUT levels after synaptic activity, which was induced by application of 60 mM KCl in HL-3 containing 1.0 mM Ca2+ for 10 min34 to examine vesicle recycling. Stimulation resulted in significant reductions in VGLUT levels in both kis mutants compared with their respective VGLUT levels at rest (Fig. 2B) suggesting that the replenishment of synaptic vesicles is impaired in kis mutants during synaptic activity.
Figure 2.
Kismet affects the readily releasable and recycling pool of vesicles. (A) Control (w1118) and kis mutant NMJs were immunolabeled for the vesicular glutamate transporter (VGLUT) at rest (Ca2+-free HL-3 with 50 mM EDTA) or after stimulation (60 mM KCl and 1.5 mM Ca2+ for 10 min). Confocal images of Drosophila 6/7 NMJ terminal boutons labeled with HRP to delineate neurons (magenta) and VGLUT (green). Scale bar = 5 µm. (B) Quantification of VGLUT fluorescence intensity normalized to controls at rest indicates that VGLUT levels are significantly reduced in kis mutants but not controls after stimulation. (C) Quantification of Rab11 fluorescence levels shows that there is a slight but significant reduction in synaptic Rab11 in kis mutants compared with controls. (D) Histogram of rab transcript levels in kis mutants normalized to controls as determined by RT-qPCR. (E) High magnification confocal micrographs of Drosophila 6/7 NMJ terminal boutons labeled with HRP to identify neurons (magenta) and Rab11 (green). Scale bar = 5 µm.
To further support the hypothesis that Kis is important for the recycling of synaptic vesicles, we examined Rab11 levels in kis mutants. Rab11 is found on synaptic vesicles35 and recycling endosomes7 and is involved in regenerating synaptic vesicles from endosomes. The levels of synaptic Rab11 were slightly but significantly decreased in kis mutants compared to w1118 controls (Fig. 2C) suggesting kis mutants may possess fewer vesicles in the recycling pool. kis mutants also exhibited reduced levels of rab11 transcripts compared to controls (Fig. 2E). Next, we used Cyclosporin A treatment to mobilize the reserve pool of vesicles36 and assess if the size of the reserve pool of synaptic vesicles is diminished in kis mutants. There were no significant differences in endocytosis as indicated by FM 1–43FX fluorescence in kis mutants compared to w1118 controls after the reserve pool was mobilized (Supplemental Fig. 2). Therefore, disruption of the recycling pool but not the reserve pool of vesicles may be responsible for the endocytosis defect observed in kis mutants.
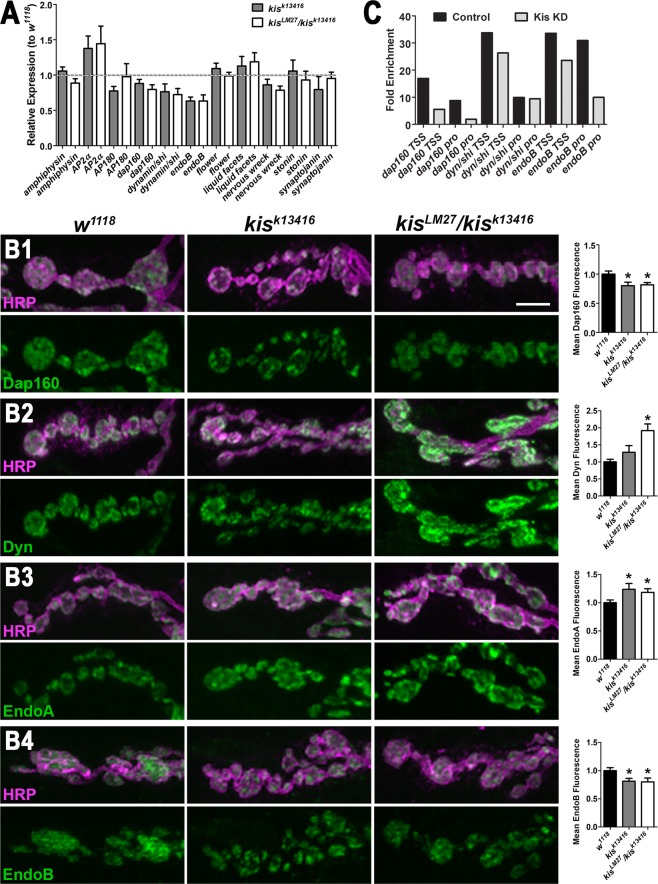
Kismet regulates transcript and synaptic levels of several endocytic proteins including Dap160, endophilin
Kis is a chromodomain helicase binding protein that influences transcription by recruiting the histone methyltransferases, ASH1 and trithorax, to active genes in salivary glands37 and trithorax-related complex in midgut intenstinal stem cells20. To determine if Kis may regulate transcription of genes important for endocytosis, we utilized RT-qPCR and ChIP-qPCR to identify potential Kis targets. We examined the levels of several transcripts that encode protein products important for endocytosis in the Drosophila third instar larval central nervous system (CNS). There were reductions in dap160, dynamin/shibire (dyn), and endophilin B (endoB) transcripts in kis mutant nervous systems relative to w1118 controls suggesting Kis functions directly or indirectly in the transcriptional regulation of these genes. To determine if the decrease in transcript levels might contribute to perturbed synaptic function, we assessed their corresponding synaptic protein levels (Fig. 3B1–4). Dap160 and EndoB levels were significantly decreased in both kis mutants compared with controls (Fig. 3B1,B2). Synaptic levels of Dyn, however, were significantly increased in kis mutants compared to w1118 controls (Fig. 3B2).
Figure 3.
Kismet mutants exhibit altered levels of transcripts and proteins involved in endocytosis. (A) Several transcripts important for clathrin-mediated endocytosis were examined in the CNS of controls and kis mutants via qRT-PCR. Transcript levels are expressed relative to w1118 controls. (B) High magnification confocal images of A3 or A4 NMJ terminal boutons labeled with HRP to denote neurons (magenta) and Dap160 (B1, green), Dynamin/Shibire (B2, Dyn, green), Endophilin A (B3, EndoA, green), or Endophilin B (B4, EndoB, green). Right histograms show quantification of fluorescence intensities relative to controls (w1118). Scale bar = 5 µm. (C) Promoter and transcription start site (TSS) occupancy was assessed for genes whose transcript and protein levels were consistently altered in kis mutants. Kismet likely binds to endoB promoters (pro) and the TSS and pro of dap160.
