Short abstract
ADAM (a disintegrin and metalloproteinase) proteins are type-1 transmembrane and secreted proteins that function in cell adhesion and signal transduction. Here we review the structural features of ADAM proteins that direct their biological functions in ectodomain shedding and cell adhesion.
Impact statement
Recent structural advances have provided a deeper appreciation for interdomain relationships that modulate the activity of ADAM proteins in ectodomain shedding and cellular adhesion. Our review covers these new findings, and places them into historical context. The new results make clear that the metalloproteinase domain works in combination with its ancillary domains to execute its biological function. The ADAM ectodomain is dynamic, and accesses conformations that require interdomain movements during its enzymatic “lifecycle.” Fundamental questions about ADAM activation and substrate selection, however, still remain unanswered. Elucidating the biochemical and structural basis for ADAM regulation will be an exciting avenue of future research that should greatly advance our understanding of ADAM function in biology and human pathogenesis.
Keywords: Metalloproteinase, structural biology, biophysics, signal transduction, notch signaling, ectodomain shedding
Introduction: History and overview of ADAMs
ADAM (a disintegrin and metalloproteinase) proteins are type-1 transmembrane and secreted proteins that function in cell adhesion and signal transduction. PH30α, the founding member of this family, was initially identified as a protein implicated in the process of sperm-egg fusion.1 It is now evident that ADAM proteins are found throughout the phylum Chordata, with expansion of the family in vertebrate evolution producing a total of 21 ADAM genes in humans.2
ADAMs are widely expressed in mammalian tissues, and exhibit a wide range of different biological functions. Knockouts of individual ADAM proteins have implicated ADAM function in fertilization, cell differentiation, immunity, angiogenesis, and development of epithelial and nervous tissue. Accordingly, dysfunctional ADAM activity is observed in a variety of pathophysiological conditions such as cancer,3 infertility,4 chronic immunity,5 Alzheimer’s disease,6 asthma, and epilepsy.7
Of the 21 human ADAM proteins, 13 are known or predicted to be catalytically active (ADAM8, -9, -10, -12, -15, -17, -19, -20, -21, -28, -30, -33, and -DEC1), whereas 8 ADAMs (ADAM2, -7, -11, -18, -22, -23, -29, and -32) are predicted to be catalytically inactive.8 Catalytically active ADAM proteins that are membrane-tethered typically result in ectodomain shedding, the irreversible processing of a membrane-associated protein to release its ectodomain into the extracellular or luminal space.9
ADAM10 and ADAM17 are the most extensively studied proteins among ADAM family members. Both of these proteins carry out critical functions in mammalian development. ADAM17 was originally identified by Moss et al. as the enzyme that catalyzes cleavage of the TNFα precursor, and dubbed “TACE,” short for TNFα-converting enzyme.10,11 Characterization of ADAM17 knockout mice, however, revealed defects in the developing epithelium and mice born with open eyelids secondary to a loss in EGFR signaling.12,13 ADAM17 was then shown also to be the primary sheddase of the epidermal growth factor precursor as well as the sheddase for transforming growth factor-alpha (TGFα) and amphregulin.14,15 ADAM10, in contrast, is the primary sheddase for Notch receptors, and ADAM10 knockout mice show characteristic Notch loss-of-function phenotypes in a variety of different tissues including the nervous and cardiovascular system.16,17 ADAM10 is also a well-characterized alpha-secretase, catalyzing cleavage of the amyloid-beta precursor protein APP, and it has been proposed that certain mutations in its prodomain are risk factors for late onset Alzheimer’s disease.18
Aberrant ADAM activity has also been linked to a number of different diseases. For example, ADAM17 has been reported to be capable of processing mutated forms of Notch1 found in T cell acute lymphoblastic leukemia.19 The ability of a single ADAM, such as ADAM17, not only to process multiple different substrates, but also to substitute for the physiologic catalyst in the cleavage of a mutated substrate of another ADAM, adds an additional layer of complexity to acquiring a complete understanding of how these molecules function in both physiology and disease.
Here we review the structural features of ADAM proteins that direct their biological functions in ectodomain shedding and cell adhesion. Table 1 lists all ADAM family proteins for which structural coordinates have been deposited, along with their PDB ID codes. The reader is referred to many other excellent reviews focused on ADAM biology in development and pathogenesis.3,8,20,21
Table 1.
ADAM structures and associated PDB ID codes.
| ADAM | Domain | PDB | References |
|---|---|---|---|
| ADAM8 | M | 4DD8 | 22 |
| ADAM10 | DC | 2AO7 | 23 |
| DC/8C7 Fab | 5L0Q | 24 | |
| MDC | 6BE6 | 25 | |
| MDC/11G2 Fab | 6BDZ | 26 | |
| ADAM17 | M | 1BKC | 27 |
| M/NT-TIMP3 | 3CKI | 28 | |
| C | 2M2F | 29 | |
| ADAM22 | MDCE | 3G5C | 30 |
| MDCE/LGI1 | 5Y2Z, 5Y31 | 31 | |
| ADAM33 | M | 1R54, 1R55 | 32 |
ADAM: a disintegrin and metalloproteinase.
Modular domain organization of ADAMs
ADAMs, ADAMTS (a disintegrin and metalloproteinase with thrombospondin repeats), and SVMP (snake venom metalloproteinase) make up the M12B adamalysin subfamily of the Metzincin superfamily of metallopeptidases. An early alternative name for the ADAM family of proteins was MDC, derived from the metalloprotease, disintegrin, and cysteine-rich modules in their ectodomains.33 Like the ADAMs, both SVMP and ADAMTS proteins have a modular domain organization, but are secreted proteins with additional structural features in the ancillary domains unique to each individual family.
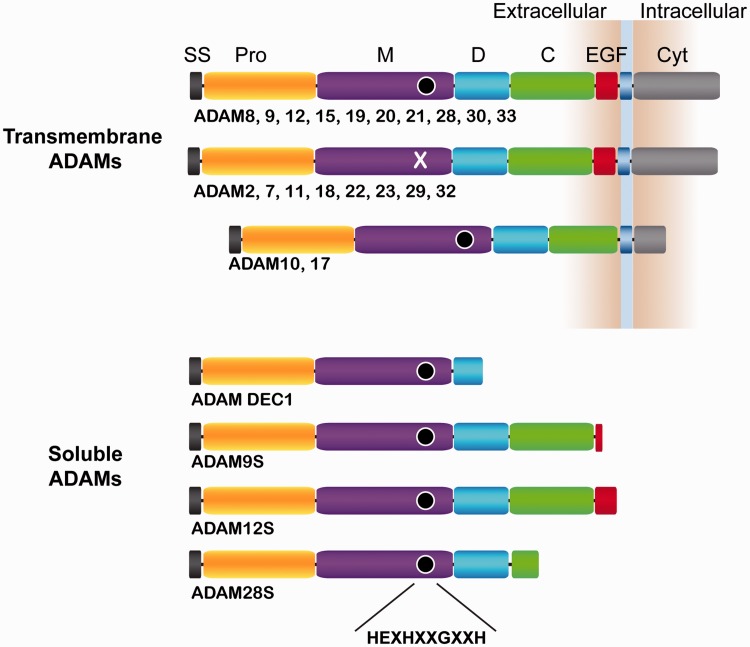
All ADAM proteins are synthesized as an inactive precursor with an N-terminal signal peptide and a prodomain immediately C-terminal to the signal peptide. Metalloproteinase (M), disintegrin (D), cysteine-rich (C), and (with the exception of ADAM10 and ADAM17) epidermal growth factor-like (EGF) domains then follow the prodomain, and precede a transmembrane segment and a cytoplasmic tail of variable length (Figure 1). Maturation of ADAMs occurs as they pass through the secretory pathway, where they are post-translationally modified with N-linked sugars and their prodomain is cleaved.
