Graphical abstract
Keywords: CRISPR, Guide RNA design, Efficiency, Specificity, Machine-learning
Abstract
The Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/ CRISPR-associated (Cas) system has emerged as the main technology for gene editing. Successful editing by CRISPR requires an appropriate Cas protein and guide RNA. However, low cleavage efficiency and off-target effects hamper the development and application of CRISPR/Cas systems. To predict cleavage efficiency and specificity, numerous computational approaches have been developed for scoring guide RNAs. Most scores are empirical or trained by experimental datasets, and scores are implemented using various computational methods. Herein, we discuss these approaches, focusing mainly on the features or computational methods they utilise. Furthermore, we summarise these tools and give some suggestions for their usage. We also recommend three versatile web-based tools with user-friendly interfaces and preferable functions. The review provides a comprehensive and up-to-date overview of computational approaches for guide RNA design that could help users to select the optimal tools for their research.
1. Introduction
Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/CRISPR-associated (Cas) systems, such as Cas9 [46] and Cas12a (formerly Cpf1) [118], are the primary tools used for genome editing due to their various abilities to manipulate, detect, and image certain DNA and RNA sequences in the cell [50]. The CRISPR/Cas system was first adapted for genome editing in 2012 [31], [46], and subsequent studies have transformed the CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) into a single guide RNA (sgRNA) that can bind to both the Cas9 protein and the target DNA sequence. Cas9 protein and sgRNA complex first scans the appropriate PAM sequence and binds to the targeted genome loci, then the activated HNH and the RuvC nuclease domain of Cas9 function to make a DNA double-strand break (DSB) in the specific region [22], [69].
The most frequently used CRISPR nuclease is type II endonuclease Cas9, which recognises the 5′-NGG-3′ PAM (SpCas9) [73]. Another popular nuclease is CRISPR type V endonuclease Cas12a (Cpf1), which recognises the 5′-TTTV-3′ PAM, and shows high efficiency in both animal and plant organisms [25], [48], [49], [71], [97], [118], [124]. Recently, several other Cas family proteins have also been discovered and adapted for DNA or RNA editing events, including Cas12b, Cas13a and Cas14 [2], [37], [94].
CRISPR-based gene editing is implemented with sequence-specific nucleases (SSNs) and a sgRNA to achieve precise gene knock-out (KO) or gene knock-in (KI). Additionally, researchers developed a catalytically inactive Cas9 (dCas9) that loses endonuclease activity, and has been adapted for gene expression regulation (CRISPRa/i) and 3D genome studies [33], [66], [67], [70], [84], [88]. Furthermore, base editing using modified nCas9 has greatly broadened application of the CRISPR system [32], [52], [123]. Compared with previous mature gene editing tools such as zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) [9], [10], [13], [100], [121], which use engineered proteins to target and cleave specific genome loci, CRISPR is lower in cost of both time and money. This advancing technology is increasingly being deployed, and has great potential for clinical detection, gene therapy and agricultural improvement [3], [21], [50], [126].
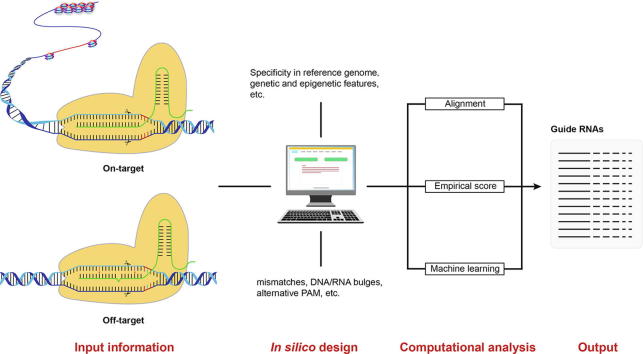
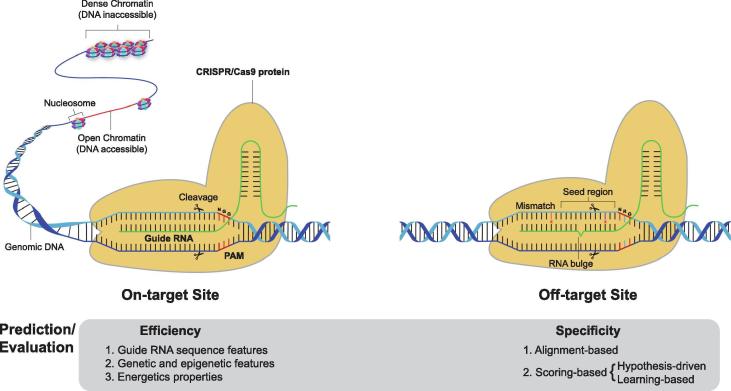
However, two major challenges hinder the development and application of the CRISPR/Cas system: potential off-target effects, and on-target efficiency (Fig. 1) [112], [120]. Successful CRISPR guide RNA (gRNA) design can resolve these issues [23], [96], and powerful computational approaches facilitate in silico gRNA design [19], [34], [104], thereby enabling the design of specific gRNAs for particular experiments.
Fig. 1.
Schematic diagram of CRISPR/Cas9 system at both on-target site and off-target site.
In this review, we summarise existing approaches for CRIPSR guide RNA design and evaluation, and assist users in choosing favourable tools for their research. Moreover, it aims to make users aware of the latest computational CRISPR tools and resources.
2. Evaluation of CRISPR cleavage efficiency
In theory, the CRISPR/Cas protein scans the PAM sequence, and sgRNA recognises target loci and activates endonuclease activity to cleave specific sites. However, cleavage efficiency varies greatly among different target sites and/or cell lines [14], [21], [27], [28], [51], [78], [87], [98], [99], [103], [115], [117], [122], [125], suggesting that several features may influence the binding and cutting efficacy of the sgRNA-Cas complex. Numerous studies have revealed that gRNA sequence features (sequence composition, nucleotide position, GC content), genetic and epigenetic features (chromatin accessibility, gene expression) and energetics properties (RNA secondary structure, melting temperature, free energy) all contribute to gRNA efficacy. Based on these features, many computational tools have been developed for designing highly efficient gRNAs. Herein, we introduce these tools based on their features, and evaluate their efficiency (Table 1).
Table 1.
Computational methods for evaluation of guide RNA efficiency.
| Tool | Enzymes | Data source | Main features | Quantitative metrics |
|---|---|---|---|---|
| E-CRISP [38] | Cas9 | – | SC, GF | – |
| CRISPRscan [78] | Cas9, Cpf1 | Zebrafish | SC | Spearman correlation = 0.309, from [36] |
| evaluateCrispr [40] | Cas9 | Drosophila | SC | Spearman correlation = 0.074, from [36] |
| sgRNAScorer [14], [15] | Cas9, Cpf1 | Human | SC, EGF | Spearman correlation = 0.225, from [36] |
| SSC [112] | Cas9 | Human, Mouse | SC | Spearman correlation = 0.274, from [36] |
| WU-CRISPR [106] | Cas9 | Human, Mouse | SC, EP | Spearman correlation = 0.215, from [36] |
| Azimuth [23], [29] | Cas9 | Human, Mouse | SC, GF, EP | Spearman correlation = 0.366, from [36] |
| CRISPRater [55] | Cas9 | Human | SC, GF | Pearson correlation = 0.399, from [55] |
| CRISPRpred [86] | Cas9 | Human, Mouse | SC, EP | ROC-AUC = 0.85, from [86] |
| CASPER [75] | Cas9, Cpf1 | – | SC | Pearson correlation = 0.443, from [75] |
| DeepCpf1 [47] | Cpf1 | Human | SC, EGF | Spearman correlation = 0.748, from [47] |
| TSAM [82] | Cas9 | Human, Mouse, Zebrafish | SC, GF, EP | Spearman correlation = 0.4, from [82] |
| TUSCAN [105] | Cas9 | Human, Mouse, Zebrafish | SC, GF | Spearman correlation = 0.12, from [105] |
| uCRISPR [119] | Cas9 | – | SC, EP | Spearman correlation = 0.3, from [119] |
SC, sequences composition; GF, genetic features; EGF, epigenetic features; EP, energetics properties.
