Summary
T cell dysfunction is a characteristic feature of chronic viral infection and cancer. Recent studies in chronic lymphocytic choriomeningitis virus (LCMV) infection have defined a PD-1+ Tcf-1+ CD8+ T cell subset capable of self-renewal and differentiation into more terminally-differentiated cells that downregulate Tcf-1 and express additional inhibitory molecules such as Tim3. Here, we demonstrated that expression of the glycoprotein CD101 divides this terminally differentiated population into two subsets. Stem-like Tcf-1+ CD8+ T cells initially differentiated into a transitory population of CD101− Tim3+ cells that later converted into CD101+ Tim3+ cells. Recently-generated CD101− Tim3+ cells proliferated in vivo, contributed to viral control, and were marked by an effector-like transcriptional signature including expression of the chemokine receptor CX3CR1, pro-inflammatory cytokines, and granzyme B. PD-1 pathway blockade increased the numbers of CD101− Tim3+ CD8+ T cells, suggesting that these newly-generated, transitional cells play a critical role in PD-1 based immunotherapy.
Keywords: CD8+ T cells, exhaustion, immunotherapy, PD-1, LCMV, chronic viral infection
Introduction
CD8+ T cells are responsible for the recognition and killing of host cells expressing intracellular non-self or mutated-self antigens arising from infection or cell transformation. Upon recognition of its cognate antigen a naive CD8+ T cell will undergo differentiation into an effector state, which is marked by high proliferative and cytotoxic capacity. After clearance of an acute viral infection, memory precursor cells present in the heterogeneous pool of effector CD8+ T cells will differentiate into long-lived memory cells in the absence of antigen(Joshi et al., 2007; Kaech et al., 2003; Sarkar et al., 2008). These memory cells undergo a slow homeostatic proliferation to maintain their numbers and remain primed for rapid effector responses upon antigen re-exposure, providing protective immunity against re-infection (Akondy et al., 2017; Kaech et al., 2002).
In conditions marked by antigen persistence such as chronic viral infection or cancer, CD8+ T cells enter an ‘exhausted’ state marked by the expression of regulatory surface proteins such as PD-1(Day et al., 2006) and progressive loss of effector functions such as cytokine production, proliferative capacity, and cytotoxicity(Barber et al., 2005; Gallimore et al., 1998; Zajac et al., 1998). These findings have led to the development of immune checkpoint inhibitors, antibodies that target negative regulatory proteins that are expressed on the surface of exhausted CD8+ T cells(Callahan and Wolchok, 2013). These drugs have proven successful at treating multiple types of cancer, although not all tumor types are susceptible nor do all patients with susceptible tumor types show clinical responses(Brahmer et al., 2012; Topalian et al., 2012). This has led to the search for additional inhibitory surface molecules expressed on CD8+ T cells for use as immunotherapy targets, often for use in combination with anti-PD-1 pathway blockade(Ott et al., 2017; Pardoll, 2012).
An additional factor complicating PD-1 therapy is the heterogeneity in exhausted CD8+ T cell populations. One key recent finding is the presence of a ‘stem-like’ population of PD-1+Tcf-1+ CD8+ T cells that sustain CD8+ T cell responses during chronic viral infection and give rise to PD-1+Tim3+ terminally differentiated cytotoxic cells(Im et al., 2016; Utzschneider et al., 2016; Wu et al., 2016). Stem-like CD8+ T cells have been found in numerous types of chronic infection and human cancers (Brummelman et al., 2018; Gettinger et al., 2018; He et al., 2016; Jiang et al., 2017; Leong et al., 2016), and it is these Tcf-1+ stem-like cells that mediate the proliferative response to PD-1 pathway blockade (Im et al., 2016). In contrast, PD-1+Tim3+ cells undergo minimal amounts of division after PD-1 blockade(Im et al., 2016). This heterogeneity again highlights the need for identification of additional surface markers of PD-1 responsive cells and targets for combination therapy in order to recruit additional antigen-specific CD8+ T cells into the immunotherapy-responsive pool.
Here, we performed a systematic approach to catalog the repertoire of surface molecules expressed by exhausted CD8+ T cells. This analysis confirmed well-known markers of exhaustion such as PD-1, Tim3, and CD244 (2B4), but also pinpointed other surface molecules with less-studied roles in T cell exhaustion such as CD7, Lax1, CD200R, and CD101. We demonstrated that this latter molecule, the type I transmembrane glycoprotein CD101, was induced specifically on CD8+ T cells in chronic viral infection and that its expression marked terminally differentiated and highly dysfunctional antigen-specific CD8+ T cells. We showed that stem-like CD8+ T cells differentiated into transitory CD101−Tim3+ cells that still retained proliferative and effector capacity and contributed to viral control before their eventual progression into an exhausted CD101+Tim3+ state. Finally, we have shown that PD-1 pathway blockade increased the proliferation and differentiation of stem-like antigen-specific CD8+ T cells to the effector-like CD101−Tim3+ state, resulting in an enlarged pool of functional cells that contribute to improved viral control.
Results
Systematic identification of CD8+ T cell exhaustion markers
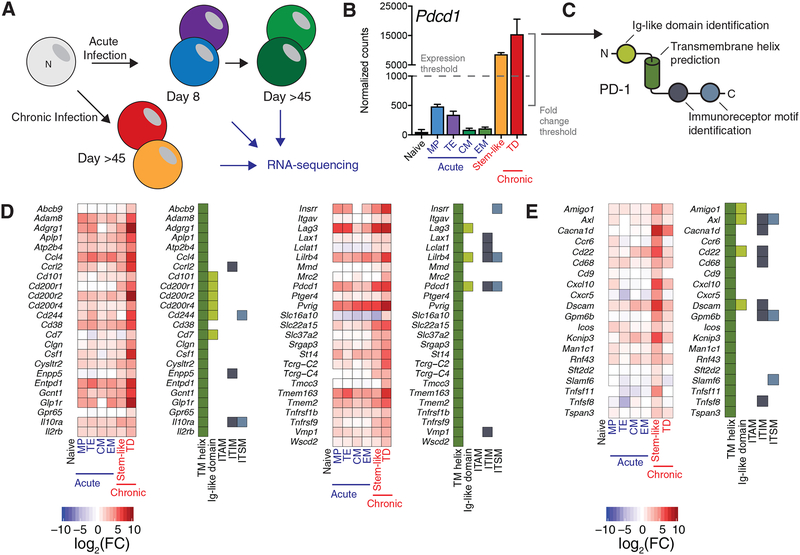
To identify markers of exhausted murine CD8+ T cells, we analyzed two RNA-sequencing datasets generated by our laboratory (Figure 1A). The first dataset consisted of RNA-sequencing data from virus-specific CD8+ T cells isolated from spleens of mice infected with LCMV Armstrong, which causes an acute infection and results in a robust, functional effector and memory CD8+ T cell response(Hudson et al., 2019). At the peak of the CD8+ T cell response to LCMV Armstrong, a heterogeneous effector cell pool is present, including Klrg1hiCD127lo terminal effector cells (TEs) and Klrg1loCD127hi memory precursor cells (MPs)(Joshi et al., 2007; Kaech et al., 2003). Both of these populations are capable of cytotoxicity and cytokine production, but long-lived memory CD8+ T cells will differentiate from the CD127+ MPs within the initially heterogeneous response(Kaech et al., 2003; Sarkar et al., 2008). To generate this first dataset, LCMV-specific MPs and TEs were isolated by fluorescence-activated cell sorting (FACS) eight days after infection. Forty-eight days post-infection, CD127+CD62L+ central memory cells (CM) and CD127+CD62L− effector memory (EM) LCMV-specific cells were isolated by FACS(Sallusto et al., 1999; Wherry et al., 2003). RNA from the aforementioned subsets as well as naive cells was isolated from sorted cells and sequenced.
Figure 1: Systematic identification of CD8+ T cell exhaustion markers.
A) Naive and virus-specific CD8+ T cells resulting from acute and chronic LCMV infection were isolated and analyzed for gene expression by RNA-sequencing. These subsets included naive CD8+ T cells, Klrg1hi CD127lo terminal effector (TE) and Klrg1lo CD127hi memory precursor (MP) cells from acute (day 8 post-infection) LCMV Armstrong and CD62L+ central (CM) and CD62L− effector (EM) memory cells from day 48 post-infection with LCMV Armstrong(Hudson et al., 2019).
C) After analysis, genes with high absolute expression in CXCR5+Tim3− stem-like CD8+ T cells or CXCR5−Tim3+ terminally differentiated (TD) CD8+ T cells from chronic infection (expression threshold) and high relative expression compared to naive CD8+ T cells and cells resulting from acute infection (fold change threshold) were identified. Expression of Pdcd1, the gene encoding PD-1, is shown as an example. See Methods for details of gene identification.
D) A bioinformatics pipeline to identify transmembrane helices, putative intracellular signaling motifs, and immunoglobulin-like domains was performed on identified genes. Expression of Pdcd1, which encodes PD-1, is shown as an example of an identified gene.
E) Heatmap of expression and protein properties of the 49 transmembrane helix-containing genes identified as highly expressed in Tim3+ CD8+ T cells from chronic infection by this algorithm. At left, log2 of the fold change from naive cells is shown. At right, a filled box indicates the presence of the indicated domain or motif.
F) Heatmap of expression and protein properties of the 20 transmembrane helix-containing genes identified as highly expressed in stem-like CD8+ T cells from chronic infection. At left, log2 of the fold change from naive cells is shown. At right, a filled box indicates the presence of the indicated domain or motif.
TD: terminally differentiated (Tcf-1−Tim3+)
The second dataset consisted of RNA-sequencing data of PD-1+ CD8+ T cells present in chronic infection(Jadhav et al., 2019). Two cell populations, PD-1+CXCR5+Tim3− cells and PD-1+CXCR5−Tim3+ cells, were isolated from spleens of mice infected with LCMV clone 13, which causes a chronic viral infection. The former subset is stem-like, expresses Tcf-1, and has the capability for self-renewal, whereas the latter subset (Tim3+) is a terminally-differentiated CD8+ T cell population that does not proliferate in response to PD-1 blockade(Im et al., 2016). In both studies, naive CD8+ T cells were sequenced, allowing a comparison with CD8+ T cells from uninfected mice and serving as a control population between the two studies.
To identify surface proteins upregulated in exhausted CD8+ T cells, we selected genes with high absolute expression and high relative expression (>3 fold) compared to naive, TE, and EM CD8+ T cells (Figure 1B). For genes with particularly high absolute expression, the relative expression threshold was relaxed to 2-fold. Genes encoding surface proteins were distinguished by the presence of a predicted transmembrane helix(Krogh et al., 2001) in their associated peptide sequences (Figure 1C). Immunoreceptor tyrosine-based motifs were determined in predicted intracellular regions and proteins with immunoglobulin-like domains were identified via the Protein Families (PFAM) annotations(Finn et al., 2016) (Figure 1C); such domains are often important for protein-protein interactions in immunological signaling pathways(Barclay, 2003). A complete and sortable list of genes, their expression, and properties of their encoded proteins are given in Table S1.
