Abstract
The emergence of carbapenem-resistant and colistin-resistant Enterobacteriaceae represents a great risk for public health. In this study, the phenotypical and genetic characteristics of eight carbapenem-resistant and colistin-resistant isolates from pig farms in China were determined by the broth microdilution method and whole genome sequencing. Antimicrobial susceptibility testing showed that the eight carbapenem-resistant and colistin-resistant strains were resistant to three aminoglycosides, twelve β-lactams, one of the phenicols, one of the tetracyclines, and one of the fluoroquinolones tested, simultaneously. The prediction of acquired resistant genes using the whole genome sequences revealed the co-existence of blaNDM-1 and mcr-1 as well as the other genes that were responsible for the multidrug-resistant phenotypes. Bioinformatics analysis also showed that the carbapenem-resistant gene blaNDM-1 was located on a putative IncFII-type plasmid, which also carried the other acquired resistant genes identified, including fosA3, blaTEM-1B and rmtB, while the colistin-resistant gene mcr-1 was carried by a putative IncX4-type plasmid. Finally, we found that these resistant genes/plasmids were conjugative, and they could be co-conjugated, conferring resistance to multiple types of antibiotics, including the carbapenems and colistin, to the recipient Escherichia coli strains.
Keywords: Escherichia coli, carbapenem-resistance, colistin-resistance, blaNDM-1, mcr-1, plasmid, co-existence
1. Introduction
Escherichia coli has become a great concern for global public health. On the one hand, the prevalence and outbreak of some intestinal pathogenic E. coli clones, particularly the well-known O157/ST11 and, more recently, the O104/ST678, has caused high levels of morbidity and mortality worldwide [1,2,3,4]; on the other hand, E. coli has had a great capacity to accumulate resistance genes, representing a natural reservoir of resistance genes, which may therefore contribute to the dissemination of antibiotic resistance and lead to treatment failures in both human and veterinary medicine [5]. It is of great importance to know how resistance genes are acquired and evaluate their capacity of dissemination between bacteria.
Recently, the emergence of the plasmid-mediated New Delhi metallo-β-lactamase-1 encoding gene blaNDM-1 and/or the plasmid-mediated colistin-resistance gene mcr-1 represents a great concern to global public health [6,7]. The plasmid-mediated blaNDM-1 confers resistance to carbapenems [6], which is considered as the last resort for treating multidrug-resistant (MDR) Enterobacteriaceae [8], while mcr-1 mediates the resistance to colistin [9], the key antibiotic used for treating carbapenem-resistant Enterobacteriaceae [8]. The emergence of carbapenems and colistin co-resistant Enterobacteriaceae means there will be little and/or no antibiotic available for the infections caused by such strains in most cases of infections caused by MDR Enterobacteriaceae. However, the co-existence of blaNDM and mcr-1 in Enterobacteriaceae originated from both humans and animals has been increasingly reported worldwide [10,11,12,13]. These isolates are involved in the co-harboring of mcr-1 and different members of blaNDM such as blaNDM-1 [11], blaNDM-4 [13], blaNDM-5 [12], and blaNDM-9 [10,14]. Of particularly concern is the recovery of an E. coli strain co-producing MCR-1 and NDM-9 from a patient with a catheter-associated urinary tract infection (UTI); the infection of such an E. coli led to the failure of antibiotic treatment and finally killed the patient [10]. Therefore, it is of great importance to monitor Enterobacteriaceae strains with co-resistance to carbapenems and colistin and understand how this resistance is accumulated. In this study, we report several carbapenem-resistant and colistin-resistant E. coli co-producing NDM-1 and MCR-1 from pig farms in China.
2. Materials and Methods
2.1. Bacterial Strains and Antimicrobial Susceptibility Testing
Fecal and environmental swabs from 7 pig farms located in Hubei Province and Henan Province in China during June 2018 and June 2019 were collected for the isolation of carbapenem-resistant and colistin-resistant E. coli, using MacConkey agar containing 4 μg/ml of imipenem and 2 μg/ml of colistin. E. coli isolates were confirmed by PCR detection of the 16S rRNA gene and the seven house-keeping genes (adk, fumC, gyrA, icd, mdh, purA, and recA) of E. coli [15]. A total of 538 samples were collected and carbapenem-resistant and colistin-resistant E. coli were only detectable on samples from one farm in Henan province (sows ≥ 4000), where eight isolates were recovered from swabs of the floor and barrier of different pigsties (RDX007, RDX012), swabs of different food troughs (RDX020, RDX024), fecal samples of different pigs suffered from diarrhea (RDX033, RDX035, RDX100), and water troughs (RDX115).
The antimicrobial susceptibility of these eight isolates was determined by testing the minimal inhibitory concentration (MIC) values of the selected antibiotics on the bacteria using the broth microdilution method, according to the protocol recommended by the United Sates Clinical and Laboratory Standards Institute (CLSI M31-S1). A total of 28 types of antibiotics were assessed for the testing, namely, amikacin (AMK), gentamicin (GEN), tobramycin (TOB), imipenem (IPM), meropenem (MRP), ertapenem (ETP), colistin (CL), cefazolin (CFZ), cefuroxime (CFX), cefoxitin (FOX), ceftazidime (CAZ), ceftriaxone (CRO), cefepime (CPM), chloramphenicol (CHL), fosfomycin (FOS), nitrofurantoin (NIT), ciprofloxacin (CIP), levofloxacin (LVX), moxifloxacin (MXF), norfloxacin (NOR), minocycline (MIN), tetracycline (TET), aztreonam (AZM), tigecycline (TGC), amoxicillin/clavulanate (AMC), ampicillin/sulbactam (AMS), and piperacillin/tazobactam (PTZ), trimethoprim/sulfamethoxazole (SXT). These 28 types of antibiotics can be assigned to several classes: aminoglycosides (AMK, GEN, TOB), β-lactams (ETP, IPM, MRP, CFZ, CFX, FOX, CAZ, CRO, CPM, AMC, AMS, PTZ, AZM), Phenicols (CHL), tetracyclines (TET, MIN, TGC), fluoroquinolones (MXF, CIP, LVX, NOR), sulfonamides (SXT), fosfomycins (FOS), nitrofurantoins (NIT), and polymyxins (CL) (Sigma-Aldrich, USA). The thirteen β-lactam class of antibiotics can be re-assigned into four sub-classes: carbapenems (ETP, IPM, MRP), cephalosporins (CFZ, CFX, FOX, CAZ, CRO, CPM), β-lactam combination agents (AMC, AMS, PTZ), and monobactams (AZM). Results were interpreted using the CLSI breakpoints (CLSI-VET06, CLSI M100, 28th Edition). If a CLSI breakpoint is not available, a European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoint is used for the interpretation. Each antibiotic was tested with three duplicates. E. coli ATCC 25922 was used as quality control.
