Short abstract
This mini review discusses the protein complexes comprised of the universal Notch signaling transcription factor, CSL (CBF1/Su(H)/Lag-1), and its activating or repressing transcriptional coregulation partners. Many of these complex structures have been solved crystallographically as well as undergoing extensive binding studies with wild-type and mutant variants. Notch signaling is critically important in a large variety of basic biological processes: cell proliferation, differentiation, cell cycle control to name a few. Aberrant Notch thus remains a coveted target for pharmaceutical intervention. To that end, we provide a molecular-level summary of the similarities and differences in the Notch coregulator complexes that ultimately govern these processes. We highlight a conserved binding motif that multiple superficially unrelated proteins have adopted to become involved in Notch target gene regulation. As CSL-interacting small molecules begin to be characterized, this review will provide insight to potential binding sites and differential complex disruption.
Impact statement
Proper Notch signaling regulation is informed by many distinct protein complexes involving a single nuclear effector. A decade of research into these protein complexes yields multiple crystal structures and a wealth of binding data to guide drug development for Notch-related diseases – cancer, cardiovascular, development disorders.
Keywords: Structural biology, X-ray crystallography, protein–protein interactions, Notch signaling, isothermal titration calorimetry, transcription
Introduction
Notch is a highly conserved juxtacrine signaling pathway in multicellular organisms that regulates many fundamental cellular processes. Specifically, Notch plays a key role in proliferation, differentiation, and cell fate determination during organismal development, and actively regulates tissue homeostasis in the adult organism.1,2 The pleiotropic effects of Notch signaling are likely due to the context-dependent activation or repression of a large repertoire of Notch target genes.3 Reported Notch targets include HES/HEY family members,4 NRARP,5 DELTEX,6 GATA3,7 c-MYC,8 and cyclinD.9 Improper Notch activity has been associated with several human diseases and mutations in Notch pathway components are responsible for some heritable conditions.10 The flagship example of a Notch-related disease is T-cell acute lymphoblastic leukemia (T-ALL), an aggressive leukemia that is characterized by aberrantly upregulated Notch signaling, which leads to a targeted increase of the oncogene c-MYC.11 With multiple critical functions and great potential for pharmaceutical intervention, many studies have sought to understand the pathway on the molecular level and identify reagents that modulate pathway activity.12,13
The canonical Notch pathway is activated at the cell membrane when a transmembrane ligand, of the Jagged (JAG) or Delta-Like (DLL) protein families, on the signal-sending cell interacts with a transmembrane Notch receptor on the signal-receiving cell.14 In mammals, there are four Notch receptors (NOTCH1-4) and five ligands (JAG1/2 and DLL1/3/4). The receptor-ligand binding event generates a pulling force from the signal-sending cell that induces a conformational opening of the negative regulatory region (NRR) of the receptor,15 exposing a cleavage site for a membrane-bound A Disintegrin and Metalloprotease (ADAM) family protease, which then leads to cleavage by the γ-secretase complex.16 This final cleavage event releases the intracellular domain of the Notch receptor (NICD) from the cell membrane, which then translocates to the nucleus and binds to the universal Notch transcription factor CSL [orthologs: CBF1/RBPJ (Homo sapiens), Su(H) (Drosophila melanogaster), Lag-1 (Caenorhabditis elegans)].17 A third protein from the Mastermind family (MAM, MAML1-3 in mammals) binds to form a ternary activation complex,17 which recruits higher order transcriptional activation molecular machinery to Notch target genes, e.g. the mediator complex and CREB-binding protein (CBP)/p300.18–20 Unlike other signaling pathways, there is no molecular cascade that amplifies the signal from the membrane to the nucleus; each ligand binding event leads to one ternary activation complex that can reside at a specific locus at a given time. Thus, the Notch pathway is very sensitive to gene dose.21 Before ligand-induced activation, or after NICD protein has been degraded, CSL can alternatively bind to one of several identified corepressors, behaving instead as a transcriptional repressor at Notch target gene loci.22 Among these direct CSL-binding, corepressors are SMRT/HDAC1-Associated Repressor Protein (SHARP, aka Msx2-interacting protein (MINT)],23,24 Hairless,25 Four and a Half LIM Domains Protein 1 (FHL1, aka KyoT2),26 RBPJ-Interacting and Tubulin-Associated Protein 1 (RITA1),27 and Lethal 3 Malignant Brain Tumor-Like Protein 3 (L3MBTL3).28
In this review, we will discuss the crystal structures and thermodynamic binding data of these CSL-coregulator complexes, with a focus on one widely conserved interaction motif used by most of the CSL-binding partners. Mutational studies have been extensively used to study the determinants of in vitro complex formation, which will be used to compare and contrast between the coregulators. Ultimately, we will highlight how multiple unique proteins have evolved to adopt a common binding mode to the versatile transcription factor CSL.
CSL and ternary activation complex
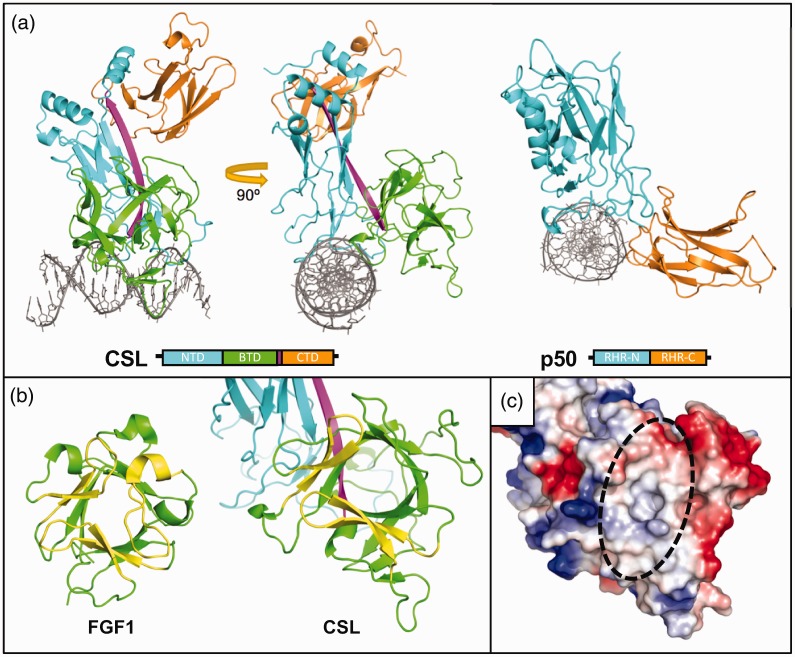
The first CSL crystal structure, Lag-1 bound to an oligonucleotide duplex, containing a binding site from the HES1 proximal promoter region, was published in 2004.29 The CSL protein is comprised of three structural domains (Figure 1(a), left, PDB: 1TTU)—the N-terminal domain (NTD), β-trefoil domain (BTD), and C-terminal domain (CTD)—which are connected by a long β-strand that integrates the individual domains into a unique singular fold. Early sequence analysis and structural studies identified similarities between CSL and the Rel homology domain (RHD) of the NF-κB and NFAT transcription factor families.29,30 The RHD has two immunoglobulin-like subdomains (Figure 1(a), right, PDB: 1NFK): the N-terminal Rel homology region (RHR-N), important for DNA binding specificity, and the C-terminal Rel homology region (RHR-C), which is associated with dimerization, other protein–protein contacts, and nonspecific DNA binding.31 Similar to RHR-N and RHR-C domains, the NTD of CSL is involved in DNA binding and the CTD is involved in protein–protein interactions. However, disparate from RHD proteins, CSL has a BTD inserted between its NTD and CTD, which is involved in both DNA binding alongside the NTD and protein–protein interactions. As we will discuss, protein partners of the BTD and/or CTD are generally distinct proteins, but some protein subdomains can bind simultaneously in a cooperative manner.
