ABSTRACT
Shigella spp. are bacterial pathogens that invade the human colonic mucosa using a type III secretion apparatus (T3SA), a proteinaceous device activated upon contact with host cells. Active T3SAs translocate proteins that carve the intracellular niche of Shigella spp. Nevertheless, the activation state of the T3SA has not been addressed in vivo. Here, we used a green fluorescent protein transcription-based secretion activity reporter (TSAR) to provide a spatio-temporal description of S. flexneri T3SAs activity in the colon of Guinea pigs. First, we observed that early mucus release is triggered in the vicinity of luminal bacteria with inactive T3SA. Subsequent mucosal invasion showed bacteria with active T3SA associated with the brush border, eventually penetrating into epithelial cells. From 2 to 8 h post-challenge, the infection foci expanded, and these intracellular bacteria displayed homogeneously high-secreting activity, while extracellular foci within the lamina propria featured bacteria with low secretion activity. We also found evidence that within lamina propria macrophages, bacteria reside in vacuoles instead of accessing the cytosol. Finally, bacteria were cleared from tissues between 8 and 24 h post-challenge, highlighting the hit-and-run colonization strategy of Shigella. This study demonstrates how genetically encoded reporters can contribute to deciphering pathogenesis in vivo.
Keywords: Shigella, type III secretion system, genetically encoded reporters, in vivo infection, fluorescence microscopy, large intestine
We mapped Shigella flexneri cells and assessed the activity of their type III secretion apparatus in the large intestine of Guinea pigs.
INTRODUCTION
Shigellosis is a human diarrheal disease induced by infection of the colonic mucosa by Shigella spp., a group of Escherichia coli pathovars that share the property of invading the epithelium and the subjacent lamina propria, thereby causing acute tissue inflammatory destruction accounting for its dysenteric symptoms (Anderson, Sansonetti and Marteyn 2016; Kang et al. 2018). The large virulence plasmid of Shigella spp. encodes a type III secretion system (T3SS) and associated effector proteins that enable host cell invasion, intracellular persistence and cell-to-cell spread (Buchrieser et al. 2000; Campbell-Valois and Pontier 2016).
The capacity of Shigella to invade host cells requires the assembly of a type III secretion apparatus (T3SA), a megadalton nanomachine composed of several proteins that oligomerize to form a syringe-like appendage. The T3SA is composed of the cytosolic sorting platform and export apparatus, inner and outer transmembrane rings and the needle that protrudes from the outer membrane surface (Hu et al. 2015). T3SA substrates, comprising 35 proteins in S. flexneri (Pinaud et al. 2017), are selected by the sorting platform and export apparatus and transit through the transmembrane rings and needle before reaching the host cell cytosol. This leads to the phagocytosis and rapid lysis of the entry vacuole. Then Shigella elicits the formation of actin comets through the action of the protein IcsA (Bernardini et al. 1989), which enables the movement of bacteria in the cytosol. Mobile bacteria eventually hit the plasma membrane, thereby reactivating their T3SAs and consequently allowing the invasion of neighboring cells through a process similar to the initial entry called cell-to-cell spread (Campbell-Valois and Pontier 2016). The iteration of cell-to-cell spread allows the expansion of the infection foci.
When T3SAs are active, the transcription activator MxiE and its co-activator IpgC upregulates the transcription of late substrate genes whose protein products are secreted after translocators and stored effectors (Campbell-Valois and Pontier 2016). Using the promoter of the late substrate ipaH7.8 and GFPsfm2, a green fluorescent protein (GFP) variant with a fast maturation rate, we developed a transcription-based secretion activity reporter (TSAR) that allows detection of the recent secretion activity of individual bacterium by measuring their fluorescent signal (Campbell-Valois et al. 2014; Campbell-Valois, Schnupf and Sansonetti 2014). The TSAR demonstrated that the T3SA cycles between on and off states when bacteria transit iteratively between vacuolar and cytosolic compartments during consecutive cell-to-cell spreading events (Campbell-Valois et al. 2014).
The study of Shigella spp. in vivo is complicated by its tropism for humans; hence, the establishment of a non-primate animal model of shigellosis has been challenging (Kang et al. 2018). Nevertheless, interest for an infection model in the Guinea pig has recently been revived (Shim et al. 2007; Arena et al. 2015; Anderson et al. 2017; Tinevez et al. 2019). In particular, quantitative analysis of fluorescence micrographs of bacteria within the colonic mucosa has indicated that Shigella invade the mucosa by targeting the apical pole of colonocytes found at the entrance of crypts (Arena et al. 2015). Nevertheless, this study did not look at stages of infection preceding invasion and those beyond 8-h post-challenge. Furthermore, it did not probe the activity of the T3SA during infection.
Herein, we have followed the invasion of the mucosa using an S. flexneri strain harboring the TSAR, which enabled us to determine the activity state of the T3SA at different time points. This allowed to study many stages of the infection of the large intestine by Shigella flexneri, ranging from events preceding the initial invasion of the mucosa to the invasion of the mucosa itself, until the final clearing of bacteria from the lamina propria by the host immune response.
MATERIALS AND METHODS
Bacteria and plasmids
In this study, we used the Shigella flexneri M90T-Sm strain resistant to streptomycin harboring the pTSAR1.1 (Sansonetti, Kopecko and Formal 1982; Campbell-Valois et al. 2014). The pTSAR1.1 allows monitoring the activity of the T3SA as previously described (Campbell-Valois et al. 2014; Campbell-Valois, Schnupf and Sansonetti 2014; Campbell-Valois et al. 2015; Campbell-Valois and Pontier 2016) pTSAR1.1 consists of: (i) the GFPsfm2 under the control of the ipaH7.8 promoter (ipaH7.8p) and (ii) a cyan fluorescent protein (CFP) variant named cerulean under the control of the strong constitutive promoter of the ribosomal protein rpsM (rpsMp). The respective fluorescence signals generated by these cistrons are referred in the figures as TSAR and rpsMp, respectively.