2.2. Detection of the Carbapenem-Resistant and Colistin-Resistant Genes
PCR assays were initially performed to determine the presence and the location of the carbapenem-resistant gene blaNDM-1 and colistin-resistant mcr-1 with primers “F: GGTTTGGCGATCTGGTTTTC; R: CGGAATGGCTCATCACGATC” (for blaNDM-1, annealing temperature 55 °C, product size: 621 bp) [16] and “F: CGGTCAGTCCGTTTGTTC; R: CTTGGTCGGTCTGTAGGG” (for mcr-1, annealing temperature 55 °C, product size: 309 bp) [9], respectively. PCR reactions were performed in a 25 μl amplification mixture containing 2 μl of template DNA, 12.5 μl of 2×Taq Master Mix (Dye Plus, Vazyme Biotech Co., Ltd; Nanjing, China), 1 μl each of 10 μM forward and reverse primer, and 8.5 μl of nuclease-free water. The reaction was performed under the following cycling conditions: an initial denaturation at 95 °C for 10 min, followed by 35 cycles of denaturation at 94 °C for 45 s, annealing at 55 °C for 45 s, extension at 72 °C for 1 min, and a final extension at 72 °C for 10 min. PCR products were analyzed by electrophoresis on 1% agarose gel. Two rounds of PCR assays were performed. The first round of PCR assays was performed using the genomic DNA isolated from the strains as the template to determine the presence of the two resistant genes. An additional round of PCR assays was performed using the plasmids isolated from the strains to determine whether the detected resistant genes were located on the plasmids.
2.3. DNA Extraction and Whole Genome Sequencing and Bioinformatic Analysis
Genomic DNA were extracted and purified using a TIANamp Bacteria DNA Kit (TIANGEN, Beijing, China). The quality and quantity of the DNA were evaluated by electrophoresis on a 1% agarose gel and using a NanoDrop2000 (Thermo Scientific, Waltham, USA). Whole genome sequencing was performed on an Illumina Hiseq Xten platform (Illumina Inc., San Diego, USA) at Guangdong Magigene Biotechnology Co. LTD (Guangzhou, China), using the pair-end 150 bp sequencing protocol. DNA libraries were constructed using a NEBNext UltraTM II DNA Library Prep Kit (New England BioLabs, Ipswich, USA). After sequencing, approximately 7,703,600~18,595,428 raw reads were yielded for the eight strains. Raw reads with low quality were filtered according to the following criteria: low quality base pairs at each terminal of the reads (Quality-Value < 20) were removed; reads with a short length (parameter setting at 50 bp), or > 15 bp overlap with Illumina TruSeq adapter sequences (parameter setting at 15 bp) were removed. Finally, approximately 7,363,540~17,856,944 clean reads (Q20% = 100, Q30% ≥ 95.59) were produced. High-quality reads were de novo assembled via SPAdes v3.9.0 [17] to generate contigs.
The assembled contigs were used for determining the genetic characteristics of the eight E. coli isolates. Serotypes and sequence types (ST) were determined by SerotypeFinder 2.0 [18] and Multi-Locus Sequence Typing (MLST) 2.0 [19], respectively. Acquired antimicrobial resistance genes and plasmids were determined using ResFinder 3.1 [20] and PlasmidFinder 2.0 [21], respectively. DNA identities between two sequences were calculated by ANI calculator [22]. A comparative genome analysis was performed and visualized using the BRIG package [23] and/or the EasyFig package [24]. Phylogenetic analysis was performed by the maximum likelihood method using the Tamura–Nei model [25] on MEGAX [26] with 1000 bootstrap iterations. The whole genome sequence of E. coli MG1655 (GenBank accession no. U00096.3) was downloaded from GenBank and used as the reference genome in the study.
The whole genome sequences of the eight E. coli isolates have been deposited into GenBank (BioProject accession no. PRJNA528830). GenBank accession numbers for each of the strains are: RXD007, SQQT00000000; RXD012: SQQW00000000; RXD020: SQQX00000000; RXD024: SQQY00000000; RXD033: SQQZ00000000; RXD035: SQRA00000000; RXD100: SQRC00000000; RXD115: SQRD00000000.
2.4. Plasmid Conjugation
Plasmid conjugation assays between the eight carbapenem-resistant and colistin-resistant E. coli (donor) and the rifampin resistant E. coli C600 (recipient) were performed on a nitrocellulose membrane, as described previously [27]. Briefly, mid-log phase donor and recipient strains (OD600 = 0.5~0.6) were mixed at a ratio of 1:3 (v/v). The bacterial mixture was then spotted on a nitrocellulose membrane which was pre-plated on the LB agar. After a 12 h of incubation at 37 °C, bacteria on the membrane were washed off using LB broth followed by being shaken at 37 °C for 4 h. Finally, the transconjugants were selected on LB agar plates laced with rifampin (1000 mgl−1) plus imipenem (20 mgl−1) plus colistin (2 mgl−1). Antimicrobial susceptibility of the transconjugants was determined using broth microdilution method as mentioned above.
3. Results
3.1. Antimicrobial Susceptibility Profile of the Eight E. coli Isolates
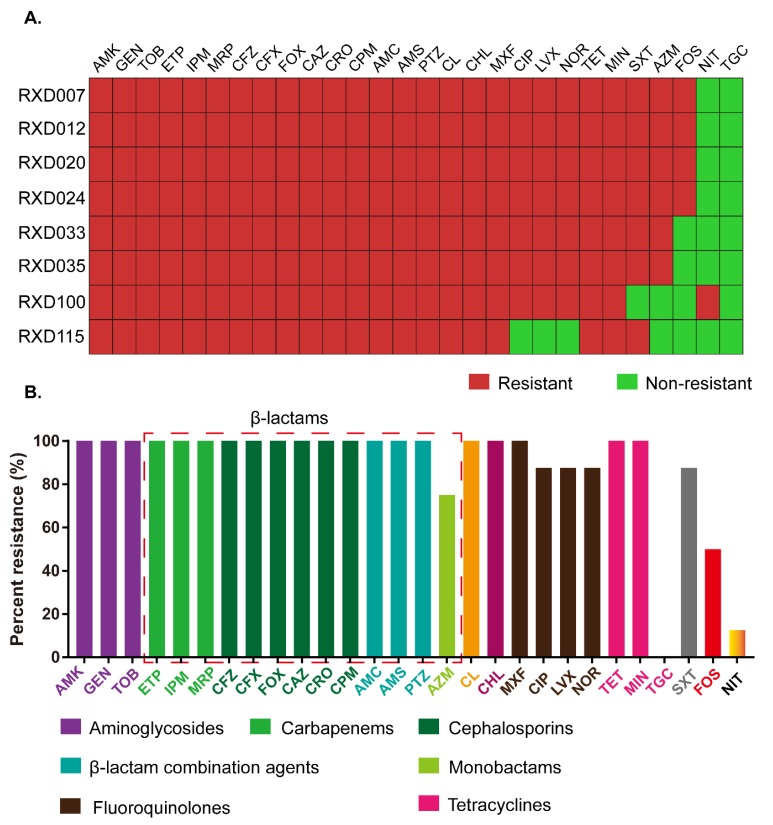
Antimicrobial susceptibility testing showed that all (100%, 8/8) of the isolates tested herein were resistant to AMK, GEN, TOB, ETP, IPM, MRP, CFZ, CFX, FOX, CAZ, CRO, CPM, AMC, AMS, PTZ, CHL, MXF, TET, and CL (Figure 1). Many of them were also resistant to CIP (75.00%, 6/8), LVX (75.00%, 6/8), NOR (75.00%, 6/8), MIN (50.00%, 4/8), SXT (87.50%, 7/8), AZM (75.00%, 6/8) and FOS (50.00%, 4/8). Only one isolate was resistant to NIT (12.5%, 1/8). All of them were susceptible to TGC (100%, 8/8) (Figure 1A).