Figure 1.
ADAM family domain organization. Domain schematics of human ADAM family members are shown. SS: signal sequence; Pro: prodomain; M: metalloproteinase; D: disintegrin-like; C: cysteine-rich; EGF: epidermal growth factor-like; Cyt: cytoplasmic tail. The transmembrane segment is indicated with a blue box. The conserved active site amino acids are collectively represented using a black circle. ADAMs which do not have the conserved active site sequence have an “X” instead of the black circle in the M domain. (A color version of this figure is available in the online journal.)
The cytosolic tails of the ADAMs differ considerably, with variations in their posttranslational modification sites and in other sequences that serve as binding motifs for proteins that control their subcellular localization. For example, ADAM10 contains an ER retention sequence, whereas ADAMs 22, 23, and 11 have PDZ binding motifs that dock onto PSD-95 scaffolding protein in the synaptic cleft.34,35 One family member, ADAMDEC-1, is secreted as a soluble protein with a short D domain, and lacks the transmembrane region and cytoplasmic tail entirely. Other ADAM family members, including ADAMs 9, 12, and 28 can undergo alternative splicing to generate secreted variants (called 9S, 12S, and 28S), but the function of these secreted forms remains poorly understood.36,37
Catalytic domain architecture
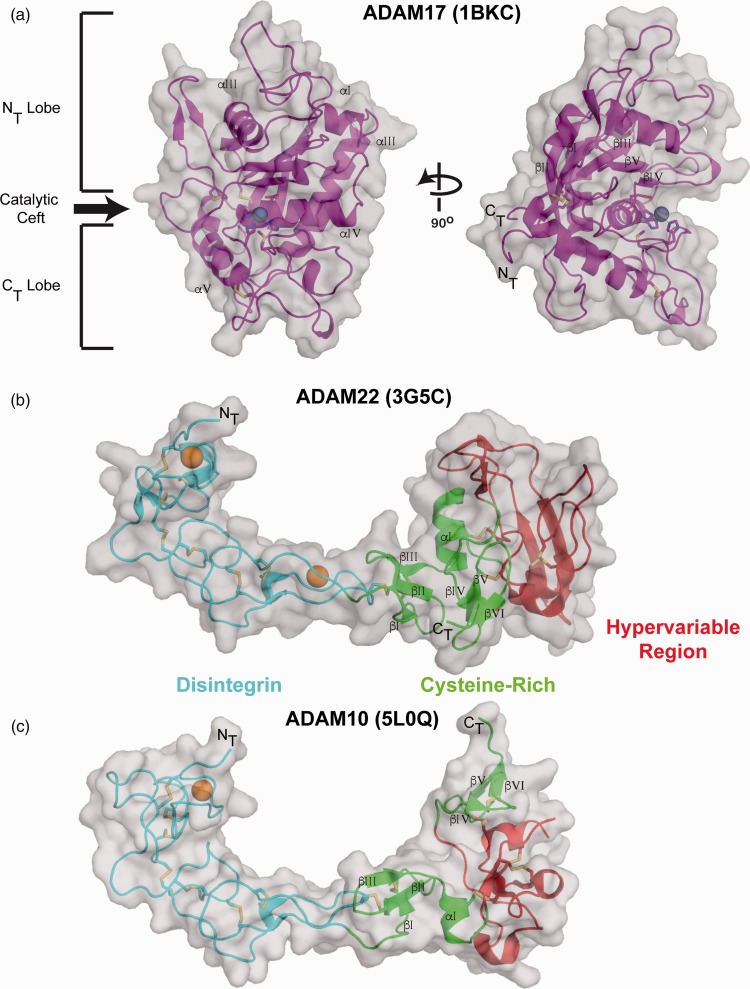
Metzincin proteins are characterized by conserved secondary structural and active site features in their M domains. The structure of the ADAM17 M domain, first determined in 1998, revealed a fold resembling a kidney bean (Figure 2), with an extended notch at the catalytic cleft dividing the domain into amino terminal (NT) and carboxyl terminal (CT) lobes.27 At the core of the NT lobe is a twisted 5-stranded antiparallel β-sheet that is enveloped by four conserved α-helices. The CT lobe contains a series of loops followed by a conserved α-helix, which is anchored to the NT lobe by a conserved disulfide bond near the C-terminus.
Figure 2.
M domain and D + C domain structures. (a) The ADAM17 M domain (PDB: 1BKC) is shown in carton representation with a transparent grey surface. The catalytic cleft and structurally conserved subdomains are indicated on the left. The active site histidine, glutamic acid, and methionine residues are represented as sticks with the bound zinc ion as a grey sphere. The bound inhibitor has been removed in these representations. The D + C domains of (b) ADAM22 (PDB: 3G5C) and (c) ADAM10 (PDB: 5L0Q) are shown in cartoon representation with a transparent grey surface. The D domain is in cyan, the C domain is in green, and the HVR is in red. Conserved structural α-helices and β-strands are labeled in the individual domains and disulfide bonds are represented as sticks. Bound calcium ions in the ADAM22 and ADAM10 D domains are represented as orange spheres. The M and EGF domains of ADAM22 have been removed from this representation. (A color version of this figure is available in the online journal.)
The catalytic cleft is formed between the NT and CT lobe along α-helix α4, generating a narrow canyon for the polypeptide substrate. Typically, metzincin family members contain a consensus HEXXHXXGXXH sequence on the fourth α-helix in the NT lobe. The three histidine residues coordinate a zinc ion, which together with the conserved glutamate residue, activates a water molecule that is the nucleophile responsible for substrate hydrolysis. Following the active site is a conserved loop in the CT lobe called the Met-turn, which wraps around the active site to position a methionine residue as a buttress beneath the zinc-coordinating cluster of histidine residues. Eight members of the ADAM family (ADAMs 2, 7, 11, 18, 22, 23, 29 and 32), however, lack the active site glutamate and have other mutations within the active site consensus sequence that almost certainly render them catalytically inactive.