In Vivo infection and tissue preparation
Female, pathogen-free Hartley Guinea pigs weighing between 120 and 250 g were obtained from Charles River Laboratories, maintained in animal care facilities of Institut Pasteur, and provided with food and water ad libitum. All animal experiments were carried out using a protocol approved by the User Committee of Institut Pasteur and by the French Ministry of Agriculture no. 2013–0113. Infection and tissue preparation were performed using intrarectal inoculation of 1 × 1010 colony forming units as described in (Arena et al. 2015). Nevertheless, a few modifications were brought to this procedure. First, we used the M90T strain harboring the pTSAR1.1 plasmid for the infection. Second, we used OCT (optimal cutting temperature) compound to embed transversal cuts of the distal part of the large intestine as opposed to using the Swiss-roll method described in (Arena et al. 2015). Thus, we did not perform extensive flushing of the lumen thereby allowing the preservation of mucus-embedded luminal bacteria. A minimum of two animals with significant levels of bacteria within the colonic mucosae were analyzed per time point. It is important to infect between two and three animals per condition or time point tested due to the inherent variability of infection observed in this animal model (Arena et al. 2015). Animals with no visible bacteria in the tissue were excluded from the analysis. To report on the limitation due to animal-to-animal variability, we selected representative images of each animal analyzed at any given timepoint and mentioned any important variation between them in the Results.
Tissue staining and microscope observation
Cut sections were transferred to slides to perform the staining as described previously (Arena et al. 2015). Staining of Goblet cells and neutrophils were realized with Wheat Germ Agglutinin (WGA) conjugated with the fluorescent dye Alexa555 (Thermo Fisher, #W32464) (Arena et al. 2015) and MUB-Cy5 (Anderson et al. 2018), respectively. Galectin-3 labeling was performed with a mouse monoclonal antibody (BD, #556 904) and a goat anti-mouse IgG secondary antibody conjugated with Alexa568 (Thermo Fisher, #A-11 004); CD68 labeling was performed with a mouse monoclonal antibody (Gentaur, KT117) and a donkey anti-mouse IgG secondary antibody conjugated with Alexa568 (Thermo Fisher, #A10037); lipopolysaccharide (LPS) labeling was performed with a rabbit polyclonal antibody raised against S. flexneri M90T LPS (Bernardini et al. 1989) and a goat anti rabbit IgG secondary antibody conjugated with Alexa488 (Thermo Fisher, #A-11 008). Tissue sections were counter stained with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma, # 32670) and phalloidin-Alexa647 (Thermo Fisher, #A22287) or cytopainter phalloidin-iFluor 405 (Abcam, #ab176752). WGA and phalloidin staining were performed as described by the supplier. Antibody labeling was performed as previously described (Arena et al. 2015). Micrographs were obtained using SP5 (Leica Microsystems) and Opterra (Bruker) confocal microscopes.
Quantification of secretion activity and bacterial location
The bacterial location within the mucosa and in the lamina propria was performed by manual observations using the staining described above. Foci at 2-h post-challenge were divided in two categories based on the presence of the bulk of the bacterial population in the lumen in the presence of mucus or in the mucosa. The pixel intensity of bacteria in the GFP (TSAR) and CFP (rpsMp) channels were obtained with ImageJ (Fiji) (Schindelin et al. 2012). First, the channels were split, and then the CFP channel was autothresholded using the MaxEntropy setting and converted into a binary mask that was next used to quantify the pixel intensity of the grayscale image. To remove signal due to noise or bacteria that were not in the center of the plane of the acquired images, objects with areas smaller than 0.5 μm2 were discarded. To remove signal due to the aggregation of individual bacteria in densely populated areas into a single object, objects with areas larger than 5 μm2 were discarded. The GFP:CFP ratio was thus obtained from all remaining objects. A minimum of four micrographs from two animals, including a total of at least 100 bacteria, were used in the quantification of each condition described.
Transmission electron microscopy
Transmission electron microscopy (TEM) images were obtained using vibratome sections of agarose-embedded tissue as described previously (Arena et al. 2015). Bacteria identified on these images were assumed to be Shigella, since previous work suggested that only this organism is found within the colonic mucosae in these conditions (Arena et al. 2015).
RESULTS
Guinea pigs were infected through the colorectal route with the invasive S. flexneri strain M90T harboring pTSAR1.1. This plasmid allows locating all living bacteria with the CFP mCerulean expressed from the constitutive rpsM promoter (rpsMp) and measuring the activity of the T3SA with a GFP TSAR (Campbell-Valois et al. 2014). To obtain this reporter, GFPsfm2, a fast-maturing variant of GFP (Campbell-Valois et al. 2014), was put under the control of the promoter of ipaH7.8 (ipaH7.8p), which is upregulated when the T3SA is active (Bongrand, Sansonetti and Parsot 2012). The maximal fluorescence signal of the TSAR is reached within 15 min following vacuole rupture and varies from bacterium-to-bacterium depending on the duration of their association with protrusions and vacuoles derived from the plasma membrane (Campbell-Valois et al. 2014). Upon inactivation of the T3SA, the stark reduction in the transcription activity leads to reduction of the fluorescent signal by dilution of the GFPsfm2 concentration due to bacterial division (Campbell-Valois et al. 2014). The intensity of the GFP signal is thus a metric of the duration and recency of the T3SA activity. It is impossible to measure membrane association and bacterial division on fixed samples; for this reason, the TSAR signal will be discussed as on or off relatively to rpsMp. Animals were sacrificed at different time points post-challenge. Tissues were cryostat-sectioned and stained with hematoxylin and eosin. Representative histology sections from time points beyond 4 h displayed the typical mucosal disruption and infiltration of immune cells when compared to non-infected tissues (Figure S1, Supporting Information). Equivalent sections from the same animals were prepared for fluorescence confocal microscopy and analyzed to determine bacterial location, fitness and the activity of their T3SA.
