Abstract
The cyclooxygenase-2 (COX-2) enzyme is considered to be an important target for developing novel anti-inflammatory agents. Selective COX-2 inhibitors offer the advantage of lower adverse effects that are commonly associated with non-selective COX inhibitors. In this work, a novel series of methyl 3-(substituted benzoyl)-7-substituted-2-phenylindolizine-1-carboxylates was synthesized and evaluated for COX-2 inhibitory activity. Compound 4e was identified as the most active compound of the series with an IC50 of 6.71 μM, which is comparable to the IC50 of indomethacin, a marketed non-steroidal anti-inflammatory drug (NSAID). Molecular modeling and crystallographic studies were conducted to further characterize the compounds and gain better understanding of the binding interactions between the compounds and the residues at the active site of the COX-2 enzyme. The pharmacokinetic properties and potential toxic effects were predicted for all the synthesized compounds, which indicated good drug-like properties. Thus, these synthesized compounds can be considered as potential lead compounds for developing effective anti-inflammatory therapeutic agents.
Keywords: COX-2 inhibition, anti-inflammatory, indolizine derivatives, 2-phenyl indolizine, molecular modeling, absorption, distribution, metabolism, excretion, toxicity [ADMET] prediction
1. Introduction
The non-steroidal anti-inflammatory drugs (NSAIDs) are considered to be one of the most frequently prescribed medications for the treatment of inflammation, pain, and fever [1]. The anti-inflammatory effects produced by the NSAIDs occur as a result of inhibition of the cyclooxygenase (COX) enzyme. This enzyme is involved in the synthesis of prostaglandin G2 (PGG2) from arachidonic acid, which is required for the inflammation process to take place [2,3]. There are mainly two important isoforms of the COX enzyme. The first one is the COX-1 isoform, which represents the constitute isoform that is distributed normally in certain tissues of the body such as the kidney and the gastrointestinal tract (GIT), this isoform carries out physiological functions such as gastric protection and maintenance of renal homeostasis [4,5,6]. The other isoform is the COX-2, which is the inducible isoform that is only expressed during exposure to certain substances such as cytokines, which are produced during injury. The use of non-selective COX inhibitors, which inhibit both the COX-1 and COX-2 isoforms usually, results in undesirable adverse effects, mainly involving the GIT. These adverse effects are caused by the inhibition of the COX-1 isoform, which is responsible for GIT protection. For instance, GIT ulceration is a common side effect of non-selective NSAIDs, which results mainly from the inhibition of the COX-1 isoform [7,8]. In order to develop anti-inflammatory agents that have an improved safety profile and reduced side effects, the discovery of selective COX-2 inhibitors is required, as these inhibitors will be devoid of the side effects observed from COX-1 isoform inhibition.
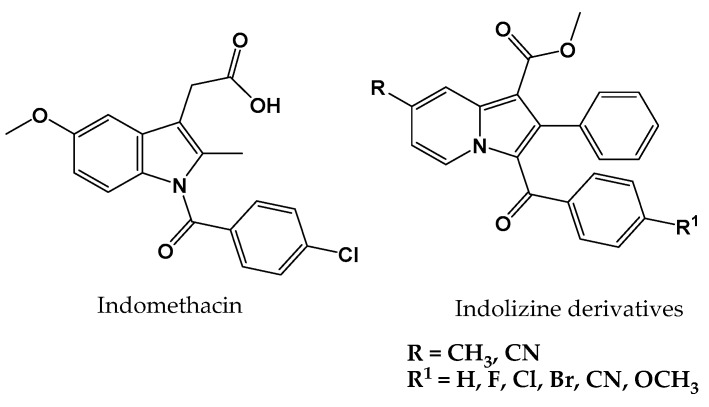
Compounds based on the indolizine scaffold (Figure 1) have been reported to possess a wide spectrum of biological activities. For instance, they have been shown to be able to bind to phospholipase A2, histamine receptors, and calcium channels, which are well-known drug targets [9,10]. In addition, indolizine derivatives were reported to have potential anti-inflammatory and anti-cancer effects [11,12,13]. Figure 1 shows the similar structures of the marketed COX-2 inhibitor indomethacin and the scaffold of the indolizine derivatives.
Figure 1.
Commercially available COX-2 enzyme inhibitor (indomethacin) and the proposed methyl 3-(substituted benzoyl)-7-substituted-2-phenylindolizine-1-carboxylate analogues for COX-2 inhibition activity.
In continuation of our effort to identify potential COX-2 inhibitors [14,15], in the present study, the anti-inflammatory activity of a series of phenylindolizine derivatives was evaluated by assessing the ability of the compounds to inhibit the COX-2 enzyme. In addition, molecular modeling studies were carried out to gain better insights into the structural requirements for achieving the high activity. The pharmacokinetic properties and toxicity of the synthesized compounds were predicted to evaluate their potential as lead compounds for developing anti-inflammatory agents.
2. Materials and Methods
2.1. General
All chemicals reported here were obtained from Sigma-Aldrich Co. (St. Louis, MO, USA), while the solvents were obtained from MilliporeSigma (Burlington, MA, USA). Thin-layer chromatography (TLC) was employed to observe chemical reactions, and this process was performed on silica gel (Sigma-Aldrich Co., St. Louis (HQ), MO, USA), on aluminum foil; n-hexane and ethyl acetate (4:6) were used as the solvent. The reactions were visualized under an ultraviolet (UV)-light/iodine chamber. A Büchi melting point B-545 apparatus was used to measure the melting points (Büchi, Labortechnik, Flawil, Switzerland). Infrared (IR) spectra were recorded on a Shimadzu IRAffinity-1S Fourier-transform infrared (FT-IR) spectrometer (Shimadzu Corporation, 1 Nishinokyo Kuwabara-cho, Nakagyo-ku, Kyoto 604-8511, Japan. Further, 1H and 13C-nuclear magnetic resonance (NMR) spectra were recorded using Bruker AVANCE III 400 MHz (Bruker Corporation, Billerica, MA, USA) with CDCl3 (solvent). Chemical shifts (δ) were indicated in ppm, with tetramethylsilane (TMS) as a reference; coupling constants (J) were recorded (Hz). The splitting pattern was documented as follows: s, singlet; d, doublet; q, quartet; and m, multiplet. Liquid chromatography–mass spectrometry (LC-MS) (Agilent 1100 series, (Agilent Technologies Agilent Technologies, 5301 Stevens Creek Blvd, Santa Clara, CA, USA) was used to measure the mass spectra, in conjunction with MSD and 0.1% aqueous trifluoroacetic acid in an acetonitrile system on the C18-BDS column. Elemental analysis was conducted using a FLASH EA 1112 CHN analyzer (Thermo Finnigan LLC, New York, NY, USA).