2.1. Guide RNA sequence features
The nucleotide composition of a target sequence is one of the most important determinants of gRNA efficiency [24], [103], [107], [112]. Large-scale CRIPSR-based screens in mammals have shown that guanines are preferred in positions 1 and 2 before the PAM sequence [103], while thymines are disfavoured within +/-4 nucleotides surrounding the PAM sequence [107]. Additionally, sequences downstream of PAMs can influence gRNA efficiency, while sequences upstream have no significant effect [24]. Cytosine is preferred at the CRISPR/Cas9 cutting site (-3 position proximal to PAM) [21], [112], and the GC content of the region 4–13 bases downstream of the PAM sequence contributes to gRNA efficiency. Based on this key information, several efficiency prediction models have been constructed.
Rule Set 1 is a predictive model built using data derived from 1,841 sgRNAs in human and mouse [24], the score is predicted by a support vector machine (SVM) model, and a supervised learning method classifies data in a generalised linear manner. Rule Set 1 is mainly used to investigate sequence features that influence CRISPR cutting efficiency. Results predicted by this model show a high correlation with experimental results. To improve the accuracy, the authors adapted more datasets and built a new model in 2016 called Rule Set 2 [23]. In this model, position-independent nucleotide counts and the location of the sgRNA target site within the gene were considered to improve predictions based on their observations. These optimised models were applied for gRNA design for both CRISPR KO and CRISPRa/i experiments. A package to predict the gRNA efficiency based on the models was also developed and implemented in Broad Institute GPP sgRNA Designer [29].
To unravel the nucleotide preference of gRNA target sites in different CRISPR-based editing events, both CRISPR KO and CRISPRa/i libraries in mammals were screened [112], and significant differences in nucleotide preference between CRISPR KO and CRISPRa/i were detected. Elastic Net is a regularised regression method for fitting and classifying data that performs better than SVM in some cases [128]. The Elastic Net algorithm was used to construct models for both CRISPR KO and CRISPRa/i, and they were applied in Spacer Scoring for CRISPR (SSC) software to predict the efficiency of gRNA. Platforms such as E-CRISP, CHOPCHOP and CRISPR-FOCUS also include this model [12], [38], [57].
Moreno-Mateos and his colleagues observed that the loading and activity of sgRNA increased with guanine enrichment and adenine depletion [78]. They measured >1,000 sgRNAs targeting 128 genes in zebrafish and used the logistical regression method to construct a predictive model that was integrated into CRISPRscan. WU-CRISPR takes advantage of this data and adds some novel features [24], [106], resulting in a model with higher precision than several other predictive models [14], [24], [112]. Labuhn et al. identified PAM-distal GC content-dependent activity and constructed a model named CRISPRater [55] that was integrated into CRISPR/Cas9 target online predictor (CCTop), a platform for CRISPR target prediction [93].
The Church laboratory developed software called sgRNA scorer to calculate sgRNA on-target scores based on their SVM model [14]. A second version of the sgRNA scorer software was proposed that improved the on-target prediction power and added prediction for other Cas systems such as SaCas9 and AsCpf1 [15]. Housden et al. then used a drug target method to screen CRISPR KO efficiency in Drosophila and developed an efficiency prediction tool [40]. CASPER integrated scores from CRISPRscan and added some new features to maximise correlations between scores and on-target experimental data [75]. This tool can also detect off-target scores and perform multipopulational analysis.
2.2. Genetic and epigenetic features
Genetic and epigenetic features like chromatin accessibility, gene position and expression are also important factors that influence sgRNA binding and Cas nucleases cleavage. Many researches have demonstrated that nucleosomes inhibit Cas9 target cleavage, and DNase I hypersensitivity and epigenomic markers alter gRNA efficacy [23], [39], [44], [101], [116]. Based on these features, several tools have been developed. By borrowing knowledge from oligonucleotide design and nucleosome occupancy models, an R package called predictSGRNA was proposed for evaluation of sgRNA efficacy [53], and this performed better than other models such as Azimuth and sgRNA scorer [14], [23], [29].
Non-homologous end joining (NHEJ) and microhomology-mediated end joining (MMEJ) are two major pathways which produce heterogeneous repair outcomes when repairing Cas9-mediated DSBs. People use CRISPR/Cas9 system to knock out genes by inducing indels into target genome location. However, CRISPR-based gene KO may induces in-frame variants in which gene functions are retained. Thus, microhomology-based prediction of CRISPR on-target efficiency should be considered. To this end, Bae et al. developed Microhomology-Predictor to improve KO efficiency by reducing in-frame editing [7]. Recent researches have showed that template-free Cas9-editing outcomes are predictable. inDelphi was the first model for precise prediction of CRISPR editing genotype [90]. Soon after, a computational predictor called FORECasT was developed using >40,000 sgRNAs in different cell lines [5]. It was shown that most reproducible mutations are single base insertion, short deletions or longer microhomology-mediated deletions, in addition, Cas9-editing outcomes were cell-line-dependent. The Shendure laboratory also built a predictive model called Lindel for prediction of the insertions and deletions of CRISPR/Cas9-mediated DSB repair based on local sequence context [16]. All these approaches are sure to assist users in guide RNAs selection for gene disruption.
CRISPR-Cpf1 achieves high efficiency and suffers minor off-targets, besides, Cpf1 prefers AT-enriched regions. Hence, more and more studies have adapted Cpf1 for KO screens. However, models for evaluation of Cpf1 cleavage efficiency are lacking. DeepCpf1 is an algorithm especially for prediction of Cpf1 activity [47]. It is implemented within the deep-learning framework and chromatin accessibility data. This program can significantly improve the accuracy of Cpf1 activity prediction. In addition, models for other Cas experiments, such as Cas9, xCas9, and base edition are also provided on the author’s GitHub (https://github.com/MyungjaeSong/Paired-Library). CRISPR-DT is a recently developed platform for prediction of Cpf1 target efficiency [127]. This SVM model displayed better performance than the deep learning-based model employed in DeepCpf1.
2.3. Energetics properties
The energetics associated with the formation of the DNA, gRNA and Cas protein complex are regular and can be analysed to eliminate bias among different models, since some energetics methods may better illustrate the Cas9 editing efficacy [95], [107], [113]. CRISPRpred includes the positions of nucleotides as well as secondary structures of sgRNAs to predict the cleavage efficiency, and this performs better than Rule Set 1 [24], [86]. Zhang et al. recently demonstrated a free energy scheme called uCRISPR for evaluating the Cas9 editing efficacy, as well as off-target effects [119]. This model is thought to apply to any cleavage-activity Cas9 dataset.
2.4. Other considerations for gRNA efficiency
Of note, these predictive models were trained by individual experiments and rules, and each model generates different on-target scores for sgRNAs, hence users must be careful when evaluating or designing guide RNAs using these models in their own experiments. However, several key features are reliable, such as guanine preference, GC content, seed region and the secondary structure of gRNAs [65], [91], [106], [112].
The accuracy of different models is controlled by their learning methods or the approaches of CRISPR activity measurement. Doench et al. tested multiple training methods and selected the best-performing one as the kernel of their model [23]. TUSCAN, a random forest-based model, outperformed models built solely by linear regression [78], [105]. A two-step averaging method (TSAM) for the regression of cleavage efficiencies also performed better than many other models [14], [23], [82], [112]. Additionally, measurement using sequencing data rather than phenotypic data may generate fewer false positive results, albeit at a cost [105].
