Abstract
Background
Androgen deprivation therapy improves the survival of castration-resistant prostate cancer (CRPC) patients, yet ultimately fails with debilitating side effects. Supraphysiological testosterone (SPT)-based therapy produces clinical responses with improved quality of life in a subset of patients. Currently, no information defines a durable response to SPT.
Objective
To identify key molecular phenotypes underlying SPT response to improve patient selection and guide combination treatment to achieve a durable response.
Design, setting, and participants
A patient-derived xenograft (PDX) preclinical trial was performed with 13 CRPC PDXs to identify molecular features associated with SPT response. Comprehensive intratumoral androgen, tumor growth, and integrated transcriptomic and protein analyses were performed in three PDXs resistant to the newer androgen receptor (AR) pathway inhibitor enzalutamide (ENZ) to define SPT response and resistance.
Intervention
Testosterone cypionate.
Outcome measurements and statistical analysis
SPT efficacy was evaluated by PDX growth, prostate-specific antigen (PSA) change, and survival. Intratumoral androgens were analyzed using mass spectrometry. Global transcriptome analysis was performed using RNA sequencing, and confirmed by quantitative real-time polymerase chain reaction and immunohistochemistry. Log-rank and Mann-Whitney tests were used for survival and molecular analyses, respectively.
Results and limitations
A durable SPT responder was identified, presenting robust repressions of ARv7 and E2F transcriptional outputs and a DNA damage response (DDR) transcriptomic program, which were altogether restored upon SPT resistance in the transient responder. ENZ rechallenge of SPT-relapsed PDXs resulted in PSA decreases but tumor progression.
Conclusions
SPT produces a durable response in AR pathway inhibitor ENZ CRPC associated with sustained suppression of ARv7 and E2F transcriptional outputs, and the DDR transcriptome, highlighting the potential of combination treatments that maintain suppression of these programs to drive a durable response to SPT.
Patient summary
Patients with enzalutamide-resistant prostate cancer have very limited treatment options. Supraphysiological testosterone presents a prominent option for improved quality of life and a potential durable response in patients with sustained suppression on ARv7/E2F transcriptional outputs and DNA repair program.
Keywords: Supraphysiological testosterone, Testosterone, Androgen deprivation therapy, Enzalutamide, Androgen receptor, Castration-resistant prostate cancer, Patient-derived xenografts
Supraphysiological testosterone (SPT) produces clinical and quality-of-life benefits in subsets of castration-resistant prostate cancer patients. Molecular characteristics associated with a response were not identified. This study indicated sustained repressions of ARv7 and E2F transcriptional outputs, and the DNA damage response program as hallmarks of a durable SPT response.
Introduction
Men with castration-resistant prostate cancer (CRPC) who progress on androgen deprivation therapy (ADT) and newer androgen receptor (AR) pathway inhibitors (ARIs) abiraterone acetate (AA) and enzalutamide (ENZ) have poor outcomes and quality of life (QoL), and limited treatment options. As CRPC develops, AR and AR variants (eg, ARv7) are often adaptively upregulated [1,2], and androgen signaling remains active even upon resistance to ARIs [2–4]. While upregulation of AR signaling supports CRPC growth, it paradoxically creates new therapeutic vulnerabilities. Supraphysiological testosterone (SPT) has shown efficacy in prostate cancer cell models [5] and in a subset of patients with improved QoL after ADT [6–8]. However, not all patients respond, and SPT resistance inevitably develops. Currently, there is no information defining a durable response to SPT.
The objectives of this study are to (1) identify molecular features associated with SPT response using a CRPC patient-derived xenograft (PDX) preclinical trial and (2) characterize SPT durable response phenotype using PDX models that exhibit de novo resistance to the newer ARI ENZ (ENZR, representing ~40% of CRPC patients [9,10]). We identified high AR and ARv7 expression, and a positive ARv7 correlation with E2F score–directed SPT response; the SPT durable response was associated with sustained repressions of the ARv7/E2F transcriptional outputs and the DNA damage response (DDR) transcriptome. Together, this work supports SPT therapy in currently incurable ENZR CRPC and highlights the opportunity for SPT-based combination therapies to achieve a durable response.
Patients and methods
2.1. Patient-derived xenografts
Animal procedures were performed in accordance with NIH guidelines and approved by University of Washington Institutional Animal Care and Use Committee. The CRPC PDX preclinical trial, ENZR PDX studies, and the ENZ rechallenge study were performed in castrated male CB17 SCID mice (Supplementary material). Testosterone (T) cypionate (1 mg) or vehicle was administered every 2 wk via intramuscular injection. Animals were sacrificed 5 d after SPT (D5) and at the end of study (EOS) for molecular analyses.
2.2. Steroid measurement
Steroids were measured using liquid chromatography-mass spectrometry as described previously [3,11]. Serum and tumors harvested at D5 and EOS were used.
2.3. RNA sequencing and analyses
RNA was sequenced using Illumina HiSeq 2500 system, and reads were aligned to the human hg38 and the mouse mm10 genome (Supplementary material). Ingenuity pathway analysis was performed on the differentially expressed genes between control and SPT-treated PDXs to identify molecular functions and upstream regulator pathways involved in the SPT response [3,12,13]. Gene set enrichment analysis (GSEA) [14] was conducted to evaluate enrichment of SPT-induced differential expression patterns in canonical signaling pathways (Supplementary material). RNA sequencing (RNA-seq) data are deposited in the GEO (GSE124704).
2.4. Quantitative real-time polymerase chain reaction
Total RNA was reverse transcribed to cDNA using the Advantage RT-for-PCR Kit (Clontech, Mountain View, CA, USA). Quantitative real-time polymerase chain reaction (qPCR) was performed with Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen, Carlsbad, CA, USA) in a 7900HT Fast Real-time PCR System (Applied Biosystems, Foster City, CA, USA) [3]. Species-specific primers are presented in Supplementary Table 1. Expression levels of the genes were normalized to human ACTB.
Flow cytometry
PDXs were dissociated using the Miltenyi human tumor dissociation kit. Cell cycle was analyzed using the BD LSR II Flow Cytometer System (Supplementary material).
2.6. Immunohistochemistry
Formalin-fixed paraffin-embedded PDXs were used for immunohistochemistry analyses (Supplementary material). All evaluations were performed in a blinded fashion using H score or percentage of cells stained.
2.7. Statistical analyses
Growth rates and serum prostate-specific antigen (PSA) levels were estimated by fitting log-linear mixed-effect models to each PDX model as described [3,15]. Kaplan-Meier survival curves were estimated and log-rank tests were used to compare survival between treatment groups [16]. Mann-Whitney tests were performed on androgen measurements and molecular analyses.
