Abstract
Ant-associated microorganisms can play crucial and often overlooked roles, and given the diversity of interactions that ants have developed, the study of the associated microbiomes is of interest. We focused here on specialist plant-ant species of the genus Allomerus that grow a fungus to build galleries on their host-plant stems. Allomerus-inhabited domatia, thus, might be a rich arena for microbes associated with the ants, the plant, and the fungus. We investigated the microbial communities present in domatia colonised by four arboreal ants: Allomerus decemarticulatus, A. octoarticulatus, A. octoarticulatus var. demerarae, and the non-fungus growing plant-ant Azteca sp. cf. depilis, inhabiting Hirtella physophora or Cordia nodosa in French Guiana. We hypothesized that the microbial community will differ among these species. We isolated microorganisms from five colonies of each species, sequenced the 16S rRNA or Internal TranscribedSpacer (ITS) regions, and described both the alpha and beta diversities. We identified 69 microbial taxa, which belong to five bacterial and two fungal phyla. The most diverse phyla were Proteobacteria and Actinobacteria. The microbial community of Azteca cf. depilis and Allomerus spp. differed in composition and richness. Geographical distance affected microbial communities and richness but plant species did not. Actinobacteria were only associated with Allomerus spp.
Keywords: microbial diversity, domatia, Allomerus decemarticulatus, Allomerus octoarticulatus, Azteca sp. cf. depilis, Cordia nodosa, Hirtella physophora
1. Introduction
The emergence of social life promoted the ecological success of the social species, but it also had profound evolutionary, ecological, and economic impacts on many other species, thus shaping life on Earth [1,2,3]. Indeed, the dominance of social species can have important consequences for the functioning of ecosystems and biodiversity in general [4]. On the other hand, sociality is associated with costs linked to the many potential risks associated with living in a group [5,6,7]. Thus, one can expect that both fitness benefits and costs shape the communities of organisms associated with social species.
Social insects, particularly ants, are good examples of these successful social organisms. They are considered as ecological engineers involved in many key ecosystem processes [2,8] and, given the high relatedness within colonies, the spread of pathogens and parasites can have strong deleterious effects both at the individual and colony levels [9]. Therefore, ant-associated microorganisms have received particular attention, mainly focusing on the complex multipartite networks of interactions among fungus-growing ants, their associated fungi, and a diversity of both detrimental and beneficial associated microorganisms [3,10,11,12,13,14,15,16,17,18]. Non fungus-growing ants also exhibit the ability to modify both abiotic and biotic characteristics, thus selecting for different microbial communities while boosting microbial diversity in their nests [19,20,21,22].
The recent discovery of bacteria and fungi associated with plant-ants (i.e., ants associated with myrmecophytes or plants providing them with a nesting place in the form of hollow structures called “domatia”) shed light on an overlooked role of microbiomes in ant-plant interactions [23,24,25,26]. It has also contributed to improving our understanding of how ants regulate their immediate environment and, thus, how they affect the diversity and functioning of the ecosystem. Moreover, myrmecophytes constitute a robust system that can be studied in order to answer such questions, since they are most often inhabited by one or a few specialized plant-ant species, with usually one colony per plant [27,28,29]. In addition, the environmental conditions provided by myrmecophytes can be considered as different from the surroundings, so that the microorganisms found inside the domatia are expected to differ according to the identity of both the associated ant species and the plant. As a consequence, both the local environment provided by the plant and the traits of the ant species might affect the diversity and composition of the associated microbial communities, which can be considered as a selection force or niche-filtering [30,31].
Here we investigated the bacterial and fungal microorganisms associated with two sympatric ant-plants, Cordia nodosa Lamarck (Boraginaceae) and Hirtella physophora Martius and Zuccharini (Chrysobalanaceae), and their associated ants at two sites in French Guiana. These myrmecophytes are mainly inhabited by ants of the genus Allomerus (Myrmicinae), but C. nodosa can also host Azteca sp. cf. depilis (Dolichoderinae) [32]. Ants of the genus Allomerus are specific plant-ants [33] that have developed a particular behaviour for prey capture, which relies on the construction of galleries on their host plants to ambush prey [34]. To this end, they have evolved the practice of a novel kind of fungal agriculture with non-nutritional purposes, which involves highly fine-tuned multipartite plant-ant-fungus-bacteria associations [26,35,36,37]. We isolated and identified bacteria and fungi present inside the domatia of the two plants inhabited by the different ant species and hypothesized that a strong niche filtering affected the microbial diversity and composition. That is, that the microbial community might be influenced by the location of the sampling site, and that both the species of the plants and the ants should influence bacteria and fungi present inside the domatia.
2. Material and Methods
2.1. Sampling
Samples were collected at two locations in French Guiana, Montagne des Singes (MdS: 5°04’21.92” N; 52°41’51.13” O) and Basevie (Bsv: 5°05’21.91” N; 53°01’28.39” O), between January and February 2009. Both sites were located about 40 km from each other. It should be noted the microbial isolation, culture and sequencing were performed just after the samples were collected (see methods below), while microbial identifications have been updated more recently, thus reflecting the potential changes in the GenBank database, which could have occurred since the sampling period. We randomly selected five colonies of each of four plant-ant species that inhabit the domatia of two myrmecophytic plant species: Cordia nodosa Lamarck (Boraginaceae) and Hirtella physophora Martius et Zuccharini (Crysobalanaceae). In the Montagne des Singes area, C. nodosa is colonized by either Allomerus octoarticulatus var. demerarae or Azteca sp. cf. depilis, and H. physophora is colonized both by A. decemarticulatus or A. octoarticulatus. Note that the two A. octoarticulatus species are different, cryptic species, and each is associated to a single host plant species, H. physophora or C. nodosa, in French Guiana. These two A. octoarticulatus species cannot be separated based on morphology alone. However, they separate into monophyletic sister clades based on the barcoding of the COI (Cytochrome c oxidase subunit I) gene fragment [38]. Allomerus octoarticulatus var. demerarae appears always and solely associated with C. nodosa, whatever its geographic origin, while A. octoarticulatus can be associated with a variety of host plants over its distribution range, although only found inhabiting H. physophora in French Guiana.
In Basevie, C. nodosa was colonized only by A. octoarticulatus var. demerarae and H. physophora only by A. decemarticulatus.
The C. nodosa domatia are located in the stems below each sympodial fork [39], while in H. physophora they result from the curling under of the leaf margin on either side of the petiole [40]. For each of the 30 plants with well-established ant colonies, we collected three leaves with domatia from the upper part of the plants to minimize the potential contamination of the leaves with soil microbiota. Leaves were collected from opposite branches. We sampled leaves of similar age based on their colouration. Then, domatia were individually transported in sterile zip bags to the laboratory. Each domatia was dissected with flame sterilized forceps and scalpel. We used 100 μL of sterile physiological saline solution (0.90% w/v NaCl) to thoroughly wash the inner walls and collect as many microorganisms as possible. Each sample was stored at 4 °C in 1.5 mL Eppendorf vials.
2.2. Microbial Isolation and Identification
Dilutions of the samples (1/106) were prepared after being gently vortexed, and 50 μL of dilution were plated on solid MYG medium (1% malt extract, 0.4% yeast extract, 0.4% glucose, 1.5% agar). Two plates were inoculated by sample, and the cultures were kept in dark conditions at 20 °C for up to 15 days. Afterward, we selected random bacterial and fungal colonies belonging to every potentially different morphospecies based on colony colour, size, and shape. Fungi were transferred to new plates to obtain pure cultures.
Fungal DNA was extracted from mycelium pieces with the Chelex® method [41], and we used direct PCR of intact bacteria as template. The 16S rRNA region of bacteria was amplified using the FD1 and RP2 primers [42] and the fungal ITS region of the rRNA with ITS1 and ITS4 primers [43]. PCR products were sequenced by Genoscreen (Lille, France), and edited with Chromas 2.6.5 (Technelysium Pty Ltd, Brisbanes, Australia). We checked for the closest sequences in GenBank [44] by following a BLAST procedure.