In total, we identified 49 transmembrane proteins markedly upregulated in PD-1+Tim3+ LCMV-specific CD8+ T cells from chronically infected mice (Figure 1D). This included a number of well-known inhibitory coregulatory molecules, such as PD-1 (Pdcd1), Lag3, and 2B4 (Cd244). Additional genes of interest included several encoding proteins with described negative effects on T cell receptor (TCR) signaling but few described roles in CD8+ T cell exhaustion, such as Lax1, Cd101, and Pvrig, which encodes a recently-described inhibitory receptor for CD112 (Murter et al., 2019; Philip et al., 2017; Soares et al., 1998; Whelan et al., 2019; Zhu et al., 2002; Zhu et al., 2016). CD7, which delivers a pro-apoptotic signal to T cells in the presence of galectin-1, as well as Gcnt1, whose genetic ablation desensitizes T cells to galectin-1-mediated apoptosis were also highly expressed in Tim3+ cells(Nguyen et al., 2001; Pace et al., 2000). An additional 20 genes encoding transmembrane proteins were identified as highly and selectively expressed in the stem-like CD8+ T cells resulting from chronic LCMV infection(Im et al., 2016). These include Slamf6, a putative stimulatory molecule (Chatterjee et al., 2011), as well as several molecules typically expressed on B cells and/or CD4+ T cells, such as Cd22 and Cxcr5 (Figure 1E). We confirmed the presence of several of these markers on antigen-specific CD8+ T cells with flow cytometry, and in this study we focused on CD101, which has allowed us to define the CD8+ T cell differentiation pathway during chronic infection.
CD101 is specifically induced on CD8+ T cells in chronic viral infection
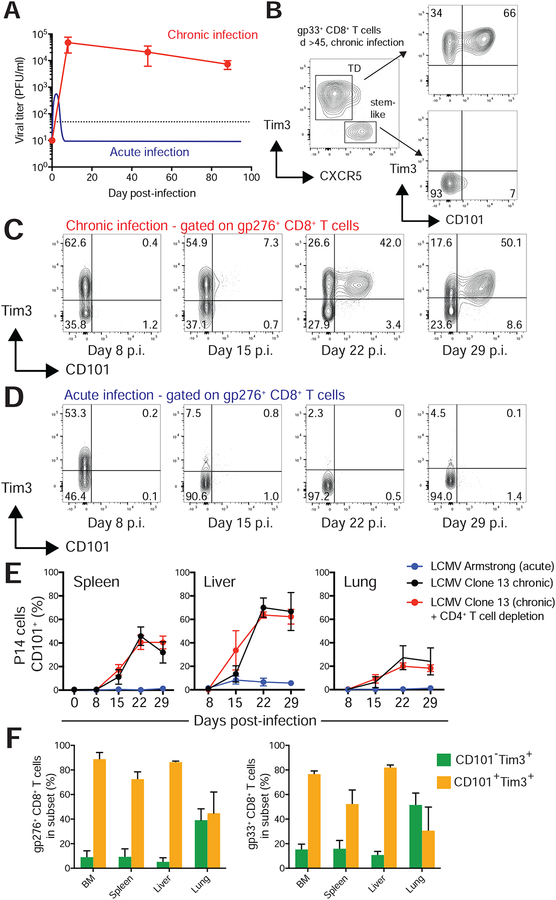
In antigen-specific CD8+ T cells from chronic LCMV infection (Figure 2A), CD101 exhibited a distinctive staining pattern; stem-like CD8+ T cells showed no expression of CD101, whereas Tim3+ cells exhibited bimodal expression of CD101 (Figure 2B). At the late stages of chronic infection (day >45 post-infection), a majority of Tim3+ cells expressed CD101. To determine kinetics of CD101 induction, we infected C57BL/6J mice with LCMV clone 13 or LCMV Armstrong, which cause chronic and acute infections respectively, and analyzed CD101 expression on splenic LCMV-specific CD8+ T cells (Figure 2C,D).
Figure 2: CD101 is induced on virus-specific CD8+ T cells during chronic infection and identifies two populations of Tim3+ cells.
A) Infection of mice with LCMV clone 13 infection with transient CD4+ T cell depletion results in high viral titers and no viral clearance for the life of the mouse. LCMV Armstrong results in an acute infection. Viral titer data from Armstrong is illustrated (Ahmed et al., 1984; Matloubian et al., 1993).
B) Flow cytometry showing CD101 expression on the stem-like and Tim3+ terminally differentiated (TD) subsets of LCMV-specific (gp33+) CD8+ T cells in chronic infection.
C) Representative CD101 and Tim3 expression on endogenous gp276-specific cells eight, 15, 22, and 29 days after induction of chronic viral infection with LCMV clone 13.
D) CD101 and Tim3 expression on endogenous Dbgp276-specific cells eight, 15, 22, and 29 days after induction of acute infection with LCMV Armstrong.
E) Percentage of gp276+ CD8+ T cells from indicated organs expressing CD101 after infection with clone 13 alone (black), clone 13 with CD4+ T cell depletion (red), or Armstrong (blue).
F) >45 days after infection with LCMV clone 13 and CD4 depletion, lymphocytes were isolated from multiple tissues. The percentage of CD101−Tim3+ and CD101+Tim3 cells among gp33-specific CD8 T cells and gp276-specific CD8 T cells were determined by flow cytometry. Cells shown in panels B-D were isolated from the spleen. Panels D and E show mean ± SEM. Panels C-D are representative flow plots from two independent experiments that both shown in panel E. Panel F is representative of three independent experiments.
Eight days after infection, expression of CD101 was not detected on LCMV-specific cells in mice with acute or chronic infection. By day 15 post-infection, CD101 was detectable on a small subset of Tim3+ LCMV-specific CD8+ T cells in chronically infected mice (Figure 2C). At 3 and 4 weeks after induction of chronic infection, most Tim3+ cells also expressed CD101. In stark contrast, no significant expression of CD101 was observed on LCMV-specific CD8+ T cells at any time point following acute infection (Figure 2D). In other organs, CD101 was induced at similar time points in chronic infection, with low but detectable expression two weeks after infection and prominent CD101+Tim3+ populations 3–4 weeks after infection (Figure 2E). As in spleen, CD101 was not detected on LCMV-specific CD8+ T cells following acute infection in spleen or liver. Late in chronic infection (day >45 after infection), the prevalence of CD101+ LCMV-specific cells differed among tissues, with lung containing the lowest frequency of CD101+Tim3+ cells, and bone marrow and liver the highest (Figure 2F). The frequencies of these populations among organs did not substantially differ between gp33-specific and gp276-specific CD8+ T cells (Figure 2F), and CD101+Tim3− cells were rarely observed. The presence or absence of CD4+ T cell help did not affect the generation of the CD8+ CD101+ subset at initial stages of infection (Figure 2E).
CD101 expression marks transcriptionally distinct antigen-specific CD8+ T cells
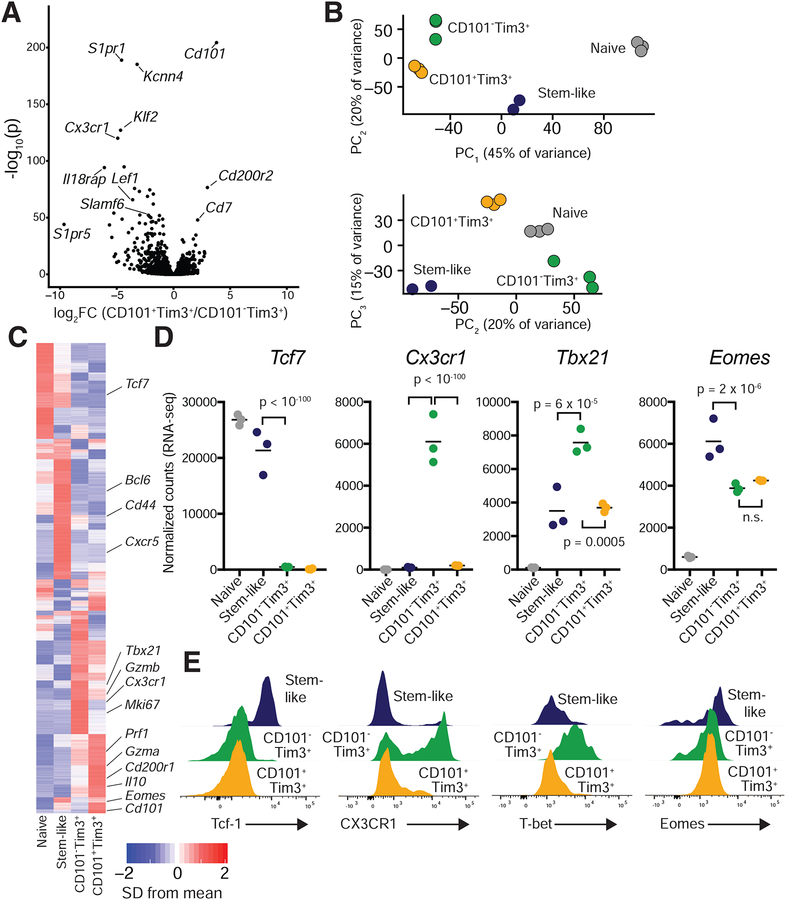
To identify transcriptional differences between stem-like (PD-1+CD101−Tim3−), PD-1+CD101−Tim3+, and PD-1+CD101+Tim3+ CD8+ T cells, we used flow cytometry to isolate these three subsets of PD-1+CD8+ T cells from spleens of chronically infected mice (>d45 post-infection; Figure S1) as well as naive CD8+ T cells from uninfected mice. 1,825 genes were differentially expressed between CD101−Tim3+ and CD101+Tim3+ PD-1+CD8+ T cells (Figure 3A, Table S2), indicating that CD101 expression markeds two transcriptionally distinct populations of Tim3+ CD8+ T cells in chronic infection. Notably, Cd101 was the most differentially upregulated gene in CD101+Tim3+ cells as measured both by adjusted p-value and fold change (Figure 3A).
Figure 3: Distinctive transcriptional signatures define stem-like, CD101−Tim3+, and CD101+Tim3+ CD8+ T cells.
RNA was isolated and sequenced from PD-1+CD8+ T cells of LCMV clone 13-infected mice (day >45). See Figure S3 for gating strategy.
A) Volcano plot showing differential gene expression between CD101+ Tim3+ and CD101−Tim3+ cells; 1,825 genes were differentially expressed (p < 0.05) between these two subsets. P values are adjusted for multiple comparisons.
B) Principal component analysis (PCA) identified three principal components that explained large amounts of transcriptional variance; samples are plotted on these components.
C) Heatmap showing mean relative expression of all differentially expressed genes across the four sequenced groups.
