Abstract
Background
The bacterial infections that prevail in the burnt patients continue to be a critical complication in the burnt patients and vary with time and place. Identification of bacterial pathogens with information of their antimicrobial susceptibility of burn wounds can help clinicians to select appropriate medication procedure as in providing them with suitable antibiotic for empirical treatment.
Methods
Retrospective study of thirty-one months (Jan 2015 to July 2017) was designed to evaluate bacteria involved in burnt wound infection and its antimicrobial susceptibilities in a Burn Intensive Care Unit of Eastern India. Pus samples were cultured on cysteine Lactose electrolyte deficient agar (Hi-Media, India). Positive bacteria cultures were identified and tested for antimicrobial susceptibility using VITEK®2 (bioMerieux, Durham, NC, USA) and interpreted according to Clinical Laboratory Standards Institute guidelines.
Results
Two hundred and seventy-two wound swabs from burnt patients were received, out of which 62.8% (n = 185) were revealed as positive for the presence of bacteria. Pseudomonas aeruginosa, Klebsiella pneumoniae, Acinetobacter baumannii and E. coli were discovered to be the most common organisms in patients. Isolated bacteria were least resistant to TIGECYCLINE and COLISTIN.
Conclusion
Data regarding the incidence of pathogens and their resistance patterns would benefit the clinicians to prescribe appropriate antibiotics, articulating policies for empirical antimicrobial therapy to control the different types of infections.
Keywords: Microbiology, Antibiotics, Antimicrobial, Antibiotic resistance, Microorganism, Pathology, Burn wound, Colistin, Tigecycline, PAN drug resistant, Extensive drug resistance
Microbiology; Antibiotics; Antimicrobial; Antibiotic resistance; Microorganism; Pathology; Burn wound; Colistin; Tigecycline; PAN drug resistant; Extensive drug resistance.
1. Introduction
Infections persist as an important complication and cause of mortality in the burn patients [1]. Disrupted skin barrier, involvement of larger burnt area, immunocompromised effects of burns and prolonged stays at the hospitals were major risk factors for initiating infection [2]. The burn wound infection is characterized by the change in the manifestation of burn wounds, such as rapid eschar separation, dark brown, black or violaceous discoloration of the eschar or edema at wound margin. It is also illustrated by the organism isolated from blood culture in the absence of other identifiable infection with following characteristic: fever (>37.5 °C) or hypothermia (<35.5 °C), hypotension (systolic pressure below 90 mmHg), oliguria (<20 mL/h), hyperglycemia or mental confusion [3]. Although a significant improvement like haemodynamic stabilization, treatment of airway and intensive care for burn victims has been established, 75% of all deaths following thermal injuries are related to infection [1].
Use of antibiotics as systemic prophylactic is a common practice with burnt patients [4]. Drug resistant bacteria with intrinsic resistance towards antibiotics, ability to survive longer in the hospital environment and hand to hand transmission of bacteria reflects their easy spread and cause outbreaks [5, 6]. Extensive drug resistance (XDR) and pandrug resistant (PDR) strains were classified as non-susceptible to at least one agent in all but two or fewer antimicrobial categories and non-susceptible to all agents in all antimicrobial categories respectively by ECDC and CDC [7]. The bacterial infections in burnt patients vary both with time and place [8, 9]. Thus, a continuous surveillance and update of antibiotic resistance pattern of micro-organisms is essential for infection control programs and accurate antibiotic treatment in the burnt patients. In the present retrospective study, we have evaluated bacterial pathogens isolated from wounds of patients admitted in Burn Care Unit and their antibiotic resistance pattern to know the trend over a period of thirty one months.
