Abstract
Activation-induced cytidine deaminase (AID) generates U:G mismatches in immunoglobulin genes that can be converted into untemplated mutations during somatic hypermutation (SHM) or DNA double-strand breaks (DSB) during class switch recombination (CSR). Null mutations in UNG and MSH2 demonstrate the complementary roles of the base excision repair (BER) and mismatch repair (MMR) pathways, respectively, in CSR. Phosphorylation of AID at serine-38 (pS38-AID) was previously hypothesized to regulate BER during CSR, as the AID phosphorylation mutant, AID(S38A), cannot interact with APE1, a BER protein. Consistent with these findings, we observe a complete block in CSR in AIDS38A/S38AMSH2−/− mouse B cells that correlates with an impaired mutation frequency at 5’Sμ. Similarly, SHM is almost negligible at the JH4 intron in AIDS38A/S38AMSH2−/− mouse B cells and, consistent with this, NP-specific affinity maturation in AIDS38A/S38AMSH2−/− mice is not significantly elevated in response to NP-CGG immunization. Surprisingly, AIDS38A/S38AUNG−/− mouse B cells also cannot complete CSR or affinity maturation despite accumulating significant mutations in 5’Sμ as well as the JH4 intron. These data identify a novel role for pS38-AID in MMR-dependent CSR and affinity maturation.
INTRODUCTION
B cells undergo three genetically programmed somatic DNA recombination and mutation events to diversify their immunoglobulin (Ig) genes – V(D)J recombination, class switch recombination (CSR), and somatic hypermutation (SHM). V(D)J recombination assembles the exons encoding the variable (V) regions of the immunoglobulin heavy (IgH) and light (IgL) chain genes [1]. CSR generates secondary Ig isotypes by replacing the IgH constant coding exon (CH) through a DNA deletional recombination reaction. The generation and subsequent ligation of DNA double-strand breaks (DSBs) in the recombining switch (S) regions, which precede each CH exon, exchanges the default Cμ for another downstream CH exon [2–6]. SHM introduces untemplated substitution mutations into V genes of both IgH and IgL chains (10−3 to10−4/bp/generation) to promote Ig affinity maturation [7].
CSR and SHM require activation-induced cytidine deaminase (AID) [8, 9], which generates deoxyuridines (U) in S or V regions that are processed either into DSBs or mutations by the base excision repair (BER) or mismatch repair (MMR) pathway [10]. During BER, uracil DNA glycosylase (UNG) recognizes and removes the uracil base to create an abasic site, which is cleaved by apurinic/apyrimidinic endonuclease 1 (APE1) to generate a single-strand break (SSB). In MMR, a heterodimer of mutS homolog 2 (MSH2) and mutS homolog 6 (MSH6) recognizes the U:G mismatch and recruits a complex of mutL homology 1 (MLH1) and PMS1 homolog 2 (PMS2) to generate a nick 5’ of the U:G mismatch. Exonuclease I (EXO1) subsequently degrades the nicked DNA to produce SSBs. Staggered SSBs generated by MMR or BER constitute the DSBs necessary for CSR. In the absence of MSH2 and UNG (MSH2−/−UNG−/−), both MMR and BER pathways are disrupted and CSR is completely blocked [11].
In V regions undergoing SHM, AID-generated U:G mismatches can be replicated to generate C:G to T:A transition mutations, which are observed in MSH2−/−UNG−/− B cells [11]. Removal of the uracil base from the U:G mismatch prior to DNA replication leaves an abasic site, which can be converted to transition or transversion mutations at C:G base pairs following replication [11]. Alternatively, mutagenic repair of the U:G mismatch by MMR generates mutations at A:T base pairs. Mice deficient in both MSH2 and Polη, the polymerase utilized in MMR, have a complete loss of mutations at A:T base pairs [12].
AID activity is regulated by phosphorylation at serine-38 (S38) by PKA (cAMP-dependent protein kinase A) [13–15]. Mice with a homozygous knock-in mutation of the phosphorylation site (AIDS38A/S38A), which changes S38 to an alanine, have significantly reduced CSR and SHM [16, 17] even though the mutant AID(S38A) enzyme retains wild-type deaminase activity [14, 18]. Similarly, B cells from mice carrying a hypomorphic mutation in the PKA regulatory subunit (RIα) display significantly reduced CSR [18]. Phosphorylation of AID mediates its interaction with APE1 [19], suggesting that pS38-AID promotes BER-dependent CSR. To determine if pS38-AID regulates MMR- or BER-dependent CSR and SHM, we bred AIDS38A/S38A mice onto either an MSH2−/− or UNG−/− background. As expected, AIDS38A/S38AMSH2−/− B cells have a complete block in CSR and reduced mutation frequency in 5’Sμ due to the absence of pS38-AID and MSH2. Similarly, almost no mutations are observed in the JH4 intron of AIDS38A/S38AMSH2−/− B cells, which correlates with a loss of affinity maturation. Interestingly, while AIDS38A/S38AUNG−/− B cells can generate mutations in S regions, AIDS38A/S38AUNG−/− mice cannot produce secondary Ig isotypes, indicating the absence of CSR. Although JH4 intron mutations are observed in AIDS38A/S38AUNG−/− B cells, no significant affinity maturation is observed. These results implicate a role for pS38-AID in MMR-dependent CSR and affinity maturation.
MATERIALS AND METHODS
Mice
C57BL/6 mice were purchased from The Jackson Laboratories. AIDS38A/S38A mice were generated as described [16], UNG−/− mice were a gift from T. Lindahl [20], MSH2−/− were a gift from H. te Riele [21], and AID−/− were a gift from T. Honjo [8]. All mutant mice were maintained on a C57BL/6 background. Age-matched male and female mice were used for all experiments. Husbandry of and experiments with mice were conducted according to protocols approved by The City College of New York IACUC.
In vitro Class Switch Recombination Assay
B cells were purified from the spleens of mice (aged 2–5 months) through negative selection with anti-CD43 magnetic beads (Miltenyi Biotec) and cultured in RPMI 1640 media (Gibco) supplemented with 10% FBS (Atlanta Biologicals), 2mM L-glutamine, 1X penicillin/streptomycin (Corning), and 47.3μM β-mercaptoethanol. Cells were stimulated in vitro with 25μg/mL LPS (Sigma-Aldrich #L7261) and 12.5ng/mL IL-4 (R&D Systems) for class switching to IgG1; 25μg/mL LPS for class switching to IgG3; or 300ng/mL anti-IgD-dextran (Fina Biosolutions), 2ng/mL TGF-β1 (R&D Systems), and 10μg/mL LPS for class switching to IgA. At 48 and 72 hours post-stimulation, the cells were passaged 1:2 with fresh stimulation media. After 96 hours, the stimulated cells were analyzed by flow cytometry using anti-IgG1 conjugated to APC (Becton Dickinson), anti-IgG3 conjugated to FITC (Becton Dickinson), or anti-IgA conjugated to FITC (Becton Dickinson). DAPI (4’,6-diamidino-2-phenylindole) was used to distinguish live and dead cells.
In vivo immunoglobulin titers (ELISA)
Mice (aged 2–4 months) were injected intraperitoneally with 50μg NP-CGG (Biosearch Technologies) in Imject Alum (Thermo Fisher) [22]. Immunized females were housed together, immunized male littermates were housed together, and immunized non-littermate males were housed individually. Mice were given a boost of NP-CGG in Imject Alum 10 days post-primary immunization (D10). Sera were obtained from blood that was collected on D0 (pre-immunization), D7, and D21 post-immunization through cheek bleeds or cardiac puncture (D21). Total immunoglobulin titers were analyzed by ELISA on anti-IgM (2 μg/mL, SouthernBiotech 1020–01), anti-IgG1 (2μg/mL, SouthernBiotech 1070–01), or anti-IgG3 (2 μg/mL, SouthernBiotech 1100–01) coated plates, which were detected using anti-IgM-HRP (1:5000, SouthernBiotech 1020–05), anti-IgG1-HRP (1:5000, SouthernBiotech 1070–05), or anti-IgG3-HRP (1:5000, SouthernBiotech 1100–05), respectively. Coating antibodies were diluted in PBS pH8. Sera and detection antibodies were diluted in 1%BSA in borate buffer (100mM boric acid, 25mM sodium borate, 75mM sodium chloride). NP-specific IgM titers were analyzed on plates coated with NP7-BSA or NP20-BSA (3μg/mL) in borate buffer and detected with anti-IgM-HRP. ELISA plates were read on SpectraMAX190 at 450nm. To determine affinity maturation, the ratio of NP7- and NP20-specific IgM concentration was calculated and this ratio was normalized to the D0 NP20 absorbance of each animal to account for variations in basal IgM titers [23].
