Abstract
Sesn3 belongs to the three-member sestrin protein family. Sestrins have been implicated in anti-oxidative stress, AMPK and mTOR signal transduction, and metabolic homeostasis. However, the role of Sesn3 in the development of nonalcoholic steatohepatitis (NASH) has not been previously studied. In this work, we generated Sesn3 whole-body knockout and liver-specific transgenic mice to investigate the hepatic function of Sesn3 in diet-induced NASH. With only 4 weeks of dietary treatment, Sesn3 knockout mice developed severe NASH phenotype as characterized by hepatic steatosis, inflammation, and fibrosis. Strikingly, after 8-week feeding with a NASH-inducing diet, Sesn3 transgenic mice were largely protected against NASH development. Transcriptomic analysis revealed that multiple extracellular matrix related processes were upregulated including TGFβ signaling and collagen production. Further biochemical and cell biological analyses have illustrated a critical control of the TGFβ-Smad pathway by Sesn3 at the TGFβ receptor and Smad3 levels. First, Sesn3 inhibits the TGFβ receptor through an interaction with Smad7; second, Sesn3 directly inhibits the Smad3 function through protein-protein interaction and cytosolic retention.
Conclusion:
Sesn3 is a critical regulator of the extracellular matrix and hepatic fibrosis by suppression of the TGFβ-Smad3 signaling.
Keywords: Sestrin, steatosis, inflammation, NASH, hepatic stellate cell
Nonalcoholic steatohepatitis (NASH) is one of the most common chronic liver diseases. The incidence and prevalence of NASH is on the rise in parallel with the increasing obesity and metabolic syndrome (1). NASH is characterized by the presence of hepatic steatosis, inflammation, and evidence of hepatocellular damage. The disease can progress to advanced fibrosis and cirrhosis (2).
Hepatic stellate cells (HSCs) are major contributors to fibrogenesis, a condition with excess accumulation of extracellular matrix in the liver, as a consequence of imbalance between its synthesis and degradation (3). Upon activation, HSCs release several profibrotic cytokines including transforming growth factor beta (TGFβ), one of the most potent inducers of fibrogenesis. Upon activation, a TGFβ dimer binds to a pair of type I and II TGFβ receptor (TGFBR1/2) homodimers. The ligand binding induces formation of a TGFBR1/2 heterotetramer and a productive conformation change that facilitates phosphorylation of TGFBR1 by TGFBR2. Subsequently, the activated TGFBR1 phosphorylates Smad family members 2 and 3 (Smad2/3) (4). The phosphorylated Smad2/3 form heterodimers or heterotrimers with Smad4 and translocate to nucleus for transcriptional activation of fibrogenic genes including collagen type I alpha 1 (COL1A1), collagen type I alpha 1 (COL3A1), smooth muscle alpha 2 actin (ACTA2), TGFB1, and tissue inhibitor of metalloproteinase 1 (TIMP1) (5). Another Smad protein — Smad7 has an inhibitory role in the regulation of the TGFBR1 activity through at least two mechanisms: 1) Smad7 can recruit ubiquitin E3 ligases such as Smurf1/2 (Smad ubiquitination-related factor 1 and 2) to TGFBR1 and that leads to the TGFBR1 degradation; 2) Smad7 can also recruit a protein phosphatase PP1C and that causes TGFBR1 inactivation via dephosphorylation (6).
Sestrin 3 (Sesn3) belongs to a small unique protein family that do not share domain structures with any other eukaryotic proteins (7). Biochemical characterization has revealed that sestrin proteins are multifunctional as they activate AMP-activated protein kinase (AMPK) and mechanistic target of rapamycin kinase complex 2 (mTORC2) but inhibit mTORC1 (7). Sesn1 and Sesn2 have been suggested to be a sensor to leucine for the regulation of mTORC1 (8, 9). Sesn1 and Sesn2 also activate the nuclear factor erythroid 2-related factor 2 (NRF2) transcriptional activity through autophagic degradation of Kelch-like ECH-associated protein 1 (Keap1) in response to oxidative stress (10). Although Sesn1/2/3 share some common functions in the regulation of AMPK and mTOR, there are also some differences. Sesn1 has the highest and Sesn3 has the lowest affinity in leucine binding (8, 9). To date, the role of Sesn3 in NASH has not been investigated. In this work, we developed Sesn3 knockout and transgenic mouse models to determine the role of Sesn3 in the pathogenesis of NASH.
Materials and Methods
Animals and diets
All animal care and experimental procedures performed in this study were approved by the Institutional Animal Care and Use Committee at Indiana University School of Medicine in accordance with National Institutes of Health guidelines for the care and use of laboratory animals. Global knockout (KO) of the Sesn3 gene was generated by crossing Sesn3 floxed mice with a CMV-Cre transgenic mouse (Jackson Laboratory, Bar Harbor, ME, USA) (11, 12). Sesn3 liver-specific knockout and transgenic (Tg) mice were generated by crossing Sesn3 floxed or conditional transgenic mice with an Albumin-Cre transgenic mouse (Jackson Laboratory, Bar Harbor, ME, USA) (13). Animals were backcrossed to C57BL6/J for six generations prior to the experiments. Animals were fed with one of the following diets: (i) regular chow (Teklad Diets 2018SX: 24% calories from protein, 18% calories from fat, and 58% calories from carbohydrate); (ii) moderately high-fat-cholesterol-cholate diet (HFCC, Research Diets D12109C, 20% calories from protein, 40% calories from fat, 40% calories from carbohydrate, and 1.25% cholesterol and 0.5% sodium cholate by weight); (iii) moderately high-fat-cholesterol diet (HFC, Research Diets D12108C, 20% calories from protein, 40% calories from fat, 40% calories from carbohydrate, and 1.25% cholesterol by weight); (iv) moderately high-fat-cholate diet (HFCA, Research Diets D17040301, 20% calories from protein, 40% calories from fat, 40% calories from carbohydrate, and 0.5% sodium cholate by weight); and (v) high-fat diet (HFD, Research Diets D12492C, 20% calories from protein, 60% calories from fat, 20% calories from carbohydrate). In the end, the animals were sacrificed for blood and tissue collection under anesthesia. As male mice had a stronger phenotype than female ones, the data presented in this report were primarily from males.
Human liver specimens
Human liver samples were obtained from control and NASH patients. Unstained liver sections from patients with simple steatosis and those with the diagnosis of NASH with different stages of fibrosis (F0-F4) were obtained under the IRB-approved protocol at the Indiana University (Supplementary Table S1).
Cell culture
Human hepatic stellate cell line LX-2 (Millipore Sigma, Burlington, MA) was cultured in DMEM (Thermo Fisher Scientific, Waltham, MA) supplemented with 2% FBS and penicillin/streptomycin (Thermo Fisher Scientific, Waltham, MA). Prior to TGFβ1 treatment cells were cultured in serum-free medium overnight. TGFβ1 (5 ng/ml) was added to the medium during the last 3 hrs of culture for Smad3 signaling analysis, or the last 24 hrs for transfection experiments. Mouse primary hepatocytes and other nonparenchymal cells such as Kupffer cells and HSCs were isolated from WT, Sesn3 KO, and TgSesn3 (Alb-Cre) mice as previously described (14, 15). Experiments were performed from 3–4 mice in each genotype. The cell line was verified in our laboratory and it did not have mycoplasma contamination.
