Abstract
BACKGROUND
Aberrant methylation in DNA regulatory regions could downregulate tumor suppressor genes without changing the sequences. However, our knowledge of secreted protein acidic and rich in cysteine (SPARC) and its aberrant methylation in gastric cancer (GC) is still inadequate. In the present research, we performed fundamental research to clarify the precise function of methylation on SPARC and its significance in GC.
AIM
To investigate promoter methylation and the effects of the SPARC gene in GC cells and tissues and to evaluate its clinical significance.
METHODS
Plasmids that overexpressed the SPARC gene were transfected into human GC BGC-823 cells; non-transfected cells were used as a control group (NC group). Quantitative real-time polymerase chain reaction and western blotting (WB) were then used to detect the expression of SPARC. Methylation-specific polymerase chain reaction was executed to analyze the gene promoter methylation status. Cell viability was measured by the cell counting kit-8 assay. The migration and invasion ability of cells were detected by scratch assays and transwell chamber assays, respectively. Cell cycle events and apoptosis were observed with a flow cytometer.
RESULTS
The expression of SPARC mRNA in GC tissues and cells was significantly lower and showed differing degrees of hypermethylation, respectively, than that in normal adjacent tissues and control cells. Treatment with 5-Aza-2’-deoxycytidine (5-Aza-Cdr) was able to restore the expression of SPARC and reverse promoter hypermethylation. Overexpression of the SPARC gene significantly inhibited proliferation, migration, and invasion of GC cells, while also causing cell cycle arrest and apoptosis; the NC group exhibited the opposite effects.
CONCLUSION
This study demonstrated that SPARC could function as a tumor suppressor and might be silenced by promoter hypermethylation. Furthermore, in GC cells, SPARC inhibited migration, invasion, and proliferation, caused cell cycle arrest at the G0/G1 phase, and promoted apoptosis.
Keywords: Gastric cancer, Secreted protein acidic and rich in cysteine, Hypermethylation, Clinicopathological features, Tumor suppressor gene
Core tip: We identified four gastric cancer (GC) cell lines and 66 paired tissues using quantitative real-time polymerase chain reaction, western blotting, and methylation-specific polymerase chain reaction. Correlation analysis between expression and clinicopathological features revealed that low expression levels of secreted protein acidic and rich in cysteine (SPARC) and high levels of methylation in GC tissues were associated with poor clinical features and a poor prognosis (high TNM stage and poor differentiation grade). The restoration of SPARC suppressed GC cell proliferation, migration, and invasion, arrested the cell cycle, and increased apoptosis. Our study found that SPARC represents a potential target for treating GC individuals.
INTRODUCTION
Worldwide, gastric cancer (GC) is one of the most common gastrointestinal cancers in humans. According to statistics, GC has the second highest global mortality rate[1] and has become a serious threat to human health. Due to the general lack of obvious symptoms and signs, patients are often identified only during progressive or late stages, with local infiltration and lymph node metastasis. Despite the use of digestive tract angiography, computed tomography, magnetic resonance imaging, endoscopy, and endoscopic ultrasound, it is still very difficult to detect and diagnose accurately GC in the early stages. The metastasis of GC is the most common phenomenon in many patients; this renders such patients non-resectable at the time of surgery, thus resulting in a low survival rate and an overall poor prognosis[2]. Consequently, there is an urgent need to conduct in-depth research on the mechanisms underlying the occurrence and development of GC and to identify an effective and minimally-invasive method for prevention and treatment.
With the development of molecular biology techniques over the last few years, many research studies have shown that the occurrence and development of GC is the result of a combination of multiple gene abnormalities and multi-step changes, including the activation of oncogenes, the inactivation of tumor suppressor genes, the loss of function of repair-related genes, and disorders in key regulatory mechanisms, including metastasis, infiltration, and signal transduction. In particular, the inactivation of tumor suppressor genes is known to play an important role. Many factors can cause the inactivation of genes, including loss of heterozygosity, haploid deficiency, polymorphism, DNA methylation, and genomic imprinting[3]. Furthermore, abnormal promoter methylation may result in the downregulation and silencing of genes related to carcinogenesis[4].
DNA methylation refers to the addition of a methyl group (CH3) to cytosines preceding guanosines (CpGs) and finally the conversion to 5-methylcytosine[5]. In the process of detecting GC cells and tissues, many cases show that reverse transcription is blocked due to abnormal promoter methylation of certain tumor suppressor genes, thereby downregulating or silencing certain genes[4]. This phenomenon may represent a new direction for studying the pathogenesis of GC.
Secreted protein acidic and rich in cysteine (SPARC), also known as BM-40, is located on human chromosome 5q31.3-q32 and has 10 exons. This protein was originally described and purified by Termine et al[6] in 1981. The gene encoding this protein is 25.9 kb in length and encodes 298-304 amino acids. SPARC belongs to a large family of proteins that also includes FSTL1, SMOC1, SMOC2, SPARCL1, SPOCK1, SPOCK2, and SPOCK2; these are located on human chromosomes 5q33.1, 3q13.33, 6q27, 4q22.1, 5q31.2, 10q22.1, and 4q32.3, respectively. Studies have shown that these proteins have very different functions in many tumors; this is due to tissue and cell heterogeneity and the fact that their relative function may be affected by their surrounding environment.
We previously published a review of how SPARC might play a suppressive role in a variety of tumors, such as ovarian cancer[7], pancreatic cancer[8], colon cancer[9], and T-cell lymphoma[10]. SPARC is also known to play a role in anti-angiogenesis, the anti-cell adhesion inhibition of cell proliferation and differentiation, regulation of the extracellular matrix, and cell cycle arrest[11]. Other researchers have shown that SPARC may promote melanoma development, invasion, metastasis, and apoptosis.
However, there is significant controversy surrounding the precise role of SPARC in GC. As previously suggested, SPARC family genes may be influenced by their surrounding microenvironment and exhibit notable tissue and cell heterogeneity. Therefore, we investigated the function of SPARC and its significance in gastric carcinomas.
MATERIALS AND METHODS
Cell lines and tumor tissue samples
A normal gastric epithelial cell line (GES-1), and four GC cell lines (AGS, MKN-45, MGC-803, and BGC-823) were purchased from Shanghai Institute of Life Sciences, Chinese Academy of Sciences. These cell lines were grown in RPMI-1640 medium (HyClone Inc., Logan, UT, United States) supplemented with 10% fetal bovine serum, and incubated in 5% CO2 at 37 °C. Sixty-six matched GC samples (50 males and 16 females) were collected from patients undergoing radical GC surgery from September 2014 to June 2016 at the Fourth Affiliated Hospital and Cancer Institute of China Medical University. All patients signed an informed consent form. The diagnosis of each patient was verified by pathological examination, and none of the patients had received any special treatment prior to surgery. All GC specimens were immediately stored at -80 °C to await further analysis. Tumor, Node, Metastasis (TNM) classification was staged according to the American Joint Committee on Cancer/Union for International Cancer Control, Eighth Edition. Histological typing was conducted according to the criteria provided by the World Health Organization.
mRNA expression analysis by quantitative real-time PCR
Total RNA was extracted from cells and tissues using a Universal RNA Extraction Kit (Takara Bio Inc., Tokyo, Japan). RNA (500 ng) was reverse transcribed to cDNA using the PrimeScript RT reagent Kit. The primer sequences designed for quantitative real-time polymerase chain reaction (qRT-PCR) were as follows: Forward primer, 5-CGAGCTGGATGAGAACAAC-3, reverse primer, 5-AAGAAGTGGCAG GAAGAGTC-3. The housekeeping gene GAPDH was used as an internal control to confirm the success of RT reaction. The primer sequences for GAPDH were as follows: Forward primer, 5-CACAAGAAGGTGGTGAAGCAG-3, reverse primer, 5’-AAAGGTGGAGGAGTGGGTCT-3. PCR amplification was carried out with an initial denaturation at 95 °C for 5 s, followed by 40 cycles of 95 °C for 4 s, 60 °C for 34 s, and a final extension step of 95 °C for 15 s, 60 °C for 1 min, and 95 °C for 15 s. The expression level of SPARC in four GC cell lines was analyzed using GES-1 cells as the relative standard. The results of qRT-PCR were subsequently analyzed by the 2-△△Ct method, and statistical tests were performed.
