Short abstract
Since their discovery just over 25 years ago, the single variable domain from heavy-chain-only antibodies plays a role in an increasing number of antibody-based applications. Structural and biophysical studies have revealed that the small, ∼15 kDa, single variable domain found in camelids displays versatility in target recognition. Such insight has served as the foundation to develop and engineer VHH domains with enhanced properties capable of targeting a range of therapeutically relevant protein antigens or low-molecular weight haptens. Furthermore, the modular nature of VHH domains allows them to be introduced into constructs that are simply not possible with conventional antibodies. Here, we review the structural and biophysical properties of VHH domains, highlight recent VHH-based therapeutics and diagnostics, and provide insight into VHH engineering that may pave the way to next-generation single domain antibody applications.
Impact statement
The development of novel antibody formats, beyond conventional antibodies, opens new possibilities in medical therapies, diagnostics, and general life science applications requiring affinity reagents. The camelid VHH domain, from heavy-chain-only antibodies, has emerged as a jack-of-all-trades module for novel affinity reagents. Applications include targeted cancer therapies, novel antimicrobial agents, conformation specific reagents, and tailor-made molecular switch entities. The breadth of unique uses for the VHH will continue to grow, opening new opportunities to treat and understand disease.
Keywords: Camelid single domain antibodies, VHH, nanobody, antibody engineering, therapeutic antibody, antibody structure
Introduction
Beyond their role in the immune response, antibodies serve as important tools for a range of life science applications. Their high affinity and specificity for a target molecule make them excellent affinity reagents for use in therapeutics and diagnostics. FDA-approved monoclonal antibodies have been used to treat a wide range of medical conditions (e.g. cancers, anthrax, asthma, Crohn’s disease, and multiple sclerosis).1 Furthermore, the relative simplicity in raising antibodies against biomarkers of interest allows the routine use of antibodies in medical diagnostics, such as point-of-care influenza testing. These applications have evolved to include non-conventional antibodies, such as antibody fragments and hybrid variants, which can provide advantages not possible with traditional antibodies.
Camelidae (e.g. camels, dromedaries, and llamas) heavy-chain-only antibodies, a class of antibodies devoid of light chains, break the mold of conventional antibody/antigen architecture. These heavy-chain-only antibodies possess a single variable domain, termed VHH, that is responsible for antigen recognition.2 As compared to conventional VH/VL antigen binding interfaces, a single VHH domain possesses a drastically reduced antigen binding interface, as only three hypervariable loops (H1-H3) are present.3 Despite the reduced interface surface area, VHH domains commonly show binding affinities that are comparable to conventional antibodies.4
Structural studies have revealed that the single variable domain from heavy-chain-only antibodies is quite versatile in target recognition. Such structural insight has served as the foundation to develop and engineer VHH domains with enhanced properties capable of targeting a range of therapeutically relevant protein antigens or low-molecular weight haptens. Here, we discuss the structural and biophysical properties of VHH domains, provide examples of recent VHH-based therapeutics and diagnostics, and review current strategies for novel VHH engineering.
Physiochemical properties of VHH domains
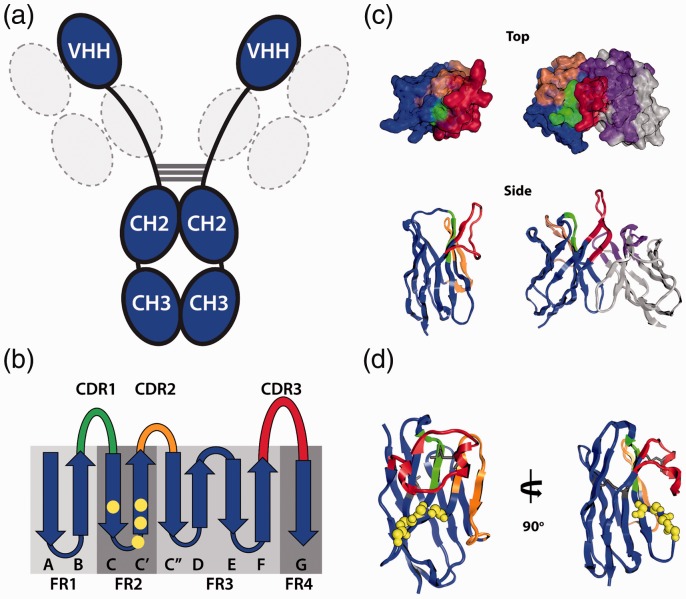
The most notable feature of the VHH domain from heavy chain-only antibodies is its small size. With a size of approximately 15 kDa, the camelid single domain represents one of the smallest, naturally occurring antibody binding units. The other example comparable in size is the IgNAR domain found in heavy chain-only antibodies from cartilaginous fish (e.g. shark), which possess similar properties to the VHH domain, likely stemming from convergent evolution.5,6 The autonomous nature of the sole VHH domain is believed to originate from several residue substitutions within framework (FW) 2 (Figure 1),7 including V37F/Y, G44E, L45R, and W47G (Kabat numbering). These residues reside within what would be the VL interface of a conventional VH domain.8 Consequently, the residue substitutions are believed to help enhance solubility/stability in the absence of a light chain.9 Another structural distinction between VHH and VH domains is the unusually long CDR3 loops typically found in VHH domains (Figure 1).10–12 The extended CDR3 found in many VHH domains has been observed to participate in intramolecular interactions with the VHH framework that would otherwise interact with VL domains. Consequently, CDR3 can serve as a surrogate for the VL domain.13–15
Figure 1.
Unique structural architecture of VHH domain antibodies. (a) Cartoon representation of a camelid immunoglobulin depicting the two heavy chains (blue) with the variable domains, VHH, the constant domains CH2 and CH3 and absence of the constant domain CH1 and the light chain (gray with dashed lines). (b) Two-dimensional representation of the VHH structure illustrating the canonical nine immunoglobulin β strands comprising the framework regions (FR1 through FR4) and CDR 1 (green), CDR2 (orange), and CDR3 (red). The locations of the four hallmark mutations in VHH domains are colored in yellow. (c) Comparison of VHH (left, PDB ID 2XA3) to a variable fragment (Fv) of conventional antibody (right, PDB ID 5IBU) with a long CDR3. The antigen binding paratope created by the CDRs (Kabat definitions) is illustrated in a surface representation of looking down (Top) and a side view ribbon diagram (Side). Light chain of the Fv is depicted in gray with the CDRs (one through three) shown in purple. (d) Ribbon representation of an anti-PD-L1 VHH (PDB ID 5JDS) with a long CDR3 loop containing an intramolecular disulfide between CDR1 and CDR3 (dark gray). CDR3 folds back along FR2 with the positions of the hallmark four mutations; backbone atoms rendered in yellow spheres. Crystal structure figures were generated using the MOE software from the Chemical Computing Group.7(A color version of this figure is available in the online journal.)
