Abstract
Primary ciliary dyskinesia (PCD) is a rare disorder that affects the biogenesis or function of motile cilia resulting in chronic airway disease. PCD is genetically and phenotypically heterogeneous, with causative mutations identified in over 40 genes; however, the genetic basis of many cases is unknown.
Using whole exome sequencing, we identified three affected siblings with clinical symptoms of PCD but normal ciliary structure, carrying compound heterozygous loss-of-function variants in CFAP221. Computational analysis suggests that these variants are the most damaging alleles shared by all three siblings. Nasal epithelial cells from one of the subjects demonstrated slightly reduced beat frequency (16.5 Hz vs 17.7 Hz, p=0.16); however, waveform analysis revealed that the CFAP221 defective cilia beat in an aberrant circular pattern.
These results show that genetic variants in CFAP221 cause PCD and that CFAP221 should be considered a candidate gene in cases where PCD is suspected but cilia structure and beat frequency appear normal.
Keywords: Primary ciliary dyskinesia, PCD, CFAP221, PCDP1, ciliary waveform
Introduction
Primary ciliary dyskinesia (PCD) is a rare genetic disease (MIM: 244400), caused by genetic lesions that impair the function of the motile cilia that line the airways, thereby reducing mucociliary clearance and resulting in chronic respiratory infections. Currently there are no specific treatments available for PCD, and early diagnosis is crucial to ensure proper disease management and minimize loss of lung function1. Advances in whole exome sequencing and analysis have contributed to the identification of PCD-causing mutations in >40 genes2. However, the underlying genetic cause is still unknown in ~30% of PCD cases, and in subjects with no obvious defects in ciliary structure or beat frequency, establishing a firm diagnosis remains challenging.
Material and Methods
Subjects
Individuals included in the study had a clinical diagnosis of PCD confirmed by standard clinical diagnostic criteria. For studies of affected individuals and their families, a signed and informed consent was obtained from all participants. All protocols involving human studies were approved by University of North Carolina Medical School and/or McGill University Health Centre Research Institute Institutional Review Boards.
Genetic Analysis
Identification of CFAP221 variants was performed by whole exome sequence analysis using the population sampling probability (PSAP) framework as previously described3, as detailed in Supplementary Information.
Airway epithelial cell cultures
Human nasal epithelial (HNE) cells from proband 2-I and controls were obtained as previously described4. The nasal cells were expanded as conditionally reprogrammed cells (CRC)5 and cultured as previously described6. Human bronchial epithelial (HBE) cells were cultured at the air/liquid interphase (ALI) as previously described7. These protocols have been approved by the University of North Carolina (UNC) Medical School Institutional Review Board.
Reverse transcription polymerase chain reaction (RT-PCR)
Semi-quantitative RT-PCR was performed using standard protocols (Takara Bio USA, Inc.), as detailed in Supplementary Information.
Waveform analysis and ciliary beat frequency measurements
To evaluate the waveform and beat direction, high resolution videos of ciliated cells were recorded and analyzed as described in Supplementary Information.
