Abstract
The intervertebral disc (IVD) is composed of three separate tissues with distinct origins and properties. Elucidating changes occurring in these tissues in response to injury or age is paramount to identify new therapies to better manage disc and spine degenerative conditions, including low back pain. Despite their small size and different mechanical load pattern compared to higher species, the use of mouse models represents a cost‐effective and powerful approach to better understand the formation, maintenance, and degeneration of the IVD. However, the isolation of the different compartments of the IVD is complicated by their diminutive size. Here, we describe a simple, step‐by‐step protocol for the isolation of the nucleus pulposus (NP) tissues that can then be processed for further analyses. Analysis from mouse NP tissues shows sufficient quantities of RNAs, purity of the NP fraction, and overall RNA quality for gene expression studies, and reveals no increase in expression of disc degeneration markers, including TNFa, IL1b, and Mmp1 up to 15 months of age in C57BL6 wildtype mice.
Keywords: aging, gene expression, intervertebral disc degeneration, nucleus pulposus
1. INTRODUCTION
The intervertebral disc (IVD) plays an important role in diffusing the various mechanical forces exerted on the spine during movement. In humans, degeneration of the IVD is associated with the onset of neck and lower back pain, a disabling, widespread and costly disease.1, 2 IVD degeneration is difficult to treat, with current treatments limited to palliative and surgical options. The fact that regenerative strategies for IVD degeneration have to date been unsuccessful reflects our incomplete understanding of the underlying etiology.3 The different cellular composition of the three distinct tissues of the IVD, the inner gelatinous nucleus pulposus (NP), the surrounding fibrous annulus fibrosus (AF), and the cartilage endplates (CEP), further complicates studies of this degenerative disease process.
Rich in water, the NP is crucial in preventing stress concentrations in the IVD by distributing forces evenly across this structure. The NP contains cells with different morphologies but all derived from the notochord4, 5: large, vacuolated notochordal cells (NC) and smaller chondrocyte‐like cells. Multiple studies support an anabolic and protective effects of NC cells on chondrocyte‐like cells and their disappearance is associated with the onset of degenerative disc disease.6, 7, 8 The collagen‐rich AF surrounds the NP in concentric laminae, providing both tensile strength and elastic rebound after movement or compression. AF cells are fibroblast‐like and still poorly characterized. Differing from the NP, where the transcription factor Brachyury (Bra) is highly and specifically expressed, the embryologic origin of AF and CEP cells is the sclerotome,9 where Paired‐box1 (Pax1) is highly expressed. Lastly, the CEP serves as a transition between the vertebral body and the IVD. It is composed of hyaline cartilage that surrounds chondrocytes morphologically similar to articular chondrocytes, although genetic expression studies have shown differences between the CEP and articular cartilage.10
Healthy, nondegenerate human samples are limited in their ease of procurement and quantity. Therefore, animal models have been utilized to better characterize healthy cell behavior and characteristics, disc formation and the degenerative process, despite the recognized morphologic differences between IVDs from small animals and humans.11 Studies in larger animals are aided in cell isolation by the gross morphological differences between the gelatinous NP and the fibrocartilaginous AF/CEP—these methods utilize gross dissection and further enzymatic digestion to ensure a high yield of cells.12 Mice present an excellent opportunity for studying disc degeneration with the wide availability of transgenic models and low overall costs, but cell isolation from each of the IVD compartments is challenging, as the size of the mouse IVD and number of cells within each IVD compartment are far less compared to larger organisms. There is no consistent method for isolating cells of the different IVD compartments in mice for either cell culture or RNA expression (Table 1). Recently, efforts have focused on cell isolation without the use of enzymes, with concerns that enzymatic digestion can alter RNA and/or cell surface expression profiles, require longer expansion times in culture and necessitate multiple passages that leads to general dedifferentiation.27 Due to the typical small NP cell yields in mice, many preclinical studies have also resolved on harvesting cells from multiple discs and pooling their contents, usually pooling discs from the same spine region, as differential composition or gene expression between IVDs from lumbar and sacral regions have been reported.28, 29, 30, 31 Lastly, because the NPs function in a unique, hypoxic, and hyperosmotic environment, gene expression is likely to change in response to enzymatic or nonenzymatic extraction due both to the time required for the procedure and the relatively dissimilar environment of cell culture conditions versus tissue. For studies focused on phenotyping IVD tissues, there is a need to prepare RNAs of the different IVD compartments in a consistent, fast, and reproductible fashion, without an extra step of ex vivo culture. Main goals of cell isolation from the mouse IVD are thus threefold: (a) preserve enough cells to allow experimental studies, (b) maintain the gene expression profile of IVD cells and limit variations and artifacts due to retrieval or culture methods, and (c) do so in an accessible and easily reproducible method with dependable results. In this report, we describe a new method to isolate pure NP RNAs from murine lumbar and thoracic IVDs, based on a simple centrifugation step.
Table 1.
Examples of methods used to prepare mouse IVD tissues (nonexhaustive)
| Purpose of study | Separation AF/NP? | Method of dissection | Vertebral level | Enzymatic digestion | Ref. |
|---|---|---|---|---|---|
| Cell culture | Mixed, NP/IAF sample | Cut was made through the middle of the AF with a scalpel blade. Exposed NP and IAF were scooped out with a needle | Lumbar | 0.01% collagenase | 13 |
| Cell culture | Yes | Identified and separated under microscope | Tail | 0.1% pronase and 0.2% collagenase type 2 | 14 |
| Cell culture | No | Microscopy was used to harvest IVD tissue completely from the end plates | Coccygeal | No | 15 |
| Cell culture | Yes | The disc was cut and bent, forcing the NP to protrude | Coccygeal | No | 16 |
| Gene expression | Yes | Dissected | Lumbar + caudal | No | 17 |
| Gene expression | Yes | NP was scraped off with a scalpel. AF was isolated by a blade under the stereo microscope. The CEP was shaved off with a scalpel | Lumbar + coccygeal | No | 18 |
| Gene expression | Yes | NP and AF from all lumbar IVD were resected separately | Lumbar | No | 19 |
| Gene expression | No | Full tissue examined together | Lumbar | No | 20 |
| Gene expression | No | Carefully dissected | Unknown | No | 21 |
| Gene expression | No | Separated from all other tissues | Tail/Lumbar | No | 22 |
| Gene expression | No | No method listed | Unknown | Unknown | 23 |
| Gene expression | No | Isolated by microdissection | Thoracic | No | 24 |
| Gene expression | No | NP isolated by microdissection | Lumbar + thoracic | No | 25 |
| Gene expression | Yes | NP and AF isolated by microdissection | Lumbar + thoracic | No | 26 |
2. MATERIALS AND METHODS
2.1. Mice
Mice were euthanized by cervical dislocation under general anesthesia induced by isoflurane inhalation. All procedures were approved by the institutional IACUC committee.