Figure 1.
Antimicrobial susceptibility profile of the eight carbapenem-resistant and colistin-resistant Escherichia coli isolates. (A) Heatmap showing resistance and multidrug resistance phenotypes of each of the E. coli isolates; (B) Percent strains resistant to the tested antibiotics. AMK: amikacin; GEN: gentamicin; TOB: tobramycin; ETP: ertapenem; IPM: imipenem; MRP: meropenem; CFZ: cefazolin; CFX: cefuroxime; FOX: cefoxitin; CAZ: ceftazidime; CRO: ceftriaxone; CPM: cefepime; AMC: amoxicillin/clavulanate; AMS: ampicillin/sulbactam; PTZ: piperacillin/tazobactam; CL: colistin; CHL: chloramphenicol; MXF: moxifloxacin; CIP: ciprofloxacin; LVX: levofloxacin; NOR: norfloxacin; TET: tetracycline; MIN: minocycline; SXT: trimethoprim/sulfamethoxazole; AZM: aztreonam; FOS: Fosfomycin; NIT: nitrofurantoin; TGC: tigecycline.
Regarding the different antibiotic classes, these eight carbapenem-resistant and colistin-resistant E. coli isolates were resistance to the three aminoglycosides tested (AMK, GEN, TOB), another nine β-lactams (CFZ, CFX, FOX, CAZ, CRO, CPM, AMC, AMS, PTZ) in addition to the three carbapenems tested, one of the phenicols tested (CHL), one of the tetracyclines tested (TET), one of the fluoroquinolones tested (MXF), simultaneously (Figure 1A). In addition, a higher proportion of these carbapenem-resistant and colistin-resistant E. coli strains showed resistance to another three fluoroquinolones tested (CIP, 75.00%; LVX, 75.00%; NOR, 75.00%), one of the β-lactams tested (AZM, 75.00%) and one of the sulfonamides tested (SXT, 87.50%). In total, 50% of the carbapenem-resistant and colistin-resistant E. coli strains were resistant to fosfomycin, and another one of the tetracyclines tested (MIN). A low proportion of the carbapenem-resistant and colistin-resistant E. coli strains displayed resistance to NIT (12.50%) and TGC (0.00%) (Figure 1B). Resistance to “AMK + GEN + TOB + ETP + IPM + MRP + CFZ + CFX + FOX + CAZ + CRO + CPM + AMC + AMS + PTZ + CL + CHL + MXF + TET” was the common characteristic for the eight carbapenem-resistant and colistin-resistant E. coli strains (Figure 1A).
3.2. Detection of the Carbapenem-Resistant Gene blaNDM-1 and Colistin-Resistant Gene mcr-1
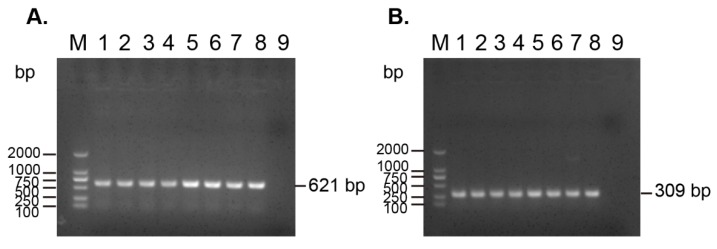
PCR assays revealed that the eight carbapenem-resistant and colistin-resistant E. coli strains contained the carbapenem-resistant gene blaNDM-1 and colistin-resistant gene mcr-1, simultaneously. In addition, PCR assays using plasmids extracted from the eight strains as template showed positive results for the detection of the two genes, suggesting that the two genes were located on plasmids (Figure 2).
Figure 2.
PCR detection of the carbapenem-resistant gene blaNDM-1 and the colistin-resistant gene mcr-1 from (A) the plasmids extracted from the eight carbapenem-resistant and (B) the colistin-resistant E. coli strains. M: DL 2000 DNA marker; 1–8: plasmid from RXD007, RXD012, RXD020, RXD024, RXD033, RXD035, RXD100, and RXD115; 9: ddH2O as negative control.