Figure 1.
Structure of CSL. (a) Left, Crystal structure of recombinant Lag-1 protein bound to 13mer DNA; PDB: 1TTU. The protein subdomains are shown schematically below. N-terminal domain (NTD, cyan), β-trefoil domain (BTD, green), and C-terminal domain (CTD, orange) are structurally connected by a single β strand (magenta). Right, NF-κB family protein p50 monomer bound to 11mer DNA; PDB: 1NFK. N-terminal Rel homology region (RHR-N, cyan) and C-terminal Rel homology region (RHR-C, orange) differ significantly structurally from the NTD and CTD of CSL due to the insertion of the BTD, with the CSL BTD adopting the DNA binding function of the RHR-C. (b) Canonical β-trefoil domain (BTD) vs. CSL. Fibroblast growth factor 1 (FGF1) has a canonical BTD with a pseudo threefold β barrel (green) capped by three hairpin motifs (yellow); PDB: 2J3P. In CSL, the BTD is missing one of the capping hairpins, exposing the interior of the protein. (c) Surface charge representation of the CSL BTD. Red is local negative charge, blue is local positive charge, and white is neutral or hydrophobic. The missing hairpin creates a hydrophobic groove in the front surface of the BTD that facilitates protein-protein interactions.
The CSL β-trefoil domain plays another major role in Notch signaling besides DNA binding, facilitated by its unique structure compared to other β-trefoil domain-containing proteins,29 including interleukin-1 and fibroblast growth factor families (Figure 1(b), PDB: 2J3P). The canonical β-trefoil fold is a β-barrel comprised of 12 β-strands with groups of 4 β-strands arranged in a pseudo threefold symmetrical arrangement, in which 3 β-hairpins form a cap structure.32 In CSL, however, two β-strands from the cap are absent on the face of the BTD that points away from the DNA, leaving a surface-exposed hydrophobic pocket that serves as a site to bind other proteins (Figure 1(c)). Importantly, early mutational studies argued for the importance of the BTD for binding to both NICD and the corepressors, specifically via its hydrophobic pocket.33,34
NICD is a large molecule that contains domains for nuclear localization, protein–protein interactions, transcriptional activation, and proteasome-targeted degradation.17 Moreover, it contains two domains important for binding to CSL and formation of the ternary activation complex with Mastermind: the RBPJ-Associated Molecule (RAM) domain and seven ankyrin repeats (ANK) domain. Originally, RAM was identified in a yeast two-hybrid screen35 as the portion of NICD that binds CSL and subsequently isothermal titration calorimetry (ITC) experiments showed a high-affinity interaction (∼20 nM Kd) between NOTCH RAM and the BTD of CSL,36,37 highlighting the importance of the RAM interaction as a first step in ternary complex formation. The isolated ANK domain of NICD in mammals shows little to no detectable binding for CSL in vitro36–38; however, ANK is absolutely required for MAM recruitment and transcriptional activation.39
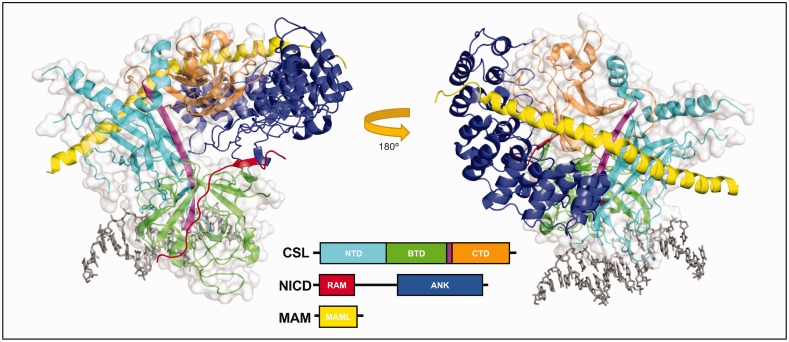
Two homologous CSL-NICD-MAM ternary activation complex crystal structures were published simultaneously, one with human proteins lacking RAM40 and the other with C. elegans proteins.41 These structures showed the overall same protein assembly and architecture (Figure 2, PDB: 2FO1), with RAM binding across the top and front of the BTD, spanning the length of the hydrophobic pocket (Figure 3(a), left). In the human protein structure containing RAM solved later,42 the RAM peptide kinks at its C-terminus, although this conformation has not been seen for any other CSL-coregulator structure and has not been functionally validated. The ANK domain of NICD binds to the CTD, with the CTD-ANK interface creating a new surface for the long α-helix of Mastermind to bind, thereby locking ANK into place and stabilizing the ternary complex.42 Consistent with the CSL-NICD-MAM ternary complex structure, site-directed mutations were introduced into the BTD (F261R, V263R, A284R, and Q333R; mouse RBPJ residue numbers) to disrupt RAM binding.43 Subsequent ITC experiments demonstrated that all of these BTD mutants significantly affect NOTCH1 and NOTCH2 RAM binding with F261 and A284 mutants having the strongest effect (see “Binding Analysis of BTD Mutants” section below).43 Moreover, these mutants have been an important tool to analyze binding and function of other coregulators that bind the BTD of CSL.
Figure 2.