Detailed step‐by‐step protocol for the isolation of NPs and CEPs/AFs:
Place the animal on its back. Douse the skin with 70% ethanol to disinfect and prevent hair from spreading during dissection (Figure 1A).
To avoid damaging the spine with too deep of a first incision, grasp and pull the skin away from the abdomen with forceps and make a small incision in the skin into the abdominal space (Figure 1B). Manually remove the pelt from the dorsal spine by either tearing or cutting the skin in the transverse plane and then pulling the two segments up to the base of the neck and below the base of the tail (Figure 1C,D).
Isolate the spinal column from the surrounding tissues—using scissors, remove the musculature and long bones, ribs, and pelvis to cut in a parallel direction to the exposed spine from the sacrum and up to the thoracic region (Figure 1E,F). Complete the isolation of the spine by making a transverse cut at the level of the femurs and at the appropriate level in the lumbar/thoracic/cervical region, depending on the region of interest (Figure 1G).
Move the excised spinal column onto an iced dissection field under a dissecting microscope (Figure 1H and 2A). The iced dissection field can be prepared by filling the cover of a Petri dish with ice and placing the other half on top, decreasing the temperature of the dissecting surface close to freezing point while avoiding contamination of the dissection field. No buffer needs to be used during the fine dissection.
With scissors or a scalpel, remove the fascia and musculature attached to the anterior aspect of the vertebral column (Figure 2B). This will expose the IVDs (Figure 2C, black arrows). Continue to remove the lateral attachments of soft tissues (Figure 2C, white arrow), as doing so will simplify the process of isolating the IVDs from the vertebral column (Figure 2D).
Cut through both pedicles to isolate the vertebral body from the other parts of the vertebrae. This can best be accomplished by inserting one blade into the spinal canal and positioning the other blade next to the vertebral body (Figure 2E). After cutting one side free, continue to the other side (Figure 2F,G). Remove any additional attachments connecting the vertebral column to the remaining posterior portion of the vertebrae (Figure 2H). Take care to remove any remaining spinal cord that can contaminate samples. Spinal cord can be visualized as a white column that extends the length of the spine.
If there is any extraneous soft tissue still attached to the isolated vertebral column, use scissors or a scalpel to remove (Figure 2I).
-
Under a dissecting microscope, examine the vertebral column, placing it with the posterior side down with the anterior aspect facing you (Figure 3A‐C). Hold the bony vertebral body with the pincers to allow proper stabilization of the specimen (Figure 3C). Isolate IVDs from lumbar and thoracic regions keeping the IVD intact (NP + AF + CEP) by placing the scalpel at the exact boundary of the CEP and the vertebral body and gently pushing down (Figure 3D,E). There will be a small color change between the two structures, and if the scalpel blade is placed correctly the structures will separate with very little force. Holding the adjacent vertebral body with the pincer allows proper control of the specimen. Repeat on the opposite aspect of the IVD (Figure 3F).
▲ Critical step—Excessive force during CEP separation from the vertebral body will force the NP out of the IVD, limiting the amount of usable sample. Be aware of the force applied to the joint and adjust the location of the blade if slight pressure is unable to separate the CEP from the vertebrae.
Place the isolated IVD (Figure 3G) on a portion of the iced dissection field and continue isolation of all other IVDs. If upon examination of the isolated IVD, marrow, soft tissue, or bone is visualized, these elements can be removed by carefully holding the IVD and separating them away from the IVD with a scalpel.
With a 28‐gauge needle, pierce the IVD from the superior to the inferior aspects (Figure 3H‐J, if NP is spared during dissection, it should be visible as a gelatinous outpouching, see Figure 3I, black arrow) and place in a previously prepared perforated tube‐within‐a‐tube, where a 0.5 mL microcentrifuge tube is punctured several times at its most inferior portion with a 25‐guage needle and placed within a 1.5 mL microcentrifuge tube (Figure 3K,L). Centrifuge at 12 000 rcf for 3 minutes at 4°C. Collect the NP cell mixture in the 1.5 mL microcentrifuge tube and deposit the AF/CEP tissue remaining in the 0.5 mL microcentrifuge tube into a new 1.5 mL tube.
-
Add TRIzol reagent to NPs immediately and thoroughly mix, proceeding then to RNA isolation or freezing.
▲ Critical step—Remember that the NP cells are enmeshed in a matrix. To better dissolve the cells in TRIzol, pipet up and down multiple times.
Snap freeze AFs/CEPs in liquid nitrogen immediately and proceed to the mechanical disruption of the AF/CEP tissues.
Figure 1.