Plasmid constructs and adenoviral vector preparation
Sesn3, SMAD3, Tgfbr1, Smad7, and GFP coding sequences were cloned into pcDNA3 (Invitrogen) with a FLAG or HA tag. Sesn3 and GFP overexpression adenoviruses were prepared in an AdEasy system (Agilent) following the manufacturer’s manual. The cloning PCR primers for coding sequence were described in Supplementary Table S2. We used a multiplicity of infection (MOI) of 20 for overexpression.
Real-time RT PCR and RNA-seq analysis
Total RNAs were isolated from tissues or cells using TRI reagent (Millipore Sigma, Burlington, MA) following the manufacturer’s instructions and converted into cDNA using a cDNA synthesis kit (Applied Biosystems, Foster City, CA). Real-time PCR analysis was performed using SYBR Green Master Mix (Promega, Madison, WI) in an Eppendorf Realplex PCR system (Hauppauge, NY). mRNA levels were analyzed and calculated with the 2−△△CT method, and all quantifications were normalized to the level of an internal control gene, peptidylprolyl isomerase A (PPIA). Primer sequences of the mouse and human genes used in this work were described in Supplementary Table S2. Liver RNA samples were submitted for RNA-seq analysis at the Center for Medical Genomics at Indiana University School of Medicine. RNA-seq data analysis was performed as previously described (16). The RNA-seq data was deposited at NCBI GEO database with an accession number GSE130642.
Immunoblot and immunoprecipitation analysis
Mouse tissues were homogenized in the lysis buffer containing 50 mM HEPES, pH 7.5, 150 mM NaCl, 10% Glycerol, 1% Triton X-100, 1.5 mM MgCl2, 1 mM EGTA, 10 mM Sodium Pyrophosphate, 100 mM Sodium Fluoride, and freshly added 100 µM Sodium Vanadate, 1 mM PMSF with cOmplete protease inhibitor cocktail tablet (Roche, Indianapolis, IN). Cells were homogenized in the NP-40 lysis buffer containing 1% NP40, 20 mM Tris, pH 7.4, 137 mM NaCl, 2 mM EDTA, 10% Glycerol, 1 mM PMSF with cOmplete protease inhibitor cocktail tablet. Equal amounts of protein lysates were resolved by SDS-PAGE and transferred to nitrocellulose for western blot analysis using specific antibodies. For quantitative analysis, enhanced chemiluminescence (Thermo Fisher Scientific, Waltham, MA) signals on immunoblots were analyzed by Gelpro32 Software (Media Cybernetics, Marlow, UK). For immunoprecipitations (IP), equal amounts of protein lysates were incubated with 1 µg of specific antibodies. After a 16-hr incubation at 4 °C, protein A/G plus Agarose (Santa Cruz Biotechnology, Dallas, TX) was added and incubated at 4 °C for 3 hrs. Normal rabbit IgG or normal mouse IgG were used as negative control. Proteins in the immunoprecipitates were analyzed by Western blot. Antibodies used in these experiments were listed in Supplementary Table S3.
Histology and immunohistochemistry analysis
Liver tissues were fixed in 10% formalin solution and processed for embedding and sectioning at the Histology Core at Indiana University School of Medicine. Liver sections (5 µm thickness) were stained with hematoxylin and eosin (H&E) or Sirius Red stain (Millipore Sigma, Burlington, MA). Immunohistochemistry analysis was performed for Sesn3, myeloperoxidase (MPO), and F4/80. Liver sections were deparaffinized, hydrated, and heated in 1 mM EDTA buffer for antigen retrieval at 100 °C for 5 min, and then treated with normal horse serum for 1 hr. Next, the slides were incubated with antibodies against MPO (1:100, Invitrogen, Carlsbad, CA), or F4/80 (1:100, Invitrogen, Carlsbad, CA) at 4 °C for overnight. After washing with PBS buffer containing 0.05% Tween-20, the tissue sections were incubated with a biotinylated universal pan-specific antibody included in a Vectastain ABC kit from Vector Laboratories (Burlingame, CA) for 2 hrs. The specimens were subsequently exposed to avidin-biotin peroxidase complexes (Vector Laboratories, Burlingame, CA) for 1 hr. Peroxidase activity was visualized using diaminobenzidine solution (Vector Laboratories, Burlingame, CA). Images were captured using a regular microscope (100X, 200X, or 400X total magnification, Leica, Buffalo Grove, IL). The areas of lipid droplets, inflammatory cells, Sirius Red-positive staining, Sesn3, MPO, and F4/80-positive signals in histological and immunohistochemical images were quantified from randomly selected sections at least five fields of each sample using Image J 1.52 software (NIH, Bethesda, MD). Antibodies used in these experiments were listed in Supplementary Table S3.
Blood chemistry and lipid analysis
Lipids were extracted from liver tissues using a chloroform-methanol extraction protocol as previously described (17). Triglyceride (TG) and cholesterol measurements were carried out using Wako L-type TG and Cholesterol E assay kits (FUJIFILM Wako Diagnostics, Richmond, VA), respectively. Serum alanine aminotransferase (ALT) was measured using an assay kit from Thermo Fisher Scientific (Waltham, MA).
Immunocytochemistry
Cells grown on a coverslip were fixed with 4% paraformaldehyde for 15 mins at room temperature, then washed 3 times with PBS and incubated overnight with primary antibodies as described in Supplementary Table S2. After washing, cells were incubated with Alexa Fluor-conjugated secondary antibodies (Invitrogen, 1:250) for 1.5 hrs, counterstained and mounted with Prolong Gold antifade mountant with DAPI (Invitrogen, Carlsbad, CA), and imaged under a fluorescent microscope (Zeiss USA, Thornwood, NY) with a total magnification of 630X.
Statistical analysis
All statistical data are expressed as mean ± SEM. Statistical analysis was performed using GraphPad prism 8 software from GraphPad (La Jolla, CA). Comparisons between two groups were performed using two‐tailed unpaired Student t-test and comparisons for more than two groups were performed using one-way ANOVA followed by Tukey post hoc test.