2.2. General Procedure for the Synthesis of Methyl 3-(Substituted Benzoyl)-7-Substituted-2-Phenylindolizine-1-Carboxylate Analogues (4a–g)
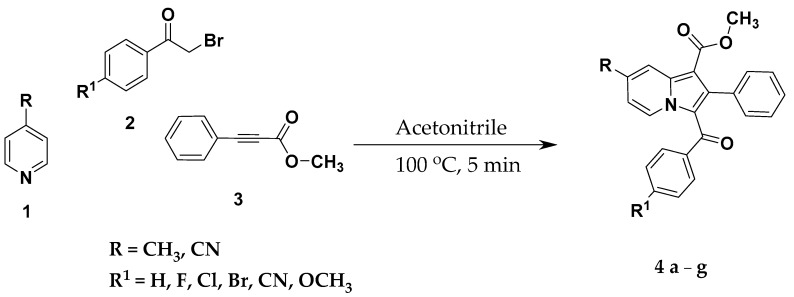
As indicated in Scheme 1, a mixture of 4-methyl/cyano pyridine (1.00 mmol), 4-substituedphenacyl bromide (1.00 mmol), and methyl phenylpropiolate (1.00 mmol) was added to 4 mL acetonitrile and triethylamine (1.00 mmol) in 10 mL of microwave tube under nitrogen atmosphere. The reaction mixture was irradiated at 100 °C in a microwave initiator up to 5 min. The reaction completion was monitored on TLC. After reaction completion, the reaction medium was evaporated under reduced pressure, the crude product obtained was diluted with water, the aqueous layer was extracted twice with ethyl acetate and the organic layer was washed with brine solution. The organic layer was evaporated under reduced pressure, and the product obtained was purified by column chromatography using 60–120 mesh silica gel with hexane and ethyl acetate solvent system to afford 76%–89% yield of methyl 3-(substituted benzoyl)-7-substituted-2-phenylindolizine-1-carboxylates.
Scheme 1.
Synthesis of methyl 3-(substituted benzoyl)-7-substituted-2-phenylindolizine-1-carboxylate analogues 4a–g. FT-IR and 1H-NMR of 4a–g see Supplementary Materials.
Methyl 3-Benzoyl-7-Methyl-2-Phenylindolizine-1-Carboxylate (4a). Appearance: yellow crystalline compound, Yield: 87%, melting point: 173–174 °C, FT-IR (KBr, cm−1): 2945.10, 1708.81 (C=O), 1591.16 (C=O), 1444.58, 1413.72, 1217.00, 1168.78, 1080.06, 821.62, 703.97, 563.18; 1H-NMR (400 MHz) δ = 9.56 (d, J = 7.2 Hz, 1H), 8.25 (s, 1H), 7.36–7.34 (d, J = 7.6 Hz, 2H), 7.16–7.09 (m, 3H), 7.03–7.00 (m, 5H), 6.92–6.90 (m, 1H), 3.70 (s, 3H), 2.53 (s, 3H); 13C-NMR (100 MHz) δ = 188.13, 164.95, 140.77, 139.68, 139.49, 138.87, 133.94, 131.44, 131.15, 130.82, 129.10, 127.65, 127.35, 127.08, 126.75, 122.08, 118.44, 117.39, 103.42, 50.77, 21.64; LC-MS (ESI, Positive): m/z: [M + H]+: 370; Anal. calculated for: C24H19NO3; C, 78.03; H, 5.18; N, 3.79; Found: C, 78.07; H, 5.19; N, 3.89.
Methyl 3-(4-Fluorobenzoyl)-7-Methyl-2-Phenylindolizine-1-Carboxylate (4b). Appearance: yellow crystalline compound, Yield: 85%, melting point: 167–168 °C, FT-IR (KBr, cm−1): 2972.10, 1685.67 (C=O), 1602.74 (C=O), 1379.01, 1217.00, 1118.64, 867.91, 783.05, 605.61, 522.67; 1H-NMR (400 MHz) δ = 9.53 (d, J = 7.2 Hz, 1H), 8.25 (s, 1H), 7.37–7.34 (m, 2H), 7.10–7.02 (m, 5H), 6.92–6.91 (d, J = 7.2Hz, 1H), 6.70–6.66 (t, J = 8.4Hz, 2H), 3.68 (s, 3H), 2.55 (s, 3H); 13C-NMR (100 MHz) δ = 186.54, 165.33, 164.88, 162.83, 140.71, 139.76, 139.03, 135.72, 135.69, 133.86, 131.52, 131.34, 131.16, 127.61, 127.25, 126.89, 121.87, 118.49, 117.48, 114.53, 114.31, 103.48, 50.81, 21.64; LC-MS (ESI, Positive): m/z: [M + H]+: 388; Anal. calculated for: C24H18FNO3; C, 74.41; H, 4.68; N, 3.62; Found: C, 74.43; H, 4.67; N, 3.63.
Methyl 3-(4-Chlorobenzoyl)-7-Methyl-2-Phenylindolizine-1-Carboxylate (4c). Appearance: yellow crystalline compound, Yield: 89%, melting point: 154–155 °C, FT-IR (KBr, cm−1): 2945.10, 1708.81 (C=O), 1593.09 (C=O), 1415.65, 1290.29, 1217.00, 918.05, 821.62, 707.83, 563.18; 1H-NMR (400 MHz) δ = 9.58 (d, J = 7.2 Hz, 1H), 8.26 (s, 1H), 7.26–7.24 (d, J = 8.0 Hz, 2H), 7.10–7.02 (m, 5H), 6.98–6.92 (m, 3H), 3.70 (s, 3H), 2.54 (s, 3H); 13C-NMR (100 MHz) δ = 186.63, 164.83, 141.02, 139.84, 139.24, 137.94, 136.78, 133.80, 131.13, 130.36, 127.71, 127.57, 127.20, 126.93, 121.83, 118.51, 117.59, 103.65, 50.82, 21.65; LC-MS (ESI, Positive): m/z: ([M + H]+: 404; Anal. calculated for: C24H18ClNO3; C, 71.38; H, 4.49; N, 3.47; Found: C, 71.33; H, 4.45; N, 3.52.
Methyl 3-(4-Bromobenzoyl)-7-Methyl-2-Phenylindolizine-1-Carboxylate (4d). Appearance: brown crystalline compound, Yield: 85%, melting point: 137–138 °C, FT-IR (KBr, cm−1): 2975.96, 1689.53 (C=O), 1614.31 (C=O), 1421.44, 1217.00, 1178.43, 1108.99, 798.47, 757.97, 636.47, 595.96; 1H-NMR (400 MHz) δ = 9.59 (d, J = 7.2 Hz, 1H), 8.25 (s, 1H), 7.18–7.16 (m, 2H), 7.13–7.02 (m, 7H), 6.94–6.92 (d, J = 7.2 Hz, 1H), 3.69 (s, 3H), 2.53 (s, 3H); 13C-NMR (100 MHz) δ = 186.72, 164.82, 141.10, 139.85, 139.28, 138.39, 133.78, 131.12, 130.53, 130.46, 127.74, 127.17, 126.95, 125.33, 121.80, 118.50, 117.62, 103.70, 50.82, 21.66; LC-MS (ESI, Positive): m/z: [M + H] +: 448; Anal. calculated for: C24H18BrNO3; C, 64.30; H, 4.05; N, 3.12; Found: C, 64.35; H, 4.06; N, 3.06.
Methyl 7-Cyano-3-(4-Cyanobenzoyl)-2-Phenylindolizine-1-Carboxylate (4e). Appearance: yellow amorphous compound, Yield: 76%, melting point: 235–236 °C, FT-IR (KBr, cm−1): 2950, 2223 (CN), 1718 (C=O), 1627 (C=O), 1375, 1226.64, 1080.06, 925.77, 702.04, 549.67; 1H-NMR (400 MHz) δ = 9.55 (d J = 7.2 Hz, 1H), 8.02 (s, 1H), 7.86–7.84 (d, J = 6.54 Hz, 2H), 7.79–7.77 (d, J = 6.54 Hz, 2H), 7.53–7.31 (m, 5H), 7.10–7.05 (m, 1H), 3.76 (s, 3H); 13C-NMR (400 MHz) δ = 184.77, 164.30, 143.63, 133.21, 132.24, 131.35, 131.09, 130.49, 129.89, 129.28, 128.99, 128.60, 127.83, 127.38, 125.50, 121.83, 121.70, 117.83, 115.51, 114.96,107.36, 52.05; LC-MS (ESI, Positive): m/z: [M + H] +: 406; Anal calculated for: C25H15N3O3: C, 74.07, H, 3.73, N, 10.36: Found: C, 73.99, H, 3.68, N, 10.42.