2.3. Alpha Diversity
Species richness was calculated as the total number of microbial species present in each community (S), and species abundance as the total number of microbial isolates of each species that appeared in our samples (N). To take into account sample size, we calculated the Margalef’s index of species richness () obtained with the ‘vegan’ R package. Then, to describe the alpha diversity, we obtained five indexes with the ‘phyloseq’ R package: Chao (± SE), Shannon, Simpson, Inversed Simpson, and Fisher. We also calculated the percentage of completeness as . All calculations were conducted for Site (Basevie and Montagne des Singes), Plant (C. nodosa and H. physophora), Ant (A. decemarticulatus, A. octoarticulatus, A. octoarticulatus var. demerarae and Azteca sp. cf. depilis), and their interactions of Site × Plant, Site × Ant, Plant × Ant, and Site × Plant × Ant. The values of the Shannon indexes between pairs of communities were compared using Student’s t-test, and p-values were corrected using a Bonferroni adjustment. Species rarefaction curves were plotted on the expected number of species, and species accumulation curves or sample rarefaction (Mao tau) were computed as a function of the six communities using PAST software [45]. For both curves, the standard errors were converted as 95% confidence intervals.
2.4. Beta Diversity
We calculated Bray-Curtis dissimilarities and, then, we clustered together the six microbial communities (Site × Plant × Ant) of domatia with the ‘picante’ R package. Furthermore, to visualize the level of similarity among microbial communities across Site × Plant × Ant and colonies, we conducted multidimensional scaling multivariate data analysis. Then, we conducted a multivariate analysis of variance (MANOVA) using the distance matrix with 999 permutations, and a multilevel pairwise comparison (permutational multivariate analysis of variance, PERMANOVA) to the Bray-Curtis distances with 40,000 permutations, using the ‘vegan’ package from R [46]. Furthermore, to validate the relevance of the pairwise comparisons after the PERMANOVA we performed a pairwise permutation MANOVA with 40,000 permutations, as found in R package ‘RVAideMemoire’.
3. Results
3.1. Microbial Isolation
A total of 67 bacteria species and two fungi species were isolated, and identified after 16S or ITS sequencing, across all ant domatia (Table 1). Bacteria belonged to the phyla Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria. Among the 11 bacterial orders, the best represented were the Rhizobiales (14 species), Enterobacterales (10), Micrococcales (9), Pseudomonadales (9) and Xanthomonadales (9). One Dothideomycetes (Ascomycota) and a Tremellomycetes (Basidiomycota) were also isolated. While Actinobacteria species (9) were not present in Azteca sp. cf. depilis nests, the two fungal species were only isolated from them. Firmicutes (class Bacilli) species were only present in A. decemarticulatus. Three bacteria were present in all ant species across all sites and plants: the insect symbiont Ochrobactrum sp. 1, Sphingomonas echinoides, and Luteibacter cf. yeojuensis st 2. Finally, Arthrobacter sp. 1 and Burkholderia sp. 9 were exclusively associated with Allomerus spp. Overall, we found 20 bacterial genera in A. decemarticulatus (18 families), A. octoarticulatus (14 families), and Azteca sp. cf. depilis (12 families) domatia, and 19 in A. octoarticulatus var. demerarae (14 families).
Table 1.
Identification of the taxa isolated from the six communities of arboreal ants and frequency of each species per community. The first column corresponds to the microbial species found in this study with their GenBank accession numbers. Second and third columns provide information about the GenBank taxid closest to our sequences and their percentage of sequence identity. Other columns represent the frequency of each microbial sequence in the studied communities. Azt: Azteca sp. cf. depilis; Ao: Allomerus octoarticulatus; Aod: A. octoarticulatus var. demerarae; Ad: A. decemarticulatus; M: Montagne des Singes; B: Basevie; C: C. nodosa; H: H. physophora.
| Microbial Species in Domatia (New GENBANK Accession) Phylum–Class–Order–Family–Species |
Closest Taxid Accession nb | Identity (%) |
Azt M_C |
Ao M_H |
Aod M_C |
Ad M_H |
Aod B_C |
Ad B_H |
|---|---|---|---|---|---|---|---|---|
| Actinobacteria–Actinobacteria–Micrococcales–Dermacoccaceae | ||||||||
| Flexivirga sp. (MN437546) | MH699193 | 97.56 | 0.018 | 0.019 | 0.039 | |||
| Actinobacteria–Actinobacteria–Micrococcales–Microbacteriaceae | ||||||||
| Arthrobacter sp. 1 (MN437547) | KX036592 | 99.00 | 0.127 | 0.027 | 0.037 | 0.023 | 0.020 | |
| Curtobacterium albidum (MN437548) | MK414948 | 99.71 | 0.027 | 0.019 | ||||
| Curtobacterium flaccumfaciens st 4 (MN437549) | KT159381 | 99.54 | 0.045 | 0.020 | ||||
| Curtobacterium luteum st 2 (MN437550) | JQ660282 | 99.70 | 0.055 | 0.019 | 0.020 | |||
| Curtobacterium sp. 1 (MN437551) | MK704290 | 99.90 | 0.023 | |||||
| Curtobacterium sp. 9 (MN437552) | MH915626 | 98.31 | 0.018 | |||||
| Microbacterium sp. 1 (MN437554) | JX566640 | 97.63 | 0.020 | |||||
| Microbacterium sp. 3 (MN437553) | MK578285 | 99.79 | 0.018 | 0.045 | ||||
| Bacteroidetes–Flavobacteriia–Flavobacteriales–Flavobacteriaceae | ||||||||
| Chryseobacterium sp. 1 (MN437556) | LT547832 | 97.00 | 0.055 | |||||
| Chryseobacterium jejuense (MN437555) | KM114947 | 98.00 | 0.018 | 0.037 | ||||
| Elizabethkingia sp. 2 (MN437557) | KP975262 | 95.30 | 0.025 | |||||
| Bacteroidetes–Sphingobacteria–Sphingobacteriales–Sphingobacteriaceae | ||||||||
| Pedobacter sp. 1 (MN437558) | KP708597 | 98.10 | 0.019 | 0.023 | ||||
| Pedobacter sp. 2 (MN437559) | AB461805 | 96.66 | 0.018 | 0.027 | 0.045 | |||
| Firmicutes–Bacilli–Bacillales–Staphylococcaceae | ||||||||
| Staphylococcus haemolyticus (MN437560) | MF157599 | 99.71 | 0.020 | |||||
| Firmicutes–Bacilli–Lactobacillales–Streptococcaceae | ||||||||
| Lactococcus lactis (MN437561) | MF972078 | 99.46 | 0.019 | |||||
| Proteobacteria–Alphaproteobacteria–Rhizobiales–Bradyrhizobiaceae | ||||||||
| Afipia sp. 1 (MN437562) | KY827230 | 99.00 | 0.019 | |||||
| Proteobacteria–Alphaproteobacteria–Rhizobiales–Brucellaceae | ||||||||
| Brucella sp. (MN437563) | CP007717 | 92.54 | 0.019 | 0.045 | ||||
| Ochrobactrum sp. 1 (MN437564) | AY914071 | 100.00 | 0.150 | 0.109 | 0.081 | 0.074 | 0.023 | 0.059 |
| Proteobacteria–Alphaproteobacteria–Rhizobiales–Hyphomicrobiaceae | ||||||||
| Devosia sp. 2 (MN437565) | LC317339 | 99.27 | 0.018 | |||||
| Proteobacteria–Alphaproteobacteria–Rhizobiales–Methylobacteriaceae | ||||||||
| Methylobacterium cf. persicinum (MN437568) | NR_041442 | 99.60 | 0.050 | 0.073 | 0.054 | 0.037 | 0.023 | |
| Methylobacterium cf. phyllostachyos (MN437567) | FR872484 | 99.80 | 0.018 | 0.019 | 0.020 | |||