It is critical for users to know which tool best suits their research. A comprehensive evaluation of different efficiency prediction tools was conducted to examine differences among models [36], and it revealed differences in the correlation between different datasets and models. Furthermore, no model performed excellently across all datasets, suggesting that a careful selection of CRISPR gRNA design tools is necessary. Users can also evaluate gRNAs using multiple models and select the best one for their experiments. Among the available tools, Rule Set 2 and DeepCpf1 are the most used and accurate scoring methods for evaluating Cas9 and Cpf1 cutting efficacy, and uCRISPR may be more accurate than some other methods, but it requires further experimental testing.
3. Prediction of CRISPR cutting specificity
The main obstacle for the application of CRISPR is off-target effects. CRISPR nucleases may cleave unintended genomic sites and cause unexpected mutations due to sgRNAs recognising DNA sequences with a few mismatches and/or DNA/RNA bulges, referred to as off-target cleavage [41], [120]. Off-target effects can be effectively relieved by predicting CRISPR cutting specificity and designing optimal gRNAs [41]. To predict the specificity of CRISPR gRNAs, two main methods have been proposed: (1) alignment-based method. Based on conventional or specialized algothrims, gRNAs are aligned to a given genome and off-target sequences and sites are returned. This method is mainly used for find out all potential off-targets in silico, (2) Scoring-based method. sgRNAs should be further scoring and ranking using identified off-targets from alignment process to select the most specific one for experiments. Two scoring approaches are shown: hypothesis-driven, where off-targets are scored based on the contribution of specific genome context factors to gRNA specificity; learning-based, where gRNAs are scored and predicted from a training model that considers the different features affecting specificity. These methods for prediction of gRNA specificity are discussed below and some of them are summarised in Table 2.
Table 2.
Computational methods for prediction of guide RNA specificity.
| Tool | Enzymes | Methods | Main features | Quantitative metrics |
|---|---|---|---|---|
| CasOT [108] | Cas9 | alignment | unlimited mismatch number, paired-gRNA mode, annotation | slow |
| Cas-OFFinder [8] | costom | alignment | unlimited mismatch number, GPU acceleration, web support | middle, fast (use GPU) |
| sgRNAcas9 [110] | Cas9 | alignment | max 5 mismatches, paired-gRNA mode, annotation, risk evaluation | slow |
| FlashFry [74] | costom | alignment | unlimited mismatch number, multiple on/off-target scores, annotation | fast |
| Crisflash [43] | Cas9 | alignment | unlimited mismatch number, variant data support | fast |
| MIT [41] | Cas9 | scoring | 20 bp sgRNA without PAM | ROC-AUC = 0.87, from [36] |
| CCTop [93] | Cas9, Cpf1 | scoring | empirically score based on number of mismatches | ROC-AUC = 0.77, from [36] |
| CFD [23] | Cas9 | scoring | 20 bp sgRNA with PAM (enable non-canonical PAM) | ROC-AUC = 0.91, from [36] |
| CRISPRoff [4] | Cas9 | scoring | energetics property and sequences composition | ROC-AUC = 0.98, from [4] |
| uCRISPR [119] | Cas9 | scoring | energetics property and sequences composition | Pearson correlation = 0.75, from [119] |
| CRISTA [1] | Cas9 | scoring | machine learning, sequences composition and epigenetic feature | ROC-AUC = 0.96, from [1] |
| Elevation [62] | Cas9 | scoring | machine learning, integrate both CFD model and epigenetic features | ROC-AUC = 0.98, from [62] |
| DeepCRISPR [18] | Cas9 | scoring | deep learning, sequences composition and epigenetic feature | ROC-AUC = 0.98, from [18] |
3.1. Alignment-based methods
In theory, potential off-target sites can be identified by aligning gRNA sequences to the reference genome based on sequence homology. Bowtie [59] and BWA [61] are traditional tools for short sequence alignment that are capable of off-target detection [36], [104]. However, there are several potential issues when using these tools. First, tools like Bowtie and BWA cannot identify small PAMs, since these alignment tools were developed for next-generation sequencing (NGS) read alignment. Second, these tools allow very limited mismatches in their seed regions, making alignment with default parameters impractical for identifying all potential off-target sites. A survey using bowtie2 [58] to detect off-targets failed to find all possible off-target sites, and could only find off-targets with up to one mismatch [23].
Despite the defects, some gRNA design methods utilise these tools, and parameters have been modified to fit the demand for off-target prediction. CCTop uses Bowtie to find off-target sites by first identifying PAM sites, and matches and mismatches in protospacer sequences are then searched for by bowtie with modified parameters [93]. Up to five mismatches are allowed in the protospacer as more mismatches may prevent DSB induction. CCTop also implements an off-target score for each candidate sgRNA. CHOPCHOP detects off-target sites using Bowtie with parameters -v and -L for searching sgRNA core regions [77]. CRISPOR uses BWA to find all potential off-target sites in iterative mode (“-N”), and can find all validated off-targets as well as Cas-OFFinder [8], [36].
In addition to these traditional tools, many other tools and algorithms have been developed for off-target site detection. Cas-OFFinder is one of the most popular tools for searching potential off-target sites, and advantages include no limit to the number of mismatches, PAM types, gRNA length or high running speed with GPUs. Cas-OFFinder can also predict off-target sites with 1 bp deletions or insertions (i.e. DNA/RNA bulges). CasOT is implemented to find Cas9 on-target sites from input sequences, as well as potential off-target sites with up to six mismatches in the seed region (12 nt adjacent to the PAM). This tool can also determine whether off-targets are within a coding exon [108]. Meanwhile, sgRNAcas9 utilises the ultrafast short sequence mapping tool SeqMap [45] to find off-targets, and classifies all sites into three categories to generate a final output of the best candidate gRNAs [110]. Recently, two new alignment-based tools have been developed. Crisflash utilises a tree-based algorithm to rapidly design CRISPR guide RNAs and optimise guide accuracy by incorporating user-supplied variant data [43]. FlashFry rapidly searches off-target sites and provides much useful information (annotation of off-target sites, on/off-target scores, GC content, etc.) for candidate gRNAs [74]. Here we still classified Crisflash and FlashFry as alignment-based methods since they both propose novel algorithms for off-target searching, whereas the scoring approaches they use are borrowed from others [23], [24], [41], [78].
Among these tools, Cas-OFFinder may be the best choice for identifying all potential off-target sites with any Cas nucleases, and FlashFry is also worth a try for its high speed and comprehensive outputs.
3.2. Scoring-based methods
3.2.1. Hypothesis-driven methods
Alignment-based methods are reliable for detection of most potential off-targets, however, not all nucleotide positions containing mismatches have the same decisive effect on off-target cleavage. Additionally, alignment-based prediction always outputs redundant off-target sites, many which are false-positives, although users can reduce the number of outputs by restricting the maximum mismatches when exploring off-target cleavage. One study compared experimentally validated off-targets and off-targets predicted by Cas-OFFinder and CCTop, and the results showed that off-targets detected by the prediction tools only covered some of the validated sites, while some off-target sites cannot be predicted solely based on sequence homology [11]. Thus, features that influence the nonspecific binding of CRISPR gRNAs need to be considered to increase the accuracy of off-target detection.
MIT (Hsu-Zhang) score was proposed for off-target evaluation during the early stages of gene editing by CRISPR. Hsu et al. evaluated >700 guide RNA variants and SpCas9-induced indel mutation levels at >100 predicted genomic off-target loci [41]. They evaluated the contributions made by different mismatch positions and numbers in the target 20 bp, and calculated a weight matrix to determine off-target efficiency for each sgRNA. The authors claimed that this score accounts for >50% of the variance in cutting frequency. The MIT score has been integrated into many CRISPR gRNA design tools such as CHOPCHOP and CRISPOR [36], [56].