Protein expression analysis by western blotting
Protein lysates from cells and samples were extracted in radioimmunoprecipitation assay buffer containing phenylmethanesulfonyl fluoride. The concentrations of protein samples were then determined using a bicinchoninic acid protein assay kit (Beyotime Bio Inc., Shanghai, China). Then, a protein standard curve was created, and sample quantities were calculated. Lysates were mixed with 6 × loading buffer, boiled for 6 min with a sealing membrane, and analyzed using 10% sodium-dodecyl sulfate polyacrylamide gel electrophoresis at 90 V for 90 min. The protein samples were then transferred to a polyvinylidene difluoride membrane (Millipore, Burlington, MA United States) at 120 mA constant current, and subsequently blocked with 5% bicinchoninic acid in phosphate-buffered saline (PBS). Membranes were incubated overnight at 4 °C with an anti-SPARC monoclonal antibody (1:1000) and an anti-GAPDH monoclonal antibody (1:10000). The next morning, the polyvinylidene difluoride membranes were washed three times in Tween tris-buffered saline prior to the application of an anti-rabbit secondary antibody for 2 h. Finally, positive protein bands were visualized using enhanced chemiluminescence developer.
DNA extraction and sodium bisulfite conversion
DNA was extracted from cells, tumors, and normal gastric mucosa samples. An EZ DNA Methylation-Gold Kit (Zymo Research, Orange, CA, United States) was used to treat extracted DNA with sodium bisulfite. The bisulfite-converted DNA was subsequently stored in 1.5 mL microcentrifuge tubes and stored at -80 °C.
Methylation-specific PCR
Methylation-specific PCR (MSP) was used to investigate SPARC gene promoter methylation. The primer sequences for methylated reactions were as follows: Forward primer, 5-GAGAGCGCGTTTTGTTTGTC-3, reverse primer, 5-AACGAC GTAAACGAAAATATCG-3’. The primer sequences designed for unmethylated reactions were as follows: Forward primer, 5-TTTTTTAGATTGTTTGGAGAGTG-3, reverse primer, 5-AACTAACAACATAAACAAAAATATC-3. The whole reaction was carried out with an initial denaturation at 94 °C for 5 min and 30 s, 58 °C for 30 s, followed by 40 cycles of 72 °C for 30 s, and a final extension step of 72 °C for 10 min. PCR products (5 μL) were loaded onto a 2% agarose gel and visualized by ethidium bromide staining.
5-Aza-2'-deoxycytidine treatment
Gastric tumor BGC-823 cells exhibiting promoter hypermethylation were incubated in culture medium with 0 μmol/L, 5 μmol/L, and 10 μmol/L of 5-Aza-2'-deoxycytidine (5-Aza-CdR), and 1 μmol/L of TSA for 72 h; the culture medium was changed every 24 h. Another group of cells was incubated in medium containing 5 μmol/L of 5-Aza-Cdr for 48 h, and 1 μmol/L of TSA for 24 h. Following treatment, we used the previously described techniques to extract RNA, DNA, and protein.
Plasmid transfection
Gastric tumor BGC-823 cells were inoculated onto 6-well plates. We also diluted Lipofectamine 3000 and prepared a master mix of DNA in Opti-MEM medium. The diluted DNA was then added to each tube of diluted Lipofectamine 3000 reagent (1:1 ratio) and incubated for 10-15 min at room temperature. The DNA-lipid complex was then added to cells and incubated for 2-4 d at 37 °C. Finally, the transfected cells were analyzed.
Cell proliferation, invasion, and migration assays
Proliferation assays were performed in a 96-well plate. Transfected BGC-823 cells were incubated in culture medium with 10 μL of cell counting kit-8. A wavelength of 490 nm was selected to measure the light absorption values of each sample every 24 h. Cell growth curves were drawn over time as the horizontal coordinate, with the light absorption value as the vertical coordinate. Invasion assays were performed in a 6-well plate containing an 8 mm pore size polycarbonate membrane that had been precoated with 50 mg/L Matrigel (BD Biosciences, Bedford, MA, United States). Transfected cells were then suspended in a serum-free medium at a concentration of 5 × 104 cells/mL. This was then seeded in the upper compartment of the chamber and incubated in the presence of medium containing 10% fetal calf serum. After re-culture with 5% CO2 at 37 °C for 24-48 h, the Transwell chambers were inverted and stained with crystal violet. The migration assay was performed in a similar manner but without coated chambers on a 6-well plate. Ten-microliter pipet tips were used to carry out a parallel scratch on the transfected cells, and the scratch observation point was recorded on the back of the 6-well plate. Then, the cells were washed off with PBS buffer; this was repeated 3-4 times. The images were then observed and photographed under an inverted imaging microscope at 0 h, 24 h, and 48 h. Proliferation, invasion, and migration assays were performed in triplicate.
Cell apoptosis and cycle assay
First, cells were digested with trypsin without ethylenediamine tetraacetic acid. Buffer solution was then added to the transfected BGC-823 cells to adjust their concentration to 1-5 × 106/mL. Five-microliters of Annexin V was then added to 100 μL of cell suspension and incubated at room temperature in the dark for 5 min. Then, we added 10 μL of propidium iodide in 400 μL of PBS solution. Cell cycle assays were performed in a similar manner, but with 400 μL of sodium citrate solution (38 mmol/L) instead of Annexin V.
Statistical analysis
The student’s T-test was used to compare the mean levels of SPARC expression. The relationship between gene expression, promoter methylation status, and clinicopathological features was analyzed by the Pearson’s χ2-test. A P value < 0.05 was regarded as being statistically significant. Measurement data were expressed as a mean ± standard deviation, and statistical analyses were carried out with SPSS 24.0 software (SPSS Inc., Armonk, NY, United States).