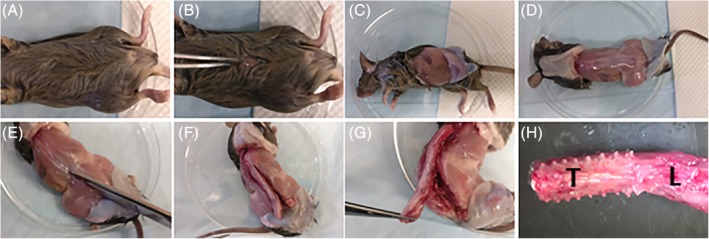
Spinal column isolation. A, Douse the fur with 70% ethanol to prevent contamination and limit spread of hair. Place the mouse on its back. B, An incision is made at level of the umbilicus on the ventral skin. C, A transverse tear is made either manually or with scissors in both directions from the incision to the dorsal spine. D, The pelt is pulled to the base of the tail and to the base of the neck. The spinous processes should now be visible, allowing visualization of the boundaries of the spine. E, With scissors, begin excising the spine from its lateral attachments of muscle, bone, and fascia in a parallel position to the spine. The process is easiest if beginning at the base of the tail and working in a superior direction, ending distal to the region of interest. F, Once one side has been cut, proceed to the opposite side, cutting in the same areas. G, Once the spine has been freed from its lateral attachments, it is still connected both superiorly and inferiorly to the cervical and sacral spine and anteriorly by viscera. Make a transverse cut below the attachment of the femurs, and, pulling the spine away from the body, begin cutting the anterior attachments. Proceed superiorly until the regions of interest have been detached anteriorly, and make another transverse cut through the superior attachment, freeing the spinal column from the body. H, The excised spine. L, lumbar region; T, thoracic region
Figure 2.
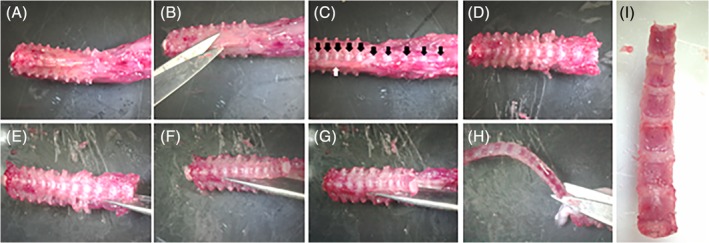
Isolation of the vertebral column from the spine. A, Display of the thoracic and lumbar segments of the spine after spine isolation. B, Using scissors or a scalpel, remove any muscles, fat, or other soft tissues adhering to the anterior part of the vertebral column. C, Removal of the soft tissue exposes the IVDs (black arrows). Tissue continues to be attached on the lateral portions of the IVD (white arrow). D, Continue removing soft tissue from the lateral portions of the vertebral column. This makes the future isolation of the IVD tissue simpler. E, Using scissors, begin cutting through the pedicles, parallel to the vertebral column, to isolate the vertebral column from the remainder of the bony spine. This is best done by inserting one tip of the scissors into the spinal canal and orienting the other tip close to the vertebral body but not on top of it. G, Continue cutting the pedicles on one side of the vertebral column. After that side is cut, proceed to the opposite side. H, Remove any adherent tissues connecting the vertebral column to the remainder of the spine. I, Isolated vertebral column, cleaned of adherent soft tissues
Figure 3.
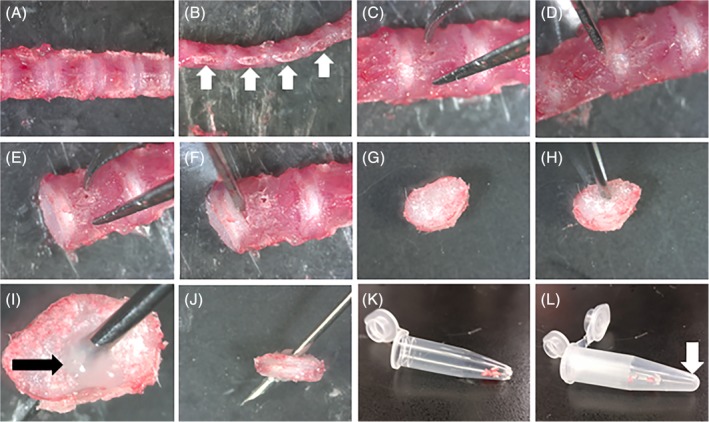
Isolation of the IVD. A, Posterior view of the isolated spine. B, Lateral view of the isolated spine, with the cut pedicles visible (white arrows). C, Proper placement of fine needle tweezers to avoid damage to the IVD. Isolation is simplified by placing the IVD with the posterior portion resting on the iced dissection field and the tweezers making contact on the anterior aspect. D, Proper placement of the scalpel blade on the boundary between the IVD and the superior vertebral body. E, Appearance of the IVD after removal of the superior vertebral body. F, Placement of the scalpel blade to remove the inferior vertebral body. G, Isolated IVD. H, Proper placement of the needle in the superior surface of the IVD. I, Presence of the NP should be confirmed by the appearance of a clear gelatinous NP (black arrow) protruding through the punctured surface. J, The needle is continued through the IVD, puncturing the inferior aspect of the IVD. K, The IVD is deposited in the 0.5 mL tube previously prepared. L, The 0.5 mL tube is placed in a larger 1.5 mL tube to be centrifuged. The NP will collect in the larger tube (white arrow) while the AF/CEP will remain in the original 0.5 mL tube
Mechanical disruption of the AF/CEP tissues:
Place the −80°C frozen tube(s) of AF/CEPs in liquid nitrogen to cool.
After cleaning the mortar and pestle with water, 70% ethanol and then RNase decontamination reagent, fill clean mortar and pestle with liquid nitrogen to cool the equipment to below freezing.
-
With liquid nitrogen still present in the mortar, deposit the AF/CEP frozen sample into the liquid nitrogen and proceed to grind the sample to a fine dust.
! Warning—It is essential that the sample remains below freezing temperature in order to reduce RNA damage.
With a cold metal spatula, scrape the fine powder down the walls of the mortar into the bottom as the liquid nitrogen evaporates. Deposit into the original labeled cold microcentrifuge tube and place back in liquid nitrogen.
-
Samples may then be stored at −80°C or processed for RNA extraction.
! Warning—Mortar and pestle must be cleaned in between each sample to remove previous tissue and prevent cross‐contamination.