Cutting frequency determination (CFD) is another popular score for off-target evaluation. It is noticeable that sgRNA can also bind genome loci with non-canonical PAMs such as NAG, NCG and NGA, which may cause off-target cleavage. Doench et al. added PAM features in their scoring metrics [23]. sgRNAs with mismatches and indels in target sequences were also included for examining correlations between sgRNAs and off-targets. CFD score was validated with GUIDE-seq and proved to perform better than MIT score, hence it was adopted by CRISPRscan [78], GuideScan [83] and CRISPOR [36].
The prediction of sgRNA specificity based on the structural features of the Cas9-sgRNA complex proved to be superior to prediction solely based on sequence features. CRISPRoff [4] and uCRISPR [119] both utilise energetics properties for off-target evaluation. Compared with other scoring methods like MIT and CFD, they both yielded better accuracy in off-target prediction. Nevertheless, neither have not been systematically evaluated by large-scale experiments, and caution should be exercised when using them.
3.2.2. Learning-based methods
Sometimes, empirical algorithms cannot effectively evaluate off-targets since they only consider minor features, whereas learning-based methods build complex models using combinations of multiple features, and they may better predict off-targets. In recent years, several new approaches for off-target prediction based on machine learning have been developed.
CRISPR Target Assessment (CRISTA) software uses BWA as the off-target search tool and implements multiple features (PAM type, nucleotide composition, GC content, chromatin structure, DNA methylation, RNA secondary structure, etc.) to predict cleavage propensity [1]. CRISTA exhibits better performance than MIT, CCTop and CFD.
The Doench laboratory also developed a linear regression model for prediction of off-target activity called Elevation that takes both sequence and chromatin accessibility features into consideration, as well as observations from others [62]. Elevation predicts individual scores for each off-target site, as well as an aggregate score for gRNAs. This method performs best among MIT, CFD and CCTop. However, this software can only compute off-target scores for the human exome (GRCh38) and cannot support other organisms on their website.
DeepCRISPR is a state-of-the-art computational platform that unifies sgRNA on-target and off-target site prediction into one framework with deep learning [18]. It identifies all possible sequence and epigenetic features that may affect sgRNA KO efficacy by learning from large data. This method was proved to surpass other available off-target prediction tools [23], [41], [92], [93].
3.3. Benchmark of scoring-based methods
Different scoring methods are based on different characteristics applied by each individual laboratory. In order to compare the accuracy of different scores, Haeussler et al. assessed four hypothesis-driven methods for off-target prediction using the same datasets [36]. CFD score exhibited the best prediction accuracy, whereas CCTop performed the worst. Data imbalance, where the number of true observed off-target sites (OTS) identified by off-target detection experiments is much less than that of all potential off-target sites recognized by alignment-based methods, is a common issue in the learning-based methods. To address the problem, a systematic survey was conducted to assess the influence of data imbalance [30]. The authors used bootstrapping sampling and PR-AUC methods to evaluate two well-established models. They emphasized the importance of using symmetric data for model construct and taking unbiased datasets for benchmark. According to current assessment results, we recommend people to use CFD score for off-target prediction. Elevation and DeepCRISPR are suitable for human genome editing, but CRISPRoff and uCRISPR may need further evaluation before use.
4. Tools for CRISPR guide RNA design
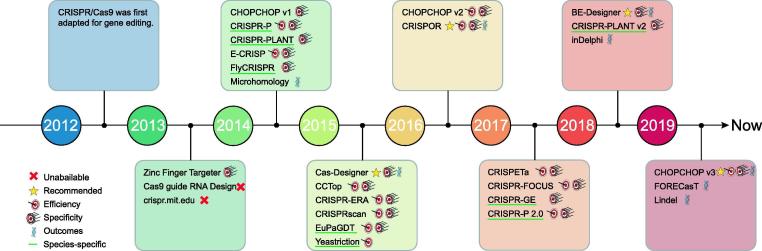
A CRISPR/Cas complex typically contains a Cas protein and a sgRNA, both of which determine the cutting activity. However, protein engineering is a complicated and uncertain strategy for most researchers, and optimising gRNAs is more feasible and efficient. A robust gRNA requires maximum on-target efficiency as well as minimum off-target activity. Basing on these two criteria, numerous computational tools have been developed for high-efficiency CRISPR gRNA design. However, each tool possesses distinct features, and user-friendliness is also important. To acquaint users with existing gRNA design tools, we provide an overview of most that are available based on the strength of their features, and this help users to make appropriate selections. Furthermore, we recommend several functional and user-friendly websites for gRNA design (Fig. 2, Table 3).
Fig. 2.
Timeline of the development of web-based tools for CRISPR guide RNA design.
Table 3.
Web-based CRISPR guide RNA design tools.
| Tool | Website |
|---|---|
| CHOPCHOP [56], [57], [77] | https://chopchop.cbu.uib.no |
| CRISPR RGEN Tools [42], [79] | http://www.rgenome.net/ |
| CRISPOR [20], [36] | http://crispor.tefor.net |
| E-CRISP [38] | http://www.e-crisp.org/E-CRISP |
| CCTop [93] | https://crispr.cos.uni-heidelberg.de |
| CRISPR-ERA [64] | http://CRISPR-ERA.stanford.edu |
| CRISPETa [85] | http://crispeta.crg.eu |
| CRISPRscan [78] | https://www.crisprscan.org |
| EuPaGDT [81] | http://grna.ctegd.uga.edu |
| CRISPR-P [60], [63] | http://crispr.hzau.edu.cn/CRISPR2/ |
| CRISPR-PLANT [76], [109] | https://www.genome.arizona.edu/crispr2 |
| CRISPR-GE [111] | http://skl.scau.edu.cn |
| inDelphi* [90] | https://www.crisprindelphi.design/ |
| FORECasT* [5] | https://partslab.sanger.ac.uk/FORECasT |
| Lindel* [16] | https://shendurelab.github.io/Lindel/ |
Tools marked by asterisk (*) are used for prediction of CRISPR editing outcomes.
4.1. A comprehensive overview of CRISPR guide RNA design tools
The CRISPR/Cas system was first adapted for gene editing in 2012, and several tools were developed immediately thereafter, such as Zinc Finger Targeter (ZiFiT) [89], Cas9 guide RNA Design [68] and CRISPR (http://crispr.mit.edu/) [41]. All these platforms have been implemented in the design of gRNAs for model organisms. ZiFiT is also used to design Zinc Finger and TALEN modules, but Cas9 guide RNA Design and CRISPR are now unavailable.
In the following years, various tools for CRISPR gRNA design emerged, due to both urgent demand for these tools, and because people have learned more and more about how the CRISPR system functions and what influences the efficiency and specificity of Cas cleavage. Many of these tools combine multiple scoring methods and provide alternative options for users. Some others have also proposed their own algorithms to rank designed sgRNAs, and this can assist users in gRNA selection. CHOPCHOP [56], [57], [77] provides alternative scores for users such as Rule Set ½ [23], [24], SSC [112], CRISPRscan [78] and deepCpf1 [47]. E-CRISP utilises its own SAE (Specificity, Annotation, Efficacy) score to determine the quality of each sgRNA, while Rule Set 1 [24] and SSC [112] are also included in E-CRISP. CCTop [93] assigns off-target scores for each sequence empirically, while the CRISPRater score [55] is also included to predict the efficiency of sgRNAs. CRISPOR [36] is a versatile platform that ranks gRNAs according to different scores for evaluating potential off-targets in the genome of interest, and for predicting on–target activity.
A large number of CRISPR/Cas-derived RNA-guided Endonucleases (RGENs) have been identified or modified to improve the cutting efficiency and enlarge the editing range. Some tools enable the design of gRNAs for RGENs. For example, Cas-Designer [79] allows users to choose 20 PAM types from different RGENs, while CRISPOR [20] also offers various PAMs from a defined list. To best serve the specialised purposes of different experiments, several web-based tools have been developed. CRISPR-ERA [64] and CHOPCHOP v3 [57] support sgRNA design for the CRISPRa/i system. CRISPETa [85] is mainly used for genome deletion with paired gRNAs, and BE-Designer [42] can be used for designing gRNA for base editing. Recently, three methods have been employed for successively predicting Cas9-editing outcomes: inDelphi [90], FORECasT [5] and Lindel [16]. These approaches can help to identify gRNAs based on forecasting results.