Structural studies have revealed details about how VHH domains remain soluble as single domains, along with their method of interacting with antigen targets. Much like their conventional antibody counterpart, VHH domains typically rely heavily on CDR3 for interactions with antigens. Notably, an elongated CDR3 loop provides VHH significant versatility in its ability to interact with target molecules. Unlike conventional antibodies, VHH domains have been observed to interact with antigens in protruding/extended conformations (Figure 2),16 which allow the antibody to bind protein clefts/pockets, including enzyme active sites.15,17,18 In addition, the CDR3 loop can produce a structurally flat paratope using CDR1/3,19 CDR1/2/3,20 and CDR2/3/FW,21 with some possessing more convex22 or concave23 paratopes (Figure 2). An analysis of 90 nonredundant VHH/protein antigen structures revealed that VHH CDRs exhibit greater structural diversity than conventional VH domains.24
Figure 2.
Versatility of the VHH antigen binding interface. VHH has been observed to possess (a) penetrative; antigen: β2 adrenoceptor-PDB ID 3P0G, (b) flat; antigen: RNase A-PDB ID 2P4A, (c) convex; antigen: Lysozyme-PDB ID 1ZVH, and (d) concave; antigen: GFP-PDB ID 3K1K paratopes. Colored surface represents VHH residues within four angstroms of antigen (displayed in white cartoon); CDR1-green; CDR2-orange, CDR3-red, framework-blue. Image generated using PyMol.16(A color version of this figure is available in the online journal.)
While protein stability and reversibility are greatly dependent on environmental conditions (e.g. pH, buffer, salt, and protein concentration), VHH domains typically possess high thermostability. In general, VHH domains exhibit melting temperatures in the range of 50–80°C25,26 and can remain functional after heating/cooling cycles,27 which makes VHH domains useful reagents for diagnostics at elevated temperatures.28 Despite having a reputation as being highly reversible after temperature melts, it is important to note that complete thermodynamic reversibility is likely a “best case” phenomena. Notably, the propensity of a given VHH clone to aggregate, an issue particularly problematic for therapeutic applications, can be easily evaluated through multi-concentration temperature melt experiments.29
Use of VHH domains in therapeutics
The use of antibodies as therapeutic agents stems from their biocompatibility and high affinity/specificity for target antigens. Since VHH domains share these properties, along with possessing additional beneficial qualities, VHH domains have been pursued as therapeutic modalities. Currently, the Therapeutic Antibody Database (https://tabs.craic.com) lists more than 31 VHH-based agents ranging from preclinical to clinical development. VHH therapeutic interests include, but are not limited to, oncology, inflammation, anti-viral, neurology, anti-infection agents, and rare hematological disorders. In February 2019, the FDA approved the first VHH-based therapy, Cablivi® (Sanofi, formally Ablynx), for the treatment of acquired thrombotic thrombocytopenic purpura.
VHH have advantages over conventional antibodies due to their small size and robustness arising from their unique physiochemical properties. Such properties include the ability to express VHH in both prokaryotic and eukaryotic expression systems, which can result in a lower cost of goods (from discovery through manufacturing). The single domain nature of VHH arises from a short encoding gene (∼360 bp). This small genetic footprint allows expanded functionality through the creation of modularity via genetic fusions to a wide-array of proteins, e.g. the creation of multispecific antibody fusions.30,31 Additionally, VHH domains possess high sequence homology to human VH3 genes,32 which may confer low immunogenicity for use in therapeutic applications. Indeed, initial human trials of an anti-Van Willebrand factor VHH for thrombotic thrombocytopenic purpura (TTP) reported low clinical immunogenicity.33 Nevertheless, despite the high sequence similarity, strategies have also been pursued to humanize camelid VHH domains.34
A caveat of using VHH domains for therapeutic applications is that the VHH hydrodynamic radius falls below the glomerular filtration limit of the kidney, which may result in rapid renal clearance, poor pharmacokinetics (PK), and potentially limit therapeutic efficacy. Moreover, VHHs lack the Fc region, which, on conventional Fc containing antibodies, facilitates extended half-lives via FcRn recycling. While in some instances this rapid clearance is a favorable behavior (e.g. diagnostic imaging reagents), several engineering strategies have been employed to elicit more favorable PK. The facile creation of genetic fusions with conventional Fc,35 VHH repeat domains (mono- or polyspecific),36 serum proteins (e.g. HSA) and anti-serum albumin VHH37 has been routinely employed to increase both the size and PK profile. As an alternative to genetic fusions and modularity, chemical approaches can also be employed including PEGylation and lipidation for improved half-life.38,39
The modular structure of VHH domains has opened new opportunities for targeting disease, such as cancer40 and viral infections.40 For example, the VHH domain has been explored in adoptive chimeric antigen receptor (CAR) T cell therapy targeting the HER2 antigen,41 as well as dual-specific HER2/CD20 CAR that takes advantage of the small modular VHH domain.42 VHH constructs targeting viral proteins, such as the influenza virus enzyme neuraminidase from H5N143 or the fusion protein F from human respiratory syncytial virus (RSV)44 have prevented viral infection in mice. Laursen et al.45 took advantage of the single VHH domain by generating multidomain VHH antibody variants that could target multiple influenza A and B hemagglutinin epitopes. These unique anti-hemagglutinin VHH domain fusion antibodies possess enhanced virus cross-reactivity/potency and provide new frameworks that may be applied to a broad range of therapeutic applications.
