Abstract
Illuminating the role of specific gene duplications within the human lineage can provide insights into human-specific adaptations. The so-called human core duplicon gene families have received particular attention in this respect, due to special features, such as expansion along single chromosomes, newly acquired protein domains and signatures of positive selection. Here, we summarize the data available for 10 such families and include some new analyses. A picture emerges that suggests broad functions for these protein families, possibly through modification of core cellular pathways. Still, more dedicated studies are required to elucidate the function of core-duplicons gene families and how they have shaped adaptations and evolution of humans.
Keywords: duplication, duplicons, gene family, human, adaptation, copy number variation
Introduction
The human genome harbors a number of rapidly evolving gene families that have been subjected to a combination of structural reorganization and bursts of segmental duplications between one to several hundred kilobases. Approximately 400 blocks of the human genome have been identified as having undergone multiple duplications during hominoid evolution. Overall, segmental duplications comprise approximately 5% of the human genome [1–3]. A detailed analysis of these segmentally duplicated regions has shown that subsets are formed around ‘core’ or ‘seed’ duplicons that are shared between all copies of the respective gene family [4–6]. Figure 1 shows the duplication structures of the Morpheus (NPIP) gene family as an example, which will be discussed in further detail below.
Figure 1.
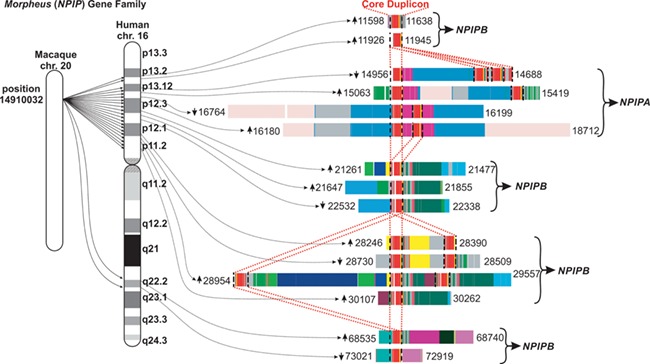
The Morpheus (NPIP) gene family as an example for human core duplicons. Figure adapted from [4, 7]. The individual members of the Morpheus core duplicons are ordered according to location on chromosome 16 (left). The mosaic structure of the complex duplication blocks is depicted on the right. The core duplicon part, which is shared by all duplication blocks, is marked by the red broken lines. Numbers next to the fragments refer to the start and end positions on the chromosome (in kb), and arrows indicate the relative orientation. Different colors refer to distinct duplication blocks, which are predicted based on reciprocal comparisons of each human subunit and its flanking sequence to outgroup mammalian genomes [4].
We focus here on 10 core duplicon gene families, which share a number of other features. (i) Their duplicates are generally confined to a single chromosome, partly in tandem but also dispersed along the chromosome. (ii) They have variable copy numbers in human populations and include some of the most variable human CNV genes (e.g. SPATA31, Morpheus (NPIP) and LRRC37A) [8–11]. (iii) Almost all of these genes show ubiquitous or at least broad patterns of expression, while their ancestral progenitor genes often exhibit tissue-specific expression, mostly in the testis [4]. Intriguingly, at least half of these core duplicon gene families (including Morpheus (NPIP), SPATA31, NBPF11, RGPD, GOLGA, PMS2P and TRIM51) evolved under positive selection (Table 1), making them among the fastest evolving genes in humans [12–17]. Hence, both the human lineage-specific expansion and the patterns of positive selection suggest that these gene families have played a direct role in the phenotypic evolution of humans.
Table 1.
Signatures of positive selection in human core duplicons
| Chr. | Name | Alternative name(s) | Ancestral gene (mouse) | Signatures of positive selection | Selection test | Literature |
|---|---|---|---|---|---|---|
| 1 | Olduvai protein domain family | Neuroblastoma breakpoint gene family, NBPF | Pde4dip | Yes | PAML, dN/dS, The likelihood ratio test, Ka/Ks, Nei-Gojobori | [13,16] |
| 2 | RGPD | RANBP2-like gene family | Ranbp2 | Yes | PAML, dN/dS, The likelihood ratio test, Ka/Ks, Nei-Gojobori | [13,15] |
| 5 | SMA-GUSBP | SMA | Gusbp2 | None described | ||
| 7 | PMS2P | PMS2 gene family | Pms2 | Yes | PAML, dN/dS, The likelihood ratio test, | [12] |
| 9 | SPATA31 | FAM75A | Spata31 | Yes | PAML, dN/dS, The likelihood ratio test, | [13] |
| 11 | TRIM51 | SPRYD5 | no homolog | Yes | PAML, dN/dS, The likelihood ratio test, | [17] |
| 15 | GOLGA8 | GOLGA, golgin | Golga2 | Yes | PAML, dN/dS, The likelihood ratio test, | [12,13] |
| 16 | Morpheus | NPIP | no homolog | Yes | PAML, dN/dS, The likelihood ratio test, Ka/Ks, Nei-Gojobori | [13,14] |
| 17 | TBC1D3 | - | Usp6nl | None described | ||
| 17 | LRRC37 | - | Lrrc37a1 | None described |
Since the initial report of human core duplicons [4], most of the genes belonging to duplicons were evolutionarily and structurally characterized. However, only three (NBPF, TBC1D3 and SPATA31) were studied functionally in more detail. Here, we provide an overview on our current knowledge on the evolution and function of gene families that are parts of the human core duplicons. We describe these gene families according to their order along the human chromosomes. The general overview is provided in Figure 2.
Figure 2.