Methyl 3-(4-Bromobenzoyl)-7-Cyano-2-Phenylindolizine-1-Carboxylate (4f). Appearance: brown crystalline compound, Yield: 84%, melting point: 208–209 °C, FT-IR (KBr, cm−1): 2950.89, 2225.70 (CN), 1706.88 (C=O), 1608.52 (C=O), 1585.36, 1514.02, 1440.51, 1419.51, 1217.00, 1143.71, 1072.35, 918.05, 705.90, 543.89; 1H-NMR (400 MHz) δ = 9.49–9.48 (m, 1H), 8.84 (s, 1H), 7.44–7.42 (d, J = 7.2 Hz, 2H), 7.23–7.13 (m, 6H), 7.12–7.08 (m, 4H), 3.77 (s, 3H); 13C-NMR (400 MHz) δ = 187.29, 163.70, 140.61, 137.01, 136.29, 132.33, 131.81, 131.10, 130.90, 129.99, 128.42, 128.14, 127.93, 127.32, 126.79, 119.90, 117.35, 115.13, 114.42, 109.35, 107.39, 51.46; LC-MS (ESI, Positive): m/z: [M + H] +: 459.2; Anal calculated for: C24H15BrN2O3: C, 62.76, H, 3.29, N, 6.10: Found: C, 62.75, H, 3.26, N, 6.12.
Methyl 7-Cyano-3-(4-Methoxybenzoyl)-2-Phenylindolizine-1-Carboxylate (4g). Appearance: yellow crystalline compound, Yield: 81%, melting point: 166–167 °C, FT-IR (KBr, cm−1): 2950.89, 2227.63, 1706.88 (C=O), 1600.81 (C=O), 1573.81, 1514.02, 1369.37, 1220.86, 1141.78, 1027.99, 802.33, 709.76, 615.25; 1H-NMR (400 MHz) δ = 9.21–9.20 (m, 1H), 8.81 (s, 1H), 7.44–7.42 (d, J = 7.2 Hz, 2H), 7.17–7.15 (m, 3H), 7.10–7.05 (m, 3H), 6.57–6.55 (d, J = 7.2 Hz, 2H), 3.78 (s, 3H), 3.72 (s, 3H); 13C-NMR (400 MHz) δ = 186.95, 163.92, 162.95, 139.14, 135.72, 132.61, 131.88, 131.14, 130.50, 130.13, 128.68, 127.75, 127.25, 126.37, 124.44, 117.63, 113.88, 113.77, 113.13, 108.34, 106.86, 55.56, 51.96; LC-MS (ESI, Positive): m/z: [M + H] +: 411; Anal calculated for: C25H18N2O4: C, 73.16, H, 4.42, N, 6.83: Found: C, 73.08, H, 4.38, N, 6.90.
2.3. Crystallography
Single crystal of compound 4a was obtained from mixture of acetone and ethyl alcohol solvent at 1:1 ratio by slow evaporation method at low temperature as block shape and brown in color. Single-crystal X-ray diffraction data were collected on a Bruker KAPPA APEX II DUO diffractometer using graphite-monochromated Mo-Kα radiation (χ = 0.71073 Å). Data collection was carried out at 173(2) K. Temperature was controlled by an Oxford Cryostream cooling system (Oxford Cryostat). Cell refinement and data reduction were performed using the program SAINT [16]. The data were scaled and absorption correction performed using SADABS [17]. The structure was solved by direct methods using SHELXS-97 [17] and refined by full-matrix least-squares methods based on F2 using SHELXL-2014 [17] and using the graphics interface program X-Seed [18,19]. Both the programs X-Seed and POV-Ray [20] were used to prepare molecular graphic images. All non-hydrogen atoms were refined anisotropically. All hydrogen atoms were placed in idealized positions and refined in riding models with Uiso assigned 1.2 or 1.5 times Ueq of their parent atoms and the C–H bond distances were constrained in the range from 0.95 Å to 0.99 Å. The structure was refined to R factor of 0.0462.
2.4. Pharmacology
2.4.1. COX-2 Inhibition Assay
In vitro, a COX-2 inhibition study of the designed methyl 3-(substituted benzoyl)-7-substituted-2-phenylindolizine-1-carboxylate analogues (4a–g) was conducted; they were screened for in vitro human recombinant COX-2 enzyme inhibitory activity as described previously [14].
2.4.2. Statistical Analysis
The one-way investigation of variance (ANOVA) was used to compare the in vitro COX 2 inhibitory activity of the designed the methyl 3-(substituted benzoyl)-7-substituted-2-phenylindolizine-1-carboxylate analogues (4a–g) with nonselective (indomethacin) and selective (celecoxib) standard substances. The steps were followed as we described in our previous communication [14].
2.5. Computational Studies
2.5.1. Molecular Docking
Computational modeling studies were conducted using Accelry’s Discovery Studio 4.0 client program, in which the algorithm used was CHARMm force fields. The X-ray co-crystal of COX-2 enzyme and indomethacin (PDB: 4COX) was used to estimate the binding mode of indolizines. The molecular interaction of indolizines and the enzyme was determined by following steps from our recently reported docking protocol [14].
2.5.2. ADME Prediction
In this study, the pharmacokinetic properties of all the synthesized compounds were predicted and analyzed. The SwissADME webserver was used for calculating and predicting various parameters related to the ADME properties of the compounds [21]. In particular, parameters related to the oral absorption property such as the number of rotatable bonds, the octanol/water partition coefficient (LogP), and the molecular weight were calculated. The ability of the compounds to cross the blood–brain barrier (BBB) was predicted as compounds entering the central nervous system (CNS) might have undesirable adverse effects that need proper considerations [22]. In addition, the probability of binding to the p-glycoprotein (P-gp) was predicted. Since this protein is known to act as an efflux pump to many xenobiotics, compounds that are substrate to this protein might be prone to resistance, hence structural modifications might be required to prevent binding to P-gp [23,24].
2.5.3. Toxicity Prediction
In this study, the SwissADME web server and the DataWarrior software were used to predict several toxic effects of the synthesized compounds [21,25]. The potential inhibition of some important cytochrome P450 (CYP 450) isoforms by the compounds was predicted. Inhibition of CYP 450 isoforms needs to be taken into consideration since it may alter the metabolism of the other drugs causing drug–drug interactions [26]. In addition, to further characterize the toxicity profiles of the synthesized compounds, the mutagenicity, tumorigenicity, and reproductive toxicity were predicted for all the compounds.