Researchers should choose suitable tools with caution since different tools are in favour of different genomes or cell types. For instance, Yeastriction [72] is specialised for yeast, FlyCRISPR [35] is specific for Drosophila, EuPaGDT [81] is recommended for eukaryotic pathogens, and CRISPR-P [60], [63], CRISPR-PLANT [76], [109] and CRISPR-GE [111] are all implemented for genome editing in plants. Also, user-friendliness is essential for these web-based design tools. Based on these considerations, we propose three comprehensive web-based platforms for CRISPR gRNA design.
4.2. Three comprehensive web platforms for CRISPR guide RNA design
4.2.1. CHOPCHOP
CHOPCHOP has a succinct interface with well-rounded functions, >200 genomes are available on the website, and users can provide gene names, genomic coordinates or target sequences as inputs. If a gene is given, users can set specific regions of the gene as targets, such as coding sequences or promoters. Before designing gRNAs, two off-target detection methods and seven efficiency scores can be chosen, which aids users in selecting optimal gRNAs for their research.
To satisfy different experimental purposes, CRISPR Cas9 nuclease/nickase, Cpf1, CasX, C2C2 and TALEN are all supported by CHOPCHOP, and various modes of DNA targeting are optional such as KO/KI, gene activation/repression, and nanopore enrichment. In activation/repression mode, gRNAs are designed to target the promoter region and its flanking sites in order to bring the activating/repressing domain into close proximity with the transcription start site [17], [54]. Meanwhile, nanopore enrichment mode is implemented to design sgRNAs for long fragment excision within 40 kb. Additionally, the inDelphi model has been integrated into CHOPCHOP for repair profile prediction by Cas9 in five cell types [90].
After clicking the ‘Find Target Sites’ button, a results table is shown, and each candidate gRNA has a rank, genomic information, GC content, self-complementarity score, mismatch number (0–3) and an efficiency score. A graphical view of all gRNAs is also provided in the UCSC Genome Browser, which helps users to select optimal gRNAs.
4.2.2. CRISPR RGEN tools
CRISPR RGEN tools, computational tools and libraries for RNA-guided endonucleases, are maintained by the Bae laboratory, and libraries comprise nine useful tools including Cas-OFFinder [8], Cas-Designer [79] and Digenome-Seq [80]. Herein, we mainly discuss the two gRNA design tools Cas-Designer and BE-Designer [42].
Compared with other designer tools, Cas-Designer allows DNA/RNA bulges when performing off-target detection. Additionally, this detection method is more rapid than others due to the Cas-OFFinder algorithm. Genomic sequences or coordinates and fasta file formats are allowed as inputs. Over 350 genomes and 20 PAM types are specified for users, and outputs include target sequences as well as out-of-frame score (calculate by microhomology), mismatch number (0–2) and off-target sites with up to 1 bp bulge. On/off-target sites are redirected to the Ensembl genome browser [26], which is capable of further inspection.
Unlike Cas-Designer, BE-Designer is primarily implemented for base editing. In this tool, four methods of base editing are specified, and the editing window region is also adjustable. After the design phase, three outcomes are based on different coding types, and gRNAs, editing window sequences and amino acids are highlighted in a user-friendly manner. This tool is a good choice for base editing.
4.2.3. CRISPOR
Since many tools have been developed for highly efficient CRISPR gRNA design, an ensemble of multiple tools can be useful, and CRISPOR does this well [20], [36]. At present, CRISPOR contains 417 genomes and 19 PAM types, and can meet the needs of most users. CRISPOR takes genome coordinates and sequences <2,000 bp as inputs and provides comprehensive information as outputs. By default, results are shown in two parts; visualisation of PAM sites along the given sequence, which allows for downloading using multiple formats including fasta, GenBank and SnapGene; a table including all information for each predicted gRNA is also generated. In the table, two specificity scores (MIT and CFD) and 10 efficiency scores (Rule Set 2, CRISPRscan, etc.) are calculated [23], [41], [78]. Furthermore, Microhomology and Lindel scores are also provided for outcome prediction [7], [16]. Warnings such as extreme GC content and poly-T values are indicated, the table is downloadable, and candidate off-target sites with up to four mismatches can be visualised and downloaded.
In addition to gRNA design, CRISPOR designs primers for each gRNA as well as off-target sites. These primers are useful when conducting on/off-target validation or in vitro expression experiments. Furthermore, CRISPOR enables filtering of gRNAs with genomic variants based on well-known variant databases or marked input sequences with N characters. Thus, CRISPOR may be the best tool for designing gRNAs.
5. Summary and perspectives
CRISPR/Cas technology has emerged as an advanced strategy for functional genomics, crop breeding and precise medicine. Guide RNAs are indispensable for CRISPR-based editing, and computational approaches provide assistance for efficient gRNA design. Numerous tools have been developed for CRISPR gRNA design and evaluation. However, many issues remain. For example, experimental datasets used to build models for predicting sgRNA specificity or efficiency are disparate; CRISPRscan score performs worse in mammals than in zebrafish, the genome the predictive model was trained in [36]. Researchers should therefore know the strengths and weaknesses of each scoring method, and select the optimal tool for their research.
As we become more aware of the mechanisms by which Cas proteins recognise gRNAs, and bind to and cleave target loci, more and more novel features contributing to cutting efficiency and specificity are being identified, including sequence composition. For instance, low chromatin accessibility may block access of Cas9, while open chromatin regions are more likely to cause undesired mutations [39], [92]. In practice, specificity is more important than efficiency for application of CRISPR. However, several off-targets remain indecipherable using current tools, and discovery of novel features may minimise off-target effects.
Too much choice is not always desirable, and an unbiased evaluation can provide guidance. To this end, the Liu laboratory has conducted the benchmarking [30], [114], and Haeussler et al. performed a comparison of different predictive scores and integrated most into CRISPOR [36]. None of the tools excel when using heterogeneous data, due to cell-specific or species-specific training models. Therefore, more cell types need to be evaluated. Also, plant and some animal genomes should be analysed thoroughly, since human or mouse have dominated to date. Moreover, it will be beneficial if useful features of multiple tools are combined in future software packages.
The major outcomes of Cas9 cleavage is non-random and predictable [102], and several tools have been created for prediction of CRISPR-Cas9 outcomes [5], [16], [90]. Such findings facilitate more accurate gene editing. However, existing tools cannot account for large indels, homozygous sites and the activity of nucleases other than SpCas9, and this should be resolved in future [6].
In summary, the development of computational approaches has revolutionised the design of effective gRNAs, and CRISPR-based gene therapies and customised gene editing may come within reach. It is likely that CRISPR technology will continue to become more powerful in the coming years.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Transgenic Major Project (2019ZX08010003-001-002, 2018ZX08020-003), Jiangsu Province Government Project (BK2018003)/The open funds of the Jiangsu Key Laboratory of Crop Genomics and Molecular Breeding (PL201801) for T.Z. and Y.Z.; the Fund of Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) for T.Z.
Contributor Information
Yong Zhang, Email: zhangyong916@uestc.edu.cn.
Tao Zhang, Email: zhangtao@yzu.edu.cn.