| Methylobacterium sp. 5 (MN437566) | KC702828 | 99.72 | 0.023 | |||||
| Proteobacteria–Alphaproteobacteria–Rhizobiales–Rhizobiaceae | ||||||||
| Agrobacterium sp. (MN437571) | EU295450 | 98.64 | 0.025 | 0.018 | 0.054 | 0.037 | 0.020 | |
| Agrobacterium tumefaciens st 2 (MN437570) | KU240580 | 99.68 | 0.020 | |||||
| Agrobacterium tumefaciens st 3 (MN437569) | KY874047 | 99.62 | 0.019 | |||||
| Rhizobium sp. 1 (MN437573) | LC385714 | 98.29 | 0.027 | |||||
| Rhizobium sp. 4 (MN437574) | KP219134 | 99.71 | 0.027 | 0.020 | ||||
| Rhizobium sp. 5 (MN437572) | MH327921 | 97.54 | 0.025 | 0.039 | ||||
| Proteobacteria–Alphaproteobacteria–Rhizobiales–Xanthobacteraceae | ||||||||
| Labrys sp. 1 (MN437575) | KR778886 | 99.72 | 0.027 | |||||
| Proteobacteria–Alphaproteobacteria–Sphingomonadales–Sphingomonadaceae | ||||||||
| Sphingobium sp. (MN437576) | HM321152 | 98.27 | 0.037 | |||||
| Sphingomonas echinoides (MN437577) | MH725538 | 98.90 | 0.125 | 0.018 | 0.054 | 0.019 | 0.114 | 0.078 |
| Sphingomonas polyaromaticivorans st 1 (MN437578) | HM241216 | 99.63 | 0.027 | |||||
| Proteobacteria–Betaproteobacteria–Burkholderiales–Burkholderiaceae | ||||||||
| Burkholderia contaminans (MN437582) | KY886142 | 99.80 | 0.050 | |||||
| Burkholderia sp. 5 (MN437583) | AB299574 | 98.98 | 0.055 | 0.068 | ||||
| Burkholderia sp. 9 (MN437580) | KX232126 | 98.78 | 0.018 | 0.027 | 0.019 | 0.045 | 0.020 | |
| Burkholderia sp. 21 (MN437581) | JN634250 | 96.51 | 0.018 | |||||
| Burkholderia tropica st 1 (MN437579) | KT390912 | 99.67 | 0.023 | 0.137 | ||||
| Paraburkholderia fungorum (MN437584) | MG576012 | 99.71 | 0.025 | 0.114 | 0.020 | |||
| Proteobacteria–Gammaproteobacteria–Enterobacterales–Enterobacteriaceae | ||||||||
| Cedecea lapagei (MN437585) | MH074798 | 99.39 | 0.039 | |||||
| Enterobacter hormaechei cf subsp xiangfangensis (MN437588) | MG928407 | 99.16 | 0.018 | 0.045 | 0.039 | |||
| Enterobacter sp. 1 (MN437587) | JQ660160 | 99.52 | 0.025 | 0.055 | 0.054 | 0.130 | 0.020 | |
| Enterobacter sp. 5 (MN437586) | KM021337 | 98.58 | 0.018 | |||||
| Enterobacteriaceae sp. 2 (MN437589) | KJ934757 | 98.55 | 0.020 | |||||
| Escherichia sp. (MN437590) | MH465145 | 99.13 | 0.027 | |||||
| Klebsiella aerogenes st 1 (MN437591) | JF494822 | 99.25 | 0.091 | 0.054 | 0.074 | 0.068 | ||
| Klebsiella oxytoca (MN437592) | KT260783 | 98.90 | 0.018 | |||||
| Proteobacteria–Gammaproteobacteria–Enterobacterales–Erwiniaceae | ||||||||
| Pantoea agglomerans (MN437593) | AF130896 | 98.90 | 0.025 | 0.054 | 0.056 | 0.023 | ||
| Pantoea dispersa st 1 (MN437594) | KC182050 | 98.11 | 0.027 | |||||
| Proteobacteria–Gammaproteobacteria–Pseudomonadales–Moraxellaceae | ||||||||
| Acinetobacter cf. bereziniae (MN437596) | MK087738 | 100.00 | 0.025 | 0.081 | 0.020 | |||
| Acinetobacter sp. 2 (MN437597) | JQ433924 | 99.27 | 0.045 | 0.020 | ||||
| Acinetobacter sp. 3 (MN437595) | KR189585 | 99.30 | 0.019 | |||||
| Proteobacteria–Gammaproteobacteria–Pseudomonadales–Pseudomonadaceae | ||||||||
| Pseudomonas cf. aeruginosa (MN437602) | KT943976 | 99.62 | 0.019 | |||||
| Pseudomonas cf. citronellolis (MN437599) | JQ659858 | 99.90 | 0.100 | |||||
| Pseudomonas fulva (MN437603) | KY511074 | 98.95 | 0.019 | |||||
| Pseudomonas nitroreducens (MN437601) | MH675504 | 98.90 | 0.045 | |||||
| Pseudomonas sp. 2 (MN437600) | KJ184870 | 99.90 | 0.037 | 0.020 | ||||
| Pseudomonas sp. 3 (MN437598) | KM187195 | 98.33 | 0.018 | |||||
| Proteobacteria–Gammaproteobacteria–Xanthomonadales–Rhodanobacteraceae | ||||||||
| Luteibacter cf. rhizovinicus st 1 (MN437606) | KY938100 | 99.90 | 0.020 | |||||
| Luteibacter cf. rhizovinicus st 2 (MN437607) | EU022023 | 99.72 | 0.027 | |||||
| Luteibacter sp.1 (MN437604) | FR714940 | 99.25 | 0.018 | 0.054 | 0.037 | |||
| Luteibacter cf. yeojuensis st 2 (MN437605) | KF668474 | 99.90 | 0.050 | 0.055 | 0.081 | 0.093 | 0.091 | 0.176 |
| Luteibacter cf. yeojuensis st 3 (MN437608) | JQ798488 | 98.74 | 0.020 | |||||
| Proteobacteria–Gammaproteobacteria–Xanthomonadales–Xanthomonadaceae | ||||||||
| Pseudoxanthomonas sp. (MN437609) | MH795540 | 99.72 | 0.018 | 0.037 | ||||
| Stenotrophomonas maltophilia st 1 (MN437612) | MK537385 | 99.61 | 0.125 | 0.027 | ||||
| Stenotrophomonas panacihumi (MN437610) | KF668484 | 99.35 | 0.025 | 0.018 | 0.054 | 0.019 | 0.059 | |
| Stenotrophomonas sp. 3 (MN437611) | JQ684520 | 99.50 | 0.025 | |||||
| Ascomycota–Dothideomycetes–Pleosporales–Didymellaceae | ||||||||
| Phoma sp. (MN435151) | KP307011 | 99.83 | 0.100 | |||||
| Basidiomycota–Tremellomycetes–Trichosporonales–Trichosporonaceae | ||||||||
| Trichosporon siamense (MN435152) | AB164370 | 99.27 | 0.025 | |||||
3.2. Alpha Diversity
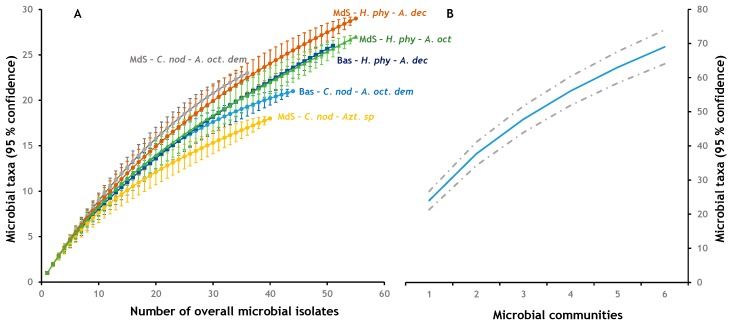
The rarefaction curve and the completeness values suggest that the microbial communities present in A. octoarticulatus var. demerarae are better characterised compared to the other four communities (Figure 1A,B; Table 2). That is, the slopes of the C. nodosa curves appear to be different from the H. physophora slopes, and indicate that total microbial diversity would likely be different between the plant species if more samples were collected. We found an overall significantly higher microbial species richness in Montagne des Singes than in Basevie (Table 2 and Table 3). There were no differences in species richness or evenness between plant species, although Chao’s abundance-based richness was higher in H. physophora than in C. nodosa (Table 2). Microbial richness associated with Azteca sp. cf. depilis (18) was more than half lower compared with A. decemarticulatus (43) or A. octoarticulatus (47), and thus exhibited significantly lower species richness and evenness (Table 3). When we analysed the diversity associated with the Site × Plant interaction, we found that C. nodosa from Basevie (21 species) had the lowest species richness and diversity, while the highest was found in H. physophora (40) from Montagne des Singes (Table 3). When investigating nest microbial diversity associated to Site × Ant communities, we found significant differences in richness in Montagne des Singes between Azteca sp. cf. depilis and A. decemarticulatus, A. octoarticulatus, and A. octoarticulatus var. demerarae (Table 3). In the interaction Plant × Ant, the only significant differences in species richness were between the microbial communities associated with Azteca sp. cf. depilis from C. nodosa, A. decemarticulatus colonising H. physophora, and A. octoarticulatus var. demerarae from C. nodosa (Table 3). Finally, in the Site × Plant × Ant interaction, the only microbial communities that exhibited significant differences in richness were those associated with Azteca sp. cf. depilis in C. nodosa and A. decemarticulatus in H. physophora, both from Montagne des Singes (Table 3).