Somatic Hypermutation Analysis
To analyze mutations in 5’Sμ and the JH4 intron, germinal center B cells (GCBC) were sorted from the Peyer’s patches of mice (aged 2–3 months) at D10 post-immunization with 50μg NP-CGG in Imject Alum. GCBC were sorted after staining with PNA-biotin (Vector Labs #B-1075–5) followed by strepavidin-APC-Cy7 (Thermo Fisher #47–4317-82) and anti-B220-APC (BD Pharmingen #553092). DAPI was used for live/dead cell exclusion. Sorted cells were lysed in lysis buffer (10mM Tris, 0.1M EDTA, 0.5% SDS) and genomic DNA was isolated by isopropanol precipitation. 5’Sμ was amplified using the following primers: Su5: 5’-GCGGCCCAGCTCATTCCAGTTCATTACAG-3’, Su3: 5’-AATGGATACCTCAGTGGTTTTTAATGGTGGGTTTA-3’. JH4 intron was amplified by a nested PCR [16, 17]. The primers for the first PCR were: J558FR3Fw: 5’-GCCTGACATCTGAGGACTCTGC-3’, VHJ558.2: 5’-CTGGACTTTCGGTTTGGTG-3’. The primers for the second PCR were: VHJ558.3: 5’-GGTCAAGGAACCTCAGTCA-3’, VHJ558.4: 5’-TCTCTAGACAGCAACTAC-3’. To analyze mutations within VH186.2, RNA was extracted from Peyer’s patch cells of D10 NP-CGG immunized mice (aged 2–3 months). cDNA was synthesized using the SuperScript III first-strand synthesis kit (Invitrogen). VH186.2 was amplified using a nested PCR as previously described [23]. All PCRs were performed with Q5 high-fidelity DNA polymerase (NEB). Amplicons were purified, ligated into pJET (ThermoFisher), and analyzed by Sanger sequencing (Genewiz). Sequences were analyzed with MegAlign (Lasergene). Reference sequences were: NCBI Ref #NG_005838 for 5’Sμ and JH4 intron; musIGHV057 on VBASE2 for VH186.2. Mutation hotspots were identified as RGYW/WRCY motifs [24, 25].
Statistical Analysis
Unless otherwise noted, a two-tailed Student’s t-test was used for statistical analysis.
RESULTS
Ablation of CSR in AIDS38A/S38AMSH2−/− and AIDS38A/S38AUNG−/− B cells
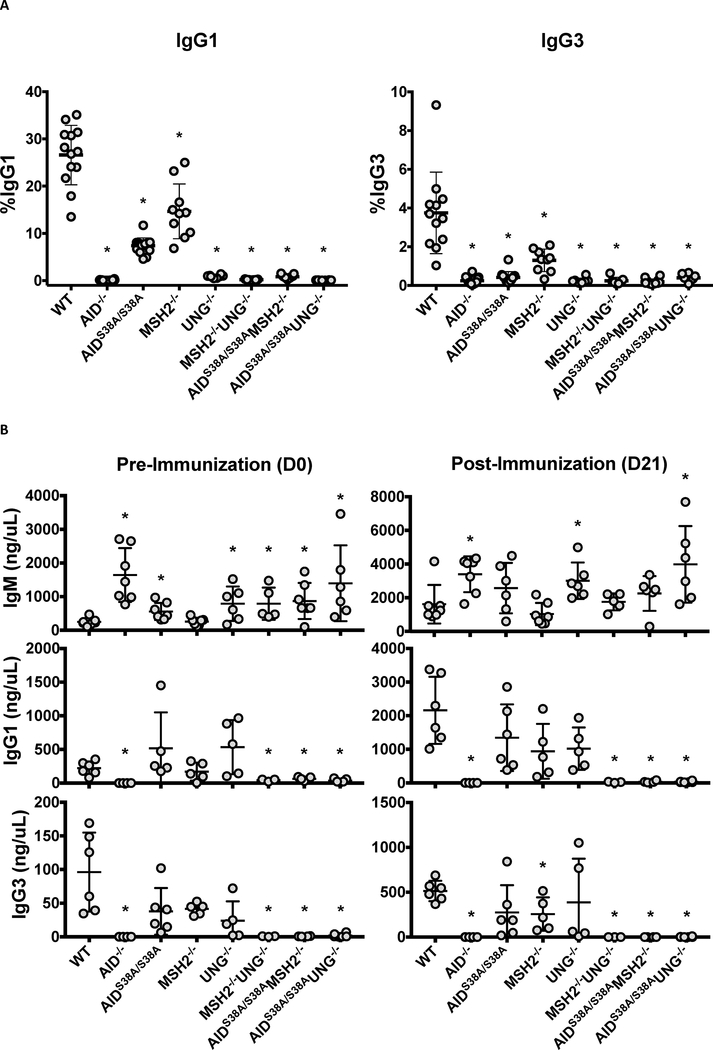
Phosphorylation of AID at serine-38 (S38) is required for wild-type (WT) levels of CSR. AID−/− B cells cannot complete CSR in vitro or in vivo [8], whereas AIDS38A/S38A B cells, which carry a serine-38 to alanine knock-in mutation of AID, complete CSR at ~25% of WT levels [16, 17] (Figures 1A and 1B). Phosphorylated AID promotes CSR in part through its interaction with APE1 [19], suggesting that the residual CSR observed in AIDS38A/S38A B cells is mediated by the MMR pathway [11]. To confirm this, we generated AIDS38A/S38AMSH2−/− mice and examined CSR to IgG1, IgG3, or IgA (Figures 1A and 1B; Supplementary Figure 1). As previously shown, AIDS38A/S38A and MSH2−/− B cells displayed a hypomorphic CSR phenotype [11, 16, 17], whereas the double mutant AIDS38A/S38AMSH2−/− B cells had a complete block in CSR in vitro and in vivo that was comparable to AID−/− and MSH2−/−UNG−/− B cells (Figures 1A and 1B). The absence of CSR was not due to reduced AID(S38A) protein expression, μ or γ1 germline transcription, cellular proliferation, or germinal center B cell formation (B220+PNAHi) (Supplementary Figures 2A–D). AIDS38A/S38AMSH2−/− mice produced IgM titers comparable to WT mice post-immunization (Figure 1B), suggesting that Ig secretion and plasma cell development were not altered. These data demonstrate that pS38-AID regulates BER-dependent CSR, as the loss of MMR and AID phosphorylation ablates CSR.
Figure 1. CSR is absent in AIDS38A/S38AMSH2−/− and AIDS38A/S38AUNG−/− B cells.