Results and Discussion
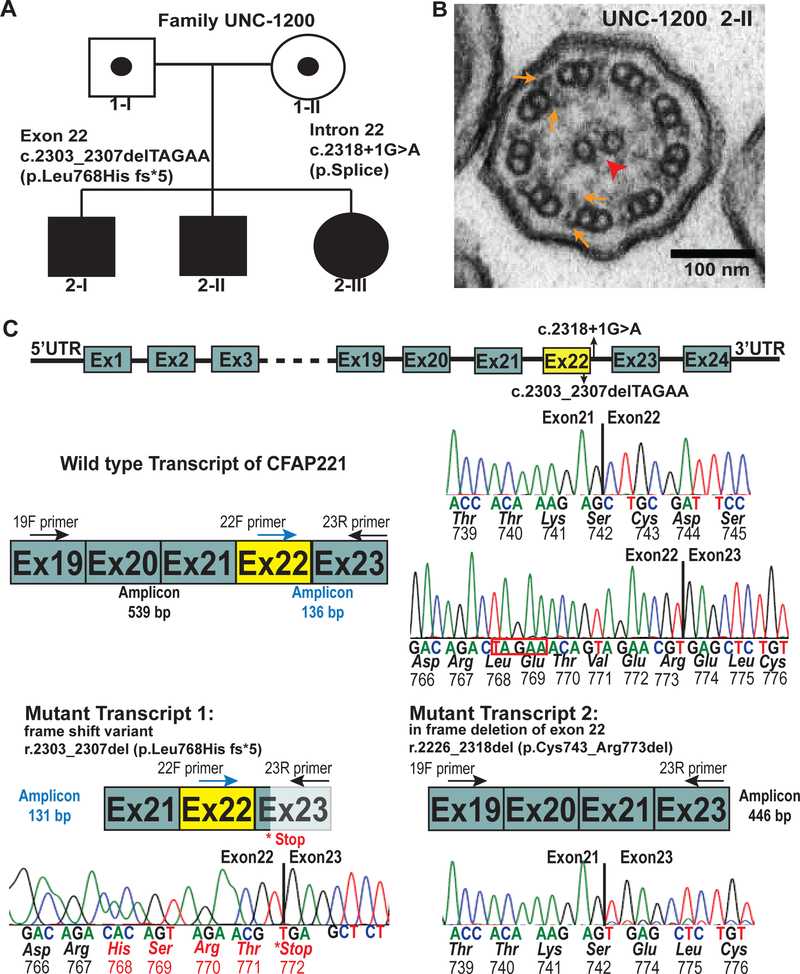
We identified three French-Canadian siblings from family UNC-1200 (Figure 1A) with phenotypes suggestive of PCD (Supplementary Table 1), including neonatal respiratory distress, chronic sinus disease, recurrent otitis media, and bronchiectasis in two individuals. None of the subjects had situs inversus. Sweat chloride testing and immunology investigations were normal in all three siblings. The rate of nasal nitric oxide production (nNO) was normal (122, 171, 189 nL/min; cut-off of 77 nL/min8), and examination of ciliary axonemes by TEM did not reveal obvious ultrastructural defects (Figure 1B).
Figure 1: CFAP221 variants in PCD subjects.
(A) Segregation analysis of CFAP221 genetic variants found in family UNC-1200 [c.2303_2307delTAGAA p.Leu768Hiffs*5] and c.2318+1G>A p. splice. Filled symbols indicate PCD affected individuals.
(B) Transmission electron micrograph of axonemal cross sections of nasal epithelium from proband 2-II showing the central pair (arrowhead) surrounded by nine microtubule doublets. Outer dynein arms and inner dynein arms (arrows) project from each doublet normally.
(C) Genomic organization of CFAP221 and sequencing of transcript variants. (top) Schematic representation of the genomic localization of the genetic variants in CFAP221 found in the PCD subjects. Solid box designates exons, horizontal lines designates introns.
(middle) Schematic showing a wild type transcript region of CFAP221 and its corresponding sequencing analysis.
(bottom) Reverse-transcriptase polymerase chain reaction with primers 19F or 22F or 23R (see Supplementary Table 3 for primer sequences) showed two major transcripts. Transcript 1 shows the deletion of 5 bases (highlighted in red box, above), leading to a frame shift of the sequence, resulting in a premature stop codon. Transcript 2 shows the in frame deletion of exon 22. Schematic of wild type and mutant transcripts and the corresponding electropherograms with exact location of the deletions are shown. Primers are designated as F (forward) and R (reverse) in their corresponding cDNA locations. Exon-exon junctions are shown by the vertical solid lines. Base sequence, amino acid sequence, and codon numbers are shown.