Notch ternary activation complex structure. The figure shows two orientations of the core Notch activation complex involving worm proteins Lag-1 (CSL), Lin-12 (Notch intracellular domain, NICD), and Sel-8 (Mastermind, MAM) bound to 13mer DNA; PDB: 2FO1. The NTD, BTD, and CTD subdomains of CSL are colored cyan, green, and orange. NICD is shown as two CSL-interacting domains: RBP-J-Associated Molecule (RAM, red) that binds along the BTD, and ankyrin repeats (ANK, blue) that bind to the NTD/CTD. RAM binds as an elongated peptide while the seven ankyrin repeats are pairs of helices that align to create a curved structure that wraps around CSL. MAM (yellow) is a single long helix that binds with an internal kink. MAM is recruited to the complex after ANK binds and thus has contacts with ANK, CTD, and NTD.
Figure 3.
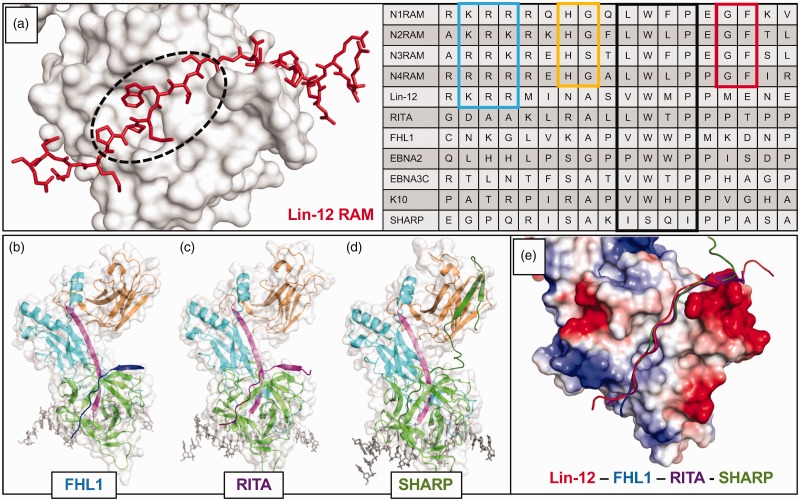
β-trefoil domain (BTD) binding notch coregulators. (a) Left, Close-up view of Lin-12 RAM domain bound to the CSL BTD; PDB: 2FO1. The dashed circle shows the “hydrophobic tetrapeptide” of the RAM domain and highlights the same hydrophobic BTD region as Figure 1(c). Right, Sequence alignment of RAM and RAM-like domains of well understood BTD binding proteins. The boxed regions of sequence are: N-terminal basic patch (light blue), H[G/S] (yellow), hydrophobic tetrapeptide (black), and GF (red). Each of these is extremely well conserved in the mammalian Notch receptor RAM domains, and they can be seen to various extents in other organisms’ Notch receptors and/or other coregulators. Specifically, the hydrophobic tetrapeptide has long served as a recognition sequence for BTD binders. (b) The crystal structure of FHL1 bound to CSL, with FHL1 peptide in blue; PDB: 4J2X. (c) The crystal structure of RITA bound to CSL, with RITA in purple; PDB: 5EG6. (d) The crystal structure of SHARP bound to CSL, with SHARP in dark green; PDB: 6DKS. SHARP binds to both the BTD (green) and the CTD (orange). (e) The similarity in BTD binding architecture of different coregulators is shown in the structural alignment of the above mentioned CSL regulation complexes. The surface charge representation of CSL illustrates how the basic N-terminus of each RAM-like domain resides near a negatively charged patch of the BTD, and the hydrophobic tetrapeptide lies along the hydrophobic patch on the front face of the BTD. Despite increasingly divergent primary sequences, the peptide backbones of BTD binders align remarkably well.
The RAM domain from all NOTCH receptor paralogs and orthologs contains several conserved features which, in some instances, are also found in other BTD-binding coregulator proteins (Figure 3(a), right). All RAM domains contain a conserved hydrophobic tetrapeptide motif (–ΦWΦP-, Φ = any hydrophobic amino acid) that makes extensive buried nonpolar contacts with the hydrophobic pocket on the BTD. Mutation of these residues results in a dramatic reduction in binding, in some cases complete loss of binding, underscoring the importance of the –ΦWΦP– in anchoring the RAM–BTD interaction.37 A stretch of basic residues upstream of the –ΦWΦP– binds a negatively charged patch on the BTD, further stabilizing this interaction. RAM domains also have two dipeptide motifs (–HG– and –GF–) flanking the –ΦWΦP– that are conserved in mammalian and fly NOTCH receptors,44 but not worms. The glycine position in the –HG– motif occupies a sterically restrictive ridge on the top of the BTD, while the histidine likely serves as another basic residue, perhaps forming a direct interaction with E260 of CSL.42 Due to the aforementioned kink of the C-terminus in the human RAM structure, there are currently no definitive binding contributions attributed to the –GF– motif. By primary sequence analysis, –ΦWΦP– motifs were readily identified in several putative BTD-binding coregulators, including EBNA2,45 EBNA3C,46 FHL1,26 K10,47 and RITA1.27 In at least two cases, the corepressors MINT/SHARP (hereafter referred to as SHARP) and L3MBTL3, there are no identifiable –ΦWΦP– motifs at all. Nonetheless, all of the mammalian corepressors that have been shown to interact with CSL, some of which the complex structures have been solved, bind along the nonpolar surface of the BTD similar to RAM rather than adopting novel binding modes.
RAM-like repression complexes (FHL1, RITA, L3MBTL3)
FHL1 represses transcription from Notch target genes by recruiting Polycomb group (PcG) proteins RING1 and HPC2.48,49 The RBPJ-FHL1-DNA crystal structure (Figure 3(b), PDB: 4J2X) was the first Notch repression complex to be determined.50 Unlike NICD, which interacts with CSL via its RAM and ANK domains, the entire interaction between FHL1 and CSL is mediated by a C-terminal peptide-like region that binds the BTD and contains the hydrophobic tetrapeptide –VWWP-.50 Structural alignment of the human CSL-NICD-MAM and mouse CSL-FHL1 complexes shows a similar backbone arrangement in the RAM and FHL1 peptides (Figure 3(e)). ITC studies determined the CSL-FHL1 binding affinity (Kd = 12 nM) to be comparable to CSL-N1RAM (Kd = 22 nM).50 When FHL1 binding was tested against the BTD mutants—F261R, V263R, A284R, and Q333R—the pattern of disrupted binding was similar, but not identical to RAM, with mutations F261R or A284R drastically reducing binding, and V263R and Q333R only modestly affecting binding.50 To further compare FHL1 and RAM interactions with CSL, ITC was performed at multiple temperatures to calculate the change in heat capacity (ΔCp) associated with binding, which correlates with the amount of buried nonpolar surface area at the binding interface. The values for CSL-RAM and CSL-FHL1 were −0.62 and −0.57 kcal/mol•K, respectively.50 These results highlight the similarity in binding modes between RAM and FHL1 and they also emphasize the critical importance of the BTD residues F261 and A284 in binding both regulators, whereas V263 and Q333 are important for RAM binding, but less so for FHL1.