Results
3.1. SPT preclinical trial using CRPC PDXs
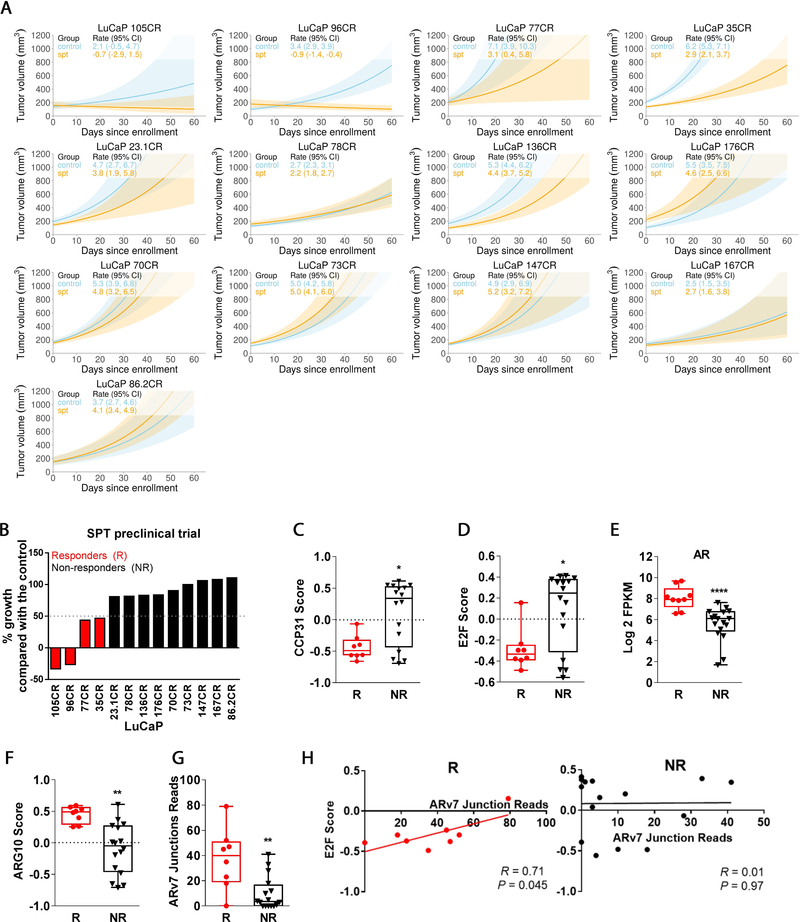
We have developed a large series of CRPC PDXs representing heterogeneity of clinical specimens [3,13,17–19]. Here, we conducted a preclinical trial of SPT using 13 CRPC PDXs (Fig. 1A and Supplementary Table 2, PDX information). Thirty-one percent of PDXs (four of 13) responded to SPT (Fig. 1B). At baseline, SPT responders exhibited lower proliferation and E2F signaling scores (Fig. 1C and 1D), and higher AR expression and AR activity score (Fig. 1E and 1F [20]). Responders also expressed higher levels of ARv7 (Fig. 1G). Notably, this E2F low/ARv7 high phenotype is characterized by a strong correlation between ARv7 and the low E2F score (Fig. 1H), suggesting that ARv7 is involved in the SPT response through cells that are slow proliferating and E2F dependent.
Fig. 1 –
SPT preclinical trial in CRPC PDX models. (A) Linear model analyses of tumor progression of 13 CRPC PDX models in the presence or absence of SPT treatment. (B) Percentage growth inhibition of SPT-treated tumors in comparison with the control tumors, based on linear model analysis of tumor volume as depicted in Figure 1A. Responders were defined as >50% inhibition of tumor growth by SPT. SPT responders demonstrated (C) lower endogenous proliferation based on CCP31 GSVA enrichment score and (D) lower E2F GSVA enrichment score, but (E) higher AR mRNA level, (F) higher AR-regulated gene set GSVA enrichment score (ARG10), and (G) higher ARv7 junction reads when compared with SPT nonresponders. (H) ARv7 expression was positively correlated with E2F GSVA enrichment score in SPT responders but not in nonresponders. N = 2–10/group. AR = androgen receptor; CI = confidence interval; CRPC = castration-resistant prostate cancer; GSVA = gene set variation analysis; PDX = patient-derived xenograft; SPT = supraphysiological testosterone.
3.2. SPT-induced growth inhibition in ENZR CRPC PDXs
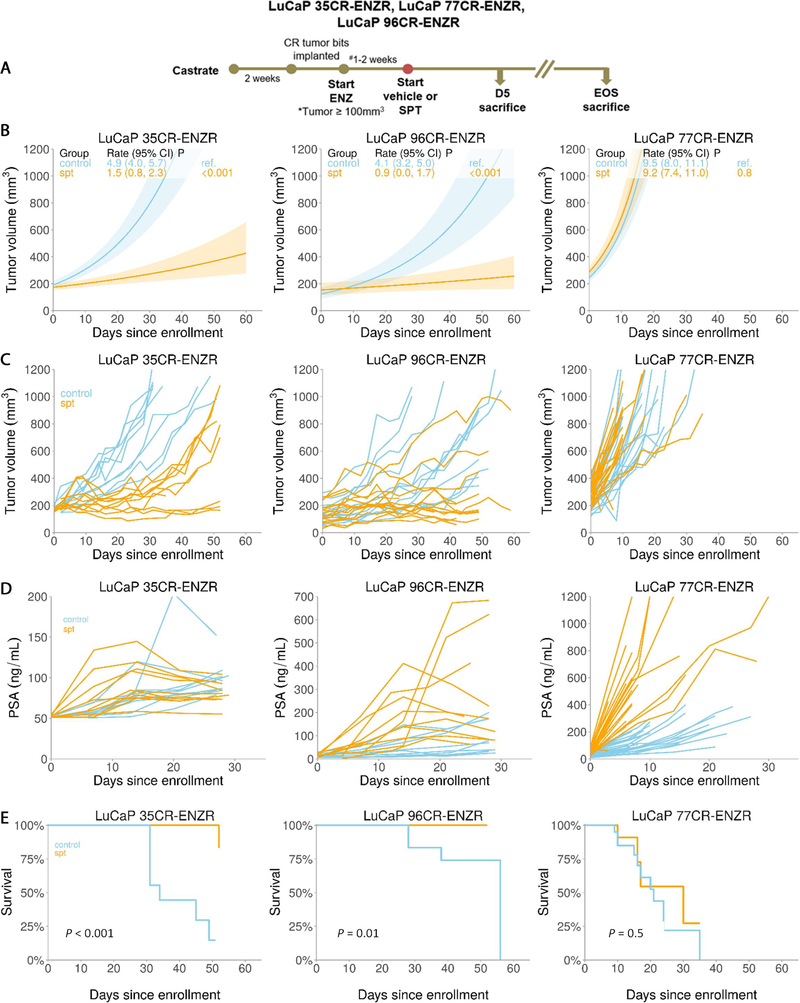
To address whether SPT is effective in CRPC failing the aggressive AR pathway repression with ARIs, we next treated CRPC PDXs with ENZ to resistance and assessed SPT efficacy in ENZR PDXs (Fig. 2A). LuCaP 35CR, LuCaP 96CR, and LuCaP 77CR were minimally responsive to both ARIs ENZ (Supplementary Fig. 1A) and AA [3]. SPT significantly suppressed LuCaP 35CR-ENZR and LuCaP 96CR-ENZR (responders), but not LuCaP 77CR-ENZR tumor growth (nonresponder; Fig. 2B and 2C, and Supplementary Fig. 1B) without apparent toxicity (Supplementary Fig. 1C and 1D). LuCaP 35CR-ENZR responded transiently to SPT, followed by the development of SPT resistance (transient responder; 30–50% increase in mean tumor volume from week 5 onwards), while LuCaP 96CR-ENZR growth suppression was durable (durable responder; Fig. 2B and 2C). SPT produced a spike in the PSA followed by a decline (Fig. 2D) and improved overall survival in responders, while no PSA decline and survival advantage were observed in the nonresponder (Fig. 2D and 2E).