We carried out whole exome sequencing in one proband (2-II) and analyzed the resulting genotype data using a statistical framework for assessing the significance of variants from n=1 cases of rare genetic disease3. This identified compound heterozygous variants in cilia and flagella associated protein 221 (CFAP221; [GenBank: NM_001271049.2, Chr.2q14.2] previously known as PCDP1) as the most damaging variants in the genome of 2-II and most plausible cause of a rare recessive disorder (Table 19, 10 and Figure 1C). Segregation analysis confirmed that both variants were present in all three affected siblings and parents were carriers for each variant (Supplementary Figure 1). A frameshift variant in exon 22 [c.2303_2307delTAGAA p.Leu768HisFs*5] is predicted to result in a premature termination codon. The other variant (c.2318+1G>A) disrupts a canonical splice donor site in intron 22 that causes aberrant splicing (Figure 1C)9. RT-PCR analysis of exon 22, using primers spanning exon 19–23, of control human nasal epithelial (HNE) cells amplified a 539 base pair product (Figure 1C). In HNE cells from subject 2-I we confirmed the five base pair deletion (r.2303_2307del) resulting in a premature stop codon (Figure 1C). Analysis of the intron 22 splice site variant revealed the in frame deletion of exon 22 (r.2226_2318del).This is predicted to result in deletion of 31 amino acid residues (p.Cys743_Arg773del) (Figure 1C).
Table 1.
List of the top genes identified in 2-II using PSAP analysis. Three genes in the entire exome contained two damaging variants after filtering.
| Gene | Transcript | Position (hg19) | Type | AAChange | Alt Read |
Total Read |
gnomAD Freq |
CADD | PSAP pvalue |
|---|---|---|---|---|---|---|---|---|---|
| PCDP1 | ENST00000413369.3 | 2:120404611_TAGAA>- | frameshift deletion | L768fs | 13 | 29 | NA | 35 | 1.00E-06 |
| PCDP1 | ENST00000413369.3 | 2:120404627_G>A | splicing | NA | 9 | 20 | 0 | 24.3 | 1.00E-06 |
| NEDD41 | ENST00000338963.2 | 15:56208554_->TA | frameshift insertion | N159fs | 36 | 82 | NA | 38 | 1.00E-06 |
| NEDD41 | ENST00000338963.2 | 15:56208558_C>A | nonsynonymous SNV | D158Y | 39 | 85 | NA | 26.4 | 1.00E-06 |
| URB12 | ENST00000382751.3 | 21:33735605_A>G | nonsynonymous SNV | S457P | 36 | 72 | 0.0001 | 26.6 | 0.000135 |
| URB12 | ENST00000382751.3 | 21:33719498_G>A | nonsynonymous SNV | P1212L | 62 | 110 | 0.0004 | 13.69 | 0.000135 |
Review of the exome sequencing data revealed that both NEDD4 variants were in cis; hence the variants in NEDD4 were excluded as a cause of PCD.
We examined the expression and localization of CFAP221 in normal human bronchial epithelial (HBE) cells (Supplementary Figure 2) cultured at an air/liquid interface (ALI)7. RT-PCR demonstrated that the expression of CFAP221 increased during ciliated cell differentiation (Supplementary Figure 2A). Western blotting and immunofluorescence showed that CFAP221 is not expressed in undifferentiated airway cells, and is localized primarily to the axoneme in differentiated ciliated cells (Supplementary Figure 2B and 2C).
To investigate the impact of the genetic variants in CFAP221 on ciliary function, a nasal biopsy from subject 2-I was expanded as conditionally reprogrammed cells11, cultured at the ALI to induce differentiation, and examined for CFAP221 expression (Supplementary Figure 3A). The localization of CFAP221 in ciliated cells was evaluated by immunofluorescence (Supplementary Figure 3B). In contrast to control cells, cells from the PCD subject showed no specific staining. The absence of CFAP221 in ciliated cells from subject 2-I probably results from the non-sense mediated decay of aberrant transcripts (Figure 1). Immunofluorescent staining for other ciliary proteins, RSPH1 and ERICH3 showed a similar pattern in the 2-I cells, as compared to control cells (Supplementary Figure 3C). This result agrees with the observation of normal ciliary ultrastructure by TEM (Figure 1B).