Dap160 helps to organize components of the endocytic machinery, directly interacts with Eps1538 and Nervous Wreck9, and promotes the localization of Endophilin A (EndoA)27,39. Therefore, we examined the levels of these proteins in kis mutants given the loss of Dap160 observed at kis mutant synapses. Synaptic levels of EndoA were increased in kis mutants compared to controls (Fig. 3B3). There were no significant differences, however, in the levels of Eps15 and Nervous Wreck (Supplemental Fig. 3). These data suggest that dap160, dyn, and endoB may be transcriptional targets of Kis. To further examine this possibility, we used ChIP-qPCR to assess Kis binding near the transcription start sites (TSS) and distal promoters (pro) of the dap160, dyn, and endoB genes because Kis is associated with both promoters and RNA polymerase II19,20. Kis binding was enriched at the dap160 TSS and pro and the endoB pro but not the dyn TSS or pro in the CNS of controls compared with animals in which Kis was knocked down in the CNS (Fig. 3C). Taken together, these data indicate that Kis regulates the levels of important endocytic proteins including Dap160 and EndoB possibly by transcriptional regulation.
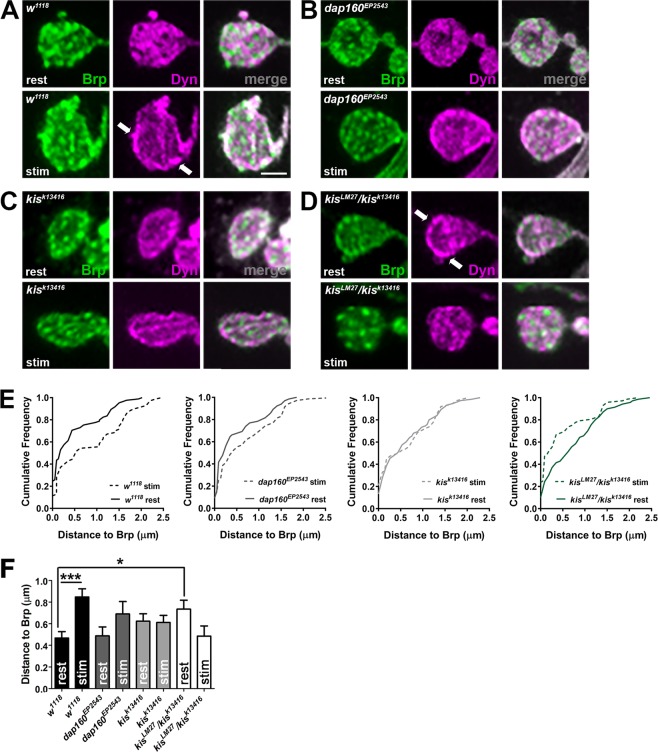
Dynamin is mislocalized in kis mutants
The localization of several proteins changes after synaptic stimulation to facilitate endocytosis. Dyn, a GTPase whose hydrolysis activity is required for membrane fission during endocytosis, clusters at active zones at resting synapses but is concentrated at periactive zones after intense synaptic activity26. To determine if Kis influences the localization of Dyn, we examined its distance from the active zone protein, Bruchpilot (Brp), in synapses at rest or after synaptic activity. Synaptic activity was induced by application of 60 mM KCl in HL-3 containing 1.0 mM Ca2+ for 10 min34. Dyn is localized within and near synaptic active zones as indicated by Brp at rest in controls (Fig. 4A). After stimulation, however, Dyn was redistributed to the periactive zone as indicated by a significant increase in the distance between Dyn and Brp (Fig. 4E,F). The redistribution of Dyn after synaptic stimulation occurred to a lesser extent in dap160EP2543 mutants but was lost in kisk13416 hypomorphs (Fig. 4B,C,E,F). This redistribution was not only lost in kisLM27/kisk13416 mutants but it was almost reversed as Dyn was localized further from active zones at rest and then was redistributed closer to active zones after synaptic stimulation (Fig. 4D,E,F). These data indicate that Kis promotes synaptic organization by influencing the localization of Dyn before and after stimulation.
Figure 4.
Kismet regulates the localization of Dynamin. (A–D) The localization of Dynamin/Shibire (Dyn) was assessed relative to the active zone protein, Bruchpilot (Brp), at rest (Ca2+-free HL-3 with 50 mM EDTA) or after stimulation (60 mM KCl and 1.5 mM Ca2+ for 10 min). Confocal images of z-projected A3 or A4 terminal NMJ boutons immunolabeled with Brp (green) and Dyn (magenta) in controls (w1118, A), dap160 mutants (B), or kis mutants (C,D). Dyn is redistributed in controls from active zones at rest to periactive zones during synaptic activity (see arrow in A). Dyn is mislocalized to periactive zones in kisLM27/kisk13416 mutants at rest (see arrow in D) and migrates toward active zones during synaptic activity. Scale bar = 2 µm. (E) Cumulative frequency histograms depicting the distance Dynamin was relative to Brp at rest and stimulated conditions in the indicated genotypes. (F) Quantification of the distance between Brp and Dyn in synaptic boutons indicating Dyn is mislocalized in kis mutants at rest.
We previously found that postsynaptic glutamate receptors are mislocalized relative to Brp at kis mutant synapses15. Thus, it is possible that, rather than Dyn and postsynaptic glutamate receptors, Brp is mislocalized at kis mutant synapses. Therefore, we examined the distance and localization of Brp relative to Synapsin (Syn) and Synaptotagmin I (SytI), which associate with the reserve and readily releasable pool of vesicles, respectively. Although Syn physically interacts with Dap16026 and its mammalian ortholog, Intersectin40, the localization of Syn in dap160 mutants at rest is unaltered26. There were no significant differences in the localization of Brp relative to Syn or SytI (Supplemental Fig. 4). These data indicate that Kis-mediated transcription influences the localization of specific synaptic proteins including Dyn rather than grossly perturbing the structure of the synapse.
