Abstract
A series of readily accessible 1-(piperidin-3-yl)thymine amides was designed, synthesised and evaluated as Mycobacterium tuberculosis TMPK (MtbTMPK) inhibitors. In line with the modelling results, most inhibitors showed reasonable MtbTMPK inhibitory activity. Compounds 4b and 4i were slightly more potent than the parent compound 3. Moreover, contrary to the latter, amide analogue 4g was active against the avirulent M. tuberculosis H37Ra strain (MIC50=35 µM). This finding opens avenues for future modifications.
Keywords: Thymidylate kinase, inhibitors, Mycobacterium tuberculosis, modelling
1. Introduction
Tuberculosis (TB) is a severe air transmitted infectious disease mainly caused by Mycobacterium tuberculosis (Mtb) and causes significant morbidity and mortality worldwide1. It is estimated that 10.4 million people fell ill with TB in 2017, and 1.6 million people succumbed the infection2. Standard treatment of drug-sensitive TB normally lasts 6 months, including a 2-month course with at least three first-line drugs and a 4-month course with at least Isoniazid (INH) and Rifampicin (RIF), resulting in poor treatment compliance and a high financial burden. Moreover, the increasing number of multidrug-resistant TB (MDR-TB), extensively drug-resistant TB (XDR-TB) and rifampicin-resistant TB (RR-TB) makes the treatment even more challenging3,4. New drugs acting on yet unexplored targets/pathways are desirable to achieve worldwide TB control.
Thymidine monophosphate kinase (TMPK), an enzyme at the junction of the de novo (involving thymidylate synthase) and salvage pathway, is responsible for the conversion of thymidine monophosphate (TMP) to thymidine diphosphate (TDP)5,6. Further phosphorylation leads to the formation of thymidine triphosphate (TTP), which is indispensable for DNA synthesis7. Mycobacterium tuberculosis TMPK (MtbTMPK) shows low (22%) sequence identity with the human isozyme and has a unique catalytic mechanism8,9, further supporting it as an attractive target for developing new anti-TB drugs. Both industrial and academic efforts have afforded several potent MtbTMPK inhibitors in the past two decennia10–19, including thymidine-like and non-nucleoside inhibitors (Figure 1)20–24.
Figure 1.
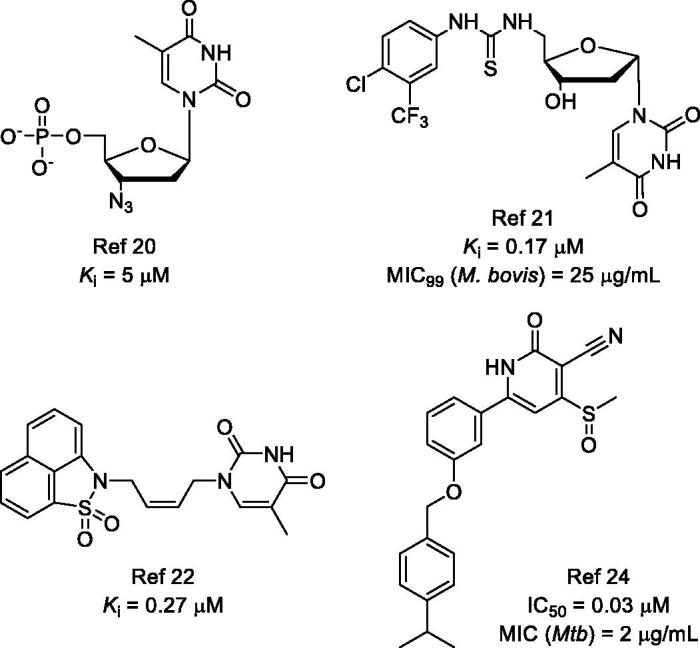
Representative thymidine-like and non-nucleoside MtbTMPK inhibitors.
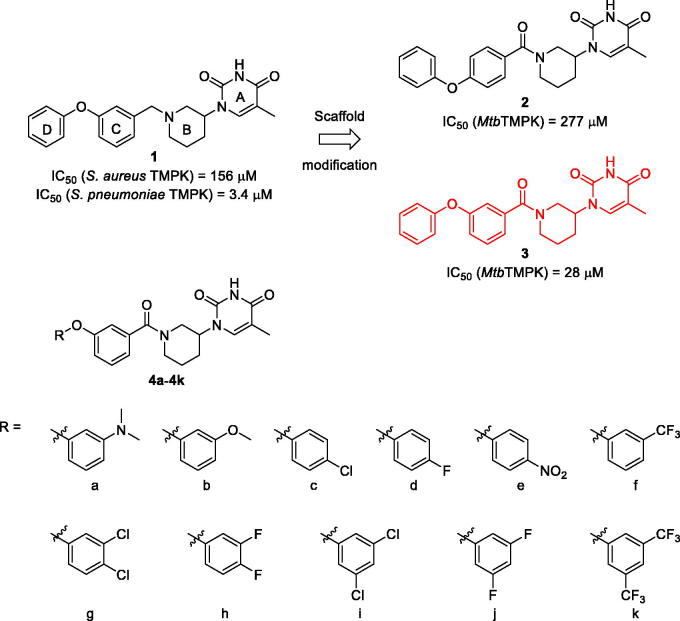
Using the Gram-positive bacterial thymidylate kinase inhibitor 1 (Figure 2) disclosed by Astra Zeneca7 as a starting point, our research group is trying to identify potent non-nucleoside MtbTMPK inhibitors25. These efforts resulted in the identification of the regiomeric racemates 2 and 3, of which the latter showed the best MtbTMPK inhibitory activity and was therefore selected for further optimisation. In this study, we synthesised a series of D-ring substituted analogues of 3 and evaluated these as MtbTMPK inhibitors (Figure 2).
Figure 2.
Structures of regiomeric racemates 2 and 3 derived from Gram-positive bacterial thymidylate kinase inhibitor 1 and amide analogues 4a–4k investigated in this study.
2. Materials and methods
2.1. Chemistry
All synthetic compounds mentioned in this study were visualised under UV light at 254 nm or stained by corresponding reagents, and purified by column chromatography (CC) on a Reveleris X2 (Grace) automated flash unit. 1H and 13C NMR spectral data were obtained from a Varian Mercury 300/75 MHz spectrometer at 300 K using TMS as an internal standard. Structural assignment was confirmed with the assistance of COSY, HSQC and HMBC. High-resolution mass spectrometry was performed on a Waters LCT Premier XETM time of flight (TOF) mass spectrometer equipped with a standard electrospray ionisation (ESI) and modular LockSpray TM interface. The purity of the tested compounds was determined by LC-MS analysis (Waters Alliance 2695 XE separation module).
3-((benzyloxy)methyl)-1–(1-(3-iodobenzoyl)piperidin-3-yl)-5-methylpyrimidine-2,4(1H,3H)-dione (6) Intermediate 8 was synthesised as previously reported25. To a solution of 8 (200.00 mg, 0.61 mmol) in dichloromethane (12.00 ml) were added N,N-bis(isopropyl) carbondiimide (153.72 mg, 1.22 mmol), 3-iodobenzoic acid (225.88 mg, 0.91 mmol) and 4-dimethylaminopyridine (7.45 mg, 0.06 mmol), the reaction mixture was stirred at room temperature for overnight. After complete consumption of 8 checked with TLC, the reaction mixture was diluted with dichloromethane (60.00 ml), and washed with 1 M HCl (60.00 ml), saturated NaHCO3 (60.00 ml) and brine (60.00 ml). The organic layer was collected and dried over anhydrous Na2SO4, followed by filtered and concentrated in vacuo. The resulting residue was purified by column chromatography (petroleum ether—EtOAc, 4: 1 v/v) to give solid 6 (230.96 mg, 68.00% yield), which was characterised by 1H and 13 C-NMR, and ESI-HRMS. 1H NMR: δH (300 MHz, CDCl3, TMS) 1.53–2.02 (m, 7 H, piperidin-4-yl, piperidin-5-yl, CH3), 2.83–3.25 (m, 2 H, piperidin-2a-yl, piperidin-6a-yl), 3.66–4.04 (m, 2 H, piperidin-2b-yl, piperidin-6b-yl), 4.34–4.50 (m, 1 H, piperidin-3-yl), 4.70 (s, 2 H, O-CH2-phenyl), 5.50 (s, 2 H, N-CH2-O), 6.93 (s, 1 H), 7.12–7.20 (m, 1 H), 7.23–7.45 (m, 6 H), 7.71–7.82 (m, 2 H). 13C NMR: δC (75 MHz, CDCl3, TMS) 13.22 (5-CH3), 24.84 (piperidin-5-yl), 29.07 (piperidin-4-yl), 53.74 (piperidin-3-yl), 60.38 (piperidin-2-yl), 70.94 (N-CH2-O), 72.33 (O-CH2-phenyl), 94.30 (I-C), 110.41 (C-5), 126.11 (Ph), 127.51 (2 C, Ph), 127.57 (Ph), 128.22 (2 C, Ph), 130.29 (Ph), 135.83 (Ph), 137.20 (Ph), 138.08 (Ph), 139.02 (2 C, C-5, Ph), 151.22 (C-2), 163.02 (C-4), 168.88 (CO, benzoyl). C (piperidin-6-yl) cannot be found. HRMS (ESI): m/z [M + H]+ Calcd. for [C25H26IN3O4+H] + 560.1041, found 560.1047.