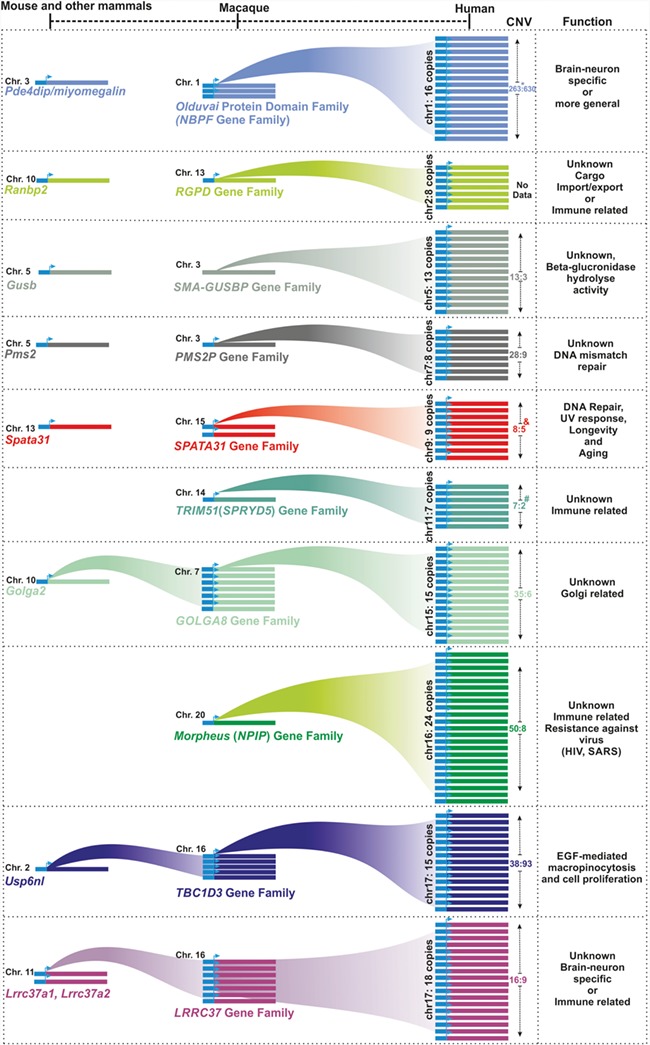
Overview of human core duplicon gene families. The figure depicts the segmentally duplicated copies of core duplicon gene families from human, macaque and mouse and includes other mammals. Functional properties of Olduvai, SPATA31 and TBC1D3, which have been studied in detail, and possible/predicted functions of other core duplicon gene families are included. CNV (median:variance) was extracted from a previous study [10]. * indicates CNV estimate based on number of Olduvai domains. # indicates CNV information for TRIM51 genes was extracted from a previous study [17], and & indicates CNV for SPATA31 genes was extracted from a previous study [18].
Chromosome 1: Olduvai protein domain (NBPF gene) family
Evolution and comparative genomics
The initial name ‘Neuroblastoma Breakpoint gene Family’ (NBPF) was given because the first identified member of the family was found to be deleted in an individual with neuroblastoma [19]. NBPF gene family members include variable numbers of tandemly repeated DUF1220 domains within their coding sequences. Since the name DUF1220 was initially a working designation assigned by the Pfam database curators, Sikela and van Roy [20] have proposed to rename the domain Olduvai. The different NBPF core duplicon genes are distributed along chromosome 1 in 16 copies, of which 6 are in tandem and 10 are dispersed [21]. The macaque genome has three copies of NBPF genes, while mice and other mammals have no clear orthologs. A possible ancestral gene is PDE4DIP, which includes a single Olduvai domain [22]. The Olduvai domain copy number has particularly expanded in humans. It increased from a single domain in mouse to 30–35 copies in new and old world monkeys and up to 300 domains in humans [22]. Based on sequence similarity, the domain structure of Olduvai can be divided into six primary subtypes including CON 1, 2 and 3 and HLS 1, 2 and 3 [16, 22, 23]. The comparative analysis of Olduvai-domain-containing NBPF genes in primates has shown that NBPF genes evolve under strong positive selection [13, 16].
Expression and subcellular localization
NBPF genes are ubiquitously expressed in all human tissues and exhibit elevated expression in the testis. The promoter region of NBPF1 genes is derived from the unrelated gene EVI5 [22, 24]. Over-expressed, myc-tagged NPBPF1 protein in human breast carcinoma cells (MCF7) was shown to be primarily localized to the cytoplasm [23].
Function
Despite its broad expression, the functional analysis of this gene family has focused on brain- and/or neuron-specific functions. This was triggered by the observation that the 1q21.1 chromosome region in humans, which is associated with brain disorders such as micro- or macrocephaly, autism and schizophrenia, includes a number of the NPBPF1 genes [25]. Further, a correlation was found between Olduvai copy number and brain size in humans. This correlation was most prominent in affected patients, but copy number was also associated with gray-matter volume in healthy controls [26]. Over-expression of Olduvai sequences in neural stem cells promotes proliferation [27–29]. Further studies also suggested a correlation between Olduvai copy number and schizophrenia risk and severity [30] as well as with IQ and total mathematical aptitude scores [31], although the breadth of this latter study is limited. While these results suggest a particularly interesting function of NBPF genes in human brain evolution with implications for the etiology of brain diseases [21], more studies are required to support this notion. Notably, copy number variations (CNVs) and deletions in the 1q21.1 region are also correlated with heart [32, 33] and kidney [34] diseases. Given that NBPF11 genes are broadly expressed, they could have a more general function. In a yeast two-hybrid system, NBPF11 was shown to interact with Chibby, which is a negative regulator of the Wnt signaling pathway [35]. There are currently no data supporting a possible specific molecular function for Olduvai domains in humans. In mouse, targeted deletion of the Olduvai domain in the Pde4dip gene resulted in not only significantly reduced fecundity and hyperactivity of mice but also in physiological changes and liver function-related effects [36].
Correlation with brain size
As discussed above, there is a strong emphasis in the literature that NBPF/Olduvai copy numbers are correlated with increasing brain size in primates and humans. However, given that the pattern of gene family expansion also holds for the other core duplicon gene families discussed here, we have re-assessed whether the same correlation is more broadly apparent. Using the brain volume measures previously provided [29] and the number of currently annotated gene copies for the various families in different genomes, we found that all these gene families show a significant correlation. In contrast, there is only a partial correlation with body mass (Table 2). Hence, one can either conclude that all human core duplicon gene families influence brain size or the correlation is incidental because these gene families are of increased interest due to their special expansion in humans.
Table 2.
Linear regression analysis of human core duplicon gene families with brain volumes and body mass across primates
| NBPF / Olduvai | RGPD | SMA-GUSB | PMS2P | SPATA31 | TRIM51 (SPRYD5) | GOLGA | MORPHEUS | TBC1D3 | LRRC37 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Brain mass (mg) | 0.90** | 0.91** | 0.90** | 0.38* | 0.72** | 0.87** | 0.68** | 0.66** | 0.82** | 0.83** |
| Brain volume (mm3) | 0.90** | 0.91** | 0.90** | 0.38* | 0.72** | 0.87** | 0.68** | 0.66** | 0.83** | 0.83** |
| Neocortex volume (mm3) | 0.92** | 0.90** | 0.92** | 0.37* | 0.72** | 0.86** | 0.68** | 0.65** | 0.81** | 0.83** |
| Cerebellum volume (mm3) | 0.83** | 0.91** | 0.81** | 0.36* | 0.72** | 0.85** | 0.64** | 0.64** | 0.81** | 0.77** |
| Body mass (g) | 0.23ns | 0.57** | 0.24ns | 0.22ns | 0.38* | 0.43* | 0.28 ns | 0.35* | 0.44* | 0.27 ns |
* P < 0.05.