Figure 1.
Rarefaction and accumulation curves of microbial species in ant nests. (A) Rarefaction curve for each bacterial community; (B) species accumulation curve.
Table 2.
Alpha diversity indexes: Margalef, Chao, Shannon, Simpson, inversed Simpson, and Fisher. S: species richness, overall number of species recorded; N: total number of microbial isolates in the community; C (%): percentage of completeness.
| Communities | S | N | C % |
Margalef’s DMG | Chao ± SE S |
Shannon’s H’ | Simpson D |
Inversed Simpson DS |
Fisher α |
|---|---|---|---|---|---|---|---|---|---|
| Site | |||||||||
| Basevie | 37 | 95 | 71.0 | 7.91 | 52.11 ± 9.73 | 3.26 | 0.959 | 18.61 | 22.27 |
| Montagne des Singes | 57 | 186 | 52.9 | 10.72 | 107.75 ± 26.46 | 3.56 | 0.946 | 24.40 | 28.05 |
| Plant species | |||||||||
| C. nodosa | 42 | 120 | 77.8 | 8.56 | 54.00 ± 7.96 | 3.43 | 0.957 | 23. 61 | 22.97 |
| H. physophora | 53 | 161 | 35.9 | 10.23 | 147.50 ± 55.56 | 3.53 | 0.958 | 23.72 | 27.56 |
| Ant species | |||||||||
| A. decemarticulatus | 43 | 106 | 73.1 | 9.01 | 58.83 ± 9.37 | 3.42 | 0.953 | 21.44 | 26.94 |
| A. octoarticulatus | 27 | 55 | 15.0 | 6.49 | 180.00 ± 74.29 | 3.02 | 0.94 | 16.01 | 20.98 |
| A. octoarticulatus demerarae | 35 | 80 | 85.4 | 7.76 | 41.00 ± 4.53 | 3.36 | 0.96 | 23.70 | 23.73 |
| Azteca sp. cf. depilis | 18 | 40 | 61.5 | 4.61 | 29.25 ± 9.53 | 2.64 | 0.9125 | 11.43 | 12.59 |
| Site × Plant | |||||||||
| Basevie × C. nodosa | 21 | 44 | 87.1 | 5.29 | 24.11 ± 3.10 | 2.89 | 0.950 | 15.61 | 15.75 |
| Basevie × H. physophora | 26 | 51 | 48.9 | 6.36 | 53.20 ± 18.16 | 2.93 | 0.946 | 13.20 | 21.24 |
| Montagne des Singes × C. nodosa | 31 | 76 | 50.8 | 6.93 | 61.00 ± 20.92 | 3.15 | 0.924 | 18.51 | 19.53 |
| Montagne des Singes × H. physophora | 40 | 110 | 65.2 | 8.30 | 61.38 ± 13.14 | 3.32 | 0.936 | 20.58 | 22.61 |
| Site × Ant | |||||||||
| Basevie × A. decemarticulatus | 26 | 51 | 48.9 | 6.36 | 53.20 ± 18.16 | 2.93 | 0.953 | 13.20 | 21.24 |
| Basevie × A. octoarticulatus demerarae | 21 | 44 | 87.1 | 5.29 | 24.11 ± 3.10 | 2.89 | 0.912 | 15.61 | 15.75 |
| Montagne des Singes × A. decemarticulatus | 29 | 55 | 68.5 | 6.99 | 42.33 ± 8.84 | 3.15 | 0.924 | 18.56 | 24.83 |
| Montagne des Singes × A. octoarticulatus | 27 | 55 | 15.0 | 6.49 | 180.00 ± 74.29 | 3.02 | 0.94 | 16.01 | 20.98 |
| Montagne des Singes × A. octoarticulatus demerarae | 23 | 36 | 77.7 | 6.14 | 29.6 ± 5.13 | 3.05 | 0.95 | 19.64 | 27.45 |
| Montagne des Singes × Azteca sp. cf. depilis | 18 | 40 | 61.5 | 4.61 | 29.25 ± 9.53 | 2.64 | 0.944 | 11.43 | 12.59 |
| Plant × Ant | |||||||||
| C. nodosa × A. octoarticulatus demerarae | 35 | 80 | 85.4 | 7.76 | 41.00 ± 4.53 | 3.36 | 0.953 | 23.70 | 23.73 |
| C. nodosa × Azteca sp. cf. depilis | 18 | 40 | 61.5 | 4.61 | 29.25 ± 9.53 | 2.64 | 0.938 | 11.43 | 12.59 |
| H. physophora × A. decemarticulatus | 43 | 106 | 73.1 | 9.01 | 58.83 ± 9.37 | 3.42 | 0.958 | 21.44 | 26.94 |
| H. physophora × A. octoarticulatus | 27 | 55 | 15.0 | 6.49 | 180.00 ± 74.29 | 3.02 | 0.913 | 16.01 | 20.98 |
| Site × Plant × Ant | |||||||||
| Basevie × C. nodosa × A. octoarticulatus demerarae | 21 | 44 | 87.1 | 5.29 | 24.11 ± 3.10 | 2.89 | 0.938 | 15.61 | 15.75 |
| Basevie × H. physophora × A. decemarticulatus | 26 | 51 | 48.9 | 6.36 | 53.20 ± 18.16 | 2.93 | 0.944 | 13.20 | 21.24 |
| Montagne des Singes × C. nodosa × A. octoarticulatus demerarae | 23 | 36 | 77.7 | 6.14 | 29. 60 ± 5.13 | 3.05 | 0.948 | 19. 64 | 27.45 |
| Montagne des Singes × C. nodosa × Azteca sp. cf. depilis | 18 | 40 | 61.5 | 4.61 | 29.25 ± 9.53 | 2.64 | 0.913 | 11.43 | 12.59 |
| Montagne des Singes × H. physophora × A. decemarticulatus | 29 | 55 | 68.5 | 6.99 | 42.33 ± 8.84 | 3.15 | 0.936 | 18.56 | 24.83 |
| Montagne des Singes × H. physophora × A. octoarticulatus | 27 | 55 | 15.0 | 6.49 | 180.00 ± 74.29 | 3.02 | 0.924 | 16.01 | 20.98 |
Table 3.