Substrate selectivity of ADAMs
In general, the substrate selectivity of metzincin family members is strongly dependent on the flexibility of the P3-P1 positions to fit into the S3-S1 pockets while permitting the extension of the side chain from the P1′ position to reach into the selective S1′ pocket.38 Analysis of the cleavage preferences of ADAMs using peptide library approaches has revealed that ADAM17 has a strong preference for small hydrophobic amino acids, such as Leu and Val, in the P1′ position, while ADAM10 can also accommodate larger, aromatic amino acids, including phenylalanine and tyrosine.39,40 The origins of this selectivity derive from a deeper S1′ pocket in ADAM10 than in ADAM1725; this deeper pocket likely contributes to the selectivity of the small molecular weight inhibitor GI254023X for ADAM10. In the ADAM10 ectodomain structure (see below), each ADAM10 molecule in the crystal has a C-terminal hydrolytic peptide product from a neighboring copy in the asymmetric unit fortuitously captured in its active site, even though the P1′ residue was glycine. This observation indicates that an extended conformation and cleavage-site accessibility also exert an important influence on substrate selection. In addition, the D and C domains of both ADAM10 and ADAM17 appear to actively participate in substrate recognition (see below), and efforts have been made to exploit these “exosites” in the design of specific ADAM10 and ADAM17 inhibitors.41,42
Disintegrin and cysteine-rich domains
The ADAM disintegrin (D) domain is named for its resemblance to the disintegrins, a family of small proteins, found in the venom of vipers and pit vipers, that block the binding of platelets to adhesion receptors and thereby suppress platelet aggregation.43 Snake venom disintegrins are classified into three groups (short, medium and long) based on the number of disulfide bonds that serve to rigidify their structures. The D domains of the ADAMs most closely resemble the largest of these snake venom disintegrins, and typically have seven conserved disulfide bonds, except for ADAM5 (a potential pseudogene) and ADAM17, which each have six. In the ADAMs, the D domain can be further rigidified by the presence of two or one structural calcium ions. The residues responsible for coordinating the second calcium ion are not conserved across all ADAM family members, and in these instances a charged side chain, such as Arg525 in ADAM10, can fill in for the calcium ion. Like the disintegrins, certain ADAMs have also been implicated in binding to adhesion receptors,44–46 though the molecular basis for this interaction has remained obscure. Whereas a conserved loop presents the RGD adhesion recognition sequence in the snake venom disintegrins, all of the ADAMs except for ADAM15 have lost this RGD sequence from their D domains. Even in ADAM15, however, this loop also contains a cysteine residue that forms a disulfide bond linking the D and C domains, likely burying the RGD in the interior of the structure.
The cysteine-rich (C) domain, consisting of 80–150 amino acids, immediately follows the disintegrin domain and exhibits poor sequence conservation across the ADAM family. This domain is primarily stabilized through a network of six disulfide bonds. The ADAM cysteine-rich domain also contains a segment dubbed the “hypervariable region” (HVR), named for the significant divergence in length and amino acid composition among the different ADAMs in this region.47 Within the ADAM family, the HVR ranges in length from 37 residues in ADAM17 to 87 residues in ADAM19. Sequence alignment of the cysteine-rich domains also shows the HVRs of ADAM10 and ADAM17 to be much shorter than in the rest of the ADAMs. This decrease in size is associated with a different disulfide bonding pattern in ADAM10 and ADAM17 when compared to ADAM22 (and likely all other ADAM family members as well).
The X-ray structure of the D + C fragment from ADAM10 revealed that the two domains actually form a continuous, elongated cup shape without a clear boundary between them (Figure 2(c)).23,24 The D domain has an arced, extended overall shape with little secondary structure, stabilized instead by a series of conserved disulfide bonds that staple the loops together and rigidify the domain. The C domain is more globular, flanked by a pair of beta-hairpins at its N- and C-terminal ends.
The C domain contributes to ADAM function by regulating subcellular localization, modulating enzymatic activity, and potentially also by influencing substrate recognition through exosites. In ADAM10, the C domain regulates subcellular localization by binding to one of a subset of tetraspanin proteins with eight cysteines in its ectodomain (tspanC8 proteins). The tspanC8 proteins not only escort ADAM10 to the cell surface but also appear to influence substrate selectivity.48–50 Whether the C domain of ADAM17 is also targeted by its escort factors, the inactive rhomboid proteinases iRhom1 and iRhom2, is not yet known. Increased surface exposure of phosphatidylserine (PS) on the cell surface, which can be induced by ionomycin stimulation of calcium influx, is associated with increased sheddase activity of ADAM10 and ADAM17 in several cell types.51,52 The working model for this observation is that the exposed PS headgroup engages the membrane proximal region of the ectodomain to optimize the positioning of the enzyme active site near the cell surface, where cleavage sites for ectodomain shedding are located. The proposed exosite function of the C domain extends influences on substrate selectivity beyond local interactions within the M domain. For ADAM10, published data suggest that an acidic pocket of three aspartic acid residues within the C domain may act as an exosite, recognizing the EphA3 receptor and stimulating the processing of EphA3 upon formation of ligand-receptor complexes.23
Interdomain relationships in mature ADAM ectodomains
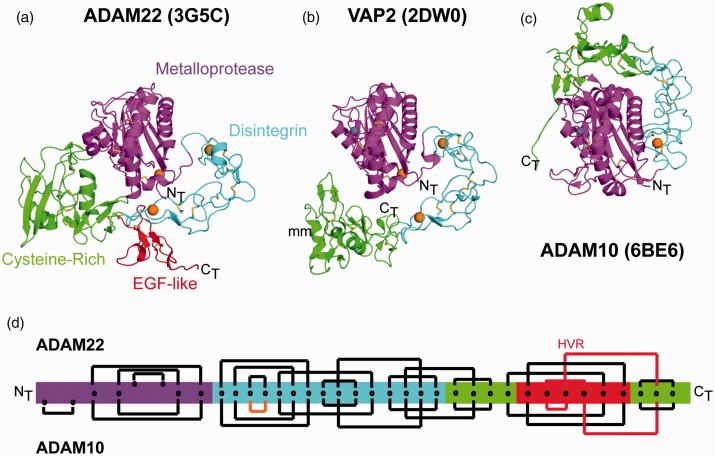
The first mammalian ADAM for which a mature, full ectodomain structure was reported was the catalytically inactive family member ADAM22.30 Its overall domain arrangement is compact, with the D + C C-shaped arm serving as an open cup into which the M domain is nestled (Figure 3(a)). This interdomain arrangement is shared with the vascular apopotosis-inducing proteins (VAPs) VAP1 and VAP2, two secreted snake venom toxins that also have the MDC architecture of the ADAMs (Figure 3(b)).47,53
Figure 3.