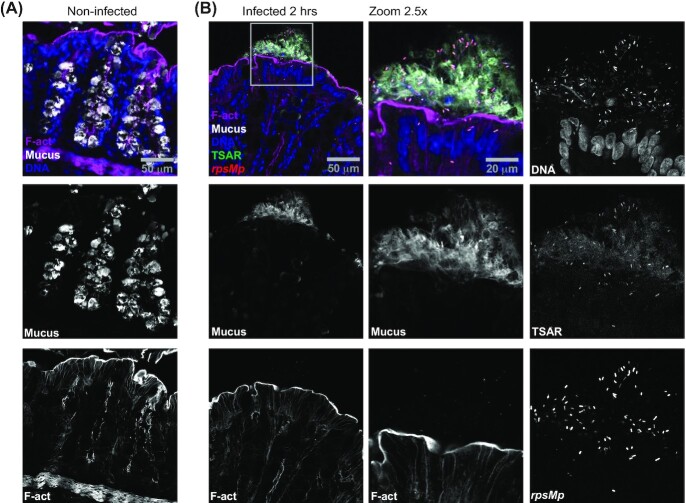
At 2 h post-challenge, we observed clusters of luminal bacteria that were rpsMp-positive (CFP-positive) and TSAR-negative (GFP-negative). They were mostly located near the surface of the mucosa, indicated by their proximity with F-actin as revealed by staining with fluorescent phalloidin. The fact that these Shigella cells were CFP-positive suggested that oxygen tension near the mucosal surface was sufficient for the maturation of its chromophore, as previously reported for GFP (Marteyn et al. 2010). In addition, these bacteria appeared to be several micrometers away from the brush border and were non-contiguous, as if an invisible web held them together. We wondered whether these clusters could be the result of interactions between bacteria and the mucus. To test this hypothesis, we stained tissue sections with fluorescently labeled WGA. Tissue from non-infected animals showed WGA-staining only within crypt goblet cells (Fig. 1A), as previously reported (Arena et al. 2015). By contrast, goblet cells in infected areas were poorly stained with WGA, while neighboring clusters of luminal bacteria were embedded in extracellular WGA-positive mucus (Fig. 1B). Interestingly, z-stacks displaying low bacterial load, appeared identical to non-infected samples, while those with high bacterial load showed massive mucus release (Figure S2, Supporting Information). This suggested that mucus release was triggered in areas, which we dubbed mucus foci, where the density of luminal bacteria was above a critical threshold. It is noteworthy that in infected stacks, luminal mucus was detectable despite PFA fixation, while it was undetectable in non-infected stacks of the same area. Finally, as indicated above, bacteria located in mucus foci were rpsMp-positive (CFP-positive) and TSAR-negative (GFP-negative), thus indicating that these bacteria were alive, though not secreting.
Figure 1.
Massive mucus release of the colonic mucosa is induced by luminal S. flexneri displaying low T3SA activity. Colonic tissue sections infected or not with WT Shigella flexneri harboring pTSAR1.1 and labeled with Wheat Germ Agglutinin (WGA), phalloidin and DAPI were imaged using confocal microscopy. A micrograph overlay of a non-infected tissue section (A). A micrograph overlay of an infected tissue section at 2 h post-challenge (left panel). The indicated channels corresponding to the boxed area are magnified by 2.5-fold and represented in the central and right panels (B). Sections are positioned with the lumen on top and are representative of infection foci observed in two animals.
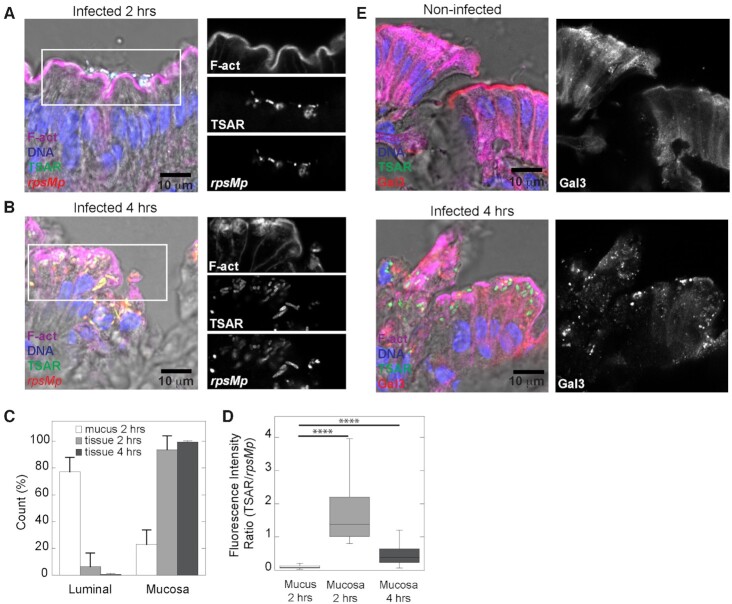
From animals infected for 2 h, we also observed tissue foci where bacteria were intimately associated with the mucosa and displayed high TSAR signals indicating that they had undergone T3SA-mediated secretion. Importantly, these bacteria were tightly associated with the brush border (Fig. 2A) or associated with the apical side of the enterocytes as indicated by their location between the F-actin of the brush border and the nearest extremity of the nucleus (Figure S3A, Supporting Information). The progression of the infection was illustrated by observation that at 4 hours, most bacteria were found within the mucosa (Fig. 2B). To quantify this, we counted the number of luminal and mucosal bacteria in mucus foci (2 h) and in tissue foci at 2–4 h (Fig. 2C). The data indicated a reduction of the number of luminal bacteria at the expense of mucosal bacteria dependent on the nature of the infection foci and the progression of the infection. As discussed in our previous study (Arena et al. 2015), bacterial density within tissue reached a maximum value at 4 h, which led to greater variability between animals at this time point. In agreement with this, we observed dense clusters of extracellular bacteria within the lamina propria of one of the animals infected for 4 h (Figure S3B, Supporting Information). Within these clusters, most bacteria were TSAR-negative and rpsMp-positive. This indicated that bacteria were not secreting in this context. The presence of this sub-population introduced higher variation in the TSAR signal of mucosal bacteria at 4 h. To quantify the T3SA activity, we measured the ratio of TSAR to rpsMp intensities for mucus-associated bacteria at 2 h and mucosa-associated bacteria at 2–4 h. The increase in the TSAR:rpsMp ratio was significant for mucosal bacteria compared to mucus-associated bacteria (P < 0.0001). Although we measured a decrease in the TSAR:rpsMp ratio for mucosal bacteria between 2 and 4 h, the variation was statistically insignificant (Fig. 2D). The migration of bacteria from the apical to the basolateral pole depends on IcsA-mediated cytosolic movement (Arena et al. 2015), which requires the prior rupture of the vacuole. In agreement with this observation, aggregation of galectin-3, a marker of vacuole rupture (Dupont et al. 2009; Paz et al. 2010), was observed in the vicinity of bacteria located on the apical pole of infected colonocytes (Fig. 2E). This suggested that these bacteria had undergone vacuole lysis at that location. In summary, mucosa-associated bacteria showed stronger secretion activity than mucus-associated bacteria. Furthermore, vacuole lysis, an important feature of cellular infection observed in cell assay systems, was reproduced in the Guinea pig model. Taken together, these observations were signs of the progression of the infection between 2 and 4 h, thus demonstrating that S. flexneri established a productive intracellular/intercellular life cycle in Guinea pigs colon.