2. Materials and methods
A retrospective study of bacteria isolates and their antibiotic susceptibility from wound swabs of patients admitted to the Burn Care Unit of a tertiary care hospital, Jharkhand, India during January 2015 to July 2017 was carried out. The pus/wound swabs obtained from individual burnt patients were processed to identify the bacteria and determine their antibiotic susceptibility. Wound swabs were collected and transported to the laboratory. Swabs were cultured aerobically in blood agar (Hi-Media, India) and CLED agar (Hi-Media, India). Identification and antibiotic susceptibility of positive cultures was done by VITEK® 2 compact system (bioMerieux, USA) using the ID-GNB, AST-N280 cards (bioMerieux, USA) for Gram's negative bacteria and ID-GPB, AST-P628 (bioMerieux, USA) for Gram's positive bacteria, in accordance with the manufacturer's instructions. Briefly, single colony was taken and made suspension in normal saline. The OD of bacterial suspensions was adjusted to 0.5. Finally the Vitek Tubes were shaking well before putting in Vitek machine to maintain homogenous suspension. The antimicrobial susceptibility testing card contained following antibiotics: AMPICILLIN, AMOXICILLIN/CLAVULANIC ACID, AMIKACIN, CEFTRIAXONE, CIPROFLOXACIN, CO-TRIMAXAZOLE, CEFOPERAZONE/SULBACTAM, COLISTIN, CEFUROXIME, GENTAMICIN, IMIPENEM, MEROPENEM, NITROFURAN, PIPERACILLIN/TAZOBACTAM and TIGECYCLINE. Escherichia coli ATCC 25922, Staphylococcus aureus ATCC 25923 and Pseudomonas aeruginosa ATCC 27853 were used as controls. Results were interpreted as recommended by the Clinical and Laboratory Standards Institute (CLSI) in 2015 and 2016 (M100-S24 and M100S, 26th Ed.). The MIC breakpoint used to identify bacteria susceptible for colistin was 2 mg/l and tigecycline was 1 or 2 mg/l [10].
3. Statistical analysis
Data was stored and managed in Microsoft Excel. All the Statistical calculations and plots were executed using R programming. The logistic regression was performed in GUIDeducerR [11].
4. Results
Two hundred and ninety nine pus samples were received from wounds of burnt patients. These samples were collected from 169 females and 130 males. The median age of the patient was 35 (range 1 year–79 years). Out of the total wound swabs collected, 61.87% (n = 185) were culture positive for bacterial infection in which 112 were from female patients and 73 were from male patients (Table 1). The distribution of patients with positive bacterial infection showed that16.76%, 35.68%, 22.7%, 15.14% and 9.73% patients were of 0–15 year's age group, 16–30 years age group, 31–45 years age group, 46–60 years age group and greater than 60 years of age respectively (Table 2). In case of female, the highest numbers of bacterial infections 41.96% (47/112) were found in the age group of 16–30 years whereas in male the highest numbers of bacterial infection 26.03% (19/73) were found in the age group of both 16–30 and 31–45 years. Logistic regression analysis was performed to evaluate the relation of infection with sex and age. The odds of getting an infection for a female burnt patients is approximately 65% higher than male burnt patients (p value = 0.045). There was no significant relationship of bacterial infection with age (p value = 0.212) was observed (Table 3).
Table 1.
Detail of burn patients.
| Infected patients N (%) | Non-infected patients N (%) | |
|---|---|---|
| Patients (n) | 185 (61.87) | 114 (38.13) |
| Male | 73 (39.46) | 57 (50) |
| Female | 112 (60.54) | 57 (50) |
| Age (in years) | ||
| Range | 1–76 years | 1–82 years |
| Median | 29 years | 30.05 years |
| Average | 32.70 years | 34.51 years |
Table 2.
Age and sex distribution of burn patients with positive bacterial wound infection.
| Age (in years) | Male N (%) | Female N (%) | Total N (%) |
|---|---|---|---|
| 0–15 | 24 (32.87) | 7 (5.73) | 31 (16.93) |
| 16–30 | 19 (26.02) | 47 (38.52) | 66 (35.67) |
| 31–45 | 19 (26.02) | 23 (18.15) | 42 (22.70) |
| 46–60 | 9 (12.32) | 19 (15.57) | 28 (15.13) |
| >61 | 2 (2.73) | 16 (13.11) | 18 (9.73) |
Table 3.
Logistic regression for presence of bacterial infection in wounds of burn patients.
| β Coefficient | St Error | Z-Value | P-Value | OR | 95% CI | |
|---|---|---|---|---|---|---|
| Age | -0.008 | 0.007 | -1.247 | 0.212 | 0.992 | 0.979–1.004 |
| Gender | 0.503 | 0.250 | 2.008 | 0.045 | 1.653 | 1.014–2.709 |
Predictor values are coded as follows: Female = 1, Male = 0; Bacteria growth in pus sample (Infection) = 1, No growth (No infection) = 0.