Troubleshooting/General caution: It is possible to include bone fragments and bone marrow with the IVD if insufficient care is not exercised during dissection. Inclusion of bone marrow into the NP fraction will be visualized after centrifugation as a dark red mass at the bottom of the tube. Younger animals are simpler to readily separate the CEP from the vertebral body.
2.2. Sample size
IVDs were isolated from a total 32 male mice (ages 1‐15 months) and 35 female mice (ages 1‐7 months) according to the method described herein. Five lumbar IVDs and seven thoracic IVDs were separated from each animal. IVDs from each region were pooled and analyses were performed separately in thoracic and lumbar IVDs.
2.3. RNA extraction
Average time for IVD isolation measured from euthanasia to freezing or Trizol addition was 17 minutes. Centrifuged NP or ground AF/CEP samples were homogenized in 500 μL of TRIzol reagent (Ambion, cat. No. 15596026). Extraction was performed according to manufacturer's directions. Gross RNA integrity was first assessed by gel electrophoresis. Yield and purity were measured by OD260/280 with a spectrophotometer (Tecan Infinite M200; Tecan Deutschland GmbH, Crailsheim, Germany). Ten RNA samples with acceptable gross RNA integrity were further assessed for RIN (RNA Integrity Number) with a bioanalyzer (Agilent 2100; Santa Clara).
2.4. cDNA synthesis and gene expression analyses
cDNAs were generated using the High‐Capacity cDNA Reverse Transcription Kit (Applied Biosystems, cat. no. 4368813). Real‐time qPCR was performed with 23 ng of starting cDNAs that were mixed with iQ SYBR Green Supermix (Bio‐Rad, cat. no. 1708882) and primers. Primer sequences can be found in Table 2. Cycling conditions consisted of an initial step at 95°C for 3 minutes followed by 40 cycles of a 10‐second period at 95°C and a 45‐second period at 60°C. Specificity of amplification was verified by the presence of a single peak on melting curves. Relative expression was determined using the 2‐ΔΔCt values with Pnn used as normalizer. Ct values are shown in Table S1.
Table 2.
qPCR primer set sequences
| Gene | Forward primer (5′→3′) | Reverse primer (5′→3′) |
|---|---|---|
| Pnn | ACCTGGAAGGGGCAGTCAGTA | ATCATCGTCTTCTGGGTCGCT |
| Pax1 | CCGCCTACGAATCGTGGAG | CCCGCAGTTGCCTACTGATG |
| Col10a1 | GCATCTCCCAGCACCAGA | CCATGAACCAGGGTCAAGAA |
| Bra | GCTCAAGGAGCTACTAACGAG | CCAGCAAGAAAGAGTACATGGC |
| Cd24 | GTTGCACCGTTTCCCGGTAA | CCCCTCTGGTGGTAGCGTTA |
| Tnfa | CCCTCACACTCAGATCATCTTCT | GCTACGACGTGGGCTACAG |
| Mmp9 | CTTCTGGCGTGTGAGTTTCC | ACTGCAGGTTGAAGCAAAGA |
| Mmp13 | CAGTCTCCGAGGAGAAACTATGAT | GGACTTTGTCAAAAAGAGCTCAG |
| Il1b | GAAATGCCACCTTTTGACAGTG | TGGATGCTCTCATCAGGACAG |
| Rankl | CAGCATCGCTCTGTTCCTGTA | CTGCGTTTTCATGGAGTCTCA |
| Rank | CACTGAGGAGACCACCCAAG | TGGCAGCCACTACTACCACA |
| Ibsp | CAGGGAGGCAGTGACTCTTC | AGTGTGGAAAGTGTGGCGTT |
| Cd31 | ACGCTGGTGCTCTATGCAAG | TCAGTTGCTGCCCATTCATCA |
2.5. Statistical analyses
Statistical tests are specified in the legend of each figure. When data were analyzed by two‐way ANOVA, tissue compartment (NP vs AF/CEP) and age were the main effects (independent variables). Levene's tests were used to confirm that the data met the equal variance assumption. Where the assumption was violated (Mmp13), the data were log transformed to correct the violation. Where the age effect was significant (and the tissue compartment by interaction was not), the age effect was further examined with trend analysis, testing for significant linear, quadratic, and cubic trends to determine if and how gene expression changed over time. Significance was defined as P < .05.
3. RESULTS
Average yield of RNA for NP and AF/CEP fractions was 817 and 778 ng/IVD, respectively, when averaging all ages, genders, and spine regions together. Further analysis for average RNA yield per IVD over different ages and per gender revealed variability in the yield of recovered RNA at each age in both NP and AF/CEP fractions, and a general trend toward a reduction in RNA yield in samples from older mice (Figure 4A,B). Significantly more RNA was procured from lumbar NP (P = .007) and AF/CEPs (P = .006) when compared to thoracic counterparts (Figure 4C). The RIN range was 4.7 to 5.8. There was no correlation between RIN number and age of the mouse.
Figure 4.