RITA1 is another Notch corepressor that contains a hydrophobic tetrapeptide (–LWTP–), albeit the third residue in its motif, threonine, is less hydrophobic and more polar than the corresponding residues in RAM and FHL1.27 RITA1 binds as a single peptide along the BTD, as shown in the RBPJ-RITA1-DNA crystal structure (Figure 3(c), PDB: 5EG6)51 with the backbone of RITA in near perfect alignment with FHL1; however, the binding affinity between RITA and RBPJ is ∼1 µM, nearly 100 times weaker than RAM or FHL1.51 Despite a much weaker interaction between RITA1 and RBPJ, mutations of F261 and A284 affected RITA1 binding the most, whereas mutations at V263 and Q333 only mildly disrupted RITA1 binding, which is a pattern consistent with FHL1, but not RAM. RITA1 has several identified post-translational modification sites in its RBPJ-interacting region, including acetylation of two lysines (K131/K136) that sit near the top of the BTD, and phosphorylation of T143 in its hydrophobic tetrapeptide. Compared to wild-type RITA1, ITC showed binding retention of only 18% for the doubly acetylated peptide and 7.5% for phosphothreonine at residue 143 of RITA1, suggesting RITA1 interactions with RBPJ may be regulated by post translational modifications.51 Note that phosphorylation, which introduces further polarity in the middle of the hydrophobic tetrapeptide via the charged phosphate, causes RITA1 to bind ∼1000-fold weaker than RAM and FHL1 under similar experimental conditions.
More recently, the malignant brain tumor (MBT) protein family member L3MBTL3 was identified as a Notch pathway regulator that directly binds to CSL and recruits the repressive lysine demethylase KDM1a to Notch loci.28 ITC experiments defined a 40-residue L3MBTL3 (31–70) peptide that binds to RBPJ with an affinity of ∼0.5 µM.28 Cellular pulldown experiments showed that this interaction required the BTD of CSL and several BTD mutants discussed previously (F261R, V263R, A284R, Q333R) dramatically reduced coimmunoprecipitation (coIP), with F261R being the most deleterious to binding.28 In addition, RBPJ-L3MBTL3 coIPs performed while titrating increasing amounts of NICD1 showed a displacement of L3MBTL3 in favor of NICD1 binding, demonstrating mutually exclusive NICD1 and L3MBTL3 binding to RBPJ.28 Taken together, these results suggest that L3MBTL3 likely binds the BTD, and given other RBPJ-coregulator complex structures, the structure of L3MBTL3 bound to RBPJ is likely similar to the RAM domain of NICD. Ongoing structural studies in the Kovall lab are focused on the structural characterization of this complex. Interestingly the L3MBTL3 construct 31–70 lacks a readily identifiable –ΦWΦP–, but it does contain a tryptophan with nonpolar residues nearby (–TWMVP–), albeit it deviates from the consensus. If this motif is involved in L3MBTL3 binding to RBPJ, it may suggest that the sequence conservation rules for binding the BTD are more flexible than previously thought.
Diverging coregulators (SHARP and hairless)
The corepressor SHARP directly binds to CSL23,24 and recruits NCoR1/2 transcriptional repression complexes via its SPOC domain.52 Prior to solving the crystal structure of the RBPJ-SHARP complex, the binding mechanism was extensively studied, and it was found that SHARP binds with high affinity to both the BTD and CTD of RBPJ,53 unlike the previously described mammalian corepressors, which only bind the BTD. Using ITC, a SHARP peptide (2776–2833) was tested for binding against RBPJ, as well as several subdomain constructs: BTD, CTD, and BTD-CTD.51 SHARP binds weakly to the isolated BTD and CTD with only 41 µM and 60 µM affinity, respectively. These individual interactions are much weaker than other CSL-coregulators complexes; however, SHARP binds to the BTD-CTD construct with ∼1 µM affinity, highlighting a striking avidity effect when both domains are available for binding. Moreover, full-length RBPJ binds SHARP (2776–2833) with 11 nM affinity, which is comparable to RAM and FHL1 binding to RBPJ. The increase in binding from the BTD-CTD construct, which does not completely recapitulate binding to full-length RBPJ, is likely due to the global fold stabilization by the NTD, keeping in mind the long β-strand that integrates and orients each domain in the context of the overall fold. The BTD-CTD construct is analogous to two domains connected by a flexible linker.
The RBPJ binding portion of SHARP does not have homology with any other BTD-binding coregulators and lacks anything resembling a –ΦWΦP– motif. This fueled speculation about a potentially new mode of BTD binding, but the recently reported RBPJ-SHARP-DNA crystal structure (Figure 3(d), PDB: 6DKS) showed that SHARP uses a bipartite binding model, in which a RAM-like peptide binds across the BTD similar to other coregulators and a β-hairpin that binds the CTD.54 Structural alignment of SHARP with other corepressors that only bind the BTD (Figure 3(e)) shows a strikingly similar peptide backbone arrangement along the BTD, with a handful of residues overlapping in unexpected positions, despite the lack of any sequence identity between SHARP and other coregulators. Within the register of –ΦWΦP–, SHARP has the sequence (–ISQI–), which deviates sharply from the –ΦWΦP– rules established from other coregulators. However, preceding the –ΦWΦP–, SHARP has an alanine residue, which is conserved in FHL1, RITA, and other coregulators, and based on previous RBPJ-coregulator structures, is constrained to residues with small side chains due to steric occlusion. Following the –ΦWΦP–, SHARP has two proline residues that structurally align with the same residues in RITA. Moreover, the first proline in this diproline motif is also conserved in EBNA2, EBNA3C, K10, LIN12, and NOTCH4 RAM. Taken together, the BTD binding region of SHARP behaves like the previous coregulators, though with much overall weaker binding affinity owing to a suboptimal hydrophobic tetrapeptide motif.