3.3. Acquired Antibiotic Resistance Genes in the Eight Carbapenem-Resistant and Colistin-Resistant E. coli
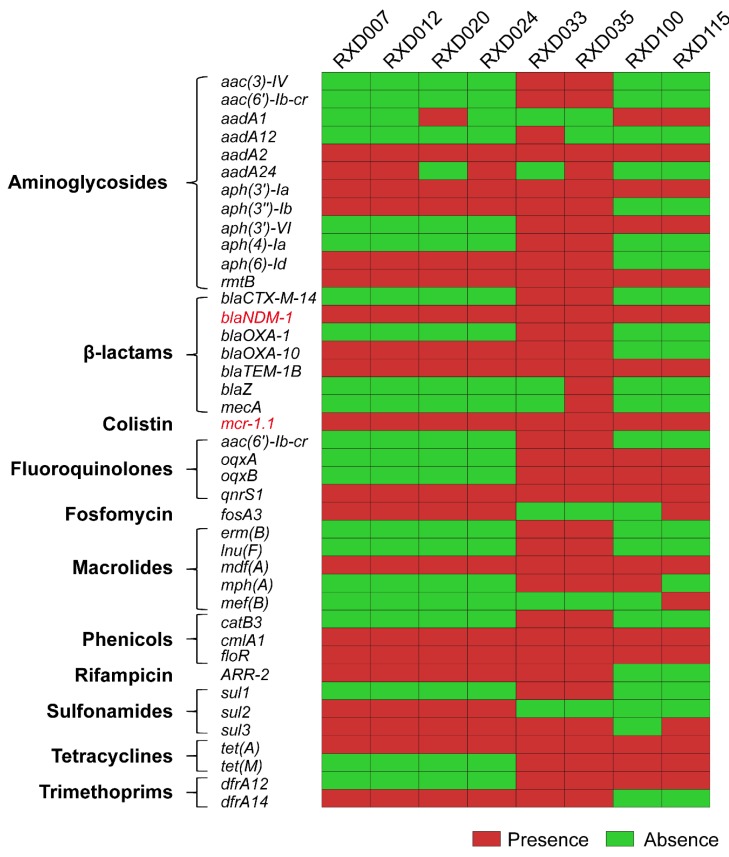
The whole genome sequencing strategy revealed that each of the eight carbapenem-resistant and colistin-resistant E. coli isolates possessed a chromosome of approximately 5.0 Mb in size and several plasmid contigs. The prediction of acquired resistance genes using whole genome sequences identified the presence of blaNDM-1 (n = 8), blaTEM-1B (n = 8), blaOXA-10 (n = 6), blaCTX-M-14 (n = 2), blaOXA-1 (n = 2), blaZ (n = 1), and mecA (n = 1), which account for the resistance to carbapenem and the other β-lactam antibiotics (Figure 2). These strains also carried aph(3′)-Ia (n = 8), aadA2 (n = 8) and rmtB (n = 8), accounting for the resistance to aminoglycosides; floR (n = 8) and cmlA1 (n = 8), accounting for the resistance to phenicols; tet(A) (n = 8) and tet(M) (n = 4), accounting for the resistance to tetracyclines; qnrS1 (n = 8), oqxA (n = 4), oqxB (n = 4), and aac(6′)-Ib-cr (n = 2), accounting for the resistance to fluoroquinolones; sul3 (n = 7), sul2 (n = 4) and sul1 (n = 2), accounting for the resistance to sulfonamides; fosA3 (n = 8), which accounts for osfomycin resistance, and mcr-1.1 (n = 8), which accounts for colistin resistance (Figure 3).
Figure 3.
Heatmap showing acquired resistant genes identified in each of the eight carbapenem-resistant and colistin-resistant E. coli isolates.
3.4. Genetic Characteristics
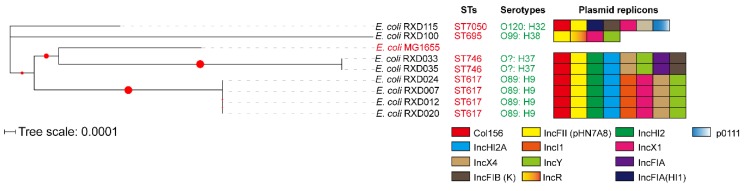
Multilocus sequence typing (MLST) identified four kinds of sequence types: ST617 (n = 4), ST746 (n = 2), ST695 (n = 1), and ST7050 (n = 1) (Figure 4). Three types of O-serotypes and four types of H-serotypes were determined: O89: H9 (n = 3), O89: H10 (n = 1), O99: H38 (n = 1), and O120: H32 (n = 1) (Figure 3). The prediction of plasmid determined four to eight replicons in each of the genomes. Among these replicons, a predicted replicon of IncFII-type plasmid pHN7A8 (GenBank accession: JN232517) was carried by all of the carbapenem-resistant and colistin-resistant E. coli strains (Figure 4).
Figure 4.
Phylogenetic analysis of the eight carbapenem-resistant and colistin-resistant E. coli isolates. Sequence types, O-serotypes, H-serotypes, and plasmid replicons are also shown. The evolutionary history was inferred by using the Maximum Likelihood method and Tamura–Nei model. The tree with the highest log likelihood (−13321.25) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved nine nucleotide sequences. There was a total of 9093 positions in the final dataset. Evolutionary analyses were conducted by using MEGA X. The circles denote bootstrap values within the range of 0.19–1.000.
3.5. Plasmid Determination
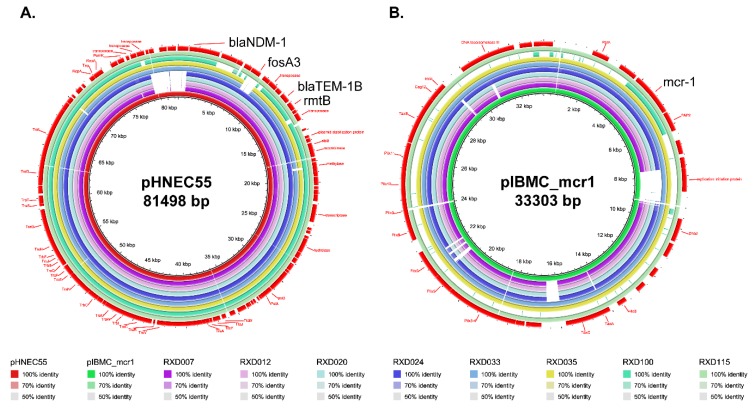
Because the results of the PCR assays revealed that the blaNDM-1 gene and the mcr-1 gene were located on plasmids (Figure 2), we used the whole genome sequences to analyze the putative blaNDM-1-carring and/or mcr-1-carrying plasmids that were harbored by the eight carbapenem-resistant and colistin-resistant E. coli isolates. Bioinformatical analysis revealed the blaNDM-1 gene was located on an IncFII-type plasmid, while the mcr-1.1 gene carried by RXD007, RXD012, RXD020, RXD024, RXD033, RXD035, and RXD115 was located on an IncX4-type plasmid. However, we did not find the putative mcr-1-carrying plasmid in RXD100 showing any homologous to any plasmids with DNA sequences available in GenBank. The IncFII-type plasmid carrying the blaNDM-1 gene was homologous to pHNEC55 (GenBank accession: KT879914), and this pHNEC55-like plasmid also carried fosA3, blaTEM-1B and rmtB (Figure 5A). An IncX4-type plasmid carrying mcr-1.1 presence in RXD007, RXD012, RXD020, RXD024, RXD033, RXD035, and RXD115 was homologous to pIBMC_mcr1 (GenBank accession: MF449287) (Figure 5B).
Figure 5.
Comparison of plasmid pHNEC55 (panel A) and pIBMC_mcr1 (panel B) with the genomes of RXD007, RXD012, RXD020, RXD024, RXD033, RXD035, RXD100, and RXD115, displayed as the outer rings inside to outside, respectively. The outmost rings (red) show the functional genes including the resistance genes as well as the structural genes carried by pHNEC55 (panel A) and pIBMC_mcr1 (panel B).