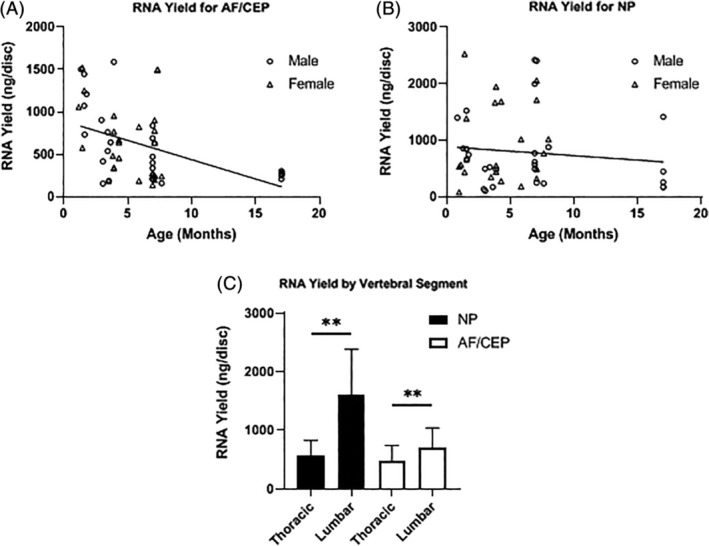
RNA yield by age, gender and IVD structure. A and B, Changing yield of RNA (ng/disc) over time by IVD compartments (n = 5 per age, linear regression for nonzero slope, *P < .05). C, RNA yield by vertebral segment (n = 5 per age, Mann‐Whitney U Test, **P < .01)
After establishing that this protocol resulted in usable quantities of RNA, samples were examined for purity. The transcription factors Brachyury and Cd24 as well as and Pax1 and Col10a1, were selected as markers of NP and AF/CEP cells, respectively.4, 32, 33 A 73‐ and 69‐fold AF/CEP enrichment vs the NP fractions for Pax1 and Col10a1 expression, respectively, across age groups indicated absence or very low AF/CEP RNA contamination in NP samples (Figure 5A,B). In contrast, Bra and Cd24 expression was not different between NP and AF/CEP fractions, thus indicating significant contamination of NP RNA in AF/CEP samples, as these latter genes are not expressed in AF and CEPs (Figure 5C, D).34 This result was confirmed histologically on IVD cryosections showing remnants of NP cells following centrifugation (Figure S1A). Compared to AF/CEP fractions, the NP fractions also showed very low (6‐ to 60‐fold weaker) expression of marker genes for mature chondrocyte (Rankl, Col10a1), monocytes (Rank), osteoblasts (Ibsp), and endothelial cells (Cd31) (Figure S1B‐E). This method, therefore, demonstrates a good capacity to isolate NP cells from the IVD, but insufficient tissue separation to ensuring an AF/CEP cell mixture free from NP contamination.
Figure 5.
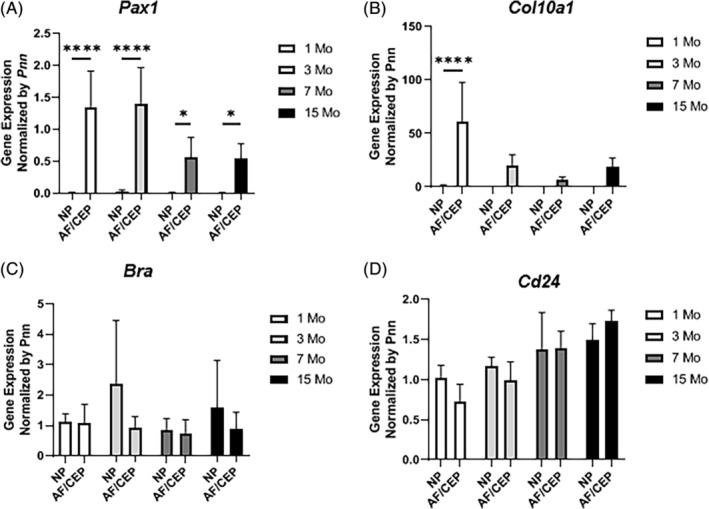
Purity of isolated samples. Gene expression (qPCR) for (A) Pax1, (B) Col10a1, (C) Bra, and (D) Cd24 in NP vs AF/CEP from 1 to 15 months of age (n = 5 per age, two‐way ANOVA with Sidak's multiple comparison test because of significant interaction for Pax1 and Col10a1. *P < .05, ****P < .0001)
Because prior studies have correlated inflammatory gene expression in mouse IVDs only in the context of disease‐causing treatments or conditions, we took advantage of this longitudinal dataset to measure the expression of inflammatory genes and extracellular remodeling enzymes in NPs and AF/CEPs, with the goal of determining if gene expression changed with age and varied between the two isolated tissue fractions. The expression of tumor necrosis factor (Tnfa) and matrix metalloproteinase‐9 (Mmp9) was similar with only tissue compartment showing a significant difference (Figure 6A,B). Expression in the NP was significantly greater than in the AF/CEP for both Tnfa (F1,31 = 5.07, P = .03, partial eta‐square = 0.14) and Mmp9 (F1,31 = 7.32, P = .01, partial eta‐square = 0.18). Age difference was nonsignificant (alpha = .05). Mmp13 expression showed no significant age‐by‐tissue compartment interaction, but difference in tissue compartment (F1,31 = 392.72, P < .01, partial eta square = .92) and age (F3,31 = 26.65, P < .01, partial eta square = .68) were both significant (Figure 6C). However, in this case, Mmp13 expression in the AF/CEP was significantly greater than in the NP. Trend analysis of the age alone indicated a significant negative linear effect (t = −1.95, P = .05, eta‐square = 0.54), indicating that Mmp13 expression significantly decreased with aging. The quadratic and cubic trends were not significant (t = 1.4, P = .16 and t = 1.11, P = .27 respectively). The expression pattern for Il1b, a key cytokine implicated in disc degeneration, was similar to that of Mmp13 with no significant age‐by‐tissue compartment interaction, but significant differences between tissue compartment (F1,31 = 46.15, P < .01, partial eta‐square = 0.62) and over time (F3,31 = 3.11, P < .04, partial eta square = 0.25) (Figure 6D). However, like Tnfa and Mmp9, Il1b expression in the NP was significantly greater than the AF/CEP. None of the trends (linear, quadratic, or cubic) were significant (alpha = .05), though Figure 6D suggests a positive linear increase in gene expression over time with a t value approaching significance (t = 1.9, P = .06, eta‐square = 0.94).
Figure 6.