Results
SESN3 is decreased in human NASH livers
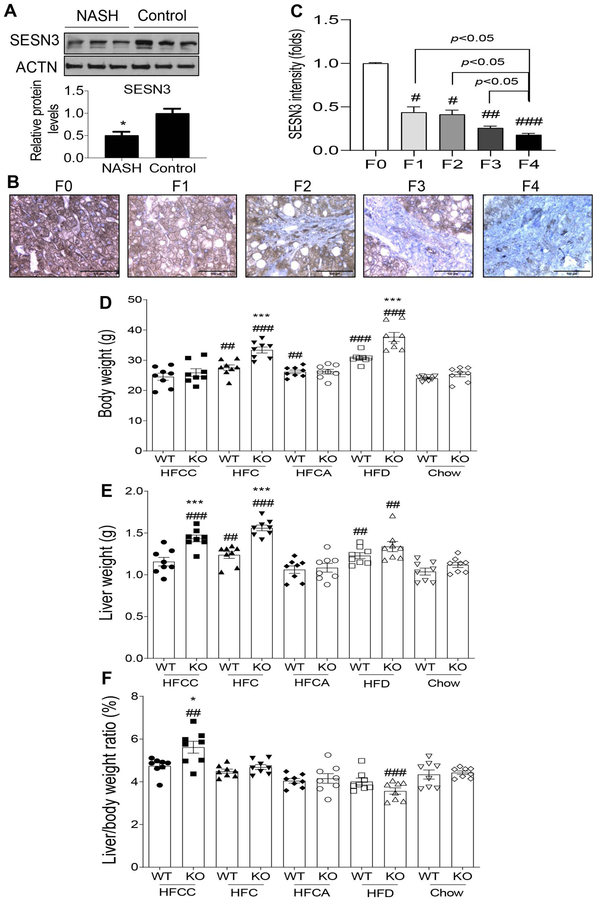
To examine whether SESN3 gene expression is altered in the liver of NASH patients, we performed immunoblot analysis of normal control and NASH liver tissue samples. Our data showed that hepatic SESN3 protein was decreased by 50% in the NASH patients compared to controls (Fig. 1A). In addition, we also performed SESN3 immunohistochemistry analysis of liver sections from NASH patients with varying degrees of hepatic fibrosis ranging from stage F0 to F4. The quantified SESN3 staining intensity per field was significantly decreased from non-fibrotic (F0) to fibrotic livers (F1-F4) (Fig. 1B and 1C), suggesting that hepatic SESN3 might be involved in the NASH pathogenesis and associated with fibrosis.
Figure 1. SESN3 protein expression in human NASH and characterization of Sesn3 knockout mice.
(A) Western blot analysis and quantification of SESN3 protein in the human liver samples from NASH patients and controls. Data are presented as mean ± SEM (n=3). *P < 0.05. (B, C) Immunohistochemistry and quantification of SESN3 expression in liver sections from human NASH patients (F0-F4). Data are presented as mean ± SEM (n=3). #P< 0.05, ##P< 0.01, ###P< 0.001 vs. F0. (D-F) Body weight, liver weight, and liver to body weight ratios in WT and Sesn3 KO male mice treated with different diets (chow groups: 5-6 months of age; special diet groups: 4-5.5 months of age). Data are presented as mean ± SEM (n=8/group). *P < 0.05 and ***P < 0.001 for Sesn3-KO vs. WT for the respective diet; ##P< 0.01 and ###P< 0.001 for the respective genotype treated with a high-fat diet vs. chow.
Mice deficient in Sesn3 are susceptible to diet-induced NASH
To investigate the pathophysiological function of Sesn3, we generated a whole-body Sesn3 knockout mouse model. Sesn3 gene KO was confirmed by immunoblot analysis of the liver and heart tissues (Supplementary Fig. 1A). To model human NASH, we challenged wild type (WT) and KO mice with four different diets in addition to regular chow as a control. Three diets (HFCC, HFC, and HFCA) have same amount of protein (20 kcal%), fat (40 kcal%), and carbohydrates (40 kcal%) but differ in either cholesterol (1.25%) or sodium cholate (0.5%). Another diet (HFD) has much higher fat content (60 kcal%) but no added cholesterol or sodium cholate. Sesn3 KO gained more weight on the HFC and HFD diets than WT controls (Fig. 1D). Liver weights were increased in the KO mice fed with HFCC or HFC and liver to body weight ratios were significantly increased in HFCC-fed WT and KO mice (Fig. 1E and 1F). Especially on the HFCC diet, KO mice had larger liver than WT mice. The Sesn3 KO mice also showed insulin resistance and glucose intolerance on either chow or HFCC diet (Supplementary Fig. 1B and 1C).
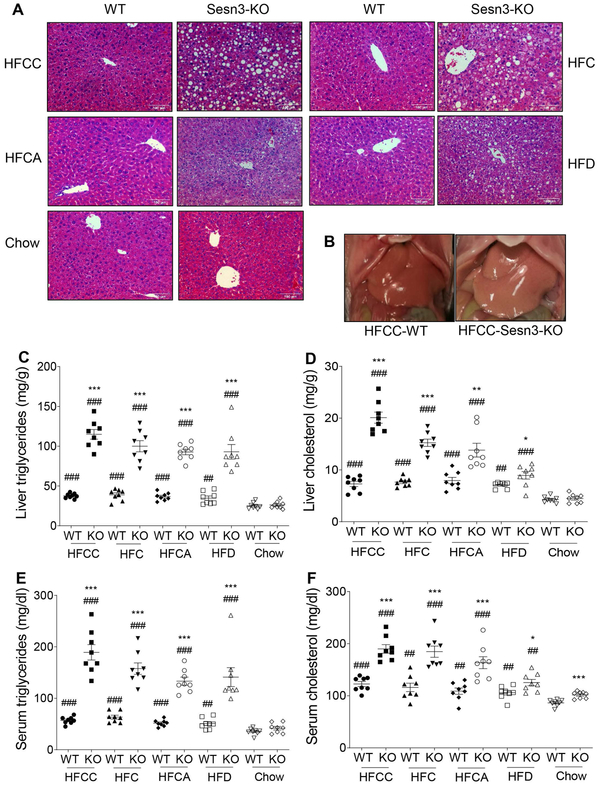
The Sesn3 KO mice developed more severe hepatic steatosis than WT controls when they were fed on four types of high-fat diet with the most severe steatosis phenotype on HFCC (Fig. 2A and 2B and Supplementary Fig. 1D). Hepatic lipid analysis also showed that liver triglyceride and cholesterol levels were significantly elevated in the Sesn3 KO mice compared to WT mice (Fig. 2C and 2D). Serum triglyceride and cholesterol levels were both markedly increased in the Sesn3 KO mice (Fig. 2E and 2F).
Figure 2. Sesn3 deficient mice are more susceptible to diet-induced hepatic steatosis.
(A) H&E staining of liver sections of WT and Sesn3 KO male mice fed with HFCC, HFC, HFCA and HFD diets for 4 weeks. (B) Macroscopic images of WT and Sesn3 KO male mouse livers. (C-F) Liver TG (C), liver cholesterol (D), serum TG (E), and serum cholesterol (F) in WT and Sesn3 KO male mice fed with HFCC, HFC, HFCA, and HFD diets for 4 weeks (n=8/group; chow groups: 5–6 months of age; special diet groups: 4-5.5 months of age). Data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 for Sesn3 KO vs. WT on the respective diet; ##P< 0.01, ###P< 0.001 for the respective genotype fed with a high-fat diet vs. chow.