Fig. 2 –
SPT response in ENZR PDX models. (A) A schema showing SPT study design and time points of tumor collection for histological and molecular analyses. (B) Linear model analyses of tumor responses of ENZR CRPC PDX in the presence and absence of SPT. (C) SPT effects on individual tumors in each arm showing heterogeneity of responses. (D) Serum PSA level upon SPT treatment in individual animals. (E) Kaplan-Meier curves showing survival benefits of SPT in the responders. N = 9–21/group. CI = confidence interval; CR = castration resistant; CRPC = castration-resistant prostate cancer; D5 = day 5 after SPT; ENZ = enzalutamide; ENZR = androgen receptor pathway inhibitor enzalutamide; EOS = end of study; PDX = patient-derived xenograft; PSA = prostate-specific antigen; Ref. = reference; SPT = supraphysiological testosterone.
3.3. Effective delivery of T to the tumor
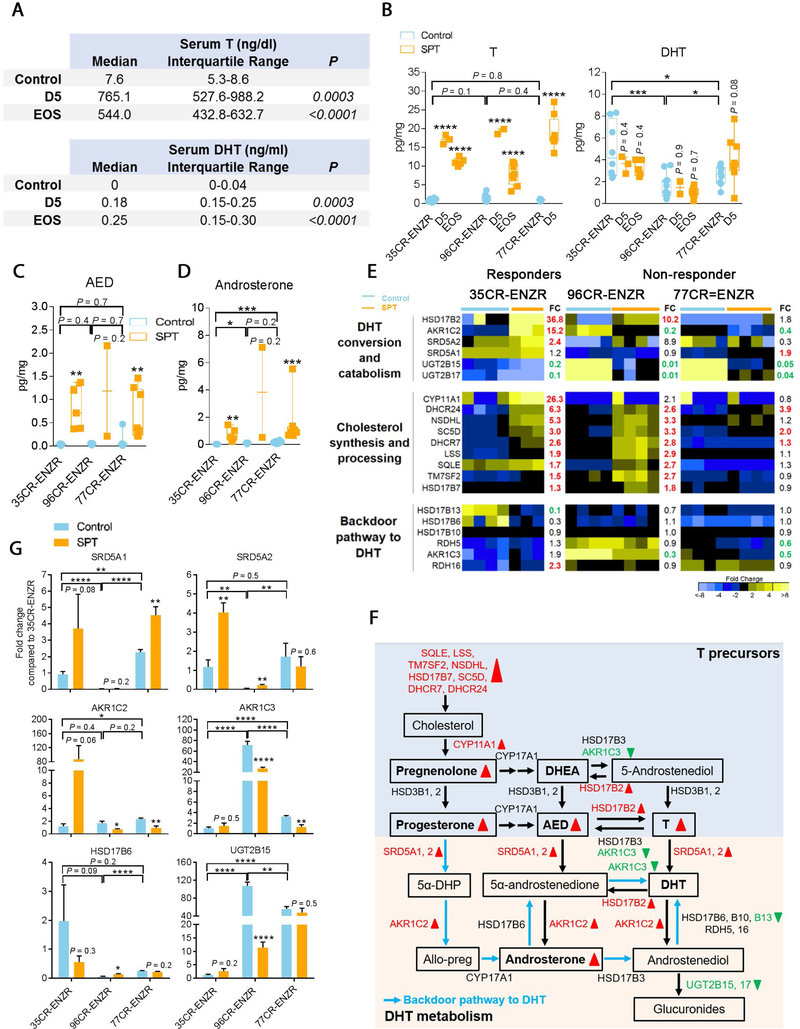
Serum T level has been the mainstay indicator of T delivery in preclinical and clinical studies [6–8,21–23]; however, the bioavailability of T in the tumor has not been examined. We confirmed that serum T and dihydrotestosterone (DHT, a potent metabolite of T) were markedly elevated upon SPT administration (Fig. 3A). Intratumorally, we detected a significant elevation of T, but a constant level of DHT, in all PDXs regardless of SPT responses (Fig. 3B), resembling stable tissue DHT in preclinical models [24] and patients receiving transdermal DHT [25].
Fig. 3 –
Effective delivery of T to the tumor. (A) Serum T and DHT levels were increased at D5 and EOS in animals bearing LuCaP 35CR-ENZR treated with SPT. (B) In all models, intratumoral T increased at D5 and EOS, while intratumoral DHT did not increase in animals treated with SPT. (C) Intratumoral AED and (D) androsterone were increased in all PDXs in comparison with levels in control tumors. (E) RNA-seq analysis of enzymes involved in steroidogenesis showing cholesterol synthesis increased upon SPT administration at D5 in responders LuCaP 35CR-ENZR and LuCaP 96CR-ENZR. Numbers in bold represent statistically significant upregulation (red) and downregulation (green) in fold change (FC) of SPT in comparison with control (p < 0.05; N = 4–7/group). (F) A summary of SPT alterations of androgen synthesis and catabolism pathways in responders LuCaP 35CR-ENZR and/or LuCaP 96CR-ENZR. Box denotes steroids and bold font represents steroids that were measured intratumorally. Steroidogenesis enzymes that showed statistically significant difference upon SPT administration (vs control) in Figure 3E are illustrated in red (upregulation) or green (downregulation). Red upward triangles represent upregulation upon SPT administration (vs control), through either intratumoral androgens or gene expression by RNA-seq analysis; green downward triangles represent downregulation. (G) Real-time PCR analysis confirmed alterations of representative steroidogenesis enzymes by SPT shown in panel F. N = 4–7/group. Mean ± SEM. AED = androstenedione; allo-preg = allopregnanolone; CR = castration resistant; D5 = day 5; DHEA = dehydroepiandrosterone; DHT = dihydrotestosterone; ENZR = androgen receptor pathway inhibitor enzalutamide; EOS = end of study; PCR = polymerase chain reaction; PDX = patient-derived xenograft; SEM = standard error of the mean; SPT = supraphysiological testosterone; T = testosterone; 5α-DHP = 5α-dihydroprogesterone. * p < 0.05 versus control unless otherwise specified. ** p < 0.01 versus control unless otherwise specified. *** p < 0.001 versus control unless otherwise specified. **** p < 0.0001 versus control unless otherwise specified.
To delineate the steroidogenic environment upon SPT therapy, we measured a comprehensive panel of steroid precursors. Intratumoral androstenedione (Fig. 3C), androsterone (Fig. 3D), pregnenolone, progesterone, but not dehydroepiandrosterone, were substantially elevated in all PDXs (Supplementary Fig. 2A), consistent with accumulation of T limiting its conversion from steroid precursors (Fig. 3F). Interestingly, SPT robustly induced androgen-regulated cholesterol processing and synthesis genes [26] exclusively in responders (Fig. 3E and 3F). The profound induction was quickly resolved upon SPT resistance in the transient responder, whereas it was sustained in the durable responder (Fig. 3E and Supplementary Fig. 2B). Despite upregulation of DHT converting enzymes SRD5A1 and SRD5A2 (Fig. 3E), upregulation of DHT catabolizing enzymes HSD17B2 and AKR1C2 (Fig. 3E–G), and downregulation of steroidogenesis enzymes in the backdoor pathway reconstituting DHT (Fig. 3D and 3F) potentially contributed to fast clearance of DHT and accumulation of androsterone in PDXs. Together, SPT administration resulted in marked upregulation of intratumoral T and steroid precursors independent of responses, but robust upregulation of cholesterol synthesis and processing genes exclusively in responders.