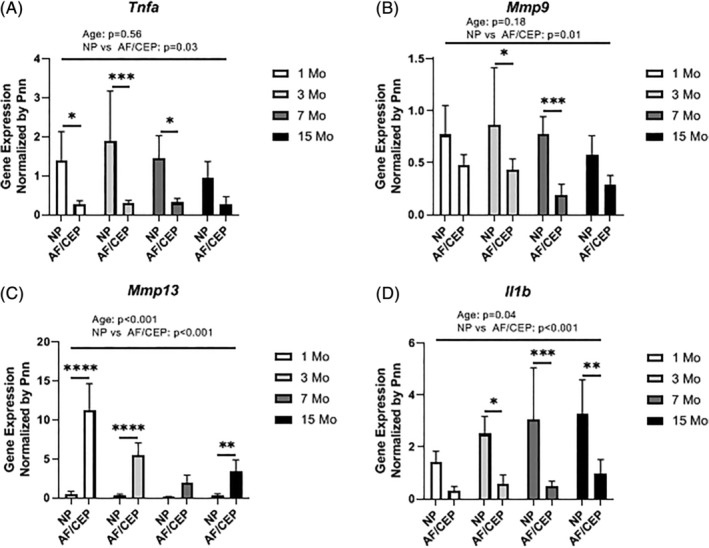
Expression of inflammatory cytokine and metalloproteinase genes. Gene expression (qPCR) for (A) Tnfa, (B) Mmp9, (C) Mmp13, and (D) Il1b (n = 5 per age, two‐way ANOVA). Interaction was nonsignificant for all genes tested. Expression differed significantly between NP and AF/CEP for each gene measured (A: P = .03, B: P = .01, C: P < .001, D: P < .001). Age was a significant factor for both Mmp13 and Il1b with P < .001 and P = .04, respectively. Changing expression over time analyzed by orthogonal polynomial contrasts, with a significant linear decrease only for Mmp13 (P < .05), with a nonsignificant linear increase in Il1b (P = .06). Post‐hoc test with Sidak's multiple comparison test was performed for analysis of NP and AF/CEP expression at each age (*P < .05, **P < .01, ***P < .001, ****P < .0001)
4. DISCUSSION
The method described herein allows multiple and reliable qPCR analyses from lumbar and thoracic mouse IVD compartments and can circumvent the need for pooling IVDs from different spine regions or mice in nonsurgical models. This is important as gene expression may differ between different levels of the spine (cervical vs thoracic vs lumbar vs tail). Likely due to their relatively larger size, lumbar IVDs yielded a greater amount of RNA than thoracic IVDs.
The NP has been implicated as an indicator and driver of IVD degeneration.35 The ability to procure a pure fraction of NP tissue allows precise investigation of molecular phenotypes, response to treatments, and the degenerative process separate from the AF/CEP. This prerequisite information can serve as a foundation for future cell‐ and stem cell‐based therapies.
A caveat of the method described herein is the low RIN numbers observed in samples examined for RNA integrity. Although inadequate sample handling or storage may cause RNA degradation, the low RIN values obtained likely result from partial RNA degradation that may occur postmortem during the dissection process, before samples are snap‐frozen or homogenized in guanidine‐containing solutions, as average dissection time to collect both lumbar and thoracic IVDs was over 15 minutes per mouse. One should note, however, that if normalized with a reference gene, RIN does not overtly affect qPCR expression profiles.36, 37 Thus, although one should always aim at obtaining RNAs with RIN > 5, the method described herein is suitable for most qPCR gene expression analyses. Reducing the time between euthanasia, dissection, and tissue freezing is likely to increase average RIN. Collecting lumbar vertebrae, which yield larger amount of RNAs than other spine regions, is also recommended. Keeping samples at low temperatures during dissection by working on a refrigerated dissection surface is also important. Mechanical disruption of the tissues while submerged in liquid nitrogen follows this principle, as disruption with a bead mill or a mechanical homogenizer exposes the samples to higher temperatures.
By comparing the expression of tissue specific transcription factors, we were able to show that the isolated NP is relatively free of AF/CEP and marrow cell contamination, but the above‐described method is insufficient to result in an AF/CEP fraction free of NP cell contamination, as shown by the detection of NP‐specific gene (Bra, Cd24) in AF/CEP fractions and the presence of NP remnants in centrifuged IVDs by histology. Therefore, the gelatinous property of the NP helps this tissue to be extracted by simple centrifugation from the IVDs, but peripheral NP cells remaining within the intervertebral discs during centrifugation contaminate AF/CEP samples.
There have been efforts made to differentiate changes due to the normal physiologic process of aging and the pathologic degeneration seen in degenerative disc disease. For instance, some studies have shown that IL1β and TNFα were key inflammatory cytokines in disc degeneration, released by IVD cells in addition to infiltrating inflammatory cells and able to induce catabolic and anti‐anabolic shifts in bovine and human IVDs.38, 39, 40 We report here no increase in expression of these key inflammatory genes, in the absence of disease‐causing conditions or degeneration‐susceptible gene knockout, up to 15 months of age in the mouse NP. The only significant change in expression with age was a decrease in Mmp13 expression, which may reflect reduced ECM remodeling. To our knowledge, this is one of the first reports where these genes were examined in the context of aging and nondegenerative state in in vivo mouse IVDs and in different IVD compartments, establishing an important foundation for future studies focused on the correlation between aging and inflammatory gene expression. Our data show that although MRI, radiographic, and histologic evidence of IVD degeneration may occur at earlier ages,41, 42, 43, 44 no significant changes in gene expression for inflammatory markers occur up to 15 months of age in mice. These data are in line with a recent study comparing global changes in gene expression during aging in the mouse NPs and AFs by microarray analysis, which did not reveal any genes related to inflammation in the top most upregulated genes.26
5. CONCLUSION
Mice are important animal models for investigating disease processes and genetic influences in spine development, aging, and degeneration. We present a reliable and reproducible method for isolating murine NP tissue from IVDs. We also show that there is no evidence of inflammation of the mouse NP up to 15 months of age at the gene expression level.
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
AUTHOR CONTRIBUTIONS
A.B., G.C., and F.E. designed experiments. A.B. and G.C. performed and analyzed measurements. A.B., G.C., and F.E. wrote the manuscript. All authors reviewed and approved the manuscript.