Interdomain relationships in the structures of ADAM22, VAP2, and ADAM10. The ectodomain of (a) ADAM22 (PDB: 3G5C), (b) VAP2 (PDB: 2DW0) and (c) ADAM10 is shown in cartoon representation with amino- (NT) and carboxy- (CT) termini labeled. The individual domains are colored according to the schematic representation in Figure 1. Bound calcium and zinc ions are represented as spheres colored orange and grey, respectively. Disulfide bonds are represented in stick form. (d) Disulfide bonding patterns in ADAM22 (Top) and ADAM10 (Bottom) are indicated with closed brackets connecting cysteine residues to their disulfide partner. Non-conserved cysteine residues that form disulfide bonds in only one of the two proteins are offset. The disulfide bond predicted to be missing from the ADAM17 D domain is colored orange. The lines for the disulfide pattern that is mismatched between ADAM22 and ADAM10 in the C domain are colored red. (A color version of this figure is available in the online journal.)
In the recently elucidated structure of the mature ADAM10 ectodomain, the overall architecture of ADAM10 is strikingly different from that of ADAM22, even though the isolated M, D, and C domains superimpose well on each other (Figure 3(c)). Superposition of the ADAM10 and ADAM22 M domains shows that the D + C region of ADAM10 wraps along the “side” of the M domain rather than along its base, resulting in the placement of the C domains on opposing surfaces of the M domains in the two structures. The different structural relationship of the D + C region with respect to the M domain in ADAM10 likely results from the shorter linker connecting the M and D domains, which places more limits on the interdomain flexibility of ADAM10.25
In the ADAM10 ectodomain structure, the C domain rests at the lip of the catalytic cleft of the M domain, and has a “fail-safe” autoinhibitory role in modulating catalytic activity.25 Accordingly, the cleavage of a fluorogenic peptide substrate by ADAM10 could be increased several-fold by addition of a monoclonal antibody, 8C7, which overcomes autoinhibition by binding to an epitope that overlaps the interdomain interface between the C and M domains.24
The high degree of sequence conservation between ADAM10 and ADAM17, together with prior studies investigating the regulation of ADAM17 activity in cells, suggests that the C domain may have a similar autoinhibitory role in ADAM17. ADAM17-catalyzed ectodomain shedding increases when cells are treated with PMA, a potent PKC pathway activator, and Blobel’s group showed that the tight binding inhibitor DPC33 only gains access to the ADAM17 active site in PMA stimulated, but not in quiescent cells.54 These data led the authors to postulate a model in which the ADAM17 active site was sterically blocked prior to stimulation with PMA, only becoming accessible to the active site inhibitor after compound treatment. In a completely independent line of investigation, the Murphy group developed a cross-domain inhibitory antibody that simultaneously recognizes the M and D + C domains of ADAM17, a finding that is also consistent with the idea that an interdomain interaction occludes the ADAM17 active site.55 Natural inhibitors targeting ADAM17 likewise show more potency toward the isolated catalytic domain than when the entire ectodomain is present.56 Based on this combination of structural information, sequence homology, and functional data, it seems likely that the active site in ADAM17 is also autoinhibited by its C domain using an autoregulatory interface analogous to that of ADAM10.
Catalytically inactive ADAMs rely on protein–protein interactions to carry out their functions. The first characterized ADAMs, fertilin-a (ADAM1a) and fertilin-b (ADAM2) are found on the surface of sperm and are required for proper sperm-egg fusion. The mature form of ADAM2 on the sperm surface lacks both its Pro and M domains, using its D + C domains to interact with surface adhesion receptors and facilitate sperm-egg binding.57
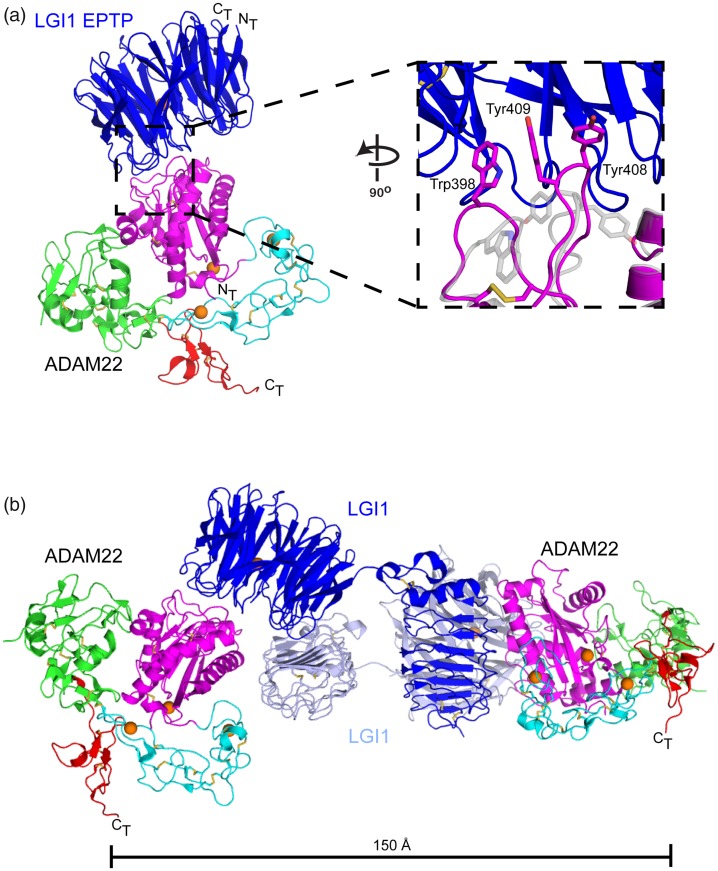
Unlike the earliest characterized non-catalytic ADAMs, ADAM22 uses its M domain to bind a secreted protein called LGI1, which is characteristically mutated in autosomal dominant partial epilepsy with auditory features (ADPEAF). These proteins appear to play an important role in neuronal development and synapse formation, as genetic disruption of lgi1, adam22, or adam23 in mice is postnatal lethal with mice suffering from severe epileptic seizures and peripheral nerve hypomyelination.26,31,58 Structures of two ADAM22-LGI1 complexes reported recently show that the epitempin-repeat of LGI1, a beta-propeller domain, uses its top face to engage the ADAM22 M domain at the distal end of the ADAM22 catalytic cleft. Structural alignment of ADAM22 in its unbound and bound conformations shows that the Cα atoms of the two states superimpose with a root mean-square deviation of only 0.5 Å. The one notable conformational change occurs in two loops of the metalloproteinase domain at the binding interface, with Trp398, Tyr408, and Tyr409 of ADAM22 flipping out to engage the LGI1 beta propeller (Figure 4(a)59). When complexes are formed with ADAM22 using a form of LGI1 that includes both its beta-propeller and leucine-rich repeat domains, the proteins form a domain-swapped 2:2 assembly, which the authors interpret as supportive of trans complex formation across the synapse (Figure 4(b)59).
Figure 4.
Structure of an ADAM22-LGI1 complex. (a) The ADAM22 ectodomain and bound LGI-1 EPTP domain (PDB: 5Y2Z) are shown in cartoon representation with disulfide bonds as sticks and bound calcium ions as orange spheres. The binding interface between the ADAM22 M domain and the LGI1 EPTP is magnified on the inset to the right. The apo ADAM22 structure (PDB: 3G5C) is superimposed on the ADAM22-LGI1 complex and shown in a transparent grey cartoon representation. The side chains of Trp398, Tyr408 and Tyr409 are represented as sticks. (b) The Cryo-EM structure of the 2:2 complex between ADAM22 and LGI1 (PDB: 5Y31) is shown in cartoon representation. (A color version of this figure is available in the online journal.)