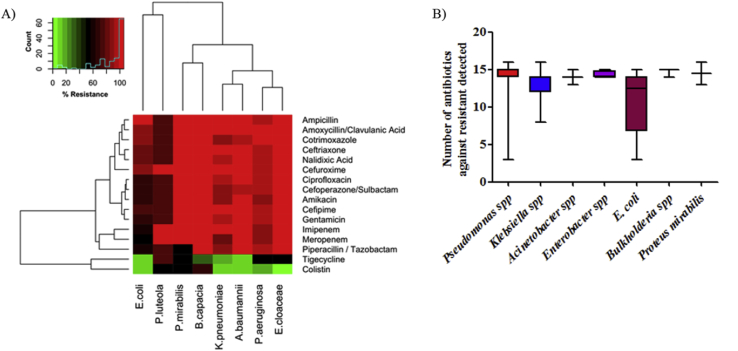
Identification of positive bacterial cultures revealed the presence of eight genera belonged to Pseudomonas, Klebsiella, Acinetobacter, Escherichia, Enterobacter, Burkholderia, Staphylococcus and Proteus (Table 4). Pseudomonas spp (43%) was the commonest pathogen isolated from wound of burnt patients followed by Klebsiella pneumoniae (28%), Acinetobacter baumannii (14.83%), E. coli (6.59%), Enterobacter cloacae (2.20%), Burkholderia cepacia (1.65%), Staphylococcus aureus (1.62%) and Proteus mirabilis (1.10%). Antimicrobial sensitivity data for the most frequently isolated organisms are shown in Figure 1/Table 5. Antibiotic susceptibility result showed that 71.25% (N = 57) and 16.25% (n = 13) Pseudomonas spp isolates were extensively drug-resistant and pandrug resistant respectively (Table 5). Extensively drug-resistant Pseudomonas spp were susceptible to COLISTIN (82.5%). All the K. pneumoniae isolates were resistant to AMPICILLIN, CEFUROXIME, CEFTRIAXONE and CEFEPIME. Out of 51 K pneumoniae isolates, 29.41% (n = 15) were labeled as multi drug-resistant and 68.63% (n = 35) were labeled as extensively drug-resistant respectively. Among XDR K. pneumoniae isolate, 17.14% (n = 6) were susceptible to only COLISTIN, 5.71% (n = 2) were susceptible to only TIGECYCLINE and 74.29% (n = 26) were susceptible to both COLISTIN/TIGECYCLINE. Except two, all Acinetobacter baumannii isolates were labeled as extensively drug-resistant and were susceptible to COLISTIN (92.59%) and TIGECYCLINE (88.89%). In E. coli isolates, the highest resistance was seen for AMPICILLIN (100%) followed by CEFUROXIME (91.67%), TRIMETHOPRIM/SULFAMETHOXAZOLE (91.67%), CEFTRIAXONE (83.33%), AMOXYCILLIN/CLAVULANIC ACID (83.33%), CEFEPIME (75%), CEFOPERAZONE/SULBACTAM (66.67%), AMINOGLYCOSIDES (66.67%), CIPROFLOXACIN (66.67%) and PIPERACILLIN/TAZOBACTAM (58.33%). Low resistance levels were seen for MEROPENEM (50%), TIGECYCLINE (8.33%) and COLISTIN (8.33%). Enterobacter cloacae isolates were resistant to all tested antibiotic except COLISTIN and TIGECYCLINE. Burkholderia cepacia and Proteus mirabilis isolates were only susceptible to TIGECYCLINE.
Table 4.
Distribution of micro-organismsisolated from burn wound.
| Species | Abundance (%) | XDR N (%) |
PDR N (%) |
|---|---|---|---|
| Pseudomonas aeruginosa | (43.0) | 56 (72.72%) | 11 (14.28%) |
| Klebsiella pneumoniae | (28.0) | 35 (68.63%) | 1 (1.96%) |
| Acinetobacter baumannii | (14.83) | 25 (92.59%) | 2 (7.41%) |
| Escherichia coli | (6.59) | 7 (58.33%) | 0 |
| Enterobacter sp. | (2.2) | 4 (100%) | 0 |
| Pseudomonas luteola | (2.18) | 1 (25.0%) | 2 (50.0%) |
| Burkholderia cepacia | (1.65) | 3 (100%) | 0 |
| Staphylococcus aureus | (1.62) | 0 | 0 |
| Proteus mirabilis | (1.1) | 2 (100%) | 0 |
Figure 1.