HDAC inhibitors suppress the endocytic phenotype in kis mutants
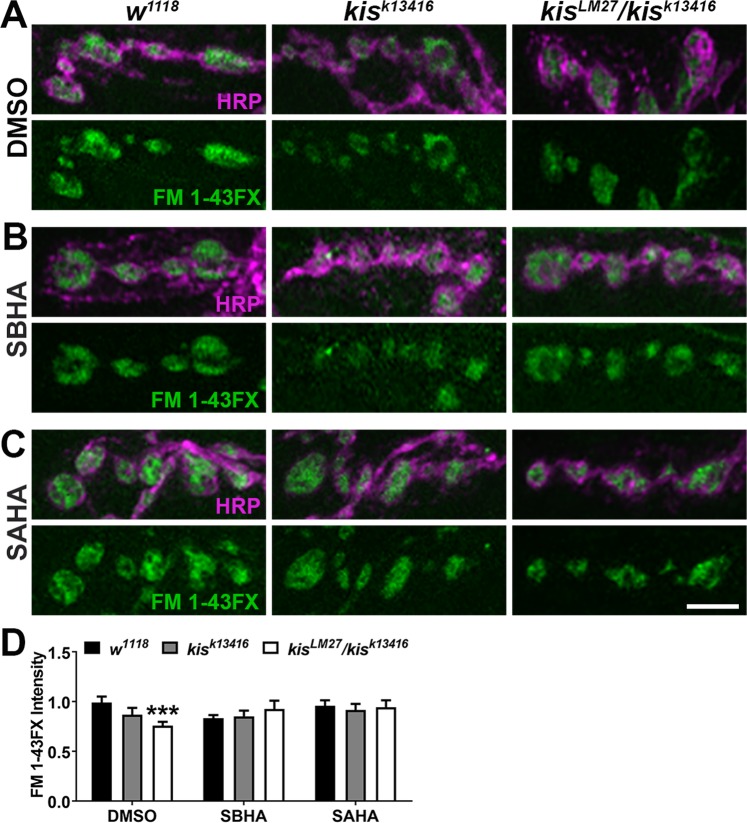
We have previously shown that histone deacetylases (HDACs) and Kis act in opposition to influence synaptic function, synaptic morphology, and motor function at third instar NMJs. The neurotransmission and synaptic morphology defects associated with kis mutants can be suppressed by the HDAC inhibitors, suberanilohydroxamic acid (SAHA) and suberoyl bis-hydroxamic acid (SBHA)16. To assess whether endocytosis is similarly regulated by HDAC and Kis activity, we used the HDAC inhibitors SAHA and SBHA to determine if these could suppress the endocytic deficiency observed in kis mutants. Endocytosis was measured after 1 min synaptic stimulation with 90 mM KCl25 in controls and kis mutants raised on DMSO or HDAC inhibitors (Fig. 5A–C). kisLM27/kisk13416 mutants raised on DMSO exhibited a significant reduction in FM 1–43FX intensity after synaptic stimulation compared with controls (Fig. 5A,C). The impaired endocytosis observed in kisLM27/kisk13416 mutants raised on DMSO was suppressed by raising the animals on either SAHA or SBHA (Fig. 5B–D). Collectively, our data indicate Kis’ regulation of endocytic transcripts, possibly antagonized by HDACs, is important for target gene expression and efficient endocytosis.
Figure 5.
The histone deacetylase inhibitors, SAHA and SBHA, suppress the deficit in endocytosis observed in kismet mutants. (A–C) High resolution confocal micrographs showing the presynaptic motor neuron (HRP, magenta) and internalization of the lipophilic dye FM 1–43FX (green) after 1 min stimulation with 90 mM KCl and 2 mM Ca2+ in controls (w1118) and kis mutants. Eggs were laid and larvae were allowed to develop on instant food containing DMSO (A), SBHA (B), or SAHA (C). Scale bar = 5 µm. (D) Quantification of FM 1–43FX fluorescence intensity relative to DMSO controls indicates that endocytosis is restored in kisLM27/kisk13416 mutants raised on either SBHA or SAHA.
Discussion
Kis is a chromatin remodeling protein homologous to CHD subfamily III proteins41 including CHD7 and CHD8. These CHD proteins are important for neurogenesis42,43 and neurodevelopment44. Their potential roles at mature synapses, however, are relatively unexplored. Given the link between chromatin remodeling proteins and neurodevelopmental disorders45, a better understanding of how these enzymes regulate synaptic function will help to gain insight into the pathology of these disorders.
We previously demonstrated that Kis positively regulated evoked and spontaneous neurotransmission and glutamate receptor localization relative to presynaptic active zones15. Here, we show that Kis is important for presynaptic endocytosis. Kis mutants exhibit significant reductions in endocytosis as evidenced by diminished internalization of FM 1–43FX (Fig. 1B,C). kKis mutants also show an increase in NMJ satellite boutons (Fig. 1A), which phenocopies endocytic mutants including endophilin, dynamin, AP180, and synaptojanin mutants10. Further, VGLUT levels (Fig. 2) were reduced after but not before synaptic stimulation suggesting that the number of synaptic vesicles is reduced in kis mutants as a result of synaptic activity. Although Kis may facilitate membrane trafficking events that promote endocytosis as evidenced by the reduction in synaptic Rab11 in kis mutants (Fig. 2C,D), Kis promotes expression of several endocytic transcripts and binds to the promoter and/or transcription start site of both dap160 and endoB (Fig. 3A,C). Based on these data, we hypothesize that Kis, similar to other chromatin remodeling proteins, alters the transcription of several genes40 important for synaptic function. These collective alterations perturb synaptic function including endocytosis.
Kismet-mediated transcription is important for maintaining the readily releasable and recycling pool of synaptic vesicles
Synaptic vesicles are grouped into “pools” based on their capacity to be released with progressively increased stimulation releasing distinct pools of vesicles. These pools are thought to be biochemically distinct but intermixed at the synapse30. Vesicles of the readily releasable pool are docked at active zones and released first after physiological stimulation. The recycling pool of vesicles is recruited to sustain neurotransmission for longer periods. Tens of seconds of high frequency stimulation, however, requires release of the reserve pool of vesicles, which is the last to be mobilized46. Our data collectively suggest that, in addition to other synaptic functions, Kis-mediated transcription may specifically influence the readily releasable and/or recycling pool of vesicles.
First, while kis mutants exhibit lower eEJC amplitudes during HFS (Fig. 1D,E), a process that mobilizes the reserve pool of vesicles, these reductions do not significantly differ from controls. Evoked responses after HFS, which require replenishment of the readily releasable pool from the recycling pool, were significantly reduced in kis mutants. Second, there were no differences in endocytosis observed in kis mutants when synapses were pretreated with Cyclosporin A (Supplemental Fig. 2), which mobilizes the reserve pool of vesicles36. This finding may be due to an increased rate of exocytosis resulting from a larger pool of vesicles available for release. The amount of membrane endocytosed is proportional to the amount of synaptic vesicle membrane added to the plasma membrane during exocytosis47. Third, the reduction in VGLUT observed at kis mutant synapses after HFS (Fig. 2A,B) is approximately proportional to the reported size of the recycling and readily releasable pools48. Finally, synaptic levels of Rab11, which is localized to recycling endosomes7, are significantly reduced in kis mutants (Fig. 2C–E).