Biological inhibition of ADAM proteins by prodomains and TIMPs
After biosynthesis, ADAMs enter the ER as inactive precursors, with latency conferred by the prodomain. The prodomains are then cleaved either by autoproteolysis or by a proprotein convertase, commonly furin, in the trans-Golgi network. ADAM prodomains are considerably larger than those of other metzincin proteins, ranging from 158 to 174 amino acids in length, and have a single conserved cysteine residue that occupies the active site in the latent state.
Although there are no atomic resolution structures yet available for an ADAM prodomain either in isolation or occupying an ADAM catalytic site, structures of matrix metalloproteases MMP1, MMP2, and MMP9 have each been solved with prodomain intact. In these structures, the free cysteine thiol protrudes from the prodomain to complete tetrahedral coordination of the zinc ion at the active site. The flanking residues of the prodomain are situated away from the active site, keeping the prodomain peptide backbone protected from autoproteolysis. Although this “cysteine switch” is considered a defining feature of metzincin family proteases, not all ADAMs contain an odd number of cysteine residues in their prodomains, and recombinant forms of the ADAM17 prodomain harboring a C184A mutation inhibits ADAM17 with comparable potency.56 These observations indicate that the prodomains use additional points of contact beyond the cysteine at the active site to confer latency. Indeed, certain ADAMs (ADAM10, ADAM17, and ADAM19) contain multiple “shadow” proprotein convertase recognition sites in predicted unstructured regions of the prodomain and are processed multiple times to enable prodomain release.60
In addition to their inhibitory properties, ADAM prodomains also serve as molecular chaperones to ensure transport of the mature ADAM through the secretory pathway to the cell surface. Two mutations in the ADAM10 prodomain, Q170H and R181G, co-segregate with late-onset Alzheimer’s disease.61 These mutations impair the maturation of ADAM10, and transgenic mice bearing forms of ADAM10 with these mutations attenuate ADAM10-dependent APP processing, leading the authors to attribute the increased disease risk from these mutations to a dysfunctional prodomain that has lost its chaperone properties.18
The human genome encodes four tissue inhibitor of metalloproteinase (TIMPs 1–4) proteins, originally identified as broad-spectrum inhibitors of MMPs.28 It has since become clear that TIMPs also inhibit ADAM metalloproteases. TIMP1 and TIMP2 show selective inhibition of ADAM10 and ADAM12, respectively, whereas TIMP3 has a broader spectrum of activity, inhibiting ADAMs 10, 17, 12, 28, and 33.62
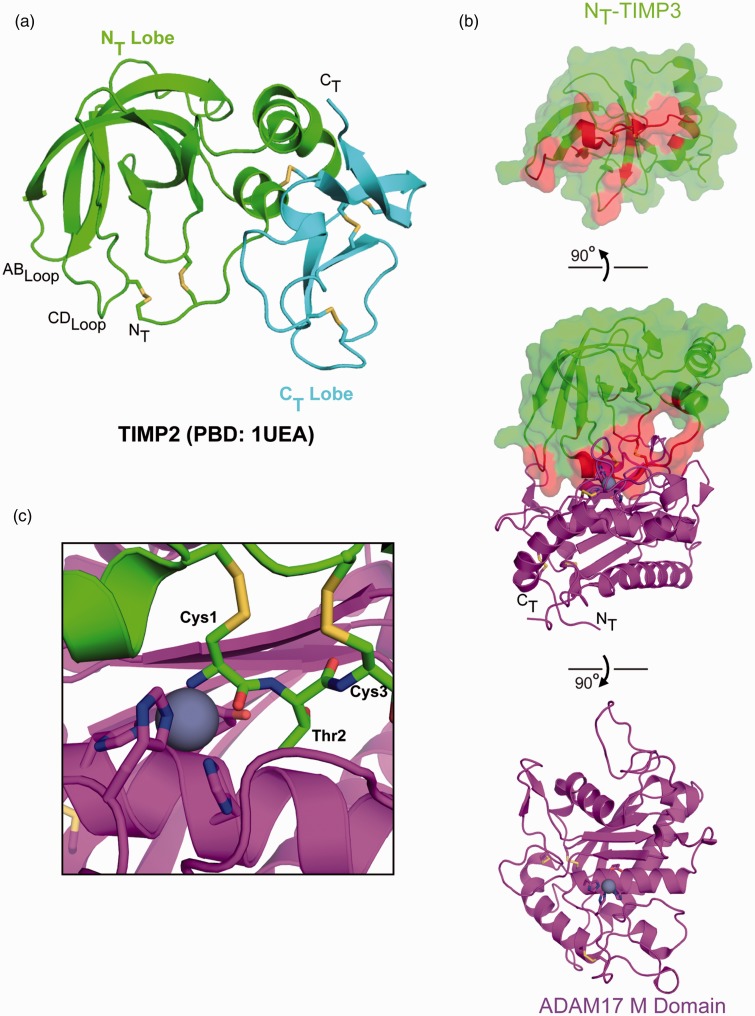
The TIMPs are small globular proteins that range from 184 to 194 amino acids in length. Structurally, they have two subdomains, referred to as NT and CT lobes.63 The NT lobe contains three α-helices and a 5-stranded β-barrel that adopts a wedge shape with its edge, consisting of the amino terminus and its AB and CD loops, binding into the catalytic cleft of the metalloproteinase (Figure 5). An intramolecular disulfide bond between the two cysteines of a conserved amino-terminal Cys-X-Cys motif places the N-terminal amine and backbone carbonyl group of Cys1 in position to coordinate the active site zinc, replacing the water molecule used in catalysis. The X-ray structure of TIMP3 in complex with the metalloprotease domain of ADAM17 also shows that the second residue, Thr2, is directed towards the S1′ pocket.64 Additional contacts from the NT lobe are observed in the catalytic cleft from the CD and AB loops protruding from the β-barrel. These additional contacts likely contribute to TIMP selectivity as engraftment of TIMP3 residues from these regions onto a TIMP1 scaffold increased the inhibitory activity of the chimeras for ADAM17.22
Figure 5.
ADAM17-TIMP3 complex. (a) The X-ray structure of full length TIMP1 (PDB: 1UEA) is shown in cartoon representation, with NT and CT subdomains are in green and cyan, respectively. Disulfide bonds are represented in stick form. (b) Contact interface of the X-ray structure of the NT lobe of TIMP3 (green) in complex with the M domain of ADAM17 (purple), shown in “open book” form. Amino acids within 4.5 Å of the M domain are highlighted red on NT-TIMP3. (c) Magnified view of the N-terminus of TIMP3 bound in the active site of the ADAM17 M domain. The conserved TIMP3 Cys-Thr-Cys sequence and ADAM17 active site amino acids are shown in stick representation. The active site zinc ion is represented as a grey sphere. (A color version of this figure is available in the online journal.)