** P < 0.001.
ns P > 0.05.
Chromosome 2: RANBP2 (RGPD) gene family
Evolution and comparative genomics
RGPD genes (RanBP2-like, GRIP domain-containing Proteins) are derived from the RANBP2 gene. RANBP2 (NUP388) is a Ran-binding protein that was shown to interact with the nuclear pore complex. The RGPD gene family has expanded to eight dispersed copies on human chromosome 2 through segmental duplication [15]. Similar to other core duplicon gene families, RGPD genes are rapidly evolving. Different exons of RGPD genes were shown to evolve under positive selection [15]. The macaque genome, as well as the mouse and other mammalian genomes, does not contain duplicates, suggesting that expansion occurred within the great apes after the separation from old world monkeys.
Expression and subcellular localization
RGPD genes are ubiquitously expressed in human tissue with elevated expression in the testis [15]. Over-expression of GFP-fused RGPD5-7 revealed its localization to the cytoplasm around the nuclear envelope [15, 37].
Function
The RANBP2 protein encoded by the progenitor gene is primarily localized within the periphery of the nuclear envelope and is thought to be required for cargo import and export [37]. Hence, the RGPD gene family members may be modifiers of this function. Interestingly, RANBP2 was also shown to be involved in resistance against Simian Immunodeficiency Virus [38]. It is thus possible that the expansion of RGPD genes is the result of an arms race between virus evolution and host resistance acquisition. The Ranbp2 knockout in mice is homozygous lethal.
Chromosome 5: SMA (GUSBP) gene family
Evolution and comparative genomics
The repetitive nature of the spinal muscular atrophy (SMA) genes was first described [39] in the context of searching for candidate genes for SMA. However, SMA is caused by mutations in the duplicated copies of the SMN1 and SMN2 genes, which are located at chromosome 5q13.3 (reviewed in detail in [40]). SMA core duplicon copies are located in close proximity both upstream and downstream (within approximately 50 kb) of the SMN2 gene. Therefore, to avoid confusion with the disease-causing genes, we will use the designation SMA-GUSBP to represent the SMA gene family. SMA-GUSBP genes are organized in 2 tandem and 10 dispersed copies along human chromosome 5; an additional copy is located on chromosome 6, and this is most slikely the ancestral copy. In the common ancestor of chimp and human, the ERV1/LTR12C retroviral element integrated upstream of SMA-GUSBP. In macaque, the GUSBP and ERV1/LTR12C elements are found in different chromosomal locations (Supplementary Fig. 1). The SMA-GUSBP gene encodes a beta-glucuronidase-like domain (Supplementary Fig. 2). No homologs of the SMA-GUSBP genes have been observed outside the great apes to date, i.e. it expanded specifically in the human lineage [10]. Therefore, SMA-GUSBP genes are the youngest expanded core duplicon family in humans.
Expression and subcellular localization
The human SMA-GUSBP mRNA transcripts include different splicing isoforms that extend more than 70 kb. The genes are broadly expressed, and the highest expression (15–20 fold) has been detected in the testis, thymus, brain and cerebellum (Supplementary Fig. 3A-B). Transiently over-expressed, C-terminally FLAG-tagged SMA-GUSBP proteins localize primarily in the vesicles of HeLa cells (Supplementary Fig. 3C).
Function
Although SMA-GUSBP genes were initially thought to be associated with SMA disease, there is no current evidence to support this, i.e. the close proximity of SMA-GUSBP genes to the SMN2 gene may not imply a functional connection. No direct functional analysis of SMA-GUSBP genes has been conducted so far.
Chromosome 7: PMS2P gene family
Evolution and comparative genomics
The PMS2P gene family is derived from the PMS2 gene, which encodes a homolog of the mutL mismatch repair gene from bacteria [41]. PMS2P genes duplicated to eight dispersed copies (PMS2P1–5, 7, 9 and 11) from the C-terminal region of the PMS2 gene located on human chromosome 7 [42]. The current marmoset and macaque genome assemblies do not include an intact PMS2P gene. However, the Orangutan genome includes three complete PMS2P copies. This suggests an expansion of PMS2P genes occurred within great apes. More work will be required to show whether they were lost in some primate genomes, or whether this is still an annotation problem. The PMS2P gene region is highly repetitive and contains multiple transposable elements (SINEs and LINEs), which make its annotation problematic.
Expression and subcellular localization
Similar to other core duplicons, PMS2P genes are expressed ubiquitously and have enhanced expression in the testis (as annotated in the NCBI description). No further data is available on the subcellular localization of these proteins.
Function
No functional analysis has been performed for PMS2P genes to date.
Chromosome 9: SPATA31 gene family
Evolution and comparative genomics
SPATA31 (formerly known as FAM75A) genes expanded from a single copy in mouse to two copies in macaque and at least nine copies on human chromosome 9 by segmental duplication. Seven of these duplicated SPATA31 gene segments are distributed on the long arm, and at least two are in tandem on the short arm. Two copies on the long arm are considered pseudogenes due to premature stop codons [18]. During the process of duplication within the primate lineage, the 5′-region of SPATA31 genes acquired an additional exon and additional protein domains, in particular a cryptochrome domain [18]. Similar to other core duplicon gene families, the SPATA31 promoter region became highly restructured before or during the duplication events. This restructuring includes the integration of a transposable element (LINE/L1, PA10-12) after the split between simians and prosimians [18].
Expression and subcellular localization
SPATA31 gene expression evolved from testis-specific expression in mice and macaques to ubiquitous expression in humans, but it still exhibits its highest expression in the testis [18]. Endogenous SPATA31 protein shows a dynamic localization between the cytoplasm and nucleus depending on fixation conditions and exposure to light [18]. In cell culture, SPATA31 protein re-localizes from the nucleolus to the nucleus upon UV exposure [18].