D-E) Confirmation of selected differentially expressed protein-coding genes by flow cytometry. D normalized counts (RNA-sequencing) of each gene. E, flow cytometry staining of each marker, gated on gp33-specific CD8+ T cells from spleens of chronically infected mice. P values shown in panel D are from Wald’s test, performed by DESeq2. Panel E are representative flow plots of at least five independent mice. Please see also Figures S1–S5.
To broadly compare the transcriptional profile of naive, stem-like, and CD101−/+ Tim3+ CD8+ T cells, we performed principal component analysis (PCA) on these samples, which identified three principal components explaining 10% or more of variance in gene expression among samples. CD101− and CD101+ Tim3+ cells were similarly situated on the first principal component (PC1), which explained 46% of transcriptional variance among all cell types measured (Figure 3B). However, the two Tim3+ subsets differed on PC2 and PC3, which collectively explain a third of transcriptional variance. Specifically, CD101−Tim3+ cells were similar to naive cells on PC2 and stem-like cells on PC3, suggesting that CD101−Tim3+ cells may retain some transcriptional programs similar to these other cell types.
Examination of individual gene expression by affinity propagation clustering revealed multiple patterns of gene expression, including many genes preferentially expressed in one of the four sequenced subsets (Figure 3C). Expression of some genes, such as Tcf7 (encoding Tcf-1) was shared by stem-like and naive cells, whereas others such as Cxcr5 were expressed only in stem-like cells. Many effector genes such as Prf1 (encoding perforin) were expressed in both CD101− and CD101+Tim3+ cells, with much lower expression in stem-like cells. Notably, a number of genes were expressed preferentially in CD101−Tim3+ cells, such as Cx3cr1, Klrg1, and Tbx21, which encodes T-bet (Figure 3D, Figure S2). Eomes expression did not differ between Tim3+ subsets, but was reduced in Tim3+ cells compared to stem-like cells. Another noticeable pattern was the very high expression of surface inhibitory molecules in CD101+ cells (Figure S2). Flow cytometric validation of selected differentially expressed proteins consistently corroborated RNA-sequencing results (Figure 3D,E).
To identify transcriptional pathways that differ between these three subsets, we performed gene set enrichment analysis (GSEA) on CD101−Tim3+cells versus CD101+Tim3+and stem-like cells (Figure S3). GSEA analysis revealed that all three CD8+ T cell subsets present in chronic infection had drastically divergent transcriptional programs: 2,078 and 2,110 gene sets were significantly enriched in CD101−Tim3+ cells versus CD101+Tim3+ cells or stem-like cells, respectively (Table S3). 1,318 of these gene sets were enriched in CD101−Tim3+ cells versus both subsets; these sets included those indicative of CD8+ T cell response to antigen and proliferation (Figure S3). Gene sets enriched in stem-like cells versus CD101−Tim3+ cells included those involved in translation, while gene sets enriched in CD101+Tim3+ cells versus CD101−Tim3+ cells were those characteristic of negative regulation of lymphocyte activation and proliferation. Only 9 gene sets were enriched in both stem-like cells and CD101+Tim3+ cells against CD101−Tim3+ cells, again illustrating that each of the three subsets express dramatically different transcriptional programs.
To compare gene expression of CD101−Tim3+ and CD101+Tim3+ cells to virus-specific CD8+ T cells following LCMV Armstrong infection, we identified genes predominately expressed in each subset (p < 0.05 and fold change ≥ 2 against all other subsets). The CD101−Tim3+ transcriptional profile resembled that of LCMV-specific Klrg1+ cells at day 8 following Armstrong infection (Figure S4A). Genes expressed in both types of cells included Zeb2 and Tbx21 (T-bet), which are responsible for effector differentiation (Dominguez et al., 2015); Cx3cr1, which may promote surveillance of non-lymphoid tissues (Gerlach et al., 2016); and S1pr1, which is required for lymphocyte migration from peripheral lymphoid organs(Matloubian et al., 2004). GSEA also revealed many shared gene sets between CD101−Tim3+ cells from chronic infection and Klrg1+ cells from acute infection (Figure S4C).
As the stem-like, CD101−Tim3+ and CD101+Tim3+ cells were first present at approximately day 15 after infection with LCMV clone 13 (Figure 2C), we also performed RNA-sequencing at this early time point to examine their transcriptional profiles and compare the subsets early and late in chronic infection. PD-1+ stem-like, CD101−Tim3+, and CD101+Tim3+ cells were sorted at day 15 post-infection as at day 45 (Figure S1). Many of the same genes identified the CD101−Tim3+ subset at day 15 and day 45 post-infection, such as Il18r1, Il18rap, Cx3cr1, Klf2, and S1pr1 (Figure S5A, Table S4). PCA analysis also showed that the three subsets were transcriptionally distinct, with little changes in their relative positions in principal component space between day 15 and 45 (Figure S5B,C). Differences in gene expression among the three subsets were highly similar at days 15 and 45, with the notable exception of higher expression of cell cycle genes in all subsets at day 15 (Figure S5D,E). However, the expression of stem-like CD8+ T cell defining genes such as Cxcr5, Tcf7 (encoding Tcf-1), and Bcl6 were fixed in the CD101−Tim3− subset at day 15 (Figure S5D), as was the expression of CD101+Tim3+-defining genes such as Cd101, Cd200r2, Cd7, and Cd160 (Figure S5E). Thus, with the exception of residual division from their initial activation, CD101+Tim3+ cells express an exhausted transcriptional signature from their earliest appearance.
Together, these results from RNA-sequencing show that stem-like, CD101−Tim3+, and CD101+Tim3+ CD8+T cells are three transcriptionally distinct populations of antigen-specific cells present in chronic infection, and we sought to determine their lineage relationship and available differentiation pathways.
CD101−Tim3+ cells are a transitory CD8+ T cell population arising from Tcf-1+ stem-like cells
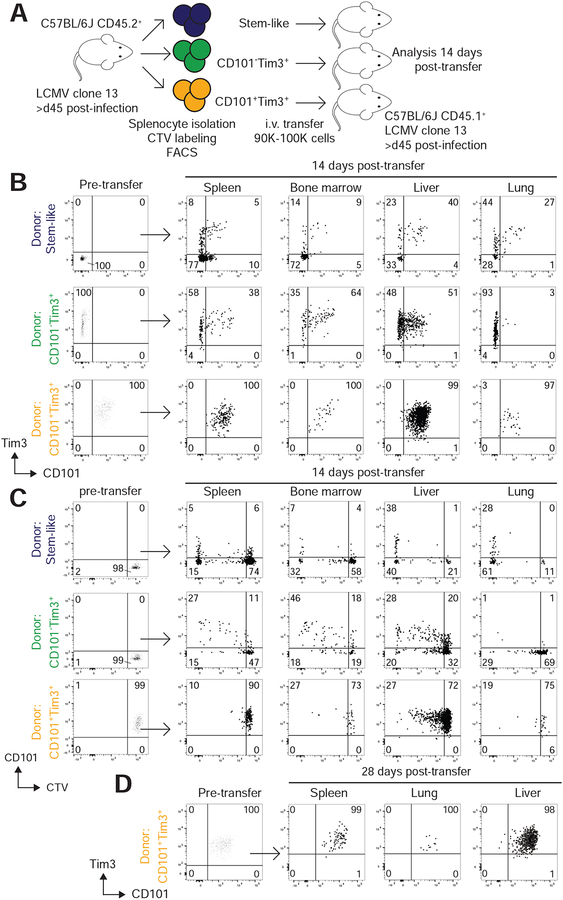
Based on the transcriptional signatures of cell division and effector function present in CD101−Tim3+ cells and the exhausted signature of CD101+ Tim3+ cells, we hypothesized that CD101−Tim3+ cells are a transitory population in the differentiation pathway between stem-like Tcf-1+ CD8+ T cells and fully exhausted CD101+Tim3+ CD8+ T cells. To test this hypothesis, we performed adoptive transfer of cells from the above subsets from chronically infected mice into congenically-distinct, infection-matched mice (Figure 4A). Splenocytes were isolated from (CD45.2+) C57BL/6J mice chronically infected with LCMV clone 13 and labeled with CellTrace Violet (CTV), a proliferation tracking dye. Using flow cytometry, we separated three populations of PD-1+CD8+ T cells from donor mice: CD101−Tim3− stem-like cells, CD101−Tim3+ cells, and CD101+Tim3+ cells. These sorted cells were then transferred intravenously into separate infection-matched, congenically distinct (CD45.1) recipient mice (Figure 4A). Two weeks post-transfer, lymphocytes were isolated from spleen, liver, lungs, and bone marrow of recipient mice and donor cells analyzed by flow cytometry for expression of Tim3, CD101, and dilution of CTV (Figure 4B,C).
Figure 4: Stem-like CD8+ T cells differentiate into transitory CD101−Tim3+ cells, followed by irreversible conversion into exhausted CD101+Tim3+ cells.
A) PD-1+ CD8+ T cells were collected from spleens of mice chronically infected with LCMV clone 13 and labeled with CellTrace Violet (CTV). From these, three subsets (CD101−Tim3−, CD101−Tim3+, and CD101+Tim3+) were isolated via FACS and transferred intravenously to infection-matched, congenically distinct mice. Recipient mice were sacrificed two weeks after transfer to analyzed phenotype of donor cells.
B-C) Phenotype (B) and CTV staining (C) of adoptively transferred cells before and 14 after transfer. All populations shown are gated on PD-1+CD45.2+CD45.1− CD8+ T cells. Cells shown are pooled from all recipient mice from one independent experiment, and results are representative of three independent experiments.
D) In one replicate, phenotype of CD101+Tim3+ donor cells was analyzed 28 days after transfer. Plots in panels A-C are pooled donor cells from one representative experiment of three independent adoptive transfer studies. Panel D shows pooled donor cell data from one independent experiment.
Stem-like donor CD8+ T cells were capable of maintaining a CD101−Tim3− phenotype or differentiating into CD101−Tim3+ or CD101+Tim3+ cells (Figure 4B). In lymphoid organs (spleen and bone marrow) most stem-like donor cells retained their pre-transfer CD101−Tim3− phenotype, whereas the majority of those recovered in lung and liver expressed Tim3. Most donor stem-like cells recovered in lung and liver had also diluted CTV, often to the maximum detectable extent, which represents eight or more rounds of division (Figure 4C). In lymphoid organs most donor cells had not divided, but those that had entered the cell cycle fully diluted CTV.
Recovered CD101−Tim3+ donor cells either retained their CD101−Tim3+ phenotype or had converted into CD101+Tim3+ cells. In bone marrow and liver, the majority of donor CD101−Tim3+ recovered cells upregulated CD101, whereas only a very small fraction of cells found in the lung had expressed CD101 (Figure 4B). These results mirror the frequency of endogenous CD101−Tim3+ cells (Figure 2F), suggesting that tissue environment may play a major role on CD8+ T cell differentiation in chronic viral infection. CD101−Tim3+ cells were also capable of division in vivo, as approximately half of recovered cells had diluted CTV, with some variation by tissue (Figure 4C). Some donor CD101−Tim3+ cells had diluted CTV to the maximum detectable extent - particularly in the liver - although this fraction was far smaller than those seen after transfer of stem-like cells.