Sustained exocytosis requires endocytosis to recycle membrane and proteins that mediate vesicle release. Endocytosis may directly reform vesicles after uncoating as occurs during clathrin-mediated endocytosis or require vesicles to pinch off from endosomes5. Indeed, endosomal recycling of vesicles is sufficiently rapid that it could allow for the rerelease of vesicles during sustained exocytosis in neuroendocrine PC12 cells49. How endosomes facilitate synaptic vesicle formation is poorly understood but it appears that endosomes sort cargo and replenish membrane pools and structures by the collective actions of several proteins including SNAREs and Rab GTPases50. Rabs are critical for vesicle trafficking and targeting8. It was recently shown that Rab11 promotes clathrin-mediated and activity-dependent bulk endocytosis51. Reduced levels of Rab11 in kis mutants suggests Kis’ transcriptional activity may affect the synaptic vesicle cycle by influencing the trafficking of endosomal compartments. Both CHD7 and CHD8 affect expression of several Rabs and Rab effectors including Rab5B52, Rab7L, Rab11B53, and Rab11FIP2, −4, and −554,55. In addition, Kis binding is enriched near several rab genes including rab5, rab7, and rab11 in Drosophila adult midgut intestinal stem cells20. Determining how Kis affects membrane trafficking will help us better understand the roles of CHD-mediated transcriptional regulation for synapse function.
Kismet regulates the levels of transcripts and synaptic proteins involved in endocytosis
Kis is a chromatin remodeling protein that associates with active sites of transcription and may promote transcriptional activation and elongation18–20. Thus, Kis regulates endocytosis by influencing the number and availability of transcripts important for endocytosis. Transcriptional regulation is accomplished by the collective actions of transcription factors, chromatin modifying enzymes, and chromatin remodeling enzymes. Chromatin modifying enzymes, including HDACs, alter the composition of functional groups on histone N-terminal tails. These enzymes work together and assemble in multi-protein complexes with chromatin remodeling enzymes to influence chromatin compaction41. Consistent with previous results16, inhibition of HDAC function by SAHA and SBHA suppresses the endocytosis defect in kis mutants (Fig. 5) suggesting Kis-mediated transcription is important for proper endocytosis. Similarly, the Class I and Class IIb HDACi, M344, strongly suppressed craniofacial cartilage defects in a zebrafish model of CHARGE syndrome56.
We identified potential Kis endocytosis target genes in the Drosophila larval central nervous system first by RT-qPCR and, for a subset of those, ChIP-qPCR. Kis influenced the transcript levels of several transcripts examined including AP2α, dynamin/shibire, and endoB (Fig. 3A). Kis was also enriched at the transcription start site and promoters for dap160 and endoB (Fig. 3C). The somewhat subtle fold changes in transcript levels are consistent with transcriptional changes in Chd7 null embryonic stem cells57 and after knock down of CHD8 in neural progenitor cells53. While our experiments used mature neurons, we would expect Kis to function similarly throughout development albeit potentially in distinct transcriptional complexes. Kis may influence the transcription of other target genes important for endocytosis as the collective actions of hundreds of genes determine endocytic trafficking58. In support of this, Gervais et al. (2019) found that Kis was enriched near the amphiphysin, AP2α, clathrin heavy chain, clathrin light chain, dynamin/shibire, flower, liquid facets, and synaptojanin in intestinal stem cells of the Drosophila midgut. We didn’t detect appreciable differences in amphiphysin, flower, liquid facets, and synaptojanin transcript levels, which may reflect tissue-specific differences in Kis target genes.
Changes in transcript levels in kis mutants sometimes corresponded to similar changes in synaptic protein levels, as was the case for Dap160 (Fig. 3B1) and EndoB (Fig. 3B4). Synaptic levels of Dyn, however, were increased in kis mutants despite dyn transcript levels being 24–29% lower than controls. Although this phenotype may be the result of Kis-mediated transcriptional changes that affect mRNA stability or translation initiation, the increase in Dyn at the synapse may occur as a result of perturbations to other synaptic proteins. There was a significant reduction in Dap160 in kis mutant synapses (Fig. 3B1). Previous work has shown that preventing the interactions between the Dap160 homolog, Intersectin, and Dyn resulted in the accumulation of Dyn at clathrin-coated pits, impaired endocytosis, and reductions in the number of synaptic vesicles at lamprey central synapses59. Synaptic levels of Dyn are, however, reduced in dap160 loss of function mutants27,39. Thus, the increased levels of Dyn may not be due to reduced levels of Dap160 in kis mutants. We previously showed that kis mutants also exhibit increased synaptic levels of the cell adhesion molecule, FasII, and the postsynaptic scaffold DLG15. Given that chromatin remodeling enzymes regulate the expression of thousands of genes20,54,55, it is likely that Kis directly or indirectly affects the levels of many synaptic proteins.
Kismet is important for proper synaptic protein localization
Dyn is redistributed after synaptic activity from active zones to periactive zones and this redistribution is dependent on interactions with Dap16026. We found that Dyn redistribution was impaired in kis mutants (Fig. 4). Unexpectedly, Dyn was mislocalized in kis mutants at rest. Dyn was diffusely localized in control NMJ boutons at rest and redistributed to the periphery of the bouton after synaptic stimulation. We observed the opposite phenotype, however, in kis mutants, which showed Dyn enriched in the periphery of boutons at rest. Dyn is recruited to phosphatidylinositol-4,5-bisphosphate (PIP2)-rich regions of the inner plasma membrane60. PIP2 membrane regions promote F-actin polymerization61 and are thought to interact with several active zone-associated proteins including Synaptotagmin, Rab3 interacting molecule, and Syntaxin62. We previously showed that Kis is important for the localization of postsynaptic glutamate receptors relative to presynaptic active zones15 and here we show that VGLUT is heterogeneously distributed in kis mutants. Thus, Kis affects target genes that promote neural function by maintaining the localization of synaptic proteins relative to PIP2 membrane domains. Future studies should investigate the role of Kis in membrane dynamics and synaptic function to gain a better picture of the basic functions that are often perturbed in neurodevelopmental disorders.