(A) Splenic B cells were purified from mice of the indicated genotypes, stimulated for CSR to IgG1 and IgG3, and analyzed for surface IgG1 and IgG3 by flow cytometry after four days in culture. For all genotypes, n ≥ 7. AIDS38A/S38AMSH2−/− and AIDS38A/S38AUNG−/− B cells had severely reduced switching compared to WT B cells (*P<0.001) (B) Serum IgM, IgG1, and IgG3 concentrations were analyzed by ELISA from mice of the indicated genotypes before and after (21 days) immunization with NP-CGG. For all genotypes, n ≥ 3. Data is displayed with error bars representing standard deviation from the mean. AIDS38A/S38AMSH2−/− and AIDS38A/S38AUNG−/− mice demonstrate increased basal IgM titers (P=0.03 and 0.04, respectively) and no IgG1 titers before (P=0.01 and 0.003, respectively) and after immunization (P=0.003 and 0.0004, respectively) when compared to WT mice. (*P<0.05; two-tailed Student’s t-test)
To examine whether pS38-AID modulates MMR-dependent CSR, AIDS38A/S38AUNG−/− mice were immunized with NP-CGG and serum Igs were analyzed by ELISA. UNG−/− B cells do not exhibit measurable levels of CSR in vitro [11] and, consistent with this, AIDS38A/S38AUNG−/− B cells could not complete CSR in vitro (Figure 1A). However, UNG−/− mice have detectable levels of serum IgG1 and IgG3 before and after immunization, indicating that UNG−/− B cells can undergo CSR in vivo likely due to compensation by the MMR pathway [11]. Surprisingly, AIDS38A/S38AUNG−/− mice had no detectable serum IgG1 and IgG3 before and after immunization (Figure 1B), indicating that AIDS38A/S38AUNG−/− B cells have a block in CSR in vivo, comparable to AIDS38A/S38AMSH2−/− and MSH2−/−UNG−/− B cells. The AIDS38A/S38AUNG−/− mice produced serum IgM that increased upon immunization with NP-CGG, suggesting that Ig secretion and plasma cell development are not impaired (Figure 1B, P=0.0032 for D0 versus D21 IgM titers in AIDS38A/S38AUNG−/− mice). AID(S38A) protein expression, μ or γ1 germline transcription, cellular proliferation, and germinal center B cell formation (B220+PNAHi) were not significantly different between WT and AIDS38A/S38AUNG−/− B cells (Supplementary Figures 2A–D). Interestingly, basal serum IgM titers in AIDS38A/S38AUNG−/− mice were increased five-fold relative to WT mice (Figure 1B, P=0.0209). Similarly, basal serum IgM titers in AIDS38A/S38AMSH2−/− mice were increased three-fold relative to WT mice (Figure 1B, P=0.0111). This hyper-IgM phenotype is also observed in MSH2−/−UNG−/− mice and suggests that IgM titers increase when CSR is blocked from the lack of MMR and BER. Therefore, the hyper-IgM and absence of CSR in AIDS38A/S38AMSH2−/− and AIDS38A/S38AUNG−/− mice demonstrates that BER and MMR, respectively, were inhibited due to the loss of phosphorylated AID.
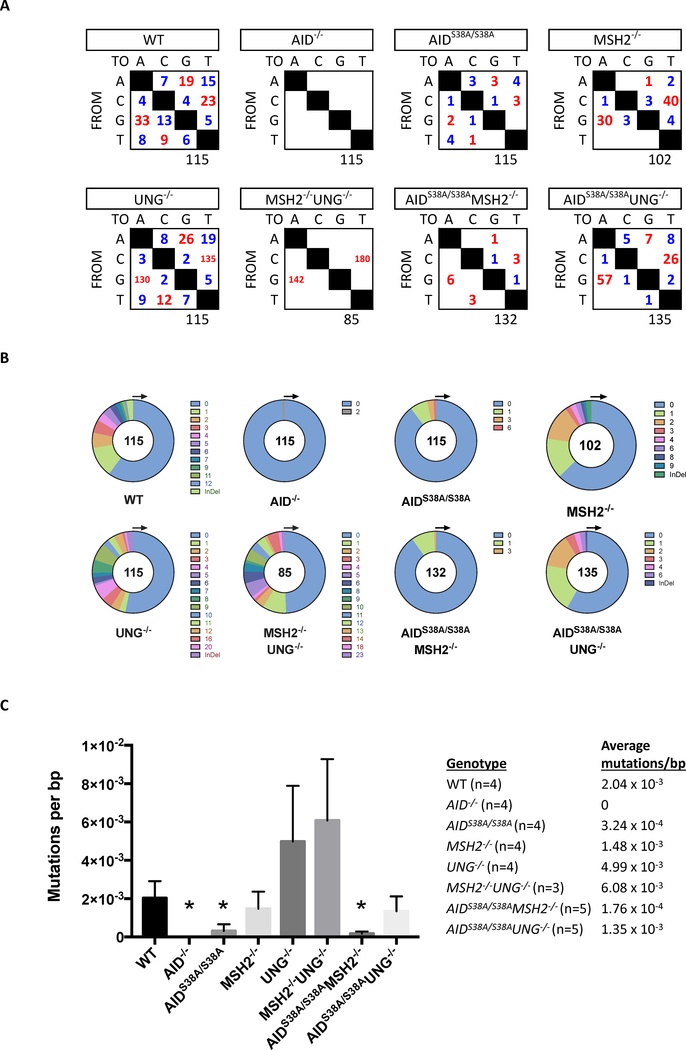
To determine if the ablation of CSR in AIDS38A/S38AMSH2−/− and AIDS38A/S38AUNG−/− B cells resulted from a lack of AID(S38A)-induced mutation of S regions, we sequenced 5’Sμ in B220+PNAhi Peyer’s patch germinal center B cells (GCBCs) that were sorted from AIDS38A/S38AMSH2−/−, AIDS38A/S38AUNG−/−, or control mice. As previously described [11], WT B cells generated transition and transversion mutations at all base pairs, MSH2−/− B cells displayed a reduction in mutations at A/T base pairs, and UNG−/− B cells showed an increase in transition mutations at G/C base pairs (Figure 2A). Mutations in 5’Sμ of AIDS38A/S38A B cells were reduced as compared to WT B cells, as previously reported [16, 17]. Similarly, the number and frequency of mutations in 5’Sμ were lower in AIDS38A/S38AMSH2−/− and AIDS38A/S38AUNG−/− B cells as compared to MSH2−/− and UNG−/− B cells, respectively (Figures 2A and 2C). AIDS38A/S38AMSH2−/− B cells had a detectable number of transition mutations and very few transversion mutations; however, the reduced mutation frequency suggests that the mutations observed in MSH2−/− B cells are pS38-AID dependent and may be mediated by the interaction of AID with RPA or APE1 [19, 26]. AIDS38A/S38AUNG−/− B cells had a higher mutation frequency at 5’Sμ as compared to AIDS38A/S38A B cells (Figure 2A) but, unlike AIDS38A/S38A B cells, AIDS38A/S38AUNG−/− B cells could not complete CSR in vivo (Figure 1B). Thus, the lack of CSR in AIDS38A/S38AUNG−/− B cells is not due to the inability of AID(S38A) to deaminate and mutate S regions, suggesting that phosphorylation of AID may regulate the generation of DSBs by MMR, the synapsis of S regions, or the ligation of recombining S regions.
Figure 2. AID-dependent DNA mutations in Sμ do not promote CSR in AIDS38A/S38AUNG−/− B cells.
Genomic DNA from B220+PNAHi GCBCs was isolated from mice of the indicated genotypes and analyzed for mutations within 5’Sμ. For all genotypes, n ≥ 3. (A) The total number of transition (red) and transversion (blue) mutations at A, C, G, and T base pairs is summarized in the tables. The total number of sequences analyzed is indicated below each table. (B) The number of mutations per 624 bp PCR amplicon is depicted in the pie charts. (C) The mean and standard deviation of the 5’Sμ mutation frequency in GCBCs of the indicated genotypes is depicted in the bar graph. The mutation frequency is calculated by dividing the total number of mutated base pairs by the total number of base pairs that was sequenced for each experimental mouse. A two-tailed Student’s t-test analyzed significant difference from WT mice (*P<0.05).