Figure 2.
In the early phase of invasion, secreting S. flexneri are associated with colonocytes. Colonic tissue sections infected or not with WT Shigella flexneri harboring pTSAR1.1 and labeled with phalloidin and DAPI were imaged using confocal microscopy. A micrograph overlay of infected tissue section at 2 h post-challenge (left panel). The right panels represent the indicated channel from the boxed area (A). A micrograph overlay of infected tissue section at 4 h post-challenge (left panel). The right panels represent the indicated channel from the boxed area (B). Count of luminal and mucosal bacteria in mucus-rich (white) and tissue foci both collected from animals infected for 2 h (light gray) and 4 h (dark grey); one way ANOVA with Bonferroni's post-tests were performed, P < 0.0001 (C). Comparison of the ratio of TSAR/rpsMp fluorescence intensity for bacteria found in the mucus at 2 h post-challenge (n = 392) or in the mucosa at 2 h (n = 103) or 4 h (n = 234) post-challenge. One way Anova with Bonferroni's post-tests were performed, P < 0.0001 (D). Micrograph overlays of infected tissue and non-infected tissue sections labeled as indicated above and with anti-galectin-3 antibody at 4 h post-challenge. The right panel show the galectin-3 channel (E). Sections are positioned with the lumen on top. Micrographs shown here together with those in Figure S3 (Supporting Information) are representative of infection foci observed in two animals.
At 8 h post-challenge, tissue damages, such as large crevices and partial effacement of the mucosa, were visible (Figure S1, Supporting Information). Interestingly, bacteria penetration into the lamina propria peaked between 6 and 8 h (Arena et al. 2015). We hypothesized that this context would favor interactions between Shigella and phagocytes. To test this, macrophages and neutrophils were, respectively, identified using the CD68 marker and MUB-Cy5 binding within tissue sections labeled with fluorescent phalloidin and the total bacterial population was revealed using an antibody recognizing the LPS of Shigella (Figure S4 and 3A, Supporting Information). Analyzes of the micrographs by confocal microscopy revealed that bacteria at 8 h were sometimes found inside colonocytes recognizable by their brush border or in cells that seemed to be extruding from the mucosal surface (Figure S4, Supporting Information). When in the lamina propria, bacteria often formed small, intracellular clusters located in the vicinity of disrupted crypts (Figure S4, Supporting Information). We also found that macrophages in less damaged sections were found in the upper part of the crypt (Figure S4A and 3A, Supporting Information), while the neutrophils were rather found in the lower part of the crypt. In damaged sections where the epithelium structure is disrupted, both cell types were more evenly distributed across the lamina propria, which may at least in part be explained by the disruption of the mucosa and the higher density of neutrophils (Figure S4B, Supporting Information). We observed the presence of bacteria in both cell types (Figure S4 and 3A, Supporting Information). Indeed, approximately 13% and 12% of the bacterial population was colocalized with neutrophils or macrophages, respectively (Fig. 3B). Some cells contained several bacteria that displayed high and homogeneous secretion activity according to the TSAR signal (Fig. 3C). Their location in the upper part of the lamina propria and the shape of their nucleus suggested that they were macrophages. To confirm the identity of these cells and the subcellular localization of internalized bacteria, we used TEM analyzes of agarose-embedded tissue sections (Fig. 3D and Figure S5, Supporting Information). Cell anatomical characteristics, such as ruffled plasma membrane, the kidney-like shape of their nucleus, the presence of vacuoles containing membrane swirls reminiscent of lysosomes or multi-vesicular bodies, and the absence of granule-containing vesicles usually encountered in neutrophils, supported the notion that these host cells were macrophages (Deane 1964; Takeuchi et al. 1965). It is noteworthy that bacteria were mostly found in vacuoles within these cells, while they were found in the cytosol or more rarely in vacuoles within neighboring epithelial cells (Fig. 3D and Figure S5, Supporting Information) (Arena et al. 2015).