T-student tests for Shannon’s H’ and Simpsons’ D. Pairwise comparisons for Site, Plant, Ant, Site × Plant, Site × Ant, Plant × Ant, and Site × Plant × Ant. Bold values denote significant pairwise comparisons. Bas.: Basevie; MdS: Montagne des Singes; C. nod.: Cordia nodosa; H. phy.: Hirtella physophora; A. dec.: Allomerus decemarticulatus; A. oct.: A. octoarticulatus; A. oct. demer.: A. octoarticulatus var. demerarae; Azteca sp.: Azteca sp. cf. depilis. * denote significant P-values after Bonferroni correction.
| Shannon’s Richness | Simpson’s Evenness | ||||||
|---|---|---|---|---|---|---|---|
| Community 1 vs | Community 2 | t | df | p-Value | t | df | p-Value |
| Basevie | Montagne des Singes | 2.399 | 203.420 | 0.017 * | −1.225 | 143.920 | 0.223 |
| C. nodosa | H. physophora | −0.829 | 274.410 | 0.408 | 0.025 | 264.410 | 0.980 |
| A. decemarticulatus | A. octoarticulatus | 2.646 | 119.85 | 0.009 | −1.085 | 104.410 | 0.281 |
| A. octoarticulatus demerarae | 0.498 | 185.1 | 0.619 | 0.423 | 185.52 | 0.673 | |
| Azteca sp. cf. depilis | 4.854 | 83.287 | <0.001 * | −2.101 | 57.704 | 0.040 | |
| A. octoarticulatus | A. octoarticulatus demerarae | −2.284 | 108.14 | 0.024 | 1.464 | 87.986 | 0.147 |
| Azteca sp. cf. depilis | 2.140 | 89.375 | 0.035 | −1.167 | 74.564 | 0.247 | |
| A. octoarticulatus demerarae | Azteca sp. cf. depilis | 4.562 | 76.011 | <0.001 * | −2.397 | 51.779 | 0.020 |
| Basevie–C. nodosa | Basevie–H. physophora | −0.225 | 92.749 | 0.823 | −0.528 | 87.968 | 0.599 |
| MdS–C. nodosa | −1.783 | 104.080 | 0.077 | 0.655 | 85.616 | 0.514 | |
| MdS–H. physophora | −3.059 | 104.050 | 0.003 * | 1.086 | 70.285 | 0.281 | |
| Basevie–H. physophora | MdS–C. nodosa | −1.313 | 98.465 | 0.192 | 1.074 | 74.704 | 0.286 |
| MdS–H. physophora | −2.387 | 95.101 | 0.019 | 1.399 | 65.154 | 0.167 | |
| MdS–C. nodosa | MdS–H. physophora | −1.286 | 172.780 | 0.200 | 0.491 | 154.160 | 0.624 |
| Basevie–A. dec. | Basevie–A. oct. demer. | 0.225 | 92.749 | 0.823 | 0.528 | 87.968 | 0.599 |
| MdS–A. dec | −1.222 | 100.810 | 0.225 | 1.018 | 86.199 | 0.311 | |
| MdS–A. oct. | −0.475 | 102.62 | 0.636 | 0.606 | 89.906 | 0.546 | |
| MdS–A. oct. demer. | −0.689 | 86.952 | 0.493 | 1.172 | 79.185 | 0.245 | |
| MdS–Azteca sp. | 1.537 | 90.593 | 0.128 | −0.463 | 90.246 | 0.644 | |
| Basevie–A. oct. demer. | MdS–A. dec | −1.635 | 98.523 | 0.105 | 0.601 | 94.668 | 0.550 |
| MdS–A. oct. | −0.780 | 98.949 | 0.437 | 0.090 | 96.931 | 0.929 | |
| MdS–A. oct. demer. | −1.032 | 78.551 | 0.305 | 0.793 | 79.779 | 0.430 | |
| MdS–Azteca sp. | 1.483 | 79.961 | 0.142 | −1.081 | 73.544 | 0.283 | |
| MdS–A. dec | MdS–A. oct. | 0.799 | 109.76 | 0.426 | −0.517 | 109.5 | 0.606 |
| MdS–A. oct. demer. | 0.596 | 87.278 | 0.553 | 0.189 | 88.710 | 0.850 | |
| MdS–Azteca sp. | 2.946 | 87.273 | 0.004 | −1.601 | 70.977 | 0.114 | |
| MdS–A. oct. | MdS–A. oct. demer. | −0.220 | 88.762 | 0.826 | 0.712 | 90.237 | 0.478 |
| MdS–Azteca sp. | 2.14 | 89.375 | 0.035 | −1.167 | 74.564 | 0.247 | |
| MdS–A. oct. demer | MdS–Azteca sp. | 2.401 | 75.388 | 0.019 | −1.767 | 65.232 | 0.082 |
| C. nodosa–A. oct. demer | C. nodosa–Azteca sp. | 4.562 | 76.011 | <0.001 * | −2.397 | 51.779 | 0.020 |
| H. physophora–A. decemarticulatus | −0.498 | 185.100 | 0.619 | −0.423 | 185.520 | 0.673 | |
| H. physophora–A. octoarticulatus | 2.284 | 108.140 | 0.024 | −1.464 | 87.986 | 0.147 | |
| C. nodosa–Azteca sp. | H. physophora–A. decemarticulatus | −4.854 | 83.287 | <0.001 * | 2.101 | 57.704 | 0.040 |
| H. physophora–A. octoarticulatus | −2.140 | 89.375 | 0.035 | 1.167 | 74.564 | 0.247 | |
| H. physophora–A. dec. | H. physophora–A. octoarticulatus | 2.646 | 119.850 | 0.009 | −1.085 | 104.410 | 0.281 |
| Bas.–C. nod.–A. oct. demer | Basevie–H. physophora–A. dec. | −0.225 | 92.749 | 0.823 | −0.528 | 87.968 | 0.599 |
| MdS–C. nodosa–A. oct. demer. | −1.032 | 78.551 | 0.305 | 0.793 | 79.779 | 0.430 | |
| MdS–C. nodosa–Azteca sp. | 1.483 | 79.961 | 0.142 | −1.081 | 73.544 | 0.283 | |
| MdS–H. physophora–A. dec. | −1.635 | 98.523 | 0.105 | 0.601 | 94.668 | 0.550 | |
| MdS–H. physophora–A. oct. | −0.780 | 98.949 | 0.437 | 0.090 | 96.931 | 0.929 | |
| Bas.–H. phy.–A. dec. | MdS–C. nodosa–A. oct. demer. | −0.688 | 86.952 | 0.493 | 1.172 | 79.185 | 0.245 |
| MdS–C. nodosa–Azteca sp. | 1.537 | 90.593 | 0.128 | −0.463 | 90.246 | 0.644 | |
| MdS–H. physophora–A. dec. | −1.222 | 100.810 | 0.225 | 1.018 | 86.199 | 0.311 | |
| MdS–H. physophora–A. oct. | −0.475 | 102.620 | 0.636 | 0.606 | 89.906 | 0.546 | |
| MdS–C. nod.–A. oct. demer | MdS–C. nodosa–Azteca sp. | 2.401 | 75.388 | 0.019 | −1.767 | 65.232 | 0.082 |
| MdS–H. physophora–A. dec. | −0.596 | 87.278 | 0.553 | −0.189 | 88.710 | 0.850 | |
| MdS–H. physophora–A. oct. | 0.220 | 88.762 | 0.826 | −0.712 | 90.237 | 0.478 | |