Methods
Fly rearing and stocks
Fly stocks were maintained at 25 °C in 12:12 h light dark cycle on Jazz Mix (Fisher Scientific) fly food. The kisk13416 and dap160EP2543 mutants and kis-eGFP flies were obtained from the Bloomington Drosophila Stock Center along with the experimental controls, w1118. UAS-kisRNAi.a flies were obtained from the Vienna Drosophila RNAi Center. The protein null kisLM27 allele is described in24.
Flies treated with histone deacetyltranferase inhibitors were raised on instant food (Nutri-Fly, 66–117, Genessee Scientific) containing 1.6% of 10% w/v Tegosept (Fisher Scientific) and either 10 µM SAHA, 10 µM SBHA (Cayman Chemicals), or 0.1% DMSO. Fly food was prepared in small batches and used within 2–3 days. The parental generation was placed on vials for 5 days and then moved new vials.
Immunohistochemistry
Third instar larvae were fillet dissected in Roger’s Ringer (135 mM NaCl, 5 mM KCl, 4 mM MgCl2*6H2O, 1.8 mM CaCl2*2H2O, 5 mM TES, 72 mM Sucrose, pH 7.15) supplemented with 2 mM glutamate on Sylgard (World Precision Instruments)-coated 60 mm plates. Dissected larvae were fixed in 3.7% paraformaldehyde (Fisher Scientific) for 30–45 min and subsequently placed in 1.5 ml centrifuge tubes. Larval pelts were washed at room temperature three times for 10 min each in PTX (PBS + 0.1% Triton, Fisher Scientific) and two times for 30 min each in PBTX (PTX + 1% Bovine Serum Albumin, Fisher Scientific). Primary antibodies were diluted in PBTX and left on overnight at 4 °C. Primary antibodies utilized included rabbit α-Brp (1:2000, Stephen Sigrist lab), mouse α-Brp (1:50, Developmental Studies Hybridoma Bank), guinea pig α-Dap160 (1:1000, Hugo Bellen lab), mouse α-Dynamin (1:300, BD Biosciences), rabbit α-Dynamin (1:2000, Hugo Bellen lab), guinea pig α-Endophilin A (1:5000, Hugo Bellen lab), rabbit α-Endophilin B (1:200, Li-Mei Pai lab), guinea pig α-Eps15 (1:2000, Hugo Bellen lab), rabbit α-Nervous Wreck (Nwk, 1:1000, Kate O’Connor-Giles lab), mouse α-Rab11 (1:50, Fisher Scientific, BDB610657), and rabbit α-VGLUT (1:5000, Aaron DiAntonio Lab). After overnight incubation in primary antibodies, larvae were washed three times for 10 min each in PBTX and then washed two times in PBTX. Larvae were next incubated in secondary antibodies diluted in PBTX for 2 h at room temperature. Secondary antibodies, including α-mouse FITC, α-mouse TRITC, α-rabbit FITC, and α-guinea pig FITC were used at 1:400 and obtained from Jackson ImmunoResearch. Cy3- and A647-HRP (Jackson ImmunoResearch) were applied at 1:125 with secondary antibodies. After 2 h, PBTX washes as described above, were performed. Larvae were mounted on slides with Vectashield (Vector Laboratories). Confocal images of the 6/7 NMJ in segments 3 or 4 were obtained using the 60x oil immersion objective on an Olympus Fluoview 1000. Imaging parameters were determined using appropriate controls for each experiment and approximately equal numbers of controls and experimental animals constituted a biological replicate. Each experiment included at least 2–3 biological replicates.
Measuring endocytosis and dynamin localization
Endocytosis was measured by quantifying the amount of internalized FM 1–43FX as described by Verstreken et al. (2008). Third instar larvae were fillet dissected in Ca2+-free HL-3 (100 mM NaCl, 5 mM KCl, 10 mM NaHCO3, 5 mM HEPES, 30 mM sucrose, 5 mM trehelose, 10 mM MgCl2, pH 7.2). After one wash with Ca2+-free HL-3 and cutting the motor neurons, larvae were treated with 4 µM FM 1–43FX (Thermo Fisher) in HL-3 containing 90 mM KCl and 1.5 mM CaCl2 for one minute. Next, larvae were washed five times over 5–10 min in Ca2+-free HL-3. Larvae were fixed in 3.7% paraformaldehyde (Fisher Scientific) in Ca2+-free HL-3 for 5 min and then placed in 1.5 ml centrifuge tubes. Larvae were washed with Ca2+-free HL-3 containing 2.5% goat serum several times and with Ca2+-free HL-3 10 times over 10–15 min. A647-HRP (1:125, Jackson ImmunoResearch) was applied for 30 min in Ca2+-free HL-3 containing 5% goat serum. Next, larvae were washed with Ca2+-free HL-3 10 times over 10–15 min and mounted on slides with Vectashield (Vector Laboratories). All larvae were imaged as described above the same day after treatment with FM 1–43FX. To label both the cycling and reserve pool of vesicles, the above protocol was executed except that larvae were incubated with 10 µM cyclosporin A (Fisher Scientific) in Ca2+-free HL-3 for 20 min prior to treatment with FM 1–43FX. Each experiment included at least 4–5 biological replicates.
Relocalization of Dyn after synaptic activity was induced as described by Winther et al. (2013, 2015). Third instar larvae were fillet dissected in Ca2+-free HL-3. The nervous system was left intact and the solution was replaced for 10 min with either Ca2+-free HL-3 containing 50 mM EDTA for controls (i.e. “rest”) or HL-3 containing 60 mM KCl and 1.0 mM CaCl2 (i.e. “stim”). After washing once with Ca2+-free HL-3 and removing the CNS, larvae were fixed with 3.7% paraformaldehyde in 1x PBS and the immunohistochemistry protocol above followed.
Quantification of Dyn relocalization and distances between Brp and Syn or SytI was accomplished using Fiji (Fiji.sc) as previously described40. Briefly, red-green-blue intensity profiles were obtained by drawing lines on z-projected images through boutons perpendicular to the NMJ branch. The distance between the maximum peaks for Brp and Dyn, Brp and SytI, or Brp and Syn were obtained. Maximum peak distances were calculated for five terminal boutons per NMJ and averaged. Pearson’s correlation coefficients were obtained by tracing around the area of the NMJ and executing the Col2 command in Fiji.