RESULTS
SPARC was expressed at low levels in gastric cell lines and tissues
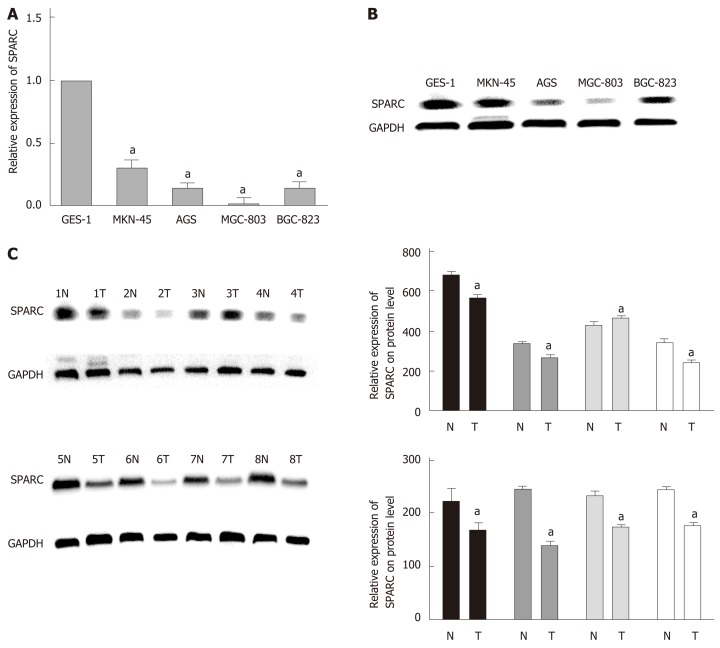
Analysis showed a significant reduction in SPARC expression in the four GC cell lines (AGS, MKN-45, MGC-803, and BGC-823) compared with the normal GES-1 cell line: AGS (0.15 ± 0.02-fold), MKN-45 (0.31 ± 0.07-fold), MGC-803 (0.03 ± 0.01-fold), and BGC-823 (0.15 ± 0.07-fold) (P < 0.05). Similar results were obtained in GC tissues when compared with paired adjacent non-tumor tissues. The representative data are separately shown in Figure 1A, B, and C. A summary of the relationships between SPARC mRNA expression and clinicopathological characteristics is given in Table 1. SPARC mRNA expression was significantly correlated with Borrmann type (P = 0.015), TNM stage (P = 0.001), and lymph node metastasis (P = 0.001) in GC patients.
Figure 1.
Expression of secreted protein acidic and rich in cysteine in gastric cancer cell lines and eight pairs of samples. A: Specific reverse transcriptase quantitative real-time PCR analyzed for secreted protein acidic and rich in cysteine (SPARC). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as loading control. Relative SPARC mRNA expression levels were analyzed by densitometry followed by quantitation relative to GAPDH; B: Western blot analysis used a rabbit monoclonal SPARC antibody (43 kDa), GAPDH was used as control (34 kDa); C: Western blot analyzed SPARC gene expression in eight cases of gastric cancer samples, and GAPDH was used as internal reference. Expression level data are presented as mean ± SE. aP < 0.05 was considered statistically significant. SPARC: Secreted protein acidic and rich in cysteine; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; N: Non-tumor tissue; T: Tumor tissue.
Table 1.
Relationship between expression levels of SPARC mRNA and clinicopathological characteristics, n (%)
| Parameters | Total, n = 66 |
Expression level of SPARC gene |
P value | |
| Low, n = 42 | High, n = 24 | |||
| Age in yr | ||||
| < 60 | 21 | 16 (76.2) | 5 (23.8) | 0.186 |
| ≥ 60 | 45 | 26 (57.8) | 19 (42.2) | |
| Gender | ||||
| Male | 51 | 32 (62.7) | 19 (37.3) | 0.348 |
| Female | 15 | 10 (66.7) | 5 (33.3) | |
| Size in cm | ||||
| < 5 | 27 | 18 (66.7) | 9 (33.3) | 0.372 |
| ≥ 5 | 39 | 24 (61.5) | 15 (38.4) | |
| Differentiation | ||||
| Good/moderate | 16 | 8 (50.0) | 8 (50.0) | 0.154 |
| Poor | 50 | 34 (68.0) | 16 (32.0) | |
| Location | ||||
| Upper | 8 | 3 (37.5) | 5 (62.5) | 0.759 |
| Middle | 18 | 8 (44.4) | 10 (56.6) | |
| Lower | 40 | 31 (77.5) | 9 (22.5) | |
| Macroscopic type | ||||
| Borrmann I + II | 19 | 6 (31.6) | 13 (68.4) | 0.015a |
| Borrmann III + IV | 47 | 36 (76.6) | 11 (23.4) | |
| TNM stage | ||||
| I + II | 27 | 7 (25.9) | 20 (74.1) | 0.001a |
| III + IV | 39 | 35 (89.8) | 4 (10.2) | |
| Lymph node metastasis | ||||
| - | 18 | 6 (33.3) | 12 (66.7) | 0.001a |
| + | 48 | 36 (75.0) | 12 (25.0) | |
P < 0.05 was considered statistically significant. SPARC: Secreted protein acidic and rich in cysteine.
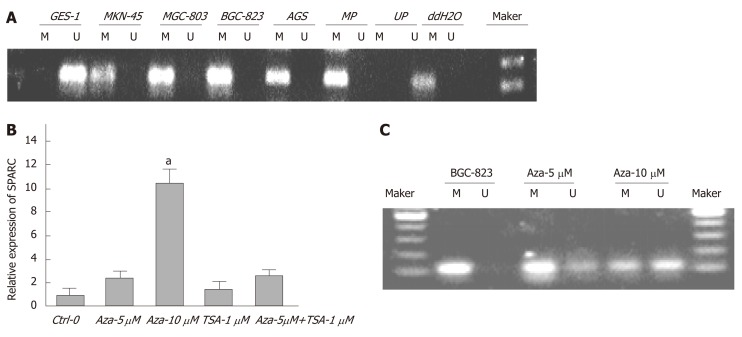
Aberrant hypermethylation in GC cell lines following 5-Aza-Cdr treatment
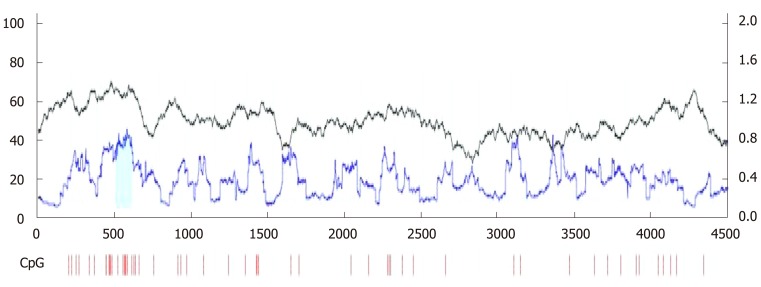
We designed suitable primers for methylation and non-methylation using Methprimer (Figure 2). Using MSP, we detected aberrant methylation of CpG islands in the SPARC gene in four GC cell lines (Figure 3A). Five-Aza-Cdr treatment was able to restore SPARC expression and reverse the hypermethylation status (Figure 3B and C). Moreover, recovery level was positively correlated with drug concentration. However, the sole application of the histone acetylation inhibitor trichostatin A (TSA) did not show this phenomenon. When the two drugs were used in combination, the outcomes were consistent with the results obtained for 5-Aza-Cdr treatment alone.
Figure 2.
Online prediction software, Methprimer, confirmed the existence of cytosines preceding guanosines island in the promoter region of SPARC gene. The green part represents the area where the cytosines preceding guanosine island is located. CpG: Cytosines preceding guanosine.
Figure 3.