| MdS–C. nod.–Azteca sp. | MdS–H. physophora–A. dec. | −2.946 | 87.273 | 0.004 | 1.601 | 70.977 | 0.114 |
| MdS–H. physophora–A. oct. | −2.140 | 89.375 | 0.035 | 1.167 | 74.564 | 0.247 | |
| MdS–H. phy.–A. dec. | MdS–H. physophora–A. oct. | 0.799 | 109.760 | 0.426 | −0.517 | 109.500 | 0.606 |
3.3. Beta Diversity
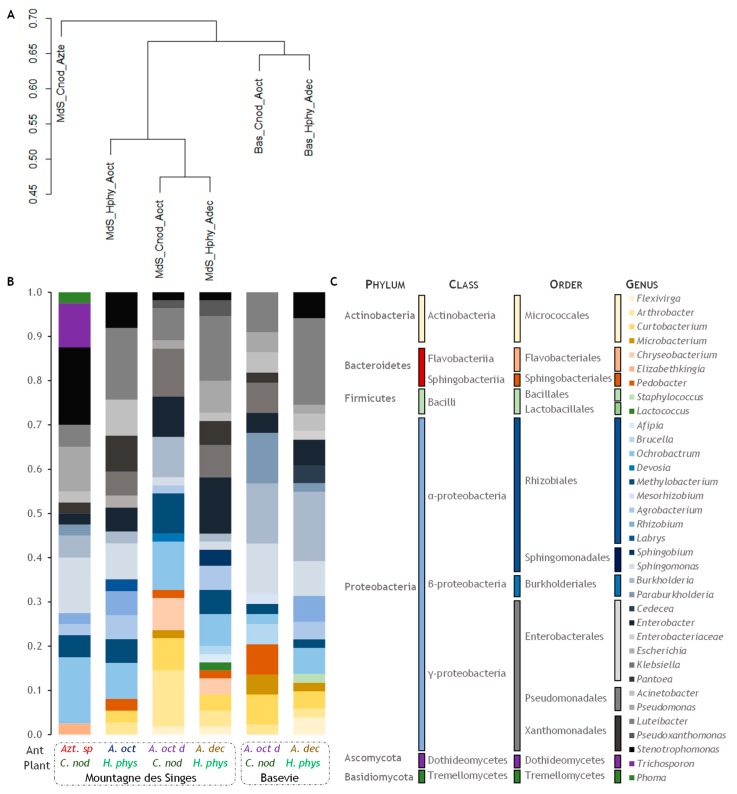
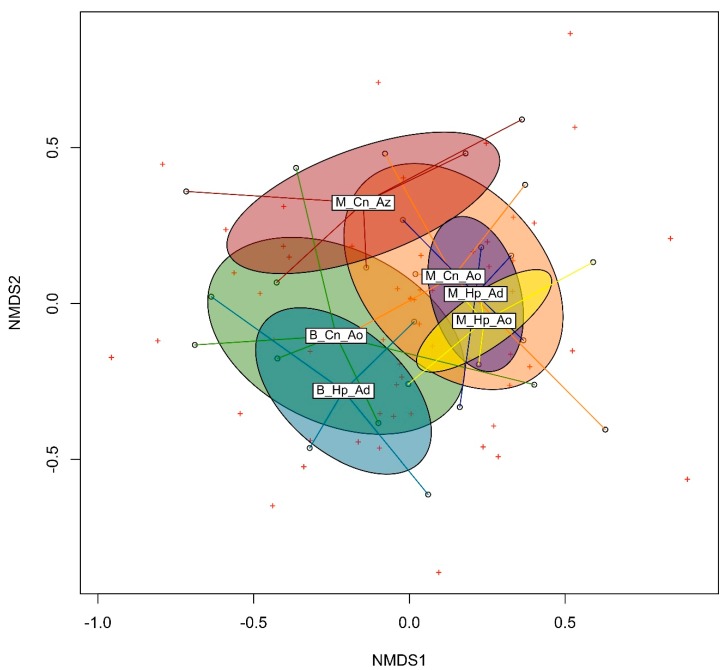
The dendrogram based on Bray-Curtis dissimilarities grouped the two microbial communities of Basevie into a clade, the microbial communities from Allomerus domatia in a second clade and the community from Azteca sp. cf. depilis into a third clade (Figure 2A). Moreover, the clades are related to the phylogenetic diversity of each microbial community (Figure 2B,C). We found overall significant differences among the six microbial communities (F5,89 = 2.6544; p-value = 0.001). The multilevel pairwise comparison detected differences for 10 pairs of communities (Table 3, which were further confirmed by the pairwise permutation MANOVA. The MDS mapping (Figure 3) was well supported by the non-metric fit of R2 = 0.948, although the linear fit (R2 = 0.708), and the stress (S = 0.228) values were slightly weak.
Figure 2.
Distribution and abundance of microbial communities in Cordia nodosa (C. nod) and Hirtella physophora (H. phys) domatia occupied by, Allomerus decemarticulatus (A. dec), A. octoarticulatus (A. oct), A. octoarticulatus var. demerarae (A. oct d), and Azteca sp. cf. depilis (Azt. sp) ant species from Montagne des Singes and Basevie. (A) Bray-Curtis dissimilarity dendrogram; (B) stackplot of bacterial taxonomic composition; (C) microbial diversity by taxonomic level (Phylum, Class, Order, and Genus).
Figure 3.
MDS mapping with two dimensions for the six microbial communities. M, Montagne des Singes; B, Basevie; Cn, Cordia nodosa; Hp, Hirtella physophora; Ad, Allomerus decemarticulatus; Ao, A. octoarticulatus; Az, Azteca sp. cf. depilis.
4. Discussion
4.1. Microbial Diversity and Composition
The microbial diversity of the studied plant-ant species is similar in genera numbers to the one found associated with fungus-growing ants in their nests [13], even if the taxonomic diversity studied here is based on the microbial ability to grow on media and, thus, the presence of more species can be expected. In another ant-plant associations between Azteca alfari and Cecropia peltata, Lucas et al. [47] found 22 bacterial phyla across internal and external structures of the plant but 90% of the microbial diversity corresponded to Proteobacteria and Actinobacteria taxa. This study also demonstrated the role that ants play in shaping the composition of microbial communities inside their nests, as it has been shown here too, which can be considered as an effect of niche filtering (see below).
That Allomerus ants were associated with the highest number of bacterial species compared to Azteca ants, which can be explained either by the fact that they cannot filter as many microbial species as Azteca spp., or because they allow and select a wider microbial community composition because of more complex functional roles. It is particularly relevant that only these ants harboured Actinobacteria species because the defensive role of some species in fungus gardens of Attine ants is well known [11,48]; and because Allomerus ants practise a particular type of fungiculture in their colonies [26]. Recent work investigating the Azteca spp. microbiome from myrmecophytic Cecropia, found different Actinobacteria species [47] from the ones identified here for Azteca sp. cf. depilis. One species of Proteobacteria, Pseudomonas citronellolis, was recorded as associated with both Azteca sp. cf. depilis (this study) and Az. alfari [47]. This species could be specifically associated with Azteca ants. Previous work on bacteria associated with both fungus-growing and arboreal ants detected bacteria from diverse genera that perform different roles, such as defence against parasites and diseases, plant substrate degradation, and nitrogen fixing bacteria from the genera Burkholderia, Curtobacterium, Enterobacter, Escherichia, Pantoea, and Rhizobium [10,13,49,50,51]. We have found bacterial species from all these genera that could perform similar roles. First, they can complement the behaviour of Allomerus ants that chew the walls of the domatia to prepare vegetal substrate and culture their mutualistic fungus Trimmatostroma cordae [26]. Second, they could act in reinforcing the active transfer of nitrogen to the plant mediated by the mutualistic fungus [36]. Third, they could play a role in the defence of the domatia against diseases by limiting pathogen proliferation.