Antibiotic resistant pattern of bacteria isolated from burn wound (A) Heat Map of antibiotic susceptibility pattern of bacteria isolated from burnt patients and (B) Box plot of number of antibiotic resistance detected in bacteria.
Table 5.
Relative frequency of resistance (%) to antibiotics in bacteria prevalent in burn wounds.
| Antibiotics | E. coli | Proteus mirabilis | Burkholderia cepacia | Pseudomonas species | Klebsiella pneumoniae | Acinetobacter baumannii | Enterobacter cloacae |
|---|---|---|---|---|---|---|---|
| Amoxycillin/Clavulanic Acid | 81.81 | 100 | 100 | 100 | 100 | 100 | 100 |
| Ceftriaxone | 81.81 | 100 | 100 | 97.37 | 100 | 100 | 100 |
| Nalidixic Acid | 81.81 | 100 | 100 | 96.05 | 92.16 | 100 | 100 |
| Cefepime | 72.72 | 100 | 100 | 94.74 | 100 | 100 | 100 |
| Gentamicin | 72.73 | 100 | 100 | 94.74 | 98.04 | 100 | 100 |
| Amikacin | 72.73 | 100 | 100 | 89.47 | 90.2 | 100 | 100 |
| Ampicillin | 100 | 100 | 100 | 98.68 | 100 | 100 | 100 |
| Cefuroxime | 90.91 | 100 | 100 | 96.05 | 100 | 100 | 100 |
| Co-trimoxazole | 90.91 | 100 | 100 | 100 | 89.8 | 96.3 | 100 |
| Ciprofloxacin | 63.64 | 100 | 100 | 96.05 | 88.24 | 100 | 100 |
| Cefoperazone/Sulbactam | 63.64 | 100 | 100 | 92.11 | 88.24 | 96.3 | 100 |
| Imipenem | 63.64 | 100 | 100 | 89.47 | 84.31 | 100 | 100 |
| Meropenem | 54.55 | 100 | 100 | 90.79 | 78.43 | 100 | 100 |
| Piperacillin/Tazobactam | 63.64 | 50 | 100 | 96.05 | 88.24 | 100 | 100 |
| Tigecycline | 9.09 | 50 | 0.00 | 50 | 11.76 | 11.11 | 25 |
| Colistin | 9.09 | 50 | 66.67 | 15.79 | 7.84 | 7.41 | 0.00 |
5. Discussion
Nosocomial infection in the burnt patients is major challenge for a clinician [12]. It has been estimated that 75% of all deaths in burnt patients were associated with infections [13]. Prolonged use of antibiotics leads to the development as well as selection of multidrug resistant (MDR) bacteria which results in treatment failure and intensifies the complications. Thus, the information of microbial flora and the current antibiotic susceptibility patterns are important for the clinician treating burn sepsis. In the present retrospective study, one hundred and eighty five (61.87%) bacteria have been isolated from the wounds of burnt patients were analyzed. Among the culture positive samples, 112 (60.54%) were from female patients and 73 (39.46%) were from male patients. The most commonly isolated organisms were Pseudomonas species (43%). Klebsiella pneumoniae and Acinetobacter baumannii were second and third predominant bacterial pathogen with a prevalence of28% and 14.83% respectively. Similar finding with P. aeruginosa a predominant isolate followed by K. pneumoniae and A. baumannii in a tertiary care hospital in India were also reported [14, 15, 16, 17]. High prevalence of these pathogens is associated with their ability to flourish well in a moist environment and persistence in hospital environment [18, 19]. A number of studies showed that Staphylococcus aureus to be a predominance etiological agent in burn wound infection [20, 21]. However, in India, incidence of S. aureus infection was quite significant but was next only to Pseudomonas spp [14, 15, 16, 17]. In contrast, in the present study we found very less number of S. aureus in burn wounds with a prevalence of 1.62% (n = 3). The variation in isolated bacteria in burn wounds has been attributed to the difference in treatment practices in the different geographical locations.