Oral-based delivery of VHH domains
While monoclonal antibody therapeutics are traditionally administered through intravenous or subcutaneous routes, opportunities exist to deliver antibodies through other routes, such as oral delivery. One such potential application includes the prevention and/or treatment of digested bacterial pathogens. For such therapeutic uses, the antibody must maintain function under gastrointestinal conditions. VHH domains targeting Escherichia coli F4 fimbriae46 and Campylobacter jejuni flagella47 have been successfully engineered to possess increased protease-resistance and stability at low pH for potential antimicrobial reagents. X-ray structures of VHH domains targeting Internalin B from Listeria monocytogenes, the organism responsible for the food-borne disease Listeriosis, suggest that VHH domains may act as competitive inhibitors preventing interactions with protein receptor c-Met, which is required for bacterial internalization.20 Finally, there are promising results in the use of orally administered VHH domains in place of conventional intravenous/subcutaneous therapeutic antibodies in addressing inflammatory bowel disease. A VHH targeting anti-tumor necrosis factor (TNF) alpha, V565, penetrates disrupted epithelium in IBD patients suggesting efficacy for IBD patients where the mucosal epithelial barrier is compromised.48
Diagnostics/small molecule detection
Antibodies generated against low molecular weight ligands (i.e. haptens) can be used to detect and quantitate molecules, such as environmental contaminants or medical biomarkers. Conventional antibodies have been effective frameworks for targeting haptens. Structural data have revealed that hapten binding pockets in conventional antibodies are typically located at the VH and VL interface, where each variable domain’s CDRs produce a binding cavity that allows significant surface area for interaction with hapten targets.49 While there is extensive structural data on VHH domains in complex with protein antigens, less is known about how VHH domains may overcome the lack of a VL domain in their interactions with haptens.
Despite only a handful of hapten/VHH structures, the single domain architecture of VHH is quite versatile in hapten recognition. The earliest structures, which involved two anti-hapten VHH domains targeting two different azo-dyes (reactive red 1 and 6), revealed that haptens interacted with the interface analogous to the VH domain from conventional VH/VL-hapten complexes.9,50 Despite the lack of an adjacent VL domain, the hapten/VHH structures revealed the extended CDR3 (and CDR2/3) are able to form sufficient binding pockets for low molecular weight hapten ligands (Figure 3). Notably, the observed binding dissociation constant for anti-RR6 VHH was ∼20 nM, although the affinity is likely enhanced due to two copper ions within RR6 (a rather rare component for a hapten), which interact with two histidine residues.50 An anti-caffeine VHH,28 which was demonstrated to bind caffeine in an unexpected 2:1 stoichiometry,51 was observed to bind caffeine in a matter analogous to conventional VH/VL hapten-antibody complexes, where caffeine is sandwiched between two VHH domains and their CDR3 loops.52
Figure 3.
Example of observed binding pockets of anti-hapten VHH complexes. The hapten interactions are found in the former VH/VL interface at the convergence of CDR3, CDR2, CDR1 or, alternatively, under CDR1. Grey sticks represent haptens from six hapten/VHH structures superimposed onto anti-methotrexate VHH complex. CDR1-green; CDR2-orange, CDR3-red; grey spheres-copper ions from reactive red dye RR6. PDB IDs (1I3U, 1QD0, 3QXT, 5VL2, 5VM0, and 5VM6). Image generated using PyMol.16(A color version of this figure is available in the online journal.)
Structural studies of an anti-methotrexate VHH53 uncovered a new binding pocket for hapten/VHH interactions.49 Structures of two different anti-methotrexate CDR graft variants revealed a noncanonical hapten recognition site, whereby methotrexate inserted itself under and through the CDR1 loop (Figure 3). This “threading” of the ligand under CDR1 resulted in significant hapten surface area burial (∼90%), which was greater than that found in many conventional anti-hapten VH/VL antibodies, likely contributing to the observed potent affinity (KD ∼5 nM) and specificity. In addition to intermolecular interactions between methotrexate and both CDR1 and VHH framework residues, “CDR4,” the non-hypervariable loop that is near the three CDRs, makes important energetic contributions towards methotrexate binding. Interestingly, structures of three different VHH domains in complex with triclocarban,54 an antibacterial/antifungal agent, as well as structures of an anti-cortisol VHH complex,55 revealed that the haptens bound the same tunnel beneath CDR1 as found in the anti-methotrexate VHH. These results suggest that the CDR1 “tunnel” may be a common mechanism for VHH interactions and may serve as a template for designing novel anti-hapten VHH domains.
Engineering a new generation of VHH domains
VHH domain development
With growing interest in the use of single domain antibodies for antibody applications, new methods to generate or enhance the VHH scaffold have emerged. Despite the single domain architecture, VHH domains have been amenable to CDR grafting, one of the frequently used antibody engineering approaches, where the binding affinity/specificity from one VHH is transferred to another VHH framework through substituting CDR loop residues.51,56 An exciting hybrid approach was successfully pursued by grafting the CDRs from a conventional antibody VH domain to a VHH domain, followed by affinity maturation against the target molecule, ultimately resulting in a nanomolar anti-fluorescein VHH.57
The tolerance of the VHH framework to accept CDR grafts, while maintaining function, is a notable characteristic for VHH development. Related efforts have explored the development of universal scaffolds/frameworks that possess high thermodynamic stability and expression levels, along with modifications that allow the VHH domain to maintain binding activity in reducing environments for intracellular use (i.e. intrabodies).56 The insertion of a non-canonical disulfide within the VHH framework can increase the protein’s melting temperature,58 and is an easy to introduce modification that may increase ambient shelf life, an important characteristic for potential anti-toxin VHH therapeutics.59 Key residues have also been identified within VHH domains for facile humanization for therapeutic use.34
A common objective after the initial generation of an antibody targeting an antigen of interest includes affinity maturation, whereby one or more protein engineering strategies are used to increase the antibody’s affinity for the antigen, often resulting in the improvement of binding constants by 10-fold or more. Typically, affinity maturation of antibodies involves modifying the gene sequences encoding the residues of the CDR loops to introduce or optimize interactions (e.g., hydrogen bonds, salt bridges, van der Waals) with the antigen. While various molecular biology methodologies can be employed for non-targeted (e.g. error prone PCR60) and targeted mutagenesis, such as degenerate oligonucleotides (e.g., hard19 or soft61 randomization), advances in DNA synthesis technologies have facilitated precise control of amino acid diversity (e.g. saturation mutagenesis). Generation of mutant sequences can be accomplished through scanning and screening individual residues one at a time or through varying multiple residues simultaneously through combinatorial libraries and in vitro display methods (e.g. phage, yeast, ribosome, etc.). More recent review of such experimental methodologies has been discussed by Tabasinezhad et al.62 and Lim et al.63 Furthermore, to complement experimental techniques, structural and computational techniques continue to emerge and evolve to aid in the affinity maturation process.