3. Results and Discussion
3.1. Chemistry
The synthetic route for the construction of target compounds, methyl 3-(substituted benzoyl)-7-substituted-2-phenylindolizine-1-carboxylate analogues (4a–g) is illustrated in Scheme 1. The treatment of 4-substituted pyridine, 4-substituedphenacyl bromide, and methyl phenylpropiolate in presence of triethylamine and acetonitrile yielded the title compounds 4a–g at 76%–89% yield by one-pot microwave-assisted synthetic approach. The purity of the compounds was confirmed by HPLC, and it was found to be over 99%. The physicochemical characteristics of methyl 3-(substituted benzoyl)-7-substituted-2-phenylindolizine-1-carboxylate analogues 4a–g are tabulated in Table 1. The chemical structure of the products 4a–g was elucidated by FT-IR, NMR (1H and 13C), mass spectrometry, and elemental analysis. The FT-IR spectra of the synthesized compounds exhibited stretching absorption band in the range of 1685–1718 cm−1 for carbonyl group (C=O). The characteristic cyano absorption peak for compounds 4e–g is found in the range of 2223–2227 cm−1. For compounds 4d and 4f carbon–bromine stretching vibration is observed in the range of 626.82–636.47 cm−1. For compounds 4c and 4b, carbon–chlorine and carbon–fluorine stretching vibrations are observed at 707.85 and 1217.00 cm−1, respectively. 1H-NMR spectra revealed the appearance of a singlet for a methyl group at the seventh position of the indolizine nucleus (4a–d) and a singlet for an ester methyl group at the first position of the indolizine nucleus (4e–g) are observed in the ranges of 3.68–3.78 and 2.53–2.55 ppm, respectively. In case of para-methoxy derivative 4g, a singlet for methoxy is observed at 3.72 ppm. The doublet peaks were commonly seen in all the derivatives 2a–g with J value at 7.12 Hz as para substituents such as fluoro, chloro, bromo, methoxy, and cyano on phenyl ring at the third position of the indolizine nucleus. In case of title compound 4b, the triplet peak was observed in the range of 6.70–6.66 ppm with J value at 8.4 Hz. The 13C-NMR spectra revealed the appearance of carbonyl carbon for compounds 4a–g in the range of 184.77–187.29 ppm. The title compounds 4a–g revealed ester alkyl carbon, which is at first position of the indolizine nucleus is found in the range of 50.77–52.05 ppm. The title compounds 4e–d exhibited a peak for a methyl group, which is at seventh position of the indolizine nucleus in the range of 21.64–21.66 ppm. The title compound 4g revealed a peak at 55.56 ppm due to the methoxy group at para position of benzoyl group, which is at third position of the indolizine nucleus. In LC-MS spectra of the compounds 4a–g the molecular ion peaks were in good agreement with molecular mass of the compounds. The results of elemental analysis were in good agreement with the calculated values of the proposed tile compounds 4a–g. The cLogP of the proposed tile compounds 4a–g was calculated using ChemDraw Professional 16.0 (PerkinElmer Informatics, Inc. 940 Winter Street Waltham, MA, USA) and the results were in the range of 4.6463–7.0802.
Table 1.
Physicochemical characteristics of methyl 3-(substituted benzoyl)-7-substituted-2-phenylindolizine-1-carboxylate analogues 4a–g.
| Compound Code | Mol Formula (Mol Weight) | R | R1 | Yield (%) a | M.P. (°C) | CLogP b |
|---|---|---|---|---|---|---|
| 4a | C24H19NO3 (369) | CH3 | H | 87 | 173–174 | 6.2094 |
| 4b | C24H18FNO3 (387) | CH3 | F | 85 | 167–168 | 6.3602 |
| 4c | C24H18ClNO3 (403) | CH3 | Cl | 89 | 154–155 | 6.9302 |
| 4d | C24H18BrNO3 (447) | CH3 | Br | 85 | 137–138 | 7.0802 |
| 4e | C25H15N3O3 (405) | CN | CN | 76 | 235–236 | 4.6463 |
| 4f | C24H15BrN2O3 (458) | CN | Br | 84 | 208–209 | 6.0300 |
| 4g | C25H18N2O4 (410) | CN | OCH3 | 81 | 166–167 | 5.3301 |
a Yields calculated after being purified using the recrystallization method; ethanol was used as the solvent. b cLogP of the compounds was calculated using ChemDraw Professional 16.
3.2. Crystallography
The parameters for crystal data collection and structure refinements, the bond lengths, angles, and torsion angles are contained in CIF file which was deposited in the Cambridge Crystallographic Data Centre (CCDC) [27] with CCDC number 1950516. The crystallographic details are listed in Table 2. Intermolecular interactions, thermal ellipsoid diagram, and packing diagrams were generated using CSD software ORTEP [28] and Mercury 3.8 [29].
Table 2.
Single crystal data collection and refinement for title compounds 4a.
| DATA | Compound 4a |
|---|---|
| Formula | C24H19 N O3 |
| Formula weight | 369.40 |
| Temperature/K | 153 (2) |
| Wavelength (Å) | 0.71073 |
| Crystal system | monoclinic |
| Space group | C 2/c |
| a (Å) | 16.550 (4) |
| b (Å) | 10.391 (2) |
| c (Å) | 22.179 (4) |
| α (°) | 90 |
| β (°) | 104.468 (4) |
| γ (°) | 90 |
| V (Å3) | 3693.1 (13) |
| Z’, Z | 1, 8 |
| Density (g cm−1) | 1.329 |
| μ (mm−1) | 0.088 |
| F (000) | 1552 |
| θ (min, max) | 2.336, 25.413 |
| hmin, max; kmin, max; lmin, max. | −19, 19; −12, 12; −26, 26 |
| No. of refl. | 3375 |
| No of unique ref./Obs. ref. | 3375, 2340 |
| No. parameters | 255 |
| Rall, Robs | 0.0462, 0.0815 |
| wRall, wRobs | 0.113, 0.099 |
| Δρmin, max (eÅ−3) | −0.272, 0.195 |
| G.O.F. | 1.020 |
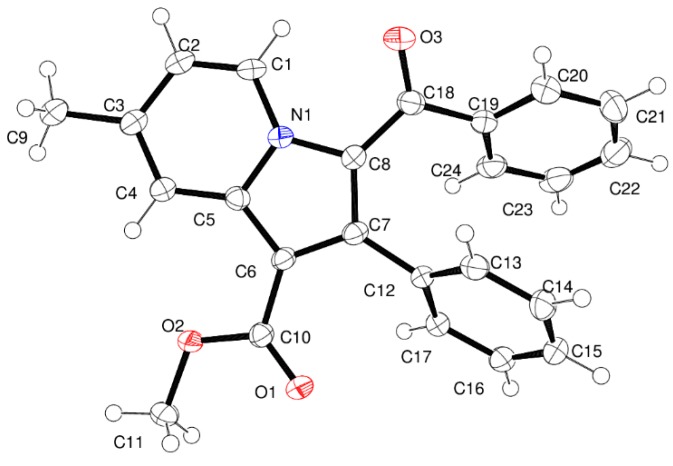
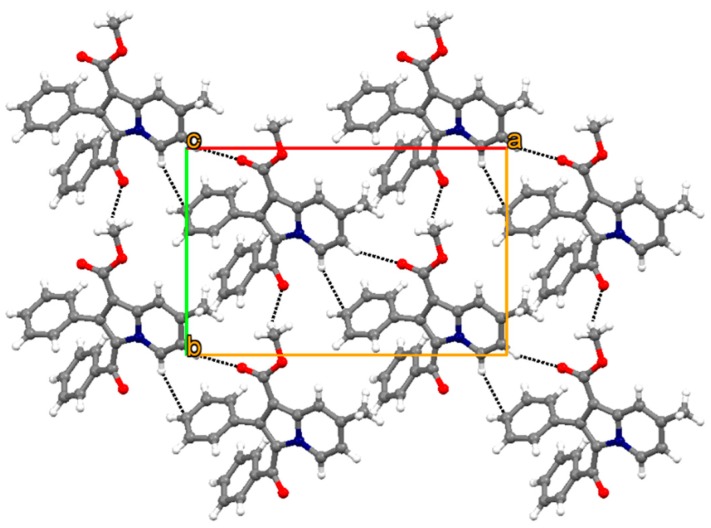
Figure 2 shows the thermal ellipsoid plot of 4a with atom labelling. Crystal structure of 4a forms molecular sheet assembly through weak C–H∙∙∙O and C–H∙∙∙π hydrogen bonds (Table 3 and Figure 3). The remaining molecules of this series of phenylindolizine are expected to have the same molecular assembly as their conformations have the same molecular core moiety and functional groups.