3-((benzyloxy)methyl)-1–(1-(3-hydroxybenzoyl)piperidin-3-yl)-5-methylpyrimidine-2,4(1H,3H)-dione (10) To a solution of 8 (1.19 g, 3.62 mmol) in dichloromethane (72 ml) were added N,N-bis(isopropyl) carbondiimide (913.60 mg, 7.24 mmol), 3-hydroxybenzoic acid (749.99 mg, 5.43 mmol) and 4-dimethylaminopyridine (43.98 mg, 0.36 mmol), the reaction mixture was stirred at room temperature for overnight. After complete consumption of 8 checked with TLC, the reaction mixture was diluted with dichloromethane (200 ml), and washed with 1 M HCl (100 ml), saturated NaHCO3 (100 ml) and brine (100 ml). The organic layer was collected and dried over anhydrous Na2SO4, followed by filtered and concentrated in vacuo. The resulting residue was purified by column chromatography (petroleum ether—EtOAc, 3: 1 v/v) to give colourless liquid 10 (1.01 g, 62.12% yield), which was characterised by 1H and 13 C-NMR, and ESI-HRMS. 1H NMR: δH (300 MHz, CDCl3, TMS) 1.59–2.10 (m, 7 H, piperidin-4-yl, piperidin-5-yl, CH3), 2.64–2.83 (m, 1 H, piperidin-6a-yl) 2.88–3.12 (m, 1 H, piperidin-2a-yl), 3.59–4.15 (m, 2 H, piperidin-2b-yl, piperidin-6b-yl), 4.32–4.53 (m, 1 H, piperidin-3-yl), 4.69 (s, 2 H, O-CH2-phenyl), 5.50 (s, 2 H, N-CH2-O), 6.74–7.08 (m, 4 H), 7.12–7.44 (m, 6 H), 7.68 (br. s., 1 H, OH). 13 C NMR: δC (75 MHz, CDCl3, TMS) 13.18 (5-CH3), 24.66 (piperidin-5-yl), 29.04 (piperidin-4-yl), 47.63 (piperidin-6-yl), 53.64 (piperidin-3-yl), 70.99 (N-CH2-O), 72.33 (O-CH2-phenyl), 110.54 (C-5), 114.32 (Ph), 117.76 (Ph), 118.47 (Ph), 127.50 (2 C, Ph), 127.59 (Ph), 128.21 (2 C, Ph), 129.97 (Ph), 135.82 (Ph), 137.97 (2 C, C-6, Ph), 151.36 (C-2), 156.70 (Ph), 163.03 (C-4), 170.91 (CO, benzoyl). C (piperidin-2-yl) cannot be found. HRMS (ESI): m/z [M + H]+ Calcd. for [C25H27N3O5+H]+ 450.2024, found 450.2036.
General procedure for substituted 3-((benzyloxy)methyl)-5-methyl-1–(1-(3 phenoxybenzoyl)piperidin-3-yl)pyrimidine-2,4(1H,3H)-dione (13b–13k). Substituted iodobenzene (0.66 mmol) was added to a stirred solution of 10 (0.44 mmol), copper iodide (0.18 mmol), potassium phosphate tribasic (0.89 mmol), picolinic acid (0.27 mmol) in DMSO under nitrogen and heated at 90 °C for overnight. The reaction mixture was cooled to room temperature, quenched with water (50 ml) and extracted with CH2Cl2 (100 ml). The separated organic layer was further washed with 1 M HCl (50 ml), saturated NaHCO3 (50 ml) and brine (50 ml), dried over anhydrous Na2SO4, and evaporated in vacuo. The concentrated crude was purified by column chromatography (petroleum ether—EtOAc, 1:1 v/v) to give colourless liquid 13a–13k (40.00–70.00% yield), which was characterised by 1H and 13C-NMR, and ESI-HRMS.
13a following the general procedure, starting with 1-iodo-3-dimethylaminobenzene (163.07 mg), 10 (197.78 mg), copper iodide (33.80 mg), potassium phosphate tribasic (189.00 mg), picolinic acid (32.90 mg) in DMSO, 13a was obtained (242.72 mg, 71.00% yield). 1H NMR: δH (300 MHz, CDCl3, TMS) 1.72–2.12 (m, 7 H, piperidin-4-yl, piperidin-5-yl, CH3), 2.68–2.97 (m, 7 H, piperidin-6a-yl, N(CH3)2), 2.97–3.23 (m, 1 H, piperidin-2a-yl,), 3.68–4.03 (m, 2 H, piperidin-2b-yl, piperidin-6b-yl), 4.32–4.51 (m, 1 H, piperidin-3-yl), 4.70 (s, 2 H, O-CH2-phenyl), 5.50 (s, 2 H, N-CH2-O) , 6.33–6.42 (m, 1 H), 6.47 (s, 1 H), 6.53–6.61 (m, 1 H), 6.94 (br. s., 1 H), 7.05 (br. s., 1 H), 7.07–7.15 (m, 2 H), 7.16–7.30 (m, 4 H), 7.31–7.45 (m, 3 H). 13 C NMR: δC (75 MHz, CDCl3, TMS) 13.60 (5-CH3), 23.86 (piperidin-5-yl), 29.54 (piperidin-4-yl), 41.16 (2 C, N(CH3)2), 54.01 (piperidin-3-yl), 71.31 (N-CH2-O), 72.71 (O-CH2-phenyl), 104.69 (Ph), 108.28 (Ph), 109.06 (Ph), 110.70 (C-5), 117.01 (Ph), 120.09 (Ph), 121.42 (Ph), 127.92 (2 C, Ph), 127.97 (Ph), 128.63 (2 C, Ph), 130.41 (Ph), 130.58 (Ph), 136.23 (Ph), 137.16 (Ph), 138.47 (C-6), 151.60 (C-2), 157.63 (Ph), 158.41 (Ph), 163.48 (C-4), 170.48 (CO, benzoyl). C (piperidin-2-yl), C (piperidin-6-yl) and C (CN(CH3)2) cannot be found. HRMS (ESI): m/z [M + H]+ Calcd. for [C33H36N4O5+H]+ 569.2758, found 569.2773.
13b following the general procedure, starting with 1-iodo-3-methoxybenzene (154.46 mg), 10 (197.78 mg), copper iodide (33.80 mg), potassium phosphate tribasic (189.00 mg), picolinic acid (32.90 mg) in DMSO, 13b was obtained (230.47 mg, 69.00% yield). 1H NMR: δH (300 MHz, CDCl3, TMS) 1.54–1.73 (m, 1 H, piperidin-5a-yl), 1.79–2.18 (m, 6 H, piperidin-4-yl, piperidin-5b-yl, CH3), 2.75–3.20 (m, 2 H, piperidin-2a-yl, piperidin-6a-yl), 3.64–4.11 (m, 5 H, piperidin-2b-yl, piperidin-6b-yl, OCH3), 4.35–4.54 (m, 1 H, piperidin-3-yl), 4.70 (s, 2 H, O-CH2-phenyl), 5.50 (s, 2 H, N-CH2-O), 6.57–6.65 (m, 2 H), 6.65–6.75 (m, 1 H), 6.94 (br. s., 1 H), 7.00–7.13 (m, 2 H), 7.16 (d, J = 6.44 Hz, 1 H), 7.20–7.47 (m, 7 H). 13 C NMR: δC (75 MHz, CDCl3, TMS) 13.64 (5-CH3), 25.27 (piperidin-5-yl), 29.56 (piperidin-4-yl), 54.04 (piperidin-3-yl), 55.80 (OCH3), 71.31 (N-CH2-O), 72.71 (O-CH2-phenyl), 105.88 (Ph), 109.96 (Ph), 110.74 (C-5), 111.87 (Ph), 117.48 (Ph), 120.47 (Ph), 121.91 (Ph), 127.92 (2 C, Ph), 127.97 (Ph), 128.63 (2 C, Ph), 130.52 (Ph), 130.70 (Ph), 137.28 (2 C, Ph), 138.47 (C-6), 151.60 (C-2), 157.93 (2 C, Ph), 161.43 (Ph), 163.46 (C-4), 170.36 (CO, benzoyl). C (piperidin-2-yl) and C (piperidin-6-yl) cannot be found. HRMS (ESI): m/z [M + H]+ Calcd. for [C32H33N3O6+H]+ 556.2442, found 556.2465.
13c following the general procedure, starting with 1-iodo-4-chlorobenzene (157.38 mg), 10 (197.78 mg), copper iodide (33.80 mg), potassium phosphate tribasic (189.00 mg), picolinic acid (32.90 mg) in DMSO, 13c was obtained (198.63 mg, 59.00% yield). 1H NMR: δH (300 MHz, CDCl3, TMS) 1.57–1.77 (m, 1 H, piperidin-5a-yl), 1.84–2.12 (m, 6 H, piperidin-4-yl, piperidin-5b-yl, CH3), 2.71–3.19 (m, 2 H, piperidin-2a-yl, piperidin-6a-yl), 3.64–4.14 (m, 2 H, piperidin-2b-yl, piperidin-6b-yl), 4.35–4.54 (m, 1 H, piperidin-3-yl), 4.70 (s, 2 H, O-CH2-phenyl), 5.49 (s, 2 H, N-CH2-O), 6.84–7.01 (m, 3 H), 7.04 (d, J = 6.15 Hz, 2 H), 7.11–7.20 (m, 1 H), 7.21–7.60 (m, 8 H). 13 C NMR: δC (75 MHz, CDCl3, TMS) 13.15 (5-CH3), 24.78 (piperidin-5-yl), 29.02 (piperidin-4-yl), 70.85 (N-CH2-O), 72.25 (O-CH2-phenyl), 110.28 (C-5), 116.92 (Ph), 119.91 (Ph), 120.60 (2 C, Ph), 121.63 (Ph), 127.45 (2 C, Ph), 127.53 (Ph), 128.17 (3 C, Ph), 129.86 (2 C, Ph), 130.17 (Ph), 136.96 (2 C, Ph), 138.00 (C-6), 151.14 (C-2), 154.96 (C, Ph), 157.35 (Ph), 163.00 (C-4), 169.80 (CO, benzoyl). C (piperidin-2-yl), C (piperidin-3-yl) and C (piperidin-6-yl) cannot be found. HRMS (ESI): m/z [M + H]+ Calcd. for [C31H30ClN3O5+H]+ 560.1947, found 560.1954.