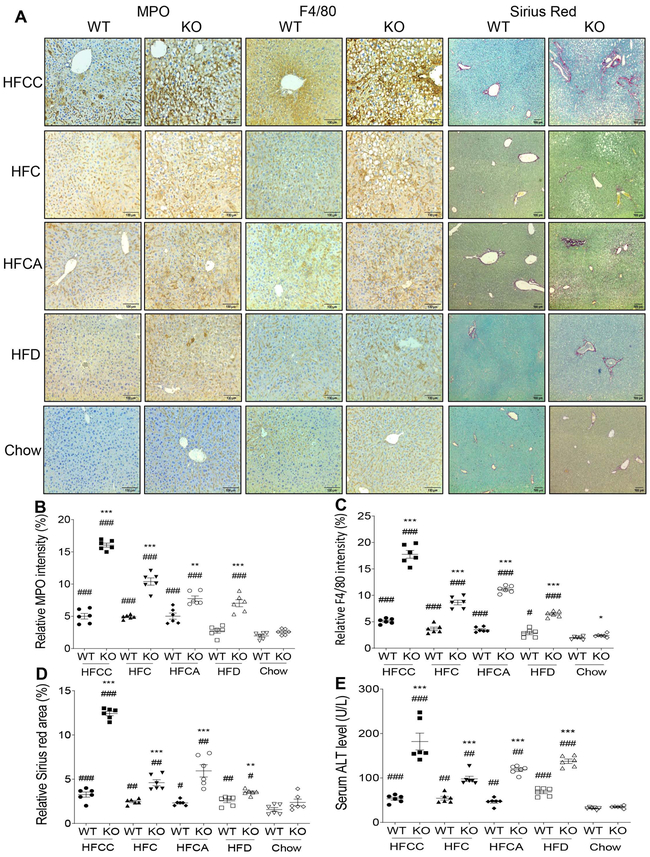
To examine the NAFLD progression, we also analyzed hepatic inflammation and fibrosis using histological staining. Neutrophil infiltration, indicated by myeloperoxidase (MPO), a marker for neutrophil, was 3-fold higher in the HFCC-treated KO mice than that in the WT mice (Fig. 3A and 3B). Similarly, the number of hepatic Kupffer cells and macrophages was also increased by more than 3-fold in the HFCC-treated KO mice than that in the WT mice (Fig. 3A and 3C). Hepatic fibrosis, indicated by Sirius Red staining, was increased 3-fold in the HFCC-treated KO mice compared to WT mice (Fig. 3A and 3D). As expected, liver injury, indicated by serum ALT, was much worse in the Sesn3 KO mice than the WT mice (Fig. 3E). To assess whether there is any gender difference with regard to the NAFLD phenotype, we also performed experiments in female WT and Sesn3 KO mice. Our data showed that when challenged with HFCC diet, the NAFLD phenotype was less severe in the female KO mice than that in the male counterparts (Supplementary Fig. 2A-F).
Figure 3. Sesn3 deficiency exacerbates the diet-induced hepatic inflammation, fibrosis and injury.
(A) Immunohistochemistry analysis of MPO (for neutrophils) and F4/80 (for macrophages) and Sirius Red staining (for fibrosis) of liver sections of WT and Sesn3 KO male mice fed with HFCC, HFC, HFCA, and HFD diets for 4 weeks. (B) Quantitative data for the MPO-positive signals in Panel A. (C) Quantitative data for the F4/80-positive signals in Panel A. (D) Quantitative data of the Sirius Red-positive signals in Panel A. (E) Serum ALT levels in WT and Sesn3 KO male mice fed with HFCC, HFC, HFCA, and HFD diets for 4 weeks (n=6/group; chow group: 5-6 months of age; special diet groups: 4-5.5 months of age). Data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 for Sesn3 KO vs. WT for the respective diet; #P< 0.05, ##P< 0.01, ###P< 0.001 for the respective genotype fed with a high-fat diet vs. chow.
Hepatic Sesn3 overexpression protects mice from diet-induced NASH
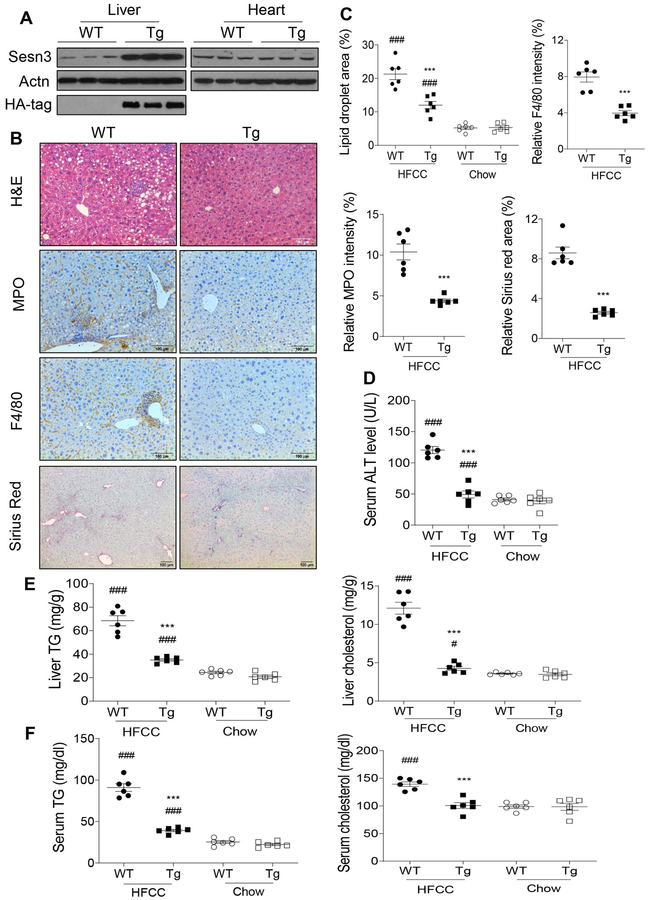
To test whether hepatic Sesn3 plays a significant role in the protection against NASH, we generated liver-specific Sesn3 Tg mice by crossing a Sesn3 conditional transgenic line and an Albumin-Cre transgenic line. Immunoblot analysis confirmed that the overexpression of Sesn3 only in the liver but not other tissues (Fig. 4A). As reported recently, Albumin-Cre also leads to a Cre-mediated recombination in both hepatocytes and HSCs (18). To verify this in our transgenic mice, we isolated primary hepatocytes, HSCs, and Kupffer cells from WT and Tg mice. Indeed, Sesn3 transgene was expressed in both hepatocytes and HSCs but not Kupffer cells after the Albumin-Cre-mediated recombination (Supplementary Fig. 3A-F). To assess the impact of Sesn3 overexpression on NASH, we treated both WT and Tg mice with the HFCC diet for 8 weeks. As expected, WT mice developed NASH on this diet with remarkable hepatic steatosis, inflammation and fibrosis whereas Tg mice were largely resistant to the development of the diet-induced NASH (Fig. 4B and 4C and Supplementary Fig. 4). In addition, the Tg mice were also protected from the HFCC diet-induced liver injury (Fig. 4D). Hepatic triglyceride and cholesterol levels were 2-fold lower in the Tg mice than that in the WT mice on the HFCC diet (Fig. 4E). Serum triglyceride and cholesterol levels were 59% and 28% lower in the Tg mice compared to the WT mice on the HFCC diet (Fig. 4F). To further verify that hepatic Sesn3 plays a major role in the regulation of triglyceride homeostasis in the liver, we also examined hepatic triglycerides in Sesn3 liver-specific knockout (LKO) and transgenic mice. Indeed, lipid droplets were increased in the liver of LKO mice but decreased in the Tg mice (Supplementary Fig. 5A-C). This was also confirmed by biochemical analysis of hepatic triglycerides (Supplementary Fig. 5D).