References
- 1.Abadi S., Yan W.X., Amar D., Mayrose I. A machine learning approach for predicting CRISPR-Cas9 cleavage efficiencies and patterns underlying its mechanism of action. PLoS Comput Biol. 2017;13 doi: 10.1371/journal.pcbi.1005807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abudayyeh O.O., Gootenberg J.S., Essletzbichler P., Han S., Joung J., Belanto J.J. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adli M. The CRISPR tool kit for genome editing and beyond. Nat Commun. 2018:9. doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alkan F., Wenzel A., Anthon C., Havgaard J.H., Gorodkin J. CRISPR-Cas9 off-targeting assessment with nucleic acid duplex energy parameters. Genome Biol. 2018;19 doi: 10.1186/s13059-018-1534-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen F., Crepaldi L., Alsinet C., Strong A.J., Kleshchevnikov V., De Angeli P. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat Biotechnol. 2019;37:64–72. doi: 10.1038/nbt.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bae S., Kim J.S. Machine learning finds Cas9-edited genotypes. Nat Biomed Eng. 2018;2:892–893. doi: 10.1038/s41551-018-0327-6. [DOI] [PubMed] [Google Scholar]
- 7.Bae S., Kweon J., Kim H.S., Kim J.S. Microhomology-based choice of Cas9 nuclease target sites. Nat Methods. 2014;11:705–706. doi: 10.1038/nmeth.3015. [DOI] [PubMed] [Google Scholar]
- 8.Bae S., Park J., Kim J.S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 10.Bogdanove A.J., Voytas D.F. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 11.Cameron P., Fuller C.K., Donohoue P.D., Jones B.N., Thompson M.S., Carter M.M. Mapping the genomic landscape of CRISPR-Cas9 cleavage. Nat Methods. 2017;14:600–606. doi: 10.1038/nmeth.4284. [DOI] [PubMed] [Google Scholar]
- 12.Cao Q., Ma J., Chen C.H., Xu H., Chen Z., Li W. CRISPR-FOCUS: a web server for designing focused CRISPR screening experiments. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0184281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chari R., Mali P., Moosburner M., Church G.M. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat Methods. 2015;12:823–826. doi: 10.1038/nmeth.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chari R., Yeo N.C., Chavez A., Church G.M. sgRNA Scorer 2.0: a species-independent model to predict CRISPR/Cas9 activity. ACS Synth Biol. 2017;6:902–904. doi: 10.1021/acssynbio.6b00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen W., McKenna A., Schreiber J., Haeussler M., Yin Y., Agarwal V. Massively parallel profiling and predictive modeling of the outcomes of CRISPR/Cas9-mediated double-strand break repair. Nucleic Acids Res. 2019;47:7989–8003. doi: 10.1093/nar/gkz487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng A.W., Wang H., Yang H., Shi L., Katz Y., Theunissen T.W. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chuai G.H., Ma H.H., Yan J.F., Chen M., Hong N.F., Xue D.Y. DeepCRISPR: optimized CRISPR guide RNA design by deep learning. Genome Biol. 2018;19 doi: 10.1186/s13059-018-1459-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuai G.H., Wang Q.L., Liu Q. In silico meets in vivo: towards computational CRISPR-based sgRNA design. Trends Biotechnol. 2017;35:12–21. doi: 10.1016/j.tibtech.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 20.Concordet J.P., Haeussler M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018;46:W242–W245. doi: 10.1093/nar/gky354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cong L., Ran F.A., Cox D., Lin S.L., Barretto R., Habib N. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y.J., Pirzada Z.A. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doench J.G., Hartenian E., Graham D.B., Tothova Z., Hegde M., Smith I. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Endo A., Masafumi M., Kaya H., Toki S. Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida. Sci Rep-Uk. 2016:6. doi: 10.1038/srep38169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flicek P., Amode M.R., Barrell D., Beal K., Brent S., Chen Y. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu Y.F., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu Y.F., Sander J.D., Reyon D., Cascio V.M., Joung J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fusi N., Smith I., Doench J., Listgarten J. In silico predictive modeling of CRISPR/Cas9 guide efficiency. bioRxiv. 2015 [Google Scholar]
- 30.Gao Y., Chuai G., Yu W., Qu S., Liu Q. Data imbalance in CRISPR off-target prediction. Brief Bioinform. 2019:bbz069. doi: 10.1093/bib/bbz069. [DOI] [PubMed] [Google Scholar]
- 31.Gasiunas G., Barrangou R., Horvath P., Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. P Natl Acad Sci USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I. Programmable base editing of A.T to G.C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilbert L.A., Horlbeck M.A., Adamson B., Villalta J.E., Chen Y., Whitehead E.H. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graham D.B., Root D.E. Resources for the design of CRISPR gene editing experiments. Genome Biol. 2015;16 doi: 10.1186/s13059-015-0823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gratz S.J., Ukken F.P., Rubinstein C.D., Thiede G., Donohue L.K., Cummings A.M. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 2014;196:961–971. doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haeussler M., Schonig K., Eckert H., Eschstruth A., Mianne J., Renaud J.B. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17 doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harrington L.B., Burstein D., Chen J.S., Paez-Espino D., Ma E., Witte I.P. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362:839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heigwer F., Kerr G., Boutros M. E-CRISP: fast CRISPR target site identification. Nat Meth. 2014;11:122–124. doi: 10.1038/nmeth.2812. [DOI] [PubMed] [Google Scholar]
- 39.Horlbeck M.A., Witkowsky L.B., Guglielmi B., Replogle J.M., Gilbert L.A., Villalta J.E. Nucleosomes impede Cas9 access to DNA in vivo and in vitro. Elife. 2016;5 doi: 10.7554/eLife.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Housden B.E., Valvezan A.J., Kelley C., Sopko R., Hu Y., Roesel C. Identification of potential drug targets for tuberous sclerosis complex by synthetic screens combining CRISPR-based knockouts with RNAi. Sci Signal. 2015:8. doi: 10.1126/scisignal.aab3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hwang G.H., Park J., Lim K., Kim S., Yu J., Yu E. Web-based design and analysis tools for CRISPR base editing. BMC Bioinf. 2018;19:542. doi: 10.1186/s12859-018-2585-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacquin A.L., Odom D.T., Lukk M. Crisflash: open-source software to generate CRISPR guide RNAs against genomes annotated with individual variation. Bioinformatics. 2019;35:3146–3147. doi: 10.1093/bioinformatics/btz019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jensen K.T., Floe L., Petersen T.S., Huang J., Xu F., Bolund L. Chromatin accessibility and guide sequence secondary structure affect CRISPR-Cas9 gene editing efficiency. FEBS Lett. 2017;591:1892–1901. doi: 10.1002/1873-3468.12707. [DOI] [PubMed] [Google Scholar]
- 45.Jiang H., Wong W.H. SeqMap: mapping massive amount of oligonucleotides to the genome. Bioinformatics. 2008;24:2395–2396. doi: 10.1093/bioinformatics/btn429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim H.K., Min S., Song M., Jung S., Choi J.W., Kim Y. Deep learning improves prediction of CRISPR-Cpf1 guide RNA activity. Nat Biotechnol. 2018;36:239–241. doi: 10.1038/nbt.4061. [DOI] [PubMed] [Google Scholar]
- 48.Kim Y., Cheong S.A., Lee J.G., Lee S.W., Lee M.S., Baek I.J. Generation of knockout mice by Cpf1-mediated gene targeting. Nat Biotechnol. 2016;34:808–810. doi: 10.1038/nbt.3614. [DOI] [PubMed] [Google Scholar]