Koide et al. provided evidence that the VHH framework and antigen interface are suitable for affinity maturation. Using a structure-guided, two-step library design/selection approach, they produced an anti-RNase A VHH variant possessing a picomolar dissociation constant with an 100-fold enhancement over the original antibody.19 Tiller et al.64 utilized natural diversity mutagenesis and yeast display combined with high-resolution crystal structure and computational modeling to improve the affinity of an anti-alpha-synuclein c-terminal peptide binding VHH by more than 5-fold. Cheng et al. employed homology modeling and molecular docking to computationally affinity mature an anti-CD47 VHH, Nb02, whereby the authors identified a series of variants that improved the affinity ranging from more than 3-fold to >87-fold. The lead VHH variant displaying the >87-fold increase in affinity simultaneously exhibited an increase in thermostability by more than 7°C.65
Development of synthetic VHH libraries
While immunization remains the most common approach to produce conventional or heavy chain-only antibodies, there is a growing interest in synthetic, or “test tube” antibody development. The ability to use synthetic libraries with in vitro selection is advantageous for target antigens that may be toxic, pathogenic, possess low immunogenicity/conserved epitopes, or are just generally difficult to work with. Synthetic antibodies and in vitro selection can even be used to target specific conformations of antigens,66 allowing not only structural insight, but energetic details of conformational transitions in proteins.67 While synthetic antibody libraries based on conventional Fab antibody fragments have become routine, recent VHH-based synthetic libraries are showing promise. While early synthetic efforts explored distinct regions of the VHH antigen interface, such as a synthetic phage display library that randomized the CDR3 loop68 of a universal VHH framework,56 more recent efforts by Zimmermann et al. developed a combined ribosome and phage display approach that mimics common VHH paratopes (e.g. concave, convex and loop/protrusive) enabling the generation of conformationally selective VHH domains targeting membrane proteins.69 A synthetic library of humanized VHH domains was developed, termed NaLi-H1, which was capable of providing highly functional VHH antibodies70; however, access to the synthetic library is only available through contract work with the commercial provider.
The Kruse and Manglik labs have taken synthetic VHH libraries a step further, with the development of a robust yeast surface display platform.71 In generating this library, the authors took a structure-based approach, using over 90 VHH structures to identify positions of variation in the CDRs. This approach appears to have diminished the population of the library that does not fold into soluble VHH domains, a known potential drawback of synthetic libraries that can reduce the total diversity of clones that can be sampled in the display approach (e.g. yeast and phage display). The synthetic library allowed the authors to identify VHH domains capable of binding a range of antigens, including soluble proteins, non-purified proteins and two human G protein-coupled receptors (GPCRs). Notably, this library has been deposited at Kerafast (Cat. No. EF0014-FP) and is available to the non-profit scientific community for a nominal fee.
Conformation specific VHH
A continually evolving application of VHH domains involves facilitating both biological and structural studies. Due to their small size, elongated H3 loops, array of paratope shape complementarity possibilities, and amenability to synthetic library development (discussed above), VHH show significant promise for their ability to stabilize antigens in different functionally relevant conformations. Consequently, the ability for VHH to stabilize GPCRs and enable conformation specific structural determination has provided invaluable insight into GPCR function.72 Rasmussen et al. identified a llama-derived VHH, termed Nb80, which stabilized the active conformation of the β2 adrenergic receptor (β2AR) and facilitated the first X-ray structure of the active conformation.18 Che et al.73 identified an active state binding VHH to κ-opioid receptor which lead to the active state crystal structure in complex with the small molecule agonist MP1104. McMahon et al. identified conformationally specific VHH domains against two different GPCRs (β2AR and A2A adenosine receptor) where the β2AR active state specific clone Nb.c200 was shown to co-crystalize in a range of conditions using lipidic cubic-phase.71 Moreover, using the same VHH library, Wingler et al.74 identified and affinity matured an active state stabilizing VHH to angiotensin II type 1 receptor, which resulted in a co-complex structure.
Conformationally selective VHH have been identified that target non-GPCR antigens, including proteases and integral membrane receptors. In one example, Kromann-Hansen et al.75 identified two VHHs to urokinase-type plasminogen activator (uPA) where one VHH is a competitive active-site inhibitor and other an allosteric modulator. Biophysical measurements combined with the VHH co-complex X-ray structures contributed to the understanding of an equilibrium between an antiparallel-to-parallel conformation of uPA. Zimmermann et al.69 identified VHHs with different paratope geometries (shape complementarity) to lock the ABC transporter TM287/288 in an outward facing, ATPase activity-inhibited, ATP bound state and also identified VHH targeting the inhibited conformation of both glycine transporter 1 and equilibrative nucleoside transporter 1.
Generation of conformation specific VHH domains has not only greatly advanced our understanding of GPCR biology, but also furthered our knowledge of integral membrane receptor biology. Such understandings can aid in the discovery of new drugs by providing both molecular details of activation states and allowing the use of VHH domains as reagents to lock receptors in druggable conformations. More thorough reviews of conformationally selective VHH have been discussed.72,76
Engineering new function (pH and ligand control)
The utility of antibodies as affinity reagents stems from their high affinity/specificity for their respective target molecules. Consequently, most antibody engineering aims to enhance affinity, specificity, or stability, and the latter extends the high affinity/specificity over elevated temperatures or adverse environmental conditions. A logical next step in engineering protein interactions is the introduction of reversible regulation. Perhaps not surprising, methods to accomplish such regulation may be modeled after commonly observed linked-equilibria found in biological systems.