Hypermethylation status of secreted protein acidic and rich in cysteine promoter in gastric cancer cells and the effects of drug intervention. A: Representative statistics of methylation-specific polymerase chain reaction (PCR) in five cell lines. PCR visualized in 2% agarose gels; B: Relative SPARC mRNA expression levels of BGC-823 cells after drug intervention were analyzed by quantitative real-time PCR followed by quantitation relative to glyceraldehyde-3-phosphate dehydrogenase; C: Methylation status of promoter region after drug intervention. Data are expressed as mean ± standard deviation (Figure 3B). aP < 0.05 was considered statistically significant. SPARC: Secreted protein acidic and rich in cysteine; M: Methylated amplified fragment; U: Unmethylated amplified fragment; MP: Methylated positive control; UP: Unmethylated positive control; ddH2O: Blank control.
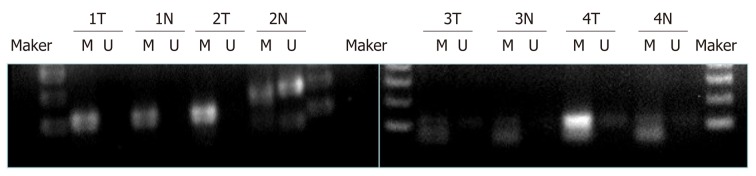
Aberrant methylation of the SPARC gene in GC tissues
MSP was used to analyze the methylation status of the SPARC gene promoter region in tumors and normal gastric mucosa samples (n = 66). The results are shown in Table 2 and Figure 4. For all GC samples, the methylation detection rate was 54.5% (n = 36), while the part methylation detection rate was 16.7% (n = 11). When comparing the methylation status of SPARC with clinicopathological characteristics, we found that there were statistically significant differences between Borrmann I + II and III and IV (P = 0.002), TNM I + II and III + IV (P = 0.001), and lymph node positive metastasis and lymph node negative metastasis (P = 0.001) in GC patients. There were no statistically significant differences with regards to gender, age, size, differentiation, or location (Table 2).
Table 2.
Relationship between methylation status of SPARC gene and clinicopathological characteristics, n (%)
| Parameters | Total, n = 66 |
Methylation status of SPARC |
P value | ||
| U, n = 19 | P, n = 11 | M, n = 36 | |||
| Age in yr | |||||
| < 60 | 21 | 6 (28.6) | 1 (4.8) | 14 (66.7) | 0.178 |
| ≥ 60 | 45 | 13 (28.9) | 10 (22.2) | 22 (48.9) | |
| Gender | |||||
| Male | 51 | 17 (33.3) | 8 (15.7) | 26 (51.0) | 0.323 |
| Female | 15 | 2 (13.3) | 3 (20.0) | 10 (66.7) | |
| Size in cm | |||||
| < 5 | 27 | 6 (22.2) | 6 (22.2) | 15 (55.6) | 0.463 |
| ≥ 5 | 39 | 13 (33.3) | 5 (12.8) | 21 (53.8) | |
| Differentiation | |||||
| Good/moderate | 16 | 6 (37.5) | 4 (25.0) | 6 (37.5) | 0.278 |
| Poor | 50 | 13 (26.0) | 7 (14.0) | 30 (60.0) | |
| Location | |||||
| Upper | 8 | 2 (25.0) | 3 (37.5) | 3 (37.5) | 0.297 |
| Middle | 18 | 6 (33.3) | 4 (22.2) | 8 (44.4) | |
| Lower | 40 | 11 (27.5) | 4 (10.0) | 25 (62.5) | |
| Macroscopic type | |||||
| Borrmann I + II | 19 | 9 (47.4) | 6 (31.6) | 4 (21.1) | 0.002a |
| Borrmann III + IV | 47 | 10 (21.3) | 5 (10.6) | 32 (68.1) | |
| TNM stage | |||||
| I + II | 27 | 14 (51.9) | 8 (29.6) | 5 (18.5) | 0.001a |
| III + IV | 39 | 5 (12.8) | 3 (7.7) | 31 (79.5) | |
| Lymph node metastasis | |||||
| - | 18 | 13 (72.2) | 2 (11.1) | 3 (16.7) | 0.001a |
| + | 48 | 6 (12.5) | 9 (18.8) | 33 (68.8) | |
P < 0.05 was considered statistically significant. SPARC: Secreted protein acidic and rich in cysteine.
Figure 4.
Methylation status of promoter region of secreted protein acidic and rich in cysteine in four matched gastric cancer tissues. T: Tumor tissue; N: Non-tumor tissue; M: Methylated amplified fragment; U: Unmethylated amplified fragment.
SPARC inhibited the proliferation, migration, and invasion of the GC cell line BGC-823
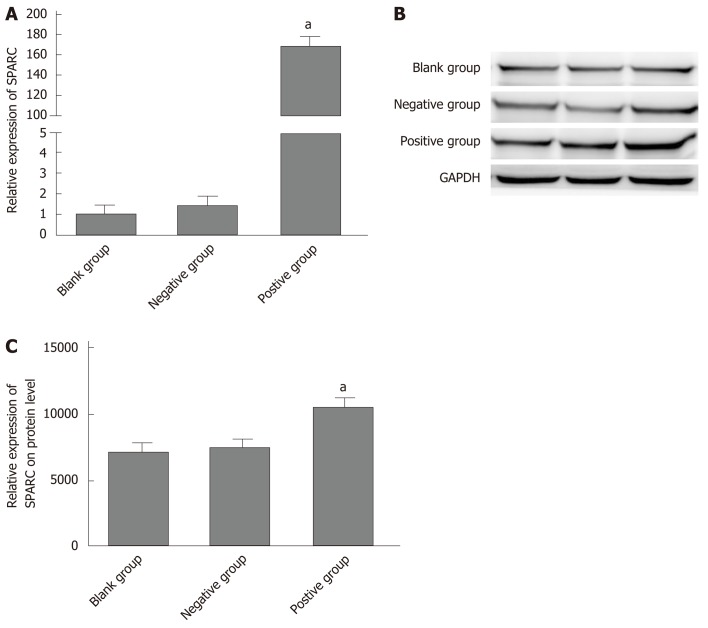
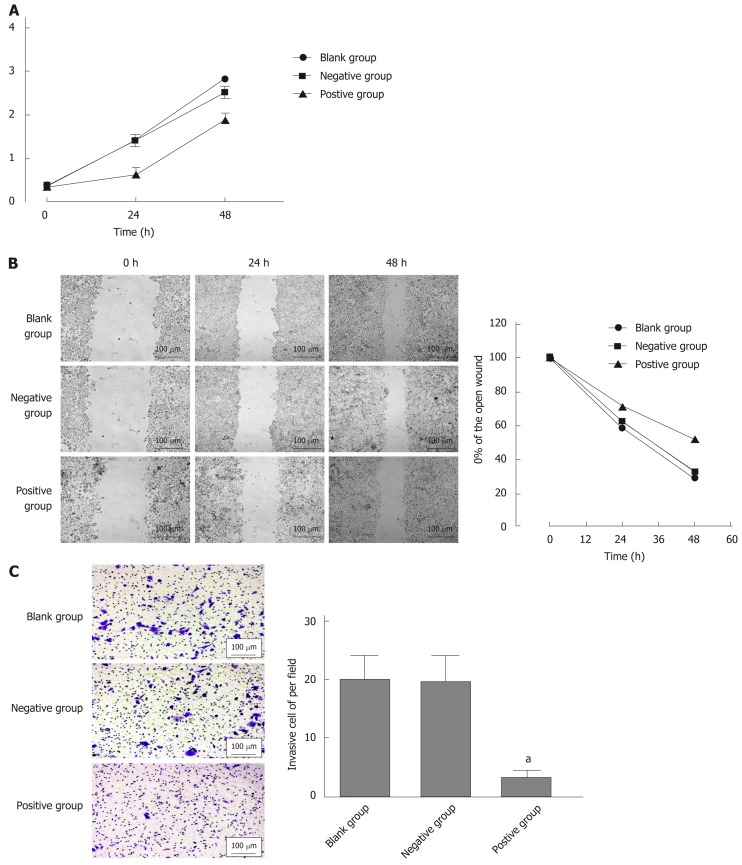
In order to verify further the biological role of SPARC in GC cell lines, we constructed a stable SPARC overexpression plasmid as a transfection model for GC BGC-823 cells. First, in order to test the transfection efficiency, we detected the mRNA and protein expression levels of SPARC in the transfected cells. This showed that the expression levels of SPARC in the positive control (PC) group was 169.6 ± 21, 4-fold higher than that in the negative control (NC) group; protein expression followed the same trend. Then, we investigated changes in the proliferation, invasion, and migration of the transfected cells (Figure 5A and B). In PC, cell proliferation and migration showed a significant decrease when compared with NC (Figure 6A and B). Compared with the NC (24 ± 9), the PC group (3 ± 1) showed a significantly lower number of cells passing through the chambers (Figure 6C). Taken together, these results suggested that the SPARC gene could significantly inhibit the proliferation, migration, and invasion of BGC-823 cells in vitro.