Figure 2.
The asymmetric unit of compound 4a with 50% probability thermal ellipsoids.
Table 3.
Intermolecular interactions of compounds 4a.
| D–X···A | D–X (Å) | X···A (Å) | D···A (Å) | <D–X···A (Å) | Symmetry Code |
|---|---|---|---|---|---|
| C2–H2···O1 | 0.95 | 2.56 | 3.450 (3) | 156 | 1/2 + x, 1/2 + y, z |
| C11–H11A···O3 | 0.98 | 2.53 | 3.383 (3) | 146 | x, −1 + y, z |
| C17–H17···Cg | 0.98 | 2.83 | 3.678 | 149 | 1/2 − x, −1/2 + y, 1/2 − z |
Cg = the centroid of six-membered ring C19/C20/C21/C22/C23/C24.
Figure 3.
Molecular assembly of 4a shown along [1 1 0] plane through weak C–H···O and C–H···π hydrogen bondings.
3.3. Pharmacology
COX-2 Inhibition
A series of methyl 3-(substituted benzoyl)-7-substituted-2-phenylindolizine-1-carboxylate analogues 4a–g were evaluated for in-vitro COX-2 inhibition activity and it was found that all the compounds displayed inhibitory effects at IC50 values between 6.71–41.59 (Table 4). From the series, compound 4e emerged as the most promising COX-2 inhibitor with a cLogP value of 4.6463 and having two cyano groups on indolizine and benzoyl moiety with IC50 of 6.71. However, compound 4g exhibited moderate inhibitory activity at IC50 9.62, having a cyano group on the indolizine ring and methoxy at the para position of the benzoyl ring, when compared to indomethacin standard. In the case of compounds 4a, 4b, 4c, and 4d, methyl functional group on the indolizine nucleus did not favor promising COX-2 inhibitory activity. The COX-2 inhibitory activity of the test compounds (4a–4d) having a methyl group at seventh position of the indolizine nucleus was in the order 4b > 4d > 4c > 4a. The COX-2 inhibitory activity of the compounds (4e–g) having a cyano group at seventh position of the indolizine nucleus was in the order 4e > 4g > 4f.
Table 4.
In vitro COX-2 inhibitory activity and docking results of methyl 3-(substituted benzoyl)-7-substituted-2-phenylindolizine-1-carboxylate scaffolds 4a–g.
| Entry | R1 | R2 | IC50 (μM)a | CDocker E. (kcal/mol) | Hydrogen Bonding (Interacting Atom, Å) | Pi Interaction |
|---|---|---|---|---|---|---|
| 4a | CH3 | H | 41.59 ± 0.03 a | −31 | ARG 120 (pi–cation) | |
| 4b | CH3 | F | 27.08 ± 0.03 c,d | −34 | HIS 90 (F, 2.09) | ARG 120 (pi–cation) |
| 4c | CH3 | Cl | 38.11 ± 0.03 b,d | −33 | ARG 120 (pi–cation) | |
| 4d | CH3 | Br | 37.66 ± 0.03 d | −32 | ARG 120 (pi–cation) | |
| 4e | CN | CN | 6.71 ± 0.03 b | −35 | HIS 90 (CN, 2.02) | |
| 4f | CN | Br | 13.55 ± 0.03 b,d | −34 | HIS 90 (CN, 2.01) | |
| 4g | CN | OCH3 | 9.62 ± 0.03 c | −35 | HIS 90 (CN, 2.07) | |
| IND | 6.84 ± 0.03 b,c | −49 | ARG 120 (anion–cation) | |||
| CLB | 0.05 ± 0.03 b | −43 | HIS 90 (SO2, 3.05) PHE 158 (SO2, 2.92) GLN 192 (NH2, 2.69) LEU 352 (NH2, 1.95) ARG120 (CF3, 2.38) |
ARG 120 (pi–cation) PHE 158 (pi–sulfur) ALA 527 (pi–amide) |
a IC50 value is the compound concentration required to produce 50% inhibition of COX-2 enzyme, expressed as means of three experimental determinations. IND: indomethacin; CLB: celecoxib. a–d Test compounds not sharing a letter differ significantly (p < 0.05).
3.4. Computational Studies
3.4.1. Molecular Modeling
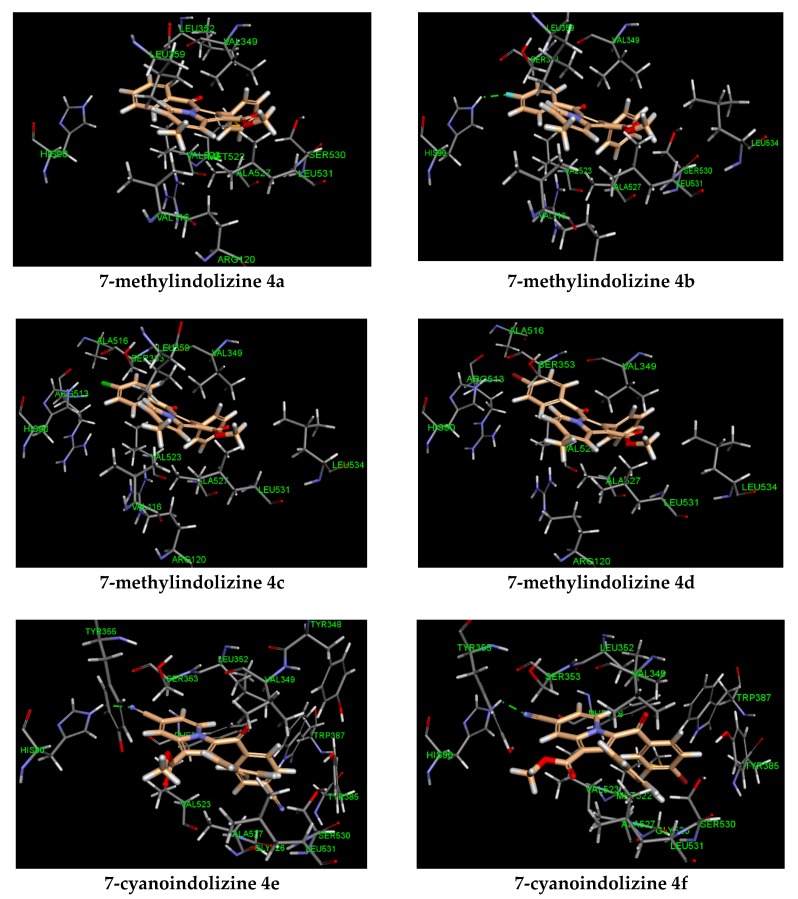
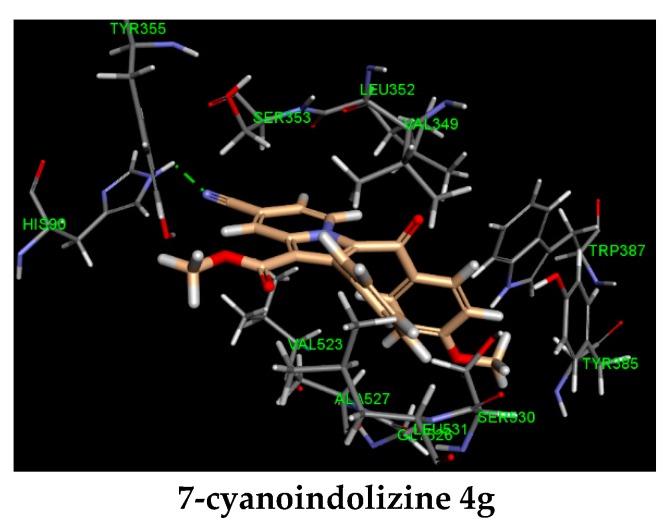
Two novel 2-phenylindolizine series, 7-methyl, and 7-cyano were investigated for COX-2 inhibitory activity. The bioactivity study revealed that 7-cyanoindolines were more active than their congener 7-methylindolizines. To gain insights into the potency of the synthesized indolizines, the key interactions between the compounds and COX-2 active site were examined through molecular docking study and reported in Table 4. Molecular modeling analysis showed two distinct binding modes for each series (Figure 4). The indolizino ring of 7-cyano series was pointed toward the residue HIS 90 participating in hydrogen bonding with cyano group at position 7 (compounds 4e–g, Figure 4), whereas the indolizino ring of 7-methyl series was projected toward the amino acid ARG 120 by interacting with the indolizino ring through pi–cation interaction (compounds 4b–d, Figure 4). The different conformation in binding site between the two series may explain the observed COX-2 activity. In the 7-methyl series, the hydrophobic interactions were mainly observed between the residues VAL 116, LEU 359, LEU 531 and methyl at position 7, and the amino acids HIS 90, ARG 513, ALA 516 and chlorine (4c) and bromine (4d) at the benzoyl ring with the exception of 3-(4-fluorobenzoyl) indolizine 4b (Figure 4). The latter exhibited strong hydrogen bonding with the residue HIS 90 and fluorine. The 7-cyanoindolizines 4e–g displayed strong H-bond with the residue HIS 90 and the cyano group at position 7 conferring greater inhibitory activity over the 7-methylindolizines 4a–d. Another interesting binding feature between the two series is the involvement of the amino acid ARG 120 in pi–cation interaction observed for only the 7-methylindolizine series. The lack of selectivity between COX-2 and COX-1 inhibitors is due to the ion pair or/and hydrogen bonding interaction with ARG 120 [30,31,32]. The non-involvement of such interaction with the 7-cyanoindolizines 4e–g may exert greater selectivity over 7-methylindolizines 4a–d.
Figure 4.