Concluding remarks
Recent structural advances have provided a deeper appreciation for interdomain relationships that modulate the activity of ADAM proteins in ectodomain shedding and cellular adhesion. It is clear the metalloproteinase domain works in combination with its ancillary domains to execute its biological function. The ADAM ectodomain is dynamic, and accesses conformations that require interdomain movements during its enzymatic “lifecycle.” Fundamental questions about ADAM activation and substrate selection, however, still remain unanswered. Maturation of ADAM10 requires association with tetraspanin family C8 proteins, which may also influence substrate selectivity for Notch or APP,48–50 yet the molecular basis for this role of the C8 tetraspanins is not understood at a structural or biophysical level. Likewise, proper ADAM17 trafficking and activation appears dependent on the iRhom family of proteins,29,32 but detailed structural information about ADAM17-iRhom complexes is also lacking. Whether other ADAM family members rely on similar maturation or regulatory factors remains unknown, but the ADAM10 and ADAM17 examples suggest that such factors may also exist. Elucidating the biochemical and structural basis for ADAM regulation will be an exciting avenue of future research that should greatly advance our understanding of ADAM function in biology and human pathogenesis.
Authors’ contributions
TCMS and SCB wrote the paper.
DECLARATION OF CONFLICTING INTERESTS
SCB is a consultant for IFM Therapeutics and for Ayala Pharmaceutical on projects unrelated to the contents of this review article. He also receives funding for an unrelated project from Novartis.
FUNDING
The authors’ work is supported by NIH grant R35 CA220340 (to S.C.B.).
References
- 1.Wolfsberg TG, Bazan JF, Blobel CP, Myles DG, Primakoff P, White JM. The precursor region of a protein active in sperm-egg fusion contains a metalloprotease and a disintegrin domain: structural, functional, and evolutionary implications. Proc Natl Acad Sci USA 1993; 90:10783–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huxley-Jones J, Clarke TK, Beck C, Toubaris G, Robertson DL, Boot-Handford RP. The evolution of the vertebrate metzincins; insights from Ciona intestinalis and Danio rerio. BMC Evol Biol 2007; 7:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy G. The ADAMs: signalling scissors in the tumour microenvironment. Nat Rev Cancer 2008; 8:929–41 [DOI] [PubMed] [Google Scholar]
- 4.Cho C, Bunch DO, Faure JE, Goulding EH, Eddy EM, Primakoff P, Myles DG. Fertilization defects in sperm from mice lacking fertilin beta. Science 1998; 281:1857–9 [DOI] [PubMed] [Google Scholar]
- 5.Lambrecht BN, Vanderkerken M, Hammad H. The emerging role of ADAM metalloproteinases in immunity. Nat Rev Immunol 2018; 18:745–58 [DOI] [PubMed] [Google Scholar]
- 6.Saftig P, Lichtenthaler SF. The alpha secretase ADAM10: a metalloprotease with multiple functions in the brain. Prog Neurobiol 2015; 135:1–20 [DOI] [PubMed] [Google Scholar]
- 7.Weskamp G, Ford JW, Sturgill J, Martin S, Docherty AJ, Swendeman S, Broadway N, Hartmann D, Saftig P, Umland S, Sehara-Fujisawa A, Black RA, Ludwig A, Becherer JD, Conrad DH, Blobel CP. ADAM10 is a principal 'sheddase' of the low-affinity immunoglobulin E receptor CD23. Nat Immunol 2006; 7:1293–8 [DOI] [PubMed] [Google Scholar]
- 8.Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol Aspects Med 2008; 29:258–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lichtenthaler SF, Lemberg MK, Fluhrer R. Proteolytic ectodomain shedding of membrane proteins in mammals-hardware, concepts, and recent developments. EMBO J 2018; 37:e99456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moss ML, Jin SL, Milla ME, Bickett DM, Burkhart W, Carter HL, Chen WJ, Clay WC, Didsbury JR, Hassler D, Hoffman CR, Kost TA, Lambert MH, Leesnitzer MA, McCauley P, McGeehan G, Mitchell J, Moyer M, Pahel G, Rocque W, Overton LK, Schoenen F, Seaton T, Su JL, Becherer JD. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-alpha. Nature 1997; 385:733–6 [DOI] [PubMed] [Google Scholar]
- 11.Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S, Nelson N, Boiani N, Schooley KA, Gerhart M, Davis R, Fitzner JN, Johnson RS, Paxton RJ, March CJ, Cerretti DP. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature 1997; 385:729–33 [DOI] [PubMed] [Google Scholar]
- 12.Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg SW, Lee DC, Russell WE, Castner BJ, Johnson RS, Fitzner JN, Boyce RW, Nelson N, Kozlosky CJ, Wolfson MF, Rauch CT, Cerretti DP, Paxton RJ, March CJ, Black RA. An essential role for ectodomain shedding in mammalian development. Science 1998; 282:1281–4 [DOI] [PubMed] [Google Scholar]
- 13.Luetteke NC, Qiu TH, Peiffer RL, Oliver P, Smithies O, Lee DC. TGF alpha deficiency results in hair follicle and eye abnormalities in targeted and waved-1 mice. Cell 1993; 73:263–78 [DOI] [PubMed] [Google Scholar]
- 14.Sunnarborg SW, Hinkle CL, Stevenson M, Russell WE, Raska CS, Peschon JJ, Castner BJ, Gerhart MJ, Paxton RJ, Black RA, Lee DC. Tumor necrosis factor-alpha converting enzyme (TACE) regulates epidermal growth factor receptor ligand availability. J Biol Chem 2002; 277:12838–45 [DOI] [PubMed] [Google Scholar]
- 15.Sahin U, Weskamp G, Kelly K, Zhou HM, Higashiyama S, Peschon J, Hartmann D, Saftig P, Blobel CP. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J Cell Biol 2004; 164:769–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartmann D, de Strooper B, Serneels L, Craessaerts K, Herreman A, Annaert W, Umans L, Lubke T, Lena Illert A, von Figura K, Saftig P. The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum Mol Genet 2002; 11:2615–24 [DOI] [PubMed] [Google Scholar]
- 17.Jorissen E, Prox J, Bernreuther C, Weber S, Schwanbeck R, Serneels L, Snellinx A, Craessaerts K, Thathiah A, Tesseur I, Bartsch U, Weskamp G, Blobel CP, Glatzel M, De Strooper B, Saftig P. The disintegrin/metalloproteinase ADAM10 is essential for the establishment of the brain cortex. J Neurosci 2010; 30:4833–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suh J, Choi SH, Romano DM, Gannon MA, Lesinski AN, Kim DY, Tanzi RE. ADAM10 missense mutations potentiate beta-amyloid accumulation by impairing prodomain chaperone function. Neuron 2013; 80:385–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bozkulak EC, Weinmaster G. Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling. Mol Cell Biol 2009; 29:5679–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber S, Saftig P. Ectodomain shedding and ADAMs in development. Development 2012; 139:3693–709 [DOI] [PubMed] [Google Scholar]