Finally, transferred CD101+Tim3+ cells were exclusively recovered in the CD101+Tim3+ state, demonstrating that these cells were incapable of interconversion to the CD101−Tim3+ state (Figure 4B). Even 28 days after transfer, CD101+Tim3+ cells did not convert to any other state, illustrating their fixed state of terminal differentiation (Figure 4D). Additionally, the vast majority of CD101+Tim3+ donor cells had not diluted CTV (Figure 4C), nor were any of these cells capable of completely diluting CTV (again, representing eight or more divisions). Together, these results confirm previous experiments demonstrating the differentiation of PD-1+ stem-like cells to a PD-1+Tim3+ state(Im et al., 2016). The data here also extend these findings by demonstrating that stem-like cells initially differentiate into transitory CD101−Tim3+ cells with proliferative capacity and an effector-like transcriptional profile. These transitory cells then convert into fully exhausted CD101+Tim3+ cells with extremely limited potential for division and no ability to revert into CD101− cells for up to 28 days.
Newly-generated CD101−Tim3+ CD8+ T cells are more functional than CD101+Tim3+ CD8+ T cells
Given the findings from the above adoptive transfer experiments, our gene expression analysis provides insight into the transcriptional changes occurring during steady-state CD8+ T cell differentiation in chronic viral infection (Figure 5A). Upon division and differentiation of stem-like cells to transitory CD101−Tim3+ cells, the expression of migration-associated (Cx3cr1, Ccr2, Itgam, Itga6, S1pr1) and division-associated (Mki67, Mcm2–7, Aurkb) genes was upregulated. Expression of genes encoding exhaustion markers such as PD-1, CD112R, and TIGIT were also upregulated, concomitant with downregulation of Cd28. This transition also involved the upregulation of the transcription factor genes Tbx21 (encoding T-bet) and Klf2 and downregulation of Tcf7 (encoding Tcf-1). Upon differentiation of transitory CD101−Tim3+ cells to exhausted CD101+Tim3+ cells, migration- and division-associated genes were downregulated. Exhausted CD101+Tim3+ downregulated Klf2 and Tbx21 and highly expressed the genes encoding Runx3, Stat3, Irf1, and Hic1. Finally, the genes encoding PD-1, CD112R, and TIGIT are further upregulated in exhausted CD101+Tim3+ cells compared to transitory cells, and other inhibitory molecules such as CD160, Lag3, and CD200R1 are exclusively upregulated in exhausted cells compared to stem-like cells (Figure 5A).
Figure 5: Transitory CD8+ T cells in chronic infection retain proliferative capacity and are more functional than CD101+Tim3+ cells.
A) Plot showing changes in gene expression during CD8+ T cell differentiation from stem-like cells to transitory CD101−Tim3+ cells and subsequent conversion to exhausted CD101+Tim3+ cells in the spleen. Selected differentially expressed genes are shown.
B) Expression of selected surface exhaustion markers in transitory (green) and exhausted CD101+Tim3+ cells (yellow). Populations are gated on gp33-specific CD8+ T cells from chronic infection.
C) Ki-67 expression in subsets of gp33-specific CD8+ T cells isolated from spleen and liver.
D) Summary data of Ki-67 staining of gp33+ CD8+ T cells from chronic infection.
E) MFI of CD101 expression on CD101+Tim3+Ki-67+ (solid circles) or CD101+Tim3+Ki-67− CD8+ T cells (open circles). Cells were gated on gp276+ CD8+ T cells.
F) P14 cells were transferred to C57BL/6J mice. Twenty-two days after infection, splenocytes were isolated and stimulated with or without gp33 peptide, and subsets analyzed by flow cytometry for cytokine expression.
G) Percentage of P14 cells expressing IFNγ by subset.
H) MFI of IFNγ staining on IFNγ+ cells within each subset.
I) MFI of granzyme B of cells within each subset.
In panel E, statistics shown are two-way ANOVA with Sidak’s test for multiple comparisons. In panels G-I, statistics shown are one-way ANOVA with Tukey’s test for multiple comparisons. In both cases, *, **, ***, and **** indicate p values less than 0.05, 0.01, 0.001, and 0.0001, respectively. Panels B-C are representative flow plots of at least five independent mice. Panels D-E are representative results from one of three independent experiments. Panels F-I are representative results from one of two independent experiments.
Flow cytometry analysis confirmed the higher expression of many inhibitory molecules in CD101+ LCMV-specific cells, including 2B4 (CD244), CD160, and CD200R(1) (Figure 5B). Ki-67 staining was most prominent in transitory cells (Figure 5C,D), as predicted by RNA-sequencing data from the spleen (Figure 5A). This is consistent with a differentiation pathway in which quiescent stem-like CD8+ T cells infrequently divide and convert into rapidly proliferating transitory CD101−Tim3+ cells. We found that Ki-67+ CD101+ CD8+ T cells expressed lower amounts of CD101 than Ki-67− CD101+ cells, suggesting that onset of dysfunction is paired with progressive upregulation of CD101, particularly in the spleen (Figure 5E).
To determine the ability of the different CD8+ T cell subsets to produce effector cytokines during chronic infection, we transferred P14 cells, which contain a transgenic TCR for the LCMV gp33 epitope, to C57BL/6J mice and infected these recipients with LCMV clone 13. Twenty-two days post-infection, approximately 70% and 60% of stem-like and transitory P14 cells, respectively, produced IFN-γ after ex vivo stimulation with gp33 peptide. In contrast, only 20% of CD101+ exhausted P14 cells were IFN-γ+ (Figure 5F,G). In addition to lower frequencies of IFN-γ-producing cells among the exhausted subset, CD101+ IFN-γ+ cells produced significantly lower amounts of IFN-γ compared to IFN-γ+ transitory or stem-like cells (Figure 5H). CD101−Tim3+ cells also expressed higher amounts of granzyme B in vivo compared to either stem-like or terminally differentiated CD101+Tim3+ cells (Figure 5I). These experiments demonstrate that during chronic viral infection, stem-like cells differentiate into transitory CD101−Tim3+ cells that retain proliferative and effector capacity before irreversible conversion into fully exhausted CD101+ cells with extremely limited capability for division and cytokine production.
CD101−Tim3+ cells contribute to viral control and are increased by PD-1 pathway blockade
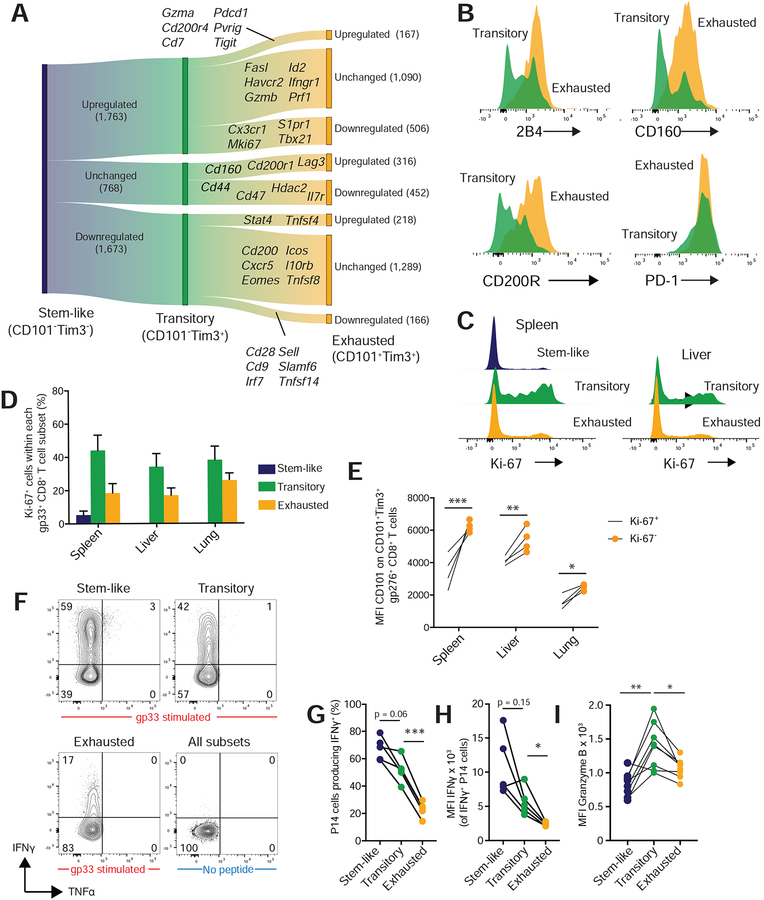
In murine LCMV infection, the CD8+ T cell response is critical for viral control, and PD-1 pathway blockade rejuvenates cell-mediated immunity in chronically infected mice (Barber et al., 2005). Given the effector capacities of CD101−Tim3+ LCMV-specific cells, we hypothesized that these transitory cells are critical for both viral control and the response to PD-1 blockade. To test the effects of CD101−Tim3+ cells on viral control, we used a diphtheria toxin (DT) receptor with a floxed stop codon driven by the Cx3cr1 promoter(Diehl et al., 2013) to deplete cells expressing CX3CR1. As very few CD4+ T cells express CX3CR1 following LCMV infection(Gerlach et al., 2016), when crossed with Lck cre mice, CX3CR1-expressing CD8+ T cells will be deleted after DT administration. CX3CR1 was exclusively expressed by CD101−Tim3+ cells (Figure 3D,E), thus DT treatment will selectively deplete the transitory CD8+ T cell subset. Fifteen days after infection with LCMV clone 13, we began DT administration (Figure 6A) and assessed LCMV-specific CD8+ T cell subsets and viral titers fifteen days later. After treatment, CX3CR1+ LCMV-specific CD8+ T cells were efficiently depleted (Figure 6B,C), resulting in a 70% decrease in splenic LCMV-specific CD101−Tim3+ CD8+ T cells (Figure 6D–F). With their CD101−Tim3+ cell population intact, LCMV titers in WT mice were 73% lower than in Lck cre Cx3cr1 DTRfl mice (Figure 6G).
Figure 6: CX3CR1+ CD101−Tim3+ cells are critical for viral control in chronic infection.
A) Lck cre+ Cx3cr1 DTR stop-floxed mice or WT controls were infected with LCMV clone 13. Fifteen days later, 250 ng of diphtheria toxin (DT) were administered every other day by intraperitoneal injection. Mice were sacrificed at day 30 post-infection to examine CD8+ T cell responses and viral titer.
B) Representative tetramer staining from WT and Lck cre+ Cx3cr1 DTR stop flox+ mice at 30 days after infection.
C-D) CD101and Tim3 and CX3CR1 and CD101 expression on tetramer positive cells at day 30 in WT and Lck cre+ Cx3cr1 DTR stop flox+ mice.
E) Total number of gp276+ CD8+ T cells in spleen at day 30.