Insignificant affinity maturation in AIDS38A/S38AMSH2−/− and AIDS38A/S38AUNG−/− B cells
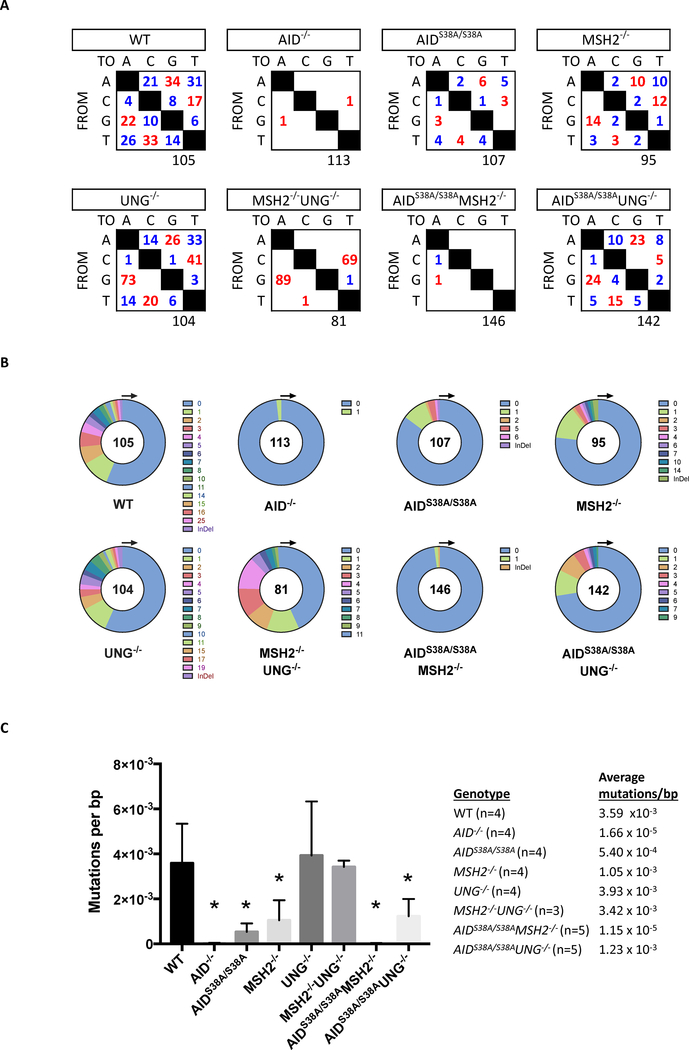
GCBCs complete SHM to generate untemplated mutations within recombined V(D)J coding segments to promote Ig affinity maturation. AID initiates SHM by deaminating deoxycytidine (C) residues in variable regions, which can be replicated across to form transition mutations at C:G base pairs [10]. Alternatively, removal of the uracil base by UNG creates an abasic site that can generate both transition and transversion mutations at C:G base pairs following DNA replication. Mutations at A:T base pairs are generated by MMR through the activity of the error prone DNA polymerase η [27]. To examine the role of AID phosphorylation in BER and MMR during affinity maturation, we analyzed mutations in the JH4 intron in B220+PNAHi Peyer’s patch GCBCs sorted from AIDS38A/S38AMSH2−/−, AIDS38A/S38AUNG−/−, or control mice [8, 28]. WT B cells had transition and transversion mutations at all base pairs, which were lost in AID−/− B cells and reduced in AIDS38A/S38A B cells (Figure 3A) [8, 16]. As previously shown, UNG−/− B cells displayed increased transition mutations at C:G base pairs, MSH2−/− B cells had reduced mutations at A:T base pairs, and MSH2−/−UNG−/− B cells had a complete loss of mutations at A:T base pairs [20]. AIDS38A/S38AMSH2−/− B cells showed mutations in the JH4 intron at the same frequency as AID−/− B cells (Figure 3A–C), which is the error rate of the DNA polymerase that was used to amplify the DNA [8, 9]. The absence of mutations in the JH4 intron of AIDS38A/S38AMSH2−/− B cells contrasts with the observed mutations in 5’Sμ (Figures 2A–C). This difference may result from the inability of AID(S38A) to deaminate V regions without RPA [26] or increased deamination within S regions, which form R-loops that expose the ssDNA substrates for AID [10]. AIDS38A/S38AMSH2−/− B cells display a VH186.2 mutation spectrum and frequency that is comparable to MSH2−/− B cells (Supplementary Figure 3), suggesting that mutations 3’ of V coding sequences in MSH2−/− B cells may be dependent on phosphorylated AID and RPA, which stabilizes V sequences for AID-mediated DNA deamination [26, 29].
Figure 3. AIDS38A/S38AUNG−/− B cells mutate the JH4 intron.
Genomic DNA from B220+PNAHi GCBCs was isolated from mice of the indicated genotypes and analyzed for mutations within the JH4 intron. For all genotypes, n ≥ 3. (A) The total number of transition (red) and transversion (blue) mutations at A, C, G, and T base pairs is summarized in the tables. The total number of sequences analyzed is indicated below each table. (B) The number of mutations per PCR amplicon is depicted in the pie charts. (C) The mean and standard deviation of the JH4 intron mutation frequency in GCBCs of the indicated genotypes is depicted in the bar graph. The mutation frequency is calculated by dividing the total number of mutated base pairs by the total number of base pairs that was sequenced for each experimental mouse. A two-tailed Student’s t-test analyzed significant difference from WT mice (*P<0.05).
In contrast, AIDS38A/S38AUNG−/− B cells can generate mutations in the JH4 intron (Figure 3A). As compared to AIDS38A/S38A B cells, the mutation spectra in AIDS38A/S38AUNG−/− B cells shifted to increased C:G transition mutations (Figure 3A), which is consistent with the observed increase in C:G transition mutations found in UNG−/− B cells [20]. However, the mutations per sequence and the mutation frequency of AIDS38A/S38AUNG−/− B cells was less than WT and UNG−/− B cells (Figures 3B and 3C), which highlights the previously described role of pS38-AID in SHM [16, 17]. Notably, the mutation spectra, the number of mutations per sequence, and the mutation frequency per base pair in AIDS38A/S38AUNG−/− B cells mirrored that of MSH2−/− B cells (Figures 2 and 3). The mutation frequency at 5’Sμ in AIDS38A/S38AUNG−/− and MSH2−/− B cells was 1.35 × 10−3 and 1.48 × 10−3 mutations/bp, respectively (Figure 2C), and the mutation frequency at the JH4 intron in AIDS38A/S38AUNG−/− and MSH2−/− B cells was 1.23 × 10−3 and 1.05 × 10−3 mutations/bp, respectively (Figure 3C). Furthermore, the frequency of mutations at A/T bases in both AIDS38A/S38AUNG−/− and MSH2−/− cells was similarly shifted. AIDS38A/S38AUNG−/− and MSH2−/− B cells showed a loss of mutations at T bases in 5’Sμ (Figure 2A) and comparable percentages of transition and transversion mutations at A bases at the JH4 intron (Figure 3A). AIDS38A/S38AUNG−/− B cells also mutated VH186.2 at frequencies comparable to MSH2−/− B cells (Supplementary Figure 3A). These data show that the SHM phenotype of the AIDS38A/S38AUNG−/− B cells closely resembles the SHM phenotype of MSH2−/− B cells and suggest that the altered SHM pattern in AIDS38A/S38AUNG−/− B cells results from a disruption of the MMR pathway due to the absence of phosphorylated AID.