One of the most common types of linked-equilibria that can regulate protein function/interactions is proton-linkage.77 Changes in protonation states of ionizable residues upon protein interactions produce pH-dependent effects, such as changes in observed binding constants. Consequently, the introduction of ionizable groups into antibody interfaces can pave the way for pH-dependent recognition. One of the earliest examples of pH-dependent engineering of an antibody/antigen interaction included the substitution of a histidine residue within a conventional antibody’s antigen binding interface, resulting in pH-dependent binding.78 Therapeutic antibodies that display pH-dependent antigen recognition have been linked with increased serum half-life, through improved antibody recycling.79,80 Murtaugh et al.81 developed a combinatorial histidine scanning library approach that enabled the generation of a highly pH-dependent anti-RNaseA VHH domains. An engineered variant is estimated as possessing an approximate 106-fold drop in the observed binding constant from pH 7 to 4 (while maintaining their native structures). While the ability to reversibly control VHH–antigen interactions may be of interest for therapeutic antibody recycling, it also opens additional application opportunities. For instance, an anti-methotrexate M36H mutant aided the use of VHH domains in immunoaffinity chromatography for the separation of methotrexate derivatives.82 It is likely that other non-pH-based methods to regulate/control VHH interactions may be of interest for life science applications. For example, combinatorial libraries allowed the generation of dual-specific VHH variants, where antigen binding could be regulated by binding of a metal ion.83,84 While in their infancy, the development and applications of tailored VHH/target regulation may open exciting possibilities for molecular-based control of antibody affinity reagents.
Summary
The camelid VHH domain, from heavy-chain-only antibodies, is a versatile tool for life science applications. Bolstered by its small size, modular shape, and diverse set of CDR interactions against protein and hapten targets, VHH domains provide opportunities not possible using conventional antibodies. VHH modularity provides opportunities for building blocks in antibody therapies, such as multi-specifics and CAR T cell therapy, as well as agile conformationally selective binding agents. The development and increasing use of synthetic VHH libraries, with user-defined diversity, along with the ability to introduce molecular switches, such as pH control, will allow VHH domains to play a pivotal role as functional “add-ons” to create next-generation therapeutics and affinity reagents.
ACKNOWLEDGMENTS
We would like to thank N. Patel and T. Laughlin for their review and comments on this manuscript.
Authors’ contributions
All authors participated in the writing and production of figures, along with the review of the manuscript.
DECLARATION OF CONFLICTING INTERESTS
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
This work was supported in part by the NIH (R15GM124607) and NSF (MCB0953323).
References
- 1.Grilo AL, Mantalaris A. The increasingly human and profitable monoclonal antibody market. Trends Biotechnol 2019; 37:9–16 [DOI] [PubMed] [Google Scholar]
- 2.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N, Hamers R. Naturally occurring antibodies devoid of light chains. Nature 1993; 363:446–8 [DOI] [PubMed] [Google Scholar]
- 3.Muyldermans S, Cambillau C, Wyns L. Recognition of antigens by single-domain antibody fragments: the superfluous luxury of paired domains. Trends Biochem Sci 2001; 26:230–5 [DOI] [PubMed] [Google Scholar]
- 4.Muyldermans S. Single domain camel antibodies: current status. J Biotechnol 2001; 74:277–302 [DOI] [PubMed] [Google Scholar]
- 5.Greenberg AS, Avila D, Hughes M, Hughes A, McKinney EC, Flajnik MF. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature 1995; 374:168–73 [DOI] [PubMed] [Google Scholar]
- 6.Flajnik MF, Deschacht N, Muyldermans S. A case of convergence: why did a simple alternative to canonical antibodies arise in sharks and camels? PLoS Biol 2011; 9:e1001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.ULC CCG. Molecular Operating Environment (MOE). 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7: Chemical Computing Group ULC, 2019
- 8.Nguyen VK, Muyldermans S, Hamers R. The specific variable domain of camel heavy-chain antibodies is encoded in the germline. J Mol Biol 1998; 275:413–8 [DOI] [PubMed] [Google Scholar]
- 9.Spinelli S, Tegoni M, Frenken L, van Vliet C, Cambillau C. Lateral recognition of a dye hapten by a llama VHH domain. J Mol Biol 2001; 311:123–9 [DOI] [PubMed] [Google Scholar]
- 10.Muyldermans S, Atarhouch T, Saldanha J, Barbosa JA, Hamers R. Sequence and structure of VH domain from naturally occurring camel heavy chain immunoglobulins lacking light chains. Protein Eng Des Sel 1994; 7:1129–35 [DOI] [PubMed] [Google Scholar]
- 11.Wu TT, Johnson G, Kabat EA. Length distribution of CDRH3 in antibodies. Proteins 1993; 16:1–7 [DOI] [PubMed] [Google Scholar]
- 12.Liang Z, Wang T, Sun Y, Yang W, Liu Z, Fei J, Guo Y, Ma Q, Pan Q, Ren L. A comprehensive analysis of immunoglobulin heavy chain genes in the Bactrian camel (Camelus bactrianus). Front Agr Sci Eng 2015; 2:249–59 [Google Scholar]
- 13.Decanniere K, Desmyter A, Lauwereys M, Ghahroudi MA, Muyldermans S, Wyns L. A single-domain antibody fragment in complex with RNase A: non-canonical loop structures and nanomolar affinity using two CDR loops. Structure 1999; 7:361–70 [DOI] [PubMed] [Google Scholar]
- 14.Desmyter A, Spinelli S, Payan F, Lauwereys M, Wyns L, Muyldermans S, Cambillau C. Three camelid VHH domains in complex with porcine pancreatic alpha-amylase – inhibition and versatility of binding topology. J Biol Chem 2002; 277:23645–50 [DOI] [PubMed] [Google Scholar]