Predicted docking pose of indolizines 4b–g (salmon-filled spheres) in the COX-2 domain (PDB 4COX). Hydrogen bonding interaction is represented as a green dotted line.
3.4.2. ADME Prediction
The prediction of the pharmacokinetic properties of lead compounds at an early stage of the drug design and development process can greatly assist in the proper selection of compounds for further development [33]. In fact, a relatively high number of compounds fail to make it to the market because of pharmacokinetic related issues that appear in later stages of the drug design and development process [33,34]. The results of the pharmacokinetic parameters prediction are listed in Table 5. In general, most of the compounds were predicted to have favorable pharmacokinetic properties. In terms of oral absorption, all the compounds fulfill the conditions of Lipinski’s rule of five criteria. The number of rotatable bonds for all the compounds is less than seven rotatable bonds, indicating a high probability of being orally bioavailable [35]. Also, The GI absorption was predicted to be high for all the compounds. Compounds 4a, 4b, 4c, and 4d were predicted to be BBB permeant, which means they have the ability to cross the BBB and enter the CNS while the remaining compounds were predicted to lack the ability to cross the BBB. All the compounds were predicted to have low probability of being P-gp substrates, which means there is low possibility of developing resistance by P-gp efflux.
Table 5.
The predicted pharmacokinetic properties of all the synthesized compounds.
| Compound ID | 4a | 4b | 4c | 4d | 4e | 4f | 4g |
|---|---|---|---|---|---|---|---|
| Rotatable bonds | 5 | 5 | 5 | 5 | 5 | 5 | 6 |
| Molecular weight | 369.41 | 387.4 | 403.86 | 448.31 | 405.4 | 459.29 | 410.42 |
| H-bond acceptors | 3 | 4 | 3 | 3 | 5 | 4 | 5 |
| H-bond donors | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| MLOGP | 3.43 | 3.8 | 3.91 | 4.01 | 1.86 | 3.11 | 2.2 |
| GI absorption | High | High | High | High | High | High | High |
| BBB permeant | Yes | Yes | Yes | Yes | No | No | No |
| P-gp substrate | No | No | No | No | No | No | No |
| Lipinski violations | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
3.4.3. Toxicity Prediction
The prediction of potential toxic effects of the compounds at an early stage of the drug design and development process is important in a similar manner to ADME prediction. The results of toxicity prediction are shown in Table 6. In general, the compounds were predicted to have a good safety profile. In terms of mutagenicity, tumorigenicity, and reproductive toxicity, all the compounds were predicted to be devoid of such major toxic effects. Also, none of the compounds was predicted to have irritant effects. In terms of CYP 450 isoforms inhibition, all the compounds except compound 4f were predicted to be potential inhibitors of the CYP1A2 isoform. On the other hand, none of the compounds was predicted to be a potential inhibitor of the CYP2D6 isoform. Compounds 4e and 4g were predicted to be CYP3A4 inhibitors. Thus, the compounds appear to have the ability to inhibit the CYP1A2 isoform, which requires consideration during further development of the compounds to avoid any possible drug–drug interactions resulting from CYP1A2 inhibition [26].
Table 6.
The predicted toxic effects of all the synthesized compounds.
| Compound ID | 4a | 4b | 4c | 4d | 4e | 4f | 4g |
|---|---|---|---|---|---|---|---|
| Mutagenic | none | none | none | none | none | none | none |
| Tumorigenic | none | none | none | none | none | none | none |
| Reproductive Effective | none | none | none | none | none | none | none |
| Irritant | none | none | none | none | none | none | none |
| CYP1A2 inhibitor | Yes | Yes | Yes | Yes | Yes | No | Yes |
| CYP2D6 inhibitor | No | No | No | No | No | No | No |
| CYP3A4 inhibitor | No | No | No | No | Yes | Yes | No |
4. Conclusions
In this study, a series of a novel series of methyl 3-(substituted benzoyl)-7-substituted-2-phenylindolizine-1-carboxylates was synthesized and assessed for COX-2 inhibition activity. All the compounds exhibited moderate to good activity as COX-2 inhibitors. The most active compound was compound 4e, which has an IC50 of 6.71 μM, similar to the IC50 of the marketed NSAID indomethacin. Molecular docking was conducted to understand the binding interactions taking place between the compounds and the amino acid residues in the active site of the COX-2 enzyme. The ADME properties, as well as the potential toxic effects of the synthesized compounds, were predicted. The results indicated a high probability for oral bioavailability for all the compounds. Also, none of the compounds was predicted to have any major toxic effects. Thus, these novel COX-2 inhibitors have the potential to be used as lead compounds for the development of improved COX-2 inhibitors.
Acknowledgments
The authors are grateful to the Deanship of Scientific Research, King Faisal University, Kingdom of Saudi Arabia for financial support and encouragement. The authors thank Dr. Hong Su, Centre for Supramolecular Chemistry Research, Department of Chemistry, University of Cape Town, Rondebosch 7701, for single-crystal X-ray data collection. S.K.N. thank DST-FIST [SR/FST/CSI-279/2016(c)] for characterization facility.