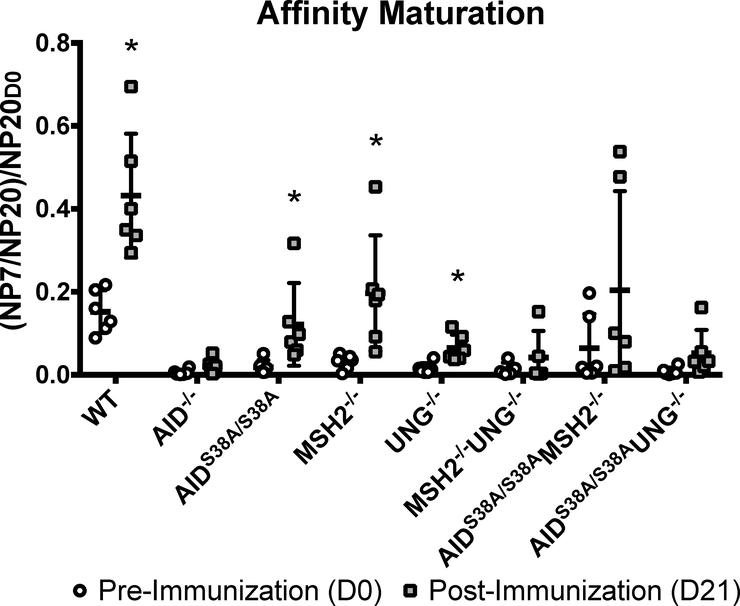
To determine if the observed JH4 and VH186.2 mutations correlate with affinity maturation, we performed NP-specific IgM ELISAs with sera from NP-CGG-immunized AIDS38A/S38AMSH2−/−, AIDS38A/S38AUNG−/−, and control mice. To quantify the change in affinity maturation, we first calculated the ratio of IgM that bound to NP7 (high affinity) and NP20 (high and low affinity) pre-immunization (D0) and post-immunization (D21). Subsequently, the NP7/NP20 ratio was normalized to the pre-immunization NP20 IgM (NP20D0) titer to account for variations in basal IgM titers, which are elevated in AID−/−, AIDS38A/S38A, MSH2−/−UNG−/−, AIDS38A/S38AMSH2−/−, and AIDS38A/S38AUNG−/− relative to WT mice (Figure 1B; P<0.05) [8, 11]. As expected, WT mice, unlike AID−/− mice, showed NP-specific IgM affinity maturation upon immunization with NP-CGG (Figure 4; P=0.006). Affinity maturation also occurred in AIDS38A/S38A, MSH2−/−, and UNG−/− mice (Figure 4; P=0.04, 0.007, 0.005, respectively) [30]. Consistent with the observed reduction in mutations at JH4 and VH186.2, AIDS38A/S38AMSH2−/− did not demonstrate significant NP-specific IgM affinity maturation (P=0.086). Surprisingly, the moderate mutation frequency in the JH4 intron and VH186.2 of AIDS38A/S38AUNG−/− mice did not translate into significant affinity maturation (P=0.105), even though these mice had a JH4 and VH186.2 mutation frequency and pattern that was comparable to MSH2−/− mice, which exhibit affinity maturation (Figures 3 and 4; Supplementary Figure 3). This observation is similar to the high mutation frequency in the 5’Sμ of AIDS38A/S38AUNG−/− B cells that did not lead to CSR, suggesting that pS38-AID is required for the recruitment of pathways downstream of AID-dependent deamination or mutation to drive affinity maturation or CSR.
Figure 4. AID-generated mutations do not promote affinity maturation in AIDS38A/S38AMSH2−/− and AIDS38A/S38AUNG−/− mice.
NP-specific serum IgM titers were measured by ELISA from mice of the indicated genotypes before and after immunization with NP-CGG. Affinity maturation was calculated by normalizing the ratio of high affinity anti-NP-IgM (NP7) and total anti-NP-IgM (NP20) at D21 post-immunization to the pre-immunization total anti-NP IgM (NP20D0) titers. Data is displayed with error bars representing standard deviation from the mean. A paired, two-tailed, Student’s t-test was used to determine statistical difference of NP affinity at D21 versus D0 of each genotype (*P<0.05). AIDS38A/S38AMSH2−/− and AIDS38A/S38AUNG−/− mice did not reach significant affinity maturation following immunization (P=0.086 and 0.105, respectively)
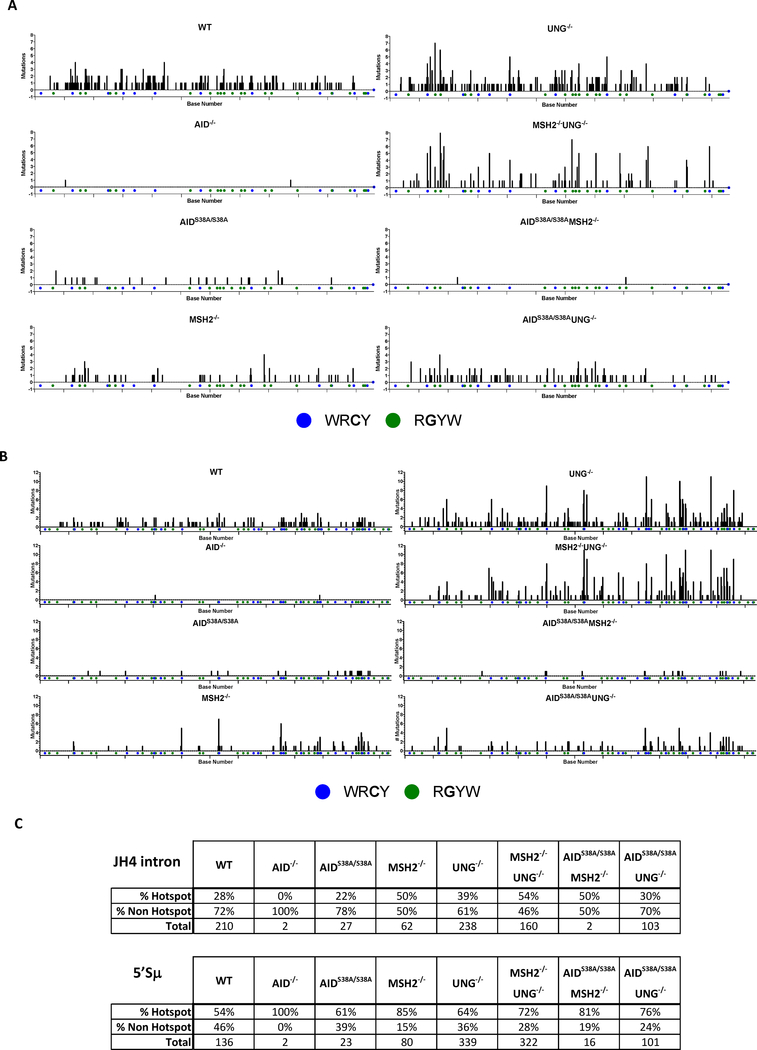
AID frequently deaminates C within RGYW/WRCY hotspot motifs in V and S regions [31]. To determine if the reduced affinity maturation in AIDS38A/S38AMSH2−/− and AIDS38A/S38AUNG−/− mice may be due to altered mutations at hotspot or non-hotspot motifs by AID(S38A), we catalogued the location and frequency of mutations at hotspot motifs in the JH4 intron and 5’Sμ for each genotype (Figure 5). Compared to WT B cells, the percentage of hotspot mutations was not reduced in the genotypes that show mutations above the background levels observed in AID−/− B cells (i.e. AIDS38A/S38A, MSH2−/−, UNG−/−, MSH2−/−UNG−/−, AIDS38A/S38AUNG−/−). Interestingly, MSH2−/−, MSH2−/−UNG−/−, and AIDS38A/S38AMSH2−/− B cells have more hotspot than non-hotspot mutations compared to WT, UNG−/−, and AIDS38A/S38A B cells, respectively (Figure 5C), consistent with the known role for MMR in generating non-hotspot mutations downstream of AID-induced deamination [32, 33]. A similar shift toward hotspot mutations was observed for UNG−/− and AIDS38A/S38AUNG−/− B cells in 5’Sμ (Figure 5C). However, at the JH4 intron, AIDS38A/S38AUNG−/− and WT B cells had similar percentages of hotspot and non-hotspot mutations, consistent with the known role for MSH2-dependent MMR in generating non-hotspot mutations at the variable region [32, 33]. Furthermore, the absence of affinity maturation in AIDS38A/S38AUNG−/− B cells was likely independent of DNA deamination by AID(S38A) because WT, AIDS38A/S38A, and AIDS38A/S38AUNG−/− B cells showed comparable percentages of JH4 hotspot mutations (Figure 5C). Thus, the decreased frequency of affinity maturation and CSR in AIDS38A/S38AUNG−/− B cells is likely due to altered processing of the AID-generated mutations by BER or MMR, rather than altered AID localization to V or S regions or reduced AID(S38A) deamination activity in vivo.
Figure 5. Mutations at the JH4 intron or 5’Sμ hotspots is unaltered in AIDS38A/S38A B cells.
The number of mutations at RGYW/WRCY hotspots within the JH4 intron (A) or 5’Sμ (B) PCR amplicon is analyzed for each genotype. On the x-axis, blue dots indicate the C in WRCY hotspots and green dots indicate the G in RGYW hotspots. Each tick mark on the x-axis demarcates a 50bp interval. (C) The percentage of mutations at RGYW/WRCY hotspots or non-hotspots within the JH4 intron or 5’Sμ is quantified for each genotype. The total number of mutations analyzed is indicated for each genotype.