3.6. Plasmid Conjugation
To assess the transferability of the plasmids and the resistance genes, conjugation experiments between the eight were performed with selective plates laced with rifampin (1000 mgl−1) plus imipenem (20 mgl−1) plus colistin (2 mgl−1). Transconjugants incubated on selective agars were selected for antimicrobial susceptibility testing. The result showed that the plasmids in seven of the eight carbapenem-resistant and colistin-resistant E. coli transferred into the recipient strain and after conjugation, the resistance phenotypes of the recipient E. coli (C600) changed remarkably (Table 1).
Table 1.
Phenotypic characteristics of the transconjugants of the eight carbapenem-resistant and colistin-resistant E. coli.
| Antibiotics tested | Transconjugants selected by rifampin plus imipenem plus colistin | E. coli C600 | ||||||
|---|---|---|---|---|---|---|---|---|
| Minimum inhibitory concentration (μg/ml) | ||||||||
| Cir007 1 | Cir012 | Cir020 | Cir033 | Cir035 | Cir100 | Cir115 | ||
| Amikacin | > 32 (R1) | > 32 (R) | > 32 (R) | > 32 (R) | > 32 (R) | > 32 (R) | > 32 (R) | ≤ 8 (S) |
| Gentamicin | > 8 (R) | > 8 (R) | > 8 (R) | > 8 (R) | > 8 (R) | > 8 (R) | > 8 (R) | ≤ 2 (S) |
| Tobramycin | > 8 (R) | > 8 (R) | > 8 (R) | > 8 (R) | > 8 (R) | > 8 (R) | > 8 (R) | ≤ 2 (S) |
| Ertapenem | > 2 (R) | > 2 (R) | > 2 (R) | > 2 (R) | > 2 (R) | > 2 (R) | 2 (R) | ≤ 0.25 (S) |
| Imipenem | > 8 (R) | > 8 (R) | > 8 (R) | > 8 (R) | > 8 (R) | 1 (S) | 8 (R) | 0.5 (S) |
| Meropenem | > 8 (R) | > 8 (R) | > 8 (R) | > 8 (R) | > 8 (R) | 1 (S) | 4 (R) | ≤ 0.13 (S) |
| Cefazolin | > 16 (R) | > 16 (R) | > 16 (R) | > 16 (R) | > 16 (R) | > 16 (R) | > 16 (R) | 4 (S) |
| Cefuroxime | > 16 (R) | > 16 (R) | > 16 (R) | > 16 (R) | > 16 (R) | > 16 (R) | > 16 (R) | 16 (R) |
| Cefoxitin | > 16 (R) | > 16 (R) | > 16 (R) | > 16 (R) | > 16 (R) | > 16 (R) | > 16 (R) | 8 (S) |
| Ceftazidime | > 32 (R) | > 32 (R) | > 32 (R) | > 32 (R) | > 32 (R) | > 32 (R) | > 32 (R) | ≤ 1 (S) |
| Ceftriaxone | > 32 (R) | > 32 (R) | > 32 (R) | > 32 (R) | > 32 (R) | > 32 (R) | > 32 (R) | ≤ 1 (S) |
| Cefepime | > 16 (R) | > 16 (R) | > 16 (R) | 16 (R) | > 16 (R) | 16 (R) | 8 (R) | ≤ 1 (S) |
| Amoxicillin/clavulanate | > 32/16 (R) | > 32/16 (R) | > 32/16 (R) | > 32/16 (R) | > 32/16 (R) | 32/16 (R) | 32/16 (R) | ≤ 8/4 (S) |
| Ampicillin/sulbactam | >16/8 (R) | >16/8 (R) | >16/8 (R) | >16/8 (R) | > 16/8 (R) | > 16/8 (R) | >16/8 (R) | 8/4 (S) |
| Piperacillin/tazobactam | >64/4 (R) | >64/4 (R) | >64/4 (R) | >64/4 (R) | >64/4 (R) | >64/4 (R) | >64/4 (R) | ≤ 4/4 (S) |
| Colistin | > 4 (R) | > 4 (R) | 4 (R) | 4 (R) | 4 (R) | > 4 (R) | > 4 (R) | ≤ 1 (S) |
| Chloramphenicol | > 16 (R) | > 16 (R) | > 16 (R) | ≤ 4 (S) | ≤ 4 (S) | > 16 (R) | ≤ 4 (S) | ≤ 4 (S) |
| Moxifloxacin | > 2 (R) | > 2 (R) | > 2 (R) | ≤ 0.5 (S) | ≤ 0.5 (S) | > 2 (R) | ≤ 0.5 (S) | ≤ 0.5 (S) |
| Ciprofloxacin | > 4 (R) | > 4 (R) | > 4 (R) | ≤ 0.5 (S) | ≤ 0.5 (S) | 2 (I) | ≤ 0.5 (S) | ≤ 0.5 (S) |
| Levofloxacin | > 8 (R) | > 8 (R) | > 8 (R) | ≤ 1 (S) | ≤ 1 (S) | 2 (S) | ≤ 1 (S) | ≤ 1 (S) |
| Norfloxacin | > 8 (R) | > 8 (R) | > 8 (R) | ≤ 2 (S) | ≤ 2 (S) | 4 (S) | ≤ 2 (S) | ≤ 2 (S) |
| Tetracycline | > 8 (R) | > 8 (R) | > 8 (R) | ≤ 2 (S) | ≤ 2 (S) | > 8 (R) | ≤ 2 (S) | ≤ 2 (S) |
| Minocycline | 4 (S) | 4 (S) | 4 (S) | ≤ 1 (S) | 2 (S) | 16 (R) | 2 (S) | ≤ 1 (S) |
| Trimethoprim/sulfamethoxazole | > 4/76 (R) | > 4/76 (R) | > 4/76 (R) | ≤1/19 (S) | ≤ 1/19 (S) | ≤ 1/19 (S) | ≤1/19 (S) | ≤ 1/19 (S) |
| Aztreonam | ≤ 2 (S) | ≤ 2 (S) | ≤ 2 (S) | ≤ 2 (S) | ≤ 2 (S) | ≤ 2 (S) | ≤ 2 (S) | ≤ 2 (S) |
| Fosfomycin | > 128 (R) | > 128 (R) | > 128 (R) | ≤ 16 (S) | ≤ 16 (S) | ≤ 16 (S) | > 128 (R) | ≤ 16 (S) |
| Nitrofurantoin | ≤ 16 (S) | 32 (S) | ≤ 16 (S) | ≤ 16 (S) | ≤ 16 (S) | > 64 (R) | ≤ 16 (S) | ≤ 16 (S) |
| Tigecycline | ≤ 1 (S) | ≤ 1 (S) | ≤ 1 (S) | ≤ 1 (S) | ≤ 1 (S) | ≤ 1 (S) | ≤ 1 (S) | ≤ 1 (S) |
1 Note: Cir007, Cir012, Cir020, Cir033, Cir035, Cir100, and Cir115 represent the transconjugant of RXD007, RXD012, RXD020, RXD033, RXD035, RXD100, and RXD115, respectively. R: Resistant; S: susceptible; I: intermediately resistant.