Supplementary Materials
The following are available online at https://www.mdpi.com/2218-273X/9/11/661/s1, Figure S1: FT-IR of methyl 3-benzoyl-7-methyl-2-phenylindolizine-1-carboxylate (4a), Figure S2: 1H-NMR of methyl 3-benzoyl-7-methyl-2-phenylindolizine-1-carboxylate (4a), Figure S3: 13C-NMR of methyl 3-benzoyl-7-methyl-2-phenylindolizine-1-carboxylate (4a), Figure S4: FT-IR of methyl 3-(4-fluorobenzoyl)-7-methyl-2-phenylindolizine-1-carboxylate (4b), Figure S5: 1H-NMR of methyl 3-(4-fluorobenzoyl)-7-methyl-2-phenylindolizine-1-carboxylate (4b), Figure S6: 13C-NMR of methyl 3-(4-fluorobenzoyl)-7-methyl-2-phenylindolizine-1-carboxylate (4b), Figure S7: FT-IR of methyl 3-(4-chlorobenzoyl)-7-methyl-2-phenylindolizine-1-carboxylate (4c), Figure S8: 1H-NMR of methyl 3-(4-chlorobenzoyl)-7-methyl-2-phenylindolizine-1-carboxylate (4c), Figure S9: 13C-NMR of methyl 3-(4-chlorobenzoyl)-7-methyl-2-phenylindolizine-1-carboxylate (4c), Figure S10: FT-IR of methyl 3-(4-bromobenzoyl)-7-methyl-2-phenylindolizine-1-carboxylate (4d), Figure S11: 1H-NMR of methyl 3-(4-bromobenzoyl)-7-methyl-2-phenylindolizine-1-carboxylate (4d), Figure S12: 13C-NMR of methyl 3-(4-bromobenzoyl)-7-methyl-2-phenylindolizine-1-carboxylate (4d), Figure S13: FT-IR of methyl 7-cyano-3-(4-cyanobenzoyl)-2-phenylindolizine-1-carboxylate (4e), Figure S14: 1H-NMR of methyl 7-cyano-3-(4-cyanobenzoyl)-2-phenylindolizine-1-carboxylate (4e), Figure S15: 13C-NMR of methyl 7-cyano-3-(4-cyanobenzoyl)-2-phenylindolizine-1-carboxylate (4e), Figure S16: FT-IR of methyl 3-(4-bromobenzoyl)-7-cyano-2-phenylindolizine-1-carboxylate (4f), Figure S17: 1H-NMR of methyl 3-(4-bromobenzoyl)-7-cyano-2-phenylindolizine-1-carboxylate (4f), Figure S18: 13C-NMR of methyl 3-(4-bromobenzoyl)-7-cyano-2-phenylindolizine-1-carboxylate (4f), Figure S19: FT-IR of methyl 7-cyano-3-(4-methoxybenzoyl)-2-phenylindolizine-1-carboxylate (4g), Figure S20: 1H-NMR of methyl 7-cyano-3-(4-methoxybenzoyl)-2-phenylindolizine-1-carboxylate (4g), Figure S21: 13C-NMR of methyl 7-cyano-3-(4-methoxybenzoyl)-2-phenylindolizine-1-carboxylate (4g).
Author Contributions
Conceptualization, K.N.V., C.T., S.K.N. and S.C.; methodology, K.N.V., O.H.A.A.-A., C.T., S.K.N., M.A.M., M.A., A.B.N., M.H., O.I.A. and S.C.; software, K.N.V., O.H.A.A.-A., C.T. and S.K.N.; validation, K.N.V., C.T., O.H.A.A.-A., S.K.N., M.A.M., M.H. and S.C.; formal analysis, K.N.V., O.H.A.A.-A., C.T., N.S. and S.C.; investigation, K.N.V., C.T., A.B.N. and S.C.; resources, K.N.V., O.H.A.A.-A., S.K.N., R.V., S.C., O.I.A. and B.O.; data curation, K.N.V., C.T. and S.C.; writing—original draft preparation, K.N.V., O.H.A.A.-A., S.K.N., M.A.M., M.A., N.S., R.V., M.H., M.B.G., O.I.A. and B.O.; writing—review and editing, K.N.V., C.T., O.H.A.A.-A., B.E.A., A.B.N., R.V. and S.C.; visualization, K.N.V. and S.K.N.; supervision, K.N.V., S.C., and B.O.; project administration, K.N.V.; funding acquisition, K.N.V., B.E.A., M.A., A.B.N., N.S.
Funding
This research was funded by the Deanship of Scientific Research at King Faisal University, Al-Ahsa, Saudi Arabia (Research Group grant number 17122011).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Lipscomb J. Management of Nonsteroidal Anti-inflammatory Drug-Induced Hypersensitivity Reactions. US Pharm. 2019;44:22–26. [Google Scholar]
- 2.Ricciotti E., Tang S.-Y., Barekat K., Veglia F., Maseda D., Bittinger K., Bushman F., FitzGerald G.A. The impact of cyclooxygenase-2 selective and non-isoform selective NSAIDs on the gut microbiota. FASEB J. 2019;33:516.511. [Google Scholar]
- 3.Moro M.G., Oliveira M.D.d.S., Oliveira L.R.d., Teixeira S.A., Muscará M.N., Spolidorio L.C., Holzhausen M. Effects of Selective Versus Non-Selective COX-2 Inhibition on Experimental Periodontitis. Braz. Dent. J. 2019;30:133–138. doi: 10.1590/0103-6440201902241. [DOI] [PubMed] [Google Scholar]
- 4.Takeuchi K., Amagase K. Roles of cyclooxygenase, prostaglandin E2 and EP receptors in mucosal protection and ulcer healing in the gastrointestinal tract. Curr. Pharm. Des. 2018;24:2002–2011. doi: 10.2174/1381612824666180629111227. [DOI] [PubMed] [Google Scholar]
- 5.Pannunzio A., Coluccia M. Cyclooxygenase-1 (COX-1) and COX-1 inhibitors in cancer: A review of oncology and medicinal chemistry literature. Pharmaceuticals. 2018;11:101. doi: 10.3390/ph11040101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mirshafiey A., Mortazavi-Jahromi S.S., Taeb M., Cuzzocrea S., Esposito E. Evaluation of the effect of α-L-guluronic acid (G2013) on COX-1, COX-2 activity and gene expression for introducing this drug as a novel NSAID with immunomodulatory property. Recent Pat. Inflamm. Allergy Drug Discov. 2018;12:162–168. doi: 10.2174/1872213X12666180607121809. [DOI] [PubMed] [Google Scholar]
- 7.Zarghi A., Zebardast T., Daraie B., Hedayati M. Design and synthesis of new 1, 3-benzthiazinan-4-one derivatives as selective cyclooxygenase (COX-2) inhibitors. Bioorg. Med. Chem. 2009;17:5369–5373. doi: 10.1016/j.bmc.2009.06.056. [DOI] [PubMed] [Google Scholar]
- 8.Omar Y.M., Abdu-Allah H.H., Abdel-Moty S.G. Synthesis, biological evaluation and docking study of 1, 3, 4-thiadiazole-thiazolidinone hybrids as anti-inflammatory agents with dual inhibition of COX-2 and 15-LOX. Bioorg. Chem. 2018;80:461–471. doi: 10.1016/j.bioorg.2018.06.036. [DOI] [PubMed] [Google Scholar]
- 9.Gubin J., de Vogelaer H., Inion H., Houben C., Lucchetti J., Mahaux J., Rosseels G., Peiren M., Clinet M. Novel heterocyclic analogs of the new potent class of calcium entry blockers: 1-[[4-(aminoalkoxy) phenyl] sulfonyl] indolizines. J. Med. Chem. 1993;36:1425–1433. doi: 10.1021/jm00062a015. [DOI] [PubMed] [Google Scholar]
- 10.Hagishita S., Yamada M., Shirahase K., Okada T., Murakami Y., Ito Y., Matsuura T., Wada M., Kato T., Ueno M. Potent inhibitors of secretory phospholipase A2: Synthesis and inhibitory activities of indolizine and indene derivatives. J. Med. Chem. 1996;39:3636–3658. doi: 10.1021/jm960395q. [DOI] [PubMed] [Google Scholar]