DISCUSSION
In this study, we describe additional data that implicates a role for phosphorylated AID in directing BER via APE1 towards CSR and affinity maturation [19]. In support of our hypothesis, mice deficient for AID phosphorylation and MMR (AIDS38A/S38AMSH2−/−) cannot complete CSR both in vitro and in vivo and have a severe decrease in SHM. Furthermore, we have uncovered a previously unknown role for AID phosphorylation in promoting MMR-dependent CSR. B cells in mice deficient for AID phosphorylation and BER (AIDS38A/S38AUNG−/−) can mutate V and S regions but cannot complete affinity maturation or CSR. These data suggest that, in the absence of UNG, pS38-AID is required for MMR-mediated processing of deaminated DNA in SHM and CSR.
We propose a model whereby phosphorylation of AID directs BER or MMR-dependent CSR or SHM in the absence of the complementary pathway [11]. The lack of CSR and reduced SHM in AIDS38A/S38AMSH2−/− B cells suggests that pS38-AID and its interaction with APE1 complements MMR-dependent CSR and SHM (Figure 1 and Figure 2) [19]. Similarly, the absence of AID phosphorylation and BER in AIDS38A/S38AUNG−/− B cells completely blocks CSR, suggesting that pS38-AID regulates the recruitment of MMR proteins to generate DSBs, synapsis of recombining S regions, or ligation of DSBs between S regions. The phosphorylated form of AID may be required to recruit PMS2 or EXO1 to S regions to generate the DNA breaks for CSR [10]. Alternatively, pS38-AID may be interacting with ATM, RPA, or another protein to tether distal, recombining S regions together and promote their subsequent ligation [34].
The lack of DSB formation in the S regions may account for the loss of CSR in AIDS38A/S38AUNG−/− B cells. MSH2−/−UNG−/− B cells cannot complete CSR likely due to the absence of DSBs in S regions and, consequently, can only produce G/C transition mutations [35–39]. AIDS38A/S38AUNG−/− B cells generate mutations at all bases in 5’Sμ (Figure 2), suggesting that SSBs are formed at sites of AID(S38A) deamination. However, the absence of CSR suggests that these SSBs do not form a sufficient number of DSBs to complete CSR. Alternatively, altered S-S junction formation may account for the lack of CSR despite the presence of 5’Sμ mutations in the AIDS38A/S38AUNG−/− B cells. AIDS38A/S38A B-cells have disrupted interaction between AID and APE1 [19] and APE1-deficient CH12 cells have reduced Sμ-Sα synapsis and CSR without affecting mutations in S regions [40]. Additionally, ATM−/− B cells have severely impaired interaction between pS38-AID and APE1 [19] and B cells from ATM−/− patients demonstrate altered S-S junctions [41]. Thus, the absence of AID phosphorylation and recruitment of APE1 may alter S-S synapsis or ligation, preventing AIDS38A/S38AUNG−/− B cells from completing CSR despite the moderate frequency of 5’Sμ mutations.
Recently, APE1 and APE2 were shown to be differentially expressed in pre-GC and GC B cells. APE1 is highly expressed in pre-GC B cells and downregulated in GC B cells, whereas APE2 expression is low in pre-GC B cells and upregulated in GC B cells [42, 43]. Furthermore, GLTs were highly expressed in pre-GC B cells and reduced in GC B cells, indicating that CSR occurred prior to GC formation [42]. Phylogenetic analysis of GC V regions indicated that CSR occurred prior to SHM of V coding sequences [42]. In our studies, the absence of CSR despite the generation of mutations in Sμ and the JH4 intron of AIDS38A/S38AUNG−/− B cells (Figures 2C and 3C) implicates differential roles for pS38-AID during CSR and SHM in the absence of UNG. Outside of GCs when APE1 and GLT are high, pS38-AID may promote the generation of a high density of DSBs at S regions by recruiting both APE1 and MMR proteins, which drives CSR [19]. The hypomorphic CSR in UNG−/− mice suggests that pS38-AID and MMR partially compensates for the absence BER. Thus, the absence of either pS38-AID or MMR completely inhibits CSR in AIDS38A/S38AUNG−/− and MSH2−/−UNG−/− mice and B cells (Figure 1) [11]. In GC B cells undergoing SHM, pS38-AID cannot recruit APE1 to V regions to create DSBs because APE1 expression is low [42, 43]. In this case, APE2 promotes mutations rather than DSB formation, possibly through a pS38-AID-dependent mechanism (Figure 3) [43]. Additional biochemical studies examining the interaction of pS38-AID with APE2 and other DNA repair proteins in pre-GC and GC B cells will clarify the differential function of AID phosphorylation in CSR and SHM.
The presence of mutations in VH186.2 and the absence of mutations in the JH4 intron was observed exclusively in AIDS38A/S38AMSH2−/− B cells (Figure 3A and Supplementary Figure 3A). We observed a decrease in mutation frequency at the JH4 intron as compared to VH186.2 in AIDS38A/S38AMSH2−/− B cells (Figure 3 and Supplementary Figure 3). This phenotype suggests pS38-AID and MSH2 are in complementary pathways for the propagation of V region mutations, which spread 3’ from the V promoter for 2 kb [29]. We hypothesize that, in the absence of MMR, pS38-AID recruits RPA to stabilize the transcription bubbles in the V region that would permit the generation of mutations downstream of the V promoter [26]. In the presence of MMR, SSBs in V sequences may recruit RPA independently of pS38-AID to propagate mutations 3’ of the V region, which is observed in AIDS38A/S38A B cells (Figure 3 and Supplementary Figure 3) [44]. In the absence of both MMR and pS38-AID, transcription of the V gene may compensate for the lack of RPA to allow AID(S38A) to generate the limited mutations observed in the V region of AIDS38A/S38AMSH2−/− B cells (i.e. mutations in VH186.2 but not the JH4 intron) (Figure 3 and Supplementary Figure 3).
The absence of CSR and the resulting hyper-IgM in AIDS38A/S38AUNG−/− mice resembles the CSR phenotype in MSH2−/−UNG−/− mice [11], suggesting a disruption of the MMR pathway due to the absence of phosphorylated AID. The similarity in the shift in SHM spectra between AIDS38A/S38AUNG−/− and MSH2−/− B cells (Figures 2 and 3; Supplementary Figure 3) further implicates a perturbation of MMR in AIDS38A/S38AUNG−/− B cells. Whether the disruption in MMR occurs as a result of disrupted interactions between MMR proteins and AID(S38A), as compared to WT AID, remains unclear. Binding assays using anti-MSH2 antibodies co-immunoprecipitated MSH6 but not AID in WT B cell lysates (Supplementary Figure 4), suggesting that pS38-AID does not actively recruit MSH2/MSH6 to sites of deamination and the disruption of MMR in AIDS38A/S38AUNG−/− mice may be downstream of MSH2/MSH6 recognition of U:G mismatches. Phosphorylated AID may promote the formation of DSBs that are required to complete CSR by promoting the recruitment or stable binding of PMS2, MLH1, or EXO1 [45]. Alternatively, the role of phosphorylated AID in MMR may be mediated by RPA, which promotes AID deamination of transcribed ssDNA [26, 39]. RPA has been shown to facilitate MMR [46, 47] and AID phosphorylation mutants, including AIDS38A/S38A, display a significantly reduced recruitment of RPA to the IgH locus [34]. Thus, the absence of RPA-dependent recruitment of MMR proteins in AIDS38A/S38AUNG−/− B cells may account for the observed block in CSR and reduced SHM.