- 15.Desmyter A, Transue TR, Ghahroudi MA, Thi MH, Poortmans F, Hamers R, Muyldermans S, Wyns L. Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nat Struct Biol 1996; 3:803–11 [DOI] [PubMed] [Google Scholar]
- 16.DeLano WL. The PyMOL molecular graphics system. San Carlos, CA, USA: DeLano Scientific, 2002 [Google Scholar]
- 17.Conrath KE, Lauwereys M, Galleni M, Matagne A, Frere JM, Kinne J, Wyns L, Muyldermans S. Beta-lactamase inhibitors derived from single-domain antibody fragments elicited in the camelidae. Antimicrob Agents Chemother 2001; 45:2807–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasmussen SGF, Choi H-J, Fung JJ, Pardon E, Casarosa P, Chae PS, DeVree BT, Rosenbaum DM, Thian FS, Kobilka TS, Schnapp A, Konetzki I, Sunahara RK, Gellman SH, Pautsch A, Steyaert J, Weis WI, Kobilka BK. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature 2011; 469:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koide A, Tereshko V, Uysal S, Margalef K, Kossiakoff AA, Koide S. Exploring the capacity of minimalist protein interfaces: interface energetics and affinity maturation to picomolar K(D) of a single-domain antibody with a flat paratope. J Mol Biol 2007; 373:941–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King MT, Huh I, Shenai A, Brooks TM, Brooks CL. Structural basis of VHH-mediated neutralization of the food-borne pathogen Listeria monocytogenes. J Biol Chem 2018; 293:13626–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubala MH, Kovtun O, Alexandrov K, Collins BM. Structural and thermodynamic analysis of the GFP:GFP-nanobody complex. Protein Sci 2010; 19:2389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Genst E, Silence K, Decanniere K, Conrath K, Loris R, Kinne J, Muyldermans S, Wyns L. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc Natl Acad Sci USA 2006; 103:4586–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirchhofer A, Helma J, Schmidthals K, Frauer C, Cui S, Karcher A, Pellis M, Muyldermans S, Casas-Delucchi CS, Cardoso MC, Leonhardt H, Hopfner KP, Rothbauer U. Modulation of protein properties in living cells using nanobodies. Nat Struct Mol Biol 2010; 17:133–8 [DOI] [PubMed] [Google Scholar]
- 24.Mitchell LS, Colwell LJ. Comparative analysis of nanobody sequence and structure data. Proteins 2018; 86:697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franco EJ, Sonneson GJ, DeLegge TJ, Hofstetter H, Horn JR, Hofstetter O. Production and characterization of a genetically engineered anti-caffeine camelid antibody and its use in immunoaffinity chromatography. J Chromatograph B 2010; 878:177–86 [DOI] [PubMed] [Google Scholar]
- 26.Goldman ER, Liu JL, Zabetakis D, Anderson GP. Enhancing stability of camelid and shark single domain antibodies: an overview. Front Immunol 2017; 8:865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Linden RH, Frenken LG, de Geus B, Harmsen MM, Ruuls RC, Stok W, de Ron L, Wilson S, Davis P, Verrips CT. Comparison of physical chemical properties of llama VHH antibody fragments and mouse monoclonal antibodies. Biochim Biophys Acta 1999; 1431:37–46 [DOI] [PubMed] [Google Scholar]
- 28.Ladenson RC, Crimmins DL, Landt Y, Ladenson JH. Isolation and characterization of a thermally stable recombinant anti-caffeine heavy-chain antibody fragment. Anal Chem 2006; 78:4501–8 [DOI] [PubMed] [Google Scholar]
- 29.Kunz P, Zinner K, Mücke N, Bartoschik T, Muyldermans S, Hoheisel JD. The structural basis of nanobody unfolding reversibility and thermoresistance. Sci Rep 2018; 8:7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steeland S, Vandenbroucke RE, Libert C. Nanobodies as therapeutics: big opportunities for small antibodies. Drug Discov Today 2016; 21:1076–113 [DOI] [PubMed] [Google Scholar]
- 31.De Vlieger D, Ballegeer M, Rossey I, Schepens B, Saelens X. Single-domain antibodies and their formatting to combat viral infections. Antibodies 2018; 8:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klarenbeek A, El Mazouari K, Desmyter A, Blanchetot C, Hultberg A, de Jonge N, Roovers RC, Cambillau C, Spinelli S, Del-Favero J, Verrips T, de Haard HJ, Achour I. Camelid Ig V genes reveal significant human homology not seen in therapeutic target genes, providing for a powerful therapeutic antibody platform. MAbs 2015; 7:693–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snoeck V. Current experience in immunogenicity assessment of next generation biologics nanobodies. In: European immunogenicity symposium, München, Germany, February 2013
- 34.Vincke C, Loris R, Saerens D, Martinez-Rodriguez S, Muyldermans S, Conrath K. General strategy to humanize a camelid single-domain antibody and identification of a universal humanized nanobody scaffold. J Biol Chem 2009; 284:3273–84 [DOI] [PubMed] [Google Scholar]
- 35.Harmsen MM, Van Solt CB, Fijten HP, Van Setten MC. Prolonged in vivo residence times of llama single-domain antibody fragments in pigs by binding to porcine immunoglobulins. Vaccine 2005; 23:4926–34 [DOI] [PubMed] [Google Scholar]
- 36.Hoefman S, Ottevaere I, Baumeister J, Sargentini-Maier ML. Pre-clinical intravenous serum pharmacokinetics of albumin binding and non-half-life extended nanobodies®. Antibodies 2015; 4:141 [Google Scholar]
- 37.Beirnaert E, Desmyter A, Spinelli S, Lauwereys M, Aarden L, Dreier T, Loris R, Silence K, Pollet C, Cambillau C, de Haard H. Bivalent llama single-domain antibody fragments against tumor necrosis factor have picomolar potencies due to intramolecular interactions. Front Immunol 2017; 8:867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding L, Tian C, Feng S, Fida G, Zhang C, Ma Y, Ai G, Achilefu S, Gu Y. Small sized EGFR1 and HER2 specific bifunctional antibody for targeted cancer therapy. Theranostics 2015; 5:378–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Witteloostuijn SB, Pedersen SL, Jensen KJ. Half-Life extension of biopharmaceuticals using chemical methods: alternatives to PEGylation. Chem Med Chem 2016; 11:2474–95 [DOI] [PubMed] [Google Scholar]