4. Discussion
Carbapenem and colistin are the last-resort antibiotics used for treating multidrug-resistant Gram-negative pathogens [8]. It is of great public health significance to monitor the carbapenem-resistant and colistin-resistant bacteria and investigate the mechanisms for the acquisition of the resistant phenotypes. In this study, we isolated eight carbapenem-resistant and colistin-resistant E. coli from a pig farm in Henan Province, China. Antimicrobial susceptibility testing revealed the eight carbapenem-resistant and colistin-resistant isolates were also resistant to aminoglycosides, cephalosporins, β-lactam combination agents, phenicols (CHL), tetracyclines, fluoroquinolones, and sulfonamides (SXT) (Figure 1 and Figure 2). Since most of these phenotypes in E. coli were conferred by resistance genes mostly through horizontal gene transfer [5], the presence of those strains in pigs/farms may represent a real public health concern: (1) these MDR isolates might transmit between pigs/environment and other animal species including humans through numerous pathways such as via direct/indirect contact and/or via food-chain, which might therefore lead to treatment failures in both human and veterinary medicine; (2) such strains might also act as a major reservoir of resistance genes, which might contribute to the spread of antimicrobial resistance.
Corresponding to the resistant phenotypes determined, prediction using the whole genome sequences identified many types of acquired genes conferring resistance to β-lactams (including cephalosporins and carbapenems), aminoglycosides, fluoroquinolones, polymyxins, tetracyclines, phenicols, sulfonamides, trimethoprim, and fosfomycin (Figure 3). Of particularly note is the blaNDM-1 gene, which encodes the New Delhi metallo-β lactamase 1 (blaNDM-1) and confers the resistance to carbapenems [28,29], and the mcr-1 gene, which encodes the phosphoethanolamine-lipid A transferase and confers the resistance to colistin [9]. Both of these two genes have substantial importance worldwide in terms of resistance [7,29,30,31] and they were identified in the eight carbapenem-resistant and colistin-resistant E. coli strains (Figure 2 and Figure 3), suggesting that the co-existence of blaNDM-1 and mcr-1 mediates the resistance phenotypes to carbapenems and colistin.
Three of the eight carbapenem-resistant and colistin-resistant E. coli were isolated from fecal samples of different diarrheal pigs. Two of them, designated RDX033 and RDX035, were found to be ST746 (Figure 4). It is worth noting that MCR-1-carrying, extended-spectrum β-lactamase (ESBL)-producing E. coli ST746 has been recovered from community-acquired urinary tract infection [32]. The human sourced E. coli ST746 contained plasmid-carrying mcr-1 as well as two kinds of β-lactam-resistant genes blaCTX-M-14 and blaTEM-1B but not blaNDM-1 gene [32]. However, the two ST746 strains recovered from diarrheal pigs carried both mcr-1 and blaNDM-1 in addition to blaCTX-M-14 and blaTEM-1B (Figure 3), and they displayed a broader spectrum of resistance compared with the human sourced ST746. The identification of such strains in pigs might represent a potential risk on public health. Another carbapenem-resistant and colistin-resistant E. coli recovered from different diarrheal pigs was RXD100, which was identified as ST695. While this isolate showed resistance to most of the antibiotics tested, it was not as resistant as the ST746 strains RDX033 and RDX035 (Figure 1). Fewer numbers of resistant genes carried by RXD100 compared to RDX033 and RDX035 might explain the difference (Figure 3).
It has been widely reported that plasmids play an important role in the dissemination of blaNDM-1 and mcr-1 [5,9,29,31]. Consistently, our PCR results using the plasmids extracted as templates from the eight strains revealed that both of the genes were located on plasmids (Figure 2). Plasmid prediction using whole genome sequences identified several putative types of plasmids harbored by the eight strains (Figure 4). Among these putative types of plasmids, IncHI2A, IncFIB(K), IncFII, IncFI1, IncR, IncHI2, and IncFIA plasmids carrying blaNDM-1 have been reported [33,34,35,36,37], while IncX4, IncFIB(K), IncFII, IncFI1, IncY, IncHI2, IncX1, and IncFIA plasmids carrying mcr-1 have been reported [38,39,40,41,42]. Although more accurate and reliable technologies such as the third-generation sequencing technologies are necessary for determining the accurate type and sequence of the plasmids, bioinformatical analysis using whole genome sequences revealed that an IncFII-type plasmid homologous to pHNEC55 (GenBank accession: KT879914) likely carried the blaNDM-1 gene, and an IncX4-type plasmid homologous to pIBMC_mcr1 (GenBank accession: MF449287) likely carried the mcr-1 gene, as the backbones of both plasmids were found in the genomes of the eight strains (Figure 5). Interestingly, the IncFII plasmid pHNEC55 carrying blaNDM-1 was also identified in a carbapenem-resistant E. coli strain isolated from pigs in Henan Province [43]. The multidrug resistance-encoding mobile elements of this plasmid contains several other resistant genes in addition to blaNDM-1, including mphA, which confers resistance to macrolides; fosA3, which confers resistance to Fosfomycin; rmtB, which confers resistance to aminoglycosides; and blaTEM, which confers resistance to β-lactams [43]. These genes were also carried by the eight carbapenem-resistant and colistin-resistant bacteria and they were also likely to be carried by the blaNDM-1-carrying plasmids (Figure 5A). In the next step, we intend to use more accurate and reliable technologies such as third-generation sequencing or a combination of second-generation with third-generation sequencing to determine the complete genome sequences and the accurate structures of the MDR plasmids in the eight E. coli strains.
Plasmid conjugation assays revealed that the MDR plasmids harbored by the eight carbapenem-resistant and colistin-resistant bacteria were transferrable, and it seems that these MDR plasmids are capable of co-conjugation at appropriate conditions, as the conjugation of the plasmids conferred both carbapenem and colistin resistance to the recipient bacteria simultaneously (Table 1). The same results have also been found in other articles [14]. In addition to carbapenems and colistin, the conjugation of these plasmids was also able to disseminate resistance to many types of antibiotics tested, including aminoglycosides, cephalosporins, β-lactam combination agents, phenicol, tetracyclines, fluoroquinolones, and sulfonamides (Table 1). Many of these antibiotics are commonly used antibiotics for bacterial infections in both human and veterinary medicine [5], and many are classified as critically important antimicrobials by the WHO and their usage should be severely restricted (https://www.who.int/foodsafety/publications/antimicrobials-fifth/en/). Therefore, the presence of these conjugative MDR plasmids might lead to the spread of the MDR plasmids and treatment failures in both human and veterinary medicine. More active actions should be taken to monitor the prevalence of such plasmids as well as their bacterial hosts. In the next step, we will evaluate the conjugative capacity and efficacy of each of the resistant plasmids after their complete genome sequences are determined by third-generation sequencing technologies.