The antibiogram studies indicate the emergence of extensively drug-resistant and pandrug resistant strains. The isolates were exhibited resistance to the commonly used antibiotics as well as new generation antibiotics. In contrast to other studies, the incidence of resistant to MEROPENEM (83.78%), IMIPENEM (85.41%) AMIKACIN (87.03%) and CIPROFLOXACIN (89.19%) are much higher in our study [8, 16, 21, 22]. COLISTIN disrupts the structure of the outer membrane and hence not required active target for antibiotic action whereas TIGECYCLINE is a broad-spectrum semisynthetic glycylcycline which is shortly introduced commercially with high susceptibility rate [23]. These two antibiotics are currently used as the last resort to the treatment of Carbapenem resistant gram-negative bacterial infections.
In this study, 12.95% and 28.64% isolates were resistant to COLISTIN and TIGECYCLINE respectively. This derivation is definitely alarming as colistin and tigecycline resistant isolates increase the catastrophic effect with reduced treatment options. Such high resistance may be because of the excessive usage of these antibiotics in the hospitals that leads to the development of multiple drug resistance pathogens.
The present study based on the automated VITEK 2 system for identification and antimicrobial susceptibility testing of the isolated that provide accurate result in tests for most of the clinical isolates and remove the requirement of human analysis and error of results [24]. However, it is mandatory to calibrate the system with ATCC strains to update the software. As there is no as well as required MIC determination of colistin by broth dilution method.
6. Conclusion
The dryness in the pipeline of new antibiotic and emergence of extensively drug-resistant as well as pandrug resistant strains pointing the current need toward active microbial surveillance in all clinical settings and prudent use of antibiotics. Data regarding the prevalence of microorganisms and their resistance patterns would definitely benefit the clinician to prescribe appropriate antibiotics, especially in resource limited countries. This also helps in formulating policies for empirical antimicrobial therapy to control infections.
Declarations
Author contribution statement
Minakshi Gupta: Performed the experiments; Contributed reagents, materials, analysis tools or data.
Aman Kumar Naik: Analyzed and interpreted the data.
Santosh Kumar Singh: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work was supported by the TATA Main Hospital.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The study design was conceived and planned by Dr. Santosh Kumar Singh and Dr. Minakshi Gupta. Dr. Minakshi Gupta performed microbiological work and Aman Kumar Naik performed statistical data analysis. Support extended by Mr. Joydeep Mukherjee (M.A in English, B. ED) for English and Grammar correction of manuscripts is sincerely acknowledged. I would like to acknowledge Mr. A. K. Biswas from TMH for his technical help to retrieve data.
References
- 1.Church D., Elsayed S., Reid O., Winston B., Lindsay R. Burn wound infections. Clin. Microbiol. Rev. 2006 Apr 1;19(2):403–434. doi: 10.1128/CMR.19.2.403-434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rowan M.P., Cancio L.C., Elster E.A., Burmeister D.M., Rose L.F., Natesan S., Chan R.K., Christy R.J., Chung K.K. Burn wound healing and treatment: review and advancements. Crit. Care. 2015 Dec;19(1):243. doi: 10.1186/s13054-015-0961-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garner J.S., Jarvis W.R., Emori T.G., Horan T.C., Hughes J.M. CDC definitions for nosocomial infections, 1988. Am. J. Infect. Contr. 1988 Jun;16(3):128–140. doi: 10.1016/0196-6553(88)90053-3. [DOI] [PubMed] [Google Scholar]