Figure 5.
Transfection efficiency of secreted protein acidic and rich in cysteine in gastric cancer cell lines BGC-823. A: Relative secreted protein acidic and rich in cysteine (SPARC) mRNA expression levels were analyzed after being transfected; B: Western blot analyzed SPARC gene protein expression after being transfected; C: Glyceraldehyde-3-phosphate dehydrogenase was used as internal reference. Data are presented as mean ± SE. aP < 0.05 was considered statistically significant. SPARC: Secreted protein acidic and rich in cysteine; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase.
Figure 6.
Effects of secreted protein acidic and rich in cysteine on cell proliferation, migration, and invasion. A: Cell proliferation rates were determined after 48 hr in complete medium by the cell counting kit-8 assay; B: The left half data represent that transfection of secreted protein acidic and rich in cysteine into cells could reduce the rate of cell scratch healing; C: Transfected cells were subjected to an assay using a two-chambered invasion apparatus as described in methods, the histogram shows the number of cells penetrating the chambers. Data are presented as mean ± SE. aP < 0.05 was considered statistically significant.
SPARC was able to cause cell cycle arrest and initiate cellular apoptosis
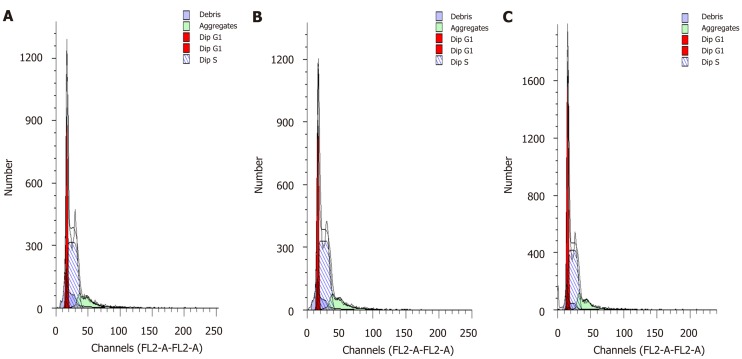
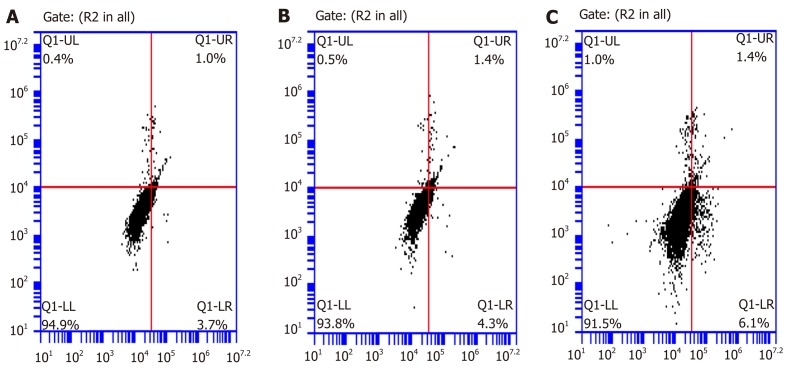
Cell cycle analyses were carried out by flow cytometry to characterize the effects of SPARC on the growth fraction distribution of BGC-823 cells. The number of cells in the G0/G1 phase in the transfected BGC-823 cells was significantly higher than that in the NC group at 72 h. This result indicated that SPARC might block the progression of the GC cell cycle from G0/G1 phase to S/G2 phase and arrest cells in the G0/G1 phase (Figure 7). In addition to this, the rate of apoptosis in PC was significantly higher (6.55% ± 0.63%) than that in NC (4.05% ± 0.35%) (P < 0.05). These results indicate that SPARC might promote apoptosis in GC cells (Figure 8).
Figure 7.
Effects of secreted protein acidic and rich in cysteine on cell cycle in BGC-823 gastric cancer cell line. Overexpressing of secreted protein acidic and rich in cysteine could change cell cycle distribution in BGC-823 gastric cancer cells at 72 h post-transfection. DNA content was measured using propidium iodide staining on flow cytometry. A: Blank control group; B: Negative control group; C: Positive control group. Data are presented as mean ± SE. aP < 0.05 was considered statistically significant.
Figure 8.
Secreted protein acidic and rich in cysteine induces apoptosis in BGC-823. For flow cytometric analysis, cells were stained with annexin V fluorescein isothiocyanate and propidium iodide. Values in histogram show increasing secreted protein acidic and rich in cysteine (SPARC) expression increased apoptosis in BGC-823 compared with negative control. A: Blank control group; B: Negative control group; C: Positive control group. Data are presented as mean ± SE. aP < 0.05 was considered statistically significant.
DISCUSSION
GC is the result of multiple genetic and epigenetic abnormalities. In contrast to genetic modifications, epigenetic modifications do not change the DNA sequence to regulate GC growth and development[12]. In our previous studies, we reported that the promoter methylation of tumor suppressor genes could inhibit their own reverse transcription and function. In the present study, we focused on the downregulation and silencing of SPARC by methylation and investigated the significance of this process in GC.
SPARC is one of the matricellular proteins; the role of these proteins is to modulate cell-matrix interactions and cell function without participating in the structural scaffold of the extracellular matrix. Many researchers have demonstrated that SPARC can inhibit the growth of tumors via anti-angiogenesis by inhibiting cell proliferation and arresting the cell cycle.