Figure 3.
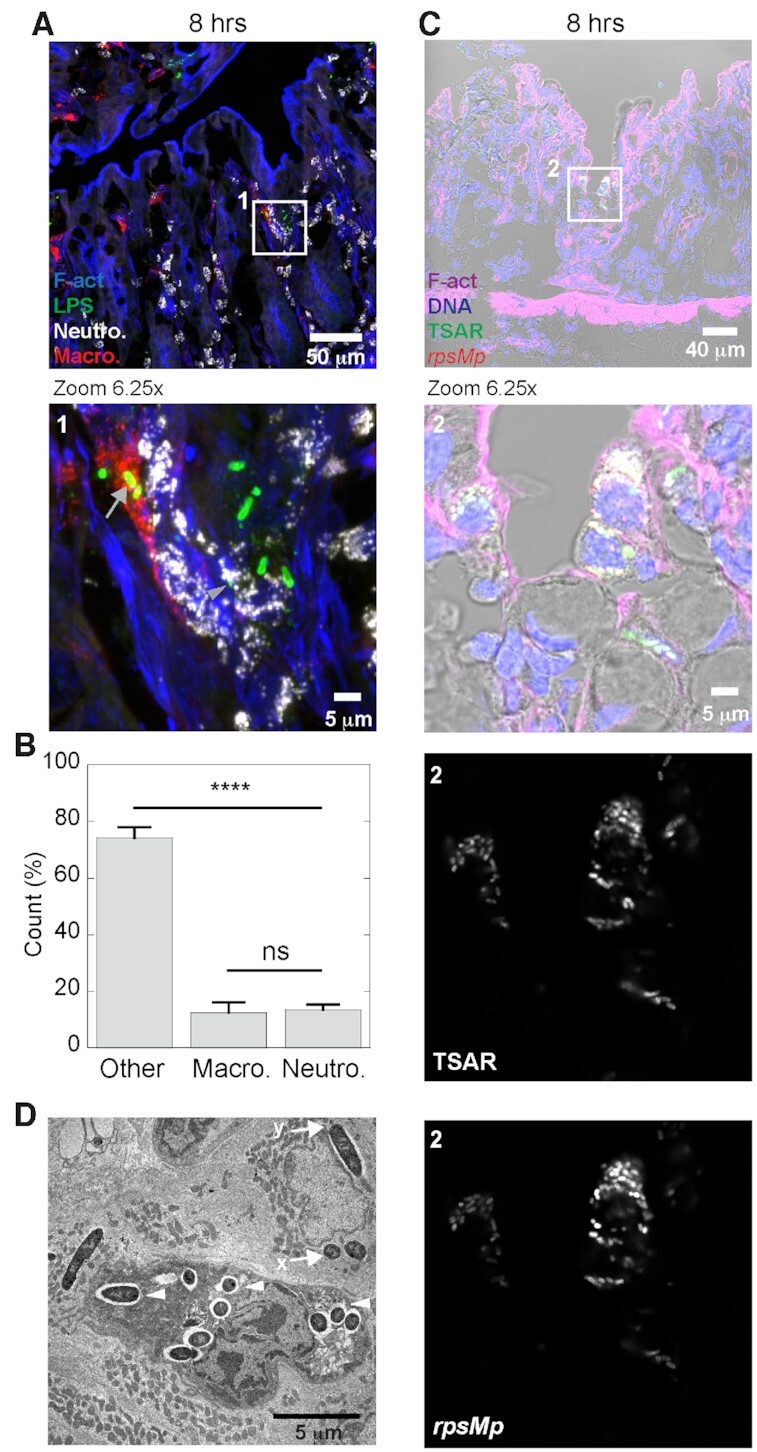
At 8 h post-challenge, secreting S. flexneri have invaded the lamina propria. Colonic tissue sections infected with WT Shigella flexneri at 8-h post-challenge were labeled with phalloidin, MUB (neutrophils), anti-LPS antibodies (S. flexneri) and anti-CD68 antibodies (macrophages), and imaged by confocal microscopy. A micrograph overlay of a maximal intensity projection of 15 z-stacks. The boxed area (1) is represented in the lower panel with a 6.25-fold magnification. Gray arrows and arrowhead show bacteria inside a macrophage or a neutrophil, respectively (A). This graph shows the fraction of bacteria (n = 760) that are associated with macrophages, neutrophils or located elsewhere in the lamina propria. Data were collected from 18 infection foci found in two animals. Error bars are SEM, and one-way Anova with Bonferroni's post-tests were performed, ****: P < 0.0001 (B). Colonic tissue section infected with WT Shigella flexneri harboring pTSAR1.1 and labeled with phalloidin and DAPI were imaged using confocal microscopy. A micrograph overlay of clusters of actively secreting bacteria present within lamina propria host cells at 8 h post-challenge. The boxed area (2) is represented in the lower panel with a 6.25-fold magnification. Bottom panels represent the indicated channel from the boxed area (C). Transmission electron microscopy images of host cells containing intracellular bacteria. Macrophages and enterocytes are respectively identified by the letter M and E. Arrowheads indicate vacuole bacteria within a macrophage. Arrows labeled with an ‘x ‘or a ‘y’ show a cytosolic or a vacuolar bacterium within a neighboring enterocyte, respectively (D). Sections in the fluorescent micrographs are positioned with the lumen on top and are representative of infection foci observed in two animals. This TEM micrograph is from a single animal, and the brush border is oriented to the right (see Figure S5, Supporting Information).
The histologic data revealed massive cellular infiltration in the lamina propria of most tissue sections from 8 h post-challenge onward (Figure S1, Supporting Information). Surprisingly, our initial observations of micrographs of tissue sections at 12–24 h showed that TSAR-positive and rpsMp-positive bacteria had disappeared from the tissue similarly to what we previously observed with bacteria expressing the GFP from a constitutive promoter (Arena et al. 2015). This suggested that fitness of this bacterial population was diminished in a coordinated fashion. We therefore used an anti-LPS antibody to locate putatively dead bacteria (Fig. 4). Using this approach, we showed that bacteria were still present in the lamina propria at 12–24 h. Nevertheless, bacteria were concentrated in small and scarcely populated clusters, and their LPS labeling was punctuated and diffused, suggesting that the integrity of their outer membrane was severely compromised. Therefore, the reduction in the TSAR and rpsMp signals was caused by the killing of bacteria through the host immune response between 8 and 24 h post-challenge.
Figure 4.
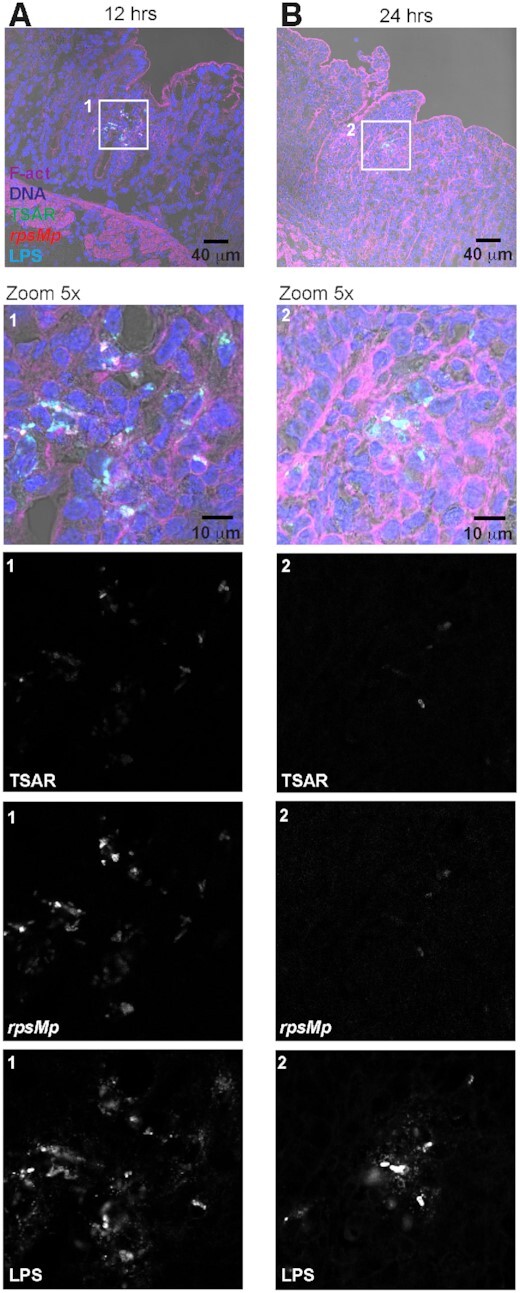
Shigella flexneri infection is controlled between 12 and 24 h post-challenge. Colonic tissue sections infected with WT Shigella flexneri harboring pTSAR1.1 and labeled with anti-lipopolysaccharide, phalloidin and DAPI were imaged using confocal microscopy. A micrograph overlay of a tissue section at 12 h post-challenge. The boxed area (1) is represented in the lower panel with a 5-fold magnification. Bottom panels represent the indicated channel from the boxed area (A). A micrograph overlay of a tissue section at 24 h post-challenge. The boxed area (2) is represented in the lower panel with a 5-fold magnification. Bottom panels represent indicated channels from the boxed area (B). Sections are positioned with the lumen on top and are representative of infection foci observed in two animals.