13d following the general procedure, starting with 1-iodo-4-fluorobenzene (146.52 mg), 10 (197.78 mg), copper iodide (33.80 mg), potassium phosphate tribasic (189.00 mg), picolinic acid (32.90 mg) in DMSO, 13d was obtained (205.87 mg, 63.00% yield). 1H NMR: δH (300 MHz, CDCl3, TMS) 1.62–1.73 (m, 1 H, piperidin-5a-yl), 1.86–1.93(m, 4 H, piperidin-5b-yl, CH3), 2.00–2.17 (m, 2 H, piperidin-4-yl), 2.68–2.94 (m, 1 H, piperidin-6a-yl), 2.99–3.13 (m, 1 H, piperidin-2a-yl), 3.63–4.12 (m, 2 H, piperidin-2b-yl, piperidin-6b-yl), 4.36–4.51 (m, 1 H, piperidin-3-yl), 4.70 (s, 2 H, O-CH2-phenyl), 5.50 (s, 2 H, N-CH2-O), 6.97–7.08 (m, 7 H), 7.10–7.17 (m, 1 H), 7.23–7.27 (m, 1 H), 7.28–7.52 (m, 5 H). 13 C NMR: δC (75 MHz, CDCl3, TMS) 13.22 (5-CH3), 24.61 (piperidin-5-yl), 29.13 (piperidin-4-yl), 70.91 (N-CH2-O), 72.33 (O-CH2-phenyl), 110.37 (C-5), 116.37 (2 C, Ph), 116.67 (Ph), 119.34 (Ph), 121.10 (Ph), 121.21 (2 C, Ph), 127.51 (2 C, Ph), 127.57 (Ph), 128.23 (2 C, Ph), 130.14 (Ph), 136.93 (2 C, Ph), 138.06 (C-6), 151.20 (C-2), 151.95 (Ph), 157.57 (Ph), 158.16 (Ph), 163.05 (C-4), 169.96 (CO, benzoyl). C (piperidin-2-yl), C (piperidin-3-yl) and C (piperidin-6-yl) cannot be found. HRMS (ESI): m/z [M + H]+ Calcd. for [C31H30FN3O5+H]+ 544.2242, found 544.2219.
13e following the general procedure, starting with 1-iodo-4-nitrobenzene (164.34 mg), 10 (197.78 mg), copper iodide (33.80 mg), potassium phosphate tribasic (189.00 mg), picolinic acid (32.90 mg) in DMSO, 13e was obtained (191.40 mg, 55.80% yield). 1H NMR: δH (300 MHz, CDCl3, TMS) 1.73–2.13 (m, 7 H, piperidin-4-yl, piperidin-5-yl, CH3), 2.67–2.90 (m, 1 H, piperidin-6a-yl), 2.95–3.12 (m, 1 H, piperidin-2a-yl), 3.57–4.17 (m, 2 H, piperidin-2b-yl, piperidin-6b-yl), 4.35–4.55 (m, 1 H, piperidin-3-yl), 4.70 (s, 2 H, O-CH2-phenyl), 5.50 (s, 2 H, N-CH2-O), 6.95 (s, 1 H), 7.01–7.10 (m, 2 H), 7.12–7.20 (m, 2 H), 7.21–7.37 (m, 6 H), 7.43–7.56 (m, 1 H), 8.16–8.30 (m, 2 H). 13 C NMR: δC (75 MHz, CDCl3, TMS) 12.81 (5-CH3), 24.44 (piperidin-5-yl), 28.63 (piperidin-4-yl), 52.91 (piperidin-3-yl), 70.48 (N-CH2-O), 72.90 (O-CH2-phenyl), 110.02 (C-5), 117.35 (2 C, Ph), 118.51 (Ph), 121.28 (Ph), 123.08 (Ph), 125.60 (2 C, Ph), 127.10 (2 C, Ph), 127.18 (Ph), 127.82 (2 C, Ph), 130.23 (Ph), 137.11 (2 C, Ph), 137.59 (C-6), 142.70 (Ph), 150.81 (C-2), 154.81 (Ph), 162.03 (Ph), 162.58 (C-4), 168.96 (CO, benzoyl). C (piperidin-2-yl) and C (piperidin-6-yl) cannot be found. HRMS (ESI): m/z [M + H]+ Calcd. for [C31H30N4O7+H]+ 571.2187, found 571.2191.
13f following the general procedure, starting with 1-iodo-3-(trifluoromethyl)benzene (179.53 mg), 10 (197.78 mg), copper iodide (33.80 mg), potassium phosphate tribasic (189.00 mg), picolinic acid (32.90 mg) in DMSO, 13f was obtained (180.56 mg, 50.60% yield). 1H NMR: δH (300 MHz, CDCl3, TMS) 1.56–2.13 (m, 7 H, piperidin-4-yl, piperidin-5-yl, CH3), 2.63–2.94 (m, 1 H, piperidin-6a-yl), 2.95–3.14 (m, 1 H, piperidin-2a-yl), 3.71–4.09 (m, 2 H, piperidin-2b-yl, piperidin-6b-yl), 4.37–4.52 (m, 1 H, piperidin-3-yl), 4.70 (s, 2 H, O-CH2-phenyl), 5.50 (s, 2 H, N-CH2-O), 6.93 (br. s., 1 H), 7.08 (d, J = 6.15 Hz, 2 H), 7.16–7.37 (m, 7 H), 7.37–7.58 (m, 4 H). 13 C NMR: δC (75 MHz, CDCl3, TMS) 13.21 (5-CH3), 23.36 (piperidin-5-yl), 29.11 (piperidin-4-yl), 53.54 (piperidin-3-yl), 70.91 (N-CH2-O), 72.31 (O-CH2-phenyl), 110.38 (C-5), 115.97 (Ph), 117.53 (Ph), 120.34 (Ph), 120.38 (Ph), 120.46 (2 C, Ph), 122.25 (Ph), 122.34 (Ph), 127.51 (2 C, Ph), 127.57 (Ph), 128.23 (2 C, Ph), 130.43 (Ph), 130.55 (Ph), 137.22 (2 C, Ph), 138.06 (C-6), 151.22 (C-2), 156.96 (Ph), 156.97 (Ph), 163.05 (C-4), 169.67 (CO, benzoyl). C (piperidin-2-yl) and C (piperidin-6-yl) cannot be found. HRMS (ESI): m/z [M + H]+ Calcd. for [C32H30F3N3O5+H]+ 594.2211, found 594.2208.
13g following the general procedure, starting with 1,2-dichloro-4-iodobenzene (180.11 mg), 10 (197.78 mg), copper iodide (33.80 mg), potassium phosphate tribasic (189.00 mg), picolinic acid (32.90 mg) in DMSO, 13 g was obtained (173.32 mg, 48.50% yield). 1H NMR: δH (300 MHz, CDCl3, TMS) 1.58–2.05 (m, 7 H, piperidin-4-yl, piperidin-5-yl, CH3), 2.57–3.08 (m, 2 H, piperidin-6a-yl, piperidin-2a-yl), 3.55–4.13 (m, 2 H, piperidin-2b-yl, piperidin-6b-yl), 4.32–4.48 (m, 1 H, piperidin-3-yl), 4.65 (s, 2 H, O-CH2-phenyl), 5.45 (s, 2 H, N-CH2-O), 6.77–6.89 (m, 1 H), 6.95 (br. s., 1 H), 6.98–7.14 (m, 3 H), 7.14–7.46 (m, 8 H). 13 C NMR: δC (75 MHz, CDCl3, TMS) 12.98 (5-CH3), 24.61 (piperidin-5-yl), 28.79 (piperidin-4-yl), 47.28 (piperidin-6-yl), 53.06 (piperidin-3-yl), 60.15 (piperidin-2-yl), 70.71 (N-CH2-O), 72.07 (O-CH2-phenyl), 109.99 (C-5), 117.28 (Ph), 118.38 (Ph), 120.20 (Ph), 120.72 (Ph), 122.20 (2 C, Ph), 126.90 (Ph), 127.27 (2 C, Ph), 127.36 (Ph), 128.00 (2 C, Ph), 130.21 (Ph), 133.01 (Ph), 137.07 (2 C, Ph), 137.86 (C-6), 151.00 (C-2), 155.51 (Ph), 156.36 (Ph), 162.82 (C-4), 169.32 (CO, benzoyl). HRMS (ESI): m/z [M + H]+ Calcd. for [C31H29Cl2N3O5+H]+ 594.1557, found 594.1588.
13h following the general procedure, starting with 1,2-difluoro-4-iodobenzene (158.39 mg), 10 (197.78 mg), copper iodide (33.80 mg), potassium phosphate tribasic (189.00 mg), picolinic acid (32.90 mg) in DMSO, 13 h was obtained (195.13 mg, 57.80% yield). 1H NMR: δH (300 MHz, CDCl3, TMS) 1.48–2.13 (m, 7 H, piperidin-4-yl, piperidin-5-yl, CH3), 2.81–3.10 (m, 2 H, piperidin-6a-yl, piperidin-2a-yl), 3.78–4.15 (m, 2 H, piperidin-2b-yl, piperidin-6b-yl), 4.30–4.37 (m, 1 H, piperidin-3-yl), 4.71 (s, 2 H, O-CH2-phenyl), 5.45 (s, 2 H, N-CH2-O), 6.98–7.06 (m, 2 H), 7.09–7.21 (m, 8 H), 7.33–7.56 (m, 3 H). 13 C NMR: δC (75 MHz, CDCl3, TMS) 12.61 (5-CH3), 24.89 (piperidin-5-yl), 28.53 (piperidin-4-yl), 53.13 (piperidin-3-yl), 70.92 (N-CH2-O), 72.19 (O-CH2-phenyl), 107.53 (Ph), 109.41 (C-5), 114.86 (Ph), 115.07 (Ph), 117.32 (Ph), 119.78 (Ph), 120.86 (Ph), 127.44 (2 C, Ph), 127.56 (Ph), 128.18 (2 C, Ph), 130.94 (Ph), 137.87 (2 C, Ph), 138.21 (C-6), 145.17 (Ph), 148.35 (Ph), 151.15 (C-2), 152.86 (Ph), 156.78 (Ph), 164.13 (C-4), 168.97 (CO, benzoyl). C (piperidin-2-yl) and C (piperidin-6-yl) cannot be found. HRMS (ESI): m/z [M + H]+ Calcd. for [C31H29F2N3O5+H]+ 562.2148, found 562.2122.