F) Numbers of CD101−Tim3+ gp276+ CD8+ T cells in spleen at day 30.
G) Spleen viral titer day 30 after infection.
In panels E-G, p values from Mann-Whitney tests are shown. Neither viral titers nor cell counts were log transformed for statistical analysis.
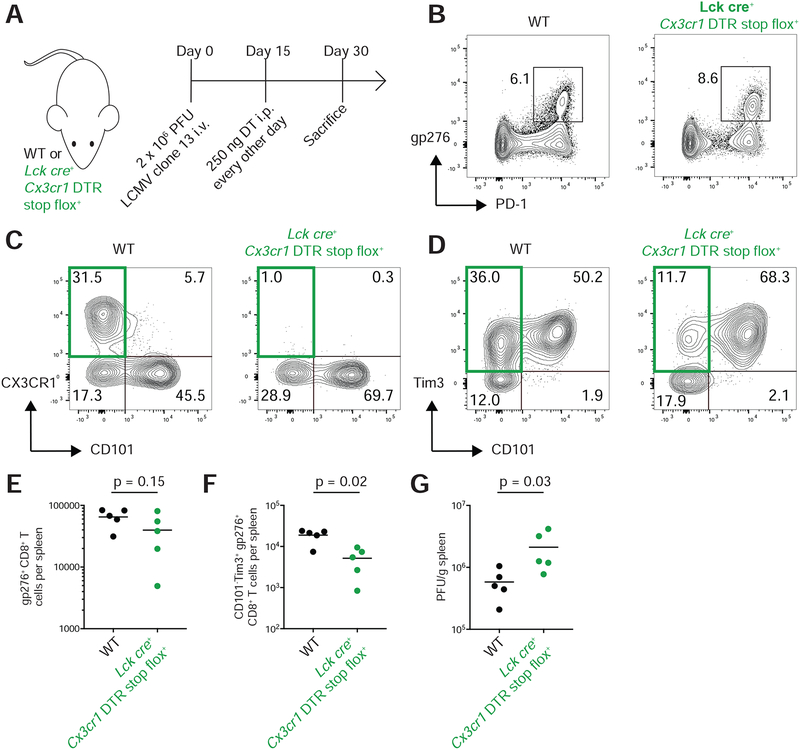
Previous work has shown that PD-1 pathway blockade increases the proliferation of PD-1+ Tcf-1+ Tim3− cells(Im et al., 2016). Here, we have shown that these stem-like cells differentiate into functional CD101−Tim3+ cells after division. These findings suggest that PD-1 pathway blockade may function by stimulating stem-like CD101−Tim3− cells to generate higher numbers of transitory CD101−Tim3+ cells. To test this hypothesis, we treated chronically infected C57BL/6J mice with ɑPD-L1 or isotype control (Figure 7A) and analyzed the number and phenotype of antigen-specific CD8+ T cells present two weeks after treatment. PD-1 pathway blockade resulted in substantial expansion of LCMV-specific cells compared to control-treated mice (Figure 7B). After treatment, the frequency of CD101−Tim3+ cells within the splenic LCMV-specific CD8+ T cell pool doubled (Figure 7C,D), driven by a 10-fold increase in the total number of these transitory cells (Figure 7E). A similar increase of transitory cell frequencies was observed in the liver after PD-1 pathway blockade compared to isotype control (Figure 7F). Again, transitory cells were present at 10-fold higher total numbers in liver, blood, and the lungs (Figure 7G–I) in ɑPD-L1 treated mice compared to controls. These results show that by increasing the generation of CD101−Tim3+ cells from CD101−Tim3− cells, checkpoint blockade enlarges the number of functional, transitory cells that contribute to improved viral control.
Figure 7: PD-1 pathway blockade increases the number of transitory antigen-specific CD8+ T cells.
A) Mice infected with LCMV clone 13 were treated with ɑPD-L1 or isotype control >45 days after infection.
B) Representative tetramer staining of splenic CD8+ T cells in isotype or ɑPD-L1-treated mice.
C) Representative staining of CD101 and Tim3 on splenic gp276+ CD8+ T cells.
D) Frequency of transitory (CD101−Tim3+) cells among CD8+ gp276+ cells in spleen.
E) Total number of transitory gp276+ CD8+ T cells in spleen.
F) Frequency of transitory (CD101−Tim3+) cells among CD8+ gp276+ cells in liver.
G) Total number of transitory gp276+ CD8+ T cells in liver.
H) Frequency of transitory gp276+ CD8+ T cells in peripheral blood.
I) Total number of transitory gp276+ CD8+ T cells in lungs.
* and ** indicate p < 0.05 and p < 0.01 by 0.01 by t-test. Data shown is representative of three independent experiments.
Discussion
In this study, we have performed a systematic analysis to define the repertoire of surface molecules that are preferentially expressed on exhausted CD8+ T cells. This analysis confirmed the presence of known checkpoint molecules, such as PD-1 and Lag3, but also identified a multitude of other, less-studied molecules present on dysfunctional CD8+ T cells. Many of these molecules such as Lax1, CD7, CD200R, CD112R, and CD101 have been shown to inhibit TCR signaling or otherwise impair T cell function(Pace et al., 2000; Rosenblum et al., 2005; Rygiel et al., 2009; Soares et al., 1998; Zhu et al., 2002; Zhu et al., 2016), but their in vivo role in exhaustion is unclear. A recent study using a liver tumor mouse model identified CD101 as a marker of antigen-specific CD8+ T cells with a dysfunctional chromatin state(Philip et al., 2017). It is unclear whether CD101 is merely a marker of dysfunctional cells or contributes to negative regulatory signaling itself, although evidence suggests an inhibitory function on T cells in vitro and in vivo (Mohammed et al., 2011; Rainbow et al., 2011; Schey et al., 2016; Soares et al., 1998). Here, we demonstrated that CD101 expression defined the terminally differentiated state of CD8+ T cells in chronic viral infection.
Initially, stem-like Tcf-1+ CD8+ T cells present in chronic viral infection differentiate into transitory CD101−Tim3+ cells, a process accelerated by PD-1 pathway blockade. Our gene expression analyses showed that this process is accompanied by downregulation of Tcf-1 and induction of transcriptional programs for cytotoxicity, cell proliferation, and migration as shown by GSEA and expression of genes encoding Ki-67, granzyme B, S1pr1, and CX3CR1. These cells also express effector- and migration-associated transcription factors, such as T-bet(Sullivan et al., 2003) and Klf2(Carlson et al., 2006). A recent report has identified increased numbers of circulating Gzmbhi CX3CR1+ CD8+ T cells in cancer patients who respond to PD-1 blockade therapy(Yan et al., 2018). We also show that transitory cells increase in numbers after PD-1 blockade and are capable of division and cytokine production in chronically infected mice, although it is unclear how analogous these transitory cells in mice are to human CX3CR1+ CD8+ T cells after PD-1 blockade. With continued persistence of antigen during chronic infection, transitory antigen-specific CD8+ T cells eventually convert into exhausted CD101+ cells. This transition involves downregulation of migratory molecules such as S1pr1 and Cx3cr1 and expression of an alternate, exhaustion-associated program. Transcription factors Klf2 and T-bet were downregulated with concomitant upregulation of Runx3, Hic1, and Nr4a2 which may program non-lymphoid residency and transcriptional repression of effector molecules (Chen et al., 2019; Liu et al., 2019; Milner et al., 2017; Seo et al., 2019; Ubaid et al., 2018).
Previous studies have postulated that T-bethi cells are precursors to Eomeshi cells in chronic viral infection (Paley et al., 2012), but we find here and previously(Im et al., 2016) that Eomes expression is highest in stem-like Tcf-1+ cells during chronic infection and is downregulated after differentiation of stem-like cells into the CD101−Tim3+ effector-like state. Additionally, we found that T-bet was highest in transitional, effector-like CD101−Tim3+ cells compared to both terminally differentiated CD101+ cells and Tcf-1+ stem-like cells. As shown by our adoptive transfer experiments, the irreversibility of Tim3 and CD101 expression on CD8+ T cells during antigen persistence made them excellent markers for the differentiation status of antigen-specific CD8+ T cells in murine models of chronic infection. In addition to the acquisition of CD101 expression, we also found that expression of well-known inhibitory receptors such as CD160, 2B4, and PD-1 as well as the dysfunction-associated markers described here (CD200Rs, CD7, Lax1, CD112R, etc.) were universally increased in exhausted terminally differentiated CD8+ T cells.
Simultaneously with our study, Zander et al. report three subsets of antigen-specific CD8+ T cells present in LCMV clone 13 infection: Slamf6+ (Ly108+), CX3CR1+, and Slamf6−CX3CR1− cells. These subsets appear largely analogous to the stem-like (CD101−Tim3−), transitory (CD101−Tim3+), and exhausted (CD101+Tim3+) populations we describe here. For example, Zander et al. find that CX3CR1+ (CD101−Tim3+) cells express higher amounts of T-bet and Zeb2 and that these cells have increased cytotoxic capacity and are crucial for viral control in LCMV clone 13 infection. Further, the Slamf6−CX3CR1− subset reported by Zander et al. express higher amounts of inhibitory molecules such as CD244, CD160, and Lag3, as do the CD101+Tim3+ cells we report in the present study. One key difference in our studies is in our conclusions regarding the differentiation pathways of the CD8+ T cell subsets. The recipient mice in our study were infection-matched, chronically infected mice whereas Zander et al. transferred the CD8+ T cell subsets into naive mice that were subsequently challenged with LCMV. These two conditions are notably different; in our experiments, the cells were transferred back into an environment of established chronic infection whereas transfer of cells into a naive mouse with subsequent infection more reflects the environment during the early stages of viral infection. Additionally, we performed transient CD4+ T cell depletion at infection, resulting in ‘helpless’ LCMV-specific donor CD8+ T cells, higher viral titers, and lifelong infection compared to the clearing infection in Zander et al.
However, together our results suggest that, in addition to canonical exhaustion-associated proteins such as PD-1 and Tim3, a second module of exhaustion-associated genes, including CD101, is upregulated on antigen-specific CD8+ T cells weeks after initial antigen exposure. PD-1 expression is seen on antigen-specific T cells following acute infection and can be observed on memory CD8+ T cells hours after antigen exposure(Hosking et al., 2013). In contrast, CD101 was not induced until 2–4 weeks after induction of chronic viral infection and was not observed on LCMV-specific cells in acute infection. Thus, CD101 and its associated surface markers such as CD7 and the CD200 receptors may constitute a ‘second wave’ of inhibitory molecules induced on antigen-specific CD8+ T cells weeks after infection. This observation also demonstrates that T cell exhaustion is a measured process, with gradual emergence of highly dysfunctional CD8+ T cells. Steady-state frequencies of CD101+ CD8+ T cells were not evident until 3–4 weeks after infection; thus, immunological models that begin checkpoint blockade less than a month after viral or tumor inoculation may adequately recapitulate neither the full extent of CD8+ T cell exhaustion present in chronic viral infection nor the presence of additional inhibitory molecules present in the CD101-associated transcriptional program.