- 40.Wu Y, Jiang S, Ying T. Single-Domain antibodies as therapeutics against human viral diseases. Front Immunol 2017; 8:1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jamnani FR, Rahbarizadeh F, Shokrgozar MA, Mahboudi F, Ahmadvand D, Sharifzadeh Z, Parhamifar L, Moghimi SM. T cells expressing VHH-directed oligoclonal chimeric HER2 antigen receptors: towards tumor-directed oligoclonal T cell therapy. Biochim Biophys Acta 2014; 1840:378–86 [DOI] [PubMed] [Google Scholar]
- 42.De Munter S, Ingels J, Goetgeluk G, Bonte S, Pille M, Weening K, Kerre T, Abken H, Vandekerckhove B. Nanobody based dual specific CARs. Int J Mol Sci 2018; 19:pii:E403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cardoso FM, Ibanez LI, Van den Hoecke S, De Baets S, Smet A, Roose K, Schepens B, Descamps FJ, Fiers W, Muyldermans S, Depicker A, Saelens X. Single-domain antibodies targeting neuraminidase protect against an H5N1 influenza virus challenge. J Virol 2014; 88:8278–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rossey I, Gilman MSA, Kabeche SC, Sedeyn K, Wrapp D, Kanekiyo M, Chen M, Mas V, Spitaels J, Melero JA, Graham BS, Schepens B, McLellan JS, Saelens X. Potent single-domain antibodies that arrest respiratory syncytial virus fusion protein in its prefusion state. Nat Commun 2017; 8:14158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laursen NS, Friesen RHE, Zhu X, Jongeneelen M, Blokland S, Vermond J, van Eijgen A, Tang C, van Diepen H, Obmolova G, van der Neut Kolfschoten M, Zuijdgeest D, Straetemans R, Hoffman RMB, Nieusma T, Pallesen J, Turner HL, Bernard SM, Ward AB, Luo J, Poon LLM, Tretiakova AP, Wilson JM, Limberis MP, Vogels R, Brandenburg B, Kolkman JA, Wilson IA. Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin. Science 2018; 362:598–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harmsen MM, van Solt CB, van Zijderveld-van Bemmel AM, Niewold TA, van Zijderveld FG. Selection and optimization of proteolytically stable llama single-domain antibody fragments for oral immunotherapy. Appl Microbiol Biotechnol 2006; 72:544–51 [DOI] [PubMed] [Google Scholar]
- 47.Hussack G, Riazi A, Ryan S, van Faassen H, MacKenzie R, Tanha J, Arbabi-Ghahroudi M. Protease-resistant single-domain antibodies inhibit Campylobacter jejuni motility. Protein Eng Des Sel 2014; 27:191–8 [DOI] [PubMed] [Google Scholar]
- 48.Crowe JS, Roberts KJ, Carlton TM, Maggiore L, Cubitt MF, Clare S, Harcourt K, Reckless J, MacDonald TT, Ray KP, Vossenkämper A, West MR. Preclinical development of a novel, orally-administered anti-tumour necrosis factor domain antibody for the treatment of inflammatory bowel disease. Sci Rep 2018; 8:4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fanning SW, Horn JR. An anti-hapten camelid antibody reveals a cryptic binding site with significant energetic contributions from a nonhypervariable loop. Protein Sci 2011; 20:1196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spinelli S, Frenken LGJ, Hermans P, Verrips T, Brown K, Tegoni M, Cambillau C. Camelid heavy-chain variable domains provide efficient combining sites to haptens. Biochemistry 2000; 39:1217–22 [DOI] [PubMed] [Google Scholar]
- 51.Sonneson GJ, Horn JR. Hapten-Induced dimerization of a Single-Domain VHH camelid antibody. Biochemistry 2009; 48:6693–5 [DOI] [PubMed] [Google Scholar]
- 52.Lesne J, Chang H-J, De Visch A, Paloni M, Barthe P, Guichou J-F, Mayonove P, Barducci A, Labesse G, Bonnet J, Cohen-Gonsaud M. Structural basis for chemically-induced homodimerization of a single domain antibody. Sci Rep 2019; 9:1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alvarez-Rueda N, Behar G, Ferre V, Pugniere M, Roquet F, Gastinel L, Jacquot C, Aubry J, Baty D, Barbet J, Birkle S. Generation of llama single-domain antibodies against methotrexate, a prototypical hapten. Mol Immunol 2007; 44:1680–90 [DOI] [PubMed] [Google Scholar]
- 54.Tabares-da Rosa S, Wogulis LA, Wogulis MD, Gonzalez-Sapienza G, Wilson DK. Structure and specificity of several triclocarban-binding single domain camelid antibody fragments. J Mol Recognit 2019; 32:e2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding L, Wang Z, Zhong P, Jiang H, Zhao Z, Zhang Y, Ren Z, Ding Y. Structural insights into the mechanism of single domain VHH antibody binding to cortisol. FEBS Lett 2019; 593:1248–56 [DOI] [PubMed] [Google Scholar]
- 56.Saerens D, Pellis M, Loris R, Pardon E, Dumoulin M, Matagne A, Wyns L, Muyldermans S, Conrath K. Identification of a universal VHH framework to graft non-canonical antigen-binding loops of camel single-domain antibodies. J Mol Biol 2005; 352:597–607 [DOI] [PubMed] [Google Scholar]
- 57.Wagner HJ, Wehrle S, Weiss E, Cavallari M, Weber W. A Two-Step approach for the design and generation of nanobodies. Int J Mol Sci 2018; 19:pii:E3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hagihara Y, Mine S, Uegaki K. Stabilization of an immunoglobulin fold domain by an engineered disulfide bond at the buried hydrophobic region. J Biol Chem 2007; 282:36489–95 [DOI] [PubMed] [Google Scholar]
- 59.Anderson GP, Liu JH, Zabetakis D, Liu JL, Goldman ER. Thermal stabilization of anti-alpha-cobratoxin single domain antibodies. Toxicon 2017; 129:68–73 [DOI] [PubMed] [Google Scholar]
- 60.Shahi B, Mousavi Gargari SL, Rasooli I, Rajabi Bazl M, Hoseinpoor R. Random mutagenesis of BoNT/E Hc nanobody to construct a secondary phage-display library. J Appl Microbiol 2014; 117:528–36 [DOI] [PubMed] [Google Scholar]
- 61.Frei JC, Lai JR. Protein and antibody engineering by phage display. Meth Enzymol 2016; 580:45–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tabasinezhad M, Talebkhan Y, Wenzel W, Rahimi H, Omidinia E, Mahboudi F. Trends in therapeutic antibody affinity maturation: from in-vitro towards next-generation sequencing approaches. Immunol Lett 2019; 212:106–13 [DOI] [PubMed] [Google Scholar]