13i following the general procedure, starting with 1,3-dichloro-5-iodobenzene (180.11 mg), 10 (197.78 mg), copper iodide (33.80 mg), potassium phosphate tribasic (189.00 mg), picolinic acid (32.90 mg) in DMSO, 13i was obtained (162.96 mg, 45.60% yield). 1H NMR: δH (300 MHz, CDCl3, TMS) 1.68–2.10 (m, 7 H, piperidin-4-yl, piperidin-5-yl, CH3), 2.67–2.90 (m, 1 H, piperidin-6a-yl), 2.95–3.17 (m, 1 H, piperidin-2a-yl), 3.63–4.14 (m, 2 H, piperidin-2b-yl, piperidin-6b-yl), 4.33–4.57 (m, 1 H, piperidin-3-yl), 4.71 (s, 2 H, O-CH2-phenyl), 5.49 (s, 2 H, N-CH2-O), 6.82–7.00 (m, 3 H) 7.05–7.17 (m, 3 H), 7.18–7.39 (m, 6 H) 7.45 (dd, J = 8.49, 7.62 Hz, 1 H). 13 C NMR: δC (75 MHz, CDCl3, TMS) 13.65 (5-CH3), 25.30 (piperidin-5-yl), 29.54 (piperidin-4-yl), 53.92 (piperidin-3-yl), 71.34 (N-CH2-O), 72.76 (O-CH2-phenyl), 110.84 (C-5), 117.91 (2 C, Ph), 118.46 (Ph), 121.36 (Ph), 123.39 (Ph), 124.26 (Ph), 127.95 (2 C, Ph), 128.02 (Ph), 128.66 (2 C, Ph), 130.95 (Ph), 136.18 (2 C, Ph), 137.66 (2 C, Ph), 138.49 (C-6), 151.65 (C-2), 156.42 (Ph), 158.47 (Ph), 163.48 (C-4), 169.96 (CO, benzoyl). C (piperidin-2-yl), C (piperidin-6-yl) cannot be found. HRMS (ESI): m/z [M + H]+ Calcd. for [C31H29Cl2N3O5+H]+ 594.2211, found 594.1580.
13j following the general procedure, starting with 1,3-difluoro-5-iodobenzene (158.39 mg), 10 (197.78 mg), copper iodide (33.80 mg), potassium phosphate tribasic (189.00 mg), picolinic acid (32.90 mg) in DMSO, 13j was obtained (154.62 mg, 45.80% yield). 1H NMR: δH (300 MHz, CDCl3, TMS) 1.75–2.10 (m, 7 H, piperidin-4-yl, piperidin-5-yl, CH3), 2.62–2.92 (m, 1 H, piperidin-6a-yl), 2.94–3.16 (m, 1 H, piperidin-2a-yl), 3.63–4.16 (m, 2 H, piperidin-2b-yl, piperidin-6b-yl), 4.34–4.55 (m, 1 H, piperidin-3-yl), 4.70 (s, 2 H, O-CH2-phenyl), 5.50 (s, 2 H, N-CH2-O), 6.45–6.62 (m, 3 H), 6.93 (br. s., 1 H), 7.05–7.17 (m, 2 H), 7.21–7.37 (m, 6 H), 7.38–7.53 (m, 1 H). 13 C NMR: δC (75 MHz, CDCl3, TMS) 13.21 (5-CH3), 24.85 (piperidin-5-yl), 29.08 (piperidin-4-yl), 53.58 (piperidin-3-yl), 70.89 (N-CH2-O), 72.30 (O-CH2-phenyl), 99.04 (Ph), 101.10 (Ph), 102.25 (Ph) 110.40 (C-5), 118.29 (Ph), 121.15 (Ph), 122.99 (Ph), 127.50 (2 C, Ph), 127.56 (Ph), 128.21 (2 C, Ph), 130.46 (Ph), 137.25 (2 C, Ph), 138.04 (C-6), 151.22 (C-2), 155.84 (Ph), 161.84 (Ph), 163.02 (C-4), 165.14 (Ph), 165.34 (Ph), 169.54 (CO, benzoyl). C (piperidin-2-yl), C (piperidin-6-yl) cannot be found. HRMS (ESI): m/z [M + H]+ Calcd. for [C31H29F2N3O5+H]+ 562.2148, found 562.2155.
13k following the general procedure, starting with 1-iodo-3,5-bis(trifluoromethyl)benzene (224.40 mg), 10 (197.78 mg), copper iodide (33.80 mg), potassium phosphate tribasic (189.00 mg), picolinic acid (32.90 mg) in DMSO, 13k was obtained (165.45 mg, 41.60% yield). 1H NMR: δH (300 MHz, CDCl3, TMS) 1.63–2.12 (m, 7 H, piperidin-4-yl, piperidin-5-yl, CH3), 2.64–2.89 (m, 1 H, piperidin-6a-yl), 2.96–3.19 (m, 1 H, piperidin-2a-yl), 3.66–4.17 (m, 2 H, piperidin-2b-yl, piperidin-6b-yl), 4.34–4.55 (m, 1 H, piperidin-3-yl), 4.70 (s, 2 H, O-CH2-phenyl), 5.49 (s, 2 H, N-CH2-O), 6.94 (s, 1 H), 7.07–7.17 (m, 2 H), 7.17–7.37 (m, 6 H), 7.43 (s, 2 H), 7.44–7.53 (m, 1 H), 7.61 (s, 1 H). 13 C NMR: δC (75 MHz, CDCl3, TMS) 13.13 (5-CH3), 24.85 (piperidin-5-yl), 29.01 (piperidin-4-yl), 53.28 (piperidin-3-yl), 70.85 (N-CH2-O), 72.25 (O-CH2-phenyl), 110.35 (C-5), 116.89 (Ph), 118.18 (Ph), 118.58 (2 C, Ph), 120.89 (2 C, Ph), 123.41 (2 C, Ph), 127.47 (2 C, Ph), 127.53 (Ph), 128.17 (2 C, Ph), 130.73 (Ph), 133.08 (Ph), 133.73 (Ph), 137.57 (2 C, Ph), 138.01 (C-6), 151.19 (C-2), 155.52 (Ph), 157.87 (Ph), 162.97 (C-4), 169.28 (CO, benzoyl). C (piperidin-2-yl) and C (piperidin-6-yl) cannot be found. HRMS (ESI): m/z [M + H]+ Calcd. for [C33H29F6N3O5+H]+ 662.2084, found 662.2086.
General procedure for substituted 5-methyl-1–(1-(3-phenoxybenzoyl)piperidin-3-yl)pyrimidine-2,4(1H,3H)-dione (4a–4b). 13a–13k was dissolved in 2–4 ml trifluoroacetic acid and heated at 73 °C for 4 h. The reaction mixture was cooled to room temperature and the trifluoroacetic acid was removed in vacuo. The residue was dissolved in 2 ml methanol, and the mixture was neutralised with saturated NaHCO3 to pH 5–6. The generated solid was filtered and the filtrate was concentrated and purified by column chromatography (CH2Cl2–MeOH, 20: 1 v/v) to give solid 4a–4k (70–85% yield), which was characterised by 1H and 13 C-NMR, and ESI-HRMS.
4a 1H NMR: δH (300 MHz, DMSO-d6, TMS) 1.47–1.60 (m, 1 H, piperidin-5a-yl), 1.74–1.93 (m, 6 H, piperidin-4-yl, piperidin-5b-yl, CH3), 2.79–2.98 (m, 7 H, piperidin-6a-yl, N(CH3)2), 3.07–3.19 (m, 1 H, piperidin-2a-yl), 3.90–4.14 (m, 2 H, piperidin-2b-yl, piperidin-6b-yl), 4.26–4.36 (m, 1 H, piperidin-3-yl), 6.27 (dd, J = 7.76, 1.90 Hz, 1 H), 6.37 (t, J = 2.34 Hz, 1 H), 6.52 (dd, J = 8.20, 2.05 Hz, 1 H), 6.92–6.96 (m, 1 H), 7.00–7.07 (m, 1 H), 7.10 (d, J = 7.62 Hz, 1 H), 7.16 (t, J = 8.20 Hz, 1 H), 7.37–7.42 (m, 1 H), 7.50 (s, 1 H), 10.92 (br. s., 1 H, 3-NH). 13 C NMR: δC (75 MHz, DMSO-d6, TMS) 12.09 (5-CH3), 24.54 (piperidin-5-yl), 28.14 (piperidin-4-yl), 50.64 (piperidin-3-yl), 103.32 (Ph), 106.57 (Ph), 108.25 (Ph), 109.03 (C-5), 115.97 (Ph), 118.98 (Ph), 120.95 (Ph), 128.40 (Ph), 130.17 (Ph), 135.54 (Ph), 137.51 (C-6), 150.71 (Ph), 151.98 (C-2), 156.77 (Ph), 157.28 (Ph), 163.58 (C-4), 168.44 (CO, benzoyl). C (N(CH3)2), C (piperidin-2-yl) and C (piperidin-2-yl) cannot be found. HRMS (ESI): m/z [M + H]+ Calcd. for [C25H28N4O4+H]+ 449.2184, found 449.2208.