Unlike AIDS38A/S38AUNG−/− B cells, which generate mutations at both 5’Sμ and the JH4 intron, AIDS38A/S38AMSH2−/− B cells generate few mutations in 5’Sμ and no mutations at the JH4 intron (Figures 2 and 3). The mutations at 5’Sμ in the AIDS38A/S38AMSH2−/− B cells may be the result of replication over abasic sites, which generates the G/C mutations, or residual BER, which generates the A/T mutations. In support of the latter hypothesis, ubiquitinated PCNA has been shown to be responsible for the generation of A/T mutation through BER in MSH2−/− B cells [48]. We speculate that the lack of mutation in the JH4 intron is due to canonical repair. In contrast to the JH4 intron, canonical DNA repair may be unable to correct the high density of deaminated DNA within the GC-rich Sμ region. Interestingly, APE1 interacts with NHEJ proteins, such as XRCC1 [49], to direct canonical DNA repair. Phosphorylation of AID and the subsequent interaction of pS38-AID with APE1 may disrupt the APE1/XRCC1 interaction at S regions, thereby preventing canonical repair mediated by XRCC1 and promoting CSR. Consistent with this hypothesis, XRCC1+/− B cells have increased SHM [50] and deletion of XRCC1 in CH12 cells increases CSR [51]. Thus, the disruption of the AID and APE1 interaction in AIDS38A/S38AMSH2−/− B cells may direct canonical DNA repair at the JH4 intron and S regions, which manifests as reduced mutation frequency.
The C-terminal domain of AID has been proposed to promote the recruitment of UNG and MSH2/MSH6 to S regions during CSR [52]. Deletion of the C-terminal ten amino acids of AID significantly reduces CSR to 10% of WT levels without altering 5’Sμ mutations [53], which almost resembles the CSR and SHM phenotype of the AIDS38A/S38AUNG−/− B cells. The similar phenotype observed in these studies suggests that the phosphorylation of AID and the C-terminal domain of AID may function cooperatively to recruit or stabilize MMR proteins at V or S regions during SHM or CSR, respectively. Alternatively, the C-terminus may bind to an unidentified factor that regulates MMR processing of deaminated DNA into DSBs. Loss of this factor could allow for mutational repair but severely limits CSR, which is consistent with the observed 5’Sμ mutations and loss of CSR in B cells expressing the AID C-terminal deletion mutant [53]. Additional biochemical studies characterizing the interaction between the C-terminal domain, pS38-AID, and MMR proteins will provide insight into the molecular pathways by which phosphorylated AID regulates CSR and SHM through MMR.
Supplementary Material
KEY POINTS.
No CSR or affinity maturation in AIDS38A/S38AMSH2−/− and AIDS38A/S38AUNG−/− mice
AID phosphorylated at serine 38 regulates BER- and MMR-dependent CSR and SHM
ACKNOWLEDGEMENTS
We thank Jayanta Chaudhuri for the anti-AID antibody; Tomas Lindahl for the UNG−/− mice; Hein te Riele for the MSH2−/− mice; Tasuku Honjo for the AID−/− mice; Simin Zheng, Montse Cols, and Emily Sible for critical reading of the manuscript; and Bharat Vaidyanathan for scientific discussions.
A. Matthews was supported by The American Association of Immunologists Careers in Immunology Fellowship
B. Q. Vuong was was supported by The American Association of Immunologists Early Career Faculty Travel Grant, the PSC-CUNY Enhanced Research Award, The National Institute on Minority Health and Health Disparities (5G12MD007603), The National Cancer Institute (2U54CA132378), and The National Institute of General Medical Sciences (1SC1GM132035-01)
REFERENCES
- 1.Murphy K, Travers P, Walport M and Janeway C (2012) Janeway’s immunobiology. Garland Science, New York. [Google Scholar]
- 2.Alt FW, Zhang Y, Meng FL, Guo C and Schwer B (2013) Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell 152:417–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kenter AL (2005) Class switch recombination: an emerging mechanism. Curr Top Microbiol Immunol 290:171–99. [DOI] [PubMed] [Google Scholar]
- 4.Xu Z, Zan H, Pone EJ, Mai T and Casali P (2012) Immunoglobulin class-switch DNA recombination: induction, targeting and beyond. Nat Rev Immunol 12:517–31. doi: 10.1038/nri3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matthews AJ, Zheng S, DiMenna LJ and Chaudhuri J (2014) Regulation of immunoglobulin class-switch recombination: choreography of noncoding transcription, targeted DNA deamination, and long-range DNA repair. Adv Immunol 122:1–57. doi: 10.1016/B978-0-12-800267-4.00001-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keim C, Kazadi D, Rothschild G and Basu U (2013) Regulation of AID, the B-cell genome mutator. Genes Dev 27:1–17. doi: 10.1101/gad.200014.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Noia JM and Neuberger MS (2007) Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem 76:1–22. [DOI] [PubMed] [Google Scholar]
- 8.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y and Honjo T (2000) Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102:553–63. [DOI] [PubMed] [Google Scholar]
- 9.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A and Durandy A (2000) Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell 102:565–75. [DOI] [PubMed] [Google Scholar]
- 10.Methot SP and Di Noia JM (2017) Molecular Mechanisms of Somatic Hypermutation and Class Switch Recombination. Adv Immunol 133:37–87. doi: 10.1016/bs.ai.2016.11.002 [DOI] [PubMed] [Google Scholar]
- 11.Rada C, Di Noia JM and Neuberger MS (2004) Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol Cell 16:163–71. [DOI] [PubMed] [Google Scholar]
- 12.Delbos F, Aoufouchi S, Faili A, Weill JC and Reynaud CA (2007) DNA polymerase eta is the sole contributor of A/T modifications during immunoglobulin gene hypermutation in the mouse. J Exp Med 204:17–23. doi: 10.1084/jem.20062131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McBride KM, Gazumyan A, Woo EM, Barreto VM, Robbiani DF, Chait BT and Nussenzweig MC (2006) Regulation of hypermutation by activation-induced cytidine deaminase phosphorylation. Proc Natl Acad Sci U S A 103:8798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Basu U, Chaudhuri J, Alpert C, Dutt S, Ranganath S, Li G, Schrum JP, Manis JP and Alt FW (2005) The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature 438:508–11. [DOI] [PubMed] [Google Scholar]
- 15.Pasqualucci L, Kitaura Y, Gu H and Dalla-Favera R (2006) PKA-mediated phosphorylation regulates the function of activation-induced deaminase (AID) in B cells. Proc Natl Acad Sci U S A 103:395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng HL, Vuong BQ, Basu U, Franklin A, Schwer B, Astarita J, Phan RT, Datta A, Manis J, Alt FW and Chaudhuri J (2009) Integrity of the AID serine-38 phosphorylation site is critical for class switch recombination and somatic hypermutation in mice. Proc Natl Acad Sci U S A 106:2717–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McBride KM, Gazumyan A, Woo EM, Schwickert TA, Chait BT and Nussenzweig MC (2008) Regulation of class switch recombination and somatic mutation by AID phosphorylation. J Exp Med 205:2585–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vuong BQ, Lee M, Kabir S, Irimia C, Macchiarulo S, McKnight GS and Chaudhuri J (2009) Specific recruitment of protein kinase A to the immunoglobulin locus regulates class-switch recombination. Nat Immunol 10:420–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vuong BQ, Herrick-Reynolds K, Vaidyanathan B, Pucella JN, Ucher AJ, Donghia NM, Gu X, Nicolas L, Nowak U, Rahman N, Strout MP, Mills KD, Stavnezer J and Chaudhuri J (2013) A DNA break- and phosphorylation-dependent positive feedback loop promotes immunoglobulin class-switch recombination. Nat Immunol 14:1183–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T and Neuberger MS (2002) Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr Biol 12:1748–55. [DOI] [PubMed] [Google Scholar]
- 21.de Wind N, Dekker M, Berns A, Radman M and te Riele H (1995) Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell 82:321–30. [DOI] [PubMed] [Google Scholar]