DISCUSSION
Herein, we have followed from onset to resolution the infection of colonic tissues by S. flexneri in the Guinea pig (Shim et al. 2007; Arena et al. 2015). Using the TSAR (Campbell-Valois et al. 2014), we mapped the bacteria inside the tissue, assessed bacterial viability at different stages of infection, and determined their T3SA activity. Our findings demonstrate the usefulness of rationally designed transcriptional reporters to understand pathogenesis (Campbell-Valois and Sansonetti 2014). We think this extensive mapping of the location of Shigella cells in parallel with the monitoring of the activity of their T3SA will facilitate future in vivo studies of shigellosis.
At 2 h post-challenge, we observed numerous clusters of luminal Shigella captured in WGA-positive mucus. These mucus embedded bacteria had low T3SA activity, suggesting that their capacity to reach this site did not require the action of T3SS effectors. Nonetheless, we cannot exclude that components of the T3SA acting as pathogen-associated molecular patterns being involved in this phenomenon. In addition, luminal WGA-positive mucus was not observed in non-infected animals. Since it is well accepted that specific fixation conditions must be used to preserve mucus (Sperandio et al. 2013), our formalin-based fixation method had most likely destroyed it in non-infected samples. Hence, the preservation of the mucus observed in luminal Shigella clusters suggests that it may have unique chemical properties, as previously suggested about mucus release observed during infection of rabbit ileal loops (Sperandio et al. 2013). Since crypt goblet cells in the vicinity of bacterial clusters had lower WGA-labeling, we reasoned that they had released the WGA-positive mucus. Goblet cells can react to microbial or pharmacological challenge by eliciting compound exocytosis (Johansson, Sjövall and Hansson 2013). The neurotransmitter acetylcholine stimulates crypt goblet cells to release their mucus (Specian 1980). Alternatively, TLR2/4/5 ligands were shown to induce mucus release by sentinel goblet cells that are located in the upper portion of the crypt (Birchenough et al. 2016). The massive coordinated release of mucus by goblet cells of crypts is therefore akin to that observed during acetylcholine treatment, as it is not limited to the upper part of the crypt. Therefore, we speculate that the release of WGA-positive mucus is due to the specific interaction of S. flexneri with areas near the entrance of crypts that are preferentially targeted in the early stages of infection (Arena et al. 2015). Further studies will be required to understand the mechanism of mucus release in the early phase of infection.
Shigella, like many other enteropathogens, is thought to invade the colonic mucosa using microfold cells (M cells) as portals (Sansonetti and Phalipon 1999). Seminal studies in Caco-2 cells also suggested that entry into colonocytes is poor from the apical pole due to the brush border, but much more favorable on the basolateral pole (Mounier et al. 1992), an entry point that may be accessed from the lamina propria through the M-cell portal (Cossart and Sansonetti 2004; Kang et al. 2018). By contrast, we rather observed that S. flexneri invades the Guinea pig colon through the apical pole of colonocytes (this work and (Arena et al. 2015)). Our attempts to stain the colon of Guinea pigs with various lectins and vimentin did not reveal the presence of M cell rich areas or of lymphoid follicles (data not shown) (Arena et al. 2015), such as those reported in humans (Fujimura, Hosobe and Kihara 1992; Mowat and Agace 2014). In non-infected murine colonic tissue, follicle M cells are also barely detectable (Bennett et al. 2016). In addition, Guinea pigs used for these experiments were young females that may have not fully developed their colonic lymphoid follicles. Although our data does not allow to discriminate between these possibilities, it is tantalizing to speculate that the large inoculum required to obtain mucosal invasion is due to the low number of M cells within distal colonic tissues of Guinea pigs. Interestingly, the number of CFU recovered from differentiated Caco-2 cells grown on a chip were as high as four orders of magnitudes larger if stretching forces mimicking peristalsis were applied (Grassart et al. 2019). Since in that model, entry was mainly through the apical pole, it suggests that peristalsis may facilitate entry through that route in vivo as well. What makes the cells invade specific regions of the mucosa is unknown. The capture of luminal Shigella in the mucus might favor entry into adjacent colonocytes. In addition, the secretion of effectors IpaJ and VirA in these cells may disrupt their polarity, thereby favoring entry of additional bacteria within the same area (Ferrari et al. 2019). Nevertheless, mucosal invasion through the brush border of colonocytes has not been reported in clinical cases to our knowledge.
Once bacteria had invaded the mucosa, we observed that their T3SA displayed high and homogenous activity. The only notable exception was observed in highly infected tissue sections where dense clusters of extracellular bacterial had formed within the mucosa; bacteria in those cases had no detectable T3SA activity. By contrast, in tissue culture cells, the TSAR signal of intracellular bacteria was heterogenous: a bacterial subpopulation displayed a high TSAR signal, while another one displayed a low TSAR signal. This was interpreted as evidence that T3SA of cytosolic bacteria might remain in an off state for long periods of time (Campbell-Valois et al. 2014; 2015). Several anatomical differences between the in vivo and in vitro experimental systems might explain this discrepancy. For example, colonocytes are small and differentiated, while Caco-2/TC7 are larger and undifferentiated colonic adenocarcinoma cells. Second, bacterial movement in tissue culture cells is constrained by their plastic substratum, while colonocytes in contact with the lamina propria have no such limitations. These factors might facilitate cell-to-cell spread in vivo, which in turn should increase and homogenize the T3SA activity across the bacterial population. Given that the half-life of the fluorescent signal emitted by the GFP of the TSAR is approximately 30 min in the absence of secretion under normal growth conditions (Campbell-Valois et al. 2014), we reasoned that variation in the TSAR signal would not be detectable across a heterogenous population if bacteria performed cell-to-cell spread on average every 60 min or so.