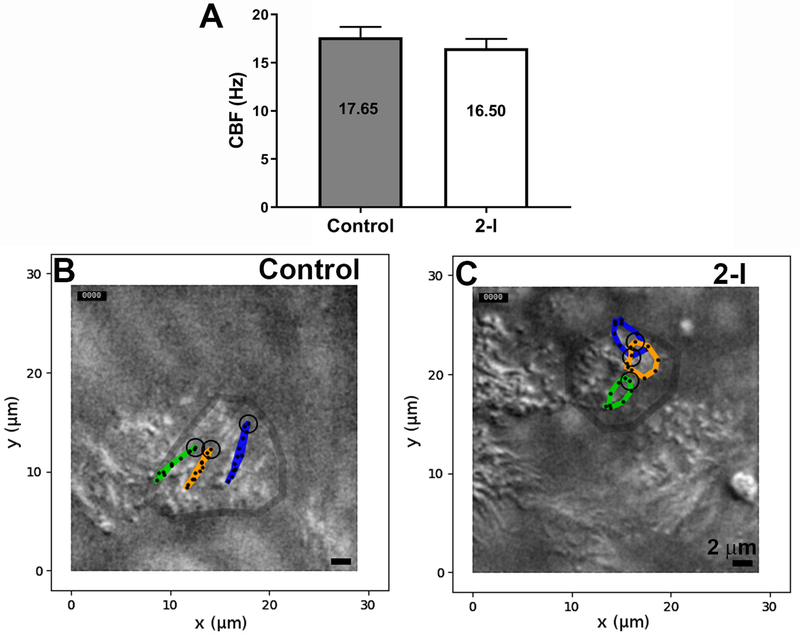
After differentiation of cells from subject 2-I, we observed vigorous ciliary activity, with a beat frequency that was only slightly reduced compared to parallel control cultures (16.5 Hz vs 17.7 Hz, p=0.16, Figure 2A). However, waveform analysis in the xy direction (top down) revealed that the CFAP221-deficient cilia beat in a slightly circular pattern, compared to the linear, back and forth motion of control cilia (Figure 2B, 2C and Supplementary Videos 1-4). This difference, when measured as the maximum deviation from a linear beat, was highly significant (control=0.27 ± 0.06 μm; 2-II=0.92 ± 0.5 μm; p<0.0015, n=24 cilia for each group).
Figure 2: Genetic defects in CFAP221 cause defective ciliary waveform.
(A) The CBF in CFAP221 deficient cells (2-II) was slightly reduced compared to control (n=6 cultures from each).
(B and C) Two-dimensional plot representing the patterns of the ciliary beat. The tips of eight cilia were traced manually in three ciliated cells from each genotype (n = 24 each). Deviation from linearity was measured as the furthest ciliary displacement from the linear axis as defined by the end-effective and end-recovery positions.
In a mouse model, mutation of Pcdp1 was reported to cause symptoms of PCD, including mucus accumulation in the nasal cavity, male infertility, and hydrocephalus12. Ciliary ultrastructure was reported as normal, however ciliary beat frequency (CBF) was reduced approximately 25% in tracheal samples of the mutant mice. In Chlamydomonas flagella, the orthologue of CFAP221, (FAP221) has been shown to be a component of the central pair projection C1d and to bind calcium13. Further, the absence of the C1d complex results in a significant impairment of mobility, including a reduced beat frequency and altered waveform. These results suggest that CFAP221 might play an important role in calcium-mediated regulation of dynein activity. This is an interesting hypothesis to explore in deciphering the underlying mechanism by which genetic variants of CFAP221 are a likely cause of PCD in humans.