4.2. Variations in Microbial Communities
The observed richness of microbial communities appears driven by both distance (geography) and ant species identity, as a result of niche filtering. That is, the effect of site location in microbial composition means horizontal acquisition of local bacterial species or strains. Furthermore, the effect of ant species in microbial richness suggest the presence of ant-engineered microbial communities. Whether these microbial communities are vertically transmitted or filtered from the environment by the ants remains unclear. Host plant species, however, do not seem to have an effect on richness or evenness of microbial species in domatia. Therefore, in the studied systems, plant species do not contribute to niche filtering. Indeed, the microbial communities associated with Azteca sp. cf. depilis appeared different from the ones associated with Allomerus ants, both in terms of richness and composition. Moreover, the pairwise comparison of the composition of bacterial communities highlighted differences between A. octoarticulatus inhabiting C. nodosa from Basevie and the other communities but A. octoarticulatus from Montagne des Singes. On the one hand, the ant species effect might be related to an active selection of potentially beneficial microorganisms by the ants; or at least the removal of pathogens. It could be also linked to differences in the environmental conditions in the domatia, not related to the plant species, but to the ants. On the other hand, the geographic effect might be driven by dispersal limitations of the microorganisms and/or differences in the local abiotic environment, surrounding plant species or even ant diet. Allomerus ants are omnivores; however, because of their particular behaviour in practising a highly specialised type of agriculture, they can be expected to exert a strong selective pressure on the composition of the microbial community such that healthy fungal symbionts are maintained [26]. The lack of bacterial communities specifically associated with host plant species is supported by previous work in acacia-ants [52]. All these results strongly suggest an active role of ants in the assemblage of their associated microbial communities.
5. Conclusions
Altogether our results show that the microbial communities present inside the domatia of ant plants are mainly influenced by ant species and, to a lower extent, by the local environment. This suggests that the ants actively select for at least part of the associated microorganisms and, thus, that the latter could have beneficial roles in the survival of the colonies and in the interaction with their host plants. Although the plant does not seem to contribute to the observed diversity in microbial communities, it could indeed benefit from their presence as already shown from the recent studies of Chaetothyriales fungi associated with plant ants. Our understanding of ant-plant-microorganism interactions and of the functioning of these interaction networks appears promising as a mean of shedding more light on the importance of biotic interactions in the evolution of biodiversity.
Acknowledgments
We thank Jérémie Lauth and Laurie Esparza for their help in the field and in the isolation and culturing of the microbial strains, to Andrea Yockey-Dejean for proof-reading the manuscript and to the Laboratoire Environnement de Petit Saut (HYDRECO) for logistical help.
Author Contributions
Conceptualization, M.X.R.-G., J.O. and C.L.; methodology, M.X.R.-G., H.G. and P.J.; software, M.X.R.-G. and A.D.A.C.; formal analysis, M.X.R.-G. and A.D.A.C.; investigation, M.X.R.-G., H.G., P.J. and C.L.; resources, J.O.; data curation, M.X.R.-G. and A.D.A.C.; writing—original draft preparation, M.X.R.G, J.O. and A.D.; writing—review and editing, all authors.; project administration, J.O.; funding acquisition, J.O and A.D.
Funding
Financial support for this study was provided by an Investissement d’Avenir grant of the Agence Nationale de la Recherche (CEBA, ANR-10-LABX-25-01) and by the PO-FEDER 2014–2020, Région Guyane (BING, GY0007194).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Hung K.-L.J., Kingston J.M., Albrecht M., Holway D.A., Kohn J.R. The worldwide importance of honey bees as pollinators in natural habitats. Proc. R. Soc. B. 2018;285:20172140. doi: 10.1098/rspb.2017.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanders D., van Veen F.J.F. Ecosystem engineering and predation: The multi-trophic impact of two ant species. J. Anim. Ecol. 2011;80:569–576. doi: 10.1111/j.1365-2656.2010.01796.x. [DOI] [PubMed] [Google Scholar]
- 3.Wilson E.O. The Insect Societies. Belknap; Cambridge, MA, USA: 1971. p. 548. [Google Scholar]
- 4.Wilson E. The effects of complex social life on evolution and biodiversity. Oikos. 1992;63:13–18. doi: 10.2307/3545511. [DOI] [Google Scholar]
- 5.Cremer S., Armitage S.A.O., Schmid-Hempel P. Social Immunity. Curr. Biol. 2007;17:693–702. doi: 10.1016/j.cub.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Meunier J. Social immunity and the evolution of group living in insects. Phil. Trans. R. Soc. B. 2015;370:20140102. doi: 10.1098/rstb.2014.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmid-Hempel P. Parasites in Social Insects. Princeton University Press; Princeton, NJ, USA: 1998. [Google Scholar]
- 8.Griffiths H.M., Ashton L.A., Walker A.E., Hasan F., Evans T.A., Eggleton P., Parr C.L. Ants are the major agents of resource removal from tropical rainforests. J. Anim. Ecol. 2018;87:293–300. doi: 10.1111/1365-2656.12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmid-Hempel P. Parasites and their social hosts. Trends Parasitol. 2017;33:453–462. doi: 10.1016/j.pt.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Aylward F.O., Burnum K.E., Scott J.J., Suen G., Tringe S.G., Adams S.M., Barry K.W., Nicora C.D., Piehowski P.D., Purvine S.O., et al. Metagenomic and metaproteomic insights into bacterial communities in leaf-cutter ant fungus gardens. ISME J. 2012;6:1688–1701. doi: 10.1038/ismej.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Currie C.R., Scott J.A., Summerbell R.C., Malloch D. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature. 1999;398:701–705. doi: 10.1038/19519. [DOI] [Google Scholar]
- 12.Currie C.R., Bot A.N.M., Boomsma J.J. Experimental evidence of a tripartite mutualism: Bacteria protect ant fungus gardens from specialized parasites. Oikos. 2003;101:91–102. doi: 10.1034/j.1600-0706.2003.12036.x. [DOI] [Google Scholar]
- 13.Kellner K., Ishak H.D., Linksvayer T.A., Mueller U.G. Bacterial community composition and diversity in an ancestral ant fungus symbiosis. FEMS Microbiol. Ecol. 2015;91:7. doi: 10.1093/femsec/fiv073. [DOI] [PubMed] [Google Scholar]
- 14.Little A.E.F., Currie C.R. Symbiotic complexity: Discovery of a fifth symbiont in the attine ant-microbe symbiosis. Biol. Lett. 2007;3:501–504. doi: 10.1098/rsbl.2007.0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mehdiabadi N.J., Hughes B., Mueller U.G. Cooperation, conflict, and coevolution in the attine ant-fungus symbiosis. Behav. Ecol. 2006;17:291–296. doi: 10.1093/beheco/arj028. [DOI] [Google Scholar]
- 16.Sanders S.B., Yek S.H., Nash D.R., Boomsma J.J. Interaction specificity between leaf-cutting ants and vertically transmitted Pseudonocardia bacteria. BMC Evol. Biol. 2015;15:27. doi: 10.1186/s12862-015-0308-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schultz T.R., Brady S.G. Major evolutionary transitions in ant agriculture. Proc. Natl. Acad. Sci. USA. 2008;105:5435–5440. doi: 10.1073/pnas.0711024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scott J.J., Budsberg K.J., Suen G., Wixon D.L., Balser T.C., Currie C.R. Microbial community structure of leaf-cutter ant fungus gardens and refuse dumps. PLoS ONE. 2010;5:e9922. doi: 10.1371/journal.pone.0009922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boots B., Clipson N. Linking ecosystem modification by the yellow meadow ant (Lasius flavus) to microbial assemblages in different soil environments. Eur. J. Soil. Biol. 2013;55:100–106. doi: 10.1016/j.ejsobi.2013.01.002. [DOI] [Google Scholar]