Figure 4. Liver-specific Sesn3 overexpression protects against diet-induced NASH.
(A) Immunoblot analysis of Sesn3 in the liver and heart tissues of WT and liver-specific Sesn3 transgenic male mice (Tg). (B) H&E staining, immunohistochemistry analysis of MPO and F4/80, and Sirius Red staining of liver sections of WT and Tg male mice fed with the HFCC diet for 8 weeks. (C) Quantitative data for lipid droplet area, MPO positive signals, F4/80 positive signals, and Sirius Red positive signals in Panel B. (D) serum ALT level in WT and Tg male mice fed with the HFCC diet for 8 weeks (n=6/group; 6-9 months of age). (E) Liver TG and cholesterol measurements in WT and Tg male mice fed with the HFCC diet for 8 weeks (n=6/group; 6-9 months of age). (F) Serum TG and cholesterol measurements in WT and Tg male mice fed with the HFCC diet for 8 weeks (n=6/group; 6-9 months of age). Data are presented as mean ± SEM. ***P < 0.001 for Tg vs. WT for the respective diet; #P< 0.05, ###P< 0.001 for the respective genotype on HFCC vs. chow.
Sesn3 deletion leads to upregulation of ECM genes
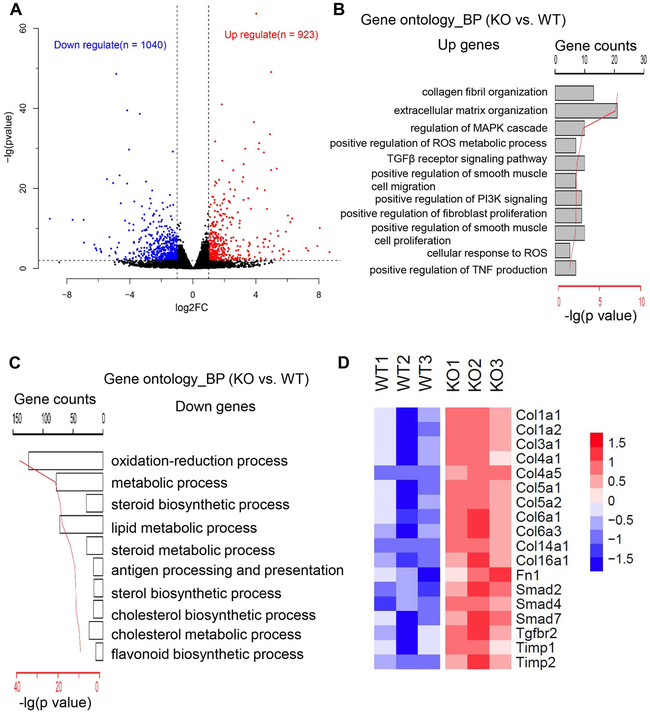
To understand the genome-wide impact of the Sesn3 gene knockout, we performed transcriptomic analysis of WT and KO liver mRNAs from the HFCC-treated mice. By filtering genes with a fold change >=2 and p value < 0.05, we found that 923 genes were significantly upregulated and 1040 genes were significantly downregulated (Fig. 5A and Supplementary Tables S4 and S5). Gene ontology analysis revealed that genes involved in the ECM, fibrosis, and inflammation were among the upregulated gene set (Fig. 5B). The downregulated genes belong to oxidation-reduction, sterol/cholesterol biosynthesis, and lipid metabolic processes (Fig. 5C). For example, a number of fibrosis-related genes including Col1a1, Col1a2, and Col3a1 were highly induced in the KO livers (Fig. 5D). We also performed qPCR to verify some of those differentially expressed genes. Indeed, a few representative genes in hepatic fibrosis and inflammation were increased in the livers of KO mice treated with HFCC but downregulated in the livers of Tg mice treated with HFCC (Fig. 6A and 6B).
Figure 5. Transcriptomic analysis reveals upregulated fibrosis genes in the liver of Sesn3 KO mice.
(A) A volcano plot representation of significantly up- and down-regulated genes in the liver of Sesn3 KO mice compared to WT mice fed with HFCC diets for 4 weeks (n=3 males/group). (B) Gene ontology analysis of significantly upregulated genes in top 11 biological processes. (C) Gene ontology analysis of significantly downregulated genes in top 10 biological processes. (D) Heatmap presentation of significantly up-regulated fibrosis related genes in the liver of Sesn3 KO mice compared to WT mice fed with an HFCC diet for 4 weeks.
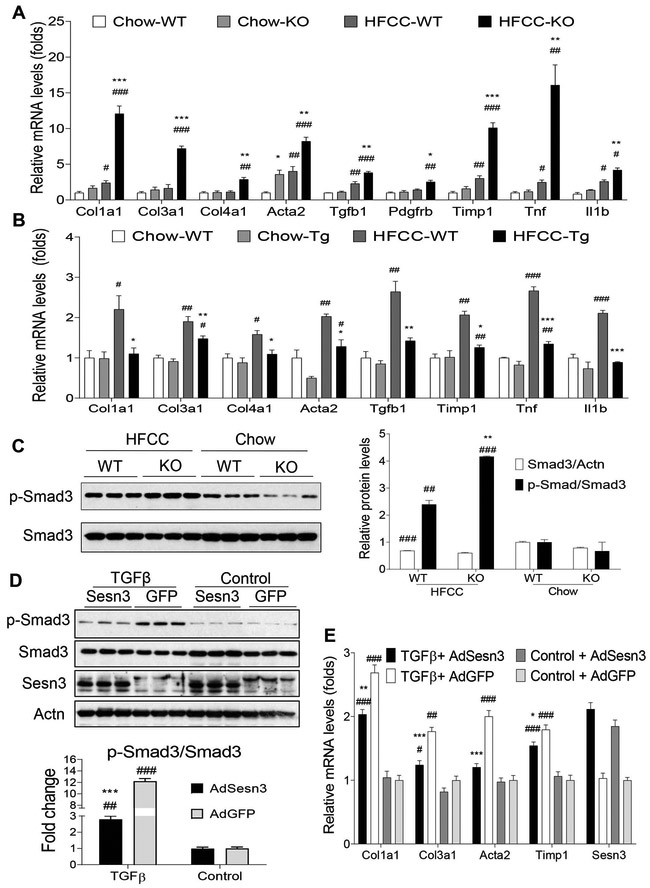
Figure 6. Sesn3 inhibits hepatic inflammation and fibrosis.