4b 1H NMR: δH (300 MHz, DMSO-d6, TMS) 1.44–1.62 (m, 1 H, piperidin-5a-yl), 1.70–2.05 (m, 6 H, piperidin-4-yl, piperidin-5b-yl, CH3), 2.80–2.96 (m, 1 H, piperidin-6a-yl), 3.09–3.24 (m, 1 H, piperidin-2a-yl), 3.73 (s, 3 H, OCH3), 3.82–4.18 (m, 2 H, piperidin-2b-yl, piperidin-6b-yl), 4.24–4.38 (m, 1 H, piperidin-3-yl), 6.53–6.63 (m, 2 H), 6.69–6.77 (m, 1 H), 6.96–7.02 (m, 1 H), 7.04–7.11 (m, 1 H), 7.11–7.18 (m, 1 H), 7.27 (t, J = 8.05 Hz, 1 H), 7.38–7.47 (m, 1 H), 7.50 (d, J = 0.88 Hz, 1 H), 10.92 (br. s., 1 H, 3-NH). 13C NMR: δC (75 MHz, DMSO-d6, TMS) 12.11 (5-CH3), 24.72 (piperidin-5-yl), 28.12 (piperidin-4-yl), 50.41 (piperidin-3-yl), 55.29 (OCH3), 105.10 (Ph), 109.04 (C-5), 109.68 (Ph), 110.95 (Ph), 116.60 (Ph), 119.50 (Ph), 121.57 (Ph), 130.26 (Ph), 130.63 (Ph), 137.63 (C-6), 150.73 (C-2), 156.67 (Ph), 157.25 (Ph), 160.76 (Ph), 163.60 (C-4), 168.33 (CO, benzoyl). C (piperidin-2-yl), C (piperidin-6-yl) and C (Ph-CO-N) cannot be found. HRMS (ESI): m/z [M + H]+ Calcd. for [C24H25N3O5+H]+ 436.1867, found 436.1899.
4c 1H NMR: δH (300 MHz, DMSO-d6, TMS) 1.45–1.66 (m, 1 H, piperidin-5a-yl), 1.74–2.02 (m, 6 H, piperidin-4-yl, piperidin-5b-yl, CH3), 2.80–2.97 (m, 1 H, piperidin-6a-yl), 3.07–3.20 (m, 1 H, piperidin-2a-yl), 3.82–4.18 (m, 2 H, piperidin-2b-yl, piperidin-6b-yl), 4.25–4.39 (m, 1 H, piperidin-3-yl), 6.98–7.02 (m, 1 H), 7.03–7.12 (m, 3 H), 7.16 (d, J = 7.61 Hz, 1 H), 7.37–7.47 (m, 3 H), 7.50 (s, 1 H), 10.93 (br. s., 1 H, 3-NH). 13 C NMR: δC (75 MHz, DMSO-d6, TMS) 12.09 (5-CH3), 24.43 (piperidin-5-yl), 28.05 (piperidin-4-yl), 50.56 (piperidin-3-yl), 109.01 (C-5), 116.78 (Ph), 119.68 (Ph), 120.66 (2 C, Ph), 121.91 (Ph), 127.60 (Ph), 129.98 (2 C, Ph), 130.38 (Ph), 137.46 (Ph), 137.80 (C-6), 150.71 (C-2), 155.19 (Ph), 156.36 (Ph), 163.58 (C-4), 168.22 (CO, benzoyl). C (piperidin-2-yl) and C (piperidin-6-yl) cannot be found. HRMS (ESI): m/z [M + H]+ Calcd. for [C23H22ClN3O4+H]+ 440.1372, found 440.1380.
4d 1H NMR: δH (300 MHz, DMSO-d6, TMS) 1.45–1.61 (m, 1 H, piperidin-5a-yl), 1.73–2.00 (m, 6 H, piperidin-4-yl, piperidin-5b-yl, CH3), 2.80–2.94 (m, 1 H, piperidin-6a-yl), 3.09–3.21 (m, 1 H, piperidin-2a-yl), 3.84–4.16 (m, 2 H, piperidin-2b-yl, piperidin-6b-yl), 4.22–4.39 (m, 1 H, piperidin-3-yl), 6.93–6.98 (m, 1 H), 7.01–7.12 (m, 3 H), 7.14 (d, J = 1.17 Hz, 1 H), 7.15–7.23 (m, 2 H), 7.41 (t, J = 11.72 Hz, 1 H), 7.50 (d, J = 0.88 Hz, 1 H), 10.93 (s, 1 H, 3-NH). 13 C NMR: δC (75 MHz, DMSO-d6, TMS) 12.12 (5-CH3), 24.69 (piperidin-5-yl), 28.15 (piperidin-4-yl), 50.56 (piperidin-3-yl), 109.06 (C-5), 116.05 (Ph), 116.57 (Ph), 116.89 (Ph), 118.89 (Ph), 121.13 (Ph), 121.25 (Ph), 121.39 (Ph), 130.31 (Ph), 137.50 (Ph), 137.69 (C-6), 150.75 (C-2), 152.07 (Ph), 156.90 (Ph), 157.26 (Ph), 163.61 (C-4), 168.35 (CO, benzoyl). C (piperidin-2-yl) and C (piperidin-6-yl) cannot be found. HRMS (ESI): m/z [M + H]+ Calcd. for [C23H22FN3O4+H]+ 424.1667, found 424.1655.
4e 1H NMR: δH (300 MHz, DMSO-d6, TMS) 1.47–1.66 (m, 1 H, piperidin-5a-yl), 1.71–2.08 (m, 6 H, piperidin-4-yl, piperidin-5b-yl, CH3), 2.76–2.99 (m, 1 H, piperidin-6a-yl), 3.08–3.22 (m, 1 H, piperidin-2a-yl), 3.80–4.23 (m, 2 H, piperidin-2b-yl, piperidin-6b-yl), 4.24–4.46 (m, 1 H, piperidin-3-yl), 7.11–7.25 (m, 4 H), 7.28 (d, J = 7.62 Hz, 1 H), 7.46–7.61 (m, 2 H), 8.18–8.32 (m, 2 H), 10.93 (br. s., 1 H, 3-NH). 13 C NMR: δC (75 MHz, DMSO-d6, TMS) 12.09 (5-CH3), 24.66 (piperidin-5-yl), 28.11 (piperidin-4-yl), 50.50 (piperidin-3-yl), 109.03 (C-5), 117.82 (2 C, Ph), 118.63 (Ph), 121.38 (Ph), 123.53 (Ph), 126.21 (2 C, Ph), 130.79 (Ph), 137.45 (Ph), 138.24 (C-6), 142.53 (Ph), 150.73 (C-2), 154.39 (Ph), 162.45 (Ph), 163.60 (C-4), 167.99 (CO, benzoyl). C (piperidin-2-yl) and C (piperidin-6-yl) cannot be found. HRMS (ESI): m/z [M + H]+ Calcd. for [C23H22N4O6+H]+ 451.1612, found 451.1636.
4f 1H NMR: δH (300 MHz, DMSO-d6, TMS) 1.44–1.65 (m, 1 H, piperidin-5a-yl), 1.69–2.07 (m, 6 H, piperidin-4-yl, piperidin-5b-yl, CH3), 2.81–2.97 (m, 1 H, piperidin-6a-yl), 3.13 (t, J = 11.86 Hz, 1 H, piperidin-2a-yl), 3.78–4.21 (m, 2 H, piperidin-2b-yl, piperidin-6b-yl), 4.23–4.43 (m, 1 H, piperidin-3-yl), 7.08 (d, J = 1.46 Hz, 1 H), 7.11–7.17 (m, 1 H), 7.22 (d, J = 7.91 Hz, 1 H), 7.29–7.37 (m, 2 H), 7.43–7.54 (m, 3 H), 7.62 (t, J = 7.83 Hz, 1 H), 10.92 (br. s., 1 H, 3-NH). 13 C NMR: δC (75 MHz, DMSO-d6, TMS) 12.11 (5-CH3), 24.72 (piperidin-5-yl), 28.12 (piperidin-4-yl), 109.04 (C-5), 115.16 (Ph), 117.38 (Ph), 120.18 (Ph), 121.86 (Ph), 122.54 (2 C, Ph), 125.48 (CF3), 130.61 (Ph), 130.99 (Ph), 131.51 (Ph), 137.37 (Ph), 137.98 (C-6), 150.73 (C-2), 155.71 (Ph), 157.00 (Ph), 163.60 (C-4), 168.13 (CO, benzoyl). C (piperidin-2-yl), C (piperidin-3-yl) and C (piperidin-6-yl) cannot be found. HRMS (ESI): m/z [M + H]+ Calcd. for [C24H22F3N3O4+H]+ 474.1635, found 474.1624.
4g 1H NMR: δH (300 MHz, DMSO-d6, TMS) 1.44–1.67 (m, 1 H, piperidin-5a-yl), 1.73–2.00 (m, 6 H, piperidin-4-yl, piperidin-5b-yl, CH3), 2.80–2.97 (m, 1 H, piperidin-6a-yl), 3.11–3.21 (m, 1 H, piperidin-2a-yl), 3.73–4.21 (m, 2 H, piperidin-2b-yl, piperidin-6b-yl), 4.25–4.38 (m, 1 H, piperidin-3-yl), 7.03 (dd, J = 8.93, 2.78 Hz, 1 H), 7.06–7.09 (m, 1 H), 7.11–7.17 (m, 1 H), 7.17–7.24 (m, 1 H), 7.29 (d, J = 2.64 Hz, 1 H), 7.45 (d, J = 7.91 Hz, 1 H), 7.48–7.53 (m, 1 H), 7.59 (d, J = 9.08 Hz, 1 H), 10.92 (s, 1 H, 3-NH). 13 C NMR: δC (75 MHz, DMSO-d6, TMS) 12.12 (5-CH3), 24.72 (piperidin-5-yl), 28.20 (piperidin-4-yl), 50.15 (piperidin-3-yl), 109.06 (C-5), 117.21 (Ph), 119.04 (Ph), 120.11 (Ph), 120.70 (Ph), 122.57 (Ph), 125.74 (Ph), 130.58(Ph), 131.69 (Ph), 132.06 (Ph), 137.50 (Ph), 137.97 (C-6), 150.75 (C-2), 155.71 (Ph), 156.01 (Ph), 163.63 (C-4), 168.15 (CO, benzoyl). C (piperidin-2-yl) and C (piperidin-6-yl) cannot be found. HRMS (ESI): m/z [M + H]+ Calcd. for [C23H21Cl2N3O4+H]+ 474.0982, found 474.0987.