Function
SPATA31’s UV-exposure response, as well as its acquired protein domains, suggests a function in DNA damage repair. Functional analysis of cells with reduced SPATA31 copy number showed increased sensitivity to UV exposure [18]. Furthermore, over-expression of SPATA31A1 in epidermal cells leads to premature senescence [43]. Interestingly, long-lived individuals (>96 years old) have significantly fewer copies of SPATA31 genes on average compared to a control group. This observation suggests that the adaptive evolution of the SPATA31 gene family is an example of antagonistic pleiotropy; it provides a fitness benefit during the reproductive phase of life (better protection against UV-light damage), but it negatively influences overall life span possibly by causing more repair-induced mutations [43]. Knockout of the progenitor gene in mice leads to spermatogenesis defects and infertility [44], which is in line with its testis-specific expression in mice.
Chromosome 11: TRIM51 (SPRYD5) gene family
Evolution and comparative genomics
A first description of the evolution of TRIM51 genes is included in a general study of TRIM genes by Han et al. [17], who showed that TRIM51 genes are recently duplicated, hominoid-specific TRIM genes. Some of the TRIM51 genes (C1,C2) were found to have CNV in human individuals and to evolve under positive selection [17]. The human genome assembly includes seven full-length, duplicated copies of TRIM51 (SPRYD5) genes, at least five fragmented duplication blocks that are expanded mainly within the centromeric region of chromosome 11 and an additional copy on chromosome 2 (TRIM51JP). Comparisons of the available assembled genomes (rheMac8, ponAbe3 and hg38) in the UCSC genome browser and Synteny in Ensembl indicate that TRIM51 genes initially evolved by segmental duplication from TRIM51EP. There is a single full-length copy in the macaque genome, which shows high similarity to human TRIM51EP. However, more detailed work is required to resolve this.
Expression and subcellular localization
Low level TRIM51 expression was detected in the developing brain and testis (https://www.ebi.ac.uk/gxa/home). Although classified as TRIM genes, members of the TRIM51 gene family lack several important motifs and domains that are otherwise common to TRIM proteins (e.g. RING and B-Box 2) [45]. TRIM51 proteins contain a coiled-coil domain and a SPRY domain. To date, no reports have shown the subcellular localization of TRIM51 proteins.
Function
In general, TRIM- or SPRYD-domain-containing genes are involved in functions related to innate immune response [17, 46]. However, there have been no studies investigating the function of TRIM51 proteins as an expanded gene family in humans because they are not classified with other TRIM- or SPRYD-containing genes [45]. But a possible involvement in immune response mechanisms could evidently explain their adaptive evolution.
Chromosome 15: GOLGA gene family
Evolution and comparative genomics
GOLGA (GOLGIN) genes encode long coiled-coil proteins associated with the Golgi apparatus. They form a large gene family distributed across different chromosomes in humans. Several of the GOLGA genes are ancient duplicates with orthologous copies in the mouse (GOLGA1, GOLGA2 on chr9, GOLGA3 on chr12, GOLGA4 on chr3, GOLGA5 on chr14, GOLGA7 on chr8 and GOLGA7B on chr10). However, there are also two subfamilies—GOLGA6 and GOLGA8—that share similarity to each other. GOLGA8 has human-specific core duplicons that are expanded along chromosome 15 by segmental duplications and are partially in tandem. The current human assembly contains 12 annotated duplicated GOLGA6 subfamily members, whereas the GOLGA8 subfamily includes 15 duplicated copies. They show the closest similarity to GM130 (GOLGA2), which is located on human chromosome 9 [47]. The macaque genome contains approximately 11 GOLGA6-like and at least 8 fragmented GOLGA8-like duplicates. The N-terminal portions of GOLGA8 genes show high variation in both macaque and human. The duplication architecture for GOLGA6 and GOLGA8 copies is complex and not yet well resolved.
Expression and subcellular localization
Both GOLGA6 and GOLGA8 are ubiquitously expressed at low levels in human tissue, and they are most highly expressed in the testis. There is no direct evidence for the subcellular localization of GOLGA6 and GOLGA8 proteins; however, based on similarity to GM130, they are predicted to localize to the Golgi apparatus, Golgi stack membrane and cytoplasm [47].
Function
The roles of the individual variants of the GOLGA proteins are not clear [47], but they may function in membrane trafficking or Golgi structure. Loss of individual GOLGA genes in mice or humans is not cell lethal, possibly due to functional redundancy between different copies. Palindromic GOLGA8 core duplicons promote recurrent chromosome 15q13.3 microdeletions that are associated with intellectual disability, schizophrenia, autism and epilepsy [48, 49] but there are currently no data that would suggest a causative role of GOLGA8 losses for the disease effects.
Chromosome 16: Morpheus (NPIP) gene family
Evolution and comparative genomics
The ‘Morpheus’ gene family, also named the nuclear pore interacting protein (NPIP) family, is one of the best studied human core duplicon gene families. It is derived from a name-giving ‘low-copy repeat sequence on chromosome 16’, LCR16a, that is approximately 20 kb long and expanded in the great ape–human lineage along chromosome 16 through segmental duplications [7, 14, 50]. It can be subdivided into two distinct subfamilies, NPIPA and NPIPB, which mostly differ with respect to exon 5 and the structure of amino acid repeats in the C-terminus [7]. In contrast to other core duplicon gene families, it has not been possible to identify paralogs outside of primates, i.e. it appears to be a newly evolved or rapidly evolving gene. In fact, the Morpheus gene family was shown to be one of the most rapidly evolving gene families during hominoid evolution [14].
Expression and subcellular localization
Morpheus genes are expressed in various tissues and are most highly expressed in the testis and thymus [7]. The 5′-exons show extensive variation in splicing [7]. Due to this alternative splicing and differences in the C-terminal repeat region, Morpheus proteins vary in size between 40 and 95 kDa. Over-expression of different types of NPIPB and NPIPA genes reveals different subcellular localizations within both the nucleus and cytoplasm [7].
Function
Although Morpheus proteins were initially suggested to interact with the nuclear pore complex [14], there has not yet been evidence to support this assumption. They were shown to be over-expressed in the retina of patients with macular degeneration [51], but the functional significance of this is unclear. Other observations indicate an involvement in innate immunity, especially with respect to viruses. Morpheus proteins were suggested to be involved in human immunodeficiency virus resistance [52, 53], were shown to interact with Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and were shown to restore the interferon-beta response in SARS-CoV cells [54]. Both Morpheus gene types (NPIPA and NPIPB) are also upregulated upon polyinosinic:polycytidylic acid (poly I:C) treatment (viral mimic), and auto-antibodies against NPIPB protein in humans have been detected [7]. It is thus possible that Morpheus genes could cause autoimmune diseases such as multiple sclerosis, systemic lupus erythematosus, myasthenia gravis and Wegener’s granulomatosis [55, 56]. Notably, newly evolved genes in mammals can generally create complications with respect to generating autoimmune responses [57].