Shigella flexneri induces pyroptosis in murine cultured macrophages through activation of the IL-1β converting enzyme (Zychlinsky et al. 1994a; Cunha and Zamboni 2013). In this model, S. flexneri rapidly lyses the phagocytic vacuole and releases the translocator IpaB, potentially other effectors, and pathogen-associated molecular patterns that induce the death of macrophages within 2 h post-challenge (Zychlinsky et al. 1994b; Navarre and Zychlinsky 2000; Suzuki et al. 2014). Neutrophils infected by Shigella in vitro undergo necrotic cell death through a process that also requires IpaB and the T3SS (François et al. 2000). In starved Guinea pigs infected by the oral ingestion of S. flexneri, TEM revealed that infiltrating neutrophils possessed a high number of dark granule-containing vesicles (Takeuchi et al. 1965). Here, we found lamina propria phagocytes that were devoid of dark vesicles and rather possessed hallmarks of macrophages (Deane 1964). TEM revealed that some macrophages found in the lamina propria contained a high proportion of vacuolar bacteria and appeared healthy. Furthermore, dense clusters of secreting bacteria within lamina propria cells matching the location and properties of macrophages were indeed observed at 8 h post-challenge. As T3SA activity reached maximal levels upon host membrane contacts (Campbell-Valois et al. 2014), we reasoned that prolonged interaction of Shigella with vacuole membranes in resident macrophages might lead to the sustained secretion activity observed. Thus, resident macrophages seem to establish a relationship with S. flexneri that differs from in vitro macrophages. This could be due to the intrinsic properties of the resident macrophages, a protective effect of the lamina propria on them, a Shigella-mediated process specifically towards resident macrophages, or a combination of these. Nevertheless, we cannot exclude that these secreting bacteria were killed in the lamina propria shortly before their phagocytosis by macrophages. Further work exploiting fluorescence-assisted cell sorting using Guinea pig specific reagents will be needed to tease these hypotheses apart.
At 12 and 24 h post-challenge, the bacteria within the tissue were in reduced number and appeared dead, as suggested by the loss of TSAR and rpsMp signals. These data suggested that the bacteria had been metabolically inactive for a while or were located in such a harsh environment that the GFP and CFP chromophore had been destroyed. This was supported by the observation that the LPS staining used to identify the remaining mucosal bacteria was punctuated, as if the integrity of their outer membrane had been compromised. It is noteworthy that this correlated with the peak of neutrophil recruitment to the mucosa (this work and (Singer and Sansonetti 2004)). Taken together, these data suggest that the clearance of the bacteria is rapid in the Guinea pig colon, similarly to what is reported in humans (Kotloff et al. 2018). This argues for the importance of studying the phenotype of Shigella strains before 12 h post-challenge if one wants to study bacterial colonization as pathogenic processes taking place beyond 24 hours are likely due to the host response. We cannot exclude the existence of Shigella persister cells at this stage of the infection (Helaine et al. 2010), although we would argue that they should emit a fluorescent signal, suggesting that if they exist at all, they are in low numbers. Further experiments are, however, required to evaluate thoroughly the existence of persisters in this context.
As illustrated here, genetically encoded reporters such as the TSAR allow to assess one or several aspects of bacterial physiology within complex environments such as host tissues (Campbell-Valois and Sansonetti 2014). In combination with relevant host staining, they may help build a holistic view of host–pathogen interactions. The extension of this concept to other host–pathogen systems and its exploitation to extract data about in vivo adaptation mechanisms of microbes are promising research avenues.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Céline Mulet for help with the hematoxylin and eosin staining and micrographs acquisition.
FUNDING
This work was supported by the European Research Council and Howard Hugues Medical Institute (PS) and Canadian Institute of Health Research Project Grant 159517 (FXCV).
Conflict of interest. None declared.
REFERENCES
- Anderson M, Sansonetti PJ, Marteyn BS. Shigella diversity and changing landscape: insights for the twenty-first century. Front Cell Infect Microbiol. 2016;6:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MC, Chaze T, Coïc Y-Met al.. MUB40 binds to lactoferrin and stands as a specific neutrophil marker. Cell Chem Biol. 2018;25:483–9. [DOI] [PubMed] [Google Scholar]
- Anderson MC, Vonaesch P, Saffarian Aet al.. Shigella sonnei encodes a functional T6SS used for interbacterial competition and niche occupancy. Cell Host Microbe. 2017;21:769–776.e3. [DOI] [PubMed] [Google Scholar]
- Arena ET, Campbell-Valois F-X, Tinevez J-Yet al.. Bioimage analysis of Shigella infection reveals targeting of colonic crypts. Proc Natl Acad Sci USA. 2015;112:E3282–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett KM, Parnell EA, Sanscartier Cet al.. Induction of colonic m cells during intestinal inflammation. Am J Pathol. 2016;186:1166–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardini ML, Mounier J, d'Hauteville Het al.. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc Nat Acad Sci. 1989;86:3867–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchenough GMH, Nyström EEL, Johansson MEVet al.. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science. 2016;352:1535–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongrand C, Sansonetti PJ, Parsot C. Characterization of the promoter, MxiE box and 5' UTR of genes controlled by the activity of the type III secretion apparatus in Shigella flexneri. Mantis NJ (ed). PLoS ONE. 2012;7:e32862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchrieser C, Glaser P, Rusniok Cet al.. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol Microbiol. 2000;38:760–71. [DOI] [PubMed] [Google Scholar]
- Campbell-Valois F-X, Pontier SM. Implications of spatiotemporal regulation of shigella flexneri type three secretion activity on effector functions: think globally, act locally. Front Cell Infect Microbiol. 2016;6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell-Valois F-X, Sachse M, Sansonetti PJet al.. Escape of actively secreting shigella flexnerifrom atg8/lc3-positive vacuoles formed during cell-to-cell spread is facilitated by icsb and virA. mBio. 2015;6:e02567–14-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell-Valois F-X, Sansonetti PJ. Tracking bacterial pathogens with genetically-encoded reporters. FEBS Lett. 2014;588:2428–36. [DOI] [PubMed] [Google Scholar]