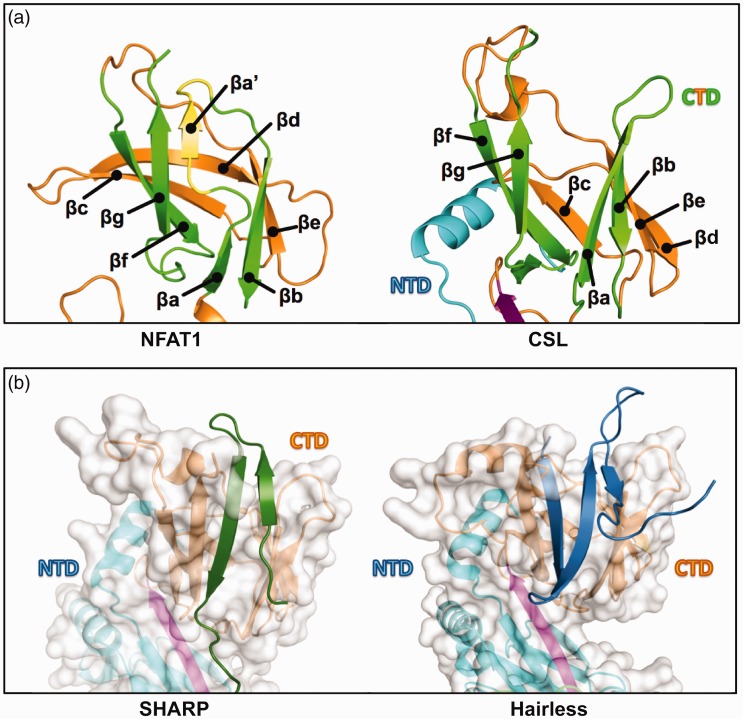
SHARP uses a β-hairpin motif to bind to the CTD of CSL (Figure 4(b), left), which is separated from the BTD-binding region by a six-residue disordered linker.54 We described above how the BTD of CSL, which is absent of two β-strands found in other canonical β-trefoil-containing proteins, has a unique coregulator binding surface. Likewise, the CSL CTD differs from canonical immunoglobulin (Ig) structures found in other RHR-C domains due to a missing βaʹ strand, which lies between strands βa and βg (Figure 4(a), PDB: 1S9K). When SHARP binds to RBPJ, a modest conformational rearrangement in the CTD accommodates the β-hairpin motif of SHARP, in which one of the SHARP β strands is positioned where βaʹ lies in a canonical RHR-C domain.54 Interestingly, the corepressor Hairless, which is the primary antagonist of Notch signaling in Drosophila and only conserved in arthropods,25 binds the CTD of CSL in a manner that is analogous to SHARP, but lacks BTD binding completely.55 Hairless interacts with CSL by inserting a three-stranded β-hairpin motif in between the two β-sheets that compose the Ig fold of the CTD (Figure 4(b), right, PDB: 5E24).55 This requires a much more dramatic structural change in CSL, which results in burial of Hairless into the hydrophobic core of CTD and facilitates high affinity (2 nM) binding.55,56 Thus, there are two different examples of how binding sites have evolved on CSL proteins, whereby loss of structural elements in canonical folds has created binding surfaces for a wide range of coregulators to interact.
Figure 4.
A second CSL binding pocket. (a) Canonical immunoglobulin (Ig) domain vs. CSL C-terminal domain (CTD) immunoglobulin fold. Left, The NFAT1 C-terminal Rel homology region (RHR-C) contains a canonical Ig fold with connected but discrete βa and βaʹ strands that form β sheet interactions with, respectively, βb and βg strands (strands are colored green for comparison, with βaʹ alone highlighted in yellow); PDB: 1S9K. Right, CSL lacks βaʹ and instead has an extended βa strand that interacts with βb but is separated from βg; PDB: 2FO1. (b) Left, the separation of βa and βg in CSL creates a protein binding cleft that SHARP (dark green) occupies for partial CSL binding. Out-of-view, SHARP extends C-terminally to bind the BTD; PDB: 6DKS. Right, Hairless (blue) also binds to the CTD cleft, but binding requires a larger conformation change to bury more protein mass; PDB: 5E24. This leads a dramatic increase in affinity (1 nM) compared to the SHARP-CTD interaction (60 µM).
Binding analysis of BTD mutants
The panoply of CSL-coregulator complex crystal structures has shown that to date all coregulators, with the exception of Hairless, incorporate a RAM-like peptide motif to bind to the BTD of CSL.22 Early structure-function studies mapped the BTD residues that were important for NICD binding, in which mutants were identified that either fully or partially disrupted CSL-coregulator complex formation.43 Subsequent ITC experiments have been performed for each CSL-coregulator complex with the following BTD variants–F261, V263, A284, and Q333—in which these residues were substituted with either an arginine residue to disrupt binding or alanine residue to remove favorable nonpolar contacts, except in the case of A284 where alanine was replaced with valine. These residues span the length of the BTD binding site (Figure 5(a) and (b), PDB: 4J2X), are surface exposed, and mutations at these sites minimally affect folding and stability of CSL. Certainly, other BTD residues contact coregulators; however, these residues also contribute to the hydrophobic core of CSL and mutating these residues are detrimental to folding/stability. Spatially, V263 sits near where the N-terminus of a coregulator would bind, followed by F261; both of these residues are part of a large β-hairpin loop that lies over the top of the binding surface, which is poorly structured in the absence of coregulator binding. A284 lies beneath this β-hairpin loop, contributing to the nonpolar BTD surface, followed by Q333, which interacts with more C-terminal regions of bound coregulators.
Figure 5.
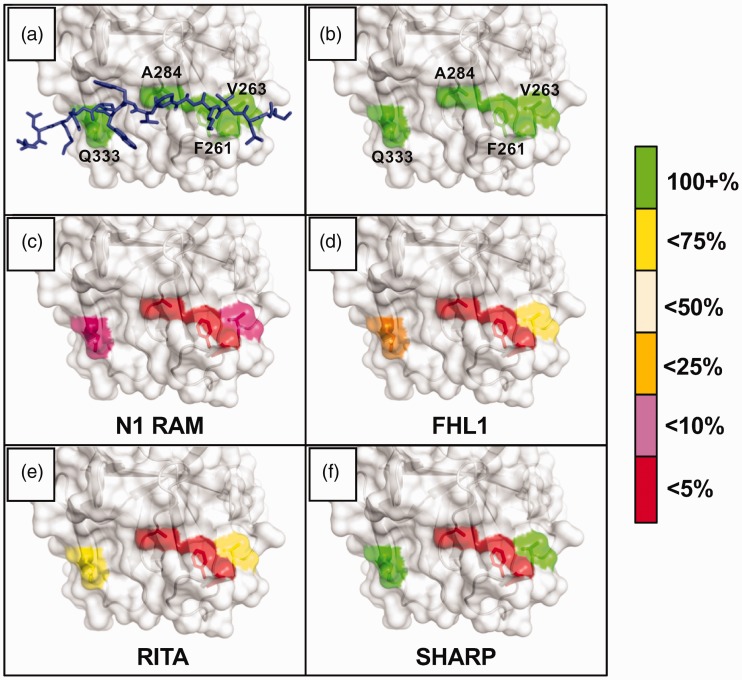
Notch coregulator binding to CSL mutants. The figure graphically depicts the results of rigorous isothermal titration calorimetry (ITC) experiments involving series of CSL BTD mutants and their effects on binding between CSL and established coregulators. The BTD residues selected for mutation—F261, V263, A284, and Q333—were chosen to disrupt RAM binding without dramatically altering CSL stability. They are shown with (a) and without (b) an overlay of the FHL1 peptide; PDB: 4J2X. The colors highlighting the individual residues reflect the binding retained after mutation, i.e. if a mutation caused the Kd to increase more than 4-fold but less than 10-fold (10–25% binding retained), the residue would be colored orange. (c) and (d) Notch1 RAM and FHL1 were tested against the arginine mutants: F261R, V263R, A284R, Q333R. Drastic decreases in binding were seen for both coregulators with the F261 and A284 mutants, illustrating the importance of the platform created by the phenylalanine sidechain, as well as the cleft afforded by the compact alanine sidechain. (e) and (f) RITA and SHARP were tested against the alanine/valine mutants: F261A, V263A, A284V, Q333A. Later experiments moved to these mutants in order to avoid introducing artifacts due to the lengthy and charged arginine sidechains.