3.4. SPT repressed ARv7 transcriptional output in ENZR PDXs
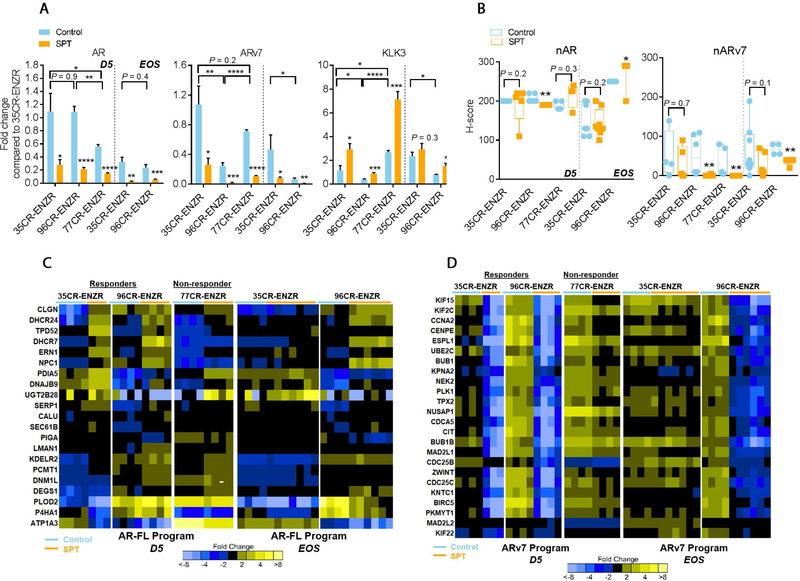
Increased levels of AR and ARv7 are evident in CRPC [27–29], promoting resistance to ENZ [2,28]. We showed that SPT downregulated both AR and ARv7 (Fig. 4A), but upregulated the AR target gene KLK3, independent of SPT responses (Fig. 4A). The elevated KLK3 expression was corroborated by abundant AR protein nuclear localization in all SPT-treated PDXs (Fig. 4B), presumably due to AR protein stabilization by the ligand. In contrast, the mean ARv7 nuclear localization H score was reduced in both responders and the nonresponder by SPT (Fig. 4B). We next examined AR full-length (AR-FL) and ARv7 transcriptional outputs [30]. Consistent with the ligand activation of AR-FL, SPT upregulated the expression of AR-FL gene set (Fig. 4C) and canonical AR gene set (Supplementary Fig. 3A) in all PDXs. In contrast, SPT induced a remarkable repression of the ARv7 program [30] only in the responders LuCaP 35CR-ENZR and LuCaP 96CR-ENZR, with a much weaker alteration in the nonresponder LuCaP 77CR-ENZR (Fig. 4D). This SPT-induced repression of the ARv7 program was mirrored by a decreased cell proliferation program (Supplementary Fig. 3B). Notably, the repressed ARv7 and proliferation programs were completely restored upon SPT resistance in the transient responder LuCaP 35CR-ENZR (Fig. 4D and Supplementary Fig. 3B), highlighting the role of ARv7 signaling underlying the SPT response.
Fig. 4 –
SPT-induced cell growth inhibition is associated with a suppressed ARv7 transcriptional program in ENZR PDXs. (A) qPCR analysis of AR, ARv7, and KLK3 at D5 and EOS in SPT-treated ENZR PDX. (B) IHC analyses of nuclear AR and ARv7 protein in SPT-treated ENZR PDX at D5 and EOS. (C) Upregulation of AR-FL target genes upon SPT treatment was independent of SPT response. (D) Marked SPT-induced downregulation of ARv7 transcriptional program at D5 was observed in responders LuCaP 35CR-ENZR and LuCaP 96CR-ENZR, but a much lesser extent of downregulation was observed in the nonresponder LuCaP 77CR-ENZR. The downregulation of genes was sustained in the durable responder LuCaP 96CR-ENZR until EOS, but was restored in the transient responder LuCaP 35CR-ENZR upon SPT resistance at EOS. N = 4–7/group. Mean ± SEM. AR = androgen receptor; AR-FL = AR full-length; D5 = day 5 after SPT; ENZR = androgen receptor pathway inhibitor enzalutamide; EOS = end of study; IHC = immunohistochemistry; PDX = patient-derived xenograft; qPCR = quantitative real-time polymerase chain reaction; SEM = standard error of the mean; SPT = supraphysiological testosterone. * p < 0.05 versus control unless otherwise specified. ** p < 0.01 versus control unless otherwise specified. *** p < 0.001 versus control unless otherwise specified. **** p < 0.0001 versus control unless otherwise specified.
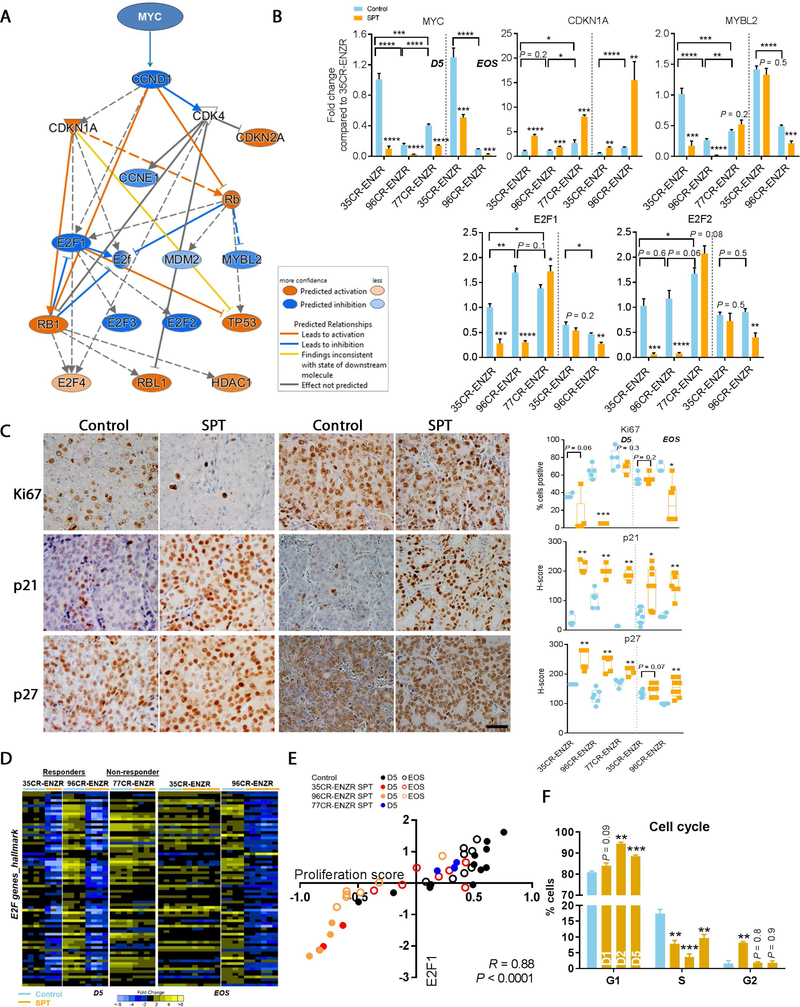
3.5. SPT treatment impaired cell cycle via robust downregulation of Myc-E2F pathway in ENZR PDXs
We next performed an unbiased pathway analysis of the global gene expression alterations at D5 to determine molecular mechanisms associated with the SPT response. SPT induced differential expression of 2998 genes in LuCaP 35CR-ENZR (false discovery rate [FDR] 0.5 and ≥2-fold SPT vs control) and significant downregulation of the Myc-E2F pathway (Fig. 5A). Proto-oncogene Myc promotes, whereas p21 inhibits, cell cycle progression [31]. SPT downregulated MYC universally and upregulated CDKN1A/p21 and p27 regardless of responses (Fig. 5B and 5C). However, downstream cell cycle activators E2F1, E2F2, and MYBL2 [32,33] were downregulated exclusively in responders (Fig. 5B), resulting in a repressed E2F program (Fig. 5D). Importantly, upon SPT resistance, repression of E2F and its transcriptional program were resolved in the transient responder (Fig. 5B and 5D) along with restored tumor proliferation (Fig. 5C). Despite the common association of MYC upregulation and cancer proliferation [34], our studies showed that upon SPT treatment, E2F1 and E2F2 were more strongly correlated with proliferation than MYC (Fig. 5E, and Supplementary Fig. 4A and 4B). Further, SPT induced G1 arrest and necrosis (Fig. 5F and Supplementary Fig. 4C). Collectively, these results provide molecular evidence to support the inhibition of Myc-E2F pathway in SPT-induced growth suppression.