ChIP-qPCR and RT-qPCR
CNS were dissected in PBS from 300–600 third instar larvae of each genotype per biological replicate. Chromatin was sheared using a Tissue Chromatin Shearing Kit with SDS Shearing Buffer (Covaris). Chromatin was sheared for 10 min by a Covaris E220 Ultrasonicator. The resulting chromatin was visualized on an agarose gel containing 1.5% Ethidium Bromide (Fisher Scientific) to confirm 100–600 bp chromatin fragments. Chromatin was then immunoprecipitated using a Magna ChIP HiSens Kit (Millipore). Sheared chromatin was incubated with either rabbit α-GFP (Abcam, ab290) or rabbit α-IgG (Abcam, ab171870) coated magnetic beads for 3 h. Next, chromatin was eluted from the magnetic beads and incubated in RNase A (10 mg/ml, Fisher Scientific) for 30 min followed by incubation overnight in Proteinase K (10 mg/ml, Millipore) at 57 °C. The next day, the Proteinase K was inactivated by incubating for 15 min at 75 °C. DNA was then isolated using the QIAquick PCR Purification Kit (Qiagen) and stored at −20 °C for qPCR.
CNS were dissected in Roger’s Ringer (135 mM NaCl, 5 mM KCl, 4 mM MgCl2*6H2O, 1.8 mM CaCl2*2H2O, 5 mM TES, 72 mM Sucrose, pH 7.15) supplemented with 2 mM glutamate from 30 third instar larvae of each genotype per biological replicate. Tissues were stored in RNALater (Fisher Scientific) at −20 °C until RNA was isolated using the Direct-zol RNA Microprep Kit (Zymo Research, R2061). qRT-PCR was performed using the iTaq Universal SYBR Green One Step Kit (Bio-Rad) and the Stratagene MX3000P thermal cycler. A minimum of three biological replicates each including three technical replicates were used for data analysis. 100 ng of RNA was used per reaction.
Electrophysiology
Third instar larvae were fillet dissected in ice cold HL-3.1 (70 mM NaCl, 5 mM KCl, 10 mM NaHCO3, 5 mM HEPES, 115 mM sucrose, 5 mM trehelose, 4 mM MgCl2 (6H2O)) containing 0.25 mM Ca2+ on Sylgard (World Precision Instruments)-coated coverslips. Recordings were performed at room temperature in HL-3.1 containing 1.5 mM Ca2+ (endocytosis) or 0.6 mM Ca2+ (paired pulse recordings). Muscle 6 in segment A3 or A4 was clamped at −60 mV using an Axoclamp 900 A amplifier (Molecular Devices). Clamp and recording electrodes were filled with 3 mM KCl and used if they exhibited 10–20 MΩ of resistance. Stimulating electrodes were filled with bath saline (HL-3.1 with Ca2+). Stimuli were delivered to segmental nerves by a Grass S88 stimulator with a SIU5 isolation unit (Grass Technologies). Endocytosis and recovery of the cycling pool of vesicles was assessed by stimulating at 0.2 Hz for 50 s, 20 Hz for 60 s, and 0.2 Hz for 50 s. Paired pulse amplitudes were measured after delivering two 10 Hz, two 20 Hz, two 50 Hz, and two 100 Hz pulses. Each stimulus pair was separated by a 20 s intertrial interval. Recordings were digitized with a Digidata 1443 digitizer (Molecular Devices). An equal number of recordings from controls and experimental animals were obtained each day. Data were analyzed in pClamp (v10.4, Molecular Devices) and GraphPad Prism (v. 5.01).
Statistical analyses
GraphPad Prism (v. 5.01) was used for all statistical analyses. A student’s t-test was used to compare two data sets. A two-way repeated measures ANOVA was used to compare evoked responses across genotypes during and after HFS. Error bars in all histograms represent the standard error of the mean. P-values in figures are represented by * = < 0.05, ** = < 0.001, and *** = < 0.0001. Summary statistics for all data can be found in Supplemental Table 1.
Supplementary information
Acknowledgements
We thank the laboratories of Drs. Hugo Bellen (Baylor College of Medicine), Aaron DiAntonio (Washington University St. Louis), Kate O’Connor-Giles (Brown University), Li-Mei Pai (Chang Gung University), and Stephan Sigrist (Freie Universitat Berlin) for generously providing antibodies used in our experiments. We also thank the Bloomington Drosophila Stock Center for fly stocks and the Developmental Studies Hybridoma Bank (University of Iowa) for additional antibodies. We thank Kelsey Lane for assistance with data analysis. Finally, we thank Dave Featherstone for his guidance and mentorship. This work was supported by SIUE’s Undergraduate Research and Creative Activities Program and by grants from the CHARGE Syndrome Foundation (DRM and FLW), NSF IOS 1256114 (DRM and FLW), NSF IOS 1856439 (DRM), and NINDS R15 NS101608-01A1 (FLW). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
N.K.L., T.L.D., D.R.M. and F.L.W. designed the experiments. N.K.L., J.M.V., R.A.S., K.B., B.H. and F.L.W. performed experiments and analyzed the data. N.K.L., T.L.D., D.R.M. and F.L.W. wrote and edited the manuscript.
Data availability
All data analyses and summary statistics are provided in Supplemental Table 1.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Nina K. Latcheva and Taylor L. Delaney.
Supplementary information
is available for this paper at 10.1038/s41598-019-55900-6.