- 20.Dauber J., Schroeter D., Wolters V. Species specific effects of ants on microbial activity and N-availability in the soil of an old-field. Eur. J. Soil. Biol. 2001;37:259–261. doi: 10.1016/S1164-5563(01)01094-9. [DOI] [Google Scholar]
- 21.Delgado-Baquerizo M., Eldridge D.J., Hamonts K., Singh B.K. Ant colonies promote the diversity of soil microbial communities. ISME J. 2019;13:1114–1118. doi: 10.1038/s41396-018-0335-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duff L.B., Urichuk T.M., Hodgins L.N., Young J.R., Untereine W.A. Diversity of fungi from the mound nests of Formica ulkei and adjacent non-nest soils. Can. J. Microbiol. 2016;62:562–571. doi: 10.1139/cjm-2015-0628. [DOI] [PubMed] [Google Scholar]
- 23.Defossez E., Selosse M.-A., Dubois M.-P., Mondolot L., Faccio A., Djieto-Lordon C., McKey D., Blatrix R. Ant-plants and fungi: A new three-way symbiosis. New Phytol. 2009;182:942–949. doi: 10.1111/j.1469-8137.2009.02793.x. [DOI] [PubMed] [Google Scholar]
- 24.Mayer V.E., Nepel M., Blatrix R., Oberhauser F.B., Fiedler K., Schönenberger J., Voglmayr H. Transmission of fungal partners to incipient Cecropia-tree ant colonies. PLoS ONE. 2018;13:e0192207. doi: 10.1371/journal.pone.0192207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayer V.E., Voglmayr H. Mycelial carton galleries of Azteca brevis (Formicidae) as a multi-species network. Proc. R. Soc. B. 2009;276:3265–3273. doi: 10.1098/rspb.2009.0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruiz-González M.X., Malé P.-J.G., Leroy C., Dejean A., Gryta H., Jargeat P., Quilichini A., Orivel J. Specific, non-nutritional association between an ascomycete fungus and Allomerus plant-ants. Biol. Lett. 2011;7:475–479. doi: 10.1098/rsbl.2010.0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janzen D.H. Coevolution of mutualism between ants and acacias in Central America. Evolution. 1966;20:249–275. doi: 10.1111/j.1558-5646.1966.tb03364.x. [DOI] [PubMed] [Google Scholar]
- 28.Solano P.J., Durou S., Corbara B., Quilichini A., Cerdan P., Belin-Depoux M., Delabie J.H.C., Dejean A. Myrmecophytes of the understory of French Guianian rainforests: Their distribution and their associated ants. Sociobiology. 2003;41:605–614. [Google Scholar]
- 29.Mayer V.E., Frederickson M.E., McKey D., Blatrix R. Current issues in the evolutionary ecology of ant-plant symbioses. New Phytol. 2014;202:749–764. doi: 10.1111/nph.12690. [DOI] [PubMed] [Google Scholar]
- 30.Ramette A., Tiedje J.M. Multiscale responses of microbial life to spatial distance and environmental heterogeneity in a patchy ecosystem. Proc. Natl. Acad. Sci. USA. 2007;104:2761–2766. doi: 10.1073/pnas.0610671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nemergut D.R., Schmidt S.K., Fukami T., O’Neill S.P., Bilinski T.M., Stanish L.F., Knelman J.E., Darcy J.L., Lynch R.C., Wickey P., et al. Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 2013;77:342–356. doi: 10.1128/MMBR.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dejean A., Azémar F., Petitclerc F., Delabie J.H.C., Corbara B., Leroy C., Céréghino R., Compin A. Highly modular pattern in ant-plant interactions involving specialized and non-specialized myrmecophytes. Sci. Nat. 2018;105:43. doi: 10.1007/s00114-018-1570-0. [DOI] [PubMed] [Google Scholar]
- 33.Fernández F. The myrmicine ant genus Allomerus Mayr (Hymenoptera: Formicidae) Caldasia. 2007;29:159–175. [Google Scholar]
- 34.Dejean A., Solano P.J., Ayroles J., Corbara B., Orivel J. Arboreal ants build traps to capture prey. Nature. 2005;434:973. doi: 10.1038/434973a. [DOI] [PubMed] [Google Scholar]
- 35.Lauth J., Ruiz-González M.X., Orivel J. New findings in insect fungiculture. Commun. Integr. Biol. 2011;4:728–730. doi: 10.4161/cib.17590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leroy C., Séjalon-Delmas N., Jauneau A., Ruiz-González M.X., Gryta H., Jargeat P., Corbara B., Dejean A., Orivel J. Trophic mediation by a fungus in an ant–plant mutualism. J. Ecol. 2011;99:583–590. doi: 10.1111/j.1365-2745.2010.01763.x. [DOI] [Google Scholar]
- 37.Seipke R.F., Barke J., Ruiz-González M.X., Orivel J., Yu D.W., Hutchings M.I. Fungus-growing Allomerus ants are associated with antibiotic-producing actinobacteria. Antonie Van Leeuwenhoek. 2012;101:443–447. doi: 10.1007/s10482-011-9621-y. [DOI] [PubMed] [Google Scholar]
- 38.Orivel J., Malé P.-J., Lauth J., Roux O., Petitclerc F., Dejean A., Leroy C. Trade-offs in an ant-plant-fungus mutualism. Proc. R. Soc. B. 2017;284:20161679. doi: 10.1098/rspb.2016.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mori S.A., Cremers G., Gracie C.A., de Granville J.-J., Heald S.V., Hoff M., Mitchell J.D. Guide to the Vascular Plants of Central French Guiana, Part 2, Dicotyledons. The New York Botanical Garden; New York, NY, USA: 2002. [Google Scholar]
- 40.Leroy C., Jauneau A., Quilichini A., Dejean A., Orivel J. Comparison between the anatomical and morphological structure of leaf blades and foliar domatia in the ant-plant Hirtella physophora (Chrysobalanaceae) Ann. Bot. 2008;101:501–507. doi: 10.1093/aob/mcm323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walsh P.S., Metzger D.A., Higuchi R. Chelex-100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 1991;10:506–513. doi: 10.2144/000114018. [DOI] [PubMed] [Google Scholar]
- 42.Weisburg W.G., Barns S.M., Pelletier D.A., Lane D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White T.J., Bruns T., Lee S., Taylor J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In: Innis N., Gelfand D., Sninsky J., White T., editors. PCR Protocols: A Guide to Methods and Applications. Academic Press; Cambridge, MA, USA: 1990. pp. 315–322. [Google Scholar]
- 44.Basic Local Alignment Search Tool (BLAST) [(accessed on 28 May 2019)]; Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi.
- 45.Hammer Ø., Harper D.A.T., Ryan P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001;4:9. [Google Scholar]
- 46.Martinez Arbizu P. PairwiseAdonis: Pairwise Multilevel Comparison Using Adonis. R Package Version 0.3. [(accessed on 17 June 2019)];2019 Available online: https://github.com/pmartinezarbizu/pairwiseAdonis.
- 47.Lucas J.M., Madden A.A., Penick C.A., Epps M.J., Marting P.R., Stevens J.L., Fergus D.J., Dunn R.R., Meineke E.K. Azteca ants maintain unique microbiomes across functionally distinct nest chambers. Proc. R. Soc. B. 2019;286:20191026. doi: 10.1098/rspb.2019.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mueller U.G., Gerardo N.M., Aanen D.K., Six D.L., Schultz T.R. The evolution of agriculture in insects. Annu. Rev. Ecol. Evol. Syst. 2005;36:563–595. doi: 10.1146/annurev.ecolsys.36.102003.152626. [DOI] [Google Scholar]
- 49.Eilmus S., Heil M. Bacterial associates of arboreal ants and their putative functions in an obligate ant-plant mutualism. Appl. Environ. Microbiol. 2009;75:4324–4332. doi: 10.1128/AEM.00455-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pinto-Tomas A.A., Anderson M.A., Suen G., Stevenson D.M., Chu F.S.T., Cleland W.W., Weimer P.J., Currie C.R. Symbiotic nitrogen fixation in the fungus gardens of leaf-cutter ants. Science. 2009;326:1120–1123. doi: 10.1126/science.1173036. [DOI] [PubMed] [Google Scholar]
- 51.Zucchi T.D., Guidolin A.S., Cônsoli F.L. Isolation and characterization of actinobacteria ectosymbionts from Acromyrmex subterraneus brunneus (Hymenoptera, Formicidae) Microbiol. Res. 2011;166:68–76. doi: 10.1016/j.micres.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 52.Rubin B.E.R., Kautz S., Wray B.D., Moreau C.S. Dietary specialization in mutualistic acacia-ants affects relative abundance but not identity of host-associated bacteria. Mol. Ecol. 2019;28:900–916. doi: 10.1111/mec.14834. [DOI] [PubMed] [Google Scholar]