Despite the gradual acquisition of this CD101-associated gene expression profile, we have shown through adoptive transfer experiments expression of CD101 is irreversible under conditions of chronic antigen exposure. Additionally, adoptive transfer of the subsets described here revealed differential migration patterns after intravenous transfer: stem-like cells were found in all tissues after transfer, with particularly high recovery in lymphoid tissues. CD101−Tim3+ CD8+ T cells were also capable of distribution to all tested tissues after transfer, with a bias to non-lymphoid tissues. CD101+Tim3+ cells, however, were nearly exclusively recovered in liver. These differences in migration could be explained by the heightened expression of chemokine receptors CCR2 and CX3CR1 in CD101−Tim3+ cells and/or the higher expression of sphingosine-1-phosphate receptors. Regardless, the unique properties and divergent transcriptional programs of each subset and their irreversible differentiation pathway demonstrates that the capabilities and immunotherapy responsiveness of the CD8+ T cell response changes with continued presence of antigen.
Finally, we have shown that PD-1 pathway blockade caused a substantial increase of antigen-specific transitory CD8+ T cells, and that these cells were critical for viral control in the LCMV model. This is consistent with recent findings that stem-like cells are responsible for the proliferative burst in response to PD-1 blockade(Im et al., 2016), as we show that these stem-like cells give rise to transitory effector cells with effector and proliferative capacity. Thus, an effective response to immunotherapy in vivo may depend not only on increased numbers of total antigen-specific cells, but also on the increased frequencies of highly functional, transitory cells within the antigen-specific CD8+ T cell population.
STAR Methods
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Rafi Ahmed (rahmed@emory.edu). This study did not generate new unique reagents.
Experimental Model and Subject Details
Mouse strains
C57BL/6J mice, Lck cre, and Cx3cr1 DTR flox mice were obtained from The Jackson Laboratory. P14 mice (on the C57BL/6J background) were bred in-house. Mice were infected at 6–8 weeks of age, and all mice in this study were females with the exception of Lck cre Cx3cr1 DTR stopfl mice and matched controls, which were of mixed sex. All mouse experiments were performed with approval of the Emory University Institutional Animal Care and Use Committee.
Method Details
Mouse infection experiments
LCMV infections were performed as previously described(Araki et al., 2017; Im et al., 2016). For chronic infections, 6–8 week old C57BL/6J mice were injected intraperitoneally (i.p.) with 300 μg of CD4+ T cell-depleting antibody GK1.5 (Bio X Cell) one and two days before intravenous (i.v.) injection with 2 × 106 pfu LCMV clone 13. CD4+ T cell depletion is required for constant viral titers in long-term experiments(Matloubian et al., 1994) and was thus used to control for reduced antigen in clearing models (such as clone 13 without CD4+ T cell depletion). For Cx3cr1 DTR experiments and those in Figure S2 only, CD4+ T cell depletion was not used, in order to measure viral clearance.
For acute infection, C57BL/6J mice were injected i.p. with 2 × 105 pfu LCMV Armstrong. Unless otherwise indicated, LCMV Armstrong-infected mice were analyzed at day 8 post-infection for the acute timepoint, and after day 45 for the memory timepoint. Unless otherwise indicated, LCMV clone 13-infected mice were analyzed after day 45 for the chronic timepoint.
C57BL/6J mice, Lck cre, and Cx3cr1 DTR flox mice were obtained from The Jackson Laboratory. P14 mice (on the C57BL/6J background) were bred in-house. All mouse experiments were performed with approval of the Emory University Institutional Animal Care and Use Committee.
RNA-sequencing and analysis of CD101 subsets from chronic LCMV infection
CD101−Tim3−, CD101−Tim3+, and CD101+Tim3+ PD-1+CD8+ T cells were isolated by FACS from three pools of ten mice each infected with LCMV clone 13 15 or >45 days prior to the sort (Figure S1). In each case, naive CD8+ T cells from three age-matched uninfected mice were also isolated by FACS. RNA was isolated using the Qiagen AllPrep Micro kit according to the manufacturer’s protocols. RNA sequencing with poly(A) selection was performed at the Yerkes Nonhuman Primate Genomics Core.
Adoptive transfer experiments
Splenocytes were isolated from LCMV clone 13 infected C57BL/6J mice >45 days after infection. CD8+ T Cells were separated with the EasySep Mouse CD8+ T Cell Isolation Kit (StemCell) and subsequently labeled with CTV (ThermoFisher) according to the manufacturer’s protocol. Using FACS, three populations of PD-1+CD8+ T cells were isolated (CD101−Tim3−, CD101−Tim3+, CD101+Tim3+). Cells were reconstituted in RPMI and 0.9–1.0 × 105 isolated cells from a single population were transferred i.v. to a single recipient CD45.1+ C57BL/6J mouse. Two (Figure 4) or four weeks (Figure S9) after transfer, cells were isolated from spleen, liver, lung, and bone marrow and stained as below.
Intracellular cytokine staining experiments
2 × 103 P14 cells (cells with a transgenic TCR for the Db-restricted gp33 LCMV epiptope) were transferred i.v. to recipient mice three days before infection with LCMV clone 13 as described above. Twenty-two days after infection, splenocytes were isolated and incubated in RPMI with 10% FBS with 0.2 μg/ml of gp33 peptide for five hours at 37 °C in the presence of BD GolgiStop and BD GolgiPlug, according to manufacturer’s protocols. After incubation, cells were stained for flow cytometry as described below.
PD-L1 blockade experiments
Mice were infected as above with LCMV clone 13. >45 days after infection, mice were injected with 200 μg of ɑPD-L1 (clone 10F.9G2) or isotype control (rat IgG2b) in PBS. Injections were given i.p. every three days. Mice were analyzed for responses 12 to 14 days after initiation of treatment.
Flow cytometry experiments
Antibodies used in flow cytometry experiments are shown in in the Key Resources Table. MHC I tetramers were prepared and used as previously described(Murali-Krishna et al., 1998). Single cell suspensions were stained with antibodies and/or tetramer in PBS with 2% FBS and 2 mM EDTA for extracellular targets. For intranuclear staining, the eBioscience Foxp3 Transcription Factor Staining Buffer Set was used for fixation, permeabilization, and intracellular staining. BD Cytofix/Cytoperm and Perm/Wash buffers were used for fixation and staining of cytoplasmic targets. BD Cytofix/Cytoperm was also used for fixation of non-intracellularly stained cells.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies (clone is given as identifier) | ||
| anti-CD101 antibody | eBioScience/ThermoFisher | Moushi101 |
| anti-CD160 antibody | Biolegend | 7H1 |
| anti-CD19 antibody | Biolegend | 6D5 |
| anti-CD200 antibody | Biolegend | OX-90 |
| anti-CD200R antibody | Biolegend | OX-110 |
| anti-CD22 antibody | Biolegend | OX-97 |
| anti-CD244/2B4 antibody | BD | 2B4 |
| anti-CD31 antibody | Biolegend | 390 |
| anti-CD38 antibody | Biolegend | 90 |
| anti-CD4 antibody | Biolegend | GK1.5 |
| anti-CD44 antibody | Biolegend | IM7 |
| anti-CD62L antibody | eBioScience/ThermoFisher | MEL-14 |
| anti-CD8 antibody | Biolegend | 53–6.7 |
| anti-CD9 antibody | Biolegend | MZ3 |
| anti-CX3CR1 antibody | Biolegend | SA011F11 |
| anti-CXCR5 antibody | Biolegend | L138D7 |
| anti-Eomes antibody | eBioScience/ThermoFisher | Dan11mag |
| anti-gp49 antibody | Biolegend | H1.1 |
| anti-Granzyme A antibody | eBioScience/ThermoFisher | GzA-3G8.5 |
| anti-IFNg antibody | Biolegend | XMG1.2 |
| anti-Ki-67 antibody | BD | B56 |
| anti-Klrg1 antibody | Southern Biotech | 2F1 |
| LIVE/DEAD Fixable Dead Cell Stain | ThermoFisher | n/a |
| anti-PD-1 antibody | Biolegend | 29F.1A12 |
| anti-T-bet antibody | Biolegend | 4B10 |
| anti-Tim3 antibody | Biolegend | RMT3–23 |
| anti-TNFa antibody | Biolegend | MP6-XT22 |
| Bacterial and Virus Strains | ||
| LCMV clone 13 | in-house | |
| LCMV Armstrong | in-house | |
| Chemicals, Peptides, and Recombinant Proteins | ||
| anti-PD-L1 antibody | 10F.9G2 | |
| isotype control | BioXCell | rat IgG2b |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J mice | Jackson Laboratories | JAX stock #000664 |
| Lck cre mice | Jackson Laboratories | JAX stock #003802 |
| DTR flox/stop mice | Jackson Laboratories | JAX stock #025629 |
Samples were acquired using a BD LSR II and analyzed using FlowJo. Summary graphs and statistics were performed in GraphPad Prism v7.
Quantification and statistical analysis
Analysis to identify markers of murine CD8+ T cell exhaustion by gene expression patterns
RNA-sequencing data from acute and chronic LCMV infection were previously described (Hudson et al., 2019). For subsets from chronic infection, tetramer-positive CXCR5+Tim3− and CXCR5−Tim3+ LCMV-specific cells were isolated by FACS from spleens of mice infected with LCMV clone 13, infected as described below. For both datasets, reads were aligned to the mm10 genome (accessed through Ensembl(Zerbino et al., 2018)) with HISAT2 version 2.0.5 ((Kim et al., 2015)). featureCounts(Liao et al., 2013) was used to assign aligned reads to genes using the Ensembl release 91 mouse genome annotation, with reads overlapping multiple features allowed assignment to more than one gene. To control for batch effects, counts were normalized with DESeq2 (ref. (Love et al., 2014)) with a design formula of ~type+batch, where ‘type’ indicates the cell type (Naive, Memory Precursor, CXCR5+, etc.) and ‘batch’ indicates the individual RNA-seq experiment (acute and chronic LCMV infection).
To identify transmembrane proteins, all peptide sequences (including splice variants) for all protein-coding genes in the Ensembl 91 genome annotation were accessed through the biomaRt R package(Durinck et al., 2005; Durinck et al., 2009) and transmembrane helices predicted by the TMHMM server version 2.0(Krogh et al., 2001). To identify ITAM, ITIM, and ITSM motifs, predicted intracellular regions were scanned for the motifs Yxx(I/L)x6–8(Y)xx(I/L), (I/L/S/V)xYxx(I/L/V), and TxYxx(A/L/V), respectively, using a custom Python script. Genes were labeled as transmembrane if any of its annotated protein sequence contained a predicted transmembrane helix, and genes were labeled as containing an ITAM, ITIM, or ITSM if any of its annotated protein sequences had the appropriate motif within a predicted intracellular region. Genes were identified as containing an immunoglobulin-like domain if the gene was annotated as containing at least one of the following PFAM domains(Finn et al., 2016) as accessed through biomaRt(Durinck et al., 2005; Durinck et al., 2009): PF00047, PF02124, PF02440, PF02480, PF03921, PF06697, PF07654, PF07679, PF07686, PF08204 PF08205, PF11465, PF13895, PF13927, PF15028, PF16680, PF16681, or PF16706.