References
- 1.Yap CC, Winckler B. Harnessing the power of the endosome to regulate neural development. Neuron. 2012;74:440–451. doi: 10.1016/j.neuron.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elkin SR, Lakoduk AM, Schmid SL. Endocytic pathways and endosomal trafficking: a primer. Wien Med Wochenschr. 2016;166:196–204. doi: 10.1007/s10354-016-0432-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 4.Kaksonen M, Roux A. Mechanisms of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2018;19:313–326. doi: 10.1038/nrm.2017.132. [DOI] [PubMed] [Google Scholar]
- 5.Saheki Y, De Camilli P. Synaptic vesicle endocytosis. Cold Spring Harb Perspect Biol. 2012;4:a005645. doi: 10.1101/cshperspect.a005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cocucci E, Aguet F, Boulant S, Kirchhausen T. The first five seconds in the life of a clathrin-coated pit. Cell. 2012;150:495–507. doi: 10.1016/j.cell.2012.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt MR, Haucke V. Recycling endosomes in neuronal membrane traffic. Biol Cell. 2007;99:333–342. doi: 10.1042/BC20070007. [DOI] [PubMed] [Google Scholar]
- 8.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 9.Rodal AA, Motola-Barnes RN, Littleton JT. Nervous wreck and Cdc42 cooperate to regulate endocytic actin assembly during synaptic growth. J Neurosci. 2008;28:8316–8325. doi: 10.1523/JNEUROSCI.2304-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dickman DK, Lu Z, Meinertzhagen IA, Schwarz TL. Altered synaptic development and active zone spacing in endocytosis mutants. Curr Biol. 2006;16:591–598. doi: 10.1016/j.cub.2006.02.058. [DOI] [PubMed] [Google Scholar]
- 11.Khodosh R, Augsburger A, Schwarz TL, Garrity PA. Bchs, a BEACH domain protein, antagonizes Rab11 in synapse morphogenesis and other developmental events. Development. 2006;133:4655–4665. doi: 10.1242/dev.02650. [DOI] [PubMed] [Google Scholar]
- 12.Meinertzhagen IA, Govind CK, Stewart BA, Carter JM, Atwood HL. Regulated spacing of synapses and presynaptic active zones at larval neuromuscular junctions in different genotypes of the flies Drosophila and Sarcophaga. J Comp Neurol. 1998;393:482–492. doi: 10.1002/(SICI)1096-9861(19980420)393:4<482::AID-CNE7>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 13.Schuster CM, Davis GW, Fetter RD, Goodman CS. Genetic dissection of structural and functional components of synaptic plasticity. II. Fasciclin II controls presynaptic structural plasticity. Neuron. 1996;17:655–667. doi: 10.1016/S0896-6273(00)80198-1. [DOI] [PubMed] [Google Scholar]
- 14.Collins CA, DiAntonio A. Synaptic development: insights from Drosophila. Curr Opin Neurobiol. 2007;17:35–42. doi: 10.1016/j.conb.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Ghosh R, et al. Kismet positively regulates glutamate receptor localization and synaptic transmission at the Drosophila neuromuscular junction. PLoS One. 2014;9:e113494. doi: 10.1371/journal.pone.0113494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Latcheva NK, et al. Epigenetic crosstalk: Pharmacological inhibition of HDACs can rescue defective synaptic morphology and neurotransmission phenotypes associated with loss of the chromatin reader Kismet. Mol Cell Neurosci. 2018;87:77–85. doi: 10.1016/j.mcn.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 17.Bouazoune K, Kingston RE. Chromatin remodeling by the CHD7 protein is impaired by mutations that cause human developmental disorders. Proc Natl Acad Sci USA. 2012;109:19238–19243. doi: 10.1073/pnas.1213825109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srinivasan S, et al. The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA Polymerase II. Development. 2005;132:1623–1635. doi: 10.1242/dev.01713. [DOI] [PubMed] [Google Scholar]
- 19.Srinivasan S, Dorighi KM, Tamkun JW. Drosophila Kismet regulates histone H3 lysine 27 methylation and early elongation by RNA polymerase II. PLoS Genet. 2008;4:e1000217. doi: 10.1371/journal.pgen.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gervais Louis, van den Beek Marius, Josserand Manon, Sallé Jérémy, Stefanutti Marine, Perdigoto Carolina N., Skorski Patricia, Mazouni Khallil, Marshall Owen J., Brand Andrea H., Schweisguth François, Bardin Allison J. Stem Cell Proliferation Is Kept in Check by the Chromatin Regulators Kismet/CHD7/CHD8 and Trr/MLL3/4. Developmental Cell. 2019;49(4):556-573.e6. doi: 10.1016/j.devcel.2019.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melicharek DJ, Ramirez LC, Singh S, Thompson R, Marenda DR. Kismet/CHD7 regulates axon morphology, memory and locomotion in a Drosophila model of CHARGE syndrome. Hum Mol Genet. 2010;19:4253–4264. doi: 10.1093/hmg/ddq348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Latcheva NK, Viveiros JM, Marenda DR. The Drosophila Chromodomain Protein Kismet Activates Steroid Hormone Receptor Transcription to Govern Axon Pruning and Memory. In Vivo. iScience. 2019;16:79–93. doi: 10.1016/j.isci.2019.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roch F, et al. Screening of larval/pupal P-element induced lethals on the second chromosome in Drosophila melanogaster: clonal analysis and morphology of imaginal discs. Mol Gen Genet. 1998;257:103–112. doi: 10.1007/pl00008620. [DOI] [PubMed] [Google Scholar]
- 24.Melicharek D, et al. Identification of novel regulators of atonal expression in the developing Drosophila retina. Genetics. 2008;180:2095–2110. doi: 10.1534/genetics.108.093302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verstreken P, Ohyama T, Bellen HJ. FM 1-43 labeling of synaptic vesicle pools at the Drosophila neuromuscular junction. Methods Mol Biol. 2008;440:349–369. doi: 10.1007/978-1-59745-178-9_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winther AM, et al. An Endocytic Scaffolding Protein together with Synapsin Regulates Synaptic Vesicle Clustering in the Drosophila Neuromuscular Junction. J Neurosci. 2015;35:14756–14770. doi: 10.1523/JNEUROSCI.1675-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koh TW, Verstreken P, Bellen HJ. Dap160/intersectin acts as a stabilizing scaffold required for synaptic development and vesicle endocytosis. Neuron. 2004;43:193–205. doi: 10.1016/j.neuron.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 28.Long AA, et al. The nonsense-mediated decay pathway maintains synapse architecture and synaptic vesicle cycle efficacy. J Cell Sci. 2010;123:3303–3315. doi: 10.1242/jcs.069468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuromi H, Kidokoro Y. Exocytosis and endocytosis of synaptic vesicles and functional roles of vesicle pools: lessons from the Drosophila neuromuscular junction. Neuroscientist. 2005;11:138–147. doi: 10.1177/1073858404271679. [DOI] [PubMed] [Google Scholar]
- 30.Denker A, Krohnert K, Rizzoli SO. Revisiting synaptic vesicle pool localization in the Drosophila neuromuscular junction. J Physiol. 2009;587:2919–2926. doi: 10.1113/jphysiol.2009.170985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuromi H, Kidokoro Y. Selective replenishment of two vesicle pools depends on the source of Ca2+ at the Drosophila synapse. Neuron. 2002;35:333–343. doi: 10.1016/S0896-6273(02)00777-8. [DOI] [PubMed] [Google Scholar]