- 22.Pucella JN, Yen WF, Kim MV, van der Veeken J, Luo CT, Socci ND, Naito Y, Li MO, Iwai N and Chaudhuri J (2015) miR-182 is largely dispensable for adaptive immunity: lack of correlation between expression and function. J Immunol 194:2635–42. doi: 10.4049/jimmunol.1402261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heise N and Klein U (2017) Somatic Hypermutation and Affinity Maturation Analysis Using the 4-Hydroxy-3-Nitrophenyl-Acetyl (NP) System. Methods Mol Biol 1623:191–208. doi: 10.1007/978-1-4939-7095-7_16 [DOI] [PubMed] [Google Scholar]
- 24.Yeap LS, Hwang JK, Du Z, Meyers RM, Meng FL, Jakubauskaite A, Liu M, Mani V, Neuberg D, Kepler TB, Wang JH and Alt FW (2015) Sequence-Intrinsic Mechanisms that Target AID Mutational Outcomes on Antibody Genes. Cell 163:1124–1137. doi: 10.1016/j.cell.2015.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu M and Schatz DG (2009) Balancing AID and DNA repair during somatic hypermutation. Trends Immunol 30:173–81. doi: S1471-4906(09)00042-8 [pii] 10.1016/j.it.2009.01.007 [DOI] [PubMed] [Google Scholar]
- 26.Chaudhuri J, Khuong C and Alt FW (2004) Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature 430:992–8. [DOI] [PubMed] [Google Scholar]
- 27.Faili A, Aoufouchi S, Weller S, Vuillier F, Stary A, Sarasin A, Reynaud CA and Weill JC (2004) DNA Polymerase {eta} Is Involved in Hypermutation Occurring during Immunoglobulin Class Switch Recombination. J Exp Med 199:265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jolly CJ, Klix N and Neuberger MS (1997) Rapid methods for the analysis of immunoglobulin gene hypermutation: application to transgenic and gene targeted mice. Nucleic Acids Res 25:1913–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maul RW and Gearhart PJ (2010) AID and somatic hypermutation. Adv Immunol 105:159–91. doi: 10.1016/S0065-2776(10)05006-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zahn A, Daugan M, Safavi S, Godin D, Cheong C, Lamarre A and Di Noia JM (2013) Separation of function between isotype switching and affinity maturation in vivo during acute immune responses and circulating autoantibodies in UNG-deficient mice. J Immunol 190:5949–60. doi: 10.4049/jimmunol.1202711 [DOI] [PubMed] [Google Scholar]
- 31.Chaudhuri J, Basu U, Zarrin A, Yan C, Franco S, Perlot T, Vuong B, Wang J, Phan RT, Datta A, Manis J and Alt FW (2007) Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv Immunol 94:157–214. doi: 10.1016/S0065-2776(06)94006-1 [DOI] [PubMed] [Google Scholar]
- 32.Roa S, Li Z, Peled JU, Zhao C, Edelmann W and Scharff MD (2010) MSH2/MSH6 complex promotes error-free repair of AID-induced dU:G mispairs as well as error-prone hypermutation of A:T sites. PLoS One 5:e11182. doi: 10.1371/journal.pone.0011182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rada C, Ehrenstein MR, Neuberger MS and Milstein C (1998) Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity 9:135–41. [DOI] [PubMed] [Google Scholar]
- 34.Yamane A, Resch W, Kuo N, Kuchen S, Li Z, Sun HW, Robbiani DF, McBride K, Nussenzweig MC and Casellas R (2011) Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat Immunol 12:62–9. doi: 10.1038/ni.1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schrader CE, Linehan EK, Mochegova SN, Woodland RT and Stavnezer J (2005) Inducible DNA breaks in Ig S regions are dependent on AID and UNG. J Exp Med 202:561–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imai K, Slupphaug G, Lee WI, Revy P, Nonoyama S, Catalan N, Yel L, Forveille M, Kavli B, Krokan HE, Ochs HD, Fischer A and Durandy A (2003) Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat Immunol 4:1023–8. [DOI] [PubMed] [Google Scholar]
- 37.Maul RW, Saribasak H, Martomo SA, McClure RL, Yang W, Vaisman A, Gramlich HS, Schatz DG, Woodgate R, Wilson DM 3rd and Gearhart PJ (2011) Uracil residues dependent on the deaminase AID in immunoglobulin gene variable and switch regions. Nat Immunol 12:70–6. doi: 10.1038/ni.1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xue K, Rada C and Neuberger MS (2006) The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2−/− ung−/− mice. J Exp Med 203:2085–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamane A, Robbiani DF, Resch W, Bothmer A, Nakahashi H, Oliveira T, Rommel PC, Brown EJ, Nussenzweig A, Nussenzweig MC and Casellas R (2013) RPA accumulation during class switch recombination represents 5’−3’ DNA-end resection during the S-G2/M phase of the cell cycle. Cell Rep 3:138–47. doi: 10.1016/j.celrep.2012.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu J, Husain A, Hu W, Honjo T and Kobayashi M (2014) APE1 is dispensable for S-region cleavage but required for its repair in class switch recombination. Proc Natl Acad Sci U S A 111:17242–7. doi: 10.1073/pnas.1420221111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peron S, Pan-Hammarstrom Q, Imai K, Du L, Taubenheim N, Sanal O, Marodi L, Bergelin-Besancon A, Benkerrou M, de Villartay JP, Fischer A, Revy P and Durandy A (2007) A primary immunodeficiency characterized by defective immunoglobulin class switch recombination and impaired DNA repair. J Exp Med 204:1207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roco JA, Mesin L, Binder SC, Nefzger C, Gonzalez-Figueroa P, Canete PF, Ellyard J, Shen Q, Robert PA, Cappello J, Vohra H, Zhang Y, Nowosad CR, Schiepers A, Corcoran LM, Toellner KM, Polo JM, Meyer-Hermann M, Victora GD and Vinuesa CG (2019) Class-Switch Recombination Occurs Infrequently in Germinal Centers. Immunity 51:337–350 e7. doi: 10.1016/j.immuni.2019.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stavnezer J, Linehan EK, Thompson MR, Habboub G, Ucher AJ, Kadungure T, Tsuchimoto D, Nakabeppu Y and Schrader CE (2014) Differential expression of APE1 and APE2 in germinal centers promotes error-prone repair and A:T mutations during somatic hypermutation. Proc Natl Acad Sci U S A 111:9217–22. doi: 10.1073/pnas.1405590111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zanotti KJ and Gearhart PJ (2016) Antibody diversification caused by disrupted mismatch repair and promiscuous DNA polymerases. DNA Repair (Amst) 38:110–6. doi: 10.1016/j.dnarep.2015.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chaudhuri J and Alt FW (2004) Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol 4:541–52. doi: 10.1038/nri1395 [DOI] [PubMed] [Google Scholar]
- 46.Genschel J and Modrich P (2003) Mechanism of 5’-directed excision in human mismatch repair. Mol Cell 12:1077–86. [DOI] [PubMed] [Google Scholar]
- 47.Lin YL, Shivji MK, Chen C, Kolodner R, Wood RD and Dutta A (1998) The evolutionarily conserved zinc finger motif in the largest subunit of human replication protein A is required for DNA replication and mismatch repair but not for nucleotide excision repair. J Biol Chem 273:1453–61. [DOI] [PubMed] [Google Scholar]
- 48.Krijger PH, Langerak P, van den Berk PC and Jacobs H (2009) Dependence of nucleotide substitutions on Ung2, Msh2, and PCNA-Ub during somatic hypermutation. J Exp Med 206:2603–11. doi: 10.1084/jem.20091707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vidal AE, Boiteux S, Hickson ID and Radicella JP (2001) XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. EMBO J 20:6530–9. doi: 10.1093/emboj/20.22.6530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saribasak H, Maul RW, Cao Z, McClure RL, Yang W, McNeill DR, Wilson DM 3rd and Gearhart PJ (2011) XRCC1 suppresses somatic hypermutation and promotes alternative nonhomologous end joining in Igh genes. J Exp Med 208:2209–16. doi: 10.1084/jem.20111135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han L, Mao W and Yu K (2012) X-ray repair cross-complementing protein 1 (XRCC1) deficiency enhances class switch recombination and is permissive for alternative end joining. Proc Natl Acad Sci U S A 109:4604–8. doi: 10.1073/pnas.1120743109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ranjit S, Khair L, Linehan EK, Ucher AJ, Chakrabarti M, Schrader CE and Stavnezer J (2011) AID binds cooperatively with UNG and Msh2-Msh6 to Ig switch regions dependent upon the AID C terminus. J Immunol 187:2464–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barreto V, Reina-San-Martin B, Ramiro AR, McBride KM and Nussenzweig MC (2003) C-terminal deletion of AID uncouples class switch recombination from somatic hypermutation and gene conversion. Mol Cell 12:501–8. doi: S1097276503003095 [pii] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.