Subjects with genetic variants in CFAP221 exhibited typical symptoms of classical PCD, but ciliary axonemes appeared normal by TEM. PCD causing mutations in other genes have also been associated with apparently normal ciliary ultrastructure14, including DNAH1115–17 (MIM:603339) and HYDIN15, 18 (MIM: 610812). A more accurate analysis of the axoneme structure of CFAP221 deficient cells could be achievable by cryo-electron microscopy. This could confirm a central pair defect, as previously shown in Chlamydomonas13. A central pair defect is consistent with a ciliary beat in a circular motion, similar to the pattern of ciliary motion in PCD-subjects carrying genetic variants in RSPH119 (MIM: 609314). In addition, the nNO levels were normal in all affected individuals, and the CBF was only slightly reduced; thus, it is clear that the diagnosis of PCD remains challenging, and a single test cannot be relied upon to detect all cases.
Our results demonstrate for the first time that genetic variants that affect the central pair protein CFAP221 cause PCD in humans. Our results suggest that in subjects with normal ultrastructure and normal or only slightly reduced CBF, CFAP221 should be considered as a candidate gene.
Supplementary Material
Acknowledgements
The authors would like to thank the PCD subjects and family members for their participation, Michele Manion (founder of US PCD Foundation), and the US PCD Foundation, and the investigators and the coordinators of the Genetic Disorders of Mucociliary Clearance Consortium, part of the Rare Disease Clinical Research Network. We thank Dr. Tony Perdue for assistance with confocal imaging; and Dr. S. H. Randell and the UNC Cell culture core for providing human airway cells. The UNC Cell Core Facility is supported by BOUCHE15R0 and P30DK065988 grants. We thank Whitney Wolf, Weining Yin, and Kimberly Burns from UNC for technical assistance and Dr. Hong Dang from UNC for bioinformatics assistance. We thank Drs. Shrikanth Mane and Francesc Loez-Giraldez and Ms. Weilai Dong from Yale Center for Mendelian Genomics for providing whole exome sequencing and bioinformatics support.
Funding:
Funding support for this research was provided to M.R.K, M.W.L. and M.A.Z by US NIH/ORDR/NHLBI grant 5U54HL096458; to M.R.K., L.E.O., and M.A.Z. by NIH-NHLBI grant R01HL071798; to L.E.O., M.A.Z., and M.R.K by NIH/NHLBI R01HL117836 and to UNC-CH by NIH/NCATS grant UL1 TR000083; to D.F.C by NIH/NICHD R01HD078641 and NIH/NIMH R01MH101810. WC is supported by NIH/NIMH T32-MH014677. Whole Exome sequencing was carried out at Yale Center for Mendelian Genomics (UM1HG006504); funded by the NHGRI and the GSP Coordinating Center (U24 HG008956) that contributes to cross-program scientific initiatives logistical and general study coordination. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Patient consent: Obtained.
Ethics approval: Provenance and peer review not commissioned; externally peer reviewed.
Web resources:
PSAP, https://github.com/awilfert/PSAP-pipeline
OMIM, http://www.omim.org
GenBank, https://www.ncbi.nlm.nih.gov/genbank/
Ensembl, http://grch37.ensembl.org/Homo_sapiens/Info/Index
1000Genome, http://www.internationalgenome.org/
gnomAD, http://gnomad.broadinstitute.org/
Uniprot, https://www.uniprot.org/
Supplementary information is available at the Journal of Human Genetics’ website.