- 49.Kleinstiver B.P., Tsai S.Q., Prew M.S., Nguyen N.T., Welch M.M., Lopez J.M. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat Biotechnol. 2016;34:869–874. doi: 10.1038/nbt.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knott G.J., Doudna J.A. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361:866–869. doi: 10.1126/science.aat5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koike-Yusa H., Li Y.L., Tan E.P., Velasco-Herrera M.D., Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 52.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuan P.F., Powers S., He S., Li K., Zhao X., Huang B. A systematic evaluation of nucleotide properties for CRISPR sgRNA design. BMC Bioinf. 2017;18 doi: 10.1186/s12859-017-1697-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.La Russa M.F., Qi L.S. The New State of the Art: Cas9 for gene activation and repression. Mol Cell Biol. 2015;35:3800–3809. doi: 10.1128/MCB.00512-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Labuhn M., Adams F.F., Ng M., Knoess S., Schambach A., Charpentier E.M. Refined sgRNA efficacy prediction improves large- and small-scale CRISPR-Cas9 applications. Nucleic Acids Res. 2018;46:1375–1385. doi: 10.1093/nar/gkx1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Labun K., Montague T.G., Gagnon J.A., Thyme S.B., Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44:W272–W276. doi: 10.1093/nar/gkw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Labun K., Montague T.G., Krause M., Torres Cleuren Y.N., Tjeldnes H., Valen E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47:W171–W174. doi: 10.1093/nar/gkz365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10 doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lei Y., Lu L., Liu H.Y., Li S., Xing F., Chen L.L. CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant. 2014;7:1494–1496. doi: 10.1093/mp/ssu044. [DOI] [PubMed] [Google Scholar]
- 61.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Listgarten J., Weinstein M., Kleinstiver B.P., Sousa A.A., Joung J.K., Crawford J. Prediction of off-target activities for the end-to-end design of CRISPR guide RNAs. Nat Biomed Eng. 2018;2:38–47. doi: 10.1038/s41551-017-0178-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu H., Ding Y., Zhou Y., Jin W., Xie K., Chen L.L. CRISPR-P 2.0: an improved CRISPR-Cas9 tool for genome editing in plants. Mol Plant. 2017;10:530–532. doi: 10.1016/j.molp.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 64.Liu H.L., Wei Z., Dominguez A., Li Y.D., Wang X.W., Qi L.S. CRISPR-ERA: a comprehensive design tool for CRISPR-mediated gene editing, repression and activation. Bioinformatics. 2015;31:3676–3678. doi: 10.1093/bioinformatics/btv423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu X., Homma A., Sayadi J., Yang S., Ohashi J., Takumi T. Sequence features associated with the cleavage efficiency of CRISPR/Cas9 system. Sci Rep. 2016;6:19675. doi: 10.1038/srep19675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lowder L.G., Zhang D., Baltes N.J., Paul J.W., 3rd, Tang X., Zheng X. A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 2015;169:971–985. doi: 10.1104/pp.15.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lowder L.G., Zhou J., Zhang Y., Malzahn A., Zhong Z., Hsieh T.F. Robust transcriptional activation in plants using multiplexed CRISPR-Act2.0 and mTALE-Act systems. Mol Plant. 2018;11:245–256. doi: 10.1016/j.molp.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 68.Ma M., Ye A.Y., Zheng W., Kong L. A guide RNA sequence design platform for the CRISPR/Cas9 system for model organism genomes. Biomed Res Int. 2013;2013 doi: 10.1155/2013/270805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Makarova K.S., Grishin N.V., Shabalina S.A., Wolf Y.I., Koonin E.V. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biology Direct. 2006;1 doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Malzahn A.A., Tang X., Lee K., Ren Q., Sretenovic S., Zhang Y. Application of CRISPR-Cas12a temperature sensitivity for improved genome editing in rice, maize, and Arabidopsis. BMC Biol. 2019;17:9. doi: 10.1186/s12915-019-0629-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mans R., van Rossum H.M., Wijsman M., Backx A., Kuijpers N.G., van den Broek M. CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res. 2015:15. doi: 10.1093/femsyr/fov004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marraffini L.A. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- 74.McKenna A., Shendure J. FlashFry: a fast and flexible tool for large-scale CRISPR target design. BMC Biol. 2018;16 doi: 10.1186/s12915-018-0545-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mendoza B.J., Trinh C.T. Enhanced guide-RNA design and targeting analysis for precise CRISPR genome editing of single and consortia of industrially relevant and non-model organisms. Bioinformatics. 2018;34:16–23. doi: 10.1093/bioinformatics/btx564. [DOI] [PubMed] [Google Scholar]
- 76.Minkenberg B., Zhang J., Xie K., Yang Y. CRISPR-PLANT v2: an online resource for highly specific guide RNA spacers based on improved off-target analysis. Plant Biotechnol J. 2019;17:5–8. doi: 10.1111/pbi.13025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Montague T.G., Cruz J.M., Gagnon J.A., Church G.M., Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42:W401–W407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moreno-Mateos M.A., Vejnar C.E., Beaudoin J.D., Fernandez J.P., Mis E.K., Khokha M.K. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Meth. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Park J., Bae S., Kim J.S. Cas-Designer: a web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics. 2015;31:4014–4016. doi: 10.1093/bioinformatics/btv537. [DOI] [PubMed] [Google Scholar]
- 80.Park J., Childs L., Kim D., Hwang G.H., Kim S., Kim S.T. Digenome-seq web tool for profiling CRISPR specificity. Nat Methods. 2017;14:548–549. doi: 10.1038/nmeth.4262. [DOI] [PubMed] [Google Scholar]
- 81.Peng D., Tarleton R. EuPaGDT: a web tool tailored to design CRISPR guide RNAs for eukaryotic pathogens. Microb Genom. 2015;1 doi: 10.1099/mgen.0.000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peng H., Zheng Y., Blumenstein M., Tao D., Li J. CRISPR/Cas9 cleavage efficiency regression through boosting algorithms and Markov sequence profiling. Bioinformatics. 2018;34:3069–3077. doi: 10.1093/bioinformatics/bty298. [DOI] [PubMed] [Google Scholar]
- 83.Perez A.R., Pritykin Y., Vidigal J.A., Chhangawala S., Zamparo L., Leslie C.S. GuideScan software for improved single and paired CRISPR guide RNA design. Nat Biotechnol. 2017;35:347–349. doi: 10.1038/nbt.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pickar-Oliver A., Gersbach C.A. The next generation of CRISPR-Cas technologies and applications. Nat Rev Mol Cell Biol. 2019;20:490–507. doi: 10.1038/s41580-019-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pulido-Quetglas C., Aparicio-Prat E., Arnan C., Polidori T., Hermoso T., Palumbo E. Scalable Design of Paired CRISPR Guide RNAs for Genomic Deletion. PLoS Comput Biol. 2017;13 doi: 10.1371/journal.pcbi.1005341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rahman M.K., Rahman M.S. CRISPRpred: a flexible and efficient tool for sgRNAs on-target activity prediction in CRISPR/Cas9 systems. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0181943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ren Q., Zhong Z., Wang Y., You Q., Li Q., Yuan M. Bidirectional promoter-based crispr-cas9 systems for plant genome editing. Front Plant Sci. 2019;10:1173. doi: 10.3389/fpls.2019.01173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sander J.D., Joung J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sander J.D., Maeder M.L., Reyon D., Voytas D.F., Joung J.K., Dobbs D. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 2010;38:W462–W468. doi: 10.1093/nar/gkq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shen M.W., Arbab M., Hsu J.Y., Worstell D., Culbertson S.J., Krabbe O. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature. 2018;563:646–651. doi: 10.1038/s41586-018-0686-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shibata M., Nishimasu H., Kodera N., Hirano S., Ando T., Uchihashi T. Real-space and real-time dynamics of CRISPR-Cas9 visualized by high-speed atomic force microscopy. Nat Commun 8. 