In each of these sets of ITC binding experiments, purified coregulator peptides were titrated into purified wild-type or mutant recombinant RBPJ, typically in phosphate buffer (50 mM sodium phosphate pH 6.5, 150 mM NaCl). The data were collected at 25°C and the binding curves were fit to a one-site binding model. Collectively, because all of these CSL-coregulator interactions were analyzed under identical experimental conditions, this provides a quantitative comparison of the relative importance of specific BTD residues for binding specific coregulators, which gives additional molecular insights into the details of how different coregulators utilize the BTD-binding surface. As summarized below, we report the changes in affinity as the percentage of wild-type binding retained after mutation, i.e. a two-fold Kd increase is 50% of wild-type binding.
The RAM domains of both Notch1 and Notch2 are highly sensitive to mutations at all four BTD residues (Figure 5(c)).43 Testing against RBPJ F261R, V263R, A284R, and Q333R, Notch1 RAM retained 0.15%, 5.1%, 3.0%, and 5.8%, respectively, of the wild-type RBPJ binding affinity (22 nM). Likewise, Notch2 RAM retained 0.34%, 0.89% 1.3%, and 16% of its wild-type RBPJ affinity (32 nM). These sharp decreases are understandable as the mutants were initially designed for their ability to disrupt RAM interactions specifically based on CSL-NICD-MAM structures.41 However, given that RAM-BTD binding is the first step toward ankyrin repeat recruitment and Mastermind binding, it is interesting to note how detrimental one of several point mutants can be to complex formation and thus Notch activation.
The RAM domains from Notch1/2 differ in only a few residues, but they still respond somewhat distinctly to the CSL mutants.43 This could be directly due to the peptide sequence near specific BTD residues; for example, the basic patch of the RAM domain aligning structurally with V263 of CSL. Another possible explanation is that sequence changes anywhere along the peptide, such as replacing glutamine in Notch1 for phenylalanine in Notch2 directly upstream of the hydrophobic tetrapeptide (Figure 3(a), right), might affect the stability and dynamics of the peptide in the given experimental conditions. This would influence not only the native binding constants but might also mitigate or amplify responses to the certain BTD mutants. We have discussed how diverging sequences have led to the wide range of affinities of CSL to its binding partners, from 12 nM (Notch1) to 41 µM (the BTD-only portion of SHARP). Now we must consider that individual molecular interactions can be more or less important for binding depending on the peptide’s overall properties.
When FHL1 was tested against the arginine mutants (Figure 5(d))—F261R, V263R, A284R, and Q333R—the wild-type binding retained was 0.19%, 63%, 0.16%, and 24%, respectively.50 F261 and A284 clearly play defining roles in coregulator binding, and a closer look at the structure explains why. All BTD binding peptides essentially sit on a surface created by the phenyl ring of F261, and the compact sidechain of A284 allows space for a cleft between the top and front face of the BTD, into which the peptides bury upon binding. In other ITC experiments, the BTD arginine mutants were changed to alanine or valine mutants—F261A, V263A, A284V, Q333A—to avoid introducing electrostatic and entropic complications with the long, guanidino-containing sidechain of arginine. The pattern of RITA binding with the new mutants nevertheless followed a similar pattern to FHL1 (Figure 5(e)): 0.36%, 58%, 2.4%, and 59% of wild-type binding (0.53 µM) was retained.51 For comparison, RITA was also tested against the corresponding arginine mutants and the retained binding was 0% (no detectable binding), 41%, 9.0%, and 25%.51
SHARP binding to the alanine mutants (Figure 5(f))—F261A, V263A, A284V, Q333A—measured 2.3%, 160%, 2.0%, and 130% of the wild-type RBPJ-SHARP affinity (5 nM).53,54 Here we see the first increases in binding caused by V263 and Q333 mutations, which have given the most variable responses among the coregulators. Located near the terminal ends of any bound peptide, it appears these residues act as auxiliary stabilizers of the overall coregulator complex, likely dependent on the local peptide sequence and conformation. As we alluded to, it is perhaps not surprising that the most sequence divergent binding partner and one that binds to multiple CSL domains, SHARP, is less affected by these mutations. It is also worth reiterating that the affinities for SHARP to wild-type RBPJ and the V263A/Q333A mutants are all still 5 nM or stronger, and even the weakened binding to F261A/A284V is significantly stronger than the wild-type RITA and L3MBTL3 binding.
Conclusions
There are currently five unique Notch coregulator complex structures solved by X-ray crystallography – those with NICD/MAML, FHL1, RITA, SHARP, and Drosophila-specific Hairless, bound to CSL.4,36,50,51,54,55 As we have shown, the four mammalian conserved CSL binders have all adopted a specific binding motif to the CSL β-trefoil domain. Likewise, several other proteins are expected to bind in a similar manner based on sequence homology (EBNA3C, K10) or experimental findings (EBNA2, L3MBTL3). Notch signaling runs entirely through the nuclear actions of CSL, so it is remarkable that this transcription factor not only binds so many distinct partner proteins, but does so in a conserved, mutually exclusive manner. There are still many outstanding questions about how the pathway is regulated in light of our current understanding. How do BTD binders compete for access to CSL, specifically when the affinities between two coregulators are comparable? What role does post translational modification of one or multiple coregulators play in determining either individual or preferential binding to CSL? How restrictive are the sequence homology rules for putative BTD binders, and are there any other predictors for finding new CSL binding partners, e.g. sidechain hydrophilicity patterns in random coil regions? Notch remains the subject of intense ongoing studies: from ligand and receptor processing and binding, novel direct CSL binding proteins and higher order complex formation, and deactivating modification and degradation of NICD.1 The list we have explored here is likely incomplete and it seems less “if” but “when” and “how many” more BTD binders will ultimately be characterized.