- Campbell-Valois F-X, Schnupf P, Nigro Get al.. A fluorescent reporter reveals on/off regulation of the shigella type iii secretion apparatus during entry and cell-to-cell spread. Cell Host and Microbe. 2014;15:177–89. [DOI] [PubMed] [Google Scholar]
- Campbell-Valois F-X, Schnupf P, Sansonetti PJ. Design of a transcription-based secretion activity reporter (TSAR) of the type III secretion apparatus of Shigella flexneri and Uses Thereof. Bio-protocol. 2014;4:e1270. [Google Scholar]
- Cossart P, Sansonetti PJ. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science. 2004;304:242–8. [DOI] [PubMed] [Google Scholar]
- Cunha LD, Zamboni DS. Subversion of inflammasome activation and pyroptosis by pathogenic bacteria. Front Cell Infect Microbiol. 2013;3:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane HW. Some electron microscopic observations on the lamina propria of the gut, with comments on the close association of macrophages, plasma cells, and eosinophils. The Anatomical Record. 1964;149:453–74. [DOI] [PubMed] [Google Scholar]
- Dupont N, Lacas-Gervais S, Bertout Jet al.. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe. 2009;6:137–49. [DOI] [PubMed] [Google Scholar]
- Ferrari ML, Malardé V, Grassart Aet al.. Shigella promotes major alteration of gut epithelial physiology and tissue invasion by shutting off host intracellular transport. Proc Natl Acad Sci USA. 2019;116:13582–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- François M, Le Cabec V, Dupont MAet al.. Induction of necrosis in human neutrophils by Shigella flexneri requires type III secretion, IpaB and IpaC invasins, and actin polymerization. Infection and Immunity. 2000;68:1289–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura Y, Hosobe M, Kihara T. Ultrastructural study of m cells from colonic lymphoid nodules obtained by colonoscopic biopsy. Dig Dis Sci. 1992;37:1089–98. [DOI] [PubMed] [Google Scholar]
- Grassart A, Malardé V, Gobba Set al.. Bioengineered human organ-on-chip reveals intestinal microenvironment and mechanical forces impacting Shigella infection. Cell Host Microbe. 2019;26:435–44..e4. [DOI] [PubMed] [Google Scholar]
- Helaine S, Thompson JA, Watson KGet al.. Dynamics of intracellular bacterial replication at the single cell level. Proc Natl Acad Sci USA. 2010;107:3746–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Morado DR, Margolin Wet al.. Visualization of the type III secretion sorting platform of Shigella flexneri. Proc Natl Acad Sci USA. 2015;112:1047–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MEV, Sjövall H, Hansson GC. The gastrointestinal mucus system in health and disease. Nature Publishing Group. 2013;10:352–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang E, Crouse A, Chevallier Let al.. Enterobacteria and host resistance to infection. Mamm Genome. 2018;29:558–76. [DOI] [PubMed] [Google Scholar]
- Kotloff KL, Riddle MS, Platts-Mills JAet al.. Shigellosis. The Lancet. 2018;391:801–12. [DOI] [PubMed] [Google Scholar]
- Marteyn B, West NP, Browning DFet al.. Modulation of Shigella virulence in response to available oxygen in vivo. Science2010;465:355–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounier J, Vasselon T, Hellio Ret al.. Shigella flexneri enters human colonic Caco-2 epithelial cells through the basolateral pole. Infection and Immunity. 1992;60:237–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol. 2014;14:667–85. [DOI] [PubMed] [Google Scholar]
- Navarre WW, Zychlinsky A. Pathogen-induced apoptosis of macrophages: a common end for different pathogenic strategies. Cellular Microbiology. 2000;2:265–73. [DOI] [PubMed] [Google Scholar]
- Paz I, Sachse M, Dupont Net al.. Galectin-3, a marker for vacuole lysis by invasive pathogens. Cell Microbiol. 2010;12:530–44. [DOI] [PubMed] [Google Scholar]
- Pinaud L, Ferrari ML, Friedman Ret al.. Identification of novel substrates of Shigella T3SA through analysis of its virulence plasmid-encoded secretome. Mantis NJ (ed). PLoS ONE. 2017;12:e0186920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansonetti PJ, Kopecko DJ, Formal SB. Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect Immun. 1982;35:852–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansonetti PJ, Phalipon A. M cells as ports of entry for enteroinvasive pathogens: mechanisms of interaction, consequences for the disease process. Semin Immunol. 1999;11:193–203. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise Eet al.. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim DH, Suzuki T, Chang SYet al.. New animal model of shigellosis in the Guinea pig: its usefulness for protective efficacy studies. J Immunol. 2007;178:2476–82. [DOI] [PubMed] [Google Scholar]
- Singer M, Sansonetti PJ. IL-8 is a key chemokine regulating neutrophil recruitment in a new mouse model of Shigella-induced colitis. J Immunol. 2004;173:4197–206. [DOI] [PubMed] [Google Scholar]
- Specian RD. Mechanism of rapid mucus secretion in goblet cells stimulated by acetylcholine. J Cell Biol. 1980;85:626–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio B, Fischer N, Chevalier-Curt MJet al.. Virulent shigella flexneri affects secretion, expression, and glycosylation of gel-forming mucins in mucus-producing cells. Baumler AJ (ed). Infect Immun. 2013;81:3632–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Mimuro H, Kim Met al.. Shigella IpaH7.8 E3 ubiquitin ligase targets glomulin and activates inflammasomes to demolish macrophages. Proc Natl Acad Sci USA. 2014;111:E4254–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi A, Sprinz H, Labrec EHet al.. An electron microscopic study of the response of the intestinal mucosa to bacterial invasion. Am J Pathol. 1965;47:1011–44. [PMC free article] [PubMed] [Google Scholar]
- Tinevez J-Y, Arena ET, Anderson Met al.. Shigella-mediated oxygen depletion is essential for intestinal mucosa colonization. Nat Microbiol. 2019;4: 2001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zychlinsky A, Fitting C, Cavaillon JMet al.. Interleukin 1 is released by murine macrophages during apoptosis induced by Shigella flexneri. J Clin Invest. 1994a;94:1328–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zychlinsky A, Kenny B, Menard Ret al.. IpaB mediates macrophage apoptosis induced by Shigella flexneri. Mol Microbiol. 1994b;11:619–27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.