Chromosome 17: TBC1D3 gene family
Evolution and comparative genomics
The TBC1D3 gene family is characterized by the TBC domain, which plays a major role in endocytosis and intracellular trafficking in other proteins. TBC1D3 genes are derived from USP6NL (alias RNTRE) by segmental duplication [58]. Comparative genomic analysis suggests that TBC1D3 genes emerged before the split of new world monkeys since the macaque genome (rheMac8) includes at least five copies of TBC1D3-like genes. TBC1D3 genes are duplicated along human chromosome 17 in 12 copies including some processed pseudogenes [59, 60]. TBC1D3 genes show extensive CNV between different populations (10 copies in Europeans and up to 50 copies in African populations) [10].
Expression and subcellular localization
TBC1D3 genes are broadly expressed in many tissues and are most highly expressed in the testis [59, 60]. Different isoforms are expressed in different tissues [60]. In HeLa cells, ectopic expression of TBC1D3 genes led to alterations of the actin cytoskeleton of HeLa cells. Over-expressed TBC1D3 protein localized in the cytoplasm, in lipid rafts and on the plasma membrane [58, 61, 62].
Function
TBC1D3 was first described as an oncogene [63] and was also identified as PRC17 during the analysis of cells derived from prostate and breast cancer patients [64]. A predicted GAP activity of the TBC domain is equivocal. While a weak GAP activity was documented in one study [64], this finding could not be confirmed by others; however, TBC1D3 is still thought to be involved in macropinocytosis [58]. TBC1D3 dysregulates the epidermal growth factor receptor signal transduction pathway and enhances cell proliferation [61]. Further studies showed that TBC1D3 protein is ubiquitinated and palmitoylated, and degradation of TBC1D3 protein is regulated by Cul7 [62, 65]. TBC1D3 genes were also shown to be involved in Insulin/IGF signaling [66]. Over-expression of TBC1D3 leads to an increase in cell proliferation in basal regions of the developing mouse cortex, as well as disruptions to adherens junctions and formation of column-like structures [67]. Similar results were also obtained in cultured human brain slices, where it was shown that TBC1D3 is critical for the generation of outer radial glial cells [67]. Finally, it was shown that TBC1D3 affects the migration of human breast cancer cells by regulating TNFα/NF-κB signaling [68].
Chromosome 17: LRRC37 gene family
Evolution and comparative genomics
The LRRC37 (leucine-rich repeat containing 37A) gene family has expanded on human chromosome 17 [4] from a single ancestral copy in other mammals [69, 70]. In humans, this family’s gene structure is highly fragmented, and only 8 of 18 copies are complete. Two of these eight complete LRRC37 genes can be classified as the LRRC37B type. The number of the duplication segments is variable within the primate lineage; there are 4, 7, 10 and 18 copies in marmoset, macaque, orangutan and human, respectively [70]. Similar to the SPATA31 genes, the N-terminal region (especially exon 1) of LRRC37A has acquired novel structures and promoters in the primate lineage [69].
Expression and subcellular localization
LRRC37 is broadly expressed in primates, partly due to the acquisition of promotor elements from unrelated genes; however, it is still most highly expressed in the testis [69, 70]. Over-expressed LRRC37A1 protein is primarily localized to the plasma membrane. Pulse-chase experiments show that it is first localized to the Golgi and then transported to the plasma membrane where it co-localizes with Ezrin [70].
Function
The LRRC37 proteins contain six leucine-rich repeat motifs (LRR), which consist of repeating 20–30 amino acid stretches. Well-known LRR domain-containing proteins include those of the innate immune system, especially in mammals and plants. For example, Toll-like receptors are single-membrane-spanning proteins, and their extracellular domains are composed of LRRs, which recognize pathogen-associated molecular patterns such as LPS, single-stranded RNA and flagellin. LRR-containing proteins may also be involved in neuron-specific functions, such as axonal guidance and neuronal migration [71, 72]. However, it is not known whether LRRC37 genes play a role in any of these functions. On the other hand, the LRRC37 gene family in humans is located at the boundary of a common inversion polymorphism of approximately 970 kb at 17q21.31 [69, 73, 74]. This has been shown to be a significant risk factor locus for the tangle diseases, including progressive supranuclear palsy [75], corticobasal degeneration [76, 77], Parkinson’s disease [78, 79] and Alzheimer’s disease [80], and it is associated with microdeletion syndromes [81–83]. LRRC37B, a member of the LRRC37 gene family is also a breakpoint for NF1 microdeletion syndrome [84].
Discussion
The evolutionary patterns of human core duplicon gene families raise a number of challenging questions. First, what is the role of the cores in the duplication process? Second, why are the non-tandem segmental duplications mostly confined to single chromosomes? Third, why are the human core duplicon genes usually derived from genes that are highly expressed in the testis? Fourth and arguably most importantly, what has the role of human core duplicon gene families been in human evolution? The data available for a number of the gene families suggest that they may not have simple single functions but instead pleiotropic effects. While it seems natural that most studies have focused on inferred specific functions, which are often related to possible disease mechanisms, we advocate for investigators to keep a broad picture in mind. For example, a number of studies have focused on possible brain expansion effects for DUF1220/Olduvai repeats in the lineage toward humans, but we show that such correlations can be drawn for all of the core duplicon gene families (Table 1). Our own work with SPATA31 had initially focused on the repair of UV-induced DNA damage because of the newly acquired protein domains. However, the incidental observation that fibroblast cells with manipulated functional copy numbers of SPATA31 exhibited altered senescence led us to study a possible connection to aging.
The most direct connections of core duplicon gene families lie in their involvement in causing chromosomal aberrations and microdeletions. But this could be due to their general repeat structure, which makes them prone to recombinational unequal cross-over mechanisms that would also affect other genes in the respective regions. Hence, the involvement in such diseases may not necessarily serve as a direct pointer toward their protein functions.