Supporting information
Supplementary Figure 1 Non‐contamination of isolated samples by marrow components. A) IVD cryosections from intact and centrifuged lumbar IVDs from 2‐month‐old mice stained by Alcian blue. Picture are representative of 6 IVD isolated from 3 mice. Bar = 50 μm. B‐E) Gene expression (qPCR) for A) Rankl, B) Rank, C) Ibsp and D) Cd31 in NP vs AF/CEP in 1‐month‐old mice (n = 5 per age, Mann‐Whitney U test, ***: P < 0.001, ****: P < 0.0001).
Supplementary Table 1 qPCR CT values.
ACKNOWLEDGMENT
We thank the expert assistance of Drs. Lisa D. White, Ph.D. and Daniel Kraushaar, Ph.D. at the Genomic and RNA Profiling Core at Baylor College of Medicine. We also thank Dr. Brian Bingham, Ph.D. for his assistance with the statistical analyses.
Bratsman A, Couasnay G, Elefteriou F. A step‐by‐step protocol for isolation of murine nucleus pulposus cells. JOR Spine. 2019;2:e1073 10.1002/jsp2.1073
REFERENCES
- 1. Juniper M, Le TK, Mladsi D. The epidemiology, economic burden, and pharmacological treatment of chronic low back pain in France, Germany, Italy, Spain and the UK: a literature‐based review. Expert Opin Pharmacother. 2009;10:2581‐2592. [DOI] [PubMed] [Google Scholar]
- 2. Hoy D, Brooks P, Blyth F, Buchbinder R. The epidemiology of low back pain. Best Pract Res Clin Rheumatol. 2010;24:769‐781. [DOI] [PubMed] [Google Scholar]
- 3. Qaseem A, Wilt TJ, McLean RM, et al. Noninvasive treatments for acute, subacute, and chronic low back pain: a clinical practice guideline from the American College of Physicians. Ann Intern Med. 2017;166:514‐530. [DOI] [PubMed] [Google Scholar]
- 4. Choi KS, Cohn MJ, Harfe BD. Identification of nucleus pulposus precursor cells and notochordal remnants in the mouse: implications for disk degeneration and chordoma formation. Dev Dyn. 2008;237:3953‐3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McCann MR, Tamplin OJ, Rossant J, et al. Tracing notochord‐derived cells using a Noto‐cre mouse: implications for intervertebral disc development. Dis Model Mech. 2012;5:73‐82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aguiar DJ, Johnson SL, Oegema TR. Notochordal cells interact with nucleus pulposus cells: regulation of proteoglycan synthesis. Exp Cell Res. 1999;246:129‐137. [DOI] [PubMed] [Google Scholar]
- 7. Boos N, Weissbach S, Rohrbach H, Weiler C, Spratt KF, Nerlich AG. Classification of age‐related changes in lumbar intervertebral discs: 2002 Volvo Award in basic science. Spine. 2002;27:2631‐2644. [DOI] [PubMed] [Google Scholar]
- 8. Hunter CJ, Matyas JR, Duncan NA. The notochordal cell in the nucleus pulposus: a review in the context of tissue engineering. Tissue Eng. 2003;9:667‐677. [DOI] [PubMed] [Google Scholar]
- 9. Christ B, Wilting J. From somites to vertebral column. Ann Anat. 1992;174:23‐32. [DOI] [PubMed] [Google Scholar]
- 10. Pattappa G, Li Z, Peroglio M, Wismer N, Alini M, Grad S. Diversity of intervertebral disc cells: phenotype and function. J Anat. 2012;221:480‐496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alini M, Eisenstein SM, Ito K, et al. Are animal models useful for studying human disc disorders/degeneration? Eur Spine J. 2008;17:2‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee JT, Cheung KM, Leung VY. Systematic study of cell isolation from bovine nucleus pulposus: improving cell yield and experiment reliability. J Orthop Res. 2015;33:1743‐1755. [DOI] [PubMed] [Google Scholar]
- 13. Cui M, Wan Y, Anderson DG, et al. Mouse growth and differentiation factor‐5 protein and DNA therapy potentiates intervertebral disc cell aggregation and chondrogenic gene expression. Spine J. 2008;8:287‐295. [DOI] [PubMed] [Google Scholar]
- 14. Kushioka J, Kaito T, Chijimatsu R, et al. A novel and efficient method for culturing mouse nucleus pulposus cells Spine J. 2019;9:1573‐1583. [DOI] [PubMed] [Google Scholar]
- 15. Ohba T, Haro H, Ando T, et al. A potential role of thymic stromal lymphopoietin in the recruitment of macrophages to mouse intervertebral disc cells via monocyte chemotactic protein 1 induction: implications for herniated discs. Arthritis Rheum. 2008;58:3510‐3519. [DOI] [PubMed] [Google Scholar]
- 16. Haro H, Crawford HC, Fingleton B, et al. Matrix metalloproteinase‐3‐dependent generation of a macrophage chemoattractant in a model of herniated disc resorption. J Clin Invest. 2000;105:133‐141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Choi H, Tessier S, Silagi ES, et al. A novel mouse model of intervertebral disc degeneration shows altered cell fate and matrix homeostasis. Matrix Biol. 2018;70:102‐122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Y, Tian Z, Ashley JW, et al. Extracellular matrix and adhesion molecule gene expression in the normal and injured murine intervertebral disc. Am J Phys Med Rehab. 2019;98:35‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alvarez‐Garcia O, Matsuzaki T, Olmer M, et al. FOXO are required for intervertebral disk homeostasis during aging and their deficiency promotes disk degeneration. Aging Cell. 2018;17:e12800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dudek M, Yang N, Ruckshanthi JP, et al. The intervertebral disc contains intrinsic circadian clocks that are regulated by age and cytokines and linked to degeneration. Ann Rheum Dis. 2017;76:576‐584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li X, Leo BM, Beck G, Balian G, Anderson GD. Collagen and proteoglycan abnormalities in the GDF‐5‐deficient mice and molecular changes when treating disk cells with recombinant growth factor. Spine. 2004;29:2229‐2234. [DOI] [PubMed] [Google Scholar]