(A) Real-time PCR analysis of fibrosis- and inflammation- related genes in the liver of WT and Sesn3 KO male mice (n = 4/group). Data are presented as mean ± SEM. *P < 0.05, **P < 0.01 and ***P < 0.001 for Sesn3 KO vs. WT; #P< 0.05, ##P< 0.01 and ###P< 0.001 for HFCC vs. chow for the same genotype. (B) Real-time PCR analysis of fibrosis- and inflammation- related genes in the livers of WT and Tg mice (n = 4/group). Data are presented as mean ± SEM. *P < 0.05, **P < 0.01 and ***P < 0.001 for Tg vs. WT; #P< 0.05, ##P< 0.01 and ###P< 0.001 for HFCC vs. chow for the same genotype. (C) Western blot analysis and quantification of total Smad3 and p-Smad3 (phosphorylated) in the livers of WT and Sesn3 KO mice. Data are presented as mean ± SEM. **P < 0.01 for Sesn3-KO vs. WT; ##P< 0.01 and ###P< 0.001 for HFCC vs. chow for the same genotype. (D, E) LX-2 cells were transduced with adenoviral AdGFP or AdSesn3 in the absence or presence of TGFβ1 (5 ng/ml) for 3 or 24 hrs. Western blot analysis and quantification (D) were performed for total Smad3 and p-Smad3; real-time PCR analysis (E) was performed for fibrosis-related genes. Data are presented as mean ± SEM. *P < 0.05 and ***P < 0.001 for AdSesn3 vs AdGFP; #P<0.05, ##P< 0.001, ###P< 0.001 for TGFβ1 treatment vs. Control.
To further characterize cell autonomous versus non-autonomous effects of Sesn3 on hepatic fibrosis, we isolated primary HSCs, hepatocytes, and Kupffer cells from WT and Sesn3 KO mice treated with chow or HFCC for 7 days and analyzed expression of a few fibrosis and inflammation related genes. Real-time PCR data showed that expression of Acta2, Col1a1, Col4a1, and Timp1 was increased in the chow KO HSCs and further induced by the HFCC diet where those genes were only modestly induced by the HFCC diet (Supplementary Fig. 6A). Expression of pro-fibrosis and pro-inflammation genes including Tgfb, Tnf, and Ccl2 was elevated in the chow KO hepatocytes and remarkably stimulated by the HFCC diet (Supplementary Fig. 6B). The Tgfb gene was modestly elevated in the chow KO Kupffer cells whereas Tnf, Il1b, Ccl2, and Tgfb were robustly induced by the HFCC diet in both WT and KO Kupffer cells (Supplementary Fig. 6C). To examine the interaction between HSCs and hepatocytes, we performed co-culture experiments by seeding HSCs in a 6-well plate and hepatocytes on an insert membrane. Sesn3-deficient HSCs were poised for transition to fibrosis even on the chow diet as indicated by elevated expression of Col1a1, Col4a1, and Timp1 genes and were further reinforced by the HFCC diet. Co-culture with Sesn3-deficient hepatocytes did not activate WT HSCs but enhanced Sesn3-deficient HSC activation on the HFCC diet (Supplementary Fig. 6D). In contrast, the effect of Sesn3-deficient HSCs on hepatocytes was modest; however, expression of Tgfb, Ccl2, and Ccl5 was highly induced by the HFCC diet in Sesn3 KO hepatocytes (Supplementary Fig. 6E).
We also analyzed multiple signaling pathways by immunoblotting. Our data showed that many of those pathways were upregulated, including NF-κB (IκBα), Jnk, p38 Mapk, Akt, Erk, and Stat3 (Supplementary Fig. 7), suggesting that Sesn3 has a broad impact on inflammation, fibrosis, and growth factor pathways.
Sesn3 inhibits the TGFβ-Smad3 pathway to block fibrogenesis
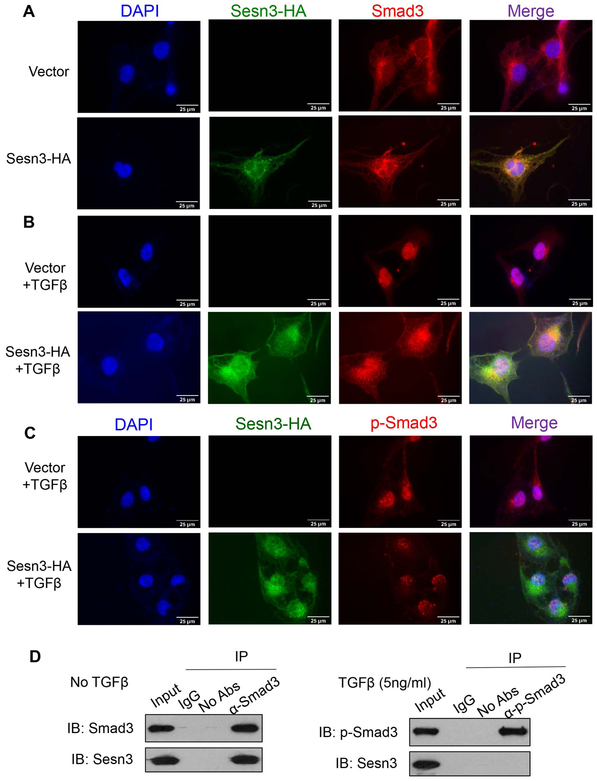
As the TGFβ signaling pathway was significantly upregulated in the HFCC-treated KO liver (Fig. 5B), we analyzed Smad3 phosphorylation at Ser465 and Ser467 residues, known substrates of TGFβ receptor 1. Our data showed that Smad3 phosphorylation was increased 74% in the KO livers compared to WT livers under the HFCC dietary conditions (Fig. 6C). To confirm the Sesn3 effect on Smad3 phosphorylation, we overexpressed either control GFP or Sesn3 in a human HSC cell line LX-2 and stimulated Smad3 phosphorylation with TGFβ. As expected, Sesn3 reduced the TGFβ-stimulated Smad3 phosphorylation by 75% (Fig. 6D). Additionally, Sesn3 also suppressed the expression of the TGFβ-stimulated fibrosis genes (Fig. 6E). To further examine the regulation of Smad3 by Sesn3, we also performed immunofluorescence microscopy. In the absence of TGFβ, there was very little nuclear Smad3 (Fig. 7A). After TGFβ stimulation, Smad3 mostly translocated to nucleus in the transfection with a vector control whereas Smad3 only partially localized to the nucleus in the Sesn3-transfected LX-2 cells and there appeared to be some colocalization of Sesn3 and Smad3 (Fig. 7B). In addition, Sesn3-transfected cells also showed less phosphorylated Smad3 in the nuclei compared to the vector-transfected cells (Fig. 7C). Co-IP analysis further confirmed that Sesn3 interacted with non-phosphorylated but not phosphorylated Smad3 (Fig. 7D). This is not surprising as Sesn3 is mostly cytoplasmic and phosphorylated Smad3 is nuclear localized (12, 19).
Figure 7. Sesn3 interacts with Smad3 and inhibits Smad3 nuclear translocation.
(A-C) LX-2 cells were transfected with Sesn3-HA or vector plasmid for 24 hrs. Immunofluorescence microscopy of Sesn3 and Smad3 in absence of TGFβ1 (A). Immunofluorescence microscopy of Sesn3 and Smad3 after treatment with TGFβ1 (5 ng/ml) for 3 hrs (B). Immunofluorescence microscopy of Sesn3 and p-Smad3 after treatment with TGFβ1 (5 ng/ml) for 3 hrs (C). (D) Co-IP analysis of interactions between Smad3 or p-Smad3 with Sesn3 in LX-2 cells with or without TGFβ1 (5 ng/ml) treatment for 3 hrs.