- 4.Avni T., Levcovich A., Ad-El D.D., Leibovici L., Paul M. Prophylactic antibiotics for burns patients: systematic review and meta-analysis. Bmj. 2010 Jan 1;340 doi: 10.1136/bmj.c241. c241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kramer A., Schwebke I., Kampf G. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect. Dis. 2006 Dec;6(1):130. doi: 10.1186/1471-2334-6-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh S.K., Mishra M., Sahoo M., Patole S., Sahu S., Misra S.R., Mohapatra H. Antibiotic resistance determinants and clonal relationships among multidrug-resistant isolates of Klebsiella pneumoniae. Microb. Pathog. 2017 Sep 1;110:31–36. doi: 10.1016/j.micpath.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Magiorakos A.P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbarth S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., Paterson D.L. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012 Mar;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 8.Mehta M., Dutta P., Gupta V. Bacterial isolates from burn wound infections and their antibiograms: a eight-year study. Indian J. Plast. Surg. 2007 Jan 1;40(1):25. [Google Scholar]
- 9.Otta S., Dash J.K., Swain B. Aerobic bacteriology of burn wound infections. CHRISMED J. Health Res. 2015 Oct 1;2(4):337. [Google Scholar]
- 10.Vasoo S. Susceptibility testing for the polymyxins: two steps back, three steps forward? J. Clin. Microbiol. 2017 Sep 1;55(9):2573–2582. doi: 10.1128/JCM.00888-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fellows I. Deducer: a data analysis GUI for R. J. Stat. Softw. 2012 Jun 30;49(8):1–5. [Google Scholar]
- 12.Ekrami A., Kalantar E. Bacterial infections in burn patients at a burn hospital in Iran. Indian J. Med. Res. 2007 Dec 1;126(6):541. [PubMed] [Google Scholar]
- 13.Srinivasan S., Vartak A.M., Patil A., Saldanha J. Bacteriology of the burn wound at the BaiJerbaiWadia hospital for children, Mumbai, India—a 13-year study, Part IBacteriological profile. Indian J. Plast. Surg. 2009 Jul;42(2):213. doi: 10.4103/0970-0358.59284. official publication of the Association of Plastic Surgeons of India. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dash M., Mishra P., Routray S. Bacteriological profile and antibiogram of aerobic burn wound isolates in a tertiary care hospital, Odisha, India. Int. J. Med. Med. Sci. 2013;3:460–463. [Google Scholar]
- 15.Bandekar N., Vinodkumar C.S., Basavarajappa K.G., Prabhakar P.J., Nagaraj P. Beta lactamases mediated resistance amongst Gram-negative bacilli in burn infection. Int. J. Biol. Med. Res. 2011;2:766–770. [Google Scholar]
- 16.Singh N.P., Goyal R., Manchanda V., Das S., Kaur I., Talwar V. Changing trends in bacteriology of burns in the burns unit, Delhi, India. Mol. Biol. Rep. 2003;29:129–133. doi: 10.1016/s0305-4179(02)00249-8. [DOI] [PubMed] [Google Scholar]
- 17.Rajput A., Singh K.P., Kumar V., Sexena R., Singh R.K. Antibacterial resistance pattern of aerobic bacteria isolates from burn patients in tertiary care hospital. Biomed. Res. 2008;19:1998–2001. [Google Scholar]
- 18.Atoyebi O.A., Sowemimo G.A., Odugbemi T. Bacterial flora of burn wounds in Lagos, Nigeria: a prospective study. Burns. 1992;18(6):448–451. doi: 10.1016/0305-4179(92)90175-t. [DOI] [PubMed] [Google Scholar]
- 19.de Abreu P.M., Farias P.G., Paiva G.S., Almeida A.M., Morais P.V. Persistence of microbial communities including Pseudomonas aeruginosa in a hospital environment: a potential health hazard. BMC Microbiol. 2014 Dec;14(1):118. doi: 10.1186/1471-2180-14-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ozumba U.C., Jiburum B.C. Bacteriology of burn wounds in Enugu, Nigeria. Burns. 2000;26:178–180. doi: 10.1016/s0305-4179(99)00075-3. [DOI] [PubMed] [Google Scholar]
- 21.Guggenheim M., Zbinden R., Handschin A.E., Gohritz A., Altintas M.A., Giovanoli P. Changes in bacterial isolates from burn wounds and their antibiograms: a 20-year study (1986-2005) Burns. 2009;35:553–560. doi: 10.1016/j.burns.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Kulkarni V., Arali S.M., Jayaraj Y.M., Shivannavar C.T., Joshi M.R. Bacterial etiology and their antibiogram in burn wound infections at Kalaburgi region (India) Indian J. Burns. 2015 Jan 1;23(1):65. [Google Scholar]
- 23.Petrosillo N., Taglietti F., Granata G. Treatment options for colistin resistant Klebsiella pneumoniae: present and future. J. Clin. Med. 2019 Jul;8(7):934. doi: 10.3390/jcm8070934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanders C.C., Peyret M., Moland E.S., Cavalieri S.J., Shubert C., Thomson K.S., Boeufgras J.M., Sanders W.E. Potential impact of the VITEK 2 system and the Advanced Expert System on the clinical laboratory of a university-based hospital. J. Clin. Microbiol. 2001 Jul 1;39(7):2379–2385. doi: 10.1128/JCM.39.7.2379-2385.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]