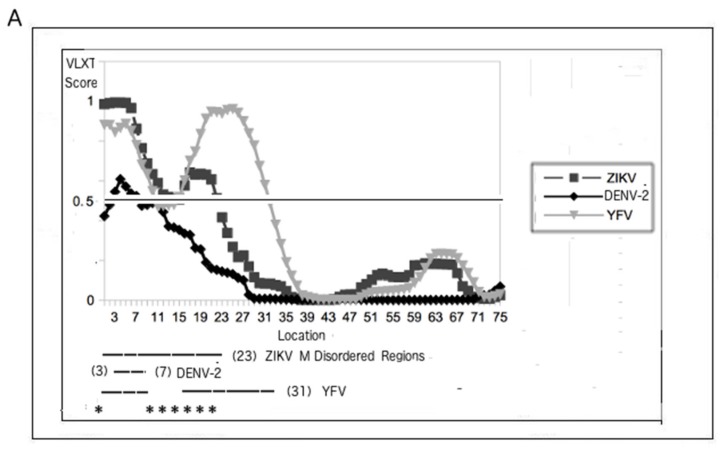
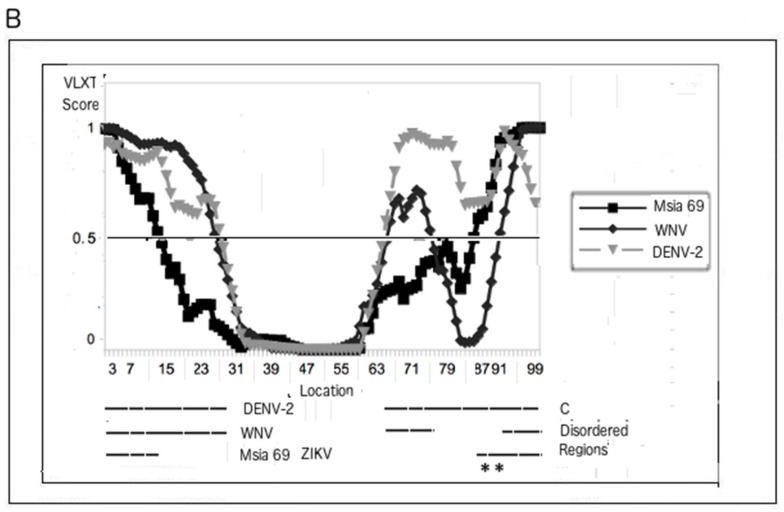
Figure 2.
Comparative PONDR VLXT plots of the M and C proteins from various flaviviruses, including ZIKV. (A) M proteins of ZIKV (UniProt: A0A127AM58), DENV-2 (UniProt: P29990), and YFV (UniProt: Q1X881). (B) C proteins of ZIKV (UniProt: H8XX11, Msia69, see Table 1), WNV (UniProt: Q9Q694), and DENV-2 (UniProt: P29990). Regions with scores of 0.5 or above represent disorder. Disorder differences between C proteins can be traced mainly to mutations and disorder near the N- and C-termini. The regions with an asterisk (*) denote potential targets for the development of a ZIKV vaccine. With the exception of Msia69 (Malaysia 1969) ZIKV, the strains were randomly chosen. Msia69 was deliberately chosen as a representative Asian strain that has a low C PID. The YFV M protein was chosen for (A), since it has one of the highest PIDs among flaviviruses. WNV was not necessary for (A), as specific strains of DENV-2 have an even lower M PID (PID: 6%) than does the ZIKV (PID: 9%). On the other hand, WNV was chosen for (B), as the low WNV C PID (among flaviviruses) allows us to suggest strategies for ZIKV vaccine development.