- 21.Reiss K, Saftig P. The “a disintegrin and metalloprotease” (ADAM) family of sheddases: physiological and cellular functions. Semin Cell Dev Biol 2009; 20:126–37 [DOI] [PubMed] [Google Scholar]
- 22.Lee MH, Maskos K, Knauper V, Dodds P, Murphy G. Mapping and characterization of the functional epitopes of tissue inhibitor of metalloproteinases (TIMP)-3 using TIMP-1 as the scaffold: a new frontier in TIMP engineering. Protein Sci 2002; 11:2493–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janes PW, Saha N, Barton WA, Kolev MV, Wimmer-Kleikamp SH, Nievergall E, Blobel CP, Himanen JP, Lackmann M, Nikolov DB. Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell 2005; 123:291–304 [DOI] [PubMed] [Google Scholar]
- 24.Atapattu L, Saha N, Chheang C, Eissman MF, Xu K, Vail ME, Hii L, Llerena C, Liu Z, Horvay K, Abud HE, Kusebauch U, Moritz RL, Ding BS, Cao Z, Rafii S, Ernst M, Scott AM, Nikolov DB, Lackmann M, Janes PW. An activated form of ADAM10 is tumor selective and regulates cancer stem-like cells and tumor growth. J Exp Med 2016; 213:1741–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seegar TCM, Killingsworth LB, Saha N, Meyer PA, Patra D, Zimmerman B, Janes PW, Rubinstein E, Nikolov DB, Skiniotis G, Kruse AC, Blacklow SC. Structural basis for regulated proteolysis by the alpha-secretase ADAM10. Cell 2017; 171:1638–48 e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fukata Y, Lovero KL, Iwanaga T, Watanabe A, Yokoi N, Tabuchi K, Shigemoto R, Nicoll RA, Fukata M. Disruption of LGI1-linked synaptic complex causes abnormal synaptic transmission and epilepsy. Proc Natl Acad Sci USA 2010; 107:3799–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maskos K, Fernandez-Catalan C, Huber R, Bourenkov GP, Bartunik H, Ellestad GA, Reddy P, Wolfson MF, Rauch CT, Castner BJ, Davis R, Clarke HR, Petersen M, Fitzner JN, Cerretti DP, March CJ, Paxton RJ, Black RA, Bode W. Crystal structure of the catalytic domain of human tumor necrosis factor-alpha-converting enzyme. Proc Natl Acad Sci USA 1998; 95:3408–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brew K, Nagase H. The tissue inhibitors of metalloproteinases (TIMPs): an ancient family with structural and functional diversity. Biochim Biophys Acta 2010; 1803:55–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McIlwain DR, Lang PA, Maretzky T, Hamada K, Ohishi K, Maney SK, Berger T, Murthy A, Duncan G, Xu HC, Lang KS, Haussinger D, Wakeham A, Itie-Youten A, Khokha R, Ohashi PS, Blobel CP, Mak TW. iRhom2 regulation of TACE controls TNF-mediated protection against Listeria and responses to LPS. Science 2012; 335:229–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H, Shim AH, He X. Structural characterization of the ectodomain of a disintegrin and metalloproteinase-22 (ADAM22), a neural adhesion receptor instead of metalloproteinase: insights on ADAM function. J Biol Chem 2009; 284:29077–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sagane K, Hayakawa K, Kai J, Hirohashi T, Takahashi E, Miyamoto N, Ino M, Oki T, Yamazaki K, Nagasu T. Ataxia and peripheral nerve hypomyelination in ADAM22-deficient mice. BMC Neurosci 2005; 6:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adrain C, Zettl M, Christova Y, Taylor N, Freeman M. Tumor necrosis factor signaling requires iRhom2 to promote trafficking and activation of TACE. Science 2012; 335:225–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blobel CP. Metalloprotease-disintegrins: links to cell adhesion and cleavage of TNF alpha and Notch. Cell 1997; 90:589–92 [DOI] [PubMed] [Google Scholar]
- 34.Fukata Y, Adesnik H, Iwanaga T, Bredt DS, Nicoll RA, Fukata M. Epilepsy-related ligand/receptor complex LGI1 and ADAM22 regulate synaptic transmission. Science 2006; 313:1792–5 [DOI] [PubMed] [Google Scholar]
- 35.Marcello E, Gardoni F, Di Luca M, Perez-Otano I. An arginine stretch limits ADAM10 exit from the endoplasmic reticulum. J Biol Chem 2010; 285:10376–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hotoda N, Koike H, Sasagawa N, Ishiura S. A secreted form of human ADAM9 has an alpha-secretase activity for APP. Biochem Biophys Res Commun 2002; 293:800–5 [DOI] [PubMed] [Google Scholar]
- 37.Roberts CM, Tani PH, Bridges LC, Laszik Z, Bowditch RD. MDC-L, a novel metalloprotease disintegrin cysteine-rich protein family member expressed by human lymphocytes. J Biol Chem 1999; 274:29251–9 [DOI] [PubMed] [Google Scholar]
- 38.Eckhard U, Huesgen PF, Schilling O, Bellac CL, Butler GS, Cox JH, Dufour A, Goebeler V, Kappelhoff R, Keller UAD, Klein T, Lange PF, Marino G, Morrison CJ, Prudova A, Rodriguez D, Starr AE, Wang Y, Overall CM. Active site specificity profiling of the matrix metalloproteinase family: proteomic identification of 4300 cleavage sites by nine MMPs explored with structural and synthetic peptide cleavage analyses. Matrix Biol 2016; 49:37–60 [DOI] [PubMed] [Google Scholar]
- 39.Caescu CI, Jeschke GR, Turk BE. Active-site determinants of substrate recognition by the metalloproteinases TACE and ADAM10. Biochem J 2009; 424:79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tucher J, Linke D, Koudelka T, Cassidy L, Tredup C, Wichert R, Pietrzik C, Becker-Pauly C, Tholey A. LC-MS based cleavage site profiling of the proteases ADAM10 and ADAM17 using proteome-derived peptide libraries. J Proteome Res 2014; 13:2205–14 [DOI] [PubMed] [Google Scholar]