Authors’ contributions
Both authors contributed to the composition and generation of this review. DPH wrote the manuscript and prepared the figures. RAK revised the manuscript and figures.
DECLARATION OF CONFLICTING INTERESTS
The author(s) declare no potential conflicts of interest in relation to the authorship and/or publication of this review.
FUNDING
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The authors are grateful for support from the NCI (NIH R01CA178974). DPH is supported by NIEHS training grant T32-ES-07250.
References
- 1.Kovall RA, Gebelein B, Sprinzak D, Kopan R. The canonical notch signaling pathway: structural and biochemical insights into shape, sugar, and force. Dev Cell 2017; 41:228–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray SJ. Notch signalling in context. Nat Rev Mol Cell Biol 2016; 17:722–35 [DOI] [PubMed] [Google Scholar]
- 3.Borggrefe T, Oswald F. The notch signaling pathway: transcriptional regulation at notch target genes. Cell Mol Life Sci 2009; 66:1631–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the notch signaling pathway. J Cell Physiol 2003; 194:237–55 [DOI] [PubMed] [Google Scholar]
- 5.Lamar E, Deblandre G, Wettstein D, Gawantka V, Pollet N, Niehrs C, Kintner C. Nrarp is a novel intracellular component of the notch signaling pathway. Genes Dev 2001; 15:1885–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Izon DJ, Aster JC, He Y, Weng A, Karnell FG, Patriub V, Xu L, Bakkour S, Rodriguez C, Allman D, Pear WS. Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch1. Immunity 2002; 16:231–43 [DOI] [PubMed] [Google Scholar]
- 7.Amsen D, Antov A, Jankovic D, Sher A, Radtke F, Souabni A, Busslinger M, McCright B, Gridley T, Flavell RA. Direct regulation of Gata3 expression determines the T helper differentiation potential of notch. Immunity 2007; 27:89–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, D, Bianco C, Rodriguez CG, Sai H, Tobias J, Li Y, Wolfe MS, Shachaf C, Felsher D, Blacklow SC, Pear WS, Aster JC. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev 2006; 20:2096–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ronchini C, Capobianco AJ. Induction of cyclin D1 transcription and CDK2 activity by notch(ic): implication for cell cycle disruption in transformation by notch(ic). Mol Cell Biol 2001; 21:5925–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siebel C, Lendahl U. Notch signaling in development, tissue homeostasis, and disease. Physiol Rev 2017; 97:1235–94 [DOI] [PubMed] [Google Scholar]
- 11.Sanchez-Martin M, Ferrando A. The NOTCH1-MYC highway toward T-cell acute lymphoblastic leukemia. Blood 2017; 129:1124–33 [DOI] [PubMed] [Google Scholar]
- 12.Ntziachristos P, Lim JS, Sage J, Aifantis I. From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell 2014; 25:318–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, Finkle D, Venook R, Wu X, Ridgway J, Schahin-Reed D, Dow GJ, Shelton A, Stawicki S, Watts RJ, Zhang J, Choy R, Howard P, Kadyk L, Yan M, Zha J, Callahan CA, Hymowitz SG, Siebel CW. Therapeutic antibody targeting of individual notch receptors. Nature 2010; 464:1052–7 [DOI] [PubMed] [Google Scholar]
- 14.Kopan R, Ilagan MX. The canonical notch signaling pathway: unfolding the activation mechanism. Cell 2009; 137:216–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon WR, Zimmerman B, He L, Miles LJ, Huang J, Tiyanont K, McArthur DG, Aster JC, Perrimon N, Loparo JJ, Blacklow SC. Mechanical allostery: evidence for a force requirement in the proteolytic activation of notch. Develop Cell 2015; 33:729–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musse AA, Meloty-Kapella L, Weinmaster G. Notch ligand endocytosis: mechanistic basis of signaling activity. Semin Cell Dev Biol 2012; 23:429–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovall RA, Blacklow SC. Mechanistic insights into notch receptor signaling from structural and biochemical studies. Curr Top Dev Biol 2010; 92:31–71 [DOI] [PubMed] [Google Scholar]
- 18.Fryer CJ, White JB, Jones KA. Mastermind recruits CycC:CDK8 to phosphorylate the notch ICD and coordinate activation with turnover. Mol Cell 2004; 16:509–20 [DOI] [PubMed] [Google Scholar]
- 19.Wallberg AE, Pedersen K, Lendahl U, Roeder RG. p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by notch intracellular domains in vitro. Mol Cell Biol 2002; 22:7812–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fryer CJ, Lamar E, Turbachova I, Kintner C, Jones KA. Mastermind mediates chromatin-specific transcription and turnover of the notch enhancer complex. Genes Dev 2002; 16:1397–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guruharsha KG, Kankel MW. Artavanis-Tsakonas S. The notch signalling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet 2012; 13:654–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oswald F, Kovall RA. CSL-Associated corepressor and coactivator complexes. Adv Exp Med Biol 2018; 1066:279–95 [DOI] [PubMed] [Google Scholar]
- 23.Kuroda K, Han H, Tani S, Tanigaki K, Tun T, Furukawa T, Taniguchi Y, Kurooka H, Hamada Y, Toyokuni S, Honjo T. Regulation of marginal zone B cell development by MINT, a suppressor of notch/RBP-J signaling pathway. Immunity 2003; 18:301–12 [DOI] [PubMed] [Google Scholar]
- 24.Oswald F, Kostezka U, Astrahantseff K, Bourteele S, Dillinger K, Zechner U, Ludwig L, Wilda M, Hameister H, Knöchel W, Liptay S, Schmid RM. SHARP is a novel component of the notch/RBP-Jkappa signalling pathway. EMBO J 2002; 21:5417–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maier D. Hairless: the ignored antagonist of the notch signalling pathway. Hereditas 2006; 143:212–21 [DOI] [PubMed] [Google Scholar]
- 26.Taniguchi Y, Furukawa T, Tun T, Han H, Honjo T. LIM protein KyoT2 negatively regulates transcription by association with the RBP-J DNA-binding protein. Mol Cell Biol 1998; 18:644–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wacker SA, Alvarado C, von Wichert G, Knippschild U, Wiedenmann J, Clauss K, Nienhaus GU, Hameister H, Baumann B, Borggrefe T, Knöchel W, Oswald F. RITA, a novel modulator of notch signalling, acts via nuclear export of RBP-J. EMBO J 2011; 30:43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu T, Park SS, Giaimo BD, Hall D, Ferrante F, Ho DM, Hori K, Anhezini L, Ertl I, Bartkuhn M, Zhang H, Milon E, Ha K, Conlon KP, Kuick R, Govindarajoo B, Zhang Y, Sun Y, Dou Y, Basrur V, Elenitoba-Johnson KS, Nesvizhskii AI, Ceron J, Lee CY, Borggrefe T, Kovall RA, Rual JF. RBPJ/CBF1 interacts with L3MBTL3/MBT1 to promote repression of notch signaling via histone demethylase KDM1A/LSD1. EMBO J 2017; 36:3232–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kovall RA, Hendrickson WA. Crystal structure of the nuclear effector of notch signaling, CSL, bound to DNA. EMBO J 2004; 23:3441–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nam Y, Weng AP, Aster JC, Blacklow SC. Structural requirements for assembly of the CSL.intracellular Notch1.Mastermind-like 1 transcriptional activation complex. J Biol Chem 2003; 278:21232–9 [DOI] [PubMed] [Google Scholar]