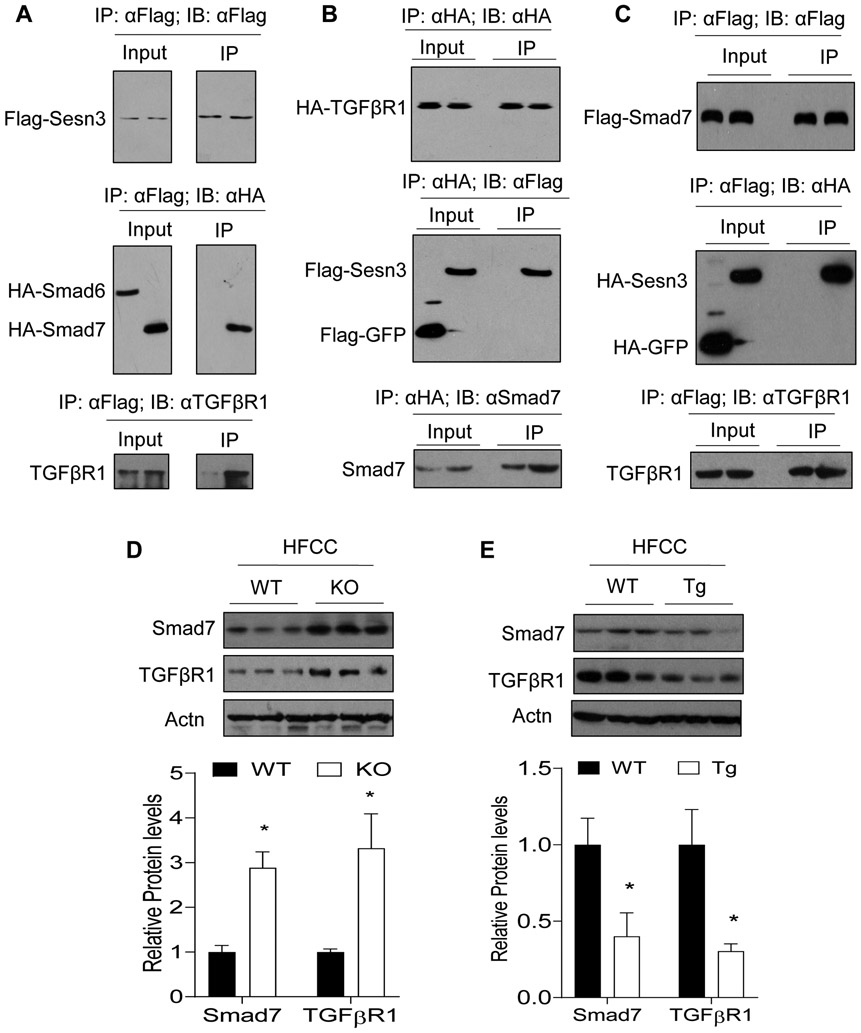
To further investigate whether Sesn3 interacts with any other Smad family members, we also performed Co-IP between Sesn3 and Smad6 or Smad7. Our data showed that Sesn3 interacted with Smad7 but not Smad6 (Fig. 8A). Since Smad7 interacts with Tgfbr1 (20), we also examined whether Sesn3/Smad7/Tgfbr1 form a protein complex. Our protein-protein interaction analysis suggested that this was the case demonstrated by using different baits (Fig. 8A-C). Tgfbr1 pulled down Sesn3 and Smad7 whereas Smad7 pulled down Sesn3 and Tgfbr1 (Fig. 8B and 8C). As Smad7 can be induced by the TGFβ receptor signaling and also feeds back on the TGFβ receptors for ubiquitin-mediated degradation, we also assessed the Sesn3 effect on Tgfbr1 and Smad7 in vivo. We found that both Tgfbr1 and Smad7 were increased in the liver of Sesn3 KO mice but decreased in the liver of Sesn3 Tg mice (Fig. 8D and 8E), suggesting that Sesn3 plays an inhibitory role in the regulation of the TGFβ receptor signaling.
Figure 8. Sesn3 interacts with Smad7 and inhibits TGFβ receptors.
(A-C) Co-IP analysis of interactions between Sesn3 and Smad7 or TGFβR1 in LX-2 cells after treatment with TGFβ1 (5 ng/ml) for 3 hrs. (D, E) Western blot analysis of Smad7 and TGFβR1 in the liver of WT, Sesn3 KO, or Tg mice fed with an HFCC diet. Data are presented as mean ± SEM. *P < 0.05 for Sesn3 KO or Tg vs. WT.
Discussion
In this work, we have reported that Sesn3 plays a critical role in the protection against the diet-induced NASH. It is known that NASH is a chronic progressive liver disease, often beginning from hepatic steatosis and then progressing to hepatic inflammation and fibrosis as an excessive accumulation of triglycerides and cholesterol leads to liver injury and inflammatory response (2). Repeated liver injury and inflammation events lead to ECM remodeling. Excessive ECM production causes fibrosis in the liver (3). Sesn3 KO mice have manifested all the characteristic features of NASH on a high-fat diet, especially the HFCC diet. It suggests that cholesterol and cholate aggravate the NASH development at least in a Sesn3-deficient state. Hepatic steatosis is much worse in the Sesn3 KO mice than WT controls for all high-fat diet treatments with ranking as HFCC > HFC > HFCA = HFD. As a signaling regulator, Sesn3 has been shown to activate AMPK and inhibit mTORC1 (12, 21-25). It is likely that Sesn3 regulates hepatic lipid homeostasis by increasing fatty acid oxidation via activation of AMPK and decreasing lipogenesis through both activation of AMPK and suppression mTORC1 (26, 27). Regarding hepatic inflammation, Sesn3 KO mice fed with the HFCC diet have shown the highest level of neutrophil and macrophage infiltration in the liver, followed by HFC, HFCA, and HFD. As expected, HFCC has also induced the most severe hepatic fibrosis in the Sesn3 KO mice even with only four weeks of the dietary treatment. The Sesn3 gene-specific effect on the NASH pathogenesis has been validated using the liver-specific Sesn3 transgenic mice. This suggests that Sesn3 function in hepatocytes and HSCs is critical for maintaining hepatic homeostasis.
According to our RNA-seq data, Sesn3 is involved in the regulation of multiple biological processes. Under Sesn3 deficient conditions, ECM and TGFβ signaling pathways are among the most significantly enriched pathways in the upregulated gene set. A number of fibrosis and TGFβ signaling genes are highly increased in the Sesn3 KO livers compared to the WT counterparts. Remarkably, these transcriptomic changes align well with the NASH phenotype manifested in the HFCC-treated Sesn3 KO mice. In this work, we have focused on the TGFβ-Smad3 pathway for the mechanistic study. Interestingly, we have identified a novel molecular interaction between Sesn3 and Smad family proteins, among them Smad3 and Smad7 have been biochemically validated (Supplementary Fig. 8). Those molecular interactions are very important for the modulation of the TGFβ-Smad signaling pathway in fibrogenesis. Our data also suggest a dual control mechanism by Sesn3. First, Smad3 nuclear translocation is blocked by Sesn3 via binding and retention of the non-phosphorylated Smad3 in the cytoplasm. Second, Sesn3 forms a tertiary protein complex with Smad7 and Tgfbr1 and by doing so it decreases the Tgfbr1 protein amount and activity through the Smad7-mediated proteasomal degradation and dephosphorylation of Tgfbr1 (Supplementary Fig. 8). Certainly, additional study will be needed to illustrate the detailed mechanism.