A gene was identified as a marker of Tim3+ CD8+ T cells from (chronic) LCMV clone 13 infection if the gene met all of the following conditions:
Expression > 1000 normalized counts in Tim3+ CD8+ T cells
Expression in CXCR5−Tim3+ CD8+ T cells > expression in CXCR5+Tim3− CD8+ T cells
log2 fold change (CXCR5−Tim3+ CD8+ T cells / naive CD8+ T cells) > 1.6 (~ 3-fold)
log2 fold change (CXCR5−Tim3+ CD8+ T cells / terminal effector CD8+ T cells) > 1.6
log2 fold change (CXCR5−Tim3+ CD8+ T cells / effector memory CD8+ T cells) > 1.6
A gene was identified as a marker of CXCR5+ CD8+ T cells from (chronic) LCMV clone 13 infection if the gene met all of the following conditions:
Expression > 1000 normalized counts in CXCR5+ CD8+ T cells
Expression in CXCR5+Tim3− CD8+ T cells > expression in CXCR5−Tim3+ CD8+ T cells
log2 fold change (CXCR5+Tim3− CD8+ T cells / naive CD8+ T cells) > 1.6
log2 fold change (CXCR5+Tim3− CD8+ T cells / terminal effector CD8+ T cells) > 1.6
log2 fold change (CXCR5+Tim3− CD8+ T cells / effector memory CD8+ T cells) > 1.6
As gene expression changes can become dampened with higher expression, the required log2 fold change in conditions 3–5 above were relaxed to 1.0 (2-fold) for genes with expression >7500 normalized counts.
Analyses described above are available in a sortable, customizable format in Table S1.
Analysis of RNA-sequencing from CD101 subsets during chronic infection
As above, reads were aligned with HISAT2 version 2.0.5 (Kim et al., 2015) to the mm10 genome (accessed through Ensembl (Zerbino et al., 2018)) and featureCounts(Liao et al., 2013) was used to assign aligned reads to genes using the Ensembl release 91 mouse genome annotation. DESeq2 (Love et al., 2014) was used to normalize for library size and calculate differential expression across groups. A gene was considered differentially expressed between two groups with an adjusted p-value of <0.05 and an average expression of >20 normalized counts across all samples.
For gene set enrichment analysis, GSEA preranked(Subramanian et al., 2005) was used against the MSigDB(Liberzon et al., 2015; Liberzon et al., 2011), with −log10(p) * sign(log2(FC)) as the ranking statistic, where p is the adjusted p-value and FC the fold change of the differential expression analysis of interest. Enrichment was considered significant with an FDR < 0.05.
Principal component analysis was performed with the FactoMineR R package(Lê et al., 2008) on all detected genes using the regularized log transformation from DESeq2. One sample of CD101−Tim3− cells was excluded from dimensionality reduction due to its status as an outlier on PCA but included on all other analyses. RNA-sequencing data was visualized with the ggplot2 R package(Wickham, 2016) and GraphPad Prism (version 7.0c).
Data and code availability
RNA-sequencing reads are available from the NCBI Sequence Read Archive under BioProject PRJNA497086. Processed RNA-sequencing data, including normalized gene counts and differential expression analyses, are available in the Supplementary Information (Tables S1–S4).
Supplementary Material
Table S1, related to Figure 1: Analysis of CD8+ T cell gene expression during acute and chronic LCMV infection, including properties of encoded proteins.
Table S2, related to Figure 2: Gene expression and differential expression testing results of CD101−Tim3−, CD101−Tim3+, and CD101+Tim3+ PD-1+ CD8+ T cells from chronic infection (day >45).
Table S3, related to Figure 2: Complete gene set enrichment analysis (GSEA) results using RNA-sequencing data of PD-1+ CD8+ T cells after chronic infection (day >45).
Table S4, related to Figure 2: Gene expression and differential expression testing results of CD101−Tim3−, CD101−Tim3+, and CD101+Tim3+ PD-1+ CD8+ T cells from chronic infection (day 15).
Chronic viral infection induces exhaustion of antigen-specific T cells. Hudson and colleagues define a transitory, effector-like population of CD8+ T cells that are recently generated from stem-like CD8+ T cells in chronic infection. These transitory cells contribute to viral control and are increased in number following PD-1 pathway blockade.
CX3CR1+ CD8+ T cells are recent progeny of stem-like cells in chronic infection
CX3CR1+ cells differentiate to a dysfunctional state marked by CD101 expression
Transitory CX3CR1+ cells express effector molecules and contribute to viral control
PD-1 pathway blockade increases the number of antigen-specific transitory cells
Acknowledgements
The authors thank Alice Kamphorst, Koichi Araki, Timothy Hoang, and Robert Karaffa and Kametha Fife of the Emory University School of Medicine Flow Cytometry Core. W.H.H. is a Cancer Research Institute Irvington Fellow supported by the Cancer Research Institute. H.T.K. is supported by grant K99CA197804 from the National Cancer Institute of the National Institutes of Health (NIH). R.A. is supported by grants R01AI030048 and P01AI056299 from the National Institute of Allergy and Infectious Diseases of the NIH. The Yerkes Nonhuman Primate Genomics Core is supported in part by ORIP/OD P51OD011132. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Declaration of Interests
Rafi Ahmed holds patents on the PD-1 pathway.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed R, Salmi A, Butler LD, Chiller JM, and Oldstone MB (1984). Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J Exp Med 160, 521–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akondy RS, Fitch M, Edupuganti S, Yang S, Kissick HT, Li KW, Youngblood BA, Abdelsamed HA, McGuire DJ, Cohen KW, et al. (2017). Origin and differentiation of human memory CD8 T cells after vaccination. Nature 552, 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, Morita M, Bederman AG, Konieczny BT, Kissick HT, Sonenberg N, and Ahmed R (2017). Translation is actively regulated during the differentiation of CD8+ effector T cells. Nature Immunology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, and Ahmed R (2005). Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 439, 682–687. [DOI] [PubMed] [Google Scholar]
- Barclay AN (2003). Membrane proteins with immunoglobulin-like domains—a master superfamily of interaction molecules. Seminars in Immunology 15, 215–223. [DOI] [PubMed] [Google Scholar]
- Brahmer JR, Tykodi SS, Chow LQM, Hwu W-J, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al. (2012). Safety and Activity of Anti–PD-L1 Antibody in Patients with Advanced Cancer. New England Journal of Medicine 366, 2455–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelman J, Mazza EMC, Alvisi G, Colombo FS, Grilli A, Mikulak J, Mavilio D, Alloisio M, Ferrari F, Lopci E, et al. (2018). High-dimensional single cell analysis identifies stem-like cytotoxic CD8+ T cells infiltrating human tumors. The Journal of Experimental Medicine 215, 2520–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan MK, and Wolchok JD (2013). At the Bedside: CTLA-4- and PD-1-blocking antibodies in cancer immunotherapy. Journal of Leukocyte Biology 94, 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CM, Endrizzi BT, Wu J, Ding X, Weinreich MA, Walsh ER, Wani MA, Lingrel JB, Hogquist KA, and Jameson SC (2006). Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature 442, 299–302. [DOI] [PubMed] [Google Scholar]
- Chatterjee M, Kis-Toth K, Thai T-H, Terhorst C, and Tsokos GC (2011). SLAMF6-driven co-stimulation of human peripheral T cells is defective in SLE T cells. Autoimmunity 44, 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, López-Moyado IF, Seo H, Lio C-WJ, Hempleman LJ, Sekiya T, Yoshimura A, Scott-Browne JP, and Rao A (2019). NR4A transcription factors limit CAR T cell function in solid tumours. Nature 567, 530–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, Mackey EW, Miller JD, Leslie AJ, DePierres C, et al. (2006). PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature 443, 350–354. [DOI] [PubMed] [Google Scholar]
- Diehl GE, Longman RS, Zhang J-X, Breart B, Galan C, Cuesta A, Schwab SR, and Littman DR (2013). Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX3CR1hi cells. Nature 494, 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez CX, Amezquita RA, Guan T, Marshall HD, Joshi NS, Kleinstein SH, and Kaech SM (2015). The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. The Journal of Experimental Medicine 212, 2041–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, and Huber W (2005). BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440. [DOI] [PubMed] [Google Scholar]
- Durinck S, Spellman PT, Birney E, and Huber W (2009). Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc 4, 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Research 44, D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallimore A, Glithero A, Godkin A, Tissot AC, Plückthun A, Elliott T, Hengartner H, and Zinkernagel R (1998). Induction and Exhaustion of Lymphocytic Choriomeningitis Virus–specific Cytotoxic T Lymphocytes Visualized Using Soluble Tetrameric Major Histocompatibility Complex Class I–Peptide Complexes. The Journal of Experimental Medicine 187, 1383–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach C, Moseman EA, Loughhead SM, Alvarez D, Zwijnenburg AJ, Waanders L, Garg R, de la Torre JC, and von Andrian UH (2016). The Chemokine Receptor CX3CR1 Defines Three Antigen-Experienced CD8 T Cell Subsets with Distinct Roles in Immune Surveillance and Homeostasis. Immunity 45, 1270–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gettinger SN, Choi J, Mani N, Sanmamed MF, Datar I, Sowell R, Du VY, Kaftan E, Goldberg S, Dong W, et al. (2018). A dormant TIL phenotype defines non-small cell lung carcinomas sensitive to immune checkpoint blockers. Nature Communications 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R, Hou S, Liu C, Zhang A, Bai Q, Han M, Yang Y, Wei G, Shen T, Yang X, et al. (2016). Follicular CXCR5-expressing CD8+ T cells curtail chronic viral infection. Nature 537, 412–416. [DOI] [PubMed] [Google Scholar]
- Hosking MP, Flynn CT, Botten J, and Whitton JL (2013). CD8+ Memory T Cells Appear Exhausted within Hours of Acute Virus Infection. The Journal of Immunology 191, 4211–4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson WH, Prokhnevska N, Gensheimer J, Akondy R, McGuire DJ, Ahmed R, and Kissick HT (2019). Expression of novel long noncoding RNAs defines virus-specific effector and memory CD8+ T cells. Nature Communications 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, Shan Q, Hale JS, Lee J, Nasti TH, et al. (2016). Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav RR, Im SJ, Hu B, Hashimoto M, Li P, Lin J-X, Leonard WJ, Greenleaf WJ, Ahmed R, and Goronzy JJ (2019). Epigenetic signature of PD-1+ TCF1+ CD8 T cells that act as resource cells during chronic viral infection and respond to PD-1 blockade. Proceedings of the National Academy of Sciences 116, 14113–14118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Li L, Han J, Sun Z, Rong Y, and Jin Y (2017). CXCR5+ CD8+ T Cells Indirectly Offer B Cell Help and Are Inversely Correlated with Viral Load in Chronic Hepatitis B Infection. DNA and Cell Biology 36, 321–327. [DOI] [PubMed] [Google Scholar]
- Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, and Kaech SM (2007). Inflammation Directs Memory Precursor and Short-Lived Effector CD8+ T Cell Fates via the Graded Expression of T-bet Transcription Factor. Immunity 27, 281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, and Ahmed R (2003). Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nature Immunology 4, 1191–1198. [DOI] [PubMed] [Google Scholar]
- Kaech SM, Wherry EJ, and Ahmed R (2002). Effector and memory T-cell differentiation: implications for vaccine development. Nature Reviews Immunology 2, 251–262. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nature Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, and Sonnhammer EL (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305, 567–580. [DOI] [PubMed] [Google Scholar]
- Lê S, Josse J, and Husson F (2008). FactoMineR: An R Package for Multivariate Analysis. Journal of Statistical Software 25. [Google Scholar]
- Leong YA, Chen Y, Ong HS, Wu D, Man K, Deleage C, Minnich M, Meckiff BJ, Wei Y, Hou Z, et al. (2016). CXCR5+ follicular cytotoxic T cells control viral infection in B cell follicles. Nature Immunology 17, 1187–1196. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2013). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov Jill P., and Tamayo P (2015). The Molecular Signatures Database Hallmark Gene Set Collection. Cell Systems 1, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, and Mesirov JP (2011). Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang Y, Lu H, Li J, Yan X, Xiao M, Hao J, Alekseev A, Khong H, Chen T, et al. (2019). Genome-wide analysis identifies NR4A1 as a key mediator of T cell dysfunction. Nature 567, 525–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matloubian M, Concepcion RJ, and Ahmed R (1994). CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J Virol 68, 8056–8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matloubian M, Kolhekar SR, Somasundaram T, and Ahmed R (1993). Molecular determinants of macrophage tropism and viral persistence: importance of single amino acid changes in the polymerase and glycoprotein of lymphocytic choriomeningitis virus. J Virol 67, 7340–7349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V, Allende ML, Proia RL, and Cyster JG (2004). Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature 427, 355–360. [DOI] [PubMed] [Google Scholar]
- Milner JJ, Toma C, Yu B, Zhang K, Omilusik K, Phan AT, Wang D, Getzler AJ, Nguyen T, Crotty S, et al. (2017). Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed JP, Fusakio ME, Rainbow DB, Moule C, Fraser HI, Clark J, Todd JA, Peterson LB, Savage PB, Wills-Karp M, et al. (2011). Identification of Cd101 as a Susceptibility Gene for Novosphingobium aromaticivorans-Induced Liver Autoimmunity. The Journal of Immunology 187, 337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murali-Krishna K, Altman JD, Suresh M, Sourdive DJD, Zajac AJ, Miller JD, Slansky J, and Ahmed R (1998). Counting Antigen-Specific CD8 T Cells: A Reevaluation of Bystander Activation during Viral Infection. Immunity 8, 177–187. [DOI] [PubMed] [Google Scholar]
- Murter B, Pan X, Ophir E, Alteber Z, Azulay M, Sen R, Levy O, Dassa L, Vaknin I, Fridman-Kfir T, et al. (2019). Mouse PVRIG Has CD8+ T Cell–Specific Coinhibitory Functions and Dampens Antitumor Immunity. Cancer Immunology Research 7, 244–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen JT, Evans DP, Galvan M, Pace KE, Leitenberg D, Bui TN, and Baum LG (2001). CD45 Modulates Galectin-1-Induced T Cell Death: Regulation by Expression of Core 2 O-Glycans. The Journal of Immunology 167, 5697–5707. [DOI] [PubMed] [Google Scholar]
- Ott PA, Hodi FS, Kaufman HL, Wigginton JM, and Wolchok JD (2017). Combination immunotherapy: a road map. Journal for ImmunoTherapy of Cancer 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace KE, Hahn HP, Pang M, Nguyen JT, and Baum LG (2000). Cutting Edge: CD7 Delivers a Pro-Apoptotic Signal During Galectin-1-Induced T Cell Death. The Journal of Immunology 165, 2331–2334. [DOI] [PubMed] [Google Scholar]
- Paley MA, Kroy DC, Odorizzi PM, Johnnidis JB, Dolfi DV, Barnett BE, Bikoff EK, Robertson EJ, Lauer GM, Reiner SL, and Wherry EJ (2012). Progenitor and Terminal Subsets of CD8+ T Cells Cooperate to Contain Chronic Viral Infection. Science 338, 1220–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardoll DM (2012). The blockade of immune checkpoints in cancer immunotherapy. Nature Reviews Cancer 12, 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip M, Fairchild L, Sun L, Horste EL, Camara S, Shakiba M, Scott AC, Viale A, Lauer P, Merghoub T, et al. (2017). Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature 545, 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainbow DB, Moule C, Fraser HI, Clark J, Howlett SK, Burren O, Christensen M, Moody V, Steward CA, Mohammed JP, et al. (2011). Evidence that Cd101 Is an Autoimmune Diabetes Gene in Nonobese Diabetic Mice. The Journal of Immunology 187, 325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum MD, Woodliff JE, Madsen NA, McOlash LJ, Keller MR, and Truitt RL (2005). Characterization of CD200-Receptor Expression in the Murine Epidermis. Journal of Investigative Dermatology 125, 1130–1138. [DOI] [PubMed] [Google Scholar]
- Rygiel TP, Rijkers ESK, de Ruiter T, Stolte EH, van der Valk M, Rimmelzwaan GF, Boon L, van Loon AM, Coenjaerts FE, Hoek RM, et al. (2009). Lack of CD200 Enhances Pathological T Cell Responses during Influenza Infection. The Journal of Immunology 183, 1990–1996. [DOI] [PubMed] [Google Scholar]
- Sallusto F, Lenig D, Förster R, Lipp M, and Lanzavecchia A (1999). Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401, 708–712. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Kalia V, Haining WN, Konieczny BT, Subramaniam S, and Ahmed R (2008). Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. The Journal of Experimental Medicine 205, 625–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schey R, Dornhoff H, Baier JLC, Purtak M, Opoka R, Koller AK, Atreya R, Rau TT, Daniel C, Amann K, et al. (2016). CD101 inhibits the expansion of colitogenic T cells. Mucosal Immunology 9, 1205–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H, Chen J, González-Avalos E, Samaniego-Castruita D, Das A, Wang YH, López-Moyado IF, Georges RO, Zhang W, Onodera A, et al. (2019). TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proceedings of the National Academy of Sciences 116, 12410–12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares LR, Tsavaler L, Rivas A, and Engleman EG (1998). V7 (CD101) ligation inhibits TCR/CD3-induced IL-2 production by blocking Ca2+ flux and nuclear factor of activated T cell nuclear translocation. J Immunol 161, 209–217. [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BM, Juedes A, Szabo SJ, von Herrath M, and Glimcher LH (2003). Antigen-driven effector CD8 T cell function regulated by T-bet. Proceedings of the National Academy of Sciences 100, 15818–15823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. (2012). Safety, Activity, and Immune Correlates of Anti–PD-1 Antibody in Cancer. New England Journal of Medicine 366, 2443–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubaid U, Andrabi SBA, Tripathi SK, Dirasantha O, Kanduri K, Rautio S, Gross CC, Lehtimäki S, Bala K, Tuomisto J, et al. (2018). Transcriptional Repressor HIC1 Contributes to Suppressive Function of Human Induced Regulatory T Cells. Cell Reports 22, 2094–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utzschneider DT, Charmoy M, Chennupati V, Pousse L, Ferreira DP, Calderon-Copete S, Danilo M, Alfei F, Hofmann M, Wieland D, et al. (2016). T Cell Factor 1-Expressing Memory-like CD8 + T Cells Sustain the Immune Response to Chronic Viral Infections. Immunity 45, 415–427. [DOI] [PubMed] [Google Scholar]
- Whelan S, Ophir E, Kotturi MF, Levy O, Ganguly S, Leung L, Vaknin I, Kumar S, Dassa L, Hansen K, et al. (2019). PVRIG and PVRL2 Are Induced in Cancer and Inhibit CD8+ T-cell Function. Cancer Immunology Research 7, 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ, Teichgräber V, Becker TC, Masopust D, Kaech SM, Antia R, von Andrian UH, and Ahmed R (2003). Lineage relationship and protective immunity of memory CD8 T cell subsets. Nature Immunology 4, 225–234. [DOI] [PubMed] [Google Scholar]
- Wickham H (2016). ggplot2: Elegant Graphics for Data Analysis (New York: Springer-Verlag; ). [Google Scholar]
- Wu T, Ji Y, Moseman EA, Xu HC, Manglani M, Kirby M, Anderson SM, Handon R, Kenyon E, Elkahloun A, et al. (2016). The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Science Immunology 1, eaai8593–eaai8593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Cao S, Liu X, Harrington SM, Bindeman WE, Adjei AA, Jang JS, Jen J, Li Y, Chanana P, et al. (2018). CX3CR1 identifies PD-1 therapy–responsive CD8+ T cells that withstand chemotherapy during cancer chemoimmunotherapy. JCI Insight 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJ, Suresh M, Altman JD, and Ahmed R (1998). Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med 188, 2205–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG, et al. (2018). Ensembl 2018. Nucleic Acids Research 46, D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Janssen E, Leung K, and Zhang W (2002). Molecular Cloning of a Novel Gene Encoding a Membrane-associated Adaptor Protein (LAX) in Lymphocyte Signaling. Journal of Biological Chemistry 277, 46151–46158. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Paniccia A, Schulick AC, Chen W, Koenig MR, Byers JT, Yao S, Bevers S, and Edil BH (2016). Identification of CD112R as a novel checkpoint for human T cells. The Journal of Experimental Medicine 213, 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, related to Figure 1: Analysis of CD8+ T cell gene expression during acute and chronic LCMV infection, including properties of encoded proteins.
Table S2, related to Figure 2: Gene expression and differential expression testing results of CD101−Tim3−, CD101−Tim3+, and CD101+Tim3+ PD-1+ CD8+ T cells from chronic infection (day >45).
Table S3, related to Figure 2: Complete gene set enrichment analysis (GSEA) results using RNA-sequencing data of PD-1+ CD8+ T cells after chronic infection (day >45).
Table S4, related to Figure 2: Gene expression and differential expression testing results of CD101−Tim3−, CD101−Tim3+, and CD101+Tim3+ PD-1+ CD8+ T cells from chronic infection (day 15).
Data Availability Statement
RNA-sequencing reads are available from the NCBI Sequence Read Archive under BioProject PRJNA497086. Processed RNA-sequencing data, including normalized gene counts and differential expression analyses, are available in the Supplementary Information (Tables S1–S4).