- 31.Phelps CB, Sengchanthalangsy LL, Malek S, Ghosh G. Mechanism of kappa B DNA binding by rel/NF-kappa B dimers. J Biol Chem 2000; 275:24392–9 [DOI] [PubMed] [Google Scholar]
- 32.Zhu X, Komiya H, Chirino A, Faham S, Fox GM, Arakawa T, Hsu BT, Rees DC. Three-dimensional structures of acidic and basic fibroblast growth factors. Science 1991; 251:90–3 [DOI] [PubMed] [Google Scholar]
- 33.Hsieh JJ, Henkel T, Salmon P, Robey E, Peterson MG, Hayward SD. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol 1996; 16:952–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsieh JJ, Hayward SD. Masking of the CBF1/RBPJ kappa transcriptional repression domain by Epstein-Barr virus EBNA2. Science 1995; 268:560–3 [DOI] [PubMed] [Google Scholar]
- 35.Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor notch and the transcription factor RBP-J kappa/Su(H). Curr Biol 1995; 5:1416–23 [DOI] [PubMed] [Google Scholar]
- 36.Friedmann DR, Wilson JJ, Kovall RA. RAM-induced allostery facilitates assembly of a notch pathway active transcription complex. J Biol Chem 2008; 283:14781–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lubman OY, Ilagan MX, Kopan R, Barrick D. Quantitative dissection of the notch:CSL interaction: insights into the notch-mediated transcriptional switch. J Mol Biol 2007; 365:577–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Del Bianco C, Aster JC, Blacklow SC. Mutational and energetic studies of notch 1 transcription complexes. J Mol Biol 2008; 376:131–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petcherski AG, Kimble J. Mastermind is a putative activator for notch. Curr Biol 2000; 10:R471–3 [DOI] [PubMed] [Google Scholar]
- 40.Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to notch transcription complexes. Cell 2006; 124:973–83 [DOI] [PubMed] [Google Scholar]
- 41.Wilson JJ, Kovall RA. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell 2006; 124:985–96 [DOI] [PubMed] [Google Scholar]
- 42.Choi SH, Wales TE, Nam Y, O'Donovan DJ, Sliz P, Engen JR, Blacklow SC. Conformational locking upon cooperative assembly of notch transcription complexes. Structure 2012; 20:340–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan Z, Friedmann DR, VanderWielen BD, Collins KJ, Kovall RA. Characterization of CSL (CBF-1, Su(H), lag-1) mutants reveals differences in signaling mediated by Notch1 and Notch2. J Biol Chem 2012; 287:34904–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson SE, Ilagan MX, Kopan R, Barrick D. Thermodynamic analysis of the CSL x notch interaction: distribution of binding energy of the notch RAM region to the CSL beta-trefoil domain and the mode of competition with the viral transactivator EBNA2. J Biol Chem 2010; 285:6681–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ling PD, Hayward SD. Contribution of conserved amino acids in mediating the interaction between EBNA2 and CBF1/RBPJk. J Virol 1995; 69:1944–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calderwood MA, Lee S, Holthaus AM, Blacklow SC, Kieff E, Johannsen E. Epstein-Barr virus nuclear protein 3C binds to the N-terminal (NTD) and beta trefoil domains (BTD) of RBP/CSL; only the NTD interaction is essential for lymphoblastoid cell growth. Virology 2011; 414:19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heinzelmann K, Scholz BA, Nowak A, Fossum E, Kremmer E, Haas J, Frank R, Kempkes B. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor 4 (vIRF4/K10) is a novel interaction partner of CSL/CBF1, the major downstream effector of notch signaling. J Virol 2010; 84:12255–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qin H, Du D, Zhu Y, Li J, Feng L, Liang Y, Han H. The PcG protein HPC2 inhibits RBP-J-mediated transcription by interacting with LIM protein KyoT2. FEBS Lett 2005; 579:1220–6 [DOI] [PubMed] [Google Scholar]
- 49.Qin H, Wang J, Liang Y, Taniguchi Y, Tanigaki K, Han H. RING1 inhibits transactivation of RBP-J by notch through interaction with LIM protein KyoT2. Nucleic Acids Res 2004;1492–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Collins KJ, Yuan Z, Kovall RA. Structure and function of the CSL-KyoT2 corepressor complex: a negative regulator of notch signaling. Structure 2014; 22:70–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tabaja N, Yuan Z, Oswald F, Kovall RA. Structure-function analysis of RBP-J-interacting and tubulin-associated (RITA) reveals regions critical for repression of notch target genes. J Biol Chem 2017; 292:10549–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ariyoshi M, Schwabe JW. A conserved structural motif reveals the essential transcriptional repression function of spen proteins and their role in developmental signaling. Genes Dev 2003; 17:1909–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.VanderWielen BD, Yuan Z, Friedmann DR, Kovall RA. Transcriptional repression in the notch pathway: thermodynamic characterization of CSL-MINT (Msx2-interacting nuclear target protein) complexes. J Biol Chem 2011; 286:14892–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan Z, VanderWielen BD, Giaimo BD, Pan L, Collins CE, Turkiewicz A, Hein K, Oswald F, Borggrefe T, Kovall RA. Structural and functional studies of the RBPJ-SHARP complex reveal a conserved corepressor binding Site. Cell Rep 2019; 26:845–54 e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuan Z, Praxenthaler H, Tabaja N, Torella R, Preiss A, Maier D, Kovall RA. Structure and function of the Su(H)-hairless repressor complex, the major antagonist of notch signaling in Drosophila melanogaster. PLoS Biol 2016; 14:e1002509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maier D, Kurth P, Schulz A, Russell A, Yuan Z, Gruber K, Kovall RA, Preiss A. Structural and functional analysis of the repressor complex in the notch signaling pathway of Drosophila melanogaster. Mol Biol Cell 2011; 22:3242–52 [DOI] [PMC free article] [PubMed] [Google Scholar]