4h 1H NMR: δH (300 MHz, DMSO-d6, TMS) 1.46–1.63 (m, 1 H, piperidin-5a-yl), 1.73–2.02 (m, 6 H, piperidin-4-yl, piperidin-5b-yl, CH3), 2.89 (t, J = 12.30 Hz, 1 H, piperidin-6a-yl), 3.10–3.22 (m, 1 H, piperidin-2a-yl), 3.82–4.17 (m, 2 H, piperidin-2b-yl, piperidin-6b-yl), 4.25–4.41 (m, 1 H, piperidin-3-yl), 6.83–6.91 (m, 1 H), 6.98–7.04 (m, 1 H), 7.05–7.21 (m, 3 H), 7.33–7.56 (m, 3 H) 10.93 (br. s., 1 H, 3-NH). 13 C NMR: δC (75 MHz, DMSO-d6, TMS) 12.14 (5-CH3), 24.73 (piperidin-5-yl), 28.20 (piperidin-4-yl), 50.53 (piperidin-3-yl), 109.10 (C-5), 109.36 (Ph), 115.59 (Ph), 116.58 (Ph), 118.19 (Ph), 118.43 (Ph), 119.47 (Ph), 122.05 (Ph), 130.47 (Ph), 137.53 (Ph), 137.83 (C-6), 148.24 (Ph), 150.78 (C-2), 152.41 (Ph), 156.52 (Ph), 163.66 (C-4), 168.27 (CO, benzoyl). C (piperidin-2-yl) and C (piperidin-6-yl) cannot be found. HRMS (ESI): m/z [M + H]+ Calcd. for [C23H21F2N3O4+H]+ 442.1573, found 442.1566.
4i 1H NMR: δH (300 MHz, DMSO-d6, TMS) 1.47–1.60 (m, 1 H, piperidin-5a-yl), 1.72–2.02 (m, 6 H, piperidin-4-yl, piperidin-5b-yl, CH3), 2.90 (t, J = 12.33 Hz, 1 H, piperidin-6a-yl), 3.87–4.12 (m, 2 H, piperidin-2b-yl, piperidin-6b-yl), 4.26–4.32 (m, 1 H, piperidin-3-yl), 7.04 (d, J = 2.05 Hz, 1 H), 7.08–7.12 (m, 1 H), 7.12–7.21 (m, 2 H), 7.21–7.27 (m, 1 H), 7.28–7.32 (m, 1 H) 7.49 (t, J = 7.98 Hz, 2 H), 10.90 (br. s., 1 H). H (piperidin-2a-yl) was hidden in water peak. 13 C NMR: δC (75 MHz, DMSO-d6, TMS) 11.48 (5-CH3), 24.03 (piperidin-5-yl), 27.82 (piperidin-4-yl), 51.28 (piperidin-3-yl), 108.81 (C-5), 117.09 (2 C, Ph), 117.36 (Ph), 120.14 (Ph), 122.66 (Ph), 122.98 (Ph), 130.26 (Ph), 134.73 (Ph), 137.08 (2 C, Ph), 137.79 (C-6), 150.38 (C-2), 154.94 (Ph), 157.97 (Ph), 163.14 (C-4), 167.92 (CO, benzoyl). C (piperidin-2-yl) and C (piperidin-6-yl) cannot be found. HRMS (ESI): m/z [M + H]+ Calcd. for [C23H21Cl2N3O4+H]+ 474.0982, found 474.0022.
4j 1H NMR: δH (300 MHz, DMSO-d6, TMS) 1.45–1.65 (m, 1 H, piperidin-5a-yl), 1.72–2.10 (m, 6 H, piperidin-4-yl, piperidin-5b-yl, CH3), 2.90 (t, J = 12.78 Hz, 1 H, piperidin-6a-yl), 3.08–3.24 (m, 1 H, piperidin-2a-yl), 3.81–4.21 (m, 2 H, piperidin-2b-yl, piperidin-6b-yl), 4.26–4.40 (m, 1 H, piperidin-3-yl), 6.65–6.79 (m, 2 H), 6.91 (tt, J = 7.59 Hz, 4.35 Hz, 1 H), 7.07–7.13 (m, 1 H), 7.13–7.21 (m, 1 H), 7.24 (dt, J = 7.62, 1.17 Hz, 1 H), 7.46 (s, 1 H), 7.49–7.53 (m, 1 H), 10.92 (br. s., 1 H, 3-NH). 13 C NMR: δC (75 MHz, DMSO-d6, TMS) 12.11 (5-CH3), 24.61 (piperidin-5-yl), 28.23 (piperidin-4-yl), 99.21 (Ph), 102.19 (Ph), 102.57 (Ph), 109.04 (C-5), 117.70 (Ph), 120.49 (Ph), 122.95 (Ph), 130.61 (Ph), 137.59 (Ph), 138.08 (C-6), 150.75 (C-2), 155.11 (Ph), 158.74 (Ph), 161.44 (Ph), 163.60 (C-4), 164.71 (Ph), 168.07 (CO, benzoyl). C (piperidin-2-yl), C (piperidin-3-yl) and C (piperidin-6-yl) cannot be found. HRMS (ESI): m/z [M + H]+ Calcd. for [C23H21F2N3O4+H]+ 442.1573, found 442.1604.
4k 1H NMR: δH (300 MHz, DMSO-d6, TMS) 1.48–1.65 (m, 1 H, piperidin-5a-yl), 1.67–2.14 (m, 6 H, piperidin-4-yl, piperidin-5b-yl, CH3), 2.80–2.96 (m, 1 H, piperidin-6a-yl), 3.11–3.23 (m, 1 H, piperidin-2a-yl), 3.74–4.20 (m, 2 H, piperidin-2b-yl, piperidin-6b-yl), 4.23–4.42 (m, 1 H, piperidin-3-yl), 7.11–7.34 (m, 3 H), 7.43–7.58 (m, 2 H), 7.63 (s, 2 H), 7.78 (s, 1 H), 10.91 (br. s., 1 H, 3- NH). 13 C NMR: δC (75 MHz, DMSO-d6, TMS) 12.16 (5-CH3), 24.47 (piperidin-5-yl), 28.23 (piperidin-4-yl), 50.62 (piperidin-3-yl), 109.18 (C-5), 116.92 (Ph), 117.85 (Ph), 119.15 (2 C, Ph), 120.75 (Ph), 121.12 (Ph), 123.45 (Ph), 124.73 (Ph), 131.02 (Ph), 131.94 (Ph), 132.38 (Ph), 137.53 (Ph), 138.18 (C-6), 150.82 (C-2), 154.97 (Ph), 158.07 (Ph), 163.74 (C-4), 168.15 (CO, benzoyl). C (piperidin-2-yl) and C (piperidin-6-yl) cannot be found. HRMS (ESI): m/z [M + H]+ Calcd. for [C25H21F6N3O4+H]+ 542.1509, found 542.1508.
2.2. Molecular modelling
The modelling was conducted using AutoDock vina and AutodockTools-1.5.626 with publicly available X-ray structure of the MtbTMPK (PDB entry 1G3U9 and 5NQ527). All PDB files of ligand were generated in ChemDraw 3 D 16.0 after being minimised the energy. Centred on MtbTMPK active site PHE70 CE2, the prepared PDBQT files of ligands and receptors were docked using a grid spacing of 0.375 and 60 × 60 × 60 number of grid points. Each ligand was docked three times in autodock vina by Lamarckian 4.2 method, affording total 60 possible conformations. All vinadock results were viewed in Chimaera, further analysed and validated through the predicted free energy in combination with possible interactions in LigPlus.
2.3. MtbTMPK assay
MtbTMPK was overexpressed in Escherichia coli and purified by a two-step procedure as previously described8. MtbTMPK activities of all compounds were measured using the spectrophotometric assay reported by Blondin et al28. All compounds were tested at varying concentration from 0.006 to 0.12 mM with constant concentrations of ATP (0.5 mM) and dTMP (0.05 mM). The studies were done at 30 °C in a medium containing 50 mM Tris–HCl, 2 mM MgCl2, pH 7.4, 50 mM KCl, 1 mM phosphoenol pyruvate, 0.2 mM NADH and two units each of coupling enzymes (lactate dehydrogenase, pyruvate kinase and nucleoside diphosphate kinase). The absorbance at 334 nm was monitored in an Eppendorf ECOM 6122 photometer. IC50 were calculated using Kaleidagraph as previously described27.
2.4. Whole cell activity against Mtb
MIC50 against mycobacteria was evaluated by a 2-fold serial dilution of the tested compounds. The in vitro antimycobacterial assay was based on a method in which a luminescent M. tuberculosis H37Ra strain Lehmann & Neumann (ATCC 25177) transformed with pSMTB1 luciferase reporter plasmid is used. Each compound was dissolved in DMSO (Sigma-Aldrich) to prepare the stock solution at concentration of 10 mM. Serial dilutions were made in Middlebrook 7H9 broth and 10% OADC (oleic acid, albumin, dextrose, catalase). Volumes of 20 μl of the serial dilutions were added in triplicate to flat-bottomed 96-well plates. By thawing and dissolving a frozen Mycobacteria pellet in 7H9-10% OADC, the bacterial suspension was made. To eliminate clumps, the dissolved pellet was passed through a 5.0 μM filter (Millipore) and left for 1 h to recover at 37 °C, 5% CO2. Subsequently, the suspension was diluted in complete 7H9 broth to obtain 50,000 Relative Light Units (RLU)/ml and a volume of 180 μl of bacteria was added to each well. After 7 days of incubation, the bacterial replication was analysed by luminometry. The bacterial suspension from each well was collected and transferred to a black 96-well plate to evade cross luminescence between wells. The luminescent signal was evoked by addition of the substrate for the bacterial luciferase, 1% n-decanal in ethanol to each well by the Discover multi-plate reader from Promega and the light emission in each well was measured. Using the plot of the % viability as a function of compound concentration, MIC50 values were determined.