All the core duplicon gene families include expansions of protein domains, of which many are known to be involved in basic cellular functions. Hence, it seems likely that they can molecularly interact with the target genes of their progenitor proteins and thus possibly modify or regulate their core functions. Future studies should focus more on this possibility.
The duplication process included the integration of repetitive transposable elements, LTRs (e.g. ERV1) or unrelated promoters (DND1 and BPTF for LRRC37A genes or EVI5 for NBPF1 genes). This integration broadened the expression to more tissues or caused ubiquitous expression. The highest expression usually remained in the testis, although the functional consequences of this expression have not been studied much to date. For example, dedicated studies should be designed to evaluate the effect of CNV of core duplicon genes on fertility phenotypes in humans.
Further, core duplicon genes commonly exist in two versions of gene structure; for example, SPATA31A versus SPATA31C, NPIPA versus NPIPB, LRRC37A versus LRRC37B, GOLGA6 versus GOLGA8 and TBC1D3 (1st cluster, A–D) versus (2nd cluster, E–H). This points to diversification of effects during the duplication process, a concept that will also require more dedicated molecular studies.
Finally, it remains to be shown whether the expansion of core duplicon gene families is a specific phenomenon in the human lineage and whether they acted as ‘game changers’ with respect to human-specific adaptations. It has been possible to readily detect duplicon gene families in humans because of the abundant high-quality data and comparative genome sequencing efforts in the primate phylogeny. However, there are now also examples of lineage-specific expansions in other clades, such as rodents [85] and elephants [86]. Still, the specific duplicates in the human lineage could evidently have contributed to human-specific phenotypes and adaptations. After all, the signatures of positive selection found around human core duplicon gene families suggest an active evolutionary role. However, these signatures would likely also arise if these genes were mere modifiers of other major changes in core pathways, i.e. if they were ‘game players’. Hence, deeper studies will be required to solve the questions raised above.
Key Points
Core duplicon gene families constitute a specific subset of gene duplicates in the human genome.
They are derived from testis-expressed genes, have acquired new promoters and have specifically expanded in the evolutionary lineage toward humans.
They express proteins with domains that suggest that they could be modifiers of general cellular pathways.
Patterns of positive selection within the genes suggest that they were important for shaping the specific adaptations of humans.
Biographical notes
The authors work at the Max-Planck Institute for Evolutionary Biology on patterns of genome evolution in mammals with a specific interest in newly evolved genes. D.T. is member of the editorial board of BFG.
Supplementary Material
Acknowledgements
This work was supported by Türkiye Bilimsel ve Teknolojik Araştırma Kurumu (TUBITAK-1001 project 112T421 to C.B.) and Max-Planck Institutional Funds (to D.T.).
References
- 1. Bailey JA, Gu Z, Clark RA, et al. Recent segmental duplications in the human genome. Science 2002;297:1003–1007. [DOI] [PubMed] [Google Scholar]
- 2. Bailey JA, Eichler EE. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet 2006;7:552–564. [DOI] [PubMed] [Google Scholar]
- 3. Zhang L, Lu HH, Chung WY, et al. Patterns of segmental duplication in the human genome. Mol Biol Evol 2005;22:135–141. [DOI] [PubMed] [Google Scholar]
- 4. Jiang Z, Tang H, Ventura M, et al. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat Genet 2007; 39:1361–1368. [DOI] [PubMed] [Google Scholar]
- 5. Marques-Bonet T, Eichler EE. The evolution of human segmental duplications and the core duplicon hypothesis. Cold Spring Harb Symp Quant Biol 2009;74:355–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dennis MY, Eichler EE. Human adaptation and evolution by segmental duplication. Curr Opin Genet Dev 2016;41:44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bekpen C, Baker C, Hebert MD, et al. Functional characterization of the morpheus gene family. biorxiv, 2017; doi: 10.1101/116087. [DOI]
- 8. Alkan C, Kidd JM, Marques-Bonet T, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet 2009;41:1061–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Conrad DF, Pinto D, Redon R, et al. Origins and functional impact of copy number variation in the human genome. Nature 2009;464:704–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sudmant PH, Kitzman JO, Antonacci F, et al. Diversity of human copy number variation and multicopy genes. Science 2010;330:641–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sudmant PH, Rausch T, Gardner EJ, et al. An integrated map of structural variation in 2,504 human genomes. Nature 2015;526:75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hahn MW, Demuth JP, Han SG. Accelerated rate of gene gain and loss in primates. Genetics 2007;177:1941–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Han MV, Demuth JP, McGrath CL, et al. Adaptive evolution of young gene duplicates in mammals. Genome Res 2009;19:859–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnson ME, Viggiano L, Bailey JA, et al. Positive selection of a gene family during the emergence of humans and African apes. Nature 2001;413:514–519. [DOI] [PubMed] [Google Scholar]
- 15. Ciccarelli FD, von C, Suyama M, et al. Complex genomic rearrangements lead to novel primate gene function. Genome Res 2005;15:343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Popesco MC, Maclaren EJ, Hopkins J, et al. Human lineage-specific amplification, selection, and neuronal expression of DUF1220 domains. Science 2006;313:1304–1307. [DOI] [PubMed] [Google Scholar]
- 17. Han K, Lou DI, Sawyer SL. Identification of a genomic reservoir for new TRIM genes in primate genomes. PLoS Genet 2011;7: e1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bekpen C, Kunzel S, Xie C, et al. Segmental duplications and evolutionary acquisition of UV damage response in the SPATA31 gene family of primates and humans. BMC Genomics 2017;18:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Laureys G, Speleman F, Opdenakker G, et al. Constitutional translocation t(1,17)(p36;q12-21) in a patient with neuroblastoma. Genes Chromosomes Cancer 1990;2:252–254. [DOI] [PubMed] [Google Scholar]