In summary, the findings from this work have demonstrated a critical function of Sesn3 in the protection of the liver from developing the diet-induced NASH. The dual inhibition of the TGFβ-Smad3 signaling by Sesn3 provides a novel regulatory mechanism against hepatic fibrosis. As NASH has become an epidemic chronic liver disease (1), identification of novel drug targets will be crucial for the NASH early intervention and treatment. Our data suggest that Sesn3 might be a potential therapeutic target for NASH.
Supplementary Material
Acknowledgments
Financial Support: This study was supported in part by the following funding sources: NIH R21AA024550 (X. Charlie Dong), R01DK107682 (Suthat Liangpunsakul and X. Charlie Dong), R01DK091592 (X. Charlie Dong), R56DK091592 (X. Charlie Dong), Showalter Scholar Award (X. Charlie Dong), Indiana Diabetes Research Center grant NIH P30DK097512, Indiana Clinical and Translational Sciences Institute funded from the NIH NCATS CTSA UL1TR002529.
Abbreviations:
- ACTA2
actin, alpha 2, smooth muscle, aorta
- ACTN
actinin
- AMPK
AMP-activated protein kinase
- CCL2
C-C motif chemokine ligand 2
- COL1A1
collagen type I alpha 1 chain
- ER
endoplasmic reticulum
- HSC
hepatic stellate cell
- MPO
myeloperoxidase
- mTORC
mechanistic target of rapamycin kinase complex
- NASH
nonalcoholic steatohepatitis
- ROS
reactive oxygen species
- Sesn3
Sestrin 3
- SMAD3
SMAD family member 3
- Smurf1/2
Smad ubiquitination-related factor 1 and 2
- TGFβ
transforming growth factor beta
- TGFBR1
TGFβ receptor type 1
Footnotes
Potential conflict of interest: Nothing to report.
References
- 1.Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, George J, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol 2018;15:11–20. [DOI] [PubMed] [Google Scholar]
- 2.Haas JT, Francque S, Staels B. Pathophysiology and Mechanisms of Nonalcoholic Fatty Liver Disease. Annu Rev Physiol 2016;78:181–205. [DOI] [PubMed] [Google Scholar]
- 3.Tsuchida T, Friedman SL. Mechanisms of hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol 2017;14:397–411. [DOI] [PubMed] [Google Scholar]
- 4.Akhurst RJ, Hata A. Targeting the TGFbeta signalling pathway in disease. Nat Rev Drug Discov 2012;11:790–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meng XM, Nikolic-Paterson DJ, Lan HY. TGF-beta: the master regulator of fibrosis. Nat Rev Nephrol 2016;12:325–338. [DOI] [PubMed] [Google Scholar]
- 6.Xu P, Liu J, Derynck R. Post-translational regulation of TGF-beta receptor and Smad signaling. FEBS Lett 2012;586:1871–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho A, Cho CS, Namkoong S, Cho US, Lee JH. Biochemical Basis of Sestrin Physiological Activities. Trends Biochem Sci 2016;41:621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu D, Shimkus KL, Lacko HA, Kutzler L, Jefferson LS, Kimball SR. Evidence for a Role for Sestrin1 in Mediating Leucine-Induced Activation of mTORC1 in Skeletal Muscle. Am J Physiol Endocrinol Metab 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolfson RL, Chantranupong L, Saxton RA, Shen K, Scaria SM, Cantor JR, Sabatini DM. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 2016;351:43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bae SH, Sung SH, Oh SY, Lim JM, Lee SK, Park YN, Lee HE, et al. Sestrins activate Nrf2 by promoting p62-dependent autophagic degradation of Keap1 and prevent oxidative liver damage. Cell Metab 2013;17:73–84. [DOI] [PubMed] [Google Scholar]
- 11.Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res 1995;23:5080–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tao R, Xiong X, Liangpunsakul S, Dong XC. Sestrin 3 Protein Enhances Hepatic Insulin Sensitivity by Direct Activation of the mTORC2-Akt Signaling. Diabetes 2015;64:1211–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Postic C, Shiota M, Niswender KD, Jetton TL, Chen Y, Moates JM, Shelton KD, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem 1999;274:305–315. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Huang X, Werner M, Broering R, Yang D, Lu M. Advanced Method for Isolation of Mouse Hepatocytes, Liver Sinusoidal Endothelial Cells, and Kupffer Cells. Methods Mol Biol 2017;1540:249–258. [DOI] [PubMed] [Google Scholar]
- 15.Mederacke I, Dapito DH, Affo S, Uchinami H, Schwabe RF. High-yield and high-purity isolation of hepatic stellate cells from normal and fibrotic mouse livers. Nat Protoc 2015;10:305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Xu H, Van der Jeught K, Li Y, Liu S, Zhang L, Fang Y, et al. Somatic mutation of the cohesin complex subunit confers therapeutic vulnerabilities in cancer. J Clin Invest 2018;128:2951–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tao R, Wei D, Gao H, Liu Y, DePinho RA, Dong XC. Hepatic FoxOs regulate lipid metabolism via modulation of expression of the nicotinamide phosphoribosyltransferase gene. J Biol Chem 2011;286:14681–14690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newberry EP, Xie Y, Lodeiro C, Solis R, Moritz W, Kennedy S, Barron L, et al. Hepatocyte and stellate cell deletion of liver fatty acid binding protein reveals distinct roles in fibrogenic injury. FASEB J 2018:fj201801976R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hata A, Chen YG. TGF-beta Signaling from Receptors to Smads. Cold Spring Harb Perspect Biol 2016;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyazawa K, Miyazono K. Regulation of TGF-beta Family Signaling by Inhibitory Smads. Cold Spring Harb Perspect Biol 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 2008;134:451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JH, Budanov AV, Talukdar S, Park EJ, Park HL, Park HW, Bandyopadhyay G, et al. Maintenance of metabolic homeostasis by Sestrin2 and Sestrin3. Cell Metab 2012;16:311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chantranupong L, Wolfson RL, Orozco JM, Saxton RA, Scaria SM, Bar-Peled L, Spooner E, et al. The Sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Rep 2014;9:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parmigiani A, Nourbakhsh A, Ding B, Wang W, Kim YC, Akopiants K, Guan KL, et al. Sestrins inhibit mTORC1 kinase activation through the GATOR complex. Cell Rep 2014;9:1281–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng M, Yin N, Li MO. Sestrins Function as Guanine Nucleotide Dissociation Inhibitors for Rag GTPases to Control mTORC1 Signaling. Cell 2014;159:122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol 2012;13:251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ricoult SJ, Manning BD. The multifaceted role of mTORC1 in the control of lipid metabolism. EMBO Rep 2013;14:242–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.