5. Conclusions
In conclusion, a total of eight carbapenem-resistant and colistin-resistant E. coli from pigs/farms in Central China were characterized in the present study. The carbapenem-resistant and colistin-resistant E. coli isolates were commonly resistant to aminoglycosides, cephalosporins, β-lactam combination agents, phenicol, tetracyclines, fluoroquinolones, and sulfonamides. A coexistence of a putative IncFII-type plasmid homologous to pHNEC55 and an IncX4-type plasmid homologous to pIBMC_mcr1 is likely to be responsible for the dissemination of the carbapenem-resistant gene blaNDM-1 and colistin-resistant gene mcr-1, and both genes/plasmids are conjugative and could be co-conjugatively transferred. In the next study, we intend to use more accurate and reliable technologies like third-generation sequencing to determine the complete genome sequences and the accurate structures of these MDR plasmids.
Author Contributions
Conceptualization, Z.P., H.C. and X.W.; methodology, Z.P., X.L., Z.H., Z.L. and Y.L.; writing—original draft preparation, Z.P.; writing—review and editing, Z.P., M.L., B.W. and X.W.; supervision, H.C. and X.W.; project administration, H.C. and X.W.; funding acquisition, H.C. and X.W.
Funding
This research was funded by the National Key Research and Development Program of China (grant numbers 2017YFC1600101 and 2017YFC1600103) and the earmarked fund for the China Agriculture Research System (grant number: CARS-35). Zhong Peng was supported in part by the China Postdoctoral Science Foundation (grant number: 2018M640719). The APC was funded by the National Key Research and Development Program of China.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Bielaszewska M., Mellmann A., Zhang W., Kock R., Fruth A., Bauwens A., Peters G., Karch H. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: A microbiological study. Lancet Infect. Dis. 2011;11:671–676. doi: 10.1016/S1473-3099(11)70165-7. [DOI] [PubMed] [Google Scholar]
- 2.Vidovic S., Korber D.R. Escherichia coli O157: Insights into the adaptive stress physiology and the influence of stressors on epidemiology and ecology of this human pathogen. Crit. Rev. Microbiol. 2016;42:83–93. doi: 10.3109/1040841X.2014.889654. [DOI] [PubMed] [Google Scholar]
- 3.Chekabab S.M., Paquin-Veillette J., Dozois C.M., Harel J. The ecological habitat and transmission of Escherichia coli O157:H7. FEMS Microbiol. Lett. 2013;341:1–12. doi: 10.1111/1574-6968.12078. [DOI] [PubMed] [Google Scholar]
- 4.Navarro-Garcia F. Escherichia coli O104:H4 Pathogenesis: An Enteroaggregative E. coli/Shiga Toxin-Producing E. coli Explosive Cocktail of High Virulence. Microbiol. Spectr. 2014;2 doi: 10.1128/microbiolspec.EHEC-0008-2013. [DOI] [PubMed] [Google Scholar]
- 5.Poirel L., Madec J.Y., Lupo A., Schink A.K., Kieffer N., Nordmann P., Schwarz S. Antimicrobial Resistance in Escherichia coli. Microbiol. Spectr. 2018;6 doi: 10.1128/microbiolspec.ARBA-0026-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wailan A.M., Paterson D.L. The spread and acquisition of NDM-1: A multifactorial problem. Expert Rev. Anti Infect. Ther. 2014;12:91–115. doi: 10.1586/14787210.2014.856756. [DOI] [PubMed] [Google Scholar]
- 7.Paterson D.L., Harris P.N. Colistin resistance: A major breach in our last line of defence. Lancet Infect. Dis. 2016;16:132–133. doi: 10.1016/S1473-3099(15)00463-6. [DOI] [PubMed] [Google Scholar]
- 8.Sun J., Yang R.S., Zhang Q., Feng Y., Fang L.X., Xia J., Li L., Lv X.Y., Duan J.H., Liao X.P., et al. Co-transfer of blaNDM-5 and mcr-1 by an IncX3-X4 hybrid plasmid in Escherichia coli. Nat. Microbiol. 2016;1:16176. doi: 10.1038/nmicrobiol.2016.176. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y.Y., Wang Y., Walsh T.R., Yi L.X., Zhang R., Spencer J., Doi Y., Tian G., Dong B., Huang X., et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 10.Lai C.C., Chuang Y.C., Chen C.C., Tang H.J. Coexistence of MCR-1 and NDM-9 in a clinical carbapenem-resistant Escherichia coli isolate. Int. J. Antimicrob. Agents. 2017;49:517–518. doi: 10.1016/j.ijantimicag.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Delgado-Blas J.F., Ovejero C.M., Abadia-Patino L., Gonzalez-Zorn B. Coexistence of mcr-1 and blaNDM-1 in Escherichia coli from Venezuela. Antimicrob. Agents Chemother. 2016;60:6356–6358. doi: 10.1128/AAC.01319-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang R.S., Feng Y., Lv X.Y., Duan J.H., Chen J., Fang L.X., Xia J., Liao X.P., Sun J., Liu Y.H. Emergence of NDM-5- and MCR-1-Producing Escherichia coli Clones ST648 and ST156 from a Single Muscovy Duck (Cairina moschata) Antimicrob. Agents Chemother. 2016;60:6899–6902. doi: 10.1128/AAC.01365-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang R., Liu Y., Zhang Q., Jin L., Wang Q., Zhang Y., Wang X., Hu M., Li L., Qi J., et al. The prevalence of colistin resistance in Escherichia coli and Klebsiella pneumoniae isolated from food animals in China: Coexistence of mcr-1 and blaNDM with low fitness cost. Int. J. Antimicrob. Agents. 2018;51:739–744. doi: 10.1016/j.ijantimicag.2018.01.023. [DOI] [PubMed] [Google Scholar]
- 14.Yao X., Doi Y., Zeng L., Lv L., Liu J.H. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect. Dis. 2016;16:288–289. doi: 10.1016/S1473-3099(16)00057-8. [DOI] [PubMed] [Google Scholar]