- 20. Sikela JM, van F. Changing the name of the NBPF/DUF1220 domain to the Olduvai domain. F1000Res 2017;6:2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sikela JM, Searles Quick VB. Genomic trade-offs: are autism and schizophrenia the steep price of the human brain? Hum Genet 2018;137:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. O'Bleness MS, Dickens CM, Dumas LJ, et al. Evolutionary history and genome organization of DUF1220 protein domains. G3 (Bethesda) 2012;2:977–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vandepoele K, Van N, Staes K, et al. A novel gene family NBPF: intricate structure generated by gene duplications during primate evolution. Mol Biol Evol 2005;22:2265–2274. [DOI] [PubMed] [Google Scholar]
- 24. Vandepoele K, Andries V, van F. The NBPF1 promoter has been recruited from the unrelated EVI5 gene before simian radiation. Mol Biol Evol 2009;26:1321–1332. [DOI] [PubMed] [Google Scholar]
- 25. Dumas L, Sikela JM. DUF1220 domains, cognitive disease, and human brain evolution. Cold Spring Harb Symp Quant Biol 2009;74:375–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dumas LJ, O'Bleness MS, Davis JM, et al. DUF1220-domain copy number implicated in human brain-size pathology and evolution. Am J Hum Genet 2012;91:444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Keeney JG, Dumas L, Sikela JM. The case for DUF1220 domain dosage as a primary contributor to anthropoid brain expansion. Front Hum Neurosci 2014;8:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Keeney JG, Davis JM, Siegenthaler J, et al. DUF1220 protein domains drive proliferation in human neural stem cells and are associated with increased cortical volume in anthropoid primates. Brain Struct Funct 2015;220:3053–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zimmer F, Montgomery SH. Phylogenetic analysis supports a link between DUF1220 domain number and primate brain expansion. Genome Biol Evol 2015;7:2083–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Searles Quick VB, Davis JM, Olincy A, et al. DUF1220 copy number is associated with schizophrenia risk and severity: implications for understanding autism and schizophrenia as related diseases. Transl Psychiatry 2015;5: e697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Davis JM, Searles VB, Anderson N, et al. DUF1220 copy number is linearly associated with increased cognitive function as measured by total IQ and mathematical aptitude scores. Hum Genet 2015;134:67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Christiansen J, Dyck JD, Elyas BG, et al. Chromosome 1q21.1 contiguous gene deletion is associated with congenital heart disease. Circ Res 2004;94:1429–1435. [DOI] [PubMed] [Google Scholar]
- 33. Greenway SC, Pereira AC, Lin JC, et al. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat Genet 2009;41:931–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Weber S, Landwehr C, Renkert M, et al. Mapping candidate regions and genes for congenital anomalies of the kidneys and urinary tract (CAKUT) by array-based comparative genomic hybridization. Nephrol Dial Transplant 2011;26:136–143. [DOI] [PubMed] [Google Scholar]
- 35. Vandepoele K, Staes K, Andries V, et al. Chibby interacts with NBPF1 and clusterin, two candidate tumor suppressors linked to neuroblastoma. Exp Cell Res 2010;316:1225–1233. [DOI] [PubMed] [Google Scholar]
- 36. Keeney JG, O'Bleness MS, Anderson N, et al. Generation of mice lacking DUF1220 protein domains: effects on fecundity and hyperactivity. Mamm Genome 2015;26:33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yokoyama N, Hayashi N, Seki T, et al. A giant nucleopore protein that binds ran/TC4. Nature 1995;376:184–188. [DOI] [PubMed] [Google Scholar]
- 38. Meyerson NR, Warren CJ, Vieira D, et al. Species-specific vulnerability of RanBP2 shaped the evolution of SIV as it transmitted in African apes. PLoS Pathog 2018;14: e1006906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Theodosiou AM, Morrison KE, Nesbit AM, et al. Complex repetitive arrangements of gene sequence in the candidate region of the spinal muscular atrophy gene in 5q13. Am J Hum Genet 1994;55:1209–1217. [PMC free article] [PubMed] [Google Scholar]
- 40. Farrar MA, Kiernan MC. The genetics of spinal muscular atrophy: Progress and challenges. Neurotherapeutics 2015;12:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Horii A, Han HJ, Sasaki S, et al. Cloning, characterization and chromosomal assignment of the human genes homologous to yeast PMS1, a member of mismatch repair genes. Biochem Biophys Res Commun 1994;204:1257–1264. [DOI] [PubMed] [Google Scholar]
- 42. Nicolaides NC, Carter KC, Shell BK, et al. Genomic organization of the human PMS2 gene family. Genomics 1995;30:195–206. [DOI] [PubMed] [Google Scholar]
- 43. Bekpen C, Xie C, Nebel A, et al. Involvement of SPATA31 copy number variable genes in human lifespan. Aging (Albany NY) 2018;10:674–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu YY, Yang Y, Xu YD, et al. Targeted disruption of the spermatid-specific gene Spata31 causes male infertility. Mol Reprod Dev 2015;82:432–440. [DOI] [PubMed] [Google Scholar]
- 45. Carthagena L, Bergamaschi A, Luna JM, et al. Human TRIM gene expression in response to interferons. PLoS One 2009;4: e4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ozato K, Shin DM, Chang TH, et al. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol 2008;8:849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Munro S. The golgin coiled-coil proteins of the Golgi apparatus. Cold Spring Harb Perspect Biol 2011;3:a005256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Antonacci F, Dennis MY, Huddleston J, et al. Palindromic GOLGA8 core duplicons promote chromosome 15q13.3 microdeletion and evolutionary instability. Nat Genet 2014;46:1293–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hassfurther A, Komini E, Fischer J, et al. Clinical and genetic heterogeneity of the 15q13.3 microdeletion syndrome. Mol Syndromol 2016;6:222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Loftus BJ, Kim UJ, Sneddon VP, et al. Genome duplications and other features in 12 Mb of DNA sequence from human chromosome 16p and 16q. Genomics 1999;60:295–308. [DOI] [PubMed] [Google Scholar]
- 51. Hornan DM, Peirson SN, Hardcastle AJ, et al. Novel retinal and cone photoreceptor transcripts revealed by human macular expression profiling. Invest Ophthalmol Vis Sci 2007;48:5388–5396. [DOI] [PubMed] [Google Scholar]