Conflict of interest
The authors declare no conflicts of interest
Referencesg
- 1.Shapiro AJ, Zariwala MA, Ferkol T, Davis SD, Sagel SD, Dell SD, et al. Diagnosis, monitoring, and treatment of primary ciliary dyskinesia: PCD foundation consensus recommendations based on state of the art review. Pediatr Pulmonol. 2016;51:115–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leigh MW, Horani A, Kinghorn B, O’Connor MG, Zariwala MA, Knowles MR. Primary ciliary dyskinesia (PCD): A genetic disorder of motile cilia. Transl Sci Rare Dis. 2019;4:51–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilfert AB, Chao KR, Kaushal M, Jain S, Zollner S, Adams DR, et al. Genome-wide significance testing of variation from single case exomes. Nat Genet. 2016;48:1455–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muller L,Brighton LE, Carson JL, Fischer WA 2nd, Jaspers I, Culturing of human nasal epithelial cells at the air liquid interface. J Vis Exp. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gentzsch M, Boyles SE, Cheluvaraju C, Chaudhry IG, Quinney NL, Cho C, et al. Pharmacological rescue of conditionally reprogrammed cystic fibrosis bronchial epithelial cells. Am J Respir Cell Mol Biol. 2017;56:568–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bustamante-Marin XM, Yin WN, Sears PR, Werner ME, Brotslaw EJ, Mitchell BJ, et al. Lack of GAS2L2 causes PCD by impairing cilia orientation and mucociliary clearance. Am J Hum Genet. 2019;104:229–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fulcher ML, Randell SH. Human nasal and tracheo-bronchial respiratory epithelial cell culture. Methods Mol Biol. 2013;945:109–21. [DOI] [PubMed] [Google Scholar]
- 8.Leigh MW, Hazucha MJ, Chawla KK, Baker BR, Shapiro AJ, Brown DE, et al. Standardizing nasal nitric oxide measurement as a test for primary ciliary dyskinesia. Ann Am Thorac Soc. 2013;10:574–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dez C, Froment C, Noaillac-Depeyre J, Monsarrat B, Caizergues-Ferrer M, Henry Y. Npa1p, a component of very early pre-60S ribosomal particles, associates with a subset of small nucleolar RNPs required for peptidyl transferase center modification. Mol Cell Biol. 2004;24:6324–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suprynowicz FA, Upadhyay G, Krawczyk E, Kramer SC, Hebert JD, Liu X, et al. Conditionally reprogrammed cells represent a stem-like state of adult epithelial cells. Proc Natl Acad Sci U S A. 2012;109:20035–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee L, Campagna DR, Pinkus JL, Mulhern H, Wyatt TA, Sisson JH, et al. Primary ciliary dyskinesia in mice lacking the novel ciliary protein Pcdp1. Mol Cell Biol. 2008;28:949–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiPetrillo CG, Smith EF. Pcdp1 is a central apparatus protein that binds Ca(2+)-calmodulin and regulates ciliary motility. J Cell Biol. 2010;189:601–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shapiro AJ, Leigh MW. Value of transmission electron microscopy for primary ciliary dyskinesia diagnosis in the era of molecular medicine: genetic defects with normal and non-diagnostic ciliary ultrastructure. Ultrastruct Pathol. 2017;41:373–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knowles MR, Leigh MW, Carson JL, Davis SD, Dell SD, Ferkol TW, et al. Mutations of DNAH11 in patients with primary ciliary dyskinesia with normal ciliary ultrastructure. Thorax. 2012;67:433–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pifferi M, Michelucci A, Conidi ME, Cangiotti AM, Simi P, Macchia P, et al. New DNAH11 mutations in primary ciliary dyskinesia with normal axonemal ultrastructure. Eur Respir J. 2010;35:1413–16. [DOI] [PubMed] [Google Scholar]
- 17.Schwabe GC, Hoffmann K, Loges NT, Birker D, Rossier C, de Santi MM, et al. Primary ciliary dyskinesia associated with normal axoneme ultrastructure is caused by DNAH11 mutations. Hum Mutat. 2008;29:289–98. [DOI] [PubMed] [Google Scholar]
- 18.Olbrich H, Schmidts M, Werner C, Onoufriadis A, Loges NT, Raidt J, et al. Recessive HYDIN mutations cause primary ciliary dyskinesia without randomization of left-right body asymmetry. Am J Hum Genet. 2012;91:672–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knowles MR, Ostrowski LE, Leigh MW, Sears PR, Davis SD, Wolf WE, et al. Mutations in RSPH1 cause primary ciliary dyskinesia with a unique clinical and ciliary phenotype. Am J Respir Crit Care Med. 2014;189:707–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.