3. Results and discussion
3.1. Chemistry
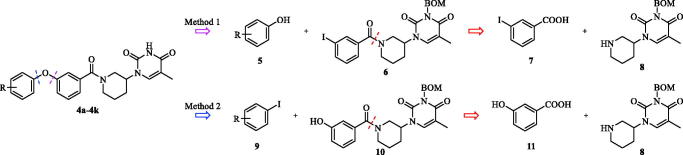
For the synthesis of the target compounds, two divergent synthesis routes were considered based on a disconnection of the ether bond between the C and D ring (Scheme 1).
Scheme 1.
Two divergent routes used for the synthesis of compounds 4a–4k.
The target compounds 4a–4k may be obtained by Ullmann coupling of either substituted phenol (5) with 3-iodobenzamide 6 (Method 1) or substituted iodobenzene (9) with phenol 10 (Method 2). We experienced that Method 1 failed to produce the coupling product in the case of electron-withdrawing substituents on the phenol ring. Except for analogue 4a, all analogues were prepared according to Method 2 as detailed Scheme 2.
Scheme 2.
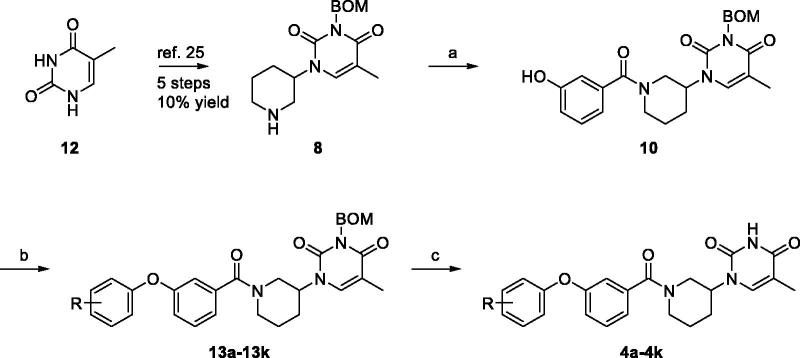
Synthesis of compound 4a–4k. Reagents and conditions: (a) N,N-bis(isopropyl) carbondiimide, DMAP, CH2Cl2, overnight, 62% yield; (b) CuI, K3PO4, DMSO, picolinic acid, 90 °C, overnight, 40.00–70.00% yield; (c) TFA, 73 °C, 4 h, 70–85% yield.
Intermediate 8 was synthesised as previously reported25. Briefly, thymine (12) was converted to N3-benzyloxymethyl-protected thymine in three steps. Next, a substitution reaction with tert-butyl 3-((methylsulfonyl)oxy)piperidine-1-carboxylate, followed by BOC-deprotection with trifluoroacetic acid afforded 3-piperidylthymine 8. Amidation of 8 with 3-hydroxybenzoic acid (11) gave compound 10. Ullmann coupling of 10 with the appropriately substituted iodobenzene, followed by BOM-deprotection afforded the desired substituted (1–(3-phenoxybenzoyl)piperidin-4-yl)thymines 4a–4k.
3.2. Docking
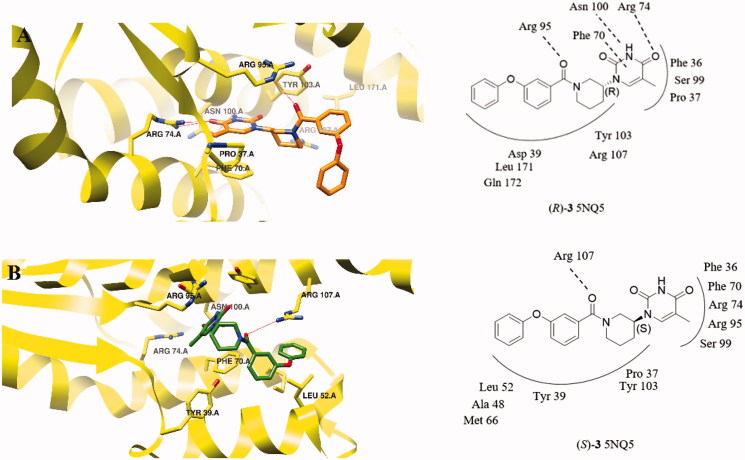
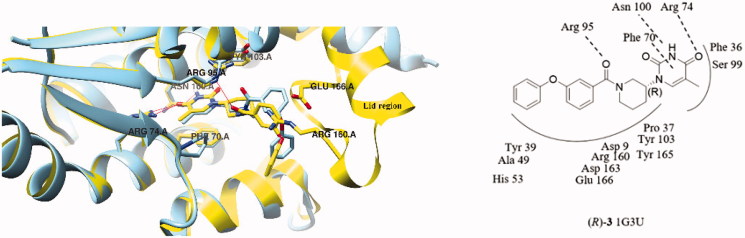
To gain insight into their putative binding mode, putative analogues were docked in the target enzyme using the crystal structure of MtbTMK in complex with (S)-1 (PDB code 5NQ5)27. Docking indicated that the (R)-3 is better accommodated than its (S)-enantiomer (Figure 3). The thymine moiety of (R)-3 adopts an identical pose as that of (S)-1 and is involved in two hydrogen bonds between its NH and C(4)O groups and the side chains of Asn100 and Arg74, as well as a π–π stacking interactions with Phe-70. By protruding out of the active site, the meta-piperidine ring forms a π-alkyl interaction with the aromatic side chain of Tyr 103. Importantly, in the (R)-enantiomer the oxygen atom of the carbonyl that connects ring B and ring C is expected to form a hydrogen bond with Arg-95 (Figure 3(A)), which justifies our choice to connect ring C to the piperidine ring B through an amide bond. Furthermore, through various hydrophobic interactions ring C and ring D contribute to the overall affinity. The putative binding mode of the (S)-enantiomer is quite different (Figure 3(B)). Although a hydrogen bond may be formed between the oxygen atom of the carbonyl connecting ring B and ring C and Arg-107, the loss of the hydrogen bonds between N3 and C4 groups of thymine ring and the side chains of Asn100 and Arg7, as well as the π–π stacking interaction between pyrimidine ring and Phe-70 results in a less promising docking score.
Figure 3.
(R)- and (S)-3 docked in the active site of MtbTMPK (PDB 5NQ5). MtbTMPK is shown in a yellow cartoon representation with selected side chains labelled and shown as sticks with carbon atoms coloured yellow. Ligands are drawn in stick representation with carbon atoms in orange ((R)-3) and green ((S)-3), hydrogen-bonding interactions are shown as red lines.
(R)-3 was also docked starting from the co-crystal structure of the natural substrate TMP (PDB code 1G3U)9. The resulting docking pose of (R)-3 in MtbTMPK is superimposed with the one described above (Figure 4). The MtbTMPK conformation is highly similar except for the flexible Lid region, leading to slightly more encouraging docking score due to extra pronounced hydrophobic interactions exhibited by the C and D ring of (R)-3. Considering the envisioned analogues are highly related to the structure of (S)-1, the co-crystal structure of MtbTMPK in complex with (S)-1 was used for modelling.
Figure 4.
Structure overlay of compound (R)-3 docked into MtbTMPK PDB 1G3U (yellow) and PDB 5NQ5 (blue), hydrogen-bonding interactions are shown as red dashed lines.
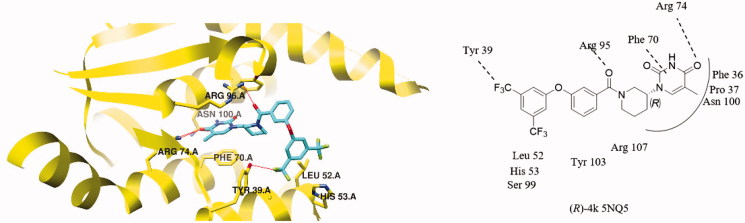
Docking analysis starting from the 5NQ5 co-crystal structure further indicated that judicious substituents of the D-ring might lead to extra interactions with the enzyme. A trifluoromethyl substituent in meta position (as in 4k) may establish a hydrogen bond with Tyr 39 (Figure 5). However, the consequent distortion of the D-ring leads to a less favourable docking score (Table 1).
Figure 5.
Compound 4k docked in the active site of MtbTMPK (PDB 5NQ5). The protein is shown in a yellow cartoon representation with selected side chains labelled and shown as sticks and 4k is drawn in stick representation with carbon atoms in cyan. Hydrogen-bonding interactions are shown as red lines.
Table 1.
Docking results of compounds 4a–4k, as well as their activities against MtbTMPK and avirulent M. tuberculosis.
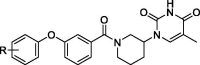 | ||||
|---|---|---|---|---|
| Compound | R | Docking score | IC50MtbTMPK (μM) | MIC50 H37Ra (μM)a |
| 4a | 3-N(CH3)2 | −8.5 | 39 ± 3 | >64 |
| 4b | 3-OCH3 | −8.5 | 15 ± 1 | >64 |
| 4c | 4-Cl | −8.8 | 39 ± 19 | >64 |
| 4d | 4-F | −8.8 | 18 ± 1 | >64 |
| 4e | 4-NO2 | −9.0 | 30 ± 3 | >64 |
| 4f | 3-CF3 | −9.2 | 53 ± 12 | >64 |
| 4g | 3,4-Cl2 | −8.8 | 18 ± 1 | 35.8 |
| 4h | 3,4-F2 | −8.8 | 23 ± 1 | >64 |
| 4i | 3,5-Cl2 | −8.7 | 13 ± 1 | >64 |
| 4j | 3,5-F2 | −8.8 | 31 ± 2 | >64 |
| 4k | 3,5-(CF3)2 | −8.2 | 158 ± 89 | >64 |
| 3 | — | −8.7 | 28 ± 2 | >64 |
Compounds were tested on H37Ra strain, measured as the half-maximal inhibitory concentration (MIC50). Using the plot of the % viability as a function of compound concentration, MIC50 values were determined.