- 52. Liu L, Oliveira NM, Cheney KM, et al. A whole genome screen for HIV restriction factors. Retrovirology 2011;8:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yeung ML, Houzet L, Yedavalli VS, et al. A genome-wide short hairpin RNA screening of jurkat T-cells for human proteins contributing to productive HIV-1 replication. J Biol Chem 2009;284:19463–19473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Huang SH, Lee TY, Lin YJ, et al. Phage display technique identifies the interaction of severe acute respiratory syndrome coronavirus open reading frame 6 protein with nuclear pore complex interacting protein NPIPB3 in modulating type I interferon antagonism. J Microbiol Immunol Infect 2017;50:277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wekerle H, Flugel A, Fugger L, et al. Autoimmunity's next top models. Nat Med 2012;18:66–70. [DOI] [PubMed] [Google Scholar]
- 56. Berer K, Wekerle H, Krishnamoorthy G. B cells in spontaneous autoimmune diseases of the central nervous system. Mol Immunol 2010;48:1332–1337. [DOI] [PubMed] [Google Scholar]
- 57. Bekpen C, Xie C, Tautz D. Dealing with the adaptive immune system during de novo evolution of genes from intergenic sequences. BMC Evol Biol 2018;18:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Frittoli E, Palamidessi A, Pizzigoni A, et al. The primate-specific protein TBC1D3 is required for optimal macropinocytosis in a novel ARF6-dependent pathway. Mol Biol Cell 2008;19:1304–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Paulding CA, Ruvolo M, Haber DA. The Tre2 (USP6) oncogene is a hominoid-specific gene. Proc Natl Acad Sci U S A 2003;100:2507–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hodzic D, Kong C, Wainszelbaum MJ, et al. TBC1D3, a hominoid oncoprotein, is encoded by a cluster of paralogues located on chromosome 17q12. Genomics 2006;88:731–736. [DOI] [PubMed] [Google Scholar]
- 61. Wainszelbaum MJ, Charron AJ, Kong C, et al. The hominoid-specific oncogene TBC1D3 activates ras and modulates epidermal growth factor receptor signaling and trafficking. J Biol Chem 2008;283:13233–13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kong C, Samovski D, Srikanth P, et al. Ubiquitination and degradation of the hominoid-specific oncoprotein TBC1D3 is mediated by CUL7 E3 ligase. PLoS One 2012;7: e46485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nakamura T, Hillova J, Mariage-Samson R, et al. A novel transcriptional unit of the tre oncogene widely expressed in human cancer cells. Oncogene 1992;7:733–741. [PubMed] [Google Scholar]
- 64. Pei L, Peng Y, Yang Y, et al. PRC17, a novel oncogene encoding a Rab GTPase-activating protein, is amplified in prostate cancer. Cancer Res 2002;62:5420–5424. [PubMed] [Google Scholar]
- 65. Kong C, Lange JJ, Samovski D, et al. Ubiquitination and degradation of the hominoid-specific oncoprotein TBC1D3 is regulated by protein palmitoylation. Biochem Biophys Res Commun 2013;434:388–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wainszelbaum MJ, Liu J, Kong C, et al. TBC1D3, a hominoid-specific gene, delays IRS-1 degradation and promotes insulin signaling by modulating p70 S6 kinase activity. PLoS One 2012;7: e31225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ju XC, Hou QQ, Sheng AL, et al. The hominoid-specific gene TBC1D3 promotes generation of basal neural progenitors and induces cortical folding in mice. Elife 2016;5:e18197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang B, Zhao H, Zhao L, et al. Up-regulation of OLR1 expression by TBC1D3 through activation of TNFalpha/NF-kappaB pathway promotes the migration of human breast cancer cells. Cancer Lett 2017;408:60–70. [DOI] [PubMed] [Google Scholar]
- 69. Bekpen C, Tastekin I, Siswara P, et al. Primate segmental duplication creates novel promoters for the LRRC37 gene family within the 17q21.31 inversion polymorphism region. Genome Res 2012;22:1050–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Giannuzzi G, Siswara P, Malig M, et al. Evolutionary dynamism of the primate LRRC37 gene family. Genome Res 2013;23:46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ko J, Kim E. Leucine-rich repeat proteins of synapses. J Neurosci Res 2007;85:2824–2832. [DOI] [PubMed] [Google Scholar]
- 72. Matsushima N, Tachi N, Kuroki Y, et al. Structural analysis of leucine-rich-repeat variants in proteins associated with human diseases. Cell Mol Life Sci 2005;62:2771–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Stefansson H, Helgason A, Thorleifsson G, et al. A common inversion under selection in Europeans. Nat Genet 2005;37:129–137. [DOI] [PubMed] [Google Scholar]
- 74. Zody MC, Jiang Z, Fung HC, et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat Genet 2008;40(9):1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Baker M, Litvan I, Houlden H, et al. Association of an extended haplotype in the tau gene with progressive supranuclear palsy. Hum Mol Genet 1999;8:711–715. [DOI] [PubMed] [Google Scholar]
- 76. Houlden H, Baker M, Morris HR, et al. Corticobasal degeneration and progressive supranuclear palsy share a common tau haplotype. Neurology 2001;56:1702–1706. [DOI] [PubMed] [Google Scholar]
- 77. Pittman AM, Myers AJ, Duckworth J, et al. The structure of the tau haplotype in controls and in progressive supranuclear palsy. Hum Mol Genet 2004;13:1267–1274. [DOI] [PubMed] [Google Scholar]
- 78. Wider C, Vilarino-Guell C, Jasinska-Myga B, et al. Association of the MAPT locus with Parkinson's disease. Eur J Neurol 2010;17:483–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Farrer M, Skipper L, Berg M, et al. The tau H1 haplotype is associated with Parkinson's disease in the Norwegian population. Neurosci Lett 2002;322:83–86. [DOI] [PubMed] [Google Scholar]
- 80. Myers AJ, Kaleem M, Marlowe L, et al. The H1c haplotype at the MAPT locus is associated with Alzheimer's disease. Hum Mol Genet 2005;14:2399–2404. [DOI] [PubMed] [Google Scholar]
- 81. Koolen DA, Sharp AJ, Hurst JA, et al. Clinical and molecular delineation of the 17q21.31 microdeletion syndrome. J Med Genet 2008;45:710–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sharp AJ, Hansen S, Selzer RR, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet 2006;38:1038–1042. [DOI] [PubMed] [Google Scholar]
- 83. Shaw-Smith C, Pittman AM, Willatt L, et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat Genet 2006;38:1032–1037. [DOI] [PubMed] [Google Scholar]
- 84. Bengesser K, Cooper DN, Steinmann K, et al. A novel third type of recurrent NF1 microdeletion mediated by nonallelic homologous recombination between LRRC37B-containing low-copy repeats in 17q11.2. Hum Mutat 2010;31:742–751. [DOI] [PubMed] [Google Scholar]
- 85. Bekpen C, Hunn JP, Rohde C, et al. The interferon-inducible p47 (IRG) GTPases in vertebrates: loss of the cell autonomous resistance mechanism in the human lineage. Genome Biol 2005;6:R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sulak M, Fong L, Mika K, et al. TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. Elife 2016;5:e11994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