- 15.Wirth T., Falush D., Lan R., Colles F., Mensa P., Wieler L.H., Karch H., Reeves P.R., Maiden M.C., Ochman H., et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Shazly S., Dashti A., Vali L., Bolaris M., Ibrahim A.S. Molecular epidemiology and characterization of multiple drug-resistant (MDR) clinical isolates of Acinetobacter baumannii. Int. J. Infect. Dis. 2015;41:42–49. doi: 10.1016/j.ijid.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joensen K.G., Tetzschner A.M., Iguchi A., Aarestrup F.M., Scheutz F. Rapid and Easy in Silico Serotyping of Escherichia coli Isolates by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2015;53:2410–2426. doi: 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larsen M.V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R.L., Jelsbak L., Sicheritz-Ponten T., Ussery D.W., Aarestrup F.M., et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F.M., Larsen M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carattoli A., Zankari E., Garcia-Fernandez A., Voldby Larsen M., Lund O., Villa L., Moller Aarestrup F., Hasman H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goris J., Konstantinidis K.T., Klappenbach J.A., Coenye T., Vandamme P., Tiedje J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 23.Alikhan N.F., Petty N.K., Ben Zakour N.L., Beatson S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sullivan M.J., Petty N.K., Beatson S.A. Easyfig: A genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamura K., Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 26.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogolsky M., Beall B.W., Wiley B.B. Transfer of the plasmid for exfoliative toxin B synthesis in mixed cultures on nitrocellulose membranes. Infect. Immun. 1986;54:265–268. doi: 10.1128/iai.54.1.265-268.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaheen B.W., Nayak R., Boothe D.M. Emergence of a New Delhi metallo-beta-lactamase (NDM-1)-encoding gene in clinical Escherichia coli isolates recovered from companion animals in the United States. Antimicrob. Agents Chemother. 2013;57:2902–2903. doi: 10.1128/AAC.02028-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y., Zhang R., Li J., Wu Z., Yin W., Schwarz S., Tyrrell J.M., Zheng Y., Wang S., Shen Z., et al. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat. Microbiol. 2017;2:16260. doi: 10.1038/nmicrobiol.2016.260. [DOI] [PubMed] [Google Scholar]
- 30.Li J., Nation R.L., Turnidge J.D., Milne R.W., Coulthard K., Rayner C.R., Paterson D.L. Colistin: The re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect. Dis. 2006;6:589–601. doi: 10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
- 31.Moellering R.C., Jr. NDM-1—A cause for worldwide concern. N. Engl. J. Med. 2010;363:2377–2379. doi: 10.1056/NEJMp1011715. [DOI] [PubMed] [Google Scholar]
- 32.Wu L., Chen J., Wang L., Wu Z. Whole genome sequence of an MCR-1-carrying, extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli ST746 isolate recovered from a community-acquired urinary tract infection. J. Glob. Antimicrob. Resist. 2018;13:171–173. doi: 10.1016/j.jgar.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 33.Cai Y., Chen C., Zhao M., Yu X., Lan K., Liao K., Guo P., Zhang W., Ma X., He Y., et al. High Prevalence of Metallo-beta-Lactamase-Producing Enterobacter cloacae from Three Tertiary Hospitals in China. Front. Microbiol. 2019;10:1610. doi: 10.3389/fmicb.2019.01610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paskova V., Medvecky M., Skalova A., Chudejova K., Bitar I., Jakubu V., Bergerova T., Zemlickova H., Papagiannitsis C.C., Hrabak J. Characterization of NDM-Encoding Plasmids from Enterobacteriaceae Recovered From Czech Hospitals. Front. Microbiol. 2018;9:1549. doi: 10.3389/fmicb.2018.01549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pedersen T., Sekyere J.O., Govinden U., Moodley K., Sivertsen A., Samuelsen O., Essack S.Y., Sundsfjord A. Spread of Plasmid-Encoded NDM-1 and GES-5 Carbapenemases among Extensively Drug-Resistant and Pandrug-Resistant Clinical Enterobacteriaceae in Durban, South Africa. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.02178-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baraniak A., Machulska M., Zabicka D., Literacka E., Izdebski R., Urbanowicz P., Bojarska K., Herda M., Kozinska A., Hryniewicz W., et al. Towards endemicity: Large-scale expansion of the NDM-1-producing Klebsiella pneumoniae ST11 lineage in Poland, 2015–2016. J. Antimicrob. Chemother. 2019 doi: 10.1093/jac/dkz315. [DOI] [PubMed] [Google Scholar]
- 37.Yang B., Feng Y., McNally A., Zong Z. Occurrence of Enterobacter hormaechei carrying blaNDM-1 and blaKPC-2 in China. Diagn. Microbiol. Infect. Dis. 2018;90:139–142. doi: 10.1016/j.diagmicrobio.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 38.Manageiro V., Clemente L., Romao R., Silva C., Vieira L., Ferreira E., Canica M. IncX4 Plasmid Carrying the New mcr-1.9 Gene Variant in a CTX-M-8-Producing Escherichia coli Isolate Recovered from Swine. Front. Microbiol. 2019;10:367. doi: 10.3389/fmicb.2019.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bai F., Li X., Niu B., Zhang Z., Malakar P.K., Liu H., Pan Y., Zhao Y. A mcr-1-Carrying Conjugative IncX4 Plasmid in Colistin-Resistant Escherichia coli ST278 Strain Isolated from Dairy Cow Feces in Shanghai, China. Front. Microbiol. 2018;9:2833. doi: 10.3389/fmicb.2018.02833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dalmolin T.V., Martins A.F., Zavascki A.P., de Lima-Morales D., Barth A.L. Acquisition of the mcr-1 gene by a high-risk clone of KPC-2-producing Klebsiella pneumoniae ST437/CC258, Brazil. Diagn. Microbiol. Infect. Dis. 2018;90:132–133. doi: 10.1016/j.diagmicrobio.2017.09.016. [DOI] [PubMed] [Google Scholar]
- 41.Xu L., Wang P., Cheng J., Qin S., Xie W. Characterization of a novel bla NDM-5-harboring IncFII plasmid and an mcr-1-bearing IncI2 plasmid in a single Escherichia coli ST167 clinical isolate. Infect. Drug Resist. 2019;12:511–519. doi: 10.2147/IDR.S192998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang C., Feng Y., Liu F., Jiang H., Qu Z., Lei M., Wang J., Zhang B., Hu Y., Ding J., et al. A Phage-Like IncY Plasmid Carrying the mcr-1 Gene in Escherichia coli from a Pig Farm in China. Antimicrob. Agents Chemother. 2017;61:e02035-16. doi: 10.1128/AAC.02035-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin D., Xie M., Li R., Chen K., Chan E.W., Chen S. IncFII Conjugative Plasmid-Mediated Transmission of blaNDM-1 Elements among Animal-Borne Escherichia coli Strains. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.02285-16. [DOI] [PMC free article] [PubMed] [Google Scholar]