Fig. 5 –
SPT-induced inhibition of proliferation via robust downregulation of MYC/p21/E2F. (A) Ingenuity pathway analysis showed SPT downregulated upstream cell cycle regulators including MYC, driving upregulation of CDKN1A (p21) and downregulation of E2F½ in LuCaP 35CR-ENZR at D5. (B) qPCR confirmed genes altered in responders LuCaP 35CR-ENZR and LuCaP 96CR-ENZR. The deregulation of genes was sustained in the durable responder LuCaP 96CR-ENZR until EOS, but was largely abolished in the transient responder LuCaP 35CR-ENZR at EOS. (C) Representative IHC images (left panel) showing reduced proliferation and increased p21 and p27 protein expression upon SPT in the responder LuCaP 35CR-ENZR, but not in the nonresponder LuCaP 77CR-ENZR at D5. Magnification: 200×, bar = 50 μm. Quantitative analyses of IHC results (right panel) demonstrated that the changes were sustained in the durable responder LuCaP 96CR-ENZR until the EOS, but were abolished in the transient responder LuCaP 35CR-ENZR at EOS. (D) E2F hallmark genes were robustly downregulated by SPT in both responders but to a much lesser extent in the nonresponder LuCaP 77CR-ENZR, and the downregulation was restored upon SPT resistance in the transient responder LuCaP 35CR-ENZ at EOS, as shown by RNA-seq analysis. (E) E2F1 gene expression was strongly correlated with the proliferation score (CCP31) in LuCaP ENZR PDXs. (F) Cell cycle analysis illustrated G1 arrest upon SPT treatment in the responder LuCaP 35CR-ENZR. N = 4–7/group. Mean ± SEM. D5 = day 5 after SPT; ENZR = androgen receptor pathway inhibitor enzalutamide; EOS = end of study; IHC = immunohistochemistry; PDX = patient-derived xenograft; qPCR = quantitative real-time polymerase chain reaction; SEM = standard error of the mean; SPT = supraphysiological testosterone. * p < 0.05 versus control unless otherwise specified. ** p < 0.01 versus control unless otherwise specified. *** p < 0.001 versus control unless otherwise specified. **** p < 0.0001 versus control unless otherwise specified.
3.6. DNA replication and damage response programs were suppressed in ENZR PDXs
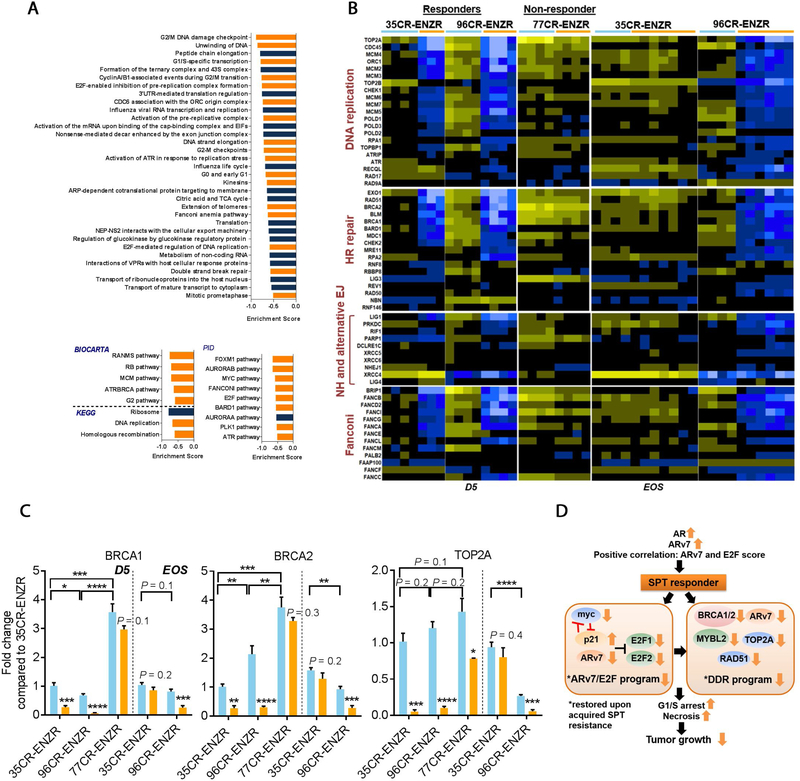
AR activation creates dsDNA breaks [35–37]. GSEA analyses of genes altered by SPT at D5 confirmed that SPT induced negative enrichment of genes involved in DNA repair, DNA replication, and cell cycle progression (FDR <0.0001; Fig. 6A). Exclusively in responders, SPT resulted in robust suppression of DNA replication and DDR programs at D5 (p < 0.05; Fig. 6B), including genes that are directly regulated by AR [38]. This suppression was largely resolved in the transient responder LuCaP 35CR-ENZR at EOS, whereas the suppression was sustained in the durable responder LuCaP 96CR-ENZR (Fig. 6B and 6C). These results indicated that SPT impaired the DNA replication and DDR programs, contributing to SPT-induced cell growth inhibition. Molecular analysis of two additional SPT nonresponders LuCaP 147CR and LuCaP 167CR (Fig. 1A) confirmed the absence of repression of DDR and E2F1 by SPT (Supplementary Fig. 4D). A summary of SPT-induced molecular alterations is illustrated in Figure 6D.
Fig. 6 –
DNA replication and DNA damage response genes were repressed exclusively in SPT responders. (A) GSEA bar plots showing SPT-induced negative enrichment of genes involved in DNA replication and repair, cell cycle, and cell division (orange bars) in LuCaP 35CR-ENZR at D5. FDR < 0.0001. (B) SPT-induced downregulation of DNA replication and DDR associated genes at D5 in responders LuCaP 35CR-ENZR and LuCaP 96CR-ENZR, but not in the nonresponder LuCaP 77CR-ENZR. The downregulation of genes was sustained in the durable responder LuCaP 96CR-ENZR until EOS, but was abolished in the transient responder LuCaP 35CR-ENZR at EOS. (C) Real-time PCR confirmation of DDR genes at D5 and the EOS. N = 4–7/group. Mean ± SEM. (D) A summary of molecular alterations underlying SPT response in ENZR CRPC PDX. AR = androgen receptor; CRPC = castration-resistant prostate cancer; D5 = day 5 after SPT; DDR = DNA damage response; EJ = end joining; ENZR = androgen receptor pathway inhibitor enzalutamide; EOS = end of study; FDR = false discovery rate; GSEA = gene set enrichment analysis; HR = homologous recombination; NH = nonhomologous; PCR = polymerase chain reaction; PDX = patient-derived xenograft; SEM = standard error of the mean; SPT = supraphysiological testosterone. * p < 0.05 versus control unless otherwise specified. ** p < 0.01 versus control unless otherwise specified. *** p < 0.001 versus control unless otherwise specified. **** p < 0.0001 versus control unless otherwise specified.