- 63.Lim CC, Choong YS, Lim TS. Cognizance of molecular methods for the generation of mutagenic phage display antibody libraries for affinity maturation. Int J Mol Sci 2019; 20:1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tiller KE, Chowdhury R, Li T, Ludwig SD, Sen S, Maranas CD, Tessier PM. Facile affinity maturation of antibody variable domains using natural diversity mutagenesis. Front Immunol 2017; 8:986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheng X, Wang J, Kang G, Hu M, Yuan B, Zhang Y, Huang H. Homology Modeling-Based in silico affinity maturation improves the affinity of a nanobody. Int J Mol Sci 2019; 20:4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rizk SS, Paduch M, Heithaus JH, Duguid EM, Sandstrom A, Kossiakoff AA. Allosteric control of ligand-binding affinity using engineered conformation-specific effector proteins. Nat Struct Mol Biol 2011; 18:437–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mukherjee S, Griffin DH, Horn JR, Rizk SS, Nocula-Lugowska M, Malmqvist M, Kim SS, Kossiakoff AA. Engineered synthetic antibodies as probes to quantify the energetic contributions of ligand binding to conformational changes in proteins. J Biol Chem 2018;293:2815–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yan J, Li G, Hu Y, Ou W, Wan Y. Construction of a synthetic phage-displayed nanobody library with CDR3 regions randomized by trinucleotide cassettes for diagnostic applications. J Transl Med 2014; 12:343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zimmermann I, Egloff P, Hutter CAJ, Arnold FM, Stohler P, Bocquet N, Hug MN, Huber S, Siegrist M, Hetemann L, Gera J, Gmür S, Spies P, Gygax D, Geertsma ER, Dawson RJP, Seeger MA. Synthetic single domain antibodies for the conformational trapping of membrane proteins. eLife 2018; 7:e34317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moutel S, Bery N, Bernard V, Keller L, Lemesre E, de Marco A, Ligat L, Rain JC, Favre G, Olichon A, Perez F. NaLi-H1: a universal synthetic library of humanized nanobodies providing highly functional antibodies and intrabodies. Elife 2016; 5:pii:e16228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McMahon C, Baier AS, Pascolutti R, Wegrecki M, Zheng S, Ong JX, Erlandson SC, Hilger D, Rasmussen SGF, Ring AM, Manglik A, Kruse AC. Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nat Struct Mol Biol 2018; 25:289–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Manglik A, Kobilka BK, Steyaert J. Nanobodies to study G protein–coupled receptor structure and function. Annu Rev Pharmacol Toxicol 2017; 57:19–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Che T, Majumdar S, Zaidi SA, Ondachi P, McCorvy JD, Wang S, Mosier PD, Uprety R, Vardy E, Krumm BE, Han GW, Lee M-Y, Pardon E, Steyaert J, Huang X-P, Strachan RT, Tribo AR, Pasternak GW, Carroll FI, Stevens RC, Cherezov V, Katritch V, Wacker D, Roth BL. Structure of the Nanobody-Stabilized active state of the kappa opioid receptor. Cell 2018; 172:55–67.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wingler LM, McMahon C, Staus DP, Lefkowitz RJ, Kruse AC. Distinctive activation mechanism for angiotensin receptor revealed by a synthetic nanobody. Cell 2019; 176:479–90.e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kromann-Hansen T, Louise Lange E, Peter Sørensen H, Hassanzadeh-Ghassabeh G, Huang M, Jensen JK, Muyldermans S, Declerck PJ, Komives EA, Andreasen PA. Discovery of a novel conformational equilibrium in urokinase-type plasminogen activator. Sci Rep 2017; 7:3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.De Groof TWM, Bobkov V, Heukers R, Smit MJ. Nanobodies: new avenues for imaging, stabilizing and modulating GPCRs. Mol Cell Endocrinol 2019; 484:15–24 [DOI] [PubMed] [Google Scholar]
- 77.Schonichen A, Webb BA, Jacobson MP, Barber DL. Considering protonation as a posttranslational modification regulating protein structure and function. Annu Rev Biophys 2013; 42:289–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ito W, Sakato N, Fujio H, Yutani K, Arata Y, Kurosawa Y. The his-probe method: effects of histidine residues introduced into the complementarity-determining regions of antibodies on antigen-antibody interactions at different pH values. FEBS Lett 1992; 309:85–8 [DOI] [PubMed] [Google Scholar]
- 79.Chaparro-Riggers J, Liang H, DeVay RM, Bai L, Sutton JE, Chen W, Geng T, Lindquist K, Casas MG, Boustany LM, Brown CL, Chabot J, Gomes B, Garzone P, Rossi A, Strop P, Shelton D, Pons J, Rajpal A. Increasing serum half-life and extending cholesterol lowering in vivo by engineering antibody with pH-sensitive binding to PCSK9. J Biol Chem 2012; 287:11090–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Igawa T, Ishii S, Tachibana T, Maeda A, Higuchi Y, Shimaoka S, Moriyama C, Watanabe T, Takubo R, Doi Y, Wakabayashi T, Hayasaka A, Kadono S, Miyazaki T, Haraya K, Sekimori Y, Kojima T, Nabuchi Y, Aso Y, Kawabe Y, Hattori K. Antibody recycling by engineered pH-dependent antigen binding improves the duration of antigen neutralization. Nat Biotechnol 2010; 28:1203–7 [DOI] [PubMed] [Google Scholar]
- 81.Murtaugh ML, Fanning SW, Sharma TM, Terry AM, Horn JR. A combinatorial histidine scanning library approach to engineer highly pH-dependent protein switches. Protein Sci 2011; 20:1619–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Davenport KR, Smith CA, Hofstetter H, Horn JR, Hofstetter O. Site-directed immobilization of a genetically engineered anti-methotrexate antibody via an enzymatically introduced biotin label significantly increases the binding capacity of immunoaffinity columns. J Chromatogr B Analyt Technol Biomed Life Sci 2016; 1021:114–21 [DOI] [PubMed] [Google Scholar]
- 83.Fanning SW, Murtaugh ML, Horn JR. A combinatorial approach to engineering a dual-specific metal switch antibody. Biochemistry 2011; 50:5093–5 [DOI] [PubMed] [Google Scholar]
- 84.Fanning SW, Walter R, Horn JR. Structural basis of an engineered dual-specific antibody: conformational diversity leads to a hypervariable loop metal-binding site. Protein Eng Des Sel 2014; 27:391–7 [DOI] [PubMed] [Google Scholar]