- 41.Minond D, Cudic M, Bionda N, Giulianotti M, Maida L, Houghten RA, Fields GB. Discovery of novel inhibitors of a disintegrin and metalloprotease 17 (ADAM17) using glycosylated and non-glycosylated substrates. J Biol Chem 2012; 287:36473–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madoux F, Dreymuller D, Pettiloud JP, Santos R, Becker-Pauly C, Ludwig A, Fields GB, Bannister T, Spicer TP, Cudic M, Scampavia LD, Minond D. Discovery of an enzyme and substrate selective inhibitor of ADAM10 using an exosite-binding glycosylated substrate. Sci Rep 2016; 6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kini RM, Evans HJ. Structural domains in venom proteins: evidence that metalloproteinases and nonenzymatic platelet aggregation inhibitors (disintegrins) from snake venoms are derived by proteolysis from a common precursor. Toxicon 1992; 30:265–93 [DOI] [PubMed] [Google Scholar]
- 44.Evans JP, Kopf GS, Schultz RM. Characterization of the binding of recombinant mouse sperm fertilin beta subunit to mouse eggs: evidence for adhesive activity via an egg beta1 integrin-mediated interaction. Dev Biol 1997; 187:79–93 [DOI] [PubMed] [Google Scholar]
- 45.Rao H, Lu G, Kajiya H, Garcia-Palacios V, Kurihara N, Anderson J, Patrene K, Sheppard D, Blair HC, Windle JJ, Choi SJ, Roodman GD. Alpha9beta1: a novel osteoclast integrin that regulates osteoclast formation and function. J Bone Miner Res 2006; 21:1657–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zigrino P, Steiger J, Fox JW, Loffek S, Schild A, Nischt R, Mauch C. Role of ADAM-9 disintegrin-cysteine-rich domains in human keratinocyte migration. J Biol Chem 2007; 282:30785–93 [DOI] [PubMed] [Google Scholar]
- 47.Takeda S, Igarashi T, Mori H, Araki S. Crystal structures of VAP1 reveal ADAMs' MDC domain architecture and its unique C-shaped scaffold. EMBO J 2006; 25:2388–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dornier E, Coumailleau F, Ottavi JF, Moretti J, Boucheix C, Mauduit P, Schweisguth F, Rubinstein E. TspanC8 tetraspanins regulate ADAM10/Kuzbanian trafficking and promote Notch activation in flies and mammals. J Cell Biol 2012; 199:481–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noy PJ, Yang J, Reyat JS, Matthews AL, Charlton AE, Furmston J, Rogers DA, Rainger GE, Tomlinson MG. TspanC8 tetraspanins and a disintegrin and metalloprotease 10 (ADAM10) interact via their extracellular regions: evidence for distinct binding mechanisms for different TspanC8 proteins. J Biol Chem 2016; 291:3145–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jouannet S, Saint-Pol J, Fernandez L, Nguyen V, Charrin S, Boucheix C, Brou C, Milhiet PE, Rubinstein E. TspanC8 tetraspanins differentially regulate the cleavage of ADAM10 substrates, Notch activation and ADAM10 membrane compartmentalization. Cell Mol Life Sci 2016; 73:1895–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bleibaum F, Sommer A, Veit M, Rabe B, Andra J, Kunzelmann K, Nehls C, Correa W, Gutsmann T, Grotzinger J, Bhakdi S, Reiss K. ADAM10 sheddase activation is controlled by cell membrane asymmetry. J Mol Cell Biol 2019;in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sommer A, Kordowski F, Buch J, Maretzky T, Evers A, Andra J, Dusterhoft S, Michalek M, Lorenzen I, Somasundaram P, Tholey A, Sonnichsen FD, Kunzelmann K, Heinbockel L, Nehls C, Gutsmann T, Grotzinger J, Bhakdi S, Reiss K. Phosphatidylserine exposure is required for ADAM17 sheddase function. Nat Commun 2016; 7:11523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Igarashi T, Araki S, Mori H, Takeda S. Crystal structures of catrocollastatin/VAP2B reveal a dynamic, modular architecture of ADAM/adamalysin/reprolysin family proteins. FEBS Lett 2007; 581:2416–22 [DOI] [PubMed] [Google Scholar]
- 54.Le Gall SM, Maretzky T, Issuree PD, Niu XD, Reiss K, Saftig P, Khokha R, Lundell D, Blobel CP. ADAM17 is regulated by a rapid and reversible mechanism that controls access to its catalytic site. J Cell Sci 2010; 123:3913–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tape CJ, Willems SH, Dombernowsky SL, Stanley PL, Fogarasi M, Ouwehand W, McCafferty J, Murphy G. Cross-domain inhibition of TACE ectodomain. Proc Natl Acad Sci USA 2011; 108:5578–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gonzales PE, Solomon A, Miller AB, Leesnitzer MA, Sagi I, Milla ME. Inhibition of the tumor necrosis factor-alpha-converting enzyme by its pro domain. J Biol Chem 2004; 279:31638–45 [DOI] [PubMed] [Google Scholar]
- 57.Blobel CP, Wolfsberg TG, Turck CW, Myles DG, Primakoff P, White JM. A potential fusion peptide and an integrin ligand domain in a protein active in sperm-egg fusion. Nature 1992; 356:248–52 [DOI] [PubMed] [Google Scholar]
- 58.Mitchell KJ, Pinson KI, Kelly OG, Brennan J, Zupicich J, Scherz P, Leighton PA, Goodrich LV, Lu X, Avery BJ, Tate P, Dill K, Pangilinan E, Wakenight P, Tessier-Lavigne M, Skarnes WC. Functional analysis of secreted and transmembrane proteins critical to mouse development. Nat Genet 2001; 28:241–9 [DOI] [PubMed] [Google Scholar]
- 59.Yamagata A, Miyazaki Y, Yokoi N, Shigematsu H, Sato Y, Goto-Ito S, Maeda A, Goto T, Sanbo M, Hirabayashi M, Shirouzu M, Fukata Y, Fukata M, Fukai S. Structural basis of epilepsy-related ligand-receptor complex LGI1-ADAM22. Nat Commun 2018; 9:1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wong E, Maretzky T, Peleg Y, Blobel CP, Sagi I. The functional maturation of a disintegrin and metalloproteinase (ADAM) 9, 10, and 17 requires processing at a newly identified proprotein convertase (PC) cleavage site. J Biol Chem 2015; 290:12135–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim M, Suh J, Romano D, Truong MH, Mullin K, Hooli B, Norton D, Tesco G, Elliott K, Wagner SL, Moir RD, Becker KD, Tanzi RE. Potential late-onset Alzheimer's disease-associated mutations in the ADAM10 gene attenuate {alpha}-secretase activity. Hum Mol Genet 2009; 18:3987–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jackson HW, Defamie V, Waterhouse P, Khokha R. TIMPs: versatile extracellular regulators in cancer. Nat Rev Cancer 2017; 17:38–53 [DOI] [PubMed] [Google Scholar]
- 63.Gomis-Ruth FX, Maskos K, Betz M, Bergner A, Huber R, Suzuki K, Yoshida N, Nagase H, Brew K, Bourenkov GP, Bartunik H, Bode W. Mechanism of inhibition of the human matrix metalloproteinase stromelysin-1 by TIMP-1. Nature 1997; 389:77–81 [DOI] [PubMed] [Google Scholar]
- 64.Wisniewska M, Goettig P, Maskos K, Belouski E, Winters D, Hecht R, Black R, Bode W. Structural determinants of the ADAM inhibition by TIMP-3: crystal structure of the TACE-N-TIMP-3 complex. J Mol Biol 2008; 381:1307–19 [DOI] [PubMed] [Google Scholar]