- 11.Chaniyara R., Tala S., Chen C.-W., Zang X., Kakadiya R., Lin L.-F., Chen C.-H., Chien S.-I., Chou T.-C., Tsai T.-H. Novel antitumor indolizino [6, 7-b] indoles with multiple modes of action: DNA cross-linking and topoisomerase I and II inhibition. J. Med. Chem. 2013;56:1544–1563. doi: 10.1021/jm301788a. [DOI] [PubMed] [Google Scholar]
- 12.Artico M., Massa S., Stefancich G., Silvestri R., Di Santo R., Corelli F. Potential antitumor agents. III. Synthesis of pyrazolo [3, 4-e] pyrrolo [3, 4-g] indolizine and 1H-pyrazolo [3, 4-e] indolizine derivatives. J. Heterocycl. Chem. 1989;26:503–507. doi: 10.1002/jhet.5570260246. [DOI] [Google Scholar]
- 13.Shrivastava S.K., Srivastava P., Bandresh R., Tripathi P.N., Tripathi A. Design, synthesis, and biological evaluation of some novel indolizine derivatives as dual cyclooxygenase and lipoxygenase inhibitor for anti-inflammatory activity. Bioorg. Med. Chem. 2017;25:4424–4432. doi: 10.1016/j.bmc.2017.06.027. [DOI] [PubMed] [Google Scholar]
- 14.Chandrashekharappa S., Venugopala K.N., Tratrat C., Mahomoodally F.M., Aldhubiab B.E., Haroun M., Venugopala R., Mohan M.K., Kulkarni R.S., Attimarad M.V. Efficient synthesis and characterization of novel indolizines: Exploration of in vitro COX-2 inhibitory activity and molecular modelling studies. New J. Chem. 2018;42:4893–4901. doi: 10.1039/C7NJ05010K. [DOI] [Google Scholar]
- 15.Sandeep C., Venugopala K., Khedr M., Padmashali B., Kulkarni R., Rashmi V., Odhav B. Design and synthesis of novel indolizine analogues as COX-2 inhibitors: Computational perspective and in vitro screening. Indian J. Pharm. Educ. Res. 2017;51:452–460. doi: 10.5530/ijper.51.3.73. [DOI] [Google Scholar]
- 16.SAINT Version 7.60a. Bruker AXS Inc.; Madison, WI, USA: 2006. [Google Scholar]
- 17.Sheldrick G.M. SHELXS-97, SHELXL-2014 and SADABS Version 2.05. University of Göttingen; Göttingen, Germany: 1997. [Google Scholar]
- 18.Barbour L.J. X-Seed—A Software Tool for Supramolecular Crystallography. Elsevier; Amsterdam, The Netherlands: 2001. [Google Scholar]
- 19.Jerry L.A., Leonard J.B. Molecular Graphics: From Science to Art. Cryst. Growth Des. 2003;3:3–8. [Google Scholar]
- 20.Barbour L. X-Seed, Graphical Interface to SHELX-97 and POV-Ray. University of Missouri–Columbia; Columbia, MO, USA: 1999. [Google Scholar]
- 21.Daina A., Michielin O., Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollastri M.P. Overview on the Rule of Five. Curr. Protoc. Pharmacol. 2010;49:9.12.11–19.12.18. doi: 10.1002/0471141755.ph0912s49. [DOI] [PubMed] [Google Scholar]
- 23.Yu D.K. The contribution of P-glycoprotein to pharmacokinetic drug-drug interactions. J. Clin. Pharmacol. 1999;39:1203–1211. doi: 10.1177/00912709922012006. [DOI] [PubMed] [Google Scholar]
- 24.Fromm M. Importance of P-glycoprotein for drug disposition in humans. Eur. J. Clin. Invest. 2003;33:6–9. doi: 10.1046/j.1365-2362.33.s2.4.x. [DOI] [PubMed] [Google Scholar]
- 25.Sander T., Freyss J., von Korff M., Rufener C. DataWarrior: An open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model. 2015;55:460–473. doi: 10.1021/ci500588j. [DOI] [PubMed] [Google Scholar]
- 26.Lin J.H. CYP induction-mediated drug interactions: In vitro assessment and clinical implications. Pharm. Res. 2006;23:1089–1116. doi: 10.1007/s11095-006-0277-7. [DOI] [PubMed] [Google Scholar]
- 27. [(accessed on 20 August 2019)]; Available online: https://www.Ccdc.Cam.Ac.Uk.
- 28.Farrugia L. WinGX suite for small-molecule single-crystal crystallography. J. Appl. Crystallogr. 1999;32:837–838. doi: 10.1107/S0021889899006020. [DOI] [Google Scholar]
- 29.Macrae C.F., Bruno I.J., Chisholm J.A., Edgington P.R., McCabe P., Pidcock E., Rodriguez-Monge L., Taylor R., van de Streek J., Wood P.A. Mercury CSD 2.0—New features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008;41:466–470. doi: 10.1107/S0021889807067908. [DOI] [Google Scholar]
- 30.Kalgutkar A.S., Crews B.C., Rowlinson S.W., Marnett A.B., Kozak K.R., Remmel R.P., Marnett L.J. Biochemically based design of cyclooxygenase-2 (COX-2) inhibitors: Facile conversion of nonsteroidal antiinflammatory drugs to potent and highly selective COX-2 inhibitors. Proc. Natl. Acad. Sci. USA. 2000;97:925–930. doi: 10.1073/pnas.97.2.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhattacharyya D.K., Lecomte M., Rieke C.J., Garavito R.M., Smith W.L. Involvement of arginine 120, glutamate 524, and tyrosine 355 in the binding of arachidonate and 2-phenylpropionic acid inhibitors to the cyclooxygenase active site of ovine prostaglandin endoperoxide H synthase-1. J. Biol. Chem. 1996;271:2179–2184. doi: 10.1074/jbc.271.4.2179. [DOI] [PubMed] [Google Scholar]
- 32.Greig G.M., Francis D.A., Falgueyret J.-P., Ouellet M., Percival M.D., Roy P., Bayly C., Mancini J.A., O’Neill G.P. The interaction of arginine 106 of human prostaglandin G/H synthase-2 with inhibitors is not a universal component of inhibition mediated by nonsteroidal anti-inflammatory drugs. Mol. Pharmacol. 1997;52:829–838. doi: 10.1124/mol.52.5.829. [DOI] [PubMed] [Google Scholar]
- 33.Lombardo F., Gifford E., Shalaeva M.Y. In silico ADME prediction: Data, models, facts and myths. Mini Rev. Med. Chem. 2003;3:861–875. doi: 10.2174/1389557033487629. [DOI] [PubMed] [Google Scholar]
- 34.Smith P., Sorich M., Low L., McKinnon R., Miners J. Towards integrated ADME prediction: Past, present and future directions for modelling metabolism by UDP-glucuronosyltransferases. J. Mol. Graph. Model. 2004;22:507–517. doi: 10.1016/j.jmgm.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 35.Veber D.F., Johnson S.R., Cheng H.-Y., Smith B.R., Ward K.W., Kopple K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.