- 22. Holguin N, Silva MJ. In‐vivo nucleus pulposus‐specific regulation of adult murine intervertebral disc degeneration via Wnt/Beta‐catenin signaling. Sci Rep. 2018;8:11191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li CG, Liang QQ, Zhou Q, et al. A continuous observation of the degenerative process in the intervertebral disc of Smad3 gene knock‐out mice. Spine. 2009;34:1363‐1369. [DOI] [PubMed] [Google Scholar]
- 24. McCann MR, Patel P, Pest MA, et al. Repeated exposure to high‐frequency low‐amplitude vibration induces degeneration of murine intervertebral discs and knee joints. Arthritis Rheumatol. 2015;67:2164‐2175. [DOI] [PubMed] [Google Scholar]
- 25. Peck SH, McKee KK, Tobias JW, et al. Whole transcriptome analysis of notochord‐derived cells during embryonic formation of the nucleus pulposus. Sci Rep. 2017;7:10504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Veras MA, McCann MR, Tenn NA, Séguin CA. Transcriptional profiling of the murine intervertebral disc and age‐associated changes in the nucleus pulposus. Connect Tissue Res. 2019;1‐19. 10.1080/03008207.2019.1665034. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 27. Tang X, Richardson WJ, Fitch RD, Brown CR, Isaacs RE, Chen J. A new non‐enzymatic method for isolating human intervertebral disc cells preserves the phenotype of nucleus pulposus cells. Cytotechnology. 2014;66:979‐986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krupkova O, Sadowska A, Kameda T, et al. p38 MAPK facilitates crosstalk between endoplasmic reticulum stress and IL‐6 release in the intervertebral disc. Front Immunol. 2018;9:1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sadowska A, Touli E, Hitzl W, et al. Inflammaging in cervical and lumbar degenerated intervertebral discs: analysis of proinflammatory cytokine and TRP channel expression. Eur Spine J. 2018;27:564‐577. [DOI] [PubMed] [Google Scholar]
- 30. Park JY, Yoon YS, Park HS, Kuh SU. Molecular response of human cervical and lumbar nucleus pulposus cells from degenerated discs following cytokine treatment. Genet Mol Res. 2013;12:838‐851. [DOI] [PubMed] [Google Scholar]
- 31. Richardson SM, Ludwinski FE, Gnanalingham KK, Atkinson RA, Freemont AJ, Hoyland JA. Notochordal and nucleus pulposus marker expression is maintained by sub‐populations of adult human nucleus pulposus cells through aging and degeneration. Sci Rep. 2017;7:1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sivakamasundari V, Kraus P, Sun W, et al. A developmental transcriptomic analysis of Pax1 and Pax9 in embryonic intervertebral disc development. Biol Open. 2017;6:187‐199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maier JA, Lo Y, Harfe BD. Foxa1 and Foxa2 are required for formation of the intervertebral discs. PLoS One. 2013;8:e55528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fujita N, Miyamoto T, Imai J, et al. CD24 is expressed specifically in the nucleus pulposus of intervertebral discs. Biochem Biophys Res Commun. 2005;338:1890‐1896. [DOI] [PubMed] [Google Scholar]
- 35. Choi H, Johnson ZI, Risbud MV. Understanding nucleus pulposus cell phenotype: a prerequisite for stem cell based therapies to treat intervertebral disc degeneration. Curr Stem Cell Res Ther. 2015;10:307‐316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schoor O, Weinschenk T, Hennenlotter J, et al. 2003. Moderate degradation does not preclude microarray analysis of small amounts of RNA. Biotechniques 35:1192–1196, 1198–1201. [DOI] [PubMed] [Google Scholar]
- 37. Fleige S, Pfaffl MW. RNA integrity and the effect on the real‐time qRT‐PCR performance. Mol Aspects Med. 2006;27:126‐139. [DOI] [PubMed] [Google Scholar]
- 38. Purmessur D, Walter BA, Roughley PJ, Laudier DM, Hecht AC, Iatridis J. A role for TNFalpha in intervertebral disc degeneration: a non‐recoverable catabolic shift. Biochem Biophys Res Commun. 2013;433:151‐156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Le Maitre CL, Freemont AJ, Hoyland JA. The role of interleukin‐1 in the pathogenesis of human intervertebral disc degeneration. Arthritis Res Ther. 2005;7:R732‐R745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Risbud MV, Shapiro IM. Role of cytokines in intervertebral disc degeneration: pain and disc content. Nat Rev Rheumatol. 2014;10:44‐56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marfia G, Campanella R, Navone SE, et al. Potential use of human adipose mesenchymal stromal cells for intervertebral disc regeneration: a preliminary study on biglycan‐deficient murine model of chronic disc degeneration. Arthritis Res Ther. 2014;16:457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Boyd LM, Richardson WJ, Allen KD, et al. Early‐onset degeneration of the intervertebral disc and vertebral end plate in mice deficient in type IX collagen. Arthritis Rheum. 2008;58:164‐171. [DOI] [PubMed] [Google Scholar]
- 43. Millecamps M, Czerminski JT, Mathieu AP, Stone LS. Behavioral signs of axial low back pain and motor impairment correlate with the severity of intervertebral disc degeneration in a mouse model. Spine J. 2015;15:2524‐2537. [DOI] [PubMed] [Google Scholar]
- 44. Ohnishi T, Sudo H, Tsujimoto T, Iwasaki N. Age‐related spontaneous lumbar intervertebral disc degeneration in a mouse model. J Orthop Res. 2018;36:224‐232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 Non‐contamination of isolated samples by marrow components. A) IVD cryosections from intact and centrifuged lumbar IVDs from 2‐month‐old mice stained by Alcian blue. Picture are representative of 6 IVD isolated from 3 mice. Bar = 50 μm. B‐E) Gene expression (qPCR) for A) Rankl, B) Rank, C) Ibsp and D) Cd31 in NP vs AF/CEP in 1‐month‐old mice (n = 5 per age, Mann‐Whitney U test, ***: P < 0.001, ****: P < 0.0001).
Supplementary Table 1 qPCR CT values.