3.3. Biological activity
Compounds 4a–4k were evaluated for their capacity to inhibit MtbTMPK and growth of an avirulent M. tuberculosis strain (Table 1). Most compounds exhibited low micromolar enzyme inhibitory activity and for most analogues the nature and the positions of the D-ring substituents have only negligible effect on the activity. This is in line with the docking scores. However, introduction of two trifluoromethyl groups in meta position of ring D (4k) significantly lowered the inhibitory potency. This may be attributed to the fact that the large trifluoromethyl groups hinder the tail of molecule to form a stacking interaction with the enzyme, or the tendency of trifluoromethyl to form a hydrogen bond by directional change of last ring, as suggested by the docking analysis. Unfortunately, only analogue 4g with a 3,4-dichlorophenyl D-ring showed moderate antimycobacterial activity, while all other compounds were virtually inactive. To further validate the antimycobacterial activity, compounds with good enzyme affinity (4b, 4g and 4i) were screened on virulent strain (H37Rv). Nevertheless, none of them displayed any whole cell antitubercular activity (MIC > 50 μM).
4. Conclusion
Based on previous findings, we report herein the synthesis and biological activities of 1-(piperidin-3-yl)thymine amides as MtbTMPK inhibitors. Modelling was conducted to understand the putative binding mode, resulting in similar docking scores with lead compound 3 for most analogues except compound 4k bearing sterically demanding substituents on the D-ring. Further MtbTMPK inhibitory activities were in line with the modelling results. Compounds 4b and 4i were slightly more potent than the parent compound 3, while compound 4k was significantly less active. Unfortunately, most analogues failed to exhibit whole cell antitubercular activity, except for 4g, which demonstrated moderate cellular activity on avirulent strain. We hypothesise that the poor permeability/uptake of these inhibitors in M. tuberculosis may be the reason for the disappointing cellular activity. Attempts to improve mycobacterial uptake will be addressed in future work.
Supplementary Material
Funding Statement
This work was supported by the China Scholarship Council (grant numbers 201607060021).
Disclosure statement
No conflict of interest was reported by the authors.
References
- 1.Giacobbo BC, Pissinate K, Rodrigues V, et al. . New insights into the SAR and drug combination synergy of 2(quinolin-4-yloxy)acetamides against Mycobacterium tuberculosis. Eur J Med Chem 2017;126:491–501. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Global Tuberculosis Report 2017. Geneva: World Health Organization; 2017. http://apps.who.int/medicinedocs/en/m/abstract/Js23360en/
- 3.Mori G, Chiarelli LR, Riccardi G, et al. . New prodrugs against tuberculosis. Drug Discov Today 2017;22:519–25. [DOI] [PubMed] [Google Scholar]
- 4.Moraski GC, Seeger N, Miller PA, et al. . Arrival of imidazo[2,1-b]thiazole-5-carboxamides: potent anti-tuberculosis agents that target QcrB. ACS Infect Dis 2016;2:393–8. [DOI] [PubMed] [Google Scholar]
- 5.Van Calenbergh S, Pochet S, Munier-Lehmann H. Drug design and identification of potent leads against Mycobacterium tuberculosis thymidine monophosphate kinase. Curr Top Med Chem 2012;12:694–705. [DOI] [PubMed] [Google Scholar]
- 6.Cui Q, Shin WS, Luo Y, et al. . Thymidylate kinase: an old topic brings new perspectives. Curr Med Chem 2013;20:1286–305. [DOI] [PubMed] [Google Scholar]
- 7.Martinez-Botella G, Breen JN, Duffy JE, et al. . Discovery of selective and potent inhibitors of gram-positive bacterial thymidylate kinase (TMK). J Med Chem 2012;55:10010–21. [DOI] [PubMed] [Google Scholar]
- 8.Munier-Lehmann H, Chaffotte A, Pochet S, et al. . Thymidylate kinase of Mycobacterium tuberculosis: a chimera sharing properties common to eukaryotic and bacterial enzymes. Protein Sci 2001;10:1195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li de la Sierra I, Munier-Lehmann H, Gilles AM, et al. . X-ray structure of TMP kinase from Mycobacterium tuberculosis complexed with TMP at 1.95 A resolution. J Mol Biol 2001;311:87–100. [DOI] [PubMed] [Google Scholar]
- 10.Vanheusden V, Munier-Lehmann H, Pochet S, et al. . Synthesis and evaluation of thymidine-5'-O-monophosphate analogues as inhibitors of Mycobacterium tuberculosis thymidylate kinase. Bioorg Med Chem Lett 2002;12:2695–8. [DOI] [PubMed] [Google Scholar]
- 11.Haouz A, Vanheusden V, Munier-Lehmann H, et al. . Enzymatic and structural analysis of inhibitors designed against Mycobacterium tuberculosis thymidylate kinase. New insights into the phosphoryl transfer mechanism. J Biol Chem 2003;278:4963–71. [DOI] [PubMed] [Google Scholar]
- 12.Vanheusden V, Munier-Lehmann H, Froeyen M, et al. . 3'-C-branched-chain-substituted nucleosides and nucleotides as potent inhibitors of Mycobacterium tuberculosis thymidine monophosphate kinase. J Med Chem 2003;46:3811–21. [DOI] [PubMed] [Google Scholar]
- 13.Vanheusden V, Van Rompaey P, Munier-Lehmann H, et al. . Thymidine and thymidine-5'-O-monophosphate analogues as inhibitors of Mycobacterium tuberculosis thymidylate kinase. Bioorg Med Chem Lett 2003;13:3045–8. [DOI] [PubMed] [Google Scholar]
- 14.Vanheusden V, Munier-Lehmann H, Froeyen M, et al. . Discovery of bicyclic thymidine analogues as selective and high-affinity inhibitors of Mycobacterium tuberculosis thymidine monophosphate kinase. J Med Chem 2004;47:6187–94. [DOI] [PubMed] [Google Scholar]
- 15.Van Daele I, Munier-Lehmann H, Hendrickx PM, et al. . Synthesis and biological evaluation of bicyclic nucleosides as inhibitors of M. tuberculosis thymidylate kinase. Chem Med Chem 2006;1:1081–90. [DOI] [PubMed] [Google Scholar]
- 16.Van Daele I, Munier-Lehmann H, Froeyen M, et al. . Rational design of 5'-thiourea-substituted alpha-thymidine analogues as thymidine monophosphate kinase inhibitors capable of inhibiting mycobacterial growth. J Med Chem 2007;50:5281–92. [DOI] [PubMed] [Google Scholar]
- 17.Gasse C, Douguet D, Huteau V, et al. . Substituted benzyl-pyrimidines targeting thymidine monophosphate kinase of Mycobacterium tuberculosis: synthesis and in vitro anti-mycobacterial activity. Bioorg Med Chem 2008;16:6075–85. [DOI] [PubMed] [Google Scholar]
- 18.Kogler M, Busson R, De Jonghe S, et al. . Synthesis and evaluation of 6-aza-2'-deoxyuridine monophosphate analogs as inhibitors of thymidylate synthases, and as substrates or inhibitors of thymidine monophosphate kinase in Mycobacterium tuberculosis. Chem Biodivers 2012;9:536–56. [DOI] [PubMed] [Google Scholar]
- 19.Adamska A, Rumijowska-Galewicz A, Ruszczynska A, et al. . Anti-mycobacterial activity of thymine derivatives bearing boron clusters. Eur J Med Chem 2016;121:71–81. [DOI] [PubMed] [Google Scholar]
- 20.Pochet S, Dugue L, Douguet D, et al. . Nucleoside analogues as inhibitors of thymidylate kinases: possible therapeutic applications. Chembiochem 2002;3:108–10. [DOI] [PubMed] [Google Scholar]
- 21.Van Poecke S, Munier-Lehmann H, Helynck O, et al. . Synthesis and inhibitory activity of thymidine analogues targeting Mycobacterium tuberculosis thymidine monophosphate kinase. Bioorg Med Chem 2011;19:7603–11. [DOI] [PubMed] [Google Scholar]
- 22.Familiar O, Munier-Lehmann H, Negri A, et al. . Exploring acyclic nucleoside analogues as inhibitors of Mycobacterium tuberculosis thymidylate kinase. Chem Med Chem 2008;3:1083–93. [DOI] [PubMed] [Google Scholar]
- 23.Familiar O, Munier-Lehmann H, Ainsa JA, et al. . Design, synthesis and inhibitory activity against Mycobacterium tuberculosis thymidine monophosphate kinase of acyclic nucleoside analogues with a distal imidazoquinolinone. Eur J Med Chem 2010;45:5910–8. [DOI] [PubMed] [Google Scholar]
- 24.Naik M, Raichurkar A, Bandodkar BS, et al. . Structure guided lead generation for M. tuberculosis thymidylate kinase (Mtb TMK): discovery of 3-cyanopyridone and 1,6-naphthyridin-2-one as potent inhibitors. J Med Chem 2015;58:753–66. [DOI] [PubMed] [Google Scholar]
- 25.Song L, Risseeuw MDP, Froeyen M, et al. . Elaboration of a proprietary thymidylate kinase inhibitor motif towards anti-tuberculosis agents. Bioorg Med Chem 2016;24:5172–82. [DOI] [PubMed] [Google Scholar]
- 26.Morris GM, Huey R, Lindstrom W, et al. . AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 2009;30:2785–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song L, Merceron R, Gracia B, et al. . Structure guided lead generation toward nonchiral M. tuberculosis thymidylate kinase inhibitors. J Med Chem 2018;61:2753–75. [DOI] [PubMed] [Google Scholar]
- 28.Blondin C, Serina L, Wiesmuller L, et al. . Improved spectrophotometric assay of nucleoside monophosphate kinase activity using the pyruvate kinase/lactate dehydrogenase coupling system. Anal Biochem 1994;220:219–21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.