Many previous studies also indicate that SPARC has a diverse array of functions, which may be dependent on the surrounding microenvironment in different tissues and cells. Therefore, whether SPARC suppresses or promotes tumors may vary from tissue to tissue. For example, SPARC can promote bone metastasis in several types of solid tumors, including glioblastoma, melanoma, and breast cancer. Furthermore, high levels of SPARC are associated with higher rates of malignancy, stronger invasiveness, and a lower overall prognosis than controls with lower levels of SPARC expression. Therefore, in such malignancies, it is possible that SPARC might serve as a marker for predicting high tumor invasion and poor prognosis. Furthermore, it may be possible to use antibodies or drugs to inhibit SPARC expression in order to weaken tumor invasion, reduce tumor metastasis, and improve the 5-year survival rate and overall prognosis of patients.
However, there is no consistent understanding with regards to the effects of the SPARC gene in GC. Nakajima et al[13] used immunohistochemistry to examine GC tissues and found that the levels of SPARC were upregulated in tumor fibroblasts but were rarely observed in the cytoplasm of GC cells. Moreover, high-matrix SPARC expression was identified as an independent predictor of a more favorable prognosis. However, laboratory studies, involving cell lines and tissues, have reported contradictory results. For instance, Gao et al[14] found that the levels of SPARC was higher in GC tissues than adjacent normal tissues; these authors also showed that increased expression levels of SPARC in GC tissues indicated that the cumulative survival time of gastric adenocarcinoma patients was shorter. Zhang et al[15] compared and analyzed the proliferation ability of SPARC between knockout cell lines and normal GC cell lines, and found that SPARC could increase cell proliferation. Consequently, further research was clearly required.
This present study used qRT-PCR to investigate the expression of SPARC mRNA in four GC cell lines, one immortalized normal gastric cell line, and 66 paired sample tissues. We showed that the SPARC gene was significantly downregulated in GC cells and tissues. Clinical data indicated that low levels of SPARC mRNA were significantly correlated with TNM stage, macroscopic type, and lymph node metastasis. Therefore, we believe that SPARC might play a role as a tumor suppressor gene in GC. SPARC has also been found to be downregulated in some other types of cancer, including neuroblastoma[16], ovarian cancer[17], pancreatic cancer[8], colon cancer[9], bladder cancer[18], prostate cancer[19], and acute leukemia[20]. When analyzing these tumors genetically, it is common to find that the promoter region of the SPARC gene in these tumors was in a state of abnormal hypermethylation. Therefore, we hypothesized that DNA methylation might be one of the mechanisms that regulates the expression of SPARC and, therefore, plays a role in tumorigenesis and development[21]. In order to verify this hypothesis, we first used the online software Methlyprimer to predict the presence of CpG islands in the promoter region of the SPARC gene. Then, we used the MSP technique to test the methylation status of the promoter region of the SPARC gene in these low-expressing GC cells and tissues. We found that the SPARC gene promoter region was not methylated in the GES-1 cell line but found that the other four GC cell lines showed methylation. Similar conclusions were previously reported for pancreatic cancer[22], prostate cancer[23], and colon cancer[24]. These results also revealed that the methylation frequency of SPARC was positively associated with TNM stage, macroscopic type, and lymph node metastasis. A high proportion of patients with Borrmann type, late TNM staging, and lymph node metastasis showed a higher proportion of SPARC promoter gene methylation than the matched control group. These results suggested that the aberrant methylation of SPARC is involved in the pathogenesis of GC and that the overall prognosis of GC may be poorer when the methylation frequency of SPARC is high. DNA methylation can be reversed by the application of demethylation drugs (e.g., 5-Aza-Cdr). We therefore selected GC BGC-823 cells, which expressed lower levels of SPARC, as a target to be treated with the demethylating agent 5-Aza-Cdr. We also treated these cells with the acetylation inhibitor drug TSA to eliminate the effects of histone acetylation on our experimental results. In the reference group, the cells were treated with TSA alone; a combination of drugs was used in another group. After treatment, the expression of the SPARC gene in the GC cell lines had increased; the extent of methylation had decreased only in cells treated with 5-Aza-Cdr alone. The application of TSA alone did not show any obvious changes. Following treatment, the restored expression level was positively related to the original drug concentration. In summary, the SPARC gene was expressed at low levels in GC cells and tissues and acts as an anti-oncogene. It is possible that DNA methylation might regulate the expression of this gene. This data provided a new basis for adjuvant treatment with clinically demethylated 5-Aza-Cdr.
Previous research has also suggested that SPARC might play a key role during the initial formation of a gastric tumor. On one hand, the SPARC gene itself could affect the proliferation, migration, and invasion of tumor cells. On the other hand, the SPARC gene could also block the progression of tumor cells from the G0/G1 phase to the S/G2 phase and initiate the process of apoptosis in tumor cells. Therefore, we further explored the effects of the SPARC gene on the biological function of GC cells from two different aspects. First, we determined the effects of increased levels of SPARC expression on tumor cell proliferation, invasion, and migration. We measured the capacity of GC cells to proliferate, invade, and migrate after transfection with a SPARC plasmid or a non-targeting control plasmid. Increased levels of SPARC expression led to the inhibition of proliferation, migration, and invasion in GC BGC-823 cells. Our results were consistent with previous results described for both ovarian cancer cells[25] and pancreatic cancer cells[26-27]. SPARC has also been shown to control the distant metastasis of ovarian cancer by regulating the expression of vascular endothelial growth factor and matrix metalloproteinases[28]. It is also possible that SPARC could alter the biological function of ovarian cancer cells by modulating signaling pathways[29]. For example, the SPARC gene could inhibit cell proliferation induced by integrin and activate growth factors such as integrin-linked kinase, focal adhesion kinase, Src, p44/42, and mitogen-activated protein kinase in downstream signaling pathways; furthermore, these signaling pathways could exert influence upon each other. SPARC has also been shown to retard angiogenesis by suppressing vascular endothelial growth factor-A via miR-410 and thus inhibit human neuroblastoma cells from proliferating[30]. Next, to elucidate the effects of SPARC on the cell cycle and rate of apoptosis in GC cells, we used flow cytometry to compare SPARC-transfected and control- transfected BGC-823 cells. Increasing levels of SPARC expression also caused delays in the cell cycle and induced apoptosis. Previous research on glioma cells by Schultz et al[31] showed that the RNA interference-mediated knock-down of SPARC could significantly change the cell cycle distribution.
DNA methylation is known to play an important role in the expression of tumor suppressor genes. Methylated genes, such as CDH11, EphA5, and HS3ST2[32], could therefore be used as biomarkers for GC. In previous research, Torres-Núñez et al[33] found that SPARC expression was regulated by DNA methylation in its core promoter region.
In conclusion, the results of our study indicated that SPARC was dramatically down-regulated in gastric carcinoma cells and tissues, and that this might be due to hypermethylation of the promoter. Furthermore, low expression and hypermethylation status of SPARC were associated with poor TNM stage, macroscopic type, and invasion into lymph nodes. Moreover, the restoration of SPARC gene expression significantly inhibited the proliferation, migration, and invasion of GC cells, caused cell cycle arrest, and promoted cellular apoptosis. Thus, we believe that our findings indicate that SPARC expression may play an important role in the regulation, development, and progression of GC.