- 32.Daniels RW, et al. Increased expression of the Drosophila vesicular glutamate transporter leads to excess glutamate release and a compensatory decrease in quantal content. J Neurosci. 2004;24:10466–10474. doi: 10.1523/JNEUROSCI.3001-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Omote H, Miyaji T, Juge N, Moriyama Y. Vesicular neurotransmitter transporter: bioenergetics and regulation of glutamate transport. Biochemistry. 2011;50:5558–5565. doi: 10.1021/bi200567k. [DOI] [PubMed] [Google Scholar]
- 34.Winther AM, et al. The dynamin-binding domains of Dap160/intersectin affect bulk membrane retrieval in synapses. J Cell Sci. 2013;126:1021–1031. doi: 10.1242/jcs.118968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 36.Kuromi H, Kidokoro Y. The optically determined size of exo/endo cycling vesicle pool correlates with the quantal content at the neuromuscular junction of Drosophila larvae. J Neurosci. 1999;19:1557–1565. doi: 10.1523/JNEUROSCI.19-05-01557.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dorighi KM, Tamkun JW. The trithorax group proteins Kismet and ASH1 promote H3K36 dimethylation to counteract Polycomb group repression in Drosophila. Development. 2013;140:4182–4192. doi: 10.1242/dev.095786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koh TW, et al. Eps15 and Dap160 control synaptic vesicle membrane retrieval and synapse development. J Cell Biol. 2007;178:309–322. doi: 10.1083/jcb.200701030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marie B, et al. Dap160/intersectin scaffolds the periactive zone to achieve high-fidelity endocytosis and normal synaptic growth. Neuron. 2004;43:207–219. doi: 10.1016/j.neuron.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 40.Gerth F, et al. Intersectin associates with synapsin and regulates its nanoscale localization and function. Proc Natl Acad Sci USA. 2017;114:12057–12062. doi: 10.1073/pnas.1715341114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tyagi M, Imam N, Verma K, Patel AK. Chromatin remodelers: We are the drivers!! Nucleus. 2016;7:388–404. doi: 10.1080/19491034.2016.1211217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Durak O, et al. Chd8 mediates cortical neurogenesis via transcriptional regulation of cell cycle and Wnt signaling. Nat Neurosci. 2016;19:1477–1488. doi: 10.1038/nn.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng W, et al. The chromatin remodeler CHD7 regulates adult neurogenesis via activation of SoxC transcription factors. Cell Stem Cell. 2013;13:62–72. doi: 10.1016/j.stem.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 44.Gompers AL, et al. Germline Chd8 haploinsufficiency alters brain development in mouse. Nat Neurosci. 2017;20:1062–1073. doi: 10.1038/nn.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopez AJ, Wood MA. Role of nucleosome remodeling in neurodevelopmental and intellectual disability disorders. Front Behav Neurosci. 2015;9:100. doi: 10.3389/fnbeh.2015.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chamberland S, Toth K. Functionally heterogeneous synaptic vesicle pools support diverse synaptic signalling. J Physiol. 2016;594:825–835. doi: 10.1113/JP270194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maritzen T, Haucke V. Coupling of exocytosis and endocytosis at the presynaptic active zone. Neurosci Res. 2018;127:45–52. doi: 10.1016/j.neures.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 48.Delgado R, Maureira C, Oliva C, Kidokoro Y, Labarca P. Size of vesicle pools, rates of mobilization, and recycling at neuromuscular synapses of a Drosophila mutant, shibire. Neuron. 2000;28:941–953. doi: 10.1016/S0896-6273(00)00165-3. [DOI] [PubMed] [Google Scholar]
- 49.Hoopmann P, et al. Endosomal sorting of readily releasable synaptic vesicles. Proc Natl Acad Sci USA. 2010;107:19055–19060. doi: 10.1073/pnas.1007037107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morgan JR, Comstra HS, Cohen M, Faundez V. Presynaptic membrane retrieval and endosome biology: defining molecularly heterogeneous synaptic vesicles. Cold Spring Harb Perspect Biol. 2013;5:a016915. doi: 10.1101/cshperspect.a016915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kokotos AC, Peltier J, Davenport EC, Trost M, Cousin MA. Activity-dependent bulk endocytosis proteome reveals a key presynaptic role for the monomeric GTPase Rab11. Proc Natl Acad Sci USA. 2018;115:E10177–E10186. doi: 10.1073/pnas.1809189115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilkinson B, et al. The autism-associated gene chromodomain helicase DNA-binding protein 8 (CHD8) regulates noncoding RNAs and autism-related genes. Transl Psychiatry. 2015;5:e568. doi: 10.1038/tp.2015.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sugathan A, et al. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc Natl Acad Sci USA. 2014;111:E4468–4477. doi: 10.1073/pnas.1405266111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katayama Y, et al. CHD8 haploinsufficiency results in autistic-like phenotypes in mice. Nature. 2016;537:675–679. doi: 10.1038/nature19357. [DOI] [PubMed] [Google Scholar]
- 55.Wang P, et al. CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in neurodevelopment. Mol Autism. 2015;6:55. doi: 10.1186/s13229-015-0048-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Asad Z, Sachidanandan C. Chemical screens in a zebrafish model of CHARGE syndrome identifies small molecules that ameliorate disease-like phenotypes in embryo. Eur J Med Genet. 2019 doi: 10.1016/j.ejmg.2019.04.018. [DOI] [PubMed] [Google Scholar]
- 57.Schnetz MP, et al. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet. 2010;6:e1001023. doi: 10.1371/journal.pgen.1001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Collinet C, et al. Systems survey of endocytosis by multiparametric image analysis. Nature. 2010;464:243–249. doi: 10.1038/nature08779. [DOI] [PubMed] [Google Scholar]
- 59.Evergren E, et al. Intersectin is a negative regulator of dynamin recruitment to the synaptic endocytic zone in the central synapse. J Neurosci. 2007;27:379–390. doi: 10.1523/JNEUROSCI.4683-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mehrotra N, Nichols J, Ramachandran R. Alternate pleckstrin homology domain orientations regulate dynamin-catalyzed membrane fission. Mol Biol Cell. 2014;25:879–890. doi: 10.1091/mbc.E13-09-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frere SG, Chang-Ileto B, Di Paolo G. Role of phosphoinositides at the neuronal synapse. Subcell Biochem. 2012;59:131–175. doi: 10.1007/978-94-007-3015-1_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koch M, Holt M. Coupling exo- and endocytosis: an essential role for PIP(2) at the synapse. Biochim Biophys Acta. 2012;1821:1114–1132. doi: 10.1016/j.bbalip.2012.02.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data analyses and summary statistics are provided in Supplemental Table 1.