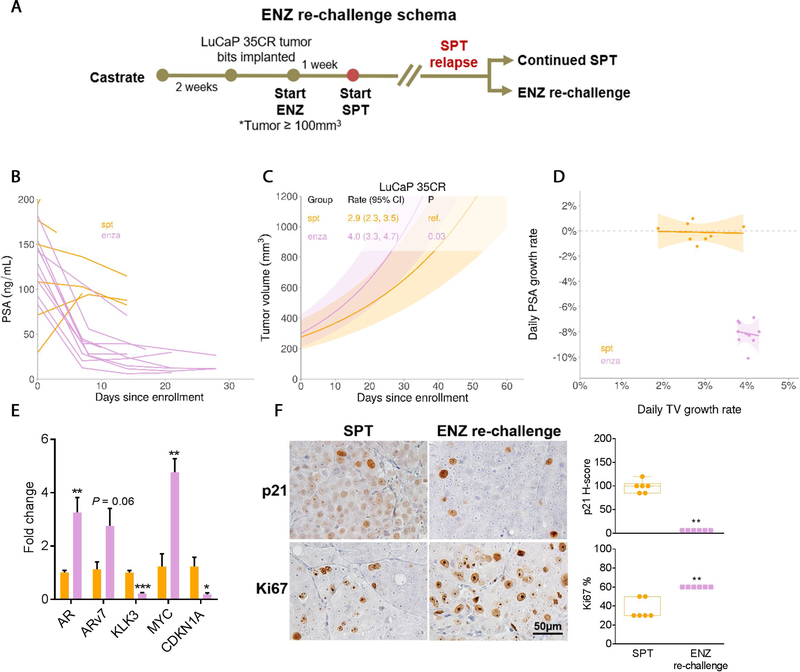
3.7. ENZ rechallenge upon SPT resistance triggered divergent PSA and tumor responses
We showed that SPT upregulated androgen signaling; therefore, we rechallenged SPT-relapsed LuCaP 35CR-ENZR PDXs with ENZ to evaluate whether SPT resensitized tumors to ENZ (Fig. 7A). Notably, serum PSA rapidly declined, but the tumor growth was significantly faster when compared with continued SPT treatment (Fig. 7B–D). Consistent with AR antagonism of ENZ, AR and ARv7 were upregulated, whereas KLK3 was suppressed (Fig. 7E). Moreover, ENZ rechallenge significantly increased MYC and downregulated the cell cycle inhibitor p21, resulting in increased proliferation (Fig. 7E and 7F). Together, our model demonstrated that ENZ rechallenge produced a favorable serum PSA response without evident tumor growth inhibition.
Fig. 7 –
ENZ rechallenge upon SPT resistance triggered divergent PSA and tumor response in LuCaP 35CR-ENZR. (A) Treatment schema of ENZ rechallenge upon SPT resistance. (B) Serum PSA levels were decreased in animals upon ENZ rechallenge. (C) Linear model analyses of tumor responses demonstrated a significant increase in tumor volume upon ENZ rechallenge than on continued SPT treatment. (D) A positive tumor growth was associated with a negative PSA growth upon ENZ rechallenge (purple data points) in LuCaP 35CR-ENZR. (E) qPCR analysis of AR, ARv7, KLK3, and cell cycle–associated MYC and CDKN1A, comparing levels in tumors upon ENZ rechallenge with those on continued SPT. (F) Representative IHC images (left panel) and scores (right panel) of p21 and Ki67 of tumors continued with SPT or upon ENZ rechallenge. Magnification: 200×, bar = 50 μm. N = 4/group. Mean ± SEM. ENZ = enzalutamide; IHC = immunohistochemistry; PSA = prostate-specific antigen; qPCR = quantitative real-time polymerase chain reaction; SEM = standard error of the mean; SPT = supraphysiological testosterone; TV = tumor volume. * p < 0.05 versus continued SPT. ** p < 0.01 versus continued SPT. *** p < 0.001 versus continued SPT.
Discussion
This study provides evidence of the preclinical efficacy of continuous SPT, and demonstrates that high AR and ARv7 signaling, and the positive association of ARv7 with the E2F score underlie SPT responsiveness. For the first time, we identify a durable response phenotype associated with sustained repressions of ARv7 and E2F transcriptional outputs, and the DDR transcriptomic program. The robust repression of ARv7/E2F/DDR transcriptional programs highlights the clinical opportunity of noninvasive monitoring of SPT early relapse and potential combination therapies for a durable SPT response.
Targeting ARv7 provides a new therapeutic angle to suppress tumor growth [39]. Despite our study and a clinical study testing cycling SPT (termed bipolar ADT [BAT]) showing no association between AR/ARv7 transcripts and SPT responses [40], we demonstrated that rather than ARv7 transcripts, repression of ARv7/E2F transcriptional outputs offer a more consistent functional readout to monitor SPT response and early relapse. While an amplified AR may indicate an SPT response [5], we argue that an amplified AR is important but not sufficient to direct an SPT response; SPT-elicited cell growth inhibition relies on the dependency of ARv7 signaling, relevant cell cycle signaling effectors such as E2F½, and the ability of a cell to repair SPT-induced DNA damage.
SPT induces dsDNA breaks [21,35], and we further demonstrate that SPT suppresses a comprehensive DDR transcriptional program involving homologous recombination (HR), nonhomologous end joining (NHEJ), and Fanconi pathways, resulting in cell cycle inhibition. HR deficiencies, such as BRCA2 and CHEK2 mutations, can shift the repair of the dsDNA breaks toward error-prone NHEJ, thereby enhancing genomic instability [41,42]. Recently, an extreme response to BAT was reported in a patient with inactivating ATM and BRCA2 mutations [43]. Similarly, LuCaP 96CR PDXs harbor inactivating BRCA2 and CHEK2 mutations, and exhibit a pronounced and sustained response to SPT. The transient responder LuCaP 35CR-ENZR has an intact DDR program, allowing the suppressed DDR program to resolve quickly. Our results along with the clinical case report provide molecular evidence to nominate patients with DDR deficiency, particularly with HR repair deficiency, to be remarkable and durable SPT responders [43]. Furthermore, our results highlight the potential inhibition of the backup DNA repair pathway, such as NHEJ, using PARP inhibitors as an unprecedented opportunity to augment SPT efficacy.
This study is based on a limited number of PDXs that exhibit de novo resistance to ENZ, and each PDX represents a different phenotype (durable vs transient vs nonresponder). Whether the ARv7, DDR, and cell cycle programs will be downregulated similarly in tumors that showed a durable response to ENZ will require further investigation. Additionally, while the study describes a molecular phenotype associated with the SPT response, corroborating studies on the detection of dsDNA breaks or ARv7/E2F knockdown warrant further investigation in other models due to the technical limitations of PDXs.