ARTICLE HIGHLIGHTS
Research background
A large body of evidence has now confirmed that secreted protein acidic and rich in cysteine (SPARC) gene plays an inhibitory role in a variety of tumors, including ovarian cancer, pancreatic cancer, and colon cancer. In addition, the SPARC gene also plays an important role in anti-angiogenesis, the inhibition of cell proliferation, and cell cycle arrest. However, little is known about the precise role of SPARC in gastric cancer (GC). In the present research, we conducted a conjoint analysis of the correlation between methylation, gene expression, and patient prognosis in GC cells and tissues.
Research motivation
SPARC is considered to represent a potential therapeutic target and biomarker of GC. Therefore, the identification and analysis of aberrant methylation of the SPARC gene, and its significance in GC, is of significant interest.
Research objectives
In this study, we aimed to investigate the expression levels of the SPARC gene in different GC cell lines and tissues and investigated how these factors were related to clinicopathological features. Further studies analyzed the correlation between methylation, gene expression, and patient prognosis. We also demonstrated potential effects on biological function.
Research methods
Quantitative real-time polymerase chain reaction (qRT-PCR) was used to detect the expression levels of SPARC in a normal gastric mucous membrane cell line, four GC cell lines, and 66 matched samples. We also used methylated-specific PCR to detect the SPARC gene promoter region and investigated the correlation between methylation and clinicopathological features. We also investigated the effects of the demethylation drug, 5-azacytidine, and the acetylated inhibitor, trichostatin A (TSA), on BGC-823 cells. By constructing a stable cell line that over-expressed SPARC, we were able to explore its effects on biological function using the cell counting kit-8 kit, the scratch method, flow cytometry, and the transwell chamber method.
Research results
qRT-PCR results showed that the expression level of SPARC gene in GC tissues and cell lines was significantly lower than controls. We also concluded that low expression levels of SPARC were related to Borrmann type, TNM staging, and lymph node metastasis. The hypermethylation of SPARC was positively correlated with Borrmann type, TNM staging, and lymph node metastasis. We transfected a plasmid vector containing the SPARC gene into BGC-823 cells and analyzed changes in biological function using the cell counting kit-8 kit, scratches, flow cytometry, and Transwell assays. Data showed that the over-expression of SPARC inhibited cell proliferation, migration, and invasion, arrested the cell cycle, and promoted cellular apoptosis.
Research conclusions
In conclusion, we provide new and reliable evidence for the role of SPARC in GC. We believe that SPARC may act as an anti-oncogene to inhibit the tumorigenesis of GC. Methylation plays a critical role in regulating SPARC gene expression and is significantly associated with GC. Furthermore, SPARC may represent a potential biomarker and therapeutic target for GC. By analyzing the effects of SPARC gene expression, it may be possible to develop better and more accurate treatment and thus improve the prognosis of patients with GC.
Research perspectives
Studying tumor inhibitor gene methylation provides a new strategy and direction for the examination, treatment, and prognosis of GC. Further studies should be conducted to analyze the regulatory mechanisms associated with the SPARC gene and potential links with the occurrence and development of GC.
Footnotes
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
Institutional review board statement: The data of gastric cancer we analyzed in this research are all from The Fourth Affiliated Hospital of China Medical University. And our research had been approved by The Fourth Affiliated Hospital of China Medical University Clinical Research Ethics Committee.
Conflict-of-interest statement: All authors declare that there is no conflict of interest related to this study.
Peer-review started: August 13, 2019
First decision: September 10, 2019
Article in press: October 22, 2019
P-Reviewer: Aoki T, Biondi A S-Editor: Wang J L-Editor: Filipodia E-Editor: Zhang YL
Contributor Information
Shuai Shao, Department of Gastrointestinal Surgery, the Fourth Affiliated Hospital of China Medical University, Shenyang 110032, Liaoning Province, China.
Nuo-Ming Zhou, Department of Gastrointestinal Surgery, the Fourth Affiliated Hospital of China Medical University, Shenyang 110032, Liaoning Province, China.
Dong-Qiu Dai, Department of Gastrointestinal Surgery, the Fourth Affiliated Hospital of China Medical University, Shenyang 110032, Liaoning Province, China. daidq63@163.com.
Data sharing statement
No additional data are available.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Maehara Y, Hasuda S, Koga T, Tokunaga E, Kakeji Y, Sugimachi K. Postoperative outcome and sites of recurrence in patients following curative resection of gastric cancer. Br J Surg. 2000;87:353–357. doi: 10.1046/j.1365-2168.2000.01358.x. [DOI] [PubMed] [Google Scholar]
- 3.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 4.Mikata R, Fukai K, Imazeki F, Arai M, Fujiwara K, Yonemitsu Y, Zhang K, Nabeya Y, Ochiai T, Yokosuka O. BCL2L10 is frequently silenced by promoter hypermethylation in gastric cancer. Oncol Rep. 2010;23:1701–1708. doi: 10.3892/or_00000814. [DOI] [PubMed] [Google Scholar]
- 5.Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis. 2000;21:461–467. doi: 10.1093/carcin/21.3.461. [DOI] [PubMed] [Google Scholar]
- 6.Termine JD, Kleinman HK, Whitson SW, Conn KM, McGarvey ML, Martin GR. Osteonectin, a bone-specific protein linking mineral to collagen. Cell. 1981;26:99–105. doi: 10.1016/0092-8674(81)90037-4. [DOI] [PubMed] [Google Scholar]
- 7.Yiu GK, Chan WY, Ng SW, Chan PS, Cheung KK, Berkowitz RS, Mok SC. SPARC (secreted protein acidic and rich in cysteine) induces apoptosis in ovarian cancer cells. Am J Pathol. 2001;159:609–622. doi: 10.1016/S0002-9440(10)61732-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Infante JR, Matsubayashi H, Sato N, Tonascia J, Klein AP, Riall TA, Yeo C, Iacobuzio-Donahue C, Goggins M. Peritumoral fibroblast SPARC expression and patient outcome with resectable pancreatic adenocarcinoma. J Clin Oncol. 2007;25:319–325. doi: 10.1200/JCO.2006.07.8824. [DOI] [PubMed] [Google Scholar]
- 9.Cheetham S, Tang MJ, Mesak F, Kennecke H, Owen D, Tai IT. SPARC promoter hypermethylation in colorectal cancers can be reversed by 5-Aza-2'deoxycytidine to increase SPARC expression and improve therapy response. Br J Cancer. 2008;98:1810–1819. doi: 10.1038/sj.bjc.6604377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan J, Zhang J, Zhang X, Li X, Li L, Li Z, Chen R, Zhang L, Wu J, Wang X, Sun Z, Fu X, Chang Y, Nan F, Yu H, Wu X, Feng X, Li W, Zhang M. SPARC is down-regulated by DNA methylation and functions as a tumor suppressor in T-cell lymphoma. Exp Cell Res. 2018;364:125–132. doi: 10.1016/j.yexcr.2017.12.022. [DOI] [PubMed] [Google Scholar]