2017 doi: 10.1038/s41467-017-01466-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Singh R., Kuscu C., Quinlan A., Qi Y., Adli M. Cas9-chromatin binding information enables more accurate CRISPR off-target prediction. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gkv575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stemmer M., Thumberger T., Del Sol Keyer M., Wittbrodt J., Mateo J.L. CCTop: an intuitive, flexible and reliable crispr/cas9 target prediction tool. PLoS One. 2015;10 doi: 10.1371/journal.pone.0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Strecker J., Jones S., Koopal B., Schmid-Burgk J., Zetsche B., Gao L.Y. Engineering of CRISPR-Cas12b for human genome editing. Nat Commun. 2019;10 doi: 10.1038/s41467-018-08224-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Szczelkun M.D., Tikhomirova M.S., Sinkunas T., Gasiunas G., Karvelis T., Pschera P. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci U S A. 2014;111:9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tang X., Liu G.Q., Zhou J.P., Ren Q.R., You Q., Tian L. A large-scale whole-genome sequencing analysis reveals highly specific genome editing by both Cas9 and Cpf1 (Cas12a) nucleases in rice. Genome Biol. 2018;19 doi: 10.1186/s13059-018-1458-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tang X., Lowder L.G., Zhang T., Malzahn A.A., Zheng X., Voytas D.F. A CRISPR–Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants. 2017;3:17018. doi: 10.1038/nplants.2017.18. [DOI] [PubMed] [Google Scholar]
- 98.Tang X., Ren Q., Yang L., Bao Y., Zhong Z., He Y. Single transcript unit CRISPR 2.0 systems for robust Cas9 and Cas12a mediated plant genome editing. Plant Biotechnol J. 2019;17:1431–1445. doi: 10.1111/pbi.13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tang X., Zheng X., Qi Y., Zhang D., Cheng Y., Tang A. A single transcript CRISPR-Cas9 system for efficient genome editing in plants. Mol Plant. 2016;9:1088–1091. doi: 10.1016/j.molp.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 100.Urnov F.D., Miller J.C., Lee Y.L., Beausejour C.M., Rock J.M., Augustus S. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 101.Uusi-Makela M.I.E., Barker H.R., Bauerlein C.A., Hakkinen T., Nykter M., Ramet M. Chromatin accessibility is associated with CRISPR-Cas9 efficiency in the zebrafish (Danio rerio) PLoS One. 2018;13 doi: 10.1371/journal.pone.0196238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.van Overbeek M., Capurso D., Carter M.M., Thompson M.S., Frias E., Russ C. DNA repair profiling reveals Nonrandom outcomes at Cas9-mediated breaks. Mol Cell. 2016;63:633–646. doi: 10.1016/j.molcel.2016.06.037. [DOI] [PubMed] [Google Scholar]
- 103.Wang T., Wei J.J., Sabatini D.M., Lander E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wilson L.O.W., O'Brien A.R., Bauer D.C. The current state and future of CRISPR-Cas9 gRNA design tools. Front Pharmacol. 2018;9 doi: 10.3389/fphar.2018.00749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wilson L.O.W., Reti D., O'Brien A.R., Dunne R.A., Bauer D.C. High activity target-site identification using phenotypic independent CRISPR-Cas9 core functionality. CRISPR J. 2018;1:182–190. doi: 10.1089/crispr.2017.0021. [DOI] [PubMed] [Google Scholar]
- 106.Wong N., Liu W., Wang X. WU-CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 2015;16:218. doi: 10.1186/s13059-015-0784-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu X.B., Scott D.A., Kriz A.J., Chiu A.C., Hsu P.D., Dadon D.B. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol. 2014;32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xiao A., Cheng Z., Kong L., Zhu Z., Lin S., Gao G. CasOT: a genome-wide Cas9/gRNA off-target searching tool. Bioinformatics. 2014;30:1180–1182. doi: 10.1093/bioinformatics/btt764. [DOI] [PubMed] [Google Scholar]
- 109.Xie K., Zhang J., Yang Y. Genome-wide prediction of highly specific guide RNA spacers for CRISPR-Cas9-mediated genome editing in model plants and major crops. Mol Plant. 2014;7:923–926. doi: 10.1093/mp/ssu009. [DOI] [PubMed] [Google Scholar]
- 110.Xie S., Shen B., Zhang C., Huang X., Zhang Y. sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS One. 2014;9 doi: 10.1371/journal.pone.0100448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xie X., Ma X., Zhu Q., Zeng D., Li G., Liu Y.G. CRISPR-GE: a convenient software toolkit for CRISPR-based genome editing. Mol Plant. 2017;10:1246–1249. doi: 10.1016/j.molp.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 112.Xu H., Xiao T.F., Chen C.H., Li W., Meyer C.A., Wu Q. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 2015;25:1147–1157. doi: 10.1101/gr.191452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xu X., Duan D., Chen S.J. CRISPR-Cas9 cleavage efficiency correlates strongly with target-sgRNA folding stability: from physical mechanism to off-target assessment. Sci Rep 7. 2017 doi: 10.1038/s41598-017-00180-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yan J., Chuai G., Zhou C., Zhu C., Yang J., Zhang C. Benchmarking CRISPR on-target sgRNA design. Brief Bioinform. 2018;19:721–724. doi: 10.1093/bib/bbx001. [DOI] [PubMed] [Google Scholar]
- 115.Yang H., Wang H.Y., Shivalila C.S., Cheng A.W., Shi L.Y., Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yarrington R.M., Verma S., Schwartz S., Trautman J.K., Carroll D. Nucleosomes inhibit target cleavage by CRISPR-Cas9 in vivo. Proc Natl Acad Sci U S A. 2018;115:9351–9358. doi: 10.1073/pnas.1810062115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.You Q., Zhong Z., Ren Q., Hassan F., Zhang Y., Zhang T. CRISPRMatch: an automatic calculation and visualization tool for high-throughput CRISPR genome-editing data analysis. Int J Biol Sci. 2018;14:858–862. doi: 10.7150/ijbs.24581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P. Cpf1 Is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang D., Hurst T., Duan D., Chen S.J. Unified energetics analysis unravels SpCas9 cleavage activity for optimal gRNA design. Proc Natl Acad Sci U S A. 2019;116:8693–8698. doi: 10.1073/pnas.1820523116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang X.H., Tee L.Y., Wang X.G., Huang Q.S., Yang S.H. Off-target effects in CRISPR/Cas9-mediated Genome Engineering. Mol Ther-Nucl Acids. 2015;4 doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang Y., Zhang F., Li X., Baller J.A., Qi Y., Starker C.G. Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol. 2013;161:20–27. doi: 10.1104/pp.112.205179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zheng X., Yang S., Zhang D., Zhong Z., Tang X., Deng K. Effective screen of CRISPR/Cas9-induced mutants in rice by single-strand conformation polymorphism. Plant Cell Rep. 2016;35:1545–1554. doi: 10.1007/s00299-016-1967-1. [DOI] [PubMed] [Google Scholar]
- 123.Zhong Z., Sretenovic S., Ren Q., Yang L., Bao Y., Qi C. Improving plant genome editing with high-fidelity xCas9 and non-canonical PAM-targeting Cas9-NG. Mol Plant. 2019;12:1027–1036. doi: 10.1016/j.molp.2019.03.011. [DOI] [PubMed] [Google Scholar]
- 124.Zhong Z., Zhang Y., You Q., Tang X., Ren Q., Liu S. Plant genome editing using FnCpf1 and LbCpf1 nucleases at redefined and altered pam sites. Mol Plant. 2018;11:999–1002. doi: 10.1016/j.molp.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 125.Zhou J., Deng K., Cheng Y., Zhong Z., Tian L., Tang X. CRISPR-Cas9 based genome editing reveals new insights into MicroRNA function and regulation in rice. Front Plant Sci. 2017;8:1598. doi: 10.3389/fpls.2017.01598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhou J., Xin X., He Y., Chen H., Li Q., Tang X. Multiplex QTL editing of grain-related genes improves yield in elite rice varieties. Plant Cell Rep. 2019;38:475–485. doi: 10.1007/s00299-018-2340-3. [DOI] [PubMed] [Google Scholar]
- 127.Zhu H., Liang C. CRISPR-DT: designing gRNAs for the CRISPR-Cpf1 system with improved target efficiency and specificity. Bioinformatics. 2019;35:2783–2789. doi: 10.1093/bioinformatics/bty1061. [DOI] [PubMed] [Google Scholar]
- 128.Zou H., Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc Ser B (Stat Methodol) 2005;67:301–320. [Google Scholar]