Cycling SPT produces clinical responses in a subset of patients with transient improved QoL [21,40]; QoL diminishes over the course of a cycle, presumably due to T levels falling below normal [23,44]. We demonstrated remarkable suppression of tumor growth without apparent toxicity by continuous SPT. So far, clinical trials on continuous SPT have not been conducted; whether such trials will offer superior QoL and treatment efficacy over cycling SPT warrant timely investigation.
Conclusions
We provided preclinical evidence highlighting ENZR CRPC addicted to ARv7- and E2F-regulated growth and/or DDR deficiency as SPT responders. We further provide tissue-based evidence for the bioavailability of intratumoral T, and identified that the SPT durable response was associated with sustained repressions of ARv7 and E2F transcriptional outputs, and an impaired DDR program. This study also provides the first-in-field rationale to support potential combination treatment with ARv7-targeting agents or DDR inhibitors to achieve a durable SPT response.
Supplementary Material
Supplementary Fig. 1. ENZ responses and toxicity assessment of CRPC PDXs. A. Individual tumor volume responses showing the minimal response of LuCaP 35CR, LuCaP 96CR, and LuCaP 77CR to ENZ. B. Estimated percentage tumor growth per day of individual ENZR LuCaP PDX tumors upon SPT treatment. Effects of SPT on C. body weight and D. serum AST level. No increase in serum AST level upon SPT treatment indicating absence of apparent damage to the heart and liver in mice for up to 81 days (LuCaP 35CR-ENZR: 34–53 days, LuCaP 96CR-ENZR: 43–81 days, LuCaP 77CR-ENZR: 16–30 days). Mean ± SEM. N=9–21/group.
Supplementary Fig. 2. Effective delivery of T to the tumor and upregulation of steroid precursors after SPT administration. A. Intra-tumoral progesterone and pregnenolone, but not DHEA, increased in all PDX models upon SPT administration. B. RNA-seq analysis of enzymes involved in DHT conversion and catabolism, and cholesterol synthesis in the transient responder LuCaP 35CR-ENZR and the durable responder LuCaP 96CR-ENZR at EOS. Numbers in bold represented statistically significant upregulation (red) or downregulation (green) in fold change (FC) comparing SPT to the control (P < 0.05). N=4–7/group.
Supplementary Fig. 3. SPT stimulated AR transcriptional program independent of growth response. A. SPT upregulated AR programs comprising of published AR-regulated genes (including genes that are regulated by both AR and AR variants) independent of SPT response. B. SPT markedly downregulated CCP31 proliferation program (GSVA enrichment score) at D5 of responders LuCaP 35CR-ENZR and LuCaP 96CR-ENZR but to a much lesser extent in the non-responder LuCaP 77CR-ENZR. The downregulation of proliferation genes was sustained in the durable responder LuCaP 96CR-ENZR until EOS, but was abolished in the transient responder LuCaP 35CR-ENZR at EOS. N=4–7/group.
Supplementary Fig. 4. Growth characteristics of ENZR PDX models. A. E2F2 and B. MYC gene expression was significantly correlated with the proliferation score in LuCaP ENZR PDX by RNA-seq analysis, with E2F2-proliferation score displaying a stronger correlation than MYCproliferation score. C. Flow analysis showed increased population of necrotic but not apoptotic cell after SPT treatment of the responder LuCaP 35CR-ENZR. N=4–7/group. D. qPCR analysis of AR, ARv7, CHEK1, RAD51, MYBL2, and E2F1 in the non-responders LuCaP 147CR and LuCaP 167CR at D5 after SPT treatment. Statistical analysis was not performed due to the small sample size of the preclinical trials.
Acknowledgments
We would like to thank the patients who generously donated tissue that made this research possible. We also thank Ms. Conner and Dalos for the assistance with the LuCaP PDX work, and Mr. Basom for the assistance in the RNA-seq analysis.
Funding/Support and role of the sponsor: This work was supported by NIH R21CA194798; the PNW Prostate Cancer SPORE P50CA097186, R50 CA221836, P30CA015704, P01CA163227, and R01CA233863; the Prostate Cancer Foundation, and the Richard M LuCas Foundation.
Financial disclosures: Eva Corey certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Chen CD, Welsbie DS, Tran C, et al. Molecular determinants of resistance to antiandrogen therapy. Nat Med 2004;10:33–9. [DOI] [PubMed] [Google Scholar]
- [2].Antonarakis ES, Lu C, Wang H, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 2014;371:1028–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lam HM, McMullin R, Nguyen HM, et al. Characterization of an abiraterone ultraresponsive phenotype in castration-resistant prostate cancer patient-derived xenografts. Clin Cancer Res 2017;23:2301–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lam HM, Corey E. Supraphysiological testosterone therapy as treatment for castration-resistant prostate cancer. Front Oncol 2018;8:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mohammad OS, Nyquist MD, Schweizer MT, et al. Supraphysiologic testosterone therapy in the treatment of prostate cancer: models, mechanisms and questions. Cancers (Basel) 2017;9:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sarosdy MF. Testosterone replacement for hypogonadism after treatment of early prostate cancer with brachytherapy. Cancer 2007;109:536–41. [DOI] [PubMed] [Google Scholar]
- [7].Leibowitz RL, Dorff TB, Tucker S, Symanowski J, Vogelzang NJ. Testosterone replacement in prostate cancer survivors with hypogonadal symptoms. BJU Int 2010;105:1397–401. [DOI] [PubMed] [Google Scholar]
- [8].Mathew P Prolonged control of progressive castration-resistant metastatic prostate cancer with testosterone replacement therapy: the case for a prospective trial. Ann Oncol 2008;19:395–6. [DOI] [PubMed] [Google Scholar]
- [9].Scher HI, Fizazi K, Saad F, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med 2012;367:1187–97. [DOI] [PubMed] [Google Scholar]
- [10].Scher HI, Beer TM, Higano CS, et al. Antitumour activity of MDV3100 in castration-resistant prostate cancer: a phase 1–2 study. Lancet 2010;375:1437–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mostaghel EA, Marck BT, Plymate SR, et al. Resistance to CYP17A1 inhibition with abiraterone in castration-resistant prostate cancer: induction of steroidogenesis and androgen receptor splice variants. Clin Cancer Res 2011;17:5913–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lam HM, Ho SM, Chen J, Medvedovic M, Tam NN. Bisphenol A disrupts HNF4α-regulated gene networks linking to prostate preneoplasia and immune disruption in noble rats. Endocrinology 2016;157:207–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ruppender N, Larson S, Lakely B, et al. Cellular adhesion promotes prostate cancer cells escape from dormancy. PLoS One 2015;10:e0130565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Nguyen HM, Ruppender N, Zhang X, et al. Cabozantinib inhibits growth of androgen-sensitive and castration-resistant prostate cancer and affects bone remodeling. PLoS One 2013;8:e78881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].TM T, PM G. Modeling survival data: extending the Cox model. New York, NY: Springer; 2000. [Google Scholar]
- [17].Nguyen HM, Vessella RL, Morrissey C, et al. LuCaP prostate cancer patient-derived xenografts reflect the molecular heterogeneity of advanced disease and serve as models for evaluating cancer therapeutics. Prostate 2017;77:654–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bluemn EG, Coleman IM, Lucas JM, et al. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell 2017;32:474–89.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Pritchard CC, Morrissey C, Kumar A, et al. Complex MSH2 and MSH6 mutations in hypermutated microsatellite unstable advanced prostate cancer. Nat Commun 2014;5:4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nelson PS, Clegg N, Arnold H, et al. The program of androgen-responsive genes in neoplastic prostate epithelium. Proc Natl Acad Sci U S A 2002;99:11890–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Schweizer MT, Antonarakis ES, Wang H, et al. Effect of bipolar androgen therapy for asymptomatic men with castration-resistant prostate cancer: results from a pilot clinical study. Sci Transl Med 2015;7:269ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Isaacs JT, D’Antonio JM, Chen S, et al. Adaptive auto-regulation of androgen receptor provides a paradigm shifting rationale for bipolar androgen therapy (BAT) for castrate resistant human prostate cancer. Prostate 2012;72:1491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Schweizer MT, Wang H, Luber B, et al. Bipolar androgen therapy for men with androgen ablation naïve prostate cancer: results from the phase II BATMAN study. Prostate 2016;76:1218–26. [DOI] [PubMed] [Google Scholar]
- [24].Zhou Y, Otto-Duessel M, He M, Markel S, Synold T, Jones JO. Low systemic testosterone levels induce androgen maintenance in benign rat prostate tissue. J Mol Endocrinol 2013;51:143–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Page ST, Lin DW, Mostaghel EA, et al. Dihydrotestosterone administration does not increase intraprostatic androgen concentrations or alter prostate androgen action in healthy men: a randomized-controlled trial. J Clin Endocrinol Metab 2011;96:430–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Han W, Gao S, Barrett D, et al. Reactivation of androgen receptor-regulated lipid biosynthesis drives the progression of castration-resistant prostate cancer. Oncogene 2018;37:710–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Watson PA, Chen YF, Balbas MD, et al. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci U S A 2010;107:16759–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Li Y, Chan SC, Brand LJ, Hwang TH, Silverstein KA, Dehm SM. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res 2013;73:483–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hu R, Dunn TA, Wei S, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res 2009;69:16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hu R, Lu C, Mostaghel EA, et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res 2012;72:3457–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Seoane J, Le HV, Massagué J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature 2002;419:729–34. [DOI] [PubMed] [Google Scholar]
- [32].Sharma N, Timmers C, Trikha P, Saavedra HI, Obery A, Leone G. Control of the p53-p21CIP1 axis by E2f1, E2f2, and E2f3 is essential for G1/S progression and cellular transformation. J Biol Chem 2006;281:36124–31. [DOI] [PubMed] [Google Scholar]
- [33].Fischer M, Quaas M, Nickel A, Engeland K. Indirect p53-dependent transcriptional repression of Survivin, CDC25C, and PLK1 genes requires the cyclin-dependent kinase inhibitor p21/CDKN1A and CDE/CHR promoter sites binding the DREAM complex. Oncotarget 2015;6:41402–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Koh CM, Bieberich CJ, Dang CV, Nelson WG, Yegnasubramanian S, De Marzo AM. MYC and prostate cancer. Genes Cancer 2010;1:617–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Haffner MC, Aryee MJ, Toubaji A, et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet 2010;42:668–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ju BG, Lunyak VV, Perissi V, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science 2006;312:1798–802. [DOI] [PubMed] [Google Scholar]
- [37].Cai C, He HH, Chen S, et al. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell 2011;20:457–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Polkinghorn WR, Parker JS, Lee MX, et al. Androgen receptor signaling regulates DNA repair in prostate cancers. Cancer Discov 2013;3:1245–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Cato L, Tribolet-Hardy Jd, Lee I, et al. ARv7 represses tumor suppressor genes in castration-resistant prostate cancer. Cancer Cell 2019;35:401–13.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Teply BA, Wang H, Luber B, et al. Bipolar androgen therapy in men with metastatic castration-resistant prostate cancer after progression on enzalutamide: an open-label, phase 2, multicohort study. Lancet Oncol 2018;19:76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Nickoloff JA, Jones D, Lee SH, Williamson EA, Hromas R. Drugging the cancers addicted to DNA repair. J Natl Cancer Inst 2017;109:djx059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Rodgers K, McVey M. Error-prone repair of DNA double-strand breaks. J Cell Physiol 2016;231:15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Teply BA, Kachhap S, Eisenberger MA, Denmeade SR. Extreme response to high-dose testosterone in BRCA2- and ATM-mutated prostate cancer. Eur Urol 2017;71:499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Teply BA, Wang H, Sullivan R, et al. Phase II study of bipolar androgen therapy (BAT) in men with metastatic castration-resistant prostate cancer (mCRPC) and progression on enzalutamide (enza). ASCO Annual Meeting, Chicago; 2017. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. ENZ responses and toxicity assessment of CRPC PDXs. A. Individual tumor volume responses showing the minimal response of LuCaP 35CR, LuCaP 96CR, and LuCaP 77CR to ENZ. B. Estimated percentage tumor growth per day of individual ENZR LuCaP PDX tumors upon SPT treatment. Effects of SPT on C. body weight and D. serum AST level. No increase in serum AST level upon SPT treatment indicating absence of apparent damage to the heart and liver in mice for up to 81 days (LuCaP 35CR-ENZR: 34–53 days, LuCaP 96CR-ENZR: 43–81 days, LuCaP 77CR-ENZR: 16–30 days). Mean ± SEM. N=9–21/group.
Supplementary Fig. 2. Effective delivery of T to the tumor and upregulation of steroid precursors after SPT administration. A. Intra-tumoral progesterone and pregnenolone, but not DHEA, increased in all PDX models upon SPT administration. B. RNA-seq analysis of enzymes involved in DHT conversion and catabolism, and cholesterol synthesis in the transient responder LuCaP 35CR-ENZR and the durable responder LuCaP 96CR-ENZR at EOS. Numbers in bold represented statistically significant upregulation (red) or downregulation (green) in fold change (FC) comparing SPT to the control (P < 0.05). N=4–7/group.
Supplementary Fig. 3. SPT stimulated AR transcriptional program independent of growth response. A. SPT upregulated AR programs comprising of published AR-regulated genes (including genes that are regulated by both AR and AR variants) independent of SPT response. B. SPT markedly downregulated CCP31 proliferation program (GSVA enrichment score) at D5 of responders LuCaP 35CR-ENZR and LuCaP 96CR-ENZR but to a much lesser extent in the non-responder LuCaP 77CR-ENZR. The downregulation of proliferation genes was sustained in the durable responder LuCaP 96CR-ENZR until EOS, but was abolished in the transient responder LuCaP 35CR-ENZR at EOS. N=4–7/group.
Supplementary Fig. 4. Growth characteristics of ENZR PDX models. A. E2F2 and B. MYC gene expression was significantly correlated with the proliferation score in LuCaP ENZR PDX by RNA-seq analysis, with E2F2-proliferation score displaying a stronger correlation than MYCproliferation score. C. Flow analysis showed increased population of necrotic but not apoptotic cell after SPT treatment of the responder LuCaP 35CR-ENZR. N=4–7/group. D. qPCR analysis of AR, ARv7, CHEK1, RAD51, MYBL2, and E2F1 in the non-responders LuCaP 147CR and LuCaP 167CR at D5 after SPT treatment. Statistical analysis was not performed due to the small sample size of the preclinical trials.