- 11.Inoue M, Senju S, Hirata S, Ikuta Y, Hayashida Y, Irie A, Harao M, Imai K, Tomita Y, Tsunoda T, Furukawa Y, Ito T, Nakamura Y, Baba H, Nishimura Y. Identification of SPARC as a candidate target antigen for immunotherapy of various cancers. Int J Cancer. 2010;127:1393–1403. doi: 10.1002/ijc.25160. [DOI] [PubMed] [Google Scholar]
- 12.Stein RA. DNA methylation profiling: a promising tool and a long road ahead for clinical applications. Int J Clin Pract. 2011;65:1212–1213. doi: 10.1111/j.1742-1241.2011.02804.x. [DOI] [PubMed] [Google Scholar]
- 13.Nakajima M, Yoshino S, Kanekiyo S, Maeda N, Sakamoto K, Tsunedomi R, Suzuki N, Takeda S, Yamamoto S, Hazama S, Hoshii Y, Oga A, Itoh H, Ueno T, Nagano H. High secreted protein acidic and rich in cysteine expression in peritumoral fibroblasts predicts better prognosis in patients with resectable gastric cancer. Oncol Lett. 2018;15:803–812. doi: 10.3892/ol.2017.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao Y, Yin SP, Xie XS, Xu DD, Du WD. The relationship between stromal cell derived SPARC in human gastric cancer tissue and its clinicopathologic significance. Oncotarget. 2017;8:86240–86252. doi: 10.18632/oncotarget.21133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang JL, Chen GW, Liu YC, Wang PY, Wang X, Wan YL, Zhu J, Gao HQ, Yin J, Wang W, Tian ML. Secreted protein acidic and rich in cysteine (SPARC) suppresses angiogenesis by down-regulating the expression of VEGF and MMP-7 in gastric cancer. PLoS One. 2012;7:e44618. doi: 10.1371/journal.pone.0044618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chlenski A, Liu S, Crawford SE, Volpert OV, DeVries GH, Evangelista A, Yang Q, Salwen HR, Farrer R, Bray J, Cohn SL. SPARC is a key Schwannian-derived inhibitor controlling neuroblastoma tumor angiogenesis. Cancer Res. 2002;62:7357–7363. [PubMed] [Google Scholar]
- 17.Said NA, Najwer I, Socha MJ, Fulton DJ, Mok SC, Motamed K. SPARC inhibits LPA-mediated mesothelial-ovarian cancer cell crosstalk. Neoplasia. 2007;9:23–35. doi: 10.1593/neo.06658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Said N, Frierson HF, Sanchez-Carbayo M, Brekken RA, Theodorescu D. Loss of SPARC in bladder cancer enhances carcinogenesis and progression. J Clin Invest. 2013;123:751–766. doi: 10.1172/JCI64782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu T, Qiu X, Zhao X, Yang R, Lian H, Qu F, Li X, Guo H. Hypermethylation of the SPARC promoter and its prognostic value for prostate cancer. Oncol Rep. 2018;39:659–666. doi: 10.3892/or.2017.6121. [DOI] [PubMed] [Google Scholar]
- 20.DiMartino JF, Lacayo NJ, Varadi M, Li L, Saraiya C, Ravindranath Y, Yu R, Sikic BI, Raimondi SC, Dahl GV. Low or absent SPARC expression in acute myeloid leukemia with MLL rearrangements is associated with sensitivity to growth inhibition by exogenous SPARC protein. Leukemia. 2006;20:426–432. doi: 10.1038/sj.leu.2404102. [DOI] [PubMed] [Google Scholar]
- 21.Goyama S, Kitamura T. Epigenetics in normal and malignant hematopoiesis: An overview and update 2017. Cancer Sci. 2017;108:553–562. doi: 10.1111/cas.13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brune K, Hong SM, Li A, Yachida S, Abe T, Griffith M, Yang D, Omura N, Eshleman J, Canto M, Schulick R, Klein AP, Hruban RH, Iacobuzio-Donohue C, Goggins M. Genetic and epigenetic alterations of familial pancreatic cancers. Cancer Epidemiol Biomarkers Prev. 2008;17:3536–3542. doi: 10.1158/1055-9965.EPI-08-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moses-Fynn E, Tang W, Beyene D, Apprey V, Copeland R, Kanaan Y, Kwabi-Addo B. Correlating blood-based DNA methylation markers and prostate cancer risk in African-American men. PLoS One. 2018;13:e0203322. doi: 10.1371/journal.pone.0203322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang E, Kang HJ, Koh KH, Rhee H, Kim NK, Kim H. Frequent inactivation of SPARC by promoter hypermethylation in colon cancers. Int J Cancer. 2007;121:567–575. doi: 10.1002/ijc.22706. [DOI] [PubMed] [Google Scholar]
- 25.Mok SC, Chan WY, Wong KK, Muto MG, Berkowitz RS. SPARC, an extracellular matrix protein with tumor-suppressing activity in human ovarian epithelial cells. Oncogene. 1996;12:1895–1901. [PubMed] [Google Scholar]
- 26.Arnold SA, Rivera LB, Miller AF, Carbon JG, Dineen SP, Xie Y, Castrillon DH, Sage EH, Puolakkainen P, Bradshaw AD, Brekken RA. Lack of host SPARC enhances vascular function and tumor spread in an orthotopic murine model of pancreatic carcinoma. Dis Model Mech. 2010;3:57–72. doi: 10.1242/dmm.003228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen G, Tian X, Liu Z, Zhou S, Schmidt B, Henne-Bruns D, Bachem M, Kornmann M. Inhibition of endogenous SPARC enhances pancreatic cancer cell growth: modulation by FGFR1-III isoform expression. Br J Cancer. 2010;102:188–195. doi: 10.1038/sj.bjc.6605440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Said N, Socha MJ, Olearczyk JJ, Elmarakby AA, Imig JD, Motamed K. Normalization of the ovarian cancer microenvironment by SPARC. Mol Cancer Res. 2007;5:1015–1030. doi: 10.1158/1541-7786.MCR-07-0001. [DOI] [PubMed] [Google Scholar]
- 29.Said N, Najwer I, Motamed K. Secreted protein acidic and rich in cysteine (SPARC) inhibits integrin-mediated adhesion and growth factor-dependent survival signaling in ovarian cancer. Am J Pathol. 2007;170:1054–1063. doi: 10.2353/ajpath.2007.060903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyineni J, Tanpure S, Gnanamony M, Antony R, Fernández KS, Lin J, Pinson D, Gondi CS. SPARC overexpression combined with radiation retards angiogenesis by suppressing VEGF-A via miR‑410 in human neuroblastoma cells. Int J Oncol. 2016;49:1394–1406. doi: 10.3892/ijo.2016.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schultz C, Lemke N, Ge S, Golembieski WA, Rempel SA. Secreted protein acidic and rich in cysteine promotes glioma invasion and delays tumor growth in vivo. Cancer Res. 2002;62:6270–6277. [PubMed] [Google Scholar]
- 32.Eyvazi S, Khamaneh AM, Tarhriz V, Bandehpour M, Hejazi MS, Sadat ATE, Sepehri B. CpG Islands Methylation Analysis of CDH11, EphA5, and HS3ST2 Genes in Gastric Adenocarcinoma Patients. J Gastrointest Cancer. 2019 doi: 10.1007/s12029-019-00290-1. [DOI] [PubMed] [Google Scholar]
- 33.Torres-Núñez E, Cal L, Suárez-Bregua P, Gómez-Marin C, Moran P, Gómez-Skarmeta JL, Rotllant J. Matricellular protein SPARC/osteonectin expression is regulated by DNA methylation in its core promoter region. Dev Dyn. 2015;244:693–702. doi: 10.1